Abstract
Rhesus macaques (Macaca mulatta) have played a valuable role in the development of human immunodeficiency virus (HIV) vaccine candidates prior to human clinical trials. However, changes and/or improvements in immunogen quality in the good manufacturing practice (GMP) process or changes in adjuvants, schedule, route, dose, or readouts have compromised the direct comparison of T-cell responses between species. Here we report a comparative study in which T-cell responses from humans and macaques to HIV type 1 antigens (Gag, Pol, Nef, and Env) were induced by the same vaccine batches prepared under GMP and administered according to the same schedules in the absence and presence of priming. Priming with DNA (humans and macaques) or alphavirus (macaques) and boosting with NYVAC induced robust and broad antigen-specific responses, with highly similar Env-specific gamma interferon (IFN-γ) enzyme-linked immunospot assay responses in rhesus monkeys and human volunteers. Persistent cytokine responses of antigen-specific CD4+ and CD8+ T cells of the central memory as well as the effector memory phenotype, capable of simultaneously eliciting multiple cytokines (IFN-γ, interleukin 2, and tumor necrosis factor alpha), were induced. Responses were highly similar in humans and primates, confirming earlier data indicating that priming is essential for inducing robust NYVAC-boosted IFN-γ T-cell responses. While significant similarities were observed in Env-specific responses in both species, differences were also observed with respect to responses to other HIV antigens. Future studies with other vaccines using identical lots, immunization schedules, and readouts will establish a broader data set of species similarities and differences with which increased confidence in predicting human responses may be achieved.
With an estimated 7,000 people acquiring human immunodeficiency virus (HIV) per day, the development of an effective HIV type 1 (HIV-1) vaccine remains a public health priority. With the failure of the first efficacy trial of a cytotoxic T-lymphocyte-based HIV-1 vaccine candidate (STEP trial) (7, 8, 22, 33), the field suffered a significant setback. The vaccine candidate evaluated in the STEP trial was based on sequential immunizations of attenuated adenovirus type 5 vectors (Ad5) expressing HIV-1 Gag, Pol, and Nef, in the absence of a previously evaluated DNA prime. The use of heterologous priming has been observed to amplify insert-specific immune responses to overcome antivector or possibly antiadenovirus immune responses (2, 6, 15, 24, 28, 29, 39). Interim analysis revealed that individuals with pre-existing immunity to the Ad5 vector were more likely to acquire HIV infection when they were immunized with this unprimed Ad5 candidate than the nonvaccinated contemporary controls. Furthermore, this vaccine candidate did not appear to reduce virus load in immunized individuals who became infected (7, 8, 21). In previous preclinical studies with human Ad5 in naive rhesus macaques, immunization with prime-boost combinations elicited a reduction in viral load after pathogenic chimeric simian/human immunodeficiency virus (SIV)-HIV (SHIV) challenge in a limited number of animals (35). Apparently, this ability to control virus load was more modest when a more stringent SIVmac239 challenge was used (6) and was more predominant in monkeys with the major histocompatibility complex haplotype Mamu-A*01, a class I molecule known to bind relatively conserved Gag epitopes (6, 23).
A debate has ensued in the field as to whether the rhesus macaque model should be considered a “gatekeeper” for promising HIV vaccine candidates as they progress toward human trials (27). Although many HIV vaccine concepts and prototypes have been evaluated in rhesus macaques, significant changes invariably occur when these vaccine candidates are refined and prepared for use in human trials. These differences have been considered minor in the past, but batch-to-batch variations of immunogens, for instance, can be considerable. Additionally, few studies have directly specifically examined human and rhesus macaque T-cell responses to multiple vaccine-encoded HIV antigens and actually compared responses using identical readouts, peptide pools, and immunization schedules.
To address this question we utilized the same good manufacturing practice (GMP) clinical lots, immunization schedule, dose, immunological readouts, and peptide pools used in matched human clinical trials. The trials EV01 and EV02 utilized EuroVacc immunogens based on HIV-1 clade C 97cn54 Gag, Pol, Nef, and Env inserted into the NYVAC pox vector administered in the presence and absence of priming with DNA (4, 12, 25). In contrast to adenovirus-based vectors, the successful eradication of smallpox through vaccination has resulted in a largely poxvirus-naive young human population, one that provided the vast majority of volunteers for these studies. In poxvirus-negative macaques, in addition to the presence or absence of DNA priming, a third group was added to determine if different priming modalities (DNA versus Semliki Forest virus [SFV]) impacted the immune responses observed following poxvirus boosting.
MATERIALS AND METHODS
Study groups.
Clinical trials were performed with low-risk counseled human volunteers as recently reported (4, 25). Preclinical trials were performed in a total of 30 mature outbred rhesus macaques (Macaca mulatta) divided into three groups of 10 animals each with similar age and sex distributions. All were housed under environmentally identical conditions at the Biomedical Primate Research Center, Rijswijk, The Netherlands. The study protocols and experimental procedures were rigorously reviewed by ethical care and use committees and approved prior to the study. All procedures were performed in accordance with Dutch law and international ethical and scientific standards and guidelines. In contrast to human volunteers, all immunizations and sample collections from macaques were performed under anesthesia.
GPN and Env immunogens.
