Abstract
Polo-like kinase 1 (Plk1) functions as a key regulator of mitotic events by phosphorylating substrate proteins on centrosomes, kinetochores, the mitotic spindle, and the midbody. Through mechanisms that are incompletely understood, Plk1 is released from and relocalizes to different mitotic structures as cells proceed through mitosis. We used fluorescence recovery after photobleaching to examine the kinetics of this process in more detail. We observed that Plk1 displayed a range of different recovery rates that differ at each mitotic substructure and depend on both the Polo-box domain and a functional kinase domain. Upon mitotic entry, centrosomal Plk1 becomes more dynamic, a process that is directly enhanced by Plk1 kinase activity. In contrast, Plk1 displays little dynamic exchange at the midbody, a process that again is modulated by the kinase activity of Plk1. Our findings suggest that the intrinsic kinase activity of Plk1 triggers its release from early mitotic structures and its relocalization to late mitotic structures. To assess the importance of Plk1 dynamic relocalization, Plk1 was persistently tethered to the centrosome. This resulted in a G2 delay, followed by a prominent prometaphase arrest, as a consequence of defective spindle formation and activation of the spindle checkpoint. The dynamic release of Plk1 from early mitotic structures is thus crucial for mid- to late-stage mitotic events and demonstrates the importance of a fully dynamic Plk1 at the centrosome for proper cell cycle progression. This dependence on dynamic Plk1 was further observed during the mitotic reentry of cells after a DNA damage G2 checkpoint, as this process was significantly delayed upon centrosomal tethering of Plk1. These results indicate that mitotic progression and control of mitotic reentry after DNA damage resides, at least in part, on the dynamic behavior of Plk1.
Polo-like kinases (Plks) are serine/threonine protein kinases that play essential roles during the cell cycle. Mammalian Plks are subdivided into four family members: Plk1, Plk2, Plk3, and Plk4 (51). Plks have a highly conserved N-terminal kinase domain and a relatively divergent C-terminal domain, called the Polo-box domain (PBD), which contains one (Plk4) or two (Plk1 to Plk3) Polo-boxes. Of the four Plks, the best characterized member is Plk1 (10). Plk1 functions in a diverse number of processes that are crucial for proper progression through multiple stages of mitosis, including mitotic entry, centrosome maturation, bipolar spindle formation, chromosome congression and segregation, cytokinesis, and mitotic exit (3, 10). The kinase domain of Plk1 is regulated in part by phosphorylation at Thr-210 within the activation loop (26, 32), likely through the actions of the upstream kinase Aurora-A in complex with the adaptor protein Bora (45, 60). Mutation of Thr-210 to Asp (T210D) mimics T-loop phosphorylation and stimulates kinase activity (26, 32, 56). Expression of an activated Plk1 T210D mutant can override the G2 arrest induced by DNA damage (62, 72) and allow cells to enter and progress completely through mitosis, albeit with a slight spindle checkpoint-dependent mitotic delay (71).
The kinase domain of Plk1, in its nonphosphorylated less active state, appears to be negatively regulated through direct interaction with the PBD (25, 48). We have shown previously that the PBDs of Plk1 function as a phosphoserine/phosphothreonine-binding module, recognizing the optimal recognition sequence motif Ser-[pSer/pThr]-[Pro/X] (15, 16). The structural basis for this phosphopeptide-specific binding has been revealed through two high-resolution X-ray structures of Plk1 PBD:phosphopeptide complexes (7, 16). In addition, we have shown that binding of phosphopeptides by the PBD in full-length Plk1 relieves its inhibitory function on kinase activity (16). These findings imply that priming phosphorylations on substrates or docking proteins by other mitotic kinases, such as cyclin-dependent kinases (Cdks), may target Plk1 to these substrates, while simultaneously activating its kinase activity. In agreement with this, a mass spectrometry study revealed that many known and potential Plk1 substrates specifically interact with the PBD of Plk1 in vitro in a phosphorylation- and mitosis-specific manner (44), while numerous studies from the Nigg, Barr, Lee, and Erikson groups, as well as others, have demonstrated that the PBD plays a crucial function in both substrate interaction and subcellular targeting of Plk1 in vivo (5, 26, 32, 33, 35, 48, 50, 54, 61).
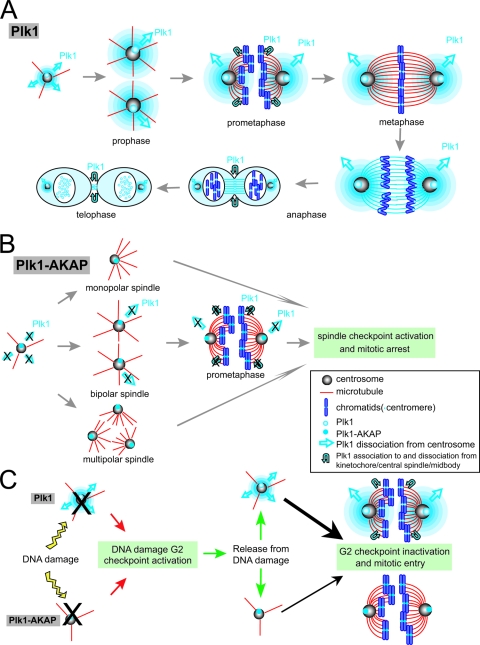
Plk1 dynamically localizes to various mitotic structures as cells progress through different stages of mitosis (3, 10) in a manner that depends on PBD function (16, 28, 33, 61, 64). During interphase and early prophase Plk1 is found at the centrosome, where it facilitates γ-tubulin recruitment (11) and centrosome maturation, separation, and microtubule nucleation during late prophase and prometaphase (2, 3, 8, 31). By metaphase, a fraction of Plk1 specifically localizes to the kinetochores, where it seems to be involved in regulating aspects of spindle checkpoint function and the metaphase-anaphase transition (1, 21). During anaphase, Plk1 is concentrated in the spindle midzone, where it likely facilitates microtubule sliding (42, 50), while after chromosome segregation in late anaphase, Plk1 remains located in the central spindle and midbody, where it participates in ingression of the cleavage furrow during cytokinesis (42, 50). Plk1 also resides at the Golgi apparatus, presumably linking Golgi fragmentation with mitotic entry by interacting with the structural Golgi protein GRASP65 (54).
Details of the movement and exchange of Plk1 between these mitotic structures remains poorly characterized, and whether Plk1 molecules exhibit similar or different dynamic behaviors at these distinct mitotic structures is largely unknown. What controls the kinetics of Plk1 localization during mitotic progression, and what is the relative importance of the PBD and kinase domains in this process? To address these questions, we analyzed the subcellular dynamics of Plk1 exchange in wild-type (WT) and mutant Plk1 proteins, at various mitotic structures, using fluorescence recovery after photobleaching (FRAP). In addition, to investigate whether dynamic relocalization of Plk1 is critical for proper mitotic progression, we examined the phenotypes of cells expressing an active but less dynamic form of Plk1, in the absence or presence of the endogenous Plk1 protein.
Intriguingly, many of the Plk1-dependent processes involved in early mitotic events appear to occur on centrosomes. Centrosomes are tiny (1 to 2 μm3) cytoplasmic organelles consisting of a pair of barrel-shaped microtubule assemblies, the centrioles, surrounded by pericentriolar material (12). In interphase cells, a single centrosome functions as the microtubule-organizing center, which then divides in S phase and separates in mitotic prophase to nucleate the bipolar mitotic spindle in prometaphase. In addition to their role in nucleating and anchoring the mitotic spindle, centrosomes are increasingly recognized as critical subcellular structures responsible for coordinately regulating the normal progression from G2 into M. For example, Jackman et al. (24) showed that the early mitotic activation of the Cdk1-cyclin B complex occurred at centrosomes, presumably through activation of the phosphatase Cdc25B, which has also been shown to localize to the centrosomes (9, 17, 24, 40). In addition to direct activators of the G2/M transition, a variety of effectors of the DNA damage-induced G2/M checkpoint have also been reported to localize to centrosomes in mammalian cells during the normal cell cycle, including the kinases Chk1, Chk2, and Plk3 and the signaling scaffold molecule BRCA1 (43). Consequently, the centrosome has now emerged as an organelle where signaling from the DNA damage checkpoint and components of the Cdk1-cyclin B activation loop are integrated during an unperturbed cell cycle to control mitotic entry. Importantly, Plk1 is involved in Cdk1-cyclin B activation, regulates DNA damage checkpoint function, and localizes to centrosomes. We therefore further investigated whether a centrosome-tethered form of Plk1 with reduced dynamic exchange would be sufficient to allow mitotic reentry after a DNA damage-induced G2 checkpoint.
MATERIALS AND METHODS
Construction of cell lines and plasmids.
Human Plk1 cDNAs were subcloned into pEGFP-C1 (Clontech, Mountain View, CA) (enhanced green fluorescent protein [EGFP]-Plk1). EGFP-Plk1-AKAP and EGFP-AKAP were generated by subcloning the PCR-derived AKAP450 fragment corresponding to residues 3644 to 3808 into EGFP-Plk1 and pEGFP-C3, respectively. Spectrin-GFP, the small interfering RNA vector pS(pSuper), pS-Plk1, and pS-Mad2 have been described previously (74). EGFP plasmids encoding Plk1 or Plk1-AKAP harboring silent mutations in the targeting region of pS-Plk1, which renders the protein insensitive to pS-Plk1-mediated degradation, have been described (71).
To isolate the human CENP-A cDNA, total RNAs purified from the human osteosarcoma U2OS cell line were transcribed to first-strand cDNAs using M-MLV reverse transcriptase (Promega, Madison, WI). The resulting products were then used as templates for PCR with a forward primer (5′-CTCTgCggCgTgTCATgg-3′) and a reverse primer (5′-TCAgCCgAgTCCCTCCTCAAgg-3′) that were designed to amplify human CENP-A. The PCR fragment was cloned into pCR2.1-TOPO (Invitrogen, Carsbad, CA) and verified as a fragment containing the CENP-A cDNA by sequencing. The human CENP-A fragment was subcloned into DsRed2-C1 vector (Clontech). DsRed2-tagged CENP-A (DsRed2-CENP-A) was amplified by PCR and subcloned into retroviral vector pLNCX2 (Invitrogen). DsRed2-CENP-A retrovirus was produced by transfection with pLNCX2-DsRed2-CENP-A, gag-pol, and VSV-g-env DNAs into 293 cells. Filtered tissue culture supernatant containing the virus was mixed with Polybrene and added to U2OS cells. The infected cells were selected by growth in the presence of 0.4 mg of G418 (Sigma, St. Louis, MO)/ml for 2 to 3 days, and U2OS cells stably expressing DsRed2-CENP-A were established. All of the constructs were confirmed by DNA sequence analysis.
Antibodies and dyes.
Thymidine, propidium iodide (PI), doxorubicin, paclitaxel, and caffeine were purchased from Sigma. Antibodies were obtained as follows: rabbit anti-phospho-Ser10-histone H3 (Upstate Biotechnology, Lake Placid, NY); monoclonal antibody cocktail against Plk1 (Zymed Laboratories, South San Francisco, CA); rabbit anti-Plk1 (Upstate Biotechnology); mouse monoclonal anti-GFP (Roche, Manheim, Germany); rabbit anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal anti-β-actin, DM1A mouse monoclonal anti-α-tubulin, and GTU88 mouse monoclonal anti-γ-tubulin (Sigma); human nuclear ANA-centromere autoantibody (CREST; Cortex Biochem, San Leandro, CA); rabbit anti-Mad2 (Covance, Princeton, NJ); mouse anti-CENP-A (Abcam, Cambridge, MA); peroxidase-conjugated goat anti-rabbit and rabbit anti-mouse (Dako, Glostrup, Denmark); goat anti-mouse Alexa Fluor 488, anti-rabbit Alexa Fluor 488, anti-mouse Alexa Fluor 594, and anti-rabbit Alexa Fluor 647 (Molecular Probes, Eugene, OR); and rhodamine-conjugated anti-human (Jackson Immunoresearch Laboratories, West Grove, PA).
Cells, transfection, synchronization, and recovery from DNA damage.
Human osteosarcoma U2OS cells were purchased from the American Type Culture Collection. U2OS cells, 293 cells, and 293T cells were grown at 37°C in 5% CO2 in Dulbecco modified Eagle medium supplemented with 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal calf serum. Transfection was performed by using Fugene 6 (Roche), Genejammer (Stratagene, Cedar Creek, TX), or electroporation (Gene Pulser II; Bio-Rad, Hercules, CA) for U2OS cells and by using Lipofectamine 2000 (Invitrogen) for 293 cells and 293T cells, according to the manufacturer's instructions. For electroporation, cells cultured in a 150-mm culture dish were transiently transfected with 20 to 40 μg of plasmid DNA/cuvette, and then the cells were allowed to adhere to dishes or plates for 24 to 48 h and used for experiments. For immunoblotting in reconstitution experiments, 10 μg of pS or pS-Plk1 was cotransfected into U2OS cells with 1 μg of pBabe-Puro combined with the indicated amounts of Plk1WT or Plk1-AKAP where indicated. Then, 2 μg of puromycin/ml was added about 8 to 16 h after transfection. After a 24-h selection, cells were harvested and lysed for immunoblotting. For flow cytometry analysis in reconstitution experiments, 10 μg of pS or pS-Plk1 was cotransfected with 1 μg of spectrin-GFP or 3 to 15 μg of the indicated Plk1 WT or mutant plasmids. Cells were synchronized in thymidine for 24 h and harvested at the indicated times after release. For recovery from DNA damage, 10 μg of pS or pS-Plk1 was cotransfected with 3 to 15 μg of the indicated Plk1 WT or mutant plasmids. Activation and inactivation of the G2 DNA damage checkpoint was performed as described previously and below (72).
Immunoblotting.
Cells were rinsed twice with ice-cold phosphate-buffered saline (PBS); solubilized in lysis buffer containing 20 mM Tris-HCl (pH 8.0), 20 mM Na4P2O7, 1 mM dithiothreitol, 1% Nonidet P-40, 1 mM MgCl2, 1 mM CaCl2, 10% glycerol, 0.5 mM Na3VO4, and 20 μM p-A-phenylmethanesulfonyl fluoride; and centrifuged at 14,000 × g for 10 min at 4°C. Protein content in the cell lysates was quantified by using a Bio-Rad protein assay, and aliquots of protein were boiled for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 2% mercaptoethanol, 10% glycerol). The solubilized proteins were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane (Bio-Rad), and detected by immunoblotting with the indicated antibodies using Western Lightning chemiluminescence reagent (Perkin-Elmer Life Sciences, Boston, MA).
Flow cytometry.
U2OS cells were transiently transfected with the indicated plasmids. At 24 to 48 h after transfection, the cells were then incubated with 2.5 mM thymidine in medium for an additional 24 h. After a thymidine block, the cells were washed three times and incubated with new medium for 20 h. The cells were harvested by trypsinization, fixed in ice-cold 70% ethanol, and stained with PI and/or anti-phospho-Ser10-histone H3 antibody in combination with anti-rabbit Alexa Fluor 647 antibody in 3% bovine serum albumin-PBS-0.05% Tween 20. Cells were analyzed by using a FACSscan (Becton Dickinson, Franklin Lakes, NJ) or a FACSCalibur (Becton Dickinson). Cell cycle profiles were generated by using CellQuest (Becton Dickinson) and FlowJo software (Tree Star, Ashland, OR).
For examining the DNA damage response, U2OS cells were transiently transfected with the indicated plasmids, and at 24 h after transfection the cells were incubated with 2.5 mM thymidine in medium for an additional 24 h. After the thymidine block, the cells were washed three times and released into new medium for 6 h to exit from S phase. Cells were then treated with 1 μM doxorubicin for 1 h, washed with drug-free medium, and incubated with medium containing 1 μM paclitaxel for 18 h. The cells were washed again with medium three times and incubated with new medium in the presence or absence of 5 mM caffeine for 7 h. The cells were harvested and stained as described above.
Immunofluorescence.
U2OS cells transfected with the indicated plasmids were seeded on 18-by-18-mm glass coverslips. At 24 to 48 h after transfection or at the indicated period after release from thymidine block, the cells were fixed in either 4% paraformaldehyde in PBS or 100% methanol for 15 min at room temperature, washed with PBS, and permeabilized 0.5% Triton X-100 in buffer (30 mM Tris buffer [pH 7.8], 75 mM NaCl, 0.3 M sucrose, 3 mM MgCl2) for 15 min at room temperature. For kinetochore staining, the cells on coverslips were extracted with PIPES buffer (20 mM PIPES, 0.3% Triton X-100, 1 mM MgCl2, 10 mM EGTA [pH 6.9]) for 5 min and then fixed with 4% paraformaldehyde for 10 min. Coverslips were blocked with 1% bovine serum albumin-1% sucrose in PