Abstract
Monoubiquitination aids in the nuclear export and entrance of proteins into the lysosomal degradative pathway, although the mechanisms are unknown. Cytidylyltransferase (CCTα) is a proteolytically sensitive lipogenic enzyme containing an NH2-terminal nuclear localization signal (NLS). We show here that CCTα is monoubiquitinated at a molecular site (K57) juxtaposed near its NLS, resulting in disruption of its interaction with importin-α, nuclear exclusion, and subsequent degradation within the lysosome. Cellular expression of a CCTα-ubiquitin fusion protein that mimics the monoubiquitinated enzyme resulted in cytoplasmic retention. A CCTα K57R mutant exhibited an extended half-life, was retained in the nucleus, and displayed proteolytic resistance. Importantly, by using CCTα-ubiquitin hybrid constructs that vary in the intermolecular distance between ubiquitin and the NLS, we show that CCTα monoubiquitination masks its NLS, resulting in cytoplasmic retention. These results unravel a unique molecular mechanism whereby monoubiquitination governs the trafficking and life span of a critical regulatory enzyme in vivo.
Protein monoubiquitination has recently emerged as an important posttranslational modification regulating transcription, endocytic vesicle trafficking, histone modification, and DNA repair (12, 26). Conjugation of one (monoubiquitination) or multiple (multiubiquitination) ubiquitin molecules to lysines within target proteins serves as an important endocytic signal for internalization and targeting of various ion channels, membrane cargo receptors, and junctional proteins to the endocytic pathway (8). Monoubiquitination of cargo receptors triggers recruitment of ubiquitin-binding proteins that bridge receptor ligands to endocytic and cell sorting elements that are further degraded or recycled to the plasma membrane (18). The addition of a monoubiquitin tag to p53, for example, is sufficient for its nuclear export; p53 monoubiquitination may cooperatively interact with sumoylation and involve its nuclear export signal (NES) for cytoplasmic targeting (5). Monoubiquitination also regulates nuclear import and export of some proteins. The tumor suppresser phosphatase and tensin homolog on chromosome ten (PTEN) requires monoubiquitination for nuclear import despite lacking a canonical nuclear localization signal (NLS) (27). Hence, in contrast to polyubiquitination of short-lived cytosolic proteins that are usually destined for rapid degradation within the proteasome, monoubiquitination appears to be a means to exquisitely regulate the availability of membrane-associated protein complexes.
CTP:phosphocholine cytidylyltransferase (CCT) is a membrane-activated enzyme that catalyzes the penultimate and rate-limiting step for biosynthesis of the major eukaryotic phospholipid, phosphatidylcholine (PtdCho) (17). CCTα, a predominant species in lung epithelia, is comprised of 367 amino acids (aa) with four functional domains: a basic residue NH2-terminal NLS, a catalytic core (C), a membrane-binding domain (M), and a carboxyl-terminal phosphorylation domain (P) (17). CCTα is an amphitrophic enzyme and thus can switch between an inactive soluble or cytoplasmic form to an active, membrane-bound species within the nucleus. In fact, the ability of CCTα to reversibly translocate to nuclear or endoplasmic membranes after stimulation by lipid activators is well established and remains central to enzyme activation (13, 17). A second, more recent mode of regulatory control involves the turnover rate of CCTα molecules. CCTα is ubiquitinated in response to cytokines, such as tumor necrosis factor alpha (TNF-α), and is a target for death effector and neutral proteinases (19, 21). However, CCTα's extended protein half-life (∼8 h) in vivo exceeds other comparable regulatory enzymes because it is stabilized, in part, by calmodulin (6). The long half-life of CCTα projects that there may be a cooperative layer of more complex regulation by additional coadaptor molecules or modificational events within its primary structure that impact the enzyme's stability. This model would strike a delicate balance between CCTα's ability to translocate to nuclear membranes to remain in an activated state and yet preserve cell membrane phospholipid homeostasis by eliminating a population of CCTα molecules when PtdCho is presented in excess.
In the present study, we demonstrate that CCTα is monoubiquitinated at a single molecular site (K57), thereby marking the enzyme for nuclear exclusion and degradation within the lysosome. A CCTα K57R mutant exhibited greater stability, was proteolytically resistant to the actions of TNF-α, and was retained in the nucleus, underscoring the functional relevance of this residue. Because K57 was in close proximity to the NLS of CCTα, we tested the hypothesis that masking of its nuclear targeting domain by monoubiquitination would impede association with importin-α and thus mislocalize the enzyme to the lysosome for degradation. This hypothesis was tested by execution of studies using CCTα-ubiquitin hybrid proteins. The results unveil a conceptually novel molecular model whereby cells regulate the constitutive behavior and levels of a membrane-activated enzyme by hindrance of a nuclear signal motif via protein monoubiquitination.
MATERIALS AND METHODS
Materials.
The sources of murine lung epithelial (MLE) cells, CCTα antibodies, TNF-α, and lactacystin were as described previously (21). A ubiquitin conjugation kit was purchased from Calbiochem (La Jolla, CA). Mouse monoclonal ubiquitin and PARP antibodies were purchased from Cell Signaling (Danvers, MA). The pAmCyan1-C1 and pZsYellow-C1 vectors were purchased from Clontech (Mountain View, CA). LysoTracker Red, mouse monoclonal V5 antibody, the To-Pro-3 nuclear staining kit, the pcDNA3.1D cloning kit, Escherichia coli OneShot competent cells, the pENTR directional TOPO cloning kits, and the Gateway mammalian expression system were purchased from Invitrogen (Carlsbad, CA). BD Talon purification and buffer kits were purchased from BD Biosciences (San Jose, CA). The QuikChange site-directed mutagenesis kit and the X-Blue cells were purchased from Stratagene (La Jolla, CA). The gel extraction kit and QIAprep spin miniprep kits were from Qiagen (Valencia, CA). FuGENE6 transfection reagent was purchased from Roche Diagnostics (Indianapolis, IN). Nucleofector transfection kits were from Amaxa (Gaithersburg, MD). Immobilized protein A/G beads were obtained from Pierce (Rockford, IL). TSG101 small interfering RNA (siRNA) and importin-2α antibody were from Santa Cruz Biotechnology (Santa Cruz, CA), and purified importin-2α was from GenWay Biotech (San Diego, CA). Mannose-6-phosphate receptor (M6PR) antibody was from Abcam (Cambridge, MA). The ubiquitinΔK plasmid was kindly provided by Peter Snyder, University of Iowa. All DNA sequencing was performed by the University of Iowa DNA Core Facility.
Cell culture.
MLE cells were cultured in Dulbecco minimum essential medium (DMEM) without fetal bovine serum (FBS) for up to 36 h with or without TNF-α (0.75 μg/ml) and with or without NH4Cl (20 mM/ml) or lactacystin (20 μM). Cell lysates were prepared by brief sonication in buffer A (150 mM NaCl, 50 mM Tris, 1.0 mM EDTA, 2 mM dithiothreitol, 0.025% sodium azide, 1 mM phenylmethylsulfonyl fluoride) at 4°C.
PtdCho synthesis and CCT activity.
PtdCho production and CCT activity were determined as described previously (32).
Immunoblot analysis.
Equal amounts of total protein in sample buffer were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and transferred to nitrocellulose, and immunoreactive CCTα or β-actin was detected as described above. CCTα was purified from rat liver as described previously (31).
Coimmunoprecipitation.
A total of 200 μg of total protein from MLE cell lysates were precleared with 20 μl of protein A/G beads for 1 h at 4°C. Then, 5 μg of CCTα antibody was added for a 3-h incubation at 4°C. Next, 40 μl of protein A/G beads was added, followed by an additional 2 h of incubation. Beads were spun down and washed five times using radioimmunoprecipitation assay buffer (50 mM HEPES, 150 mM NaCl, 0.5 mM EGTA, 50 mM NaF, 10 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 20 μM leupeptin, 1% [vol/vol] Triton X-100) as described previously (2). Beads were heated at 100°C for 5 min with 80 μl of protein sample buffer prior to SDS-PAGE and immunoblotting.
Construction of tagged CCTα mutants.
Glutathione S-transferase (GST)-CCTFL, GST-CCTMEM, GST-CCTCAT, GST-CCTΔN40, GST-CCT315, and GST-CCT210 were constructed as described previously (6). GST-CCTFL, harboring point mutations within the catalytic domain, was constructed by mutating Lys57, Lys100, Lys122, Lys183, and Lys186 to Arg using a QuikChange site-directed mutagenesis kit. A full-length CCTα template (GST-CCTFL) plasmid DNA was used as a template. A ubiquitin-His vector was constructed using primary rat alveolar epithelial cell cDNA as a template with appropriate primers in a PCR, generating a 228-bp ubiquitin fragment. The PCR product was gene cleaned, followed by directional cloning into a pDEST26 His-tagged expression vector. V5-CCTFL and V5-CCTK57R were constructed using GST-CCTFL and GST-CCTK57R as templates in PCR; amplified fragments were directionally cloned into a pcDNA3.1D/V5-His expression vector.
Construction of CFP-tagged CCTα mutants.
CFP-CCT (full-length CCTα) was constructed as described previously (6). A CFP-CCTK57R mutant was constructed using PCR with CFP-CCT as a template.
The CFP-ubiquitin-CCT fusion construct comprised of full-length CCTα with an NH2-terminal ubiquitin tag (CFP-UbWT-CCT and CFP-UbΔK-CCT) was constructed as follows. pcDNA-ubiquitinWT or ubiquitinΔK was used as the template, using the primers 5′-ACTCTCGAGATGCAGATCTTCGTGAAGACTCT-3′ (forward) and 5′-ACTGTCGACCCCACCTCTGAGACGGAG-3′ (reverse) to amplify the 228-bp wild-type (WT) or lysine knockout ubiquitin fragment. The forward primer contains a XhoI site and a 3-bp overhang; the reverse primer contains a SalI site and a 3-bp overhang. CFP-CCT was used as the template, using the primers 5′-ACTGTCGACATGGATGCACAGAGTTCAGCT-3′ (forward) and 5′-ACTGGATCCGTCCTCTTCATCCTCGCTGA-3′ (reverse) to amplify the 1,100-bp CCT fragment. The forward primer contains a SalI site and a 3-bp overhang; the reverse primer contains a BamHI site and a 3-bp overhang. Both PCR products were gel purified and digested with the enzymes described above prior to directional cloning into the CFP construct.
CFP-CCTΔN40 mutant lacking the first 40 residues of CCTα was constructed as follows. GST-CCT was used as the template utilizing the primers 5′-ACTACTAGATCTTTACGGCAGCCAGCTCCT-3′ (forward) and 5′-ACTGTCGACGTCCTCTTCATCCTCGCTGA-3′ (reverse) to amplify the 980-bp CCT fragment. The forward primer contains a BglII site and a 3-bp overhang; the reverse primer contains a SalI site and a 3-bp overhang. The PCR product was gel purified, followed by digestion with the enzymes described above, prior to directional cloning into the CFP vector.
A CFP-CCTK57-Ub mutant where ubiquitin was fused internally and in frame between CCTα Ser56 and Lys57 was constructed by double SOE-PCR. Six CCTUbi primers were designed: CCTUbi1, 5′-ACTCTCGAGATGGATGCACAGAGTTCAGCT-3′; CCTUbi2, 5′-AGTCTTCACGAAGATCTGCATACTAAAGTCAACTTCAATTTC-3′; CCTUbi3, 5′-GAAATTGAAGTTGACTTTAGTATGCAGATCTTCGTGAAGACT-3′; CCTUbi4, 5′-AGTCACCCTGACATAGGGCTTCCCACCTCTGAGACGGAGCAC-3′; CCTUbi5, 5′-GTGCTCCGTCTCAGAGGTGGGAAGCCCTATGTCAGGGTGACT-3′; and CCTUbi6, 5′-ACTGTCGACGTCCTCTTCATCCTCGC-3′). In the first step, the primers CCTUbi1 and CCTUbi2 were used to amplify an NH2-terminal 1- to 56-aa fragment of CCTα. In the second step, primers CCTUbi3 and CCTUbi4 were used to amplify the ubiquitin fragment. In the third step, primers CCTUbi5 and CCTUbi6 were used to amplify a carboxyl-terminal 57- to 367-aa fragment of CCTα. In the last step, the three gel-purified fragments from the steps above were used as a template in the final PCR using the primers CCTUbi1 and CCTUbi6 to amplify a desired 1,329-bp product with the ubiquitin insertion. The CCTUbi1 primer contains an XhoI site and a 3-bp overhang; the CCTUbi6 primer contains a SalI site and a 3-bp overhang. The PCR product was gel purified, followed by digestion with the above enzymes, prior to directional cloning into the CFP vector.
A CFP-CCTΔN40NLSCTerm mutant where an NH2-terminally truncated CCTα harbored a carboxyl-terminal NLS was constructed as follows. GST-CCT was used as the template using the primers 5′-ACTGTCGACATGGATGCACAGAGTTCAGCT-3′ (forward) and 5′-ACTGGATCCACCAACTGCACAGCGCTG-3′ (reverse) to amplify the 120-bp NH2-terminal CCT fragment. The forward primer and reverse primers contain a SalI site and a BamHI site, respectively, each with 3-bp overhangs. The PCR product was gel purified, followed by digestion with the above enzymes, prior to directional cloning into the CFP-CCTN40 vector.
A CFP-CCTΔN40K57-Ub-NLSCTerm mutant where the NH2-terminally truncated CCTα also contained an internal ubiquitin signal and a carboxyl-terminal NLS was constructed as follows. CFP-CCTK57-ubiquitin was used as the template utilizing the primers 5′-ACTCTCGAGTTACGGCAGCCAGCTCCT-3′ (forward) and 5′-ACTGTCGACGTCCTCTTCATCCTCGC-3′ (reverse) to amplify the 1,209-bp CCTΔN40Ubi fragment. The forward primer contains an XhoI site and a 3-bp overhang; the reverse primer contains a SalI site and a 3-bp overhang. The PCR product was gel purified, followed by digestion with the enzymes described above. In another reaction, GST-CCT was used as the template utilizing the primers 5′-ACTGTCGACATGGATGCACAGAGTTCAGCT-3′ (forward) and 5′-ACTGGATCCACCAACTGCACAGCGCTG-3′ (reverse) to amplify the 120-bp NH2-terminal CCTα fragment. The forward primer contains a SalI site and a 3-bp overhang; the reverse primer contains a BamHI site and a 3-bp overhang. The PCR product was gel purified, followed by digestion with the enzymes described above. The two PCR products were ligated into the CFP vector in a three-fragment ligation. All of the PCR conditions were as follows: 98°C for 30 s and then 35 cycles of 98°C for 15 s, 62°C for 30 s, and 72°C for 30 s.
Protein expression and siRNA.
Transfection of all protein constructs was conducted using FuGENE6 using a modified protocol. MLE cells were suspended in DMEM plus F-12 medium containing 2% FBS, 18 μl of FuGENE6 reagent, and 6 μg of the desired plasmid/dish. Using this protocol, cellular expression of green fluorescence-tagged plasmids was achieved at 60 to 80% in MLE cells. In some studies, at 24 h after transfection, TNF-α (0.75 μg/ml) was added for an additional 36 h. For TSG101 siRNA studies, 106 cells were transfected with 0.2 pmol of scramble RNA or TSG101 siRNA for 24 h. Cells were then treated with or without TNF-α (0.75 μg/ml) for an additional 24 h prior to harvest.
In vitro ubiquitin conjugation assay.
Ubiquitin conjugation reaction mixtures were prepared before the addition of purified CCTα substrate. A 1× reaction mixture contains 0.5 μg of conjugation fraction I and 0.5 μg of conjugation fraction II, 1 μg of ubiquitin, and energy solution containing ATP that was provided with the ubiquitin conjugation kit. A total of 0.1 μg of purified rat liver CCTα was added to individual reactions containing increasing concentrations of the ubiquitin conjugation reaction mixtures; reactions were incubated at 30°C for 120 min. As a negative control, 0.1 μg of CCTα was incubated with the reaction mixture, without ubiquitin. In other studies, 0.1 μg of purified rat liver CCTα was added to a 9× reaction mixture, followed by incubation at 30°C for up to 120 min. Reactions were then stopped by heating at 100°C for 5 min with 80 μl of protein sample buffer, and products were subsequently processed for immunoblotting. In other studies, 2 to 4 μg of purified His-CCT was used as a substrate in the ubiquitin conjugation reaction with or without PtdCho/oleic acid vesicles.
CCTα degradation.
CCTα turnover was determined by incubating MLE cells with cycloheximide. Cells were first transfected with V5-CCT or V5-CCTK57R, followed by exposure of the cells to cycloheximide (15 μg/ml) in DMEM without FBS for various times prior to harvesting. In other studies, MLE cells were first transfected with CFP-CCT or CFP-CCTK57R, followed by exposure of the cells to cycloheximide (15 μg/ml), in DMEM without FBS, for various times prior to fixing in paraformaldehyde and immunostaining.
Immunostaining.
MLE cells were plated and transfected with different CCTα constructs. Immunofluorescence cell imaging was performed on a Zeiss LSM 510 confocal microscope using a 458-, 568-, or 615-nm wavelength. All experiments were done with a Zeiss 63× oil differential interference contrast objective lens. MLE cells were pretreated with 0.001% digitonin-phosphate-buffered saline (PBS) for 2 min to partially permeabilize cell membranes, washed with PBS, fixed with 4% paraformaldehyde for 20 min, and then exposed to 15% bovine serum albumin, 1:200 M6PR primary antibody, and 1:200 Alexa 568-labeled goat anti-rabbit secondary antibody sequentially for immunostaining. In other studies, MLE cells were washed with PBS, fixed with 4% paraformaldehyde for 20 min, and blocked by 15% bovine serum albumin for 1 h. A 1:2,000 dilution of To-Pro-3 nuclear staining solution was used to visualize nuclei. In separate studies, MLE cells were incubated with 1:10,000 LysoTracker Red, followed by PBS washing and 4% paraformaldehyde fixation. A 458-nm wavelength was used to excite CFP-CCT fusion protein, with fluorescence emission collected through a 475- to 500-nm filter. A 568-nm wavelength was used to excite the Alexa 568 dye, with fluorescence emission collected through a 585-nm filter. A 568-nm wavelength was used to excite LysoTracker Red, with fluorescence emission collected through a 585-nm filter. A 613-nm wavelength was used to excite the To-Pro-3 dye, with fluorescence emission collected through a 633-nm filter. Scanning was bidirectional at the highest possible rate measurement using a 1× zoom lens. Images were exported as 12-bit lsm files. For quantitation of fluorescence, 10 individual cells for each condition were analyzed. After subtraction of background, the regions of interest were selected in the nucleus and in the cytosol of each cell, and the average intensity was measured and calculated by using ImageJ analysis software.
His pulldown assay.
Transfected His-tagged constructs were extracted in lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 10 mM imidazole, 1% Triton X-100; pH 7.4) and purified and eluted from Talon beads according to the manufacturer's instructions.
His pulldown importin binding.
Purified CCTWT or CCTK57R (4 μg) was used as a substrate for in vitro ubiquitination reactions. After 4 h of incubation at 30°C, the reaction mixtures and untreated controls were incubated with 40 μl of Talon cobalt beads at 4°C for 2 h. Beads were washed thoroughly with PBS containing 0.2% NP-40 and 0.2% Triton X-100. After a washing step, 2 μg of purified importin-2α was added to the beads for an additional 1 h of incubation. The beads were washed thoroughly again, followed by elution with 250 mM imidazole and concentration with Centricon, and the products were subsequently processed for immunoblotting. Densitometry was performed on control and monoubiquitinated CCTαs, and their relative association with importin-2α was assessed.