The construction of the immunogens is described in detail elsewhere (12, 26). In brief, clade C/B′ Gag-Pol-Nef (GPN) and Env (gp120) codon-optimized sequences were designed by Geneart GmbH, based on sequence information derived from a 97cn54 provirus clone (38). PCRScript-based vectors were used to generate the DNA vaccine candidate plasmids comprising pORT1aGPN and pORT1agp120 and were produced in accordance with GMP standards using Escherichia coli strain DH1 lac dapD (Cobra Therapeutics Inc.). The genetic stability of both plasmids was evaluated in a DH1 lac dapD host strain up to 39 cell generations and controlled by double-strand DNA sequencing (Geneart GmbH, Regensburg, Germany). Upscaling and production of clinical GMP-grade material were performed by Cobra Biomanufacturing Plc, Keele, Staffordshire, United Kingdom. Therapeutics Inc. Sanofi Pasteur, Lyon, France, generated the NYVAC vector expressing GPN and Env of clade C HIV-1 97cn54 in a back-to-back orientation (11) under GMP conditions. Expression of the GPN polygene and the Env gene was checked by Western blot analysis (data not shown). The infectious-virus concentration (107.7 50% cell culture infective doses/ml) of the final product was determined by titration on QT35 cells (continuous cell line derived from a chemically induced tumor in quail).
pSFV4.2-HIVC-Env/syngp120 (clade C, 97cn54) and pSFV4.2-HIVC-Gag-Pol-Nef (clade C, 97cn54) SFV vectors were constructed at the Karolinska Institute, Stockholm, Sweden, according to the following methods. The sequence encoding the HIV-1 clade C syngp120 was isolated from pCR-Script-syngp120 obtained from Geneart GmbH (Regensburg, Germany) as a NotI-ApaI fragment and ligated into pSFV4.2 expression vector. For production of pSFV4.2-HIVC-Gag-Pol-Nef, the sequence encoding GPN was isolated as a KpnI-XhoI fragment from pScript-synGag-Pol-Nef, also provided by Geneart. The fragment was first inserted into the pET43 transfer plasmid and thereafter excised as an XhoI-SmaI fragment for insertion into the pSFV4.2 vector. For generation of recombinant particles, RNAs from the two SFV recombinant plasmids were synthesized and packaged into SFV particles using the two-helper RNA system described elsewhere (37). The recombinant SFV particles were harvested and purified by ultracentrifugation through a 20% sucrose cushion. Indirect immunofluorescence of infected BHK cells was performed to determine the titer of the recombinant virus stocks (19, 20). Antigen expression was verified in infected BHK-21 cells by metabolic labeling with [35S]methionine and further confirmed by immunoprecipitation as described previously (10). Immunoprecipitation and indirect immunofluorescence assays for analysis of GPN were performed with monoclonal antibodies (MAbs) against p24 (EVA repository reagent MAb HIV-1 p55/p24 ARP313 [EH12E1]). For the analysis of expression of syngp120, polyclonal antibodies against envelope clade C (kindly supplied by M. Esteban, Centro Nacional de Biotecnologia, Consejo Superior de Investigaciones Científicas, Madrid, Spain) were used.
Immunization schedule.
In accordance to the human clinical trial schedules (4, 25), at weeks 0 and 4, groups of 10 animals each were primed with either DNA or SFV. A third group of 10 animals did not receive any priming. At weeks 20 and 24, all animals were immunized (“boosted”) with NYVAC. The DNA pORT-gp120 and pORT-GPN plasmids were mixed prior to administration in an equimolar fashion (final total DNA concentration, 1.05 mg/ml). Each animal received 2 ml DNA intramuscularly (i.m.) in each upper leg (4.2 mg/4 ml total per monkey). Similarly, for the alphavirus-primed group, two constructs, pSFV4.2-HIVC-Env/syngp120 and pSFV4.2-HIVC-Gag-Pol-Nef SFV, were mixed prior to administration to give a final concentration of 109 PFU/ml of each construct. This mixture (500 μl) was administered i.m. in the left upper arm (5 × 108 PFU of each construct). The NYVAC vector expressing Gag-Pol-Nef and Env was later administered i.m. in the left upper arm (107.7 PFU/ml total).
ELISA.
Antibodies to HIV-1 Env (gp140 97cn54) in serum were measured by enzyme-linked immunosorbent assay (ELISA). Plates were coated overnight with Env in 100 mM NaHCO3 and were blocked for 1 h with 1% nonfat milk before application of serum, serially diluted in 1% bovine serum albumin-containing phosphate-buffered saline buffer. After 1 h incubation, 1 μg/ml anti-human immunoglobulin G-horseradish peroxidase conjugate was added for 1 h before the addition of ultra-tetramethylbenzidine ELISA development reagent. The reaction was stopped by addition of 0.5 M H2SO4. Results were expressed as immunoglobulin G endpoint dilution titers.
Enumeration of peptide-specific T-cell responses.
Quantification of antigen-specific cytokine-secreting cells in macaques was performed by gamma interferon (IFN-γ), interleukin 2 (IL-2), and IL-4 enzyme-linked immunospot (ELISpot) assays on freshly isolated peripheral blood mononuclear cells (PBMC), according to the manufacturer's instructions (U-Cytech, Utrecht, The Netherlands). The positive control was staphylococcal endotoxin B (1 μg/ml), and the negative control was medium alone. Details of human assays are reported elsewhere (12). For both human and macaque assays, antigen-specific responses were measured against 5-μg/ml peptide pools: 15-mers with an 11-amino-acid overlap spanning the entire GPN polygene and the Env clade C of HIV-1 97cn54 (Synpep Corporation, Dublin, CA). Eight peptide pools were made as follows: Gag1 60 peptides (Cg1-Cg240), Gag2 61 peptides (Cg244-Cg486), Pol1 60 peptides (Cgp485-Cp721), Pol2 61 peptides (Cp725-Cpn817, Cnp1017-Cp1161), Pol3 61 peptides (Cp1165-Cp1403), Nef 49 peptides (Cn838-Cnp1030), Env1 49 peptides (CN9-CN249), and Env2 63 peptides (CN253-CN485). Results are expressed as the mean number of spot-forming cells (SFC) per 106 PBMC from triplicate assays minus background values (mean number of SFC plus two standard deviations [SD] of triplicate assays with medium alone). Mean numbers of spots for each animal or individual per antigen are reported (responses to two Gag pools, three Pol pools, and two Env pools were combined).
Intracellular cytokine analysis.
The phenotypes of responding T cells were analyzed by intracellular cytokine staining and fluorescence-activated cell sorting (FACS) analysis as described elsewhere (3), with minor modifications. PBMC were incubated for 2 h at 37°C and 5% CO2 in the presence of the costimulatory molecules CD28 and CD49d (2 μg/ml each), with medium alone (negative control), with 1 μg/ml staphylococcal endotoxin B (positive control), or with 5 μg/ml of the eight peptide pools used in the ELISpot assay as described above. To inhibit cytokine secretion, Golgi Plug (Becton Dickinson) was added to a final dilution of 1:1,000 from the stock, and cells were further incubated overnight at 37°C and 5% CO2. After stimulation, cells were stained with directly conjugated antibodies (Becton Dickinson, Pharmingen, CA) for cell surface markers to CD3 (clone SP34, allophycocyanin labeled), CD4 (L200, peridinin-chlorophyll-protein complex labeled), and CD8 (SK1, allophycocyanin 7 labeled). Memory phenotyping was possible by staining with antibodies to CCR7 (150503, fluorescein isothiocyanate labeled; R&D Systems) and CD45RA (5H9, custom biotin labeled by Becton-Dickinson, followed by streptavidin Pacific Orange; Molecular Probes). The cells were fixed and permeabilized with Cytofix/Cytoperm buffer (Becton Dickinson) and stained intracellularly with IFN-γ (B27, Alexa-700 labeled), IL-2 (MQ1-17H12, phycoerythrin labeled), and tumor necrosis factor alpha (TNF-α) (MAb 11, phycoerythrin-cyanin 7 labeled) in Perm/wash solution (Becton Dickinson). A total of 300,000 to 800,000 events were acquired on a FACS Aria (Becton Dickinson, CA) and analyzed using FACS Diva software.
Data were analyzed by comparing proportions using a one-tailed z test, to test if the antigen-specific response was higher than the negative control response (response with medium alone). The value for α was set at 0.05. When the antigen-specific response was significantly higher than the background, the percentage of cytokine producing CD4+ or CD8+ T cells was calculated. The range of cytokine secretion for unstimulated cells or background responses in unvaccinated animals (data not shown) was 0 to 0.05%. In human volunteers, background responses never exceeded 0.02%. The reported values of cytokine-producing cells of each functional subset have had the background subtracted.
Statistical analysis.
Statistical analysis between immunization groups or between rhesus and human data was performed by the Wilcoxon rank sum test or the Mann-Whitney test. A P value of <0.05 was considered significant.
RESULTS
Safety and tolerability.
All animals remained healthy following immunizations and gained weight similarly to nonstudy peers. None of the immunizations induced measurable adverse effects, confirming data from humans indicating that the vaccine immunogens are well tolerated (4, 25).
Comparison of macaque and human vaccine-induced HIV-specific IFN-γ ELISpot assay responses.
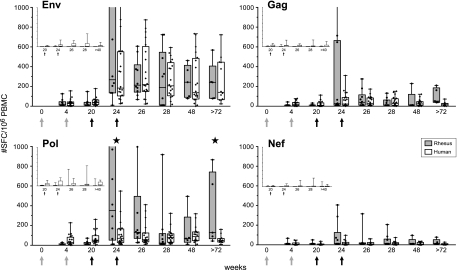
Details of the clinical trial have been published separately (4, 12, 25). Vaccine-induced HIV-specific IFN-γ responses in macaques were highly similar to responses measured in human volunteers (Fig. 1). In the absence of DNA priming, responses were low and infrequent in both species (Fig. 1, insets). However, when DNA priming preceded NYVAC immunizations, responses increased dramatically (Fig. 1). In both species, dominant responses were directed to Env, with the same relative magnitude and kinetics. In contrast to the response in humans, the IFN-γ response to Pol in macaques had a significantly higher peak and was more persistent (P = 0.045 [week 24 human versus week 24 rhesus] and P = 0.037 [week 72 human versus week 82 rhesus]). Again, IFN-γ responses to Gag and Nef were similar (low) in both species, with the occasional individual exception in some macaques in which a Gag response was observed (week 24).
FIG. 1.
Direct comparison of vaccine-induced antigen-specific IFN-γ ELISpot assay responses in rhesus monkeys and human volunteers. IFN-γ secretion by PBMC of individual DNA/NYVAC-immunized animals (n = 10) and human volunteers (n = 20) to HIV-1 clade C Env (two peptide pools), Gag (two peptide pools), Pol (three peptide pools), and Nef (one peptide pool) overlapping peptides was measured by ELISpot assay. Immunizations were given at weeks 0 and 4 (DNA; gray arrows) and weeks 20 and 24 (NYVAC; black arrows). Box-whisker plots show the interquartile ranges and medians (horizontal lines) of the groups at each time point. The last time point is week 82 for rhesus monkeys and week 72 for humans. Background responses (mean number of SFC plus 2 SD of triplicate assays with medium alone) were subtracted. Statistically significant differences (P < 0.05; Wilcoxon's rank sum test) between humans and rhesus monkeys are indicated by stars. (Insets) Antigen-specific IFN-γ responses of rhesus monkeys (first bars) and human volunteers (second bars) that were not primed with DNA but only immunized at week 20 and 24 (black arrows) with NYVAC expressing HIV-1 clade C Env, Gag, Pol, and Nef. The last time point is week 40 for rhesus monkeys and week 48 for humans. Values on the y axes range from 0 to 1,000 SFC/106 PBMC.
Influence of priming on NYVAC-boosted HIV-specific immune responses.
As the vaccine vector strategy used was developed to induce T-cell responses, it was not surprising that low antibody titers were observed (data not shown). The preclinical macaque study was expanded by an additional group to determine if priming with alphavirus particles (SFV) differed from DNA priming, in terms of the cumulative immune response following boosting. Intramuscular administration of either DNA or SFV alone induced low antigen-specific T-cell responses. DNA priming did induce low Env-specific IL-2 responses (50 to 150 SFC/106 PBMC) that were not detected after SFV priming (P = 0.0043) (Fig. 2, week 20).
FIG. 2.
Vaccine-induced antigen-specific T-cell responses in time. IFN-γ, IL-2, and IL-4 secretion by PBMC of individual animals (dots) to HIV-1 clade C Env (two peptide pools), Gag (two peptide pools), Pol (three peptide pools), and Nef (one peptide pool) overlapping peptides was measured by ELISpot assay. Immunizations were given at weeks 0 and 4 (DNA or SFV priming) and weeks 20 and 24 (NYVAC boosting). Box-whisker plots show the interquartile ranges and medians (horizontal lines) of the groups at each time point. Background responses (mean number of SFC plus 2 SD of triplicate assays with medium alone) were subtracted. Responses of animals that were not primed (monitored from weeks 20 to 40), primed with DNA, or primed with SFV are presented in one graph for direct comparison. Statistically significant differences (P < 0.05; Wilcoxon's rank sum test) between the two priming groups are indicated by stars. (DNA priming > SFV priming) and diamonds (SFV priming > DNA priming).
In the absence of priming, IFN-γ, IL-2, and IL-4 peptide-specific responses were either extremely low or negligible (Fig. 2). When groups were primed by either DNA or alphavirus, boosting by NYVAC dramatically increased the magnitude of clade C HIV-specific cytokine responses (more than 1,000 IFN-γ SFC/106 PBMC) (Fig. 2) and increased the frequency of responders (up to 100%). Importantly, priming facilitated the diversity of the HIV-specific cytokine (IFN-γ, IL-2, and IL-4) responses, which were all markedly increased. Antigen-specific cytokine responses were very similar between DNA- and SFV-primed groups, with only subtle differences in ELISpot assay responses. All DNA-primed animals demonstrated antigen-specific IFN-γ, IL-2, and IL-4 responses at least at one time point, while the percentages of antigen-specific IL-2 and IL-4 responders in the SFV-primed animals were lower (75% and 62.5%, respectively). With respect to antigen (Env, Gag, Pol, or Nef)-specific responses, animals that received the DNA prime-NYVAC boost combination developed generally higher Env-specific cytokine responses than SFV-primed animals (Fig. 2). Interestingly, in contrast, alphavirus-primed animals developed higher Gag- and Nef-specific IFN-γ peak responses as observed at week 26 (P = 0.045 and P = 0.007) (Fig. 2). The antigen-specific IL-2 responses were low but persistent after the poxvirus boosts (Fig. 2, middle), and although they were highly comparable following DNA and SFV priming, they had a tendency to be more robust in the DNA-primed group. Again, responses to the Nef insert were poorest. Very interestingly, IL-4 responses showed a strong peak after the first NYVAC boost; however, these declined rapidly, independently of the type of priming (Fig. 2, right).
Functional and phenotypic profile of vaccine-induced peptide-specific cytokine-producing T cells elicited by different priming strategies in macaques.
The preclinical macaque study was performed to determine the best priming agent to take forward for further clinical evaluation. To determine if the responses observed by ELISpot assay were mediated by either CD4+ or CD8+ T cells, multiparameter FACS analysis was performed. The HIV-specific peptide-stimulated cytokine responses in CD4+ and CD8+ T-cell populations from five animals of each group were analyzed at week 26 (peak response) and week 82 (memory) for intracellular production of IFN-γ, IL-2, and/or TNF-α (Fig. 3). Interestingly, data revealed that vaccine-induced T-cell responses were mediated by both CD4+ and CD8+ T cells and were multifunctional (producing more than one cytokine simultaneously), persistent, and directed to all of the four vaccine-encoded HIV antigens (Fig. 4).
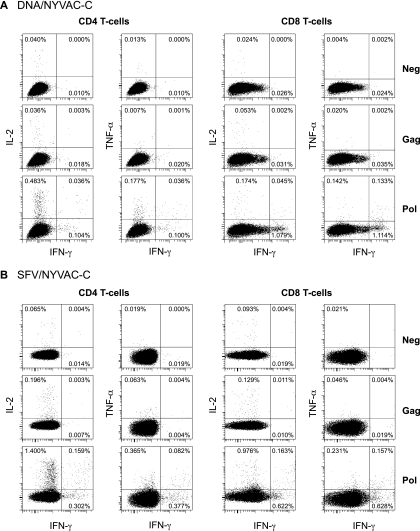
FIG. 3.
Vaccine-induced antigen-specific intracellular cytokine production as measured by FACS analysis at week 82. Flow cytometry profiles of Gag- and Pol-specific CD4+ (left) and CD8+ (right) T cells able to secrete IFN-γ, IL-2, and TNF-α of a representative DNA-primed animal (A) and an SFV-primed animal (B) are shown.
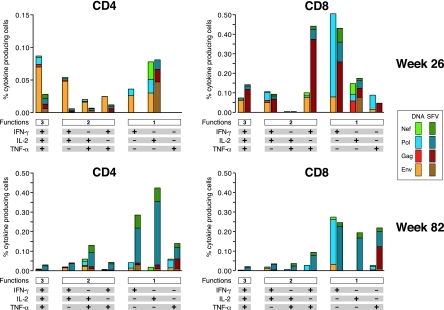
FIG. 4.
Functional profiles of vaccine-induced antigen-specific CD4+ and CD8+ T cells. The results shown are from five animals of each group (DNA/NYVAC-C and SFV/NYVAC-C) at week 26 and week 82. Responses are grouped on the basis of the number of functions (producing one, two, or three cytokines simultaneously). All the possible combinations of cytokine responses are shown on the x axis. The responses to vaccine antigens (Env, Gag, Pol, and Nef) are color coded. Bars correspond to the mean number of different functionally distinct antigen-specific CD4+ or CD8+ T cells per group. Background responses have been subtracted.
When antigen specificity at each time point was examined, subtle differences were found between primed groups. DNA-primed animals showed an Env-dominated (P < 0.01) peak response (week 26) that elicited higher responses than SFV-primed animals (P = 0.04), consistent with ELISpot assay data. The Env response was mediated by only CD4+ T cells in three of five animals and by both CD4+ and CD8+ T cells in two of five animals (data not shown). These results were very consistent with the Env response in human volunteers; 50% of the human responders to NYVAC boosting possessed CD4+-T-cell-mediated responses (12). In NYVAC-boosted macaques, Env-specific T cells showed a polyfunctional profile, with 65% of CD4+ and CD8+ T cells exhibiting two or three functions. Interestingly, it was by this multiparameter analysis that differences were observed, not only in terms of HIV antigen specificity but also in the nature of the population of antigen-specific T cells. SFV-primed animals responded relatively more strongly to Pol and Nef antigens, and this response was mediated by both CD4+ and CD8+ T-cell populations in this group. Similarly, the CD4+ T-cell-mediated Pol- and Nef-specific responses were significantly higher than those elicited by DNA priming in macaques (P = 0.046). Additionally, two out of five SFV-primed animals possessed a strong Gag-specific cytokine response mediated by CD8+ T cells. Although the Gag-specific CD8+ T-cell response in DNA-primed animals tended to be lower, the difference did not reach statistical significance. The percentage of antigen-specific T cells with a polyfunctional profile was lower at week 82 than at week 26 (Fig. 4). However, it must be pointed out that the Env-dominated response shifted toward a different antigen (Pol) over time, which was mediated by more single-cytokine-producing cells than the Env-specific response. As we measured responses only in circulation, we cannot rule out the possibility that different memory kinetics may exist in secondary lymphoid tissues.
Comparison of macaque and human functional profiles and memory phenotypes of vaccine-induced Env-specific CD4+ and CD8+ T cells.
Given the general similarity with respect to the magnitude of the ELISpot assay responses in DNA- and SFV-primed groups and difficulties with clinical-scale production of SFV, the human trial was carried out with (and without) DNA priming only.
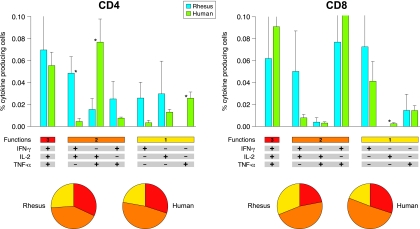
Cytokine profiles of Env-specific CD4+ and CD8+ T cells in DNA-primed rhesus monkeys (Fig. 4) were highly similar to the functional profiles of Env-specific human T cells (Fig. 5) (12). Direct comparison and detailed analysis showed differences between some T-cell populations, revealing a statistically significantly higher percentage of IFN-γ- and IL-2-producing CD4+ T cells in rhesus macaques, while humans showed more IL-2- and TNF-α-producing CD4+ T cells. Furthermore, TNF-α-producing CD4+ and IL-2-producing CD8+ T cells were detected only in humans, not in rhesus macaques. However, the relative proportions of CD4+ and CD8+ T cells producing either one, two, or three cytokines simultaneously were the same for rhesus macaques and human volunteers (Fig. 5).
FIG. 5.
Direct comparison of rhesus and human functional profiles of vaccine-induced Env-specific CD4+ and CD8+ T cells. The results shown were generated from five animals and eight human volunteers immunized with DNA/NYVAC-C at the peak of the response. Responses are grouped on the basis of the number of functions (producing one, two, or three cytokines simultaneously). All the possible combinations of cytokine responses are shown on the x axis. Bars correspond to the mean number of different functionally distinct Env-specific CD4+ or CD8+ T cells per group. Standard errors of the means are also presented. Each slice of the pie charts corresponds to the fraction of CD4+ or CD8+ T cells with a given number of functions (as shown below the graphs) within the total CD4+ and CD8+ T-cell populations. Statistically significant differences between rhesus macaques and human are indicated by asterisks (Mann-Whitney). Background responses have been subtracted.
CD45RA and CCR7 define functionally distinct populations of memory T cells (31, 32). The memory phenotype was measured in T cells of 10 animals (5 of each primed group). Since rhesus CD4+ T cells have much lower expression of the CD45RA molecule on their surfaces than CD8+ T cells or human T cells, it was difficult to detect antigen-specific cytokine-producing CD4+ CD45RA+ T cells in this study. However, at the peak (week 26) of the T-cell response, we were able to detect Env-specific cytokine (triple [IFN-γ, IL-2, TNF-α], dual, and single)-producing CD4+ T cells that were predominantly CCR7− (data not shown), indicating that the cells were of either the effector memory (EM; CCR7− CD45RA−) or the effector (EFF; CCR7− CD45RA+) phenotype, consistent with the human response to Env (12). Similarly, the responses to other antigens (Gag, Pol, and Nef) were also mediated by CCR7− (EM or EFF) CD4+ T cells in animals from both primed (DNA and SFV) groups.
The Env-specific cytokine-producing rhesus monkey CD8+ T-cell population possessed mainly an EM (CD45RA− CCR7−) and EFF (CD45RA+ CCR7−) phenotype (Fig. 6). This phenotypic profile did not change over time (data not shown), while the Env-specific human CD8+ T cells shifted from the EM phenotype to the EFF phenotype (12). Also, the CD8+ T cells specific for other antigens (Gag, Pol, or Nef) in rhesus macaques from either group (DNA or SFV primed) were of the EM and EFF phenotypes and remained unchanged in time (data not shown).
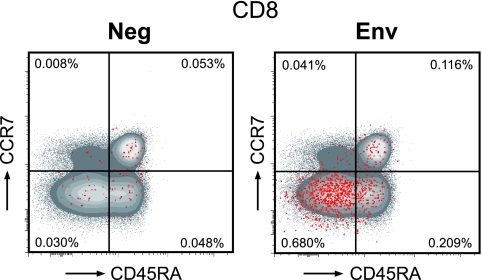
FIG. 6.
