Abstract
The adapter protein CRKL is required for the normal development of multiple tissues that rely on fibroblast growth factor 8 (FGF8). The precise role of CRKL in receptor signaling has been unclear, however. To address this issue, we first modeled the three-dimensional structure of CRKL by molecular dynamics. By taking advantage of structural simulations, we performed in silico analysis of the interactions of the autophosphorylation sites of FGR receptor 1 (FGFR1) with the SH2 domain of CRKL or a highly related protein, CRK. As predicted by simulations, we confirm the specific physical interaction of phosphorylated Y463 (pY463) in FGFR1 with the CRKL SH2 domain at an affinity ∼30-fold stronger than that of CRK. We also provide evidence that interactions outside of the core YXXP motif have a significant impact on physical association, which is consistent with predictions from molecular-dynamics simulations. Furthermore, we identify CRKL as an essential component of an FGF8-induced feed-forward loop permissive for efficient activation of the mitogen-activated protein kinase Erk1/2, as well as FGF8-induced anchorage-independent cell growth, using Crkl-deficient cells or a pY463 synthetic peptide. Although many cells generally require cell-matrix adhesion, our results demonstrate that CRKL permits cells to bypass the strict need for adhesion in response to FGF8 through direct interaction with receptor.
The 22q11 gene CRKL encodes an adapter protein consisting of one SH2 and two SH3 domains (39). Mouse models of 22q11 deletion (del22q11) syndrome, clinically known as DiGeorge syndrome, predict that a reduced gene dosage of CRKL significantly contributes to the pathogenesis of this contiguous gene syndrome (10, 11). While the entire spectrum of the physiological functions that CRKL mediates during development is unclear, our previous studies indicated an involvement of CRKL protein in pathways induced by fibroblast growth factor 8 (FGF8) and cell-matrix adhesion (19, 24).
FGF8 is required for many critical processes in vertebrate development (21). The FGF8 splice isoform b (FGF8b) displays more potent biological activity than the a-isoform in vivo and is essential for normal vertebrate gastrulation and heart development (6, 9, 13). Recent structural and functional studies indicate that FGF8b is a high-affinity ligand for the c-isoform of FGFR1, FGFR2, and FGFR3, as well as FGFR4 (26, 44). It has been a challenge to precisely map physiologically relevant interactions between ligand and receptor isoforms during pharyngeal development affected by del22q11 syndrome. Close correlation between phenotypes resulting from a deficiency of Fgf8 and mutations in Fgfr1 or Fgfr2, however, suggests that these receptors at least partly mediate Fgf8-induced signaling events in vivo (27, 28, 30, 33, 40, 43).
A recent study of FGF receptor 1 (FGFR1) revealed a regulated sequence of autophosphorylation (7). One of the tyrosine residues autophosphorylated at an early stage of receptor activation is Y463. This tyrosine residue when phosphorylated was initially reported as an SH2 domain binding site for CRK, a protein closely related to CRKL (18). Previous studies suggest that CRK and CRKL SH2 domains may associate with a similar spectrum of tyrosine phosphorylated proteins (reviewed in reference 5). In contrast to this common view, however, we found that FGF8-stimulated FGFR1 and FGFR2 associate preferentially with the SH2 domain of CRKL rather than CRK (24).
In order to reconcile previous findings and to provide novel insight into the mechanism by which FGF8 exerts its functions through CRKL, we have taken advantage of both structural and functional approaches. We used molecular-dynamics (MD) simulations to explore potential interactions of the SH2 domain of CRK and CRKL with the autophosphorylation sites of FGFR1. As predicted by MD simulations, saturation binding experiments confirm specific binding between the CRKL SH2 domain and a phosphorylated Y463 peptide at an affinity more than 1 order of magnitude stronger than that of CRK. Furthermore, we demonstrate that FGF8 initiates a CRKL-dependent pathway to activate the small G proteins Rac1 and Cdc42, as well as their effector PAK. This pathway is a feed-forward loop that feeds into Raf1 and Mek1 downstream of Ras, whereas Ras is activated by FGF8 through a CRKL independent pathway. We show that disruption of CRKL abrogates the ability of FGF8 to sustain anchorage-independent growth. The unique ability of FGF receptors to recruit CRKL therefore bypasses the critical need for cell-matrix adhesion. Our findings importantly imply that CRKL-dependent pathways may permit cell migration or tissue remodeling upon activation of FGFR1 and/or FGFR2 by allowing the cell to leave the site where it has relied upon previously established stable cell-matrix adhesion without compromising function or survival.
MATERIALS AND METHODS
MD simulations.
MD simulations were performed by updated software and hardware, while the methodology was described previously (37). The nuclear magnetic resonance (NMR) structure of the full-length human CRK (isoform a; also known as CRK-II) in nonphosphorylated form, as well as that of a partial protein (residues 1 to 228 of isoform a) in phosphorylated form, was obtained from the RCSB Protein Data Bank (PDB ID = 2EYZ and 2DVJ, respectively [http://www.rcsb.org/]) (15). All MD simulations were performed by Amber8.0 on a system equipped with the RIKEN MDGRAPE-3 special purpose computer for MD simulations (38). The all-atom point charge force field AMBER ff03 was chosen to represent the protein in the present study. All other parameters were used as described previously (37). After 10-ns MD simulations, the average structures of the full-length CRK and CRKL were calculated based on snapshots every 10 ps between 7 and 10 ns.
Free energy calculation.
To evaluate potential intramolecular interaction between the SH2 domain and phosphotyrosine containing sequences, we first modeled interaction between the structure of the SH2 domain obtained above and the amino acid sequence that includes a phosphotyrosine residue. The averaged structures of phosphorylated CRK and CRKL were obtained during the last 3 ns of MD simulations performed for a period of 10 ns and used as template structures. For simulation of the interaction between the SH2 domain to a pY peptide, the amino acid residues surrounding the pY221 or pY207 for CRK or CRKL in the structure obtained above were replaced with appropriate residues in pY peptide being tested (Table 1), and the structure of the pY peptide is energy minimized with atom position fixing of the SH2 domain. These model structures were optimized by MD simulations for a period of 5 ns. The trajectories were recorded every 10 ps during the last 1 ns period (during ca. 4 to 5 ns). We then applied the molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) method to estimate the binding free energy as previously described (37). The dielectric constants inside and outside the molecule were 4.0 and 80.0, respectively.
TABLE 1.
Calculated binding free energy
| Potential binding sitea | Avg free energy (ΔG) ± SDb
|
|
|---|---|---|
| CRK SH2 | CRKL SH2 | |
| FGFR1 autophosphorylation sites | ||
| pY463 (GVSEpYELPEDPRWELPR) | −231.44 ± 21.90 | −343.99 ± 32.63 |
| pY583/pY585 (PGLEpYCpYNPSHNPEEQLSS) | −113.38 ± 39.06 | −220.74 ± 27.41 |
| pY653/pY654* (HHIDpYpYKKTTNGRLPVKW) | ND | ND |
| pY730 (TNELpYMMMRDCWHAVPS) | −268.08 ± 29.44 | −379.22 ± 18.95 |
| pY766 (SNQEpYLDLSMPLDQYSP) | −167.49 ± 24.39 | −247.40 ± 17.28 |
| CRK/CRKL internal sites | ||
| p221 in CRK (EPGPpYAQPSVNTPLPNL) | −262.85 ± 39.02 | ND |
| p207 in CRKL (PAHApYAQPQTTTPLPAV) | ND | −268.40 ± 13.56 |
Amino acid sequences used for simulation are given in single-letter code in parentheses; pY (indicated in boldface) denotes a phosphotyrosine. *, Y653/Y654 locate within the activation loop of the catalytic domain and are therefore omitted in the current analysis.
The values are shown as averages from structures extracted every 10 ps between 4 and 5 ns of simulations in kcal/mol. ND, not determined.
Peptides and recombinant protein synthesis.
Peptides were purchased from GenScript or synthesized and purified by liquid chromatography-mass spectrometry. The purity of all peptides used was >95%. Peptides used in saturation binding experiments were labeled with biotin at the N terminus.
