Abstract
Hypertonicity (most often present as high salinity) is stressful to the cells of virtually all organisms. Cells survive in a hypertonic environment by increasing the transcription of genes whose products catalyze cellular accumulation of compatible osmolytes. In mammals, the kidney medulla is normally hypertonic because of the urinary concentrating mechanism. Cellular accumulation of compatible osmolytes in the renal medulla is catalyzed by the sodium/myo-inositol cotransporter (SMIT), the sodium/chloride/betaine cotransporter, and aldose reductase (synthesis of sorbitol). The importance of compatible osmolytes is underscored by the necrotic injury of the renal medulla and subsequent renal failure that results from the inhibition of SMIT in vivo by administration of a specific inhibitor. Tonicity-responsive enhancers (TonE) play a key role in hypertonicity-induced transcriptional stimulation of SMIT, sodium/chloride/betaine cotransporter, and aldose reductase. We report the cDNA cloning of human TonE binding protein (TonEBP), a transcription factor that stimulates transcription through its binding to TonE sequences via a Rel-like DNA binding domain. Western blot and immunohistochemical analyses of cells cultured in hypertonic medium reveal that exposure to hypertonicity elicits slow activation of TonEBP, which is the result of an increase in TonEBP amount and translocation to the nucleus.
Because cells are permeable to water, exposure to a hypertonic environment causes cell to shrink and elevates the concentration of intracellular ions, primarily potassium salts. Elevated cell potassium perturbs cell proteins, imposing considerable stress. Bacteria (except for halobacteria), plant, and animal cells alleviate the stress by accumulating compatible osmolytes, small organic solutes such as myo-inositol, betaine, sorbitol, taurine, and glycerol, which do not perturb macromolecules (1). The protective effect of accumulation of compatible osmolytes is believed to be attributable to the substitution of their osmotic activity for that of the high concentration of potassium, which typically returns to near normal concentrations.
Stimulation of transcription of specific genes plays a key role in the cellular adaptation to hypertonicity because it leads to an increase in the activity of transporters and enzymes that catalyze the accumulation of compatible osmolytes. Examples are the pro U operon in Escherichia coli and Salmonella typhimurium that code for an ATP-consuming betaine transporter (2); GPD1 in Saccharomyces cerevisiae codes for a key enzyme in the biosynthesis of glycerol (3); and betaine aldehyde dehydrogenase in beet and spinach code for synthesis of betaine (4). In mammals, the kidney medulla is normally hypertonic because of the urinary concentrating mechanism. Cells in the renal medulla accumulate compatible osmolytes to high concentrations as a result of elevated transcription of genes for the sodium/myo-inositol cotransporter (SMIT; ref. 5), the sodium/chloride/betaine cotransporter (BGT1; ref. 6), and aldose reductase (for synthesis of sorbitol; ref. 7). The importance of compatible osmolytes is underscored by the necrotic injury of the renal medulla and renal failure that result from inhibition of SMIT in vivo by administration of a specific inhibitor (8). This injury is caused by the prevention of accumulation of myo-inositol as a compatible osmolyte because the injury can be prevented by the simultaneous administration of an excess of myo-inositol (to overcome the inhibition of SMIT by the competitor) or betaine (another compatible osmolyte that compensates for the lack of myo-inositol). The protective effect of compatible osmolytes is universal in that, during systemic hypertonicity, an abnormal state, nonrenal tissues such as brain (9, 10) and eye (11) accumulate myo-inositol as a result of increased transcription of SMIT.
Although the sensors of hypertonicity are well understood in bacteria (12) and yeast (13), the cis- and trans-acting factors involved in hypertonicity-induced transcription are best understood in mammals. Tonicity-responsive enhancer (TonE), whose putative consensus sequence is TGGAAANN(C/T)N(C/T) (14), regulates genes for SMIT (14), BGT1 (15), and aldose reductase (16, 17). Electrophoretic mobility-shift assays (EMSAs) of nuclear extracts and in vivo footprinting reveal that exposure of cells to hypertonicity results in activation of TonE binding protein (TonEBP), which binds to TonE sites, leading to stimulation of transcription (14, 15). Thus, activation of TonEBP is a key event in hypertonicity-induced transcription. Here, we report the molecular cloning of TonEBP. The sequence of TonEBP reveals that it is a Rel-like activator of transcription.
MATERIALS AND METHODS
Cloning of TonEBP.
Yeast one-hybrid screening (18) was performed by using a commercial system (Match-Maker One-Hybrid System, CLONTECH). A TonE reporter strain of yeast was generated by inserting four tandem copies of the TonE sequence from the canine BGT1 gene (15) (TGGTGGAAAAGTCCAGCT; TonE is underlined) into the genome immediately upstream of a minimal promoter and HIS3. A mutant TonE reporter strain was made the same way by using an inactive mutant TonE sequence (TGGTGtccccGTCCAGCT). A human kidney cDNA library (CLONTECH) that directs expression of the cDNAs fused to the transactivation domain of yeast transcription factor GAL4 was transformed into the TonE reporter strain. Of five million transformants, one colony consistently grew in the absence of histidine. This colony contained a cDNA clone with a 4.1-kilobase insert. The mutant TonE reporter strain transformed with this clone did not grow in the absence of histidine. A HeLa cell cDNA library in λgt10 (CLONTECH) was screened by using the 4.1-kilobase cDNA probe to obtain additional overlapping cDNA clones covering a total of 6.0 kilobases.
Antibodies, Western Blot Analysis, and Immunostaining.
The TonEBP cDNA corresponding to amino acids 2 to 472 was subcloned into pQE30 (Qiagen, Chatsworth, CA). The truncated protein now fused to six histidine residues at its N terminus was expressed in E. coli and was purified by using a Ni+-agarose column. The purified protein was used to raise antibodies in rabbits by using a commercial service (Covance, Denver, PA). For Western blot analysis, whole cell lysates or nuclear extracts of MDCK cells were separated on an 8% SDS-polyacrylamide gel and were transferred onto a nitrocellulose membrane. The membrane was probed with the TonEBP antiserum and was visualized by using chemiluminescence reagents (NEN). To quantify the difference in TonEBP amount, semiquantitative Western blot analysis was performed by comparing the isotonic sample with serially diluted hypertonic samples. For immunostaining of TonEBP, MDCK cells were fixed for 15 min in PBS containing 3.7% formaldehyde, were permeabilized for 5 min with 0.1% Triton X-100 in PBS, and were washed with Tris-buffered saline. The cells were incubated for 1 hour at 37°C with the TonEBP antiserum (1:50 dilution) and then were incubated as above in a 1:100 dilution of anti-rabbit IgG conjugated with rhodamine (Zymed). After washing with PBS, cells were observed by fluorescence microscopy. Preimmune serum did not yield significant staining (data not shown).
Cell Transfection.
Transfection and analysis of luciferase activity were performed as described (15). One of the two luciferase constructs used contains two copies of “hTonE” in front of the SV40 promoter and the luciferase gene (15). The other contains three copies of κB sequence from the immunoglobin κ light chain gene (19). Medium was made hypertonic by adding 200 mM raffinose.
RESULTS
Cloning of TonEBP.
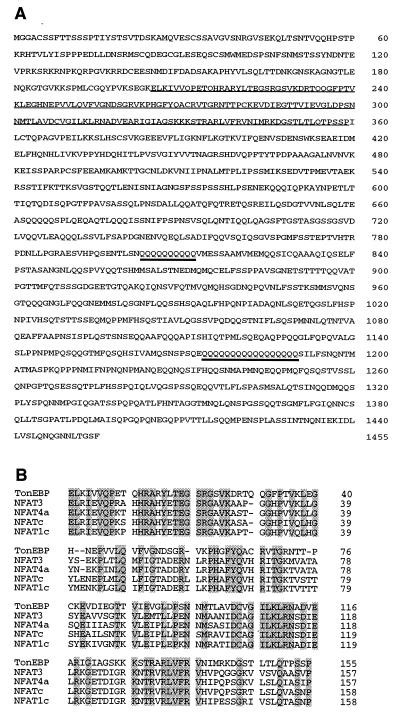
To learn more about regulation of TonEBP by hypertonicity, we cloned its cDNA by using a yeast one-hybrid strategy (18). Sequencing of the positive clone revealed that it contains a large ORF (encoding the N-terminal 1,182 amino acids shown in Fig. 1A) that extends to the end of the cDNA without a stop codon, indicating that this is a partial clone missing the C-terminal sequence. This ORF is not in frame with that of GAL4, suggesting that the cDNA encodes a transactivating domain that is functional in yeast. It appears that the glutamine-rich region in the C-terminal half is responsible for the transactivation (see more below). Overlapping clones were obtained from the HeLa library by conventional hybridization with the cloned cDNA probe. All of the clones cover a sequence of 6,045 bp (the nucleotide sequence is available from the GenBank database, with accession no. AF089824), in which there is a large ORF of 1,455 amino acids (Fig. 1A). Each of the nucleotides of the ORF is represented by at least two independent cDNA clones. All of the overlapping sequences of the independent cDNA clones are identical, indicating that these clones are from the same mRNA. Because the TonEBP mRNA is >12 kilobases (Fig. 4), the cloned region represents less than half of the mRNA sequence. Sizes of the 5′- and 3′-untranslated regions are not known at this time because none of the cDNA clones have poly(A) tails.
Figure 1.
TonEBP is a Rel-like protein. (A) The predicted amino acid sequence of human TonEBP. The NFAT-like Rel homology domain and two stretches of glutamine residues are underlined. (B) Sequence alignment of Rel-homology domains in TonEBP and NFAT isoforms (21). Amino acids conserved in all of the members are shaded.
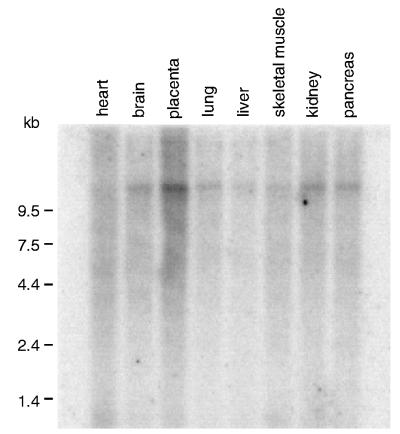
Figure 4.
Expression of TonEBP mRNA in human tissues. A blot containing poly(A) RNA from human tissues (CLONTECH) was probed with TonEBP cDNA. All of the tissues shown express TonEBP mRNA. Similar results (not shown) also were obtained with RNA from spleen, thymus, prostate, testis, ovary, small intestine, and colon.
Analysis of the deduced amino acid sequence reveals a stretch of ≈160 amino acids near the N terminus that displays significant similarity to the Rel-like DNA binding domain (20) of the nuclear factor of activated T cells (NFAT) transcription factor family (Fig. 1B). In this region, 88% of the amino acids are conserved among the NFAT isoforms; 45% are identical between the NFAT isoforms and TonEBP. On the other hand, TonEBP does not have a sequence similar to the highly conserved N-terminal region among the NFAT isoforms that functions as an interface for calcineurin and kinases that control nuclear import or export (21), suggesting that the signaling pathways for regulation are different for TonEBP and the NFATs. We conclude that TonEBP is a more distantly related member of the NFAT family.
Confirmation of the Cloned TonEBP.