Phenotypic analysis of vaccine-induced CD8+ T cells. The memory phenotype of antigen-specific T cells was determined in 10 animals (5 animals from each primed group). PBMC were unstimulated (Neg) or stimulated with Env peptides for 16 h and stained with IFN-γ, IL-2, TNF-α, CD3, CD4, CD8, CCR7, and CD45RA antibodies. A flow cytometry scatter plot of CD8+ T cells from animal Ri508 (DNA primed) at week 26 (peak) is shown. The red dots indicate the cytokine-producing vaccine-induced CD8+ T cells. Env-specific CD8+ T cells are of the EM (CD45RA− CCR7−) and EFF (CD45RA+ CCR7−) phenotypes.
DISCUSSION
Differences between preclinical immunogen batches, routes, doses, and vaccine schedules are variables that are often overlooked as confounding factors when vaccine-specific responses in macaques and humans are being compared. This issue has recently been recognized as one for which there is a lack of data and knowledge in the HIV vaccine development field (9, 27). Here we provide such data in the context of clade C HIV-1 immunogens administered by DNA priming and NYVAC boosting using identical vaccine lots and protocols in both species.
The direct comparison of vaccine induced IFN-γ responses in rhesus and humans clearly confirmed that priming is essential and has an important impact on the quantity and quality as well as the persistence of insert-specific immune responses. Both humans and rhesus macaques showed a relative predominant response to Env, with similar magnitude and kinetics. The reason for the relative predominance of Env over the other three HIV antigens encoded by these vectors is most likely related to the level of antigen expression at the cellular level. When the immunogenicity of these vaccine vectors was evaluated in HLA-A2 class I transgenic mice, a similar Env-dominated response was observed (11).
While immune responses in humans and macaques were similar in terms of Env-, Gag-, and Nef-specific IFN-γ ELISpot assay results, it should also be noted where the two primate species differed. Macaques generated significantly higher Pol-specific IFN-γ responses. This discrepancy may possibly be explained by differences in antigen processing, such as presentation or epitope binding by certain human major histocompatibility complex and rhesus Mamu molecules. The presence of different endogenous retroviruses in rhesus monkeys (for instance, foamy virus) might possibly explain the higher Pol-specific responses in this species.
The polyfunctional profiles of Env-specific T-cell populations in rhesus monkeys showed strong similarities with those in immunized human volunteers (12), having the same proportions of CD4+ and CD8+ T cells producing either three, two, or one cytokine simultaneously. However, rhesus macaques showed more IFN-γ- and IL-2-producing CD4+ T cells, while humans had more IL-2- and TNF-α-producing CD4+ T cells. Furthermore, TNF-α-producing CD4+ and IL-2-producing CD8+ T cells were detected only in humans, not in rhesus macaques. Some differences in the memory phenotype of CD8+ T cells were also observed between species. The Env-specific human CD8+ T cells shifted from the EM phenotype to the EFF phenotype over time (12), while in rhesus macaques the antigen-specific CD8+ T cells of the EM and EFF phenotypes remained unchanged.
It can only be speculated that these subtle differences are related to actual species differences. In some instances, differences may possibly be attributable to differences in avidity or specificity of the MAbs used to detect human and rhesus macaque cytokines. Alternatively, there may be different levels of expression of surface molecules on rhesus macaque and human T cells. Vaccine doses in both species were the same, but responses could have been different if the doses had been adjusted per kilogram of body weight.
Vaccine-induced polyfunctional responses in both CD4+ and CD8+ T-cell subsets in the rhesus macaques suggests the potential to recruit robust CD8+ cytotoxic T lymphocytes, which may then facilitate the clearance of HIV-infected cells (13, 16, 17, 34, 36). The presence of polyfunctional phenotypes has been shown to be beneficial and to correlate with a long-term-nonprogressor status in rhesus macaques (1, 30) and humans (5, 18, 40). In the current setting with the existing GMP HIV-1 immunogens, it was not possible to show protection against SIV or SHIV challenge without the corresponding SIV inserts. Subsequent challenge studies of the same DNA-NYVAC strategy matched to SHIV inserts (with matching HIV-1 Envs) have provided evidence of vaccine efficacy (26).
The ability to directly compare macaque to human immune responses in studies with identical immunogens, doses, and schedules will provide us with a more complete comparative immunology database with which to improve our understanding of the similarities but also the differences between these species. Therefore, the direct extrapolation of rhesus data to humans should be made with due consideration. This understanding may facilitate more confidence in the use of these valuable models in the iterative process of assembling a strong clinical trial pipeline of HIV vaccine candidates (14, 27).
Acknowledgments
We are indebted to S. Ding, T. de Koning and H. van Westbroek for coordination and documentation. We thank E. Remarque for valuable statistical advice.
This study was supported by funding of the EuroVacc (European Vaccine Effort Against HIV/AIDS) project QLK2-CT-1999-01321.
Footnotes
Published ahead of print on 25 March 2009.
REFERENCES
- 1.Amara, R. R., C. Ibegbu, F. Villinger, D. C. Montefiori, S. Sharma, P. Nigam, Y. Xu, H. M. McClure, and H. L. Robinson. 2005. Studies using a viral challenge and CD8 T cell depletions on the roles of cellular and humoral immunity in the control of an SHIV-89.6P challenge in DNA/MVA-vaccinated macaques. Virology 343246-255. [DOI] [PubMed] [Google Scholar]
- 2.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 29269-74. [DOI] [PubMed] [Google Scholar]
- 3.Balla-Jhagjhoorsingh, S. S., G. Koopman, P. Mooij, W. Koornstra, S. McCormack, J. Weber, G. Pantaleo, and J. L. Heeney. 2004. Long-term persistence of HIV-1 vaccine-induced CD4+CD45RA−CD62L−CCR7− memory T-helper cells. AIDS 18837-848. [DOI] [PubMed] [Google Scholar]
- 4.Bart, P. A., R. Goodall, T. Barber, A. Harari, A. Guimaraes-Walker, M. Khonkarly, N. C. Sheppard, Y. Bangala, M. J. Frachette, R. Wagner, P. Liljestrom, J. P. Kraehenbuhl, M. Girard, J. Goudsmit, M. Esteban, J. Heeney, Q. Sattentau, S. McCormack, A. Babiker, G. Pantaleo, and J. Weber. 2008. EV01: A phase I trial in healthy HIV negative volunteers to evaluate a clade C HIV vaccine, NYVAC-C undertaken by the EuroVacc Consortium. Vaccine 263153-3161. [DOI] [PubMed] [Google Scholar]
- 5.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 1074781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casimiro, D. R., F. Wang, W. A. Schleif, X. Liang, Z. Q. Zhang, T. W. Tobery, M. E. Davies, A. B. McDermott, D. H. O'Connor, A. Fridman, A. Bagchi, L. G. Tussey, A. J. Bett, A. C. Finnefrock, T. M. Fu, A. Tang, K. A. Wilson, M. Chen, H. C. Perry, G. J. Heidecker, D. C. Freed, A. Carella, K. S. Punt, K. J. Sykes, L. Huang, V. I. Ausensi, M. Bachinsky, U. Sadasivan-Nair, D. I. Watkins, E. A. Emini, and J. W. Shiver. 2005. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with DNA and recombinant adenoviral vaccine vectors expressing Gag. J. Virol. 7915547-15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J. 2007. AIDS research. Did Merck's failed HIV vaccine cause harm? Science 3181048-1049. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, J. 2007. AIDS research. Promising AIDS vaccine's failure leaves field reeling. Science 31828-29. [DOI] [PubMed] [Google Scholar]
- 9.Fauci, A. S., M. I. Johnston, C. W. Dieffenbach, D. R. Burton, S. M. Hammer, J. A. Hoxie, M. Martin, J. Overbaugh, D. I. Watkins, A. Mahmoud, and W. C. Greene. 2008. HIV vaccine research: the way forward. Science 321530-532. [DOI] [PubMed] [Google Scholar]
- 10.Garoff, H., J. Wilschut, P. Liljestrom, J. M. Wahlberg, R. Bron, M. Suomalainen, J. Smyth, A. Salminen, B. U. Barth, H. Zhao, et al. 1994. Assembly and entry mechanisms of Semliki Forest virus. Arch. Virol. Suppl. 9329-338. [DOI] [PubMed] [Google Scholar]
- 11.Gomez, C. E., J. L. Najera, V. Jimenez, K. Bieler, J. Wild, L. Kostic, S. Heidari, M. Chen, M. J. Frachette, G. Pantaleo, H. Wolf, P. Liljestrom, R. Wagner, and M. Esteban. 2007. Generation and immunogenicity of novel HIV/AIDS vaccine candidates targeting HIV-1 Env/Gag-Pol-Nef antigens of clade C. Vaccine 251969-1992. [DOI] [PubMed] [Google Scholar]
- 12.Harari, A., P. A. Bart, W. Stohr, G. Tapia, M. Garcia, E. Medjitna-Rais, S. Burnet, C. Cellerai, O. Erlwein, T. Barber, C. Moog, P. Liljestrom, R. Wagner, H. Wolf, J. P. Kraehenbuhl, M. Esteban, J. Heeney, M. J. Frachette, J. Tartaglia, S. McCormack, A. Babiker, J. Weber, and G. Pantaleo. 2008. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J. Exp. Med. 20563-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heeney, J. L. 2002. The critical role of CD4+ T-cell help in immunity to HIV. Vaccine 201961-1963. [DOI] [PubMed] [Google Scholar]
- 14.Heeney, J. L., and S. A. Plotkin. 2006. Immunological correlates of protection from HIV infection and disease. Nat. Immunol. 71281-1284. [DOI] [PubMed] [Google Scholar]
- 15.Hel, Z., W. P. Tsai, A. Thornton, J. Nacsa, L. Giuliani, E. Tryniszewska, M. Poudyal, D. Venzon, X. Wang, J. Altman, D. I. Watkins, W. Lu, A. von Gegerfelt, B. K. Felber, J. Tartaglia, G. N. Pavlakis, and G. Franchini. 2001. Potentiation of simian immunodeficiency virus (SIV)-specific CD4+ and CD8+ T cell responses by a DNA-SIV and NYVAC-SIV prime/boost regimen. J. Immunol. 1677180-7191. [DOI] [PubMed] [Google Scholar]
- 16.Janssen, E. M., E. E. Lemmens, T. Wolfe, U. Christen, M. G. von Herrath, and S. P. Schoenberger. 2003. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 421852-856. [DOI] [PubMed] [Google Scholar]
- 17.Kaech, S. M., and R. Ahmed. 2003. Immunology. CD8 T cells remember with a little help. Science 300263-265. [DOI] [PubMed] [Google Scholar]
- 18.Kannanganat, S., B. G. Kapogiannis, C. Ibegbu, L. Chennareddi, P. Goepfert, H. L. Robinson, J. Lennox, and R. R. Amara. 2007. Human immunodeficiency virus type 1 controllers but not noncontrollers maintain CD4 T cells coexpressing three cytokines. J. Virol. 8112071-12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson, G. B., and P. Liljestrom. 2004. Delivery and expression of heterologous genes in mammalian cells using self-replicating alphavirus vectors. Methods Mol. Biol. 246543-557. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson, G. B., and P. Liljestrom. 2003. Live viral vectors: Semliki Forest virus. Methods Mol. Med. 8769-82. [DOI] [PubMed] [Google Scholar]
- 21.Kresge, K. J. 2008. AIDS vaccine researchers STEP up to the challenge. IAVI Report September-October. 121-7. [PubMed] [Google Scholar]
- 22.Ledford, H. 2007. HIV vaccine may raise risk. Nature 450325. [DOI] [PubMed] [Google Scholar]
- 23.Liang, X., D. R. Casimiro, W. A. Schleif, F. Wang, M. E. Davies, Z. Q. Zhang, T. M. Fu, A. C. Finnefrock, L. Handt, M. P. Citron, G. Heidecker, A. Tang, M. Chen, K. A. Wilson, L. Gabryelski, M. McElhaugh, A. Carella, C. Moyer, L. Huang, S. Vitelli, D. Patel, J. Lin, E. A. Emini, and J. W. Shiver. 2005. Vectored Gag and Env but not Tat show efficacy against simian-human immunodeficiency virus 89.6P challenge in Mamu-A*01-negative rhesus monkeys. J. Virol. 7912321-12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, J., B. A. Ewald, D. M. Lynch, M. Denholtz, P. Abbink, A. A. Lemckert, A. Carville, K. G. Mansfield, M. J. Havenga, J. Goudsmit, and D. H. Barouch. 2008. Magnitude and phenotype of cellular immune responses elicited by recombinant adenovirus vectors and heterologous prime-boost regimens in rhesus monkeys. J. Virol. 824844-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormack, S., W. Stohr, T. Barber, P. A. Bart, A. Harari, C. Moog, D. Ciuffreda, C. Cellerai, M. Cowen, R. Gamboni, S. Burnet, K. Legg, E. Brodnicki, H. Wolf, R. Wagner, J. Heeney, M. J. Frachette, J. Tartaglia, A. Babiker, G. Pantaleo, and J. Weber. 2008. EV02: a phase I trial to compare the safety and immunogenicity of HIV DNA-C prime-NYVAC-C boost to NYVAC-C alone. Vaccine 263162-3174. [DOI] [PubMed] [Google Scholar]
- 26.Mooij, P., S. S. Balla-Jhagjhoorsingh, G. Koopman, N. Beenhakker, P. van Haaften, I. Baak, I. G. Nieuwenhuis, I. Kondova, R. Wagner, H. Wolf, C. E. Gomez, J. L. Najera, V. Jimenez, M. Esteban, and J. L. Heeney. 2008. Differential CD4+ versus CD8+ T-cell responses elicited by different poxvirus-based human immunodeficiency virus type 1 vaccine candidates provide comparable efficacies in primates. J. Virol. 822975-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan, C., M. Marthas, C. Miller, A. Duerr, C. Cheng-Mayer, R. Desrosiers, J. Flores, N. Haigwood, S. L. Hu, R. P. Johnson, J. Lifson, D. Montefiori, J. Moore, M. Robert-Guroff, H. Robinson, S. Self, and L. Corey. 2008. The use of nonhuman primate models in HIV vaccine development. PLoS Med. 5e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson, C., B. Makitalo, P. Berglund, F. Bex, P. Liljestrom, G. Sutter, V. Erfle, P. ten Haaft, J. Heeney, G. Biberfeld, and R. Thorstensson. 2001. Enhanced simian immunodeficiency virus-specific immune responses in macaques induced by priming with recombinant Semliki Forest virus and boosting with modified vaccinia virus Ankara. Vaccine 193526-3536. [DOI] [PubMed] [Google Scholar]
- 29.Robinson, H. L., D. C. Montefiori, R. P. Johnson, K. H. Manson, M. L. Kalish, J. D. Lifson, T. A. Rizvi, S. Lu, S. L. Hu, G. P. Mazzara, D. L. Panicali, J. G. Herndon, R. Glickman, M. A. Candido, S. L. Lydy, M. S. Wyand, and H. M. McClure. 1999. Neutralizing antibody-independent containment of immunodeficiency virus challenges by DNA priming and recombinant pox virus booster immunizations. Nat. Med. 5526-534. [DOI] [PubMed] [Google Scholar]
- 30.Sadagopal, S., R. R. Amara, D. C. Montefiori, L. S. Wyatt, S. I. Staprans, N. L. Kozyr, H. M. McClure, B. Moss, and H. L. Robinson. 2005. Signature for long-term vaccine-mediated control of a simian and human immunodeficiency virus 89.6P challenge: stable low-breadth and low-frequency T-cell response capable of coproducing gamma interferon and interleukin-2. J. Virol. 793243-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sallusto, F., J. Geginat, and A. Lanzavecchia. 2004. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22745-763. [DOI] [PubMed] [Google Scholar]
- 32.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401708-712. [DOI] [PubMed] [Google Scholar]
- 33.Sekaly, R. P. 2008. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J. Exp. Med. 2057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shedlock, D. J., and H. Shen. 2003. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 300337-339. [DOI] [PubMed] [Google Scholar]
- 35.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415331-335. [DOI] [PubMed] [Google Scholar]
- 36.Slifka, M. K., and J. L. Whitton. 2000. Activated and memory CD8+ T cells can be distinguished by their cytokine profiles and phenotypic markers. J. Immunol. 164208-216. [DOI] [PubMed] [Google Scholar]
- 37.Smerdou, C., and P. Liljestrom. 1999. Two-helper RNA system for production of recombinant Semliki forest virus particles. J. Virol. 731092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su, L., M. Graf, Y. Zhang, H. von Briesen, H. Xing, J. Kostler, H. Melzl, H. Wolf, Y. Shao, and R. Wagner. 2000. Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B′) recombinant strain in China. J. Virol. 7411367-11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xin, K. Q., N. Jounai, K. Someya, K. Honma, H. Mizuguchi, S. Naganawa, K. Kitamura, T. Hayakawa, S. Saha, F. Takeshita, K. Okuda, M. Honda, D. M. Klinman, and K. Okuda. 2005. Prime-boost vaccination with plasmid DNA and a chimeric adenovirus type 5 vector with type 35 fiber induces protective immunity against HIV. Gene Ther. 121769-1777. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerli, S. C., A. Harari, C. Cellerai, F. Vallelian, P. A. Bart, and G. Pantaleo. 2005. HIV-1-specific IFN-γ/IL-2-secreting CD8 T cells support CD4-independent proliferation of HIV-1-specific CD8 T cells. Proc. Natl. Acad. Sci. USA 1027239-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]