Recombinant glutathione S-transferase (GST) fusion proteins were induced with IPTG (isopropyl-β-d-thiogalactopyranoside) in the Escherichia coli strain BL21(DE3) (for human CRKL or CRK SH2 domain) or DH5α (for Raf1-RBD or Pak1-RBD). After induction, bacterial cells were resuspended and sonicated in phosphate-buffered saline (PBS) containing 0.5 mM dithiothreitol, 10 μg of lysozyme (Sigma)/ml, and protease inhibitor cocktail (Sigma). After Triton X-100 extract, bacterial lysate was cleared by centrifugation at 12,000 × g for 10 min at 4°C. GST fusion proteins were coupled to glutathione-Sepharose 4B (GE Life Sciences) and used for pulldown experiments immediately. For saturation binding experiments, GST fusion proteins were purified using a glutathione-Sepharose 4B column with elution buffer (10 mM reduced glutathione in 50 mM Tris [pH 8.0]) and dialyzed against conjugate buffer (0.1 M sodium carbonate [pH 9.3]). The purity of GST fusion proteins was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by Coomassie blue staining. In order to remove GST from the SH2 domain, we used immobilized thrombin (Sigma) according to the manufacturer's protocol. Cleaved GST and undigested GST fusion proteins were removed by glutathione-Sepharose 4B columns. Since lyophilized proteins gave higher nonspecific backgrounds in our experiements, protein preparations were divided into aliquots and stored at −70°C.
Saturation binding assays.
Saturation binding experiments were designed to estimate the affinity of interaction between the SH2 domain and immobilized peptide. The CRKL SH2 or CRK SH2 domain (with or without GST) was labeled using Cy3 monoreactive dye (protein array grade; GE Life Sciences) according to the manufacturer's protocol. Labeled proteins were separated from unincorporated dye and eluted in 20 mM HEPES (pH 7.4)-100 mM KCl using Sephadex G-25 columns (Illustra NAP-10; GE Life Sciences). The Cy3-labeled protein concentration was calculated from the A280 by a predicted molecular extinction coefficient for each protein with Cy3 absorbance at 552 nm taken into account using the following formula: [Cy3-bound protein] (M) = (A280 - 0.08A552)/ɛ, where the molar extinction coefficient of the protein is expressed as ɛ (M− cm−). The dye-to-protein ratio was 0.8.
For binding experiments, 96-well microtiter plates coated with streptavidin (Nunc Immobilizer) were used to immobilize 20 pmol of biotin-labeled peptide per well. Cy3-labeled GST-SH2 fusion proteins or cleaved SH2 domains were diluted at increasing concentrations in binding buffer (20 mM HEPES [pH 7.4], 100 mM KCl, 0.1% Tween 20, 1% bovine serum albumin [BSA], 1 mM dithiothreitol) and then added to the plate. After 1 h of incubation, followed by three washes with binding buffer, the amount of bound protein was determined in a fluorescent plate reader (Perkin-Elmer VICTOR2) using a Cy3-compatible excitation/emission filter set. Binding data were analyzed in the computer program Prism 5 (GraphPad Software) by global nonlinear regression for total and nonspecific binding data with the assumption of 1:1 binding. In experiments using GST-SH2 domains, the affinity was calculated by using half the total monomer concentration as the dimer concentration based on two assumptions: all GST fusion molecules are dimerized (assumption 1) and, at a sufficiently low density of immobilized peptide, one dimer binds to a single peptide to negate the possibility of avidity effects (assumption 2) (17). These assumptions seemed to hold true when we compared the dissociation constant (KD) values obtained with GST fusion or thrombin-cleaved SH2 domains in our experimental conditions (data not shown). However, the values presented here are from experiments using thrombin-cleaved SH2 domains.
Cells, growth factors, and suspension culture.
The mouse embryonic fibroblasts (MEFs) used in the present study have been described previously (19, 24). Cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% bovine calf serum in 5% CO2 in a humidified incubator at 37°C. Cells were starved for serum for 16 h and then harvested by trypsin-EDTA treatment, followed by the use of soybean trypsin inhibitor (Sigma). Cells were then suspended in serum-free DMEM (1.5 × 107 cells in 30 ml) and plated onto a 150-mm plate coated with 1.2% agarose to keep them in suspension in a CO2 incubator for 1 h prior to treatment with either mouse Fgf8b (R&D Systems) or human epidermal growth factor (EGF; Sigma). After 10 min of incubation with growth factor, cells were lysed in lysis buffer (15% glycerol, 1% NP-40, 50 mM Tris [pH 7.4], 0.2 M NaCl, 2.5 mM MgCl2), including protease inhibitor cocktail (Roche Applied Science), 1 mM sodium orthovanadate, and 10 mM NaF. Cell lysates were used immediately for the biochemical experiments described below. For some experiments, PEP-1 (a 21-amino-acid peptide carrier) was used to deliver pYELP or control peptides into the cell (25). The phosphorylated or control peptide was preincubated with PEP-1 for 30 min at a molar ratio of 1:20. The peptide mix was then added to the cell at the start of the suspension culture.
In some experiments, the 293 human embryonic kidney cells were used for transient expression of the wild type and FGFR1 Y463F mutant. Expression vectors of these FGFR1 are generous gifts from Moosa Mohammadi (2). Transfection was carried out with Effectene (Qiagen) according to the manufacturer's protocol, using 0.5 g of DNA per 60-mm culture plate 24 h before serum starvation. Serum starvation, suspension culture, and Fgf8 stimulation were carried out as described above.
Coimmunoprecipitation, pulldown assays, and immunoblot analysis.
Coimmunoprecipitation, pulldown assays, SDS-PAGE, and immunoblot analysis were performed as previously described (19, 24). Rabbit polyclonal antiactin (C-11), anti-Cdc42 (P1), anti-CRKL (C-20), anti-DOCK180 (C-19), anti-FGFR1 (C-15), and anti-FGFR2 (C-17) antibodies, as well as antiphosphotyrosine monoclonal antibody (PY99), were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-phospho-Erk p44/42 (pTEpY) (E10), anti-Pak, anti-phospho-S338 Raf-1, rabbit polyclonal anti-Erk, anti-phospho-T423/403 Pak, and anti-Raf1 antibodies were obtained from Cell Signaling Technology. Mouse monoclonal anti-Ras, anti-Rac1, and anti-MEK were purchased from BD Transduction Laboratories. Rabbit polyclonal anti-phospho-S222 MEK1, anti-phospho-S298 polyclonal antibody were purchased from Invitrogen. Mouse monoclonal anti-p130Cas (BD Biosciences), anti-CRKL (Upstate Biotechnology), and anti-FGFR1 (Zymed) antibodies were used for compatible combinations of antibody host species between immunoprecipitations and immunoblots.
In vitro kinase assays.
Specific antibodies against Pak, Raf1, or Mek1 were added to cell lysate containing 0.5 mg of total proteins. The kinase-antibody complex was precipitated with protein A-agarose beads. Beads were washed once with lysis buffer, twice with PBS containing 2 mM sodium orthovanadate and 1% Triton X-100, once with 100 mM HEPES (pH 7.4), and once with kinase buffer (12.5 mM HEPES [pH 7.4], 12.5 mM β-glycerophosphate, 7.5 mM MgCl2, 0.5 mM EGTA, 0.5 mM NaF, 0.5 mM sodium orthovanadate). A total of 40 μl of kinase buffer containing 4 μg of bovine myelin basic protein (MBP; Sigma) and 4 μCi of [γ-32P]ATP as substrates were added, and the mixtures were incubated at 30°C for 15 min. Kinase reactions were terminated by the addition of an equal volume of 2× Laemmli sample butter, and proteins were separated by SDS-PAGE. The gel was dried, and incorporation of radioactivity on to MBP was quantitated in a phosphorimager.
Colony-forming assays.
To evaluate the ability of cells to grow without cell-matrix adhesion, cells were plated in soft low-melting-point agarose (SeaPlaque). A base layer was prepared with 1.4% agarose (2 ml per well in a six-well plate) in DMEM containing 10% fetal bovine serum with or without 10 ng of Fgf8/ml. Cells were suspended into 0.3% top agarose in DMEM plus serum with or without Fgf8 at a final density of 105 cells/ml of top agarose/well. At 14 days after plating, colonies were stained with ethidium bromide for 15 min at 37°C and photographed on a UV illuminator. Experiments were repeated four times, each time in triplicate. In some experiments, pYELP and carrier peptide PEP-1 were included in both base and top agarose.
RESULTS
MD simulations for the protein structure of CRK and CRKL.
A previous study reported an NMR structure of the full-length nonphosphorylated CRK (CRK isoform a [also known as CRK-II]) and a partial structure (residue 1 to 228) of phosphorylated CRK (15). The entire structure of CRKL has not been solved, while a crystal structure of the second SH3 domain has been reported (12). In order to provide structural insight into the interaction between CRKL and the FGF receptor, we first calculated the three-dimensional structures of full-length CRK and CRKL in their nonphospholyrated and phosphorylated forms by MD using the NMR structure recently determined for nonphosphorylated and phosphorylated CRK as templates.
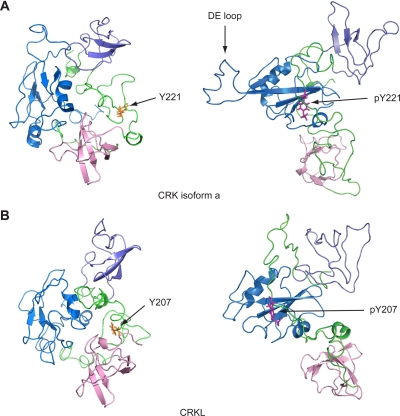
Over the course of MD simulations for 10 ns, the root mean square deviation of the whole protein, as well as that of each domain, stabilized, thus indicating that we have obtained stable structures for both CRK and CRKL (results not shown). The predicted structure of CRKL is in general agreement with features in the NMR structures of CRK in nonphosphorylated and phosphorylated forms (Fig. 1). Some structural differences between CRK and CRKL are notable in linker regions between the SH domains, as well as in the DE loop region (which exists only in CRK SH2 domain, not in CRKL), while the core structure of each domain and relative positions of domains are maintained (Fig. 1; also see the supplemental material). The conformational change in the DE loop in relation to intramolecular association between pY221 and the CRK SH2 domain is consistent with previous reports (1a, 3).
FIG. 1.
Full-length structures of CRK and CRKL predicted by MD simulation. (A) Structure of full-length CRK (isoform a) with or without phosphorylation at Y221. (B) Structure of CRKL with or without phosphorylation at Y207. These structures were obtained by MD simulations as the average structure of 7- to 10-ns simulations (shown in ribbon representations). The SH2, SH3n, and SH3c domains are highlighted by blue, pink, and slate, respectively. The linker sequences between domains are shown in green. The internal phosphotyrosine residue (pY221 and pY207 in CRK and CRKL, respectively) is shown in magenta; the tyrosine residues without phosphorylation are shown in yellow.
Simulated interactions of CRK/CRKL SH2 domains with FGFR1 autophosphorylation sites.
The three-dimensional structures of CRK and CRKL in their phosphorylated forms obtained above have provided us with useful templates to further simulate interactions between the SH2 domain and its potential target sequences. Previous mapping studies identified seven autophosphorylation sites: Y463, Y583, Y585, Y653, Y654, Y730, and Y766 (22). In order to predict the possible interaction of the CRK or CRKL SH2 domain with these autophosphorylation sites, we calculated the free energy (ΔG) in silico (Table 1). The binding free energy is calculated as −344 kcal/mol for binding between the CRKL SH2 domain and the peptide including pY463, whereas it is −231 kcal/mol for binding between the CRK SH2 domain and the same amino acid sequence. Likewise, the ΔG was estimated as −379 kcal/mol for binding between the pY730 sequence and the CRKL SH2 domain, while it was −268 kcal/mol for the CRK SH2 domain. These results therefore predict strong interaction of pY463 and pY730 with the CRKL SH2 domain, rather than that of CRK.
A recent study demonstrated that autophosphorylation occurs precisely in the order of pY653, pY583, pY463, pY585, and pY654 before the receptor is fully activated, whereas phosphorylation at Y730 is not detected (7). Autophosphorylation at Y730 may still occur at low stoichiometry as reported previously (22). However, the crystal structure of the FGFR1 dimer (PDB identification, 1fgk) indicates that Y730 is hidden between two receptor monomers juxtaposed, while Y463 is easily accessible (23). In addition, the local residues that surrounds Y463, but not Y730, is in agreement with the previously reported amino acid consensus for CRK and CRKL SH2 domain binding, YXXP (5). These observations suggest that it is more likely that pY463, rather than pY730, is a high-affinity binding site for the CRKL SH2 domain.
Simulated interactions of CRK/CRKL SH2 domains with the internal tyrosine phosphorylation site.
The internal tyrosine residue Y221 in CRK becomes a binding site for the SH2 domain when phosphorylated (3, 15, 31). Consistent with a similarity between the amino acid sequences surrounding Y221 in CRK and Y207 in CRKL, our MD simulations demonstrate that intramolecular interaction between pY207 and the SH2 domain is also possible (Fig. 1). The ΔG values calculated by the MM-PBSA method were −263 kcal/mol and −268 kcal/mol for CRK-pY221 versus CRKL-pY207 binding, respectively (Table 1). Since these ΔG values predict weaker interactions than that of phosphorylated Y463 in FGFR1 noted above, recruitment of CRKL to ligand-stimulated FGFR1 may still take place at a higher affinity even when the internal Y207 is phosphorylated.
Characterization of interactions of CRK/CRKL SH2 domains with FGFR1 pY463 in silico.
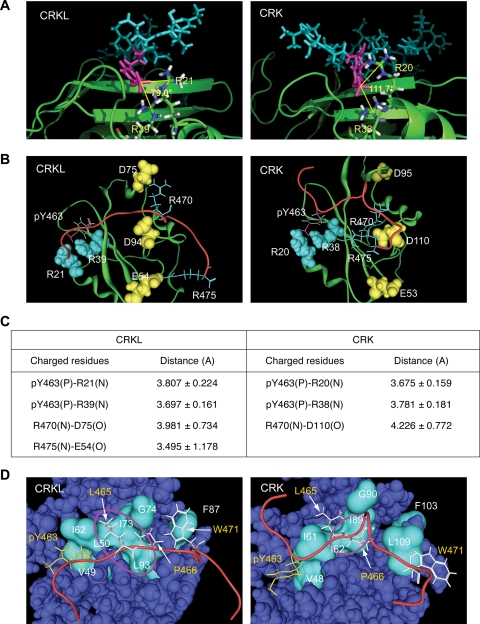
Component analysis indicates that a major determinant of binding between pY463 sequence and the SH2 domain is electrostatic attraction (data not shown). In general agreement with phosphotyrosine-SH2 domain interactions, two arginine residues R21 and R39 in CRKL interact with phosphorylated Y463, while R20 and R38 in CRK have similar electrostatic interaction with pY463 (Fig. 2A; also see the supplemental material). We have noted that two arginine residues, R470 and R475, in the proximity of pY463 participate in peptide binding to the CRKL SH2 domain through their interaction with two negatively charged residues in the SH2 domain, D75 and E54, respectively (Fig. 2B). In contrast, although R470 seems to interact with D110 in the CRK SH2 domain, the distance of this interaction is greater than that of the CRKL SH2 domain, thus electrostatic force is weaker (Fig. 2B; Fig. 2C summarizes the distance between charged residues that interact).
FIG. 2.
Structural basis of interaction between CRKL SH2 domain and pY peptide. (Structural information is available in the supplemental material.) (A) Electrostatic interaction between pY463 in the peptide sequence and two arginine residues in the SH2 domain. pY463 is shown in magenta. SH2 domains are shown in ribbon representations in green, except that two arginine residues interacting with the phosphotyrosine are shown in a stick model. The number shown between the two lines is the angle formed by the central carbon atom in the guanidinium group in the two arginine residues and the phosphorus atom in the phosphate group in pY463 to illustrate qualitative differences in the way that CRKL and CRK SH2 domains interact with the phosphotyrosine in the peptide. (B) Interaction of charged residues between the SH2 domain and FGFR1 peptide sequence. The SH2 domain of CRK and CRKL are colored in green in a ribbon representation. The backbone of the phosphorylated Y463 peptide sequence is shown in red. Charged residues in the peptide sequence and SH2 domain are shown in stick or space fill models, respectively. The phosphate group in pY463 is shown in red. Negatively and positively charged amino acids are shown in yellow or light blue, respectively. (C) Summary table for the distance between two atoms involved in electrostatic interactions. (D) Hydrophobic interactions between the SH2 domain and FGFR1 peptide sequence. The backbone of the pY peptide is shown in red. Hydrophobic residues in the peptide sequence are shown in stick models; pY463 in yellow and the others in white. Hydrophobic residues along the peptide binding area in the SH2 domain are shown in cyan in the surface model. Hydrophobic pockets are highlighted by a circle in magenta.
We have also noted a hydrophobic pocket in the CRKL SH2 domain that fits two hydrophobic residues L465 and P466 in the pY463 peptide (Fig. 2C). The position of this hydrophobic pocket seems to allow additional hydrophobic interactions between the aromatic ring of pY463 and V49, as well as between W471 and F87 (Fig. 2C). Although the CRK SH2 domain has a similar hydrophobic pocket, it is smaller than that of CRKL, and the two hydrophobic residues L465 and P466 of the FGFR1 peptide do not fit to the hydrophobic surface of the CRK SH2 domain. Moreover, unlike the CRKL SH2 domain, the aromatic ring of pY463 or W471 does not appear to participate in hydrophobic attractions in the CRK SH2 domain. These qualitative and quantitative differences noted above in electrostatic and hydrophobic attractions at least partly explain the overall differences in calculated binding free energy of the pY463 sequence with CRKL SH2 versus that of CRK.
Physical interaction of CRKL and CRK SH2 domains with FGFR1 pY463 peptide.
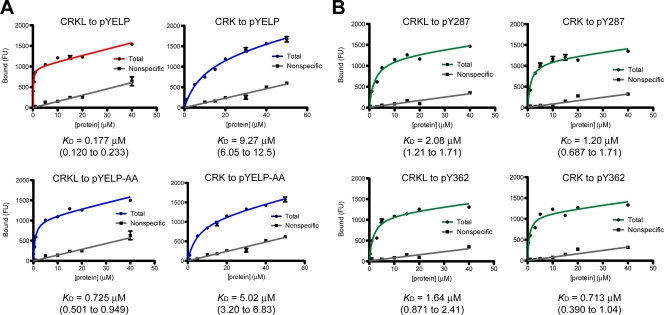
In order to provide evidence for physical interaction between the CRKL SH2 domain and pY463 predicted above, we conducted saturation binding experiments between the SH2 domain of CRK or CRKL and a synthetic 17-amino-acid peptide including pY463 at the +5 position, GVSEpYELPEDPRWELPR (abbreviated as pYELP peptide). In our initial experiments, we used tetramethylrhodamine-labeled pYELP peptide to determine its affinity to immobilized GST-SH2 domains without success due partly to high nonspecific binding (not shown). We then used immobilized pYELP peptide on the surface and incubation with increasing concentrations of Cy3-labeled CRKL or CRK SH2 domain (Fig. 3). We have conducted several experiments using GST fusion of these domains or SH2 domains after thrombin cleavage. While GST fusion proteins exist as dimers, it is assumed that each dimer binds to a single phosphopeptide when the density of immobilized peptide is sufficiently low (17). Consistent with this assumption, removal of GST from SH2 domains did not affect the KD values in our experimental conditions (not shown). These results indicate that pY463 is a high-affinity binding site for the SH2 domain of CRKL, while the CRK SH2 domain exhibits more than an order of magnitude weaker affinity to the same phosphopeptide (Fig. 3A). From five independent repeats of this experiment, we obtained mean (± the standard error) KD values of 0.235 (± 0.033) and 6.91 (± 1.13) μM for the CRKL and CRK SH2 domains, respectively (repeat data not shown). As expected, the CRKL or CRK SH2 domain did not show specific binding to a control peptide without phosphorylation at Y463 (not shown).
FIG. 3.
Saturation binding experiments for interactions between the SH2 domain of CRKL or CRK and phosphotyrosyl peptides. (A) Representative plots of saturation binding data between the SH2 domain of CRKL or CRK and a peptide corresponding to FGFR1 pY463 (pYELP peptide) or a modified pYELP peptide (pYELP-AA), in which R470/475 are replaced with alanine residues. (B) Representative plots of saturation binding data between CRKL or CRK SH2 domain and a peptide corresponding to BCAR1 pY287 and pY362 peptides. Experiments were carried out using Cy3-labeled SH2 domains after cleavage of GST by thrombin. The x axis shows the concentration of Cy3-labeled SH2 domain, and the y axis shows the arbitrary fluorescent unit (FU) that corresponds to the amount of bound protein. The KD values are shown below the plots. The 95% confidence interval is shown in parentheses.
MD simulations suggested that two arginine residues, R470/475, may provide additional electrostatic force for the association of the CRKL SH2 domain to autophosphorylated Y463 in FGFR1 (Fig. 2). To validate this prediction, we compared the affinity of CRKL and CRK SH2 domains to a mutated pYELP peptide in which the two arginine residues, R470/475, were replaced with alanine residues (Fig. 3A). The replacement made the affinity of the pYELP peptide significantly weaker (KD = 0.177 versus 0.725 μM, with 95% confidence intervals of 0.120 to 0.233 versus 0.501 to 0.949 μM, respectively). Although the alanine replacement may have an opposite effect for the CRK SH2 domain, the results were not statistically different (KD = 9.27 versus 5.02 μM, with 95% confidence intervals of 6.05 to 12.5 versus 3.20 to 6.83 μM, respectively). Nevertheless, these results provide evidence that amino acid residues downstream of the core binding motif YXXP contribute to the specificity of phosphotyrosine-mediated interactions of two proteins.
Although unlikely, the greater affinity of the CRKL SH2 domain compared to that of CRK could be explained by the possibility that Cy3 labeling may selectively affect the ability of CRK SH2 domain (rather than CRKL SH2 domain) to bind phosphorylated peptides. To address this issue, we conducted control experiments using the same preparations of Cy3-labeled SH2 domains for their binding to phosphorylated peptides from BCAR1 (p130CAS: CRK-associated substrate) (Fig. 3B). p130CAS is a scaffold protein that associates with both CRK and CRKL when phosphorylated (5) and has multiple YXXP motifs phosphorylated by SRC family tyrosine kinases (32, 34). When the same SH2 domain preparations were used in the experiment shown in Fig. 3A, the pY287 peptide (PLLEVpYDVPPSVEKG) exhibited KD values of 2.08 and 1.20 μM, and the pY362 peptide (AEDVpYDVPPPA) showed KD values of 1.64 and 0.713 μM to the CRKL or CRK SH2 domain, respectively (Fig. 3B). These results demonstrated that two phosphorylated p130CAS peptides corresponding to Y287 and Y362 showed similar affinity values for both CRKL and CRK SH2 domains, thereby contrasting the high-affinity interaction of CRKL SH2 domain to pY463 in FGFR1.
Y463 is essential for the physical association of CRKL to FGF8-stimulated FGFR1.
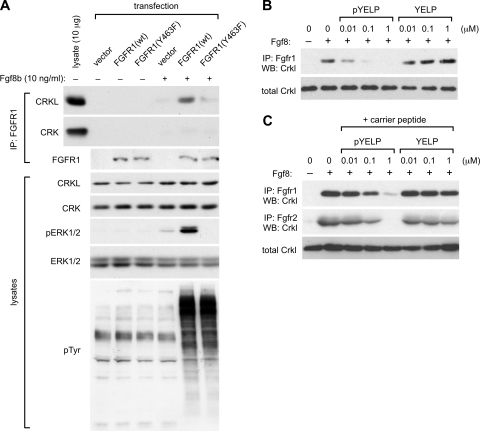
In order to provide further evidence that pY463 in FGFR1 is a binding site for the CRKL SH2 domain rather than CRK, we determined whether a mutant FGFR1 lacking Y463 could physically associate with CRKL in the cell. In this experiment, we transfected human embryonic kidney 293 cells transiently with expression vectors of FGFR1. When we transfected cells with high levels of FGFR1, cells detached from the plate either with wild-type or mutant FGFR1 even without Fgf8 stimulation (data not shown). Thus, we used half the dose of plasmid recommended in the manufacturer's transfection manual in our experiments. With this dosage, we observed comparable levels of Fgf8 induced an increase in the overall cellular phosphotyrosine either with wild-type or mutated FGFR1 (Fig. 4A). Conveniently, 293 cells express little endogenous FGFR1 at levels difficult to detect in immunoblots and respond poorly to Fgf8b (Fig. 4A). This cell line therefore seemed ideal for our experiments utilizing expression of exogenous FGFR1 which can be stimulated by Fgf8b. In addition, we minimized the effects of the adhesion-dependent pathways that CRKL mediates by stimulating cells in suspension.
FIG. 4.
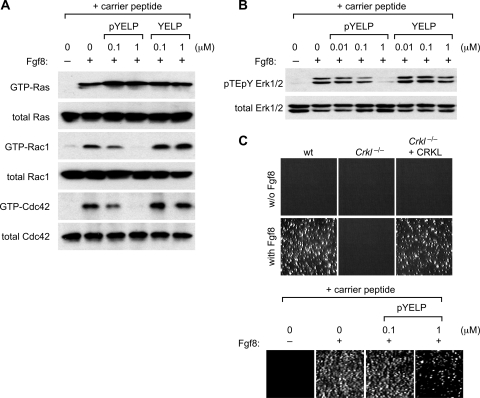
The physical association of CRKL to Fgf8-stimulated FGFR1 depends on Y463. (A) Coimmunoprecipitation of CRKL and FGFR1. Human embryonic kidney 293 cells were transfected transiently with expression vectors of wild-type (wt) or mutant FGFR1 lacking Y463 (Y463F). Suspension culture was stimulated with 10 ng of Fgf8/ml for 10 min, and then cell lysates were prepared. Fgfr1 was immunoprecipitated (IP: FGFR1). Coimmunoprecipitation with CRKL or CRK was assessed by immunoblotting. The leftmost lane includes 10 μg of total protein in lysate as a control in order to estimate the amount of associated CRKL or CRK protein relative to the total amount in lysates. In parallel experiments, cell lysates (lysates) were evaluated by SDS-PAGE and immunoblotted with the antibody indicated. (B) pYELP peptide inhibits association of Crkl with Fgfr1. A suspension culture of wild-type MEFs was stimulated with 10 ng of Fgf8/ml for 10 min, and then cell lysates were prepared. Peptide was added to cell lysates at different concentrations, and Fgfr1 was immunoprecipitated. Coimmunoprecipitation with Crkl was assessed by immunoblotting. (C) pYELP peptide inhibits in vivo association of Crkl with Fgf8-stimulated Fgfr1. Suspension cultures of wild-type MEFs were pretreated with either pYELP or YELP peptide mixed with the carrier peptide PEP-1 and then stimulated with 10 ng of Fgf8/ml for 10 min. Cell lysates were prepared, and Fgfr1 or Fgfr2 was immunoprecipitated. Coimmunoprecipitation of Fgfr1 with Crkl was assessed by immunoblotting.
Using these conditions, we found that Y463F mutant receptor showed poor CRKL association after receptor stimulation with Fgf8b at a level comparable to that of the vector control, whereas CRKL was coimmunoprecipitated efficiently with wild-type FGFR1 (Fig. 4A). Although both CRKL and CRK were easily detectable in 293 cell lysates, CRK did not seem to form a stable complex with FGFR1 with or without Y463F mutation (Fig. 4A). Although we could detect CRK association with a longer exposure, consistent with a weaker affinity of the CRK SH2 domain with pY463 shown above, there was little difference between wild-type and Y463F mutant receptors (data not shown). These results are consistent with the high-affinity interaction between the CRKL SH2 domain and pYELP peptide. Interestingly, Fgf8-induced phosphorylation of ERK1 and ERK2 was also dependent on Y463 after Fgf8b stimulation (Fig. 4A). While there is a relatively low level of response to Fgf8 in vector control, expression of the Y463F mutant may be inhibitory.
To provide further evidence that pY463 is important for association of CRKL to the receptor complex, we determined whether pYELP peptide could inhibit their association. In the first experiment, MEFs were starved for serum and stimulated with Fgf8b for 10 min. Each peptide was added in vitro to cell lysates prepared at the end of Fgf8 stimulation. Consistent with our results noted above, pYELP peptide blocked association of Crkl to the Fgf8-stimulated Fgfr1 in a dosage-dependent manner in vitro (Fig. 4B). In the second experiment, pYELP or control peptide was applied to MEFs in culture using the carrier peptide PEP-1 (25) before cells were stimulated with Fgf8b. The cell lysates were then prepared, and FGF receptors were immunoprecipitated. pYELP peptide blocked association of endogenous Crkl with Fgfr1 and Fgfr2 after Fgf8 stimulation in the cell (Fig. 4C). These results demonstrate that pYELP peptide inhibits association of CRKL with FGFR1 and FGFR2 not only in vitro but also in live cells at relatively low concentrations.
Fgf8 activates the small G-proteins Ras, Rac, and Cdc42 without adhesion.
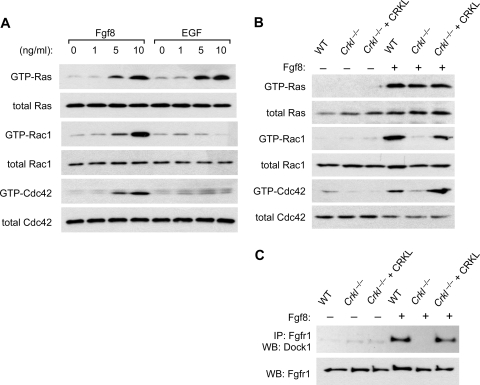
Crkl participates in not only FGF8-induced pathways but also cell adhesion-associated signaling pathways (19, 24). We previously found that localization of Crkl to focal adhesions is sufficient to activate the small G proteins Rac1 and Cdc42, but not Ras. In order to minimize the effects of such adhesion-dependent pathways that CRKL mediates, we evaluated FGF8 or EGF-induced signals while MEFs are suspended in tissue culture medium (Fig. 5). As expected, EGF activated Ras, while it had little effect on GTP-loading of Rac1 or Cdc42 without adhesion (Fig. 5A). In contrast, Fgf8 efficiently stimulated GTP loading of not only Ras but also Rac1 and Cdc42. Fgf8-induced activation of Rac1 and Cdc42 was dependent on Crkl, while Ras was similarly activated in the presence or absence of Crkl in MEFs (Fig. 5B). Dock1 (also known as Dock180) is a member of the CZH family of unconventional guanine nucleotide exchange factors for Rac and Cdc42 lacking the Dbl domain (reviewed in reference 20). We found that association of endogenous Dock1 with the receptor complex is dependent on Crkl in MEFs stimulated with Fgf8 (Fig. 5C). Therefore, our results indicate that CRKL is essential for the assembly of signaling components immediately adjacent to Fgf8-stimulated Fgfr1, leading to activation of the small G proteins Rac1 and Cdc42.
FIG. 5.
Crkl is essential for Fgf8-induced activation of Rac1 and Cdc42, but not Ras. (A) Pulldown assays for GTP-bound small G proteins. Wild-type MEFs were stimulated with either Fgf8 or EGF, while cells were kept in suspension. Cell lysates were prepared after 10 min of stimulation. The levels of total Ras, Rac1, and Cdc42 are shown in the lower panel for each GTP-bound G protein pulldown. (B) Pulldown assays for GTP-bound small G proteins after 10 min of stimulation with 10 ng of Fgf8/ml in suspension culture of MEFs isolated from wild-type or Crkl−/− embryos. An additional control was prepared by introducing a CRKL transgene into Crkl−/− MEFs (Crkl−/− + CRKL). A pulldown assay of GTP-bound small G-proteins was carried out as described above. (C) Coimmunoprecipitation of Fgfr1 with Dock1. Cell lysates were prepared as described above to immunoprecipitate Fgfr1. Dock1 protein was probed in the immunoprecipitate by immunoblotting.
Crkl is required for optimal activity of the serine/threonine kinases Raf1 and Mek induced by Fgf8.
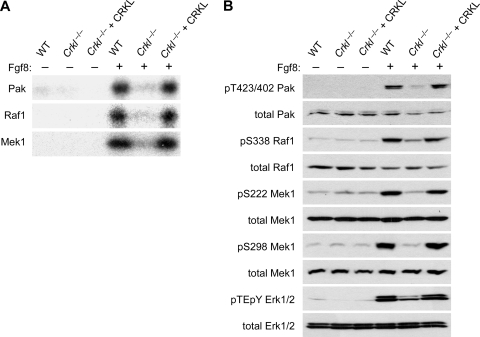
Two classes of protein serine/threonine kinases, RAF (mitogen-activated protein [MAP] kinase kinase kinase) and MEK (MAP kinase kinase), lie directly downstream of the small G-protein RAS and form a pathway to activate the MAP kinases ERK1 and ERK2 (reviewed in reference 16). It has been shown that another class of serine/threonine kinases, PAKs, positively regulate RAF1 and MEK1 through adhesion-dependent phosphorylation at S338 and S298, respectively (4, 14, 35). We therefore determined whether Fgf8 promotes the activity of Pak, Raf1, and Mek1 through Crkl, while cells are kept in suspension in order to dissociate the effect of cell-matrix adhesion from that of receptor-induced pathways. In these conditions, we found that Crkl is indeed required for optimal activity of these kinases when judged by in vitro kinase assays using MBP as an exogenous substrate (Fig. 6A). The profile of kinase activity was also consistent with the phosphorylation status of these kinases as probed by phospho-specific antibodies (Fig. 6B). We found that Fgf8-induced phosphorylation of Pak at T423/T402/T421 (on Pak1, Pak2, and Pak3, respectively), as well as that of Raf1 and Mek1 at S338 and S298, respectively, is dependent on Crkl in the cell (Fig. 6B). Since phosphorylation of these sites is necessary for full activation of these serine/threonine kinases, our result explains why Erk1/Erk2 are not fully activated without Crkl (Fig. 6B), despite the fact that Fgf8 can induce comparable levels of GTP-loaded Ras in the presence or absence of Crkl (Fig. 6B).
FIG. 6.
Crkl is essential for Fgf8-induced activation of the cascades of serine/threonine kinases. (A) In vitro kinase assays for Pak, Raf1, and Mek1. Suspension culture of MEFs was stimulated with 10 ng of Fgf8/ml for 10 min, and each protein was immunoprecipitated from cell lysates for in vitro kinase assays. (B) Immunoblot analysis for Fgf8-induced site specific phosphorylation of Pak, Raf1, Mek1, and Erk1/2. Cell lysates were prepared as described above, and phospho-specific antibodies were used to probe phosphorylation events. Total protein levels of each protein are shown in the lower panel for each protein.
Consistent with the results from our analysis of Crkl-deficient MEFs, pYELP peptide efficiently inhibited Fgf8-induced GTP loading of Rac1 and Cdc42 in the cell without inhibitory effects on Fgf8-induced Ras activation (Fig. 7A). pYELP peptide, but not YELP peptides, was also capable of inhibiting Fgf8-induced Erk activation in wild-type MEFs (Fig. 7B).
FIG. 7.
pYELP inhibits cellular responses to Fgf8. (A) pYELP peptide inhibits Fgf8-induced activation of Rac1 and Cdc42, but not Ras. A suspension culture of wild-type MEFs was pretreated with either pYELP or YELP peptide mixed with the carrier peptide PEP-1, and then cells were stimulated with 10 ng of Fgf8/ml for 10 min. Cell lysates were prepared, and pulldown assays were performed. (B) pYELP peptide inhibits Fgf8-induced Erk1/2 activation. Peptide and Fgf8 treatment was done as described above. Cell lysates were analyzed by immunoblots for phospho-Erk. (C) CRKL is essential for anchorage-independent cell growth induced by Fgf8. Colony-forming assays were performed using MEFs as described in Materials and Methods. Photographs of the colonies are shown.
Fgf8-induced anchorage-independent cell growth depends on Crkl.
Our results presented above suggest that the unique ability of Crkl to mediate Fgf8-induced signals toward Rac1 and Cdc42 activation may be similar to what adhesion signals can provide to the cell. If so, Fgf8 may support anchorage independent cell growth in a manner dependent on Crkl. Indeed, we have found that Crkl−/− MEFs did not form colonies in soft agar in the presence of Fgf8, whereas wild-type MEFs and Crkl−/− MEFs with CRKL transgene expression formed a number of colonies (Fig. 7C). Consistent with the results with Crkl-deficient MEFs, treatment of wild-type MEFs with pYELP peptide also inhibited Fgf8-induced anchorage-independent growth. These results demonstrate that Crkl plays a specific and essential role in mediating Fgf8-induced receptor signals that provide cells with the capacity to bypass the critical need for cell-matrix adhesion.
DISCUSSION
It is an important challenge for biologists to understand the complexity of interactions that evolutionally, genetically, and structurally related proteins mediate with their upstream and downstream targets, particularly when such proteins coexist in the same cell. The 22q11 gene CRKL (CRK-like) is closely related to the 17p13 gene CRK. CRKL is broadly expressed with high levels in neural crest derivatives during mouse development (11). Likewise, CRK is detectable in all tissues thus far examined (Mouse Genome Informatics [http://www.informatics.jax.org]). Despite largely overlapping expression patterns, experimental disruptions of the mouse homologue of either CRKL or CRK results in a homozygous lethal phenotype (11, 29), indicating that the loss of either CRKL or CRK cannot be fully compensated for by the presence of the other gene. Although these observations do not directly contradict the widely accepted notion that these proteins have redundant functions, our previous study demonstrated a specific role for CRKL in FGF8 signaling in the pathogenesis of del22q11 syndrome in mouse models (24). In the present study, we used a multiprong approach to shed light on the mechanisms of signaling specificity that CRKL can provide to FGF8-induced signaling cascades and cellular responses.
Interactions between CRKL SH2 domain and FGFR1 autophosphorylation sites.
Using MD simulations, we demonstrated that the SH2 domain of CRKL has unique structural properties compared to that of CRK. These differences became noticeable when the SH2 domain was simulated to bind the amino acid sequence, including pY463. Predictions from these structures led us to further investigate differences between the SH2 domains of CRKL and CRK. The MM-PBSA method used in the present study has been tested extensively, although it tends to overestimate free energy compared to the affinity experimentally determined (reviewed in reference 8). Nonetheless, this method provides a reliable rank order within a given set of interactions (37); thus, these simulations have provided useful predictions for our purposes to identify likely binding sites for the SH2 domain of CRKL. While other methods are being developed to bring predicted values closer to experimental values, the applicability of such methods to a broader collection of molecules remains to be explored (41, 42).
Despite general similarities between the three-dimensional structures of CRKL and CRK, we noted some features in the SH2 domains when they interact with the FGFR1 amino acid sequence surrounding phosphorylated Y463. Albeit similar, distinct distributions of charged or hydrophobic residues on the surface of the SH2 domain in CRKL and CRK contribute significantly to their binding specificities. It is noteworthy that such atomic attractions are mediated by not only the phosphotyrosine residue but additional residues outside of the core YXXP motif on the C-terminal side of the peptide sequence. Our biochemical data further confirmed influence from two arginine residues R470/475 for the affinity of protein-protein interaction (Fig. 3).
One interesting prediction we can make from our observations is that due to a larger hydrophobic pocket, the CRKL SH2 domain may accommodate a residue with a larger hydrophobic side chain at the +2 position (relative to the phosphotyrosine) in addition to a proline residue at the +3 position. In contrast, the CRK SH2 domain may prefer a sequence with a hydrophobic residue with a smaller side chain at this position. In this regard, it is noteworthy that p130CAS (BCAR1) has 15 YXXP motifs in its substrate domain, many of which have a valine residue at the +2 position. Although our saturation binding experiments indicate a moderate affinity of CRK and CRKL SH2 domains of ∼1 μM to two of these YXXP motifs, the tandem positioning of binding motifs in a single protein molecule likely generates switch like on-off overall binding pattern. Since the side chain of valine occupies a smaller space than that of leucine, it is plausible that YXVP motifs may be compatible with both CRKL and CRK SH2 domains, whereas YXLP motifs such as YELP in FGFR1 may be accommodated more preferentially by the CRKL SH2 domain. Future studies of CRK specific binding proteins, once identified, will provide a more precise structural basis for the selectivity of CRKL- and CRK-mediated pathways in the cell.
As predicted from MD simulations, pYELP peptide corresponding to phosphorylated Y463 offered a high-affinity binding site for CRKL SH2 domain (Fig. 3). Furthermore, effective peptide concentrations found in other experiments correlate closely with our KD estimates (Fig. 4). While these results are consistent with direct binding of the CRKL SH2 domain to the receptor through pY463, it remains possible that other proteins may also bind to phosphorylated Y463. The SH2 domain of NCK1/2 also prefers a proline residue at the +3 position similar to CRK and CRKL SH2 domains (36). However, endogenous Nck did not form stable association with Fgf8-stimulated Fgfr1 in MEFs, although Nck proteins were easily detectable in these cells (data not shown). In addition, our current results do not exclude the possibility that CRKL may associate with the receptor complex indirectly through other pY463 binding proteins. Either directly or indirectly, however, our results indicate that CRKL associates with FGFR1 through autophosphorylation at Y463 and that this residue and the presence of CRKL are essential for full activation of the MAP kinases ERK1 and ERK2 induced by Fgf8.
Essential role of CRKL in an FGF8-induced feed-forward loop.
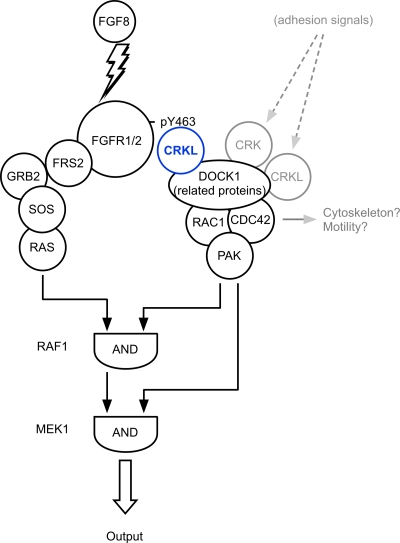
We have demonstrated in the present study that Crkl plays a unique role in a feed-forward loop for Fgf8-induced MAP kinase activation through Dock1, Rac1, and Cdc42 and then Pak (Fig. 8). It has been shown that adhesion-dependent PAK activation is critical for RAF1 and MEK to mediate EGF-induced ERK activation (4, 35). PAK phosphorylates RAF1 and MEK1 at S338 and S298, respectively, both of which are essential for the competence of these kinases to mediate the signal from Ras (14, 35). Therefore, cell-matrix adhesion provides permissive conditions for the cell to respond to growth factors such as EGF. Our study has revealed that CRKL can provide similar permissive conditions for the cell stimulated by FGF8 without relying on cell-matrix adhesion. It is tempting to compare these pathways with a binary logic network in which tandem AND gates can regulate the output (Fig. 8) (1). These results provide insight into the mechanism of genetic interaction between Fgf8 and Crkl that we previously reported in mice (24). Many other modifiers also contribute to regulation of the MAP kinases through this Raf1-Mek1 axis (reviewed in reference 16), although the effects of modifiers could be context dependent in different cell types and with different stimuli.
FIG. 8.
Model of the network that activates the MAP kinase by FGF8. The figure illustrates a role for the feed-forward loop mediated by interaction of CRKL and FGFR1 in organizing the output through the MAP kinase ERK1/2. Note that RAF1 and MEK1 function as tandem AND-gates that integrate signals effectively from FGFR1. Although cell-matrix adhesion can also participate in this network (shaded in gray), cells stimulated by FGF8 can escape from this requirement. This is a simplified model that does not show additional regulatory pathways mediated by other signaling modifiers.
We have found that Crkl is essential for Fgf8 to sustain growth and survival without cell adhesion. It might be interpreted that Fgf8 and Crkl may be important for morphogenetic processes such as cell migration, vascular sprouting/remodeling, and/or epithelial mesenchymal transformation. Previously, we showed that Crkl is also required for Fgf8-induced chemotaxis in culture, as well as expression of the ETS class of genes Pea3 and Erm in mouse embryos (24). Crkl-dependent activation of small G proteins such as Rac1 and Cdc42 triggered by Fgf8 is likely to be important for cytoskeletal rearrangements and cell migration in addition to their contributions to the activation of the MAP kinases Erk1/2. These possibilities are consistent with increased numbers of apoptotic cells in pharyngeal epithelia, splanchnic mesoderm, and decreased neural crest cells observed in mouse embryos deficient for Crkl, particularly when combined with a reduced gene dosage of Fgf8 (24). Future studies will address more precise tissue-specific and context-dependent functions of CRKL during development, as well as in the abnormal cell behavior of cancer cells. Quantitative analysis of protein interactions that CRKL and a related protein CRK provide to the cell will aid future endeavors.
Supplementary Material
Acknowledgments
We thank M. Mohammadi for reagents; M. R. Rosner for sharing protocols for in vitro kinase assays; W.-J. Rhee for peptide synthesis and purification; and B. Liu, P. D. Nash, R. B. Jones, and D. L. Guris for discussion and critical reading of the manuscript.
This study was supported in part by grants from the National Institutes of Health (R01 DE015883) and the American Heart Association (0640109N) to A.I. and by the contracted research Protein 3000 Project by the Ministry of Education, Culture, Sports, Science and Technology of Japan to M.T. A.I. is an Established Investigator of the American Heart Association. J.S. is a Postdoctoral Fellow supported by the Greater Midwest Affiliate of the American Heart Association (0720073Z).
We declare that we have no competing financial interests.
Footnotes
Published ahead of print on 23 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alon, U. 2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8450-461. [DOI] [PubMed] [Google Scholar]
- 1a.Anafi, M., M. K. Rosen, G. D. Gish, L. E. Kay, and T. Pawson. 1996. A potential SH3 domain-binding site in the Crk SH2 domain. J. Biol. Chem. 27121365-21374. [DOI] [PubMed] [Google Scholar]
- 2.Dell'Era, P., M. Mohammadi, and M. Presta. 1999. Different tyrosine autophosphorylation requirements in fibroblast growth factor receptor-1 mediate urokinase-type plasminogen activator induction and mitogenesis. Mol. Biol. Cell 1023-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donaldson, L. W., G. Gish, T. Pawson, L. E. Kay, and J. D. Forman-Kay. 2002. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proc. Natl. Acad. Sci. USA 9914053-14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edin, M. L., and R. L. Juliano. 2005. Raf-1 serine 338 phosphorylation plays a key role in adhesion-dependent activation of extracellular signal-regulated kinase by epidermal growth factor. Mol. Cell. Biol. 254466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feller, S. M. 2001. Crk family adaptors-signalling complex formation and biological roles. Oncogene 206348-6371. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher, R. B., J. C. Baker, and R. M. Harland. 2006. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development 1331703-1714. [DOI] [PubMed] [Google Scholar]
- 7.Furdui, C. M., E. D. Lew, J. Schlessinger, and K. S. Anderson. 2006. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell 21711-717. [DOI] [PubMed] [Google Scholar]
- 8.Gilson, M. K., and H. X. Zhou. 2007. Calculation of protein-ligand binding affinities. Annu. Rev. Biophys. Biomol. Struct. 3621-42. [DOI] [PubMed] [Google Scholar]
- 9.Guo, Q., and J. Y. Li. 2007. Distinct functions of the major Fgf8 spliceform, Fgf8b, before and during mouse gastrulation. Development 1342251-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guris, D. L., G. Duester, V. E. Papaioannou, and A. Imamoto. 2006. Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev. Cell 1081-92. [DOI] [PubMed] [Google Scholar]
- 11.Guris, D. L., J. Fantes, D. Tara, B. J. Druker, and A. Imamoto. 2001. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat. Genet. 27293-298. [DOI] [PubMed] [Google Scholar]
- 12.Harkiolaki, M., R. J. Gilbert, E. Y. Jones, and S. M. Feller. 2006. The C-terminal SH3 domain of CRKL as a dynamic dimerization module transiently exposing a nuclear export signal. Structure 141741-1753. [DOI] [PubMed] [Google Scholar]
- 13.Hutson, M. R., P. Zhang, H. A. Stadt, A. K. Sato, Y. X. Li, J. Burch, T. L. Creazzo, and M. L. Kirby. 2006. Cardiac arterial pole alignment is sensitive to FGF8 signaling in the pharynx. Dev. Biol. 295486-497. [DOI] [PubMed] [Google Scholar]
- 14.King, A. J., H. Sun, B. Diaz, D. Barnard, W. Miao, S. Bagrodia, and M. S. Marshall. 1998. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature 396180-183. [DOI] [PubMed] [Google Scholar]
- 15.Kobashigawa, Y., M. Sakai, M. Naito, M. Yokochi, H. Kumeta, Y. Makino, K. Ogura, S. Tanaka, and F. Inagaki. 2007. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat. Struct. Mol. Biol. 14503-510. [DOI] [PubMed] [Google Scholar]
- 16.Kolch, W. 2005. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell. Biol. 6827-837. [DOI] [PubMed] [Google Scholar]
- 17.Ladbury, J. E., M. A. Lemmon, M. Zhou, J. Green, M. C. Botfield, and J. Schlessinger. 1995. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc. Natl. Acad. Sci. USA 923199-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsson, H., P. Klint, E. Landgren, and L. Claesson-Welsh. 1999. Fibroblast growth factor receptor-1-mediated endothelial cell proliferation is dependent on the Src homology (SH) 2/SH3 domain-containing adaptor protein Crk. J. Biol. Chem. 27425726-25734. [DOI] [PubMed] [Google Scholar]
- 19.Li, L., D. L. Guris, M. Okura, and A. Imamoto. 2003. Translocation of CrkL to focal adhesions mediates integrin-induced migration downstream of Src family kinases. Mol. Cell. Biol. 232883-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meller, N., S. Merlot, and C. Guda. 2005. CZH proteins: a new family of Rho-GEFs. J. Cell Sci. 1184937-4946. [DOI] [PubMed] [Google Scholar]
- 21.Meyers, E. N., M. Lewandoski, and G. R. Martin. 1998. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 18136-141. [DOI] [PubMed] [Google Scholar]
- 22.Mohammadi, M., I. Dikic, A. Sorokin, W. H. Burgess, M. Jaye, and J. Schlessinger. 1996. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol. Cell. Biol. 16977-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohammadi, M., J. Schlessinger, and S. R. Hubbard. 1996. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell 86577-587. [DOI] [PubMed] [Google Scholar]
- 24.Moon, A. M., D. L. Guris, J. H. Seo, L. Li, J. Hammond, A. Talbot, and A. Imamoto. 2006. Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev. Cell 1071-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris, M. C., J. Depollier, J. Mery, F. Heitz, and G. Divita. 2001. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat. Biotechnol. 191173-1176. [DOI] [PubMed] [Google Scholar]
- 26.Olsen, S. K., J. Y. Li, C. Bromleigh, A. V. Eliseenkova, O. A. Ibrahimi, Z. Lao, F. Zhang, R. J. Linhardt, A. L. Joyner, and M. Mohammadi. 2006. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 20185-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, E. J., L. A. Ogden, A. Talbot, S. Evans, C. L. Cai, B. L. Black, D. U. Frank, and A. M. Moon. 2006. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development 1332419-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, E. J., Y. Watanabe, G. Smyth, S. Miyagawa-Tomita, E. Meyers, J. Klingensmith, T. Camenisch, M. Buckingham, and A. M. Moon. 2008. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development 1353599-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, T. J., K. Boyd, and T. Curran. 2006. Cardiovascular and craniofacial defects in Crk-null mice. Mol. Cell. Biol. 266272-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riley, B. M., M. A. Mansilla, J. Ma, S. Daack-Hirsch, B. S. Maher, L. M. Raffensperger, E. T. Russo, A. R. Vieira, C. Dode, M. Mohammadi, M. L. Marazita, and J. C. Murray. 2007. Impaired FGF signaling contributes to cleft lip and palate. Proc. Natl. Acad. Sci. USA 1044512-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosen, M. K., T. Yamazaki, G. D. Gish, C. M. Kay, T. Pawson, and L. E. Kay. 1995. Direct demonstration of an intramolecular SH2-phosphotyrosine interaction in the Crk protein. Nature 374477-479. [DOI] [PubMed] [Google Scholar]
- 32.Sakai, R., T. Nakamoto, K. Ozawa, S. Aizawa, and H. Hirai. 1997. Characterization of the kinase activity essential for tyrosine phosphorylation of p130Cas in fibroblasts. Oncogene 141419-1426. [DOI] [PubMed] [Google Scholar]
- 33.Scholpp, S., C. Groth, C. Lohs, M. Lardelli, and M. Brand. 2004. Zebrafish fgfr1 is a member of the fgf8 synexpression group and is required for Fgf8 signalling at the midbrain-hindbrain boundary. Dev. Genes Evol. 214285-295. [DOI] [PubMed] [Google Scholar]
- 34.Shin, N. Y., R. S. Dise, J. Schneider-Mergener, M. D. Ritchie, D. M. Kilkenny, and S. K. Hanks. 2004. Subsets of the major tyrosine phosphorylation sites in Crk-associated substrate (CAS) are sufficient to promote cell migration. J. Biol. Chem. 27938331-38337. [DOI] [PubMed] [Google Scholar]
- 35.Slack-Davis, J. K., S. T. Eblen, M. Zecevic, S. A. Boerner, A. Tarcsafalvi, H. B. Diaz, M. S. Marshall, M. J. Weber, J. T. Parsons, and A. D. Catling. 2003. PAK1 phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. J. Cell Biol. 162281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Songyang, Z., S. E. Shoelson, M. Chaudhuri, G. Gish, T. Pawson, W. G. Haser, F. King, T. Roberts, S. Ratnofsky, R. J. Lechleider, et al. 1993. SH2 domains recognize specific phosphopeptide sequences. Cell 72767-778. [DOI] [PubMed] [Google Scholar]
- 37.Suenaga, A., N. Takada, M. Hatakeyama, M. Ichikawa, X. Yu, K. Tomii, N. Okimoto, N. Futatsugi, T. Narumi, M. Shirouzu, S. Yokoyama, A. Konagaya, and M. Taiji. 2005. Novel mechanism of interaction of p85 subunit of phosphatidylinositol 3-kinase and ErbB3 receptor-derived phosphotyrosyl peptides. J. Biol. Chem. 2801321-1326. [DOI] [PubMed] [Google Scholar]
- 38.Taiji, M., T. Narumi, Y. Ohno, N. Futatsugi, A. Suenaga, N. Takada, and A. Konagaya. 2003. Protein Explorer: a Petaflops special-purpose computer system for molecular dynamics simulations, p. 15. In Proceedings of the 2003 ACM/IEEE Conference on Supercomputing. IEEE Computer Society, Washington, DC.
- 39.ten Hoeve, J., C. Morris, N. Heisterkamp, and J. Groffen. 1993. Isolation and chromosomal localization of CRKL, a human Crk-like gene. Oncogene 82469-2474. [PubMed] [Google Scholar]
- 40.Trokovic, N., R. Trokovic, P. Mai, and J. Partanen. 2003. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 17141-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, J., Y. Deng, and B. Roux. 2006. Absolute binding free energy calculations using molecular dynamics simulations with restraining potentials. Biophys. J. 912798-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woo, H. J., and B. Roux. 2005. Calculation of absolute protein-ligand binding free energy from computer simulations. Proc. Natl. Acad. Sci. USA 1026825-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, K., and D. M. Ornitz. 2008. FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development 135483-491. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, X., O. A. Ibrahimi, S. K. Olsen, H. Umemori, M. Mohammadi, and D. M. Ornitz. 2006. Receptor specificity of the fibroblast growth factor family: the complete mammalian FGF family. J. Biol. Chem. 28115694-15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.