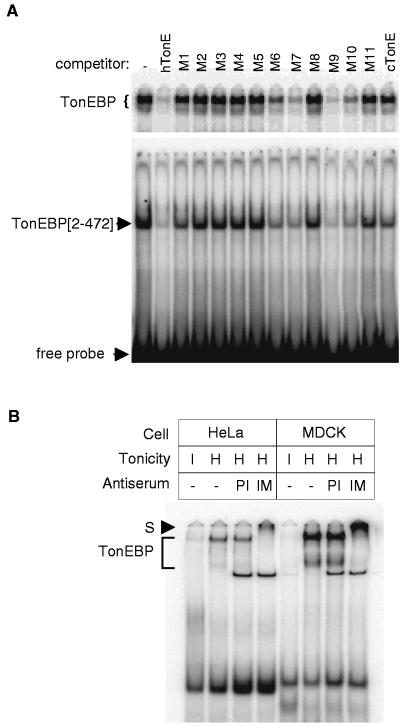
The identity of the cloned TonEBP was confirmed by analysis of its DNA binding and the effect of specific antibodies raised to it. To examine DNA binding activity, EMSA was performed on a truncated TonEBP containing the Rel-like domain (N-terminal 472 amino acids). As shown in Fig. 2A, the truncated TonEBP forms a complex with the TonE DNA probe that migrates faster than the native TonEBP, as expected. The competition profile of a panel of TonE sequences for binding to the truncated protein is indistinguishable from that for binding to the native TonEBP of HeLa cells, demonstrating that the cloned TonEBP binds DNA with correct specificity. Next, we performed “super-shift” analysis of TonEBP by using antibodies raised against the N-terminal portion of TonEBP. The antibodies specifically retard mobility of the TonEBP bands from HeLa and MDCK cells (Fig. 2B), indicating that they bind the native TonEBP in human and canine cells. Western blot analysis of MDCK cells (Fig. 5A) and HeLa cells (not shown) detects a single band of 200 kDa that appears to be the same 200-kDa polypeptide that is specifically labeled when TonE is UV cross-linked to nuclear extracts of MDCK cells (15). This is in reasonable agreement with the calculated molecular mass of 160 kDa. Collectively, these results establish that the cloned protein is TonEBP.
Figure 2.
(A) DNA binding profile of the recombinant TonEBP. The N-terminal 472 amino acids of TonEBP (amino acids 2–472—TonEBP[2–472]), including the Rel-like domain, were bacterially expressed and purified. EMSA was performed by using 0.3 nM 32P-hTonE without or with 10 nM competitors as indicated on top: double-stranded DNA probes hTonE, hTonE mutants—M1 to M11—and cTonE were described previously (15). Each binding reaction includes 10 μg of nuclear extract from HeLa cells cultured in hypertonic medium (Upper) or 0.05 μg of TonEBP[2–472] (Lower). The reactions were separated on nondenaturing gels, and autoradiograms of the gels are shown. Only the top portion of the gel is shown for the HeLa nuclear extracts. Bands representing the native TonEBP (from HeLa cells), TonEBP[2–472], and free probe are marked at left. (B) Super-shift analysis of TonEBP. Each lane contains 10 μg of nuclear extract prepared from HeLa or MDCK cells cultured in hypertonic medium for 18 hours (H) or in isotonic medium (I). The antiserum raised against TonEBP[2–472] (IM) and preimmune serum (PI) were added to binding mixtures—0.1 μl or serum added undiluted per reaction—as indicated. The gel was run longer than those in A to allow the super-shifted bands to move into the gel. The TonEBP bands and the super-shifted TonEBP band (S) are marked at left.
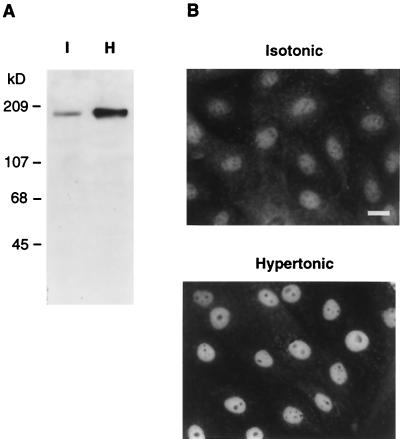
Figure 5.
Regulation of TonEBP by hypertonicity. (A) Western blot analysis of whole cell lysates prepared from MDCK cells cultured in isotonic (I) or hypertonic (H) medium for 18 hours by using the antiserum raised against TonEBP[2–472] (Fig. 2). (B) Distribution of TonEBP in isotonic and hypertonic MDCK cells. MDCK cells were grown on glass coverslips and were treated with either isotonic or hypertonic medium for 18 hours. Cells then were fixed and stained with the TonEBP antiserum. (Bar = 20 μm.)
Dominant Negative Form of TonEBP.
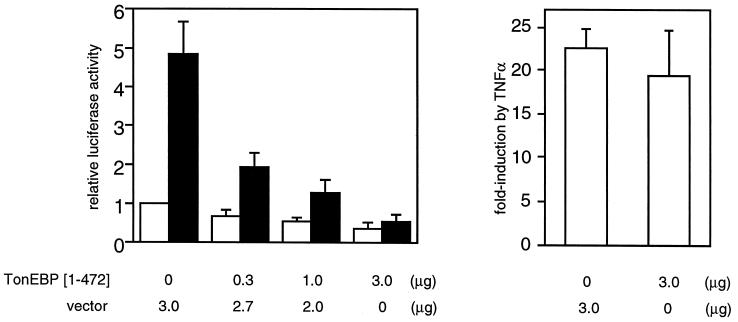
The large region (>1,000 amino acids) downstream of the DNA-binding N-terminal region has two long stretches (10 or more) of glutamine residues and overall is rich (18%) for this amino acid (Fig. 1A). Because glutamine-rich regions are involved in activation of transcription (22), it appears that the TonEBP has a bipartite structure—DNA binding domain in the N terminus and transactivation domain toward the C terminus. Based on this prediction, we asked the question of whether the N-terminal truncated protein that includes the DNA-binding domain but not the glutamine-rich region could function as a dominant negative TonEBP. To address this question, we studied the effects of expressing the truncated TonEBP on TonE-mediated stimulation of reporter gene expression. As shown in Fig. 3, expression of the truncated TonEBP suppresses stimulation of transcription by hypertonicity in a dose-dependent manner. These effects are specific for the TonE/TonEBP system in that NF-κB-mediated regulation of transcription is not affected (Fig. 3 Right). These data provide in vivo evidence that TonEBP is the transcription factor interacting with TonE.
Figure 3.
TonEBP[1–472] inhibits stimulation of transcription by hypertonicity. (Left) MDCK cells were transiently transfected with 2 μg of a plasmid containing a TonE-driven luciferase gene (15) along with plasmid pcDNA3.1(+) (Invitrogen) directing expression of TonEBP[1–472] or pcDNA3.1(+) by itself (vector) in amounts indicated at the bottom. Transfected cells were cultured in isotonic (open bars) or hypertonic medium (filled bars) for 18 hours, and the luciferase activity was measured. Luciferase activity was normalized to the control cells [transfected with the pcDNA3.1(+) and the luciferase construct] cultured in isotonic medium. Results are mean ± SEM; n = 4. (Right) Lack of effect of TonEBP[1–472] on tumor necrosis factor α-induced NF-κB activity. MDCK cells were transfected with 2 μg of a plasmid containing the luciferase gene under the control of κB sequence from the immunoglobin κ light chain gene (19) along with 3 μg of TonEBP[1–472] in pcDNA3.1(+) or pcDNA3.1(+) by itself as indicated at the bottom. The transfected cells were treated without or with tumor necrosis factor α (20 ng/ml) for 6 hours and were analyzed for luciferase activity. Fold-induction of luciferase by tumor necrosis factor α treatment is shown. Results are mean ± SEM; n = 4.
TonEBP Is Ubiquitously Expressed.
Northern blot analysis of human tissues reveals that TonEBP mRNA is expressed in all tissues examined, including kidney and brain (Fig. 4). Such widespread TonEBP expression in tissues that normally are not exposed to a hypertonic environment raises the possibility that TonEBP may be a general safety system that protects against pathologic hypertonicity, as demonstrated for brain (9, 10) and eye (11). Alternatively, TonEBP may play an unknown, widely distributed role other than mediating the transcriptional response to hypertonicity.
Hypertonicity Increases the Amount and Nuclear Distribution of TonEBP.
We have shown that activation of TonEBP is the key step in stimulation of transcription of the BGT1 (15) and SMIT genes (14) in response to hypertonicity. To investigate the mechanism of activation of TonEBP, we performed semiquantitative Western blot analysis and immunocytochemistry on MDCK cells. Western blot analysis on three independent pairs of whole cell extracts consistently showed a 4-fold increase in TonEBP when cells were cultured in hypertonic medium for 18 hours (Fig. 5A). A similar analysis of nuclear extracts showed 10× more TonEBP in nuclei of hypertonic cells compared with isotonic cells (data not shown). In addition, the rise in amount of nuclear TonEBP is slow; it takes >10 hours to complete (data not shown). These observations are in agreement with the EMSA data (15), which demonstrated a 9-fold increase in TonEBP activity in nuclei with a similar time course in response to hypertonicity treatment. Immunostaining of isotonic cells shows that TonEBP is present in both cytoplasm and nucleus (Fig. 5B). In contrast, when MDCK cells are cultured in hypertonic medium, the nuclear staining increases while the cytoplasmic staining decreases. In summary, hypertonicity-induced activation of nuclear TonEBP is achieved by a combination of an increase in the amount of TonEBP and an increase in nuclear distribution of TonEBP.