FRET.
Importin-2α was cloned using PCR with mouse type II cell cDNA as a template. Primers contained convenient restriction sites for ligation of importin-2α into the YFP vector. For fluorescence resonance energy transfer (FRET), cells were cotransfected with YFP-importin and one of six other plasmids (CFP-CCT, CFP-CCTK57R, CFP-Ub-CCT, CFP-CCTK57Ub, CFP-CCTΔN40K57-Ub-NLSCTerm, and CFP alone; 2 μg of plasmid/chamber), and importin-CCT interaction was detected at the single cell level by using a combination laser-scanning microscope system as described previously (6). The average fluorescence intensities/per pixel were calculated following background subtraction. FRET efficiency (EFRET) was calculated as follows: EFRET = (1 − CFPbefore/CFPafter) × 100. Three experiments were performed, and >12 randomly selected cells for each condition were analyzed.
Statistical analysis.
Statistical analysis was performed by using one-way analysis of variance with a Bonferroni adjustment or a Student unpaired t test. The data are expressed as means ± the standard errors of the mean.
RESULTS
CCTα is monoubiquitinated in vitro and in vivo.
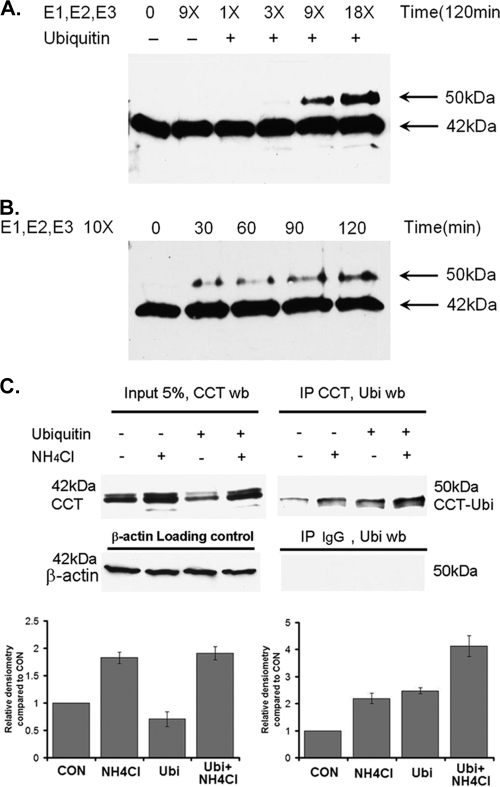
Purified CCTα was incubated with increasing concentrations of the full complement of purified conjugation enzymes (E1, E2s, and E3s), plus ubiquitin (∼8.5 kDa) and ATP. Reactions were resolved by SDS-PAGE and analyzed by immunoblotting. After 120 min., CCTα antibodies recognized both the 42-kDa purified form of CCTα, plus an additional species of ∼50 kDa, a predicted size consistent with monoubiquitinated CCTα. The proportion of this 50-kDa species relative to the 42-kDa species increased with increasing concentrations (Fig. 1A) and duration (Fig. 1B) of reaction with the ubiquitin-conjugating enzymes. As a negative control, CCTα was incubated with an intermediate concentration of the conjugation enzyme mixture, but in the absence of ubiquitin; these control reactions did not generate the 50-kDa immunoreactive species (Fig. 1A). These data strongly suggest that purified CCTα is ubiquitinated in vitro. Moreover, the presence of only one additional slower-migrating species at 50 kDa suggests that CCTα is subject to monoubiquitination.
FIG. 1.
CCTα is monoubiquitinated in vitro and in vivo. (A) Purified CCTα (0.1 μg) was incubated with increasing amounts of conjugation enzyme mixture for 120 min. Reactions were terminated by adding 20 μl of SDS loading buffer, boiled for 5 min, and then resolved by SDS-PAGE prior to CCTα immunoblotting. (B) Purified CCTα (0.1 μg) was incubated with 10 μg of conjugation enzyme mixture for 0 to 120 min. Reactions were terminated and processed by SDS-PAGE prior to CCTα immunoblotting as described above. (C) On the left, MLE cells transfected with ubiquitin (Ubi) were incubated for 24 h and then exposed to 20 mM NH4Cl for additional 24 h to impair endosomal function, thereby allowing monoubiquitinated protein to accumulate. Cells were lysed, and 10 μg of lysate was resolved by SDS-PAGE prior to CCTα immunoblotting. The presence of faint upper bands represents CCT phosphorylation variants. Alternatively 200-μg lysates were immunoprecipitated with a CCTα rabbit polyclonal antibody, as shown on the right. Immunoprecipitants were resolved by SDS-PAGE prior to ubiquitin immunoblotting. As a negative control, 200-μg lysates were immunoprecipitated with a rabbit immunoglobulin G. Immunoprecipitants were resolved by SDS-PAGE prior to ubiquitin immunoblotting. Below are the results of a densitometric analysis of bands on immunoblots from four independent experiments.
To evaluate whether monoubiquitinated CCTα could be detected in cells, MLE cells were first transfected with ubiquitin and, after 24 h, exposed to 20 mM NH4Cl. NH4Cl impairs acidification in endocytic vesicles, thereby attenuating degradative capacity, allowing ubiquitinated proteins to be better visualized (18). Cells were then lysed and immunoprecipitated with a rabbit polyclonal antibody recognizing CCTα, or a rabbit immunoglobulin G as a negative control; immunoprecipitates were then resolved by SDS-PAGE and probed with a ubiquitin antibody. Typically, CCTα appears as two or three bands clustered at ∼42 kDa, a finding indicative of phosphorylation variants (Fig. 1C, left). Monoubiquitinated proteins are very difficult to detect in cells because the subpopulation of select proteins that are ubiquitinated is extremely small (16). However, after immunoprecipitation, immunoblots identified a 50-kDa immunoreactive band, which is consistent with the presence of monoubiquitinated CCTα (Fig. 1C, right panel). Cells exposed to NH4Cl with transfected ubiquitin plasmid displayed the highest levels of the 50-kDa (monoubiquitinated) CCTα protein (Fig. 1C, right panel, lower graph). Thus, CCTα may be naturally targeted by monoubiquitination for subsequent degradation via the lysosomal pathway. Polyubiquitinated CCTα species were not detected using these approaches. These results suggest that CCTα undergoes monoubiquitination in vitro and in vivo.
TNF-α induces CCTα monoubiquitination and degradation.
In our prior studies, TNF-α stimulates protein ubiquitination and decreases CCTα activity (21). We exploited the observation that monoubiquitinated proteins are generally translocated and degraded in the lysosome, whereas polyubiquitinated proteins are degraded in the 26S proteasome (5). To distinguish between monoubiquitination and polyubiquitination of CCTα after TNF-α stimulation, cells were treated with NH4Cl or the proteasomal inhibitor, lactacystin. Compared to control cells, TNF-α with or without lactacystin treatment reduced CCT activity by 50% (Fig. 2A). Unlike the effects of lactacystin, inclusion of 20 mM NH4Cl in the medium totally blocked TNF-α inhibition of CCT activity. Immunoblotting experiments revealed that CCTα was degraded in cells exposed to TNF-α, an effect totally blocked with NH4Cl; similar effects were not observed with lactacystin (Fig. 2B). Further, in cells exposed to NH4Cl and exposed to TNF-α, a 50-kDa band was also detected in cell lysates, which is consistent with monoubiquitinated CCTα (Fig. 2B, right side). This band was only seen after larger amounts of cellular protein (50 μg) were loaded onto gels and the films were exposed extensively (Fig. 2B, right side). Densitometric analysis indicates that endogenous CCTα-ubiquitin only represents about 1 to 2% of the total CCTα in MLE cells, whereas ubiquitinated CCTα can accumulate up to 5% of this total when cells are exposed to TNF-α and NH4Cl.
FIG. 2.
TNF-α degradation of CCTα is mediated by the endosome-lysosomal pathway. (A and B) MLE cells were exposed to TNF-α (0.75 μg/ml) with or without NH4Cl (20 mM) or lactacystin (20 μM) treatment for 24 h. Cells were then harvested for analysis of CCT activity (A) or for CCTα and β-actin immunoblotting (B). (C and D). MLE cells were transfected with 0.2 pmol of scramble siRNA or TSG101 siRNA for 24 h. Cells were then treated with or without TNF-α (0.75 μg/ml) for an additional 24 h. Cells were then harvested for analysis of CCT activity (C) or for CCTα, TSG101, and β-actin immunoblotting (D). The data represent the results of three independent experiments. *, P < 0.05 versus the control.
To examine the molecular machinery that mediates monoubiquitinated CCT lysosomal degradation, we used siRNA against the ESCORT I (for endosomal sorting complex required for transport) subunit, TSG101. ESCORT I complex proteins bind ubiquitinated proteins on endosomal membranes and assist in lysosomal degradation (16). TSG101 is a key subunit of ESCORT, which directly interacts with ubiquitinated proteins (16). Loss-of-function studies of TSG101 impairs targeting of ubiquitin proteins to the endocytic pathway (16). Cells were transfected with scrambled RNA or TSG101 siRNA for 24 h and then treated with or without TNF-α for an additional 24 h. TNF-α reduced CCT activity by 50% (Fig. 2C) and produced a significant decrease in CCTα protein in cells transfected with scrambled RNA. TSG101 siRNA produced an 80% knockdown of TSG101 expression (Fig. 2D). Further, in cells transfected with TSG101 siRNA, CCT activity and immunoreactive levels were significantly restored despite TNF-α treatment. These data suggest that TNF-α utilizes the ESCORT I complex to degrade ubiquitinated CCTα within the endosomal-lysosomal degradation pathway.
Monoubiquitinated CCTα is localized to the endosome and/or lysosome.
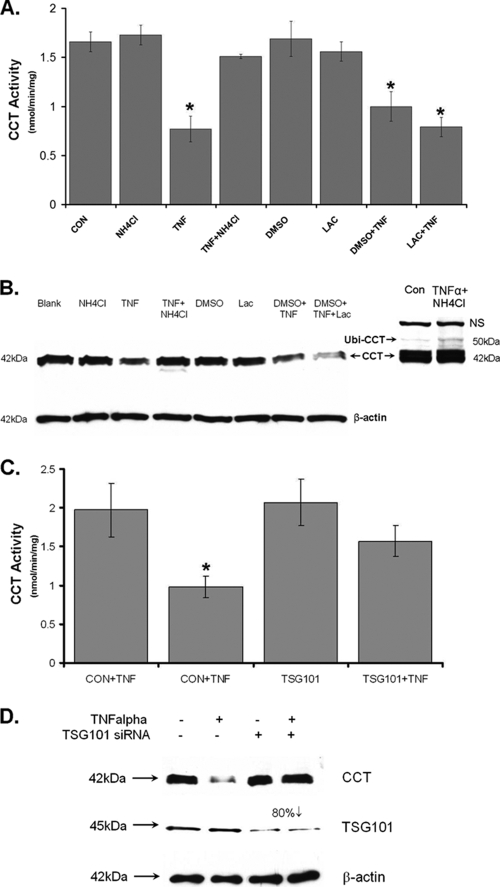
To evaluate whether ubiquitinated CCTα is targeted to the endosomal-lysosomal pathway, cells were transfected with CFP-CCT prior to exposure to NH4Cl with or without TNF-α or vehicle. As previously shown, overexpressed CFP-CCT was detected within the nucleus (Fig. 3A, top panels) (6). Next, upon selective permeabilization of the plasma membrane, cytosolic signal intensities were reduced to optimize colocalization with subcellular markers. Cells were permeabilized with 0.001% digitonin, fixed, and immunostained with antibody to the late endosomal marker, M6PR (Fig. 3A, lower left panels). Under these conditions, treatment with NH4Cl effectively increased expression of CFP-CCT within the cytoplasm, a finding consistent with enzyme accumulation secondary to the inhibition of lysosomal degradation. The addition of TNF-α alone or in combination with NH4Cl resulted in a robust accumulation of CFP-CCT in the cytoplasmic compartment. Further, a pronounced punctate signal was observed in all cells visualized with M6PR antibody, and these signals colocalized with CCTα (Fig. 3A, lower left panels [Merge]). In other experiments, cells were treated with LysoTracker Red for 30 min to stain the lysosome. Similarly, NH4Cl, TNF-α, or both agents significantly induced CCTα cytosolic accumulation in cells (Fig. 3A, lower right panels). TNF-α alone was not able to induce strong CCTα colocalization with the lysosomal marker, suggesting that CCTα rapidly degrades within these organelles. However, TNF-α in combination with NH4Cl resulted in intense punctate signals that colocalized with LysoTracker Red. These results provide the first evidence that TNF-α promotes the trafficking and disposal of CCTα within the endosomal-lysosomal pathway.
FIG. 3.
Monoubiquitinated CCTα accumulates in the cytosol after lysosomal inhibition and effects of a CCTα-ubiquitin mimic. (A) MLE cells were first transfected with CFP-CCTα for 12 h and then exposed to NH4Cl (20 mM/ml) with or without TNF-α (0.75 μg/ml) or vehicle for an additional 36 h. Cells were permeabilized with 0.001% digitonin-PBS and then fixed and counterstained with To-Pro-3 (to visualize the nucleus, top panel). Cells were also immunostained with an antibody recognizing M6PR (late endosome marker, left panel). Cells were incubated with a 1:10,000 dilution of LysoTracker Red (lysosomal marker), followed by PBS washing and 4% paraformaldehyde fixation (right panel). (B) Expression of CCTα-ubiquitin fusion proteins. WT CCTα (CFP-CCT) and two monoubiquitinated CCTα mimics, where one copy of WT ubiquitin or ubiquitin with all six lysines mutated (CFP-UbiΔK-CCT) was fused to the CCTα NH2 terminus (CFP-Ubi-CCT) and was transfected into MLE cells. At 24 h after transfection, cells were analyzed as in panel A. The CCTα-ubiquitin fusion proteins that mimic monoubiquitinated CCTα accumulate predominantly in the cytoplasm and colocalize with M6PR and LysoTracker Red. The data represents the results of three independent experiments.
To investigate whether monoubiquitination is sufficient for cytoplasmic translocation of CCTα, we constructed fusion proteins that mimic the monoubiquitinated form of CCTα. These proteins, CFP-CCTα and CFP-Ubi-CCT (where one copy of the ubiquitin sequence was fused in frame to the NH2 terminus of CCTα), were functional and exhibited appropriate membrane targeting when expressed in cells (data not shown). CFP-CCTα expressed nuclear localization, whereas immunostaining showed both nuclear and cytosolic localization of the CFP-Ubi-CCT chimera, with significant colocalization with late endosomes or lysosomes (Fig. 3B). Thus, NH2-terminal fusion of CCTα with monoubiquitin is sufficient to sequester the enzyme within the cytoplasm of lung epithelia. Last, because the fused ubiquitin moiety to CCTα contains several lysine residues that could potentially be ubiquitinated forming polyubiquitin chains, we tested a CFP-UbΔK-CCT chimera where all six lysines within ubiquitin were substituted with arginines. Immunostaining of cells after expression of this construct indicated similar cytoplasmic colocalization with the late endosomal marker as with CFP-Ubi-CCT. These data suggest that monoubiquitination is sufficient for the lysosomal degradation of CCTα.
Mapping the CCTα ubiquitination site.
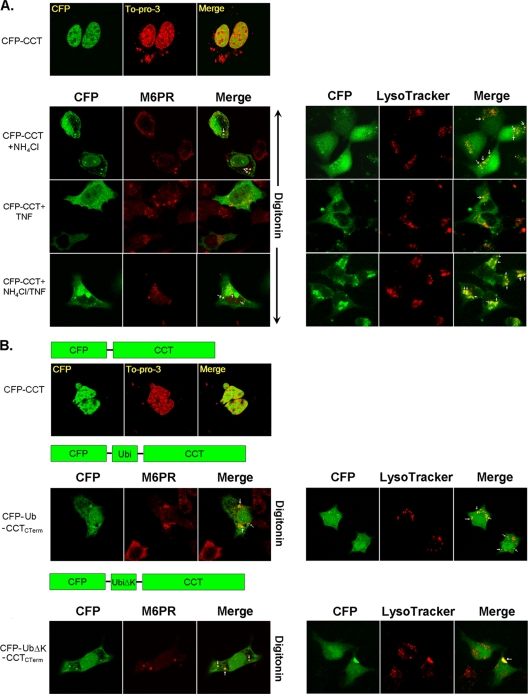
We expressed various GST-tagged CCTα proteins harboring mutations in known functional domains and coexpressed them with His-tagged ubiquitin (Fig. 4A). Cells were then harvested, and proteins were purified on cobalt beads. After stringent washing, products were eluted and resolved by SDS-PAGE prior to GST immunoblotting. As a negative control, cells were transfected with each GST-tagged CCTα construct alone (without His-ubiquitin plasmid), and cells were processed as described above (lanes 2, 5, 8, 11, 14, and 17 on each gel [Fig. 4B]). Under these conditions, the individual constructs were not detected using GST-immunoblotting after cobalt bead purification. Each of these CCTα constructs was sufficiently expressed and fusion proteins exhibited the expected mobilities on immunoblots (lanes 1, 4, 7, 10, 13, and 16 on each gel [Fig. 4B]). Full-length GST-CCT and GST-CCT mutants devoid of the membrane-binding domain (aa 236 to 315; CCTMEM), NH2-terminal residues harboring a PEST sequence (aa 1 to 40, CCTΔN40), phosphorylation (P) domain (aa 315 to 367; CCT315), and membrane-binding and P domain (aa 210 to 367; CCT210) all bound ubiquitin (Fig. 4B, lanes 3, 6, 12, 15, and 18 on each gel). However, a GST-CCTα fusion protein devoid of the catalytic region (including the catalytic core aa 73 to 236 and the NH2-terminal linker region [aa 40 to 72]; CCTCAT) did not bind ubiquitin (Fig. 4B). Thus, the acceptor site for monoubiquitination resides within the CCTα catalytic region.
FIG. 4.
Mapping of a ubiquitination site within CCTα. (A) Map illustrating full-length GST-CCTα and GST-CCTα mutants devoid of the NH2-terminal sequence (aa 1 to 40) (CCTΔN40), the membrane-binding domain (aa 236 to 315) (CCTMEM), the catalytic region (aa 40 to 236) (CCTCAT), the carboxyl-terminal phosphorylation domain (aa 315 to 367) (CCT315), or the membrane and carboxyl terminus (aa 210 to 367) (CCT210). (B) GST-tagged CCT mutants were copurified by using His-tagged ubiquitin in pulldown assays. Thus, GST-tagged CCT mutants were coexpressed in MLE cells, with or without His-tagged ubiquitin. At 24 h after transfection, cells were harvested with lysis buffer. Cell lysate was used for His purification; after stringent washing, the products were eluted and resolved by SDS-PAGE prior to GST immunoblotting. (C) The CCTα primary sequence within the catalytic core, which indicates (by vertical arrows) all of the candidate ubiquitin acceptor sites that were mutated. (D) GST-tagged CCT variants harboring point mutations within the catalytic core were copurified by His-tagged ubiquitin in pull-down assays as in panel B. GST-CCTK57R, GST-CCTK100R, GST-CCTK122R, and GST-CCTK183/186R were cotransfected with His-tagged ubiquitin. At 24 h after transfection, the cells were harvested with lysis buffer, and cell lysates were used for His pull-down assays. After stringent washing, products were eluted and resolved by SDS-PAGE prior to GST immunoblotting. The data in panels B and D represent the results of three independent experiments.
We next tested additional GST-CCT mutants harboring point mutations on each of five lysine sites within the catalytic region: GST-CCTK57R, GST-CCTK100R, GST-CCTK122R, and a double mutant, GST-CCTK183/186R (Fig. 4C). Each mutant was cotransfected with His-tagged ubiquitin and processed by purification and immunoblotting as described above. His pull-down assays clearly indicate that full-length GST-CCT and GST-CCTK100R, GST-CCTK122R, GST-CCTK183/6R mutants all bound ubiquitin (Fig. 4D). However, GST-CCTK57R was not detected by GST immunoblotting. The results indicate that K57 is the primary acceptor site for CCTα monoubiquitination.
A CCTα K57R mutant is proteolytically resistant.
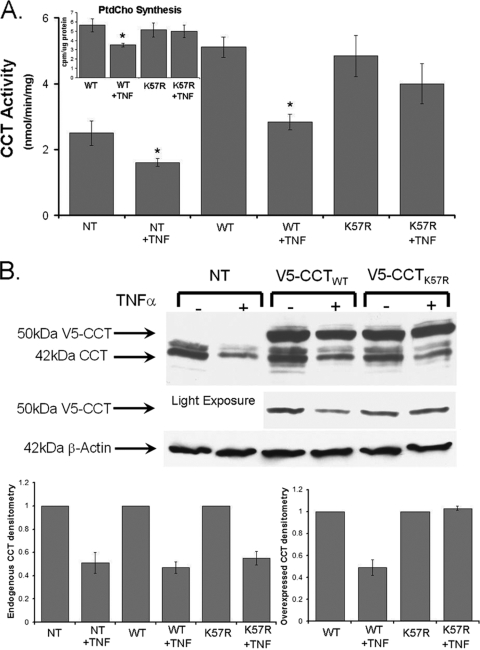
To assess the functionality of the CCT K57 site, cells were transfected with V5-CCTWT and V5-CCTK57R and exposed to TNF-α. TNF-α decreased the activity of both endogenous and overexpressed WT CCTα to 50% of control values (Fig. 5A). However, TNF-α did not significantly decrease CCT activity in cells expressing V5-CCTK57R (Fig. 5A). Importantly, these changes in enzyme activity by TNF-α were mirrored in measurements of PtdCho synthesis, where phospholipid synthesis was preserved in cells expressing the CCTK57R mutant (Fig. 5A [inset]). Immunoblotting experiments revealed that both endogenous 42-kDa CCTα and V5-CCTWT were degraded by TNF-α, whereas the levels of the 50-kDa V5-CCTK57R fusion product displayed considerable stability compared to untreated controls (Fig. 5B [light exposure and right lower graph]). Thus, K57 likely serves as the key monoubiquitination site regulating CCTα turnover in response to TNF-α since mutagenesis of this residue significantly blocks TNF-α activity in vivo.
FIG. 5.
The K57R ubiquitin acceptor site within CCTα is functionally relevant. (A) MLE cells were transfected with V5-CCTWT or V5-CCTK57R to assess the physiologic relevance of the ubiquitin acceptor site (K57). At 12 h after transfection, cells were exposed to TNF-α (0.75 μg/ml) for an additional 36 h. In some studies, cells were pulsed with [3H]choline (1 μCi/dish) for the final 3 h of incubation. Cells were then harvested for analysis of CCT activity (A), PtdCho synthesis (inset), or for CCTα and β-actin immunoblotting (B). In panel B, the overexpressed CCT mutant levels are better visualized after light exposure of immunoblots. The data in each panel represents the results of three independent experiments, and endogenous and overexpressed bands from the upper part of panel B are quantitated densitometrically below. *, P < 0.05 versus the control. NT, not transfected.
Mutating CCTα K57 significantly prolongs protein half-life and nuclear retention.
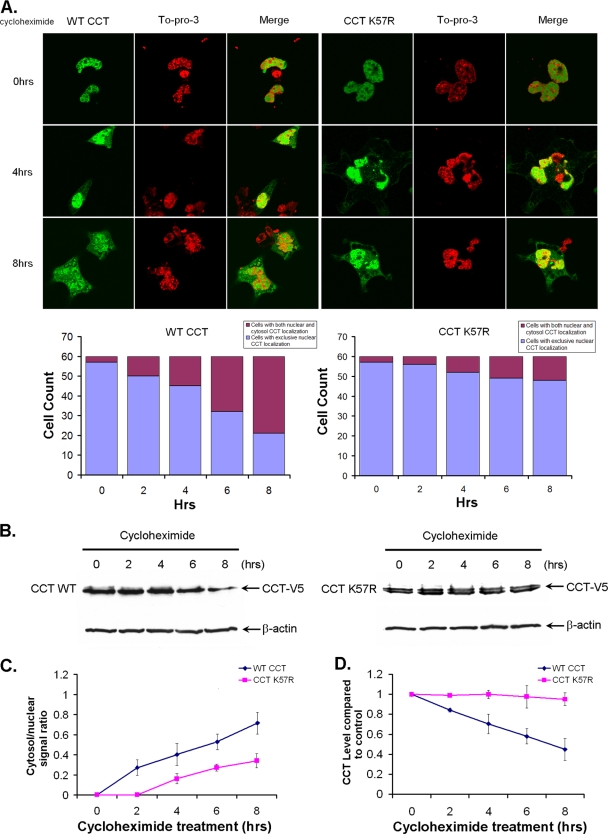
MLE cells were transfected with CFP-CCT or CFP-CCTK57R and then exposed to cycloheximide (15 μg/ml) in serum-free DMEM for 2 to 8 h prior to paraformaldehyde fixing. Sixty random cells were counted at each time point and analyzed for trafficking of each CCTα construct from the nucleus to the cytoplasm, and fluorescent signals were calculated and graphed. As shown in Fig. 6A, at baseline (t = 0 h), ∼97% of cells after expressing each construct displayed exclusive nuclear expression (Fig. 6A, graph). CFP-CCT readily translocates to the cytosol by 4 h. The cytoplasmic/nuclear ratio of the CCT fluorescent signal increases from 2 to 8 h in a time-dependent manner (Fig. 6C, graph). Compared to the WT, however, CFP-CCTK57R behaved differently in that the majority of cells still exhibited an exclusive CCTα fluorescent signal in the nucleus, and the amount of cytosolic fluorescent signal was significantly less than for the cells transfected with WT CCT over 4 to 8 h (Fig. 6A).
FIG. 6.
A K57R CCTα mutant exhibits greater protein stability and nuclear retention. (A) MLE cells were transfected with CFP-CCTWT CFP-CCTK57R and then exposed to cycloheximide (15 μg/ml) in DMEM with 0% FBS for 2 to 8 h prior to paraformaldehyde fixing. Cells were also counterstained with TOPO-3 to visualize the nucleus. A total of 60 random cells were counted at each time point, and CFP fluorescent signals within the cytosol and nucleus were calculated and plotted below in panel A and also in panel C. (B) MLE cells were transfected with V5-CCTWT (left) or V5-CCTK57R (right) and then exposed to cycloheximide (15 μg/ml) in DMEM with 0% FBS for 2 to 8 h prior to harvesting for V5 immunoblotting. (D) The densitometric results of CCTα protein were calculated and plotted. The data in each panel represents the results of three independent experiments. *, P < 0.05 versus the control.
Next, we tested whether nuclear retention of CCTK57R could extend the half-life of this functional protein. Cells were transfected with V5-CCT or V5-CCTK57R, exposed to cycloheximide (15 μg/ml) as described above, and then harvested for V5 immunoblotting. Using these methods, the levels of native CCTα enzyme decreased in a time-dependent manner when exposed to cycloheximide (Fig. 6B). In addition, the ratio of cytosolic/nuclear CCTα was greater over time for native enzyme versus the CCTK57R mutant (Fig. 6C). The densitometry results in Fig. 6D show that CCTα exhibited a t1/2 of 6 to 8 h, which is consistent with prior studies (6). In contrast, CCTK57R turnover was significantly decreased in cells exposed to cycloheximide. Together, these observations indicate that a population of CCTα molecules must be exported from the nucleus to be degraded.
Monoubiquitination of CCTα at K57 masks the NLS.
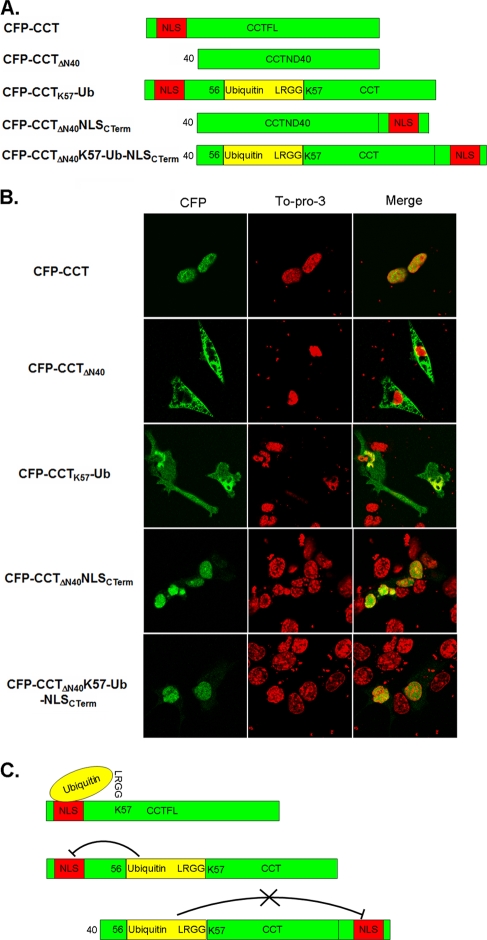
Within the first 40 NH2-terminal residues, CCTα contains an NLS necessary and sufficient for CCTα nuclear localization (9, 30). To test the mechanism of CCTα nuclear export by monoubiquitination several CFP-CCT deletion and hybrid constructs were expressed in cells. These mutants varied in the position of the NLS relative to a one-copy fusion of the ubiquitin moiety within CCTα. First, a CFP-CCTΔN40 construct that lacks the NLS localized to the cytosol exclusively (Fig. 7). We similarly tested a CFP-CCTK57-Ub mutant where ubiquitin was inserted in frame between residues Ser56 and Lys57. Since Lys57 is the major monoubiquitination site of CCTα, this fusion protein should mimic the monoubiquitinated form of CCTα. Interestingly, this fusion protein localized to both the cytosolic and the nuclear compartments. We next examined whether the inserted ubiquitin sequence masks the NLS, thus hindering the ability of CCTα to import to the nuclear membrane. As a positive control, we shifted the 40-residue stretch of the CCTα NH2 terminus containing its nuclear signal motif to the carboxyl terminus of the protein. This mutant, CFP-CCTΔN40NLSCTerm, was localized to the cell nucleus (Fig. 7B). Another mutant, CFP-CCTΔN40K57-Ub-NLSCTerm, where the internal ubiquitin signal was displaced from the carboxyl-terminal NLS, localized to the nucleus. Taken together, these novel observations strongly suggest that when CCTα is monoubiquitinated at a site near the NLS, the NLS is masked and CCTα is unable to translocate to the nucleus. The net effect is that CCTα is retained in the cytoplasm, where it may enter the endosomal-lysosomal pathway. However, if the CCTα NLS is sufficiently distanced from a putative monoubiquitination site, nuclear import and CCTα stability are preserved.
FIG. 7.
Monoubiquitination of CCTα K57 masks its NLS. (A) Schematic diagram of five CCTα constructs: CFP-CCT, CFP-CCTΔN40, CFP-CCTK57-Ub, CFP-CCTΔN40NLSCTerm, and CFP-CCTΔN40K57-Ub-NLSCTerm. (B) All constructs shown were transfected into MLE cells. At 24 h after transfection, cells were fixed by paraformaldehyde and counterstained with TOPO-3 to visualize the nucleus. (C) Diagram illustration of the proposed mechanism of CCTα nuclear export.
Monoubiquitination disrupts CCTα-importin-α interaction.
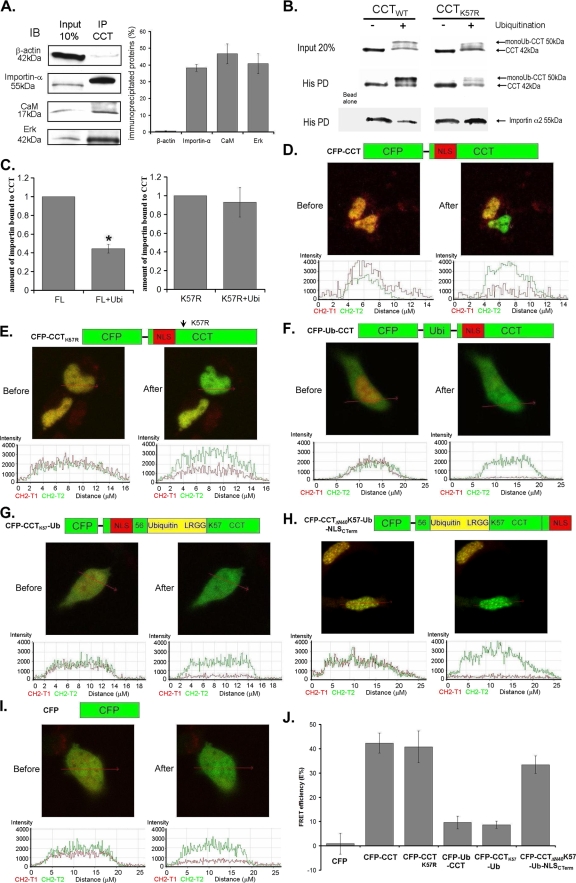
Nuclear import of NLS-containing proteins involves binding with importin-α, resulting in protein complex assembly that transits through the nuclear pore for further processing (11, 26). Disruption of cargo-importin-α association impairs nuclear entry. Thus, we tested whether monoubiquitination of CCTα impairs its ability to bind importin-α. First, by coimmunoprecipitation, importin-α was bound to CCTα (Fig. 8A). As positive controls, extracellular signal-regulated protein kinase (Erk) and calmodulin were also associated with CCTα (2, 6). His-CCTWT or CCTK57R were next monoubiquitinated in vitro, and Talon cobalt beads were added. The preparations were rinsed and then incubated with importin-α; after washing, the preparations were processed for His-CCTα pull-downs and importin-α immunoblotting. As shown in Fig. 8B, CCTWT was effectively monoubiquitinated prior to incubation with importin-α. In contrast, the CCTK57R mutant exhibited a significantly reduced ability to be monoubiquitinated (Fig. 8B, far right, upper lane). After monoubiquitination and the first purification of His-CCTα, equal amounts of enzyme were detected by CCTα immunoblotting (Fig. 8B, middle lane). Last, these preparations were incubated with importin-α and probed for CCTα association with the nuclear chaperone (Fig. 8B, lower lane). As can be seen, WT CCTα-importin-α complex formation was remarkably reduced after in vitro monoubiquitination of purified enzyme compared to unmodified substrate (Fig. 8B, lower lane, right panel). This association between importin-α and the CCTK57R mutant, however, was better preserved (Fig. 8C).
FIG. 8.
Monoubiquitination of CCTα at K57 disrupts its interaction with importin-α. (A) Coimmunoprecipitation. CCTα was immunoprecipitated from MLE cells, and samples were processed for immunoblotting for β-actin (negative control), importin-α, calmodulin (CaM), or Erk. The relative association is presented densitometrically (right graph). (B) Importin interaction assay. Purified CCTWT or CCTK57R (4 μg) were reacted in an in vitro ubiquitination reaction or incubated in buffer (top row). After incubation on cobalt beads and washing (middle row), products were reincubated with purified importin-α (2 μg), washed, and processed for CCTα or importin-α immunoblotting (lower row). (C) Densitometric analysis of immunoblots was performed showing relative amounts of importin-α associated with unmodified or monoubiquitinated CCTα after correcting for loading. (D to J) FRET analysis. Cells were cotransfected with YFP-importin-α and one of six constructs: CFP-CCT, CFP-CCTK57R, CFP-Ub-CCT, CFP-CCTK57Ub, CFP-CCTΔN40K57-Ub-NLSCTerm, and CFP alone. Importin-α-CCT interaction at the single cell level was imaged by using laser scanning microscopy before and after photobleaching. Shown in the upper sets of panels is single cell imaging showing that, after acceptor photobleaching, the fluorescence intensity of YFP decreases, and CFP increases in panels D, E, and H, confirming the protein interaction between importin-α and CCTα. In panels F, G, and I, after acceptor photobleaching, the fluorescence intensity of YFP decreases, and yet CFP does not change, indicating a lack of protein interaction between importin-α and CFP-Ub-CCT or CFP-CCTK57Ub. (J) The same FRET in each panel was confirmed quantitatively by graphing of fluorescence intensities. FRET efficiencies (E%) were calculated and graphed in panel J from three experiments and >12 randomly selected cells for each condition that was analyzed.
To assess CCTα-importin-α interaction in vivo, cells were analyzed by FRET by cotransfecting YFP-importin-α with one of six CCTα plasmids: CFP-CCT, CFP-CCTK57R, CFP-Ub-CCT, CFP-CCTK57Ub, CFP-CCTΔN40K57-Ub-NLSCTerm, and CFP alone. In FRET, energy is transferred from the donor fluorophore (CFP-CCT variant) to an acceptor fluorophore (YFP-importin-α) when proteins are in close proximity. If FRET is observed using the acceptor photobleaching method indicative of protein interaction between binding partners, the donor emission (CFP) signal increases after a nearby acceptor fluorophore (YFP) is inactivated by irreversible photobleaching. The emission fluorescence of both the donor CFP-CCT and acceptor YFP-importin-α before and after acceptor photobleaching in the region of interest in nuclei (red arrow) are shown (Fig. 8D to I, upper photos and lower plots). The data from three independent experiments and >12 randomly selected cells for each condition were analyzed and are shown graphically (Fig. 8J). As can be seen in Fig. 8D and E, cotransfection of YFP-importin-α with either CFP-CCT or CFP-CCTK57R show that, after bleaching, there was decreased acceptor fluorescence (YPF) coupled with an increase in donor emission fluorescence (CFP) because the acceptor cannot take in energy after its photobleaching. Thus, the data support CCT and importin-α interaction in lung epithelia. These results are similar to findings using FRET with importins and their binding to nuclear pore proteins (7). In contrast, neither CFP-Ub-CCT, CFP-CCTK57Ub, nor the negative control CFP alone exhibited FRET when cotransfected with YFP-importin-α (Fig. 8F, G, and I). Specifically, CFP emission fluorescence of the donor did not increase despite marked reduction in acceptor (YFP) emission after photobleaching. Importantly, the CFP-CCTΔN40K57-Ub-NLSCTerm construct, where the internal ubiquitin signal was displaced from the carboxyl-terminal NLS within CCTα, showed a positive FRET result with importin-α, indicating that the distal carboxy-terminal NLS is able to interact with importin-α (Fig. 8H). The data are consistent with the importin binding assays and indicate that monoubiquitinated CCTα variants do not interact robustly with importin-α in vivo.
DISCUSSION
Monoubiquitination has emerged as a means to target resident nuclear proteins for export into the cytoplasm (5); however, the mechanisms have not been elucidated. In the present study, we provide the first evidence that protein ubiquitination masks the NLS of a key regulatory enzyme, CCTα, thereby leading to abrogation of its binding to importin-α, its cytosolic retention, and lysosomal degradation. This was demonstrated using positional hybrid mutant constructs of CCTα that varied in intermolecular spacing between the NLS and ubiquitin. We further demonstrate that CCTα is not polyubiquitinated but undergoes site-specific monoubiquitination as a means to regulate enzyme half-life in cells. Evidence in support of CCTα monoubiquitination includes the observations that (i) CCTα was effectively conjugated with single ubiquitin in vitro; (ii) the enzyme was detected in cells in a monoubiquitin-CCTα complex; and (iii) cellular levels accumulated after treatment with NH4Cl, which impairs the clearance of monoubiquitinated substrates. The data also suggest that juxtapositioning of a ubiquitin acceptor site (K57) near an exposed nuclear targeting sequence is efficiently exploited by cells to mislocalize an enzyme that is normally targeted for nuclear membrane activation. Thus, monoubiquitination may be an important homeostatic signal by which cells brand CCTα for degradation to maintain an optimal balance of membrane phospholipid.
The present study differs from our previous work, where we showed that TNF-α-induced CCTα degradation is attenuated with lactacystin, suggesting that the enzyme is also polyubiquitinated and processed within the proteasome (21). However, that earlier study differs significantly from the present study with regard to experimental design, the concentrations and kinetics of several reagents used, and the culture conditions. Despite using clasto-lactacystin in the present study, an active metabolite of lactacystin that is severalfold more active than the lactacystin used previously, we were not able to abrogate TNF-α-induced CCTα degradation. Our results do not exclude the possibility that CCTα is monoubiquitinated, multiubiquitinated, or even polyubiquitinated depending on experimental conditions as observed for other regulatory proteins (e.g., p53) or hormone receptors that are channeled via different degradative pathways (15, 28, 29). TNF-α causes both polyubiquitination and monoubiquitination of IκB kinase proteins (4, 10), and the cytokine's ability to differentially regulate CCTα ubiquitination would not be surprising given its pleiotropic effects. Although we identified a physiologically relevant ubiquitin acceptor site (K57) within CCTα, the CCTK57R mutant significantly, but not completely, displayed reduced ability to be monoubiquitinated (Fig. 8A). Thus, there may be a second ubiquitination acceptor site within the enzyme. Preliminary mass spectrometry data showing that CCTK341 may be ubiquitinated is consistent with the detection of two immunoreactive CCTα bands at 50 and 58 kDa at times in cell lysates (data not shown). Thus, CCTα might be multiubiquitinated in vivo.
The CCTα NLS harbors a stretch of basic residues typical of nuclear signal motifs that might interact with the karyopherin, importin-α, that after forming a complex with CCT translocates through the nuclear pore complex; once within the nucleus, the karyopherin-cargo complex dissociates by the action of RanGTPase (26). CCTα cytoplasmic-nuclear shuttling also appears to utilize this pathway since importin-α was bound to unmodified enzyme in vitro and in vivo, but less so with monoubiquitinated enzyme (Fig. 8). Preliminary fluorescence recovery after photobleaching studies show that CFP-CCTα nuclear import is accelerated after cotransfection of cells with importin-α (see Movies S1 and S2 in the supplemental material). Moreover, posttranslational modifications, masking of cargo nuclear transport signals by conformational changes, and protein-protein interaction can prevent karyopherin-cargo binding and disrupt the trafficking of NLS-containing proteins (1, 14, 22, 25, 26). The masking of nuclear transport signals by monoubiquitination, however, represents a previously unrecognized mechanism whereby proteins are retained within the cytoplasm.
Thus far, the most well-understood monoubiquitinated protein is p53, a protein ubiquitinated by the RING domain E3 ligase, MDM2; this monoubiquitination event causes p53 nuclear export (20). The detailed mechanism of p53 nuclear export, however, remains largely unknown. A recent study hypothesized, but did not test, the possibility that p53 monoubiquitination within its carboxyl terminus alters its three-dimensional structure, unmasking a hidden carboxyl-terminal NES (5). Unlike p53, CCTα does not harbor a canonical leucine-rich NES, nor does leptomycin, an inhibitor of exportin-α, lead to nuclear enzyme accumulation (24). Thus, it would appear that the molecular model for CCTα trafficking by monoubiquitination would be NES independent and might involve factors that lead to its nuclear exclusion by functional disruption of its NLS. To address this, a ubiquitin-CCTα fusion protein (CFP-CCTK57-Ub) was constructed. When a single ubiquitin copy is linked to Lys57, the carboxyl-terminal ubiquitin Gly will form an isopeptide bond with the Lys ɛ-amino group of CCTα. Because of the flexibility of the Gly residue, the CCTα-ubiquitin fusion protein will mimic monoubiquitinated CCTα. However, there are two caveats using this approach. First, ubiquitin fusions can themselves serve as ubiquitins and conjugate to other molecules (5). Second, spacing between Ser56 and Lys57 within the CCTα amino terminus would normally be in the order of only 1 to 2 Å but, based on the crystal structure of ubiquitin, its insertion between these residues is predicted to increase this distance to 20 to 30 Å, thus potentially altering CCTα conformation. Nevertheless, since residues 8 to 28 representing the CCTα NLS would be in close proximity to the ubiquitinated moiety, we hypothesized that in the CFP-CCTK57-Ub fusion protein the NLS is masked. To verify this, we shifted the NH2-terminal segment (aa 1 to 40) of CCTα to the carboxyl-terminal end to increase the intermolecular distance between the NLS and ubiquitin (Fig. 7). Indeed, expression of this ubiquitin fusion construct (CFP-CCTΔN40K57-Ub-NLSCTerm) was sufficient to trigger nuclear import of CCTα by exposure of its NLS. These intermolecular spatial features may be important since the nuclear targeting motif consists mostly of basic residues that, in the cytosol, would be predicted to have a net positive charge at a pH range of ∼7.40. The isoelectric point of ubiquitin is 6.79, giving ubiquitin a net negative charge under physiologic conditions. Thus, electrostatic interactions would promote ubiquitin binding to the NLS, thereby neutralizing its charge and potentially impairing the assembly of a CCTα-karyopherin cargo complex.
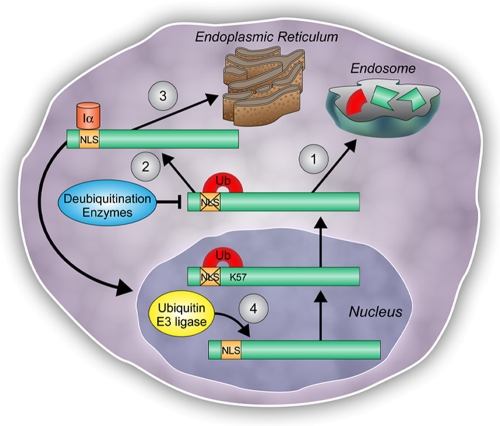
Nuclear import of CCTα is also essential for the generation of the nuclear membrane network (13). A CCTα mutant devoid of a 21-residue stretch within its NH2 terminus lacked nuclear import ability and yet remained functional (30). This NH2-truncated CCTα mutant perhaps retains activity by relocating and binding to other membrane structures via the M domain, presumably within the cytoplasm. Still other data suggest that CCTα is stored within the nucleus when cellular requirements for PtdCho are low (24). Upon increased demand for PtdCho synthesis, CCTα is recruited to the endoplasmic reticulum. Our model of CCTα monoubiquitination is compatible with each of these findings. When cell needs for PtdCho synthesis are low, nuclear CCTα containing an intact NLS that is disassembled by RanGTPase may be rapidly exported, monoubiquitinated, and targeted for lysosomal disposal (Fig. 9). In this way, soluble, but not membrane-associated CCTα would be more prone to ubiquitination. However, in the setting of increased demand for membrane biogenesis, the levels of monoubiquitinated CCTα may be low to facilitate nuclear translocation and expansion of the nuclear reticulum (13). Here, more CCTα would be membrane associated and relatively protected from ubiquitin modification. In support of this model, inclusion of lipid vesicles in the in vitro ubiquitination conjugation reaction or exposure of cells to oleic acid (to induce membrane binding of CCTα) reduces the pool of ubiquitinated CCTα (data not shown). Thus, perhaps CCTα membrane association and ubiquitination are linked, thereby regulating cellular PtdCho homeostasis. Last, monoubiquitinated CCTα may be modified in the cytoplasm as a mechanism to augment phospholipid synthesis. In the cytosol, sequential ubiquitin editing by actions of a deubiquitinating enzyme (DUB) could cleave ubiquitin from the CCTα-ubiquitin complex, thus preventing endosomal degradation of the enzyme (Fig. 9). This scenario occurs for membrane recycling of the epithelial sodium channel (3). Deubiquitinated or free CCTα molecules could then bind ER membranes to help catalyze the formation of PtdCho or reenter the nucleus.
FIG. 9.
Model for CCTα nuclear activation, exclusion, and proteolytic processing mediated by protein monoubiquitination. A schematic diagram illustrates the proposed pathways by which CCTα is regulated by monoubiquitination. (Arrow 1) CCTα monoubiquitination at K57 leads to masking of its NLS, thus mislocalizing the enzyme to the lysosome for degradation. (Arrow 2) Under conditions when the demand for PtdCho synthesis is high, editing of monoubiquitinated CCTα by the action of DUBs may result in free CCTα. Deubiquitinated CCTα can either dock to endoplasmic reticulum membranes (arrow 3) or translocate to the nuclear envelope by NLS-directed recruitment of karyopherins, such as importin-α (Iα), to augment PtdCho synthesis (arrow 4). Finally, when the demand for membrane phospholipid synthesis is low, nuclear CCTα can undergo monoubiquitination and export for proteolytic processing via the lysosomal pathway outlined in the present study.
When analyzed by immunoblotting, considerably little CCTα is monoubiquitinated compared to the levels of deubiquitinated CCTα. The endogenous 50-kDa CCTα-ubiquitin complex is extremely difficult to detect by standard chemiluminescent analysis unless the film is exposed extensively or cells are exposed to NH4Cl (Fig. 2). These results likely reflect either very rapid translocation of CCTα-ubiquitin complexes to the lysosome coupled with efficient degradation, that monoubiquitination of CCTα occurs very slowly, or that there exists competition of substrate between ubiquitin ligases and DUBs. The latter might also explain why CCTα exhibits a relatively long t1/2 (6 to 8 h). Interestingly, the CCTK57R mutant was retained in the nucleus and had an extremely long half-life. These results suggest that translocation of CCTα from the nucleus to the cytosol is required for its turnover and that proteasomes or proteinases within the nucleus play a lesser role in regulating enzyme stability. The translocation of CCTα might also have additional roles similar to that of other membrane proteins that require sorting into the multivesicular bodies prior to degradation in the lysosome. In this process, ubiquitin serves as a sorting signal (23).
Supplementary Material
Acknowledgments
We thank Chantal Allamargot for helping with the fluorescence confocal imaging.
This study was supported by an American Heart Association Predoctoral Fellowship Award (B.B.C.), a Merit Review Award from the Department of Veteran's Affairs, and NIH R01 grants HL081784, HL097376, and HL068135 (to R.K.M.). B.B.C. is a recipient of the 2008 ASBMB Graduate Student Travel Award for presentation of this study.
Footnotes
Published ahead of print on 30 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abrams, C. C., D. A. Chapman, R. Silk, E. Liverani, and L. K. Dixon. 2008. Domains involved in calcineurin phosphatase inhibition and nuclear localization in the African swine fever virus A238L protein. Virology 374477-486. [DOI] [PubMed] [Google Scholar]
- 2.Agassandian, M., J. Zhou, L. A. Tephly, A. J. Ryan, A. B. Carter, and R. K. Mallampalli. 2005. Oxysterols Inhibit phosphatidylcholine synthesis via ERK docking and phosphorylation of CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 28021577-21587. [DOI] [PubMed] [Google Scholar]
- 3.Butterworth, M. B., R. S. Edinger, H. Ovaa, D. Burg, J. P. Johnson, and R. A. Frizzell. 2007. The deubiquitinating enzyme UCH-L3 regulates the apical membrane recycling of the epithelial sodium channel. J. Biol. Chem. 28237885-37893. [DOI] [PubMed] [Google Scholar]
- 4.Carter, R. S., K. N. Pennington, P. Arrate, E. M. Oltz, and D. W. Ballard. 2005. Site-specific monoubiquitination of IκB kinase IKKβ regulates its phosphorylation and persistent activation. J. Biol. Chem. 28043272-43279. [DOI] [PubMed] [Google Scholar]
- 5.Carter, S., O. Bischof, A. Dejean, and K. H. Vousden. 2007. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 9428-435. [DOI] [PubMed] [Google Scholar]
- 6.Chen, B. B., and R. K. Mallampalli. 2007. Calmodulin binds and stabilizes the regulatory enzyme, CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 28233494-33506. [DOI] [PubMed] [Google Scholar]
- 7.Damelin, M., and P. A. Silver. 2000. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol. Cell 5133-140. [DOI] [PubMed] [Google Scholar]
- 8.d'Azzo, A., A. Bongiovanni, and T. Nastasi. 2005. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6429-441. [DOI] [PubMed] [Google Scholar]
- 9.DeLong, C. J., L. Qin, and Z. Cui. 2000. Nuclear localization of enzymatically active green fluorescent protein-CTP:phosphocholine cytidylyltransferase alpha fusion protein is independent of cell cycle conditions and cell types. J. Biol. Chem. 27532325-32330. [DOI] [PubMed] [Google Scholar]
- 10.Ea, C. K., L. Deng, Z. P. Xia, G. Pineda, and Z. J. Chen. 2006. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22245-257. [DOI] [PubMed] [Google Scholar]
- 11.Faul, C., S. Huttelmaier, J. Oh, V. Hachet, R. H. Singer, and P. Mundel. 2005. Promotion of importin α-mediated nuclear import by the phosphorylation-dependent binding of cargo protein to 14-3-3. J. Cell Biol. 169415-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedberg, E. C. 2006. Reversible monoubiquitination of PCNA: a novel slant on regulating translesion DNA synthesis. Mol. Cell 22150-152. [DOI] [PubMed] [Google Scholar]
- 13.Gehrig, K., R. B. Cornell, and N. D. Ridgway. 2008. Expansion of the nucleoplasmic reticulum requires the coordinated activity of lamins and CTP:phosphocholine cytidylyltransferase α. Mol. Biol. Cell 19237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harreman, M. T., T. M. Kline, H. G. Milford, M. B. Harben, A. E. Hodel, and A. H. Corbett. 2004. Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J. Biol. Chem. 27920613-20621. [DOI] [PubMed] [Google Scholar]
- 15.Huang, F., D. Kirkpatrick, X. Jiang, S. Gygi, and A. Sorkin. 2006. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21737-748. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda, H., and T. K. Kerppola. 2008. Lysosomal localization of ubiquitinated jun requires multiple determinants in a lysine-27 linked polyubiquitin conjugate. Mol. Biol. Cell. 194588-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackowski, S., and P. Fagone. 2005. CTP:phosphocholine cytidylyltransferase: paving the way from gene to membrane. J. Biol. Chem. 280853-856. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, K. G., H. Barriere, C. J. Carbone, J. Liu, G. Swaminathan, P. Xu, Y. Li, D. P. Baker, J. Peng, G. L. Lukacs, and S. Y. Fuchs. 2007. Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis. J. Cell Biol. 179935-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagace, T. A., J. R. Miller, and N. D. Ridgway. 2002. Caspase processing and nuclear export of CTP:phosphocholine cytidylyltransferase alpha during farnesol-induced apoptosis. Mol. Cell. Biol. 224851-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, M., C. L. Brooks, F. Wu-Baer, D. Chen, R. Baer, and W. Gu. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 3021972-1975. [DOI] [PubMed] [Google Scholar]
- 21.Mallampalli, R. K., A. J. Ryan, R. G. Salome, and S. Jackowski. 2000. Tumor necrosis factor-alpha inhibits expression of CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 2759699-9708. [DOI] [PubMed] [Google Scholar]
- 22.Moorthy, A. K., and G. Ghosh. 2003. p105. IκBγ and prototypical IκBs use a similar mechanism to bind but a different mechanism to regulate the subcellular localization of NF-κB. J. Biol. Chem. 278556-566. [DOI] [PubMed] [Google Scholar]
- 23.Nikko, E., and B. Andre. 2007. Evidence for a direct role of the Doa4 deubiquitinating enzyme in protein sorting into the MVB pathway. Traffic 8566-581. [DOI] [PubMed] [Google Scholar]
- 24.Northwood, I. C., A. H. Tong, B. Crawford, A. E. Drobnies, and R. B. Cornell. 1999. Shuttling of CTP:phosphocholine cytidylyltransferase between the nucleus and endoplasmic reticulum accompanies the wave of phosphatidylcholine synthesis during the G0 → G1 transition. J. Biol. Chem. 27426240-26248. [DOI] [PubMed] [Google Scholar]
- 25.Okamura, H., J. Aramburu, C. Garcia-Rodriguez, J. P. Viola, A. Raghavan, M. Tahiliani, X. Zhang, J. Qin, P. G. Hogan, and A. Rao. 2000. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell 6539-550. [DOI] [PubMed] [Google Scholar]
- 26.Pemberton, L. F., and B. M. Paschal. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6187-198. [DOI] [PubMed] [Google Scholar]
- 27.Trotman, L. C., X. Wang, A. Alimonti, Z. Chen, J. Teruya-Feldstein, H. Yang, N. P. Pavletich, B. S. Carver, C. Cordon-Cardo, H. Erdjument-Bromage, P. Tempst, S. G. Chi, H. J. Kim, T. Misteli, X. Jiang, and P. P. Pandolfi. 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128141-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varghese, B., H. Barriere, C. J. Carbone, A. Banerjee, G. Swaminathan, A. Plotnikov, P. Xu, J. Peng, V. Goffin, G. L. Lukacs, and S. Y. Fuchs. 2008. Polyubiquitination of prolactin receptor stimulates its internalization, postinternalization sorting, and degradation via the lysosomal pathway. Mol. Cell. Biol. 285275-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walrafen, P., F. Verdier, Z. Kadri, S. Chretien, C. Lacombe, and P. Mayeux. 2005. Both proteasomes and lysosomes degrade the activated erythropoietin receptor. Blood 105600-608. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Y., J. I. MacDonald, and C. Kent. 1995. Identification of the nuclear localization signal of rat liver CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 270354-360. [DOI] [PubMed] [Google Scholar]
- 31.Weinhold, P. A., M. E. Rounsifer, and D. A. Feldman. 1986. The purification and characterization of CTP:phosphorylcholine cytidylyltransferase from rat liver. J. Biol. Chem. 2615104-5110. [PubMed] [Google Scholar]
- 32.Zhou, J., A. J. Ryan, J. Medh, and R. K. Mallampalli. 2003. Oxidized lipoproteins inhibit surfactant phosphatidylcholine synthesis via calpain-mediated cleavage of CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 27837032-37040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.