DISCUSSION
We conclude that the cloned protein is indeed TonEBP, which initially was identified from nuclear extracts based on a correlation between its binding to a panel of wild-type and mutant TonE sequences and their enhancer activity (15): in EMSA, TonEBP binds active TonEs with high affinity whereas it binds inactive TonEs with much lower affinity. Evidence for this conclusion includes: (i) the Rel-like domain of the cloned protein binds DNA with a specificity indistinguishable from the native TonEBP (Fig. 2A); (ii) antibodies raised to this protein recognize the native TonEBP in EMSA (Fig. 2B) as well as immobilized TonEBP in Western blot analysis (Fig. 5A); and (iii) the Rel-like DNA binding domain without the putative transactivation domain functions as a dominant negative form of TonEBP (Fig. 3).
We believe that TonEBP is probably the DNA-binding subunit of a larger complex. Although TonEBP appears as a single polypeptide of 200 kDa in UV crosslinking (15) and Western blot analysis (Fig. 5A), the native TonEBP appears as two bands in EMSA gels (Fig. 2). In gel-filtration analyses of nuclear extracts assayed by EMSA, the native TonEBP behaves as a complex >1,000 kDa (data not shown), far exceeding the apparent size of TonEBP: 200 kDa. Our preliminary data show that immunoprecipitation of TonEBP from MDCK cells labeled with [35S]methionine brings down several proteins (data not shown), supporting the idea that TonEBP forms stable interactions with other proteins.
Throughout the biologic spectrum, transcriptional stimulation of proteins that catalyze the cellular accumulation of compatible osmolytes is a key event in adaptation to hypertonicity. Despite more than a decade of intense research, cis-elements and proteins regulating the pro U operon in bacteria have not been defined clearly (23). In yeast, even though the kinase signaling pathways for stimulation of transcription of GPD1 have been identified (ref. 13 and see below), the cis- and trans-acting factors are unknown (24). TonEBP is a transcription factor that regulates hypertonicity-induced cellular accumulation of compatible osmolytes. TonEBP regulates SMIT (catalyzes myo-inositol accumulation; ref. 14), BGT1 (betaine; ref. 15), and aldose reductase (sorbitol; refs. 16 and 17). It is possible that TonEBP is involved in the regulation of the sodium/chloride/taurine cotransporter, whose mRNA is increased by hypertonicity (25). Thus, TonEBP regulates accumulation of most of the major compatible osmolytes in mammalian kidney, with the clear exception of glycerophosphocholine, whose cellular accumulation is achieved by inhibition of an enzyme that degrades glycerophosphocholine (26).
Many transcription factors are activated by hypertonicity. Activation of Janus kinases in response to hypertonicity leads to activation of transcription factors STAT1 and STAT3 (27). Hypertonicity also activate three different isoforms of mitogen-activated protein kinases: extracellular regulated kinases, c-Jun N-terminal kinase, and p38 (reviewed in ref. 28). These kinases in turn activate transcription factors, including c-Myc, Elk-1, c-Jun, ATF-2, and Max (28). The role of mitogen-activated protein kinases in activation of TonEBP has been addressed indirectly by measuring the induction of SMIT and BGT1 mRNA or TonE-regulated reporter gene expression. Preventing activation of extracellular regulated kinases (29) or c-Jun N-terminal kinases (30) does not affect the induction by hypertonicity of BGT1 and SMIT mRNA or a TonE regulated reporter gene, respectively, indicating that these kinases are not involved in signaling to TonEBP. The role of p38 is disputed. Prevention of p38 activation by expressing a dominant negative form of MKK-3, an upstream kinase, does not prevent TonE-mediated increase in transcription in response to hypertonicity (30). On the other hand, treatment with a specific inhibitor of p38 prevents induction of BGT1 mRNA by hypertonicity (31). Because p38 is a functional homolog of Hog1p (32) that mediates induction by hypertonicity of GPD1 in yeast (13), it is of particular interest to ask the question of whether p38 is involved in activation of TonEBP. The TonEBP antibodies described here should provide a critical probe to address this question directly.
Regardless of the role of p38 in TonEBP activation, the time course of activation of TonEBP is much slower than other transcription factors that are activated by hypertonicity. Although STATs (27) and c-Jun (33) are fully activated within half an hour, it takes >3 hours for a detectable increase in TonEBP activity and >10 hours for full activation (ref. 15, and see above). In fact, activation of TonEBP in response to hypertonicity is one of the slowest among known stress responses: full activation of heat shock factor 1 is reached within 10 min of heat shock (34); c-fos induction by UV or oxidative stress is complete in 1 hour (35); and activation of hypoxia-inducible factor 1 is complete in 4 hours (36). Of the two different modes of TonEBP activation—increase in TonEBP amount and nuclear redistribution (Fig. 5)—kinetics of the former is primarily responsible for the slow overall activation of TonEBP. The slow kinetics and dual modes of regulation (amount and distribution) of TonEBP suggest that signaling pathways for hypertonicity would be different from those for the other stresses mentioned above.
Acknowledgments
We thank Randall Reed and Gregg Semenza for advice and Ulrich Siebenlist for the κB-luciferase construct. This work was supported by National Institutes of Health Grant PO1 DK44484 and Grant-in-Aid 96014490 from the American Heart Association.
ABBREVIATIONS
- TonE
tonicity-responsive enhancer
- TonEBP
TonE binding protein
- SMIT
sodium/myo-inositol cotransporter
- BGT1
sodium/chloride/betaine cotransporter
- EMSA
electrophoretic mobility-shift assays
- NFTA
nuclear factor of activated T cells
Footnotes
References
- 1.Yancey P H, Clark M E, Hand S C, Bowlus R D, Somero G N. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 2.Lucht J M, Bremer E. FEMS Microbiol Rev. 1994;14:3–20. doi: 10.1111/j.1574-6976.1994.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 3.Albertyn J, Hohmann S, Thevelein J M, Prior B A. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishitani M, Nakamura T, Han S Y, Takabe T. Plant Mol Physiol. 1995;27:307–315. doi: 10.1007/BF00020185. [DOI] [PubMed] [Google Scholar]
- 5.Yamauchi A, Uchida S, Preston A S, Kwon H M, Handler J S. Am J Physiol. 1993;264:F20–F23. doi: 10.1152/ajprenal.1993.264.1.F20. [DOI] [PubMed] [Google Scholar]
- 6.Uchida S, Yamauchi A, Preston A S, Kwon H M, Handler J S. J Clin Invest. 1993;91:1604–1607. doi: 10.1172/JCI116367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smardo F L, Burg M B, Garcia-Perez A. Am J Physiol. 1992;262:C776–C782. doi: 10.1152/ajpcell.1992.262.3.C776. [DOI] [PubMed] [Google Scholar]
- 8.Kitamura H, Yamauchi A, Sugiura T, Matsuoka Y, Horio M, Tohyama M, Shimada S, Imai E, Hori M. Kidney Int. 1998;53:146–153. doi: 10.1046/j.1523-1755.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 9.Ibsen L, Strange K. Am J Physiol. 1996;271:F877–F885. doi: 10.1152/ajprenal.1996.271.4.F877. [DOI] [PubMed] [Google Scholar]
- 10.Minami Y, Inoue K, Shimada S, Mormura H, Miyai A, Yamauchi A, Matsunaga T, Tohyama M. Mol Brain Res. 1996;40:64–70. doi: 10.1016/0169-328x(96)00034-4. [DOI] [PubMed] [Google Scholar]
- 11.Morimura H, Shimada S, Otori Y, Yamauchi A, Minami Y, Inoue K, Miyai A, Ishimoto I, Tano Y, Tohyama M. Mol Brain Res. 1996;35:333–338. doi: 10.1016/0169-328x(95)00245-n. [DOI] [PubMed] [Google Scholar]
- 12.Mizuno T, Mizushima S. Mol Microbiol. 1990;4:1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 13.Maeda T, Takekawa M, Saito H. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 14.Rim J S, Atta M G, Dahl S C, Berry G T, Handler J S, Kwon H M. J Biol Chem. 1998;273:20615–20621. doi: 10.1074/jbc.273.32.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyakawa H, Woo S K, Chen C P, Dahl S C, Handler J S, Kwon H M. Am J Physiol. 1998;274:F753–F761. doi: 10.1152/ajprenal.1998.274.4.F753. [DOI] [PubMed] [Google Scholar]
- 16.Ferraris J D, Williams C K, Jung K, Bedford J J, Burg M B, Garcia-Perez A. J Biol Chem. 1996;271:18318–18321. doi: 10.1074/jbc.271.31.18318. [DOI] [PubMed] [Google Scholar]
- 17.Ko B C B, Ruepp K M, Bohren K M, Gabbay K H, Chung S S M. J Biol Chem. 1997;272:16431–16437. doi: 10.1074/jbc.272.26.16431. [DOI] [PubMed] [Google Scholar]
- 18.Wang M M, Reed R R. Nature (London) 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 19.Fujita T, Nolan G P, Liou H C, Scott M L, Baltimore D. Genes Dev. 1993;7:1354–1363. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- 20.Wolfe S A, Zhou P, Dotsch V, Chen L, You A, Ho S N, Crabtree G R, Wagner G, Verdine G L. Nature (London) 1997;385:172–176. doi: 10.1038/385172a0. [DOI] [PubMed] [Google Scholar]
- 21.Rao A, Luo C, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 22.Gill G, Pascal E, Tseng Z H, Tjian R. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordi B J A M, Fielder A E, Burns C M, Hinton J C D, Dover N, Ussery D W, Higgins C F. J Biol Chem. 1997;272:12083–12090. doi: 10.1074/jbc.272.18.12083. [DOI] [PubMed] [Google Scholar]
- 24.Marquez J A, Pascual-Ahuir A, Proft M, Serrano R. EMBO J. 1998;17:2543–2553. doi: 10.1093/emboj/17.9.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchida S, Kwon H M, Yamauchi A, Preston A S, Marumo F, Handler J S. Proc Natl Acad Sci USA. 1992;89:8230–8234. doi: 10.1073/pnas.89.17.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon E D, Zablocki K, Jung K Y, Peters E M, Garcia-Perez A, Burg M B. Am J Physiol. 1995;269:C35–C41. doi: 10.1152/ajpcell.1995.269.1.C35. [DOI] [PubMed] [Google Scholar]
- 27.Gatsios P, Terstegen L, Schliess F, Haussinger D, Kerr I M, Heinrich P C, Graeve L. J Biol Chem. 1998;273:22962–22968. doi: 10.1074/jbc.273.36.22962. [DOI] [PubMed] [Google Scholar]
- 28.Whitmarsh A J, Davis R J. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 29.Kwon H M, Itoh T, Rim J S, Handler J S. Biochem Biophys Res Commun. 1995;213:975–979. doi: 10.1006/bbrc.1995.2224. [DOI] [PubMed] [Google Scholar]
- 30.Kultz D, Garcia-Perez A, Ferraris J D, Burg M B. J Biol Chem. 1997;272:13166–13170. doi: 10.1074/jbc.272.20.13165. [DOI] [PubMed] [Google Scholar]
- 31.Sheikh-Hamad D, Mari J D, Suki W N, Safirstein R, Watts B A, Rouse D. J Biol Chem. 1998;273:1832–1837. doi: 10.1074/jbc.273.3.1832. [DOI] [PubMed] [Google Scholar]
- 32.Han J, Lee J D, Bibbs L, Ulevitch R J. Science. 1994;265:808–810. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 33.Raingeaud J, Gupta S, Rogers J S, Dickens M, Han J, Ulevitch R J, Davis R J. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 34.Kline M P, Morimoto R I. Mol Cell Biol. 1997;17:2107–2115. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devary Y, Gottlieb R A, Lau L F, Karin M. Mol Cell Biol. 1991;11:2804–2811. doi: 10.1128/mcb.11.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G L, Jiang B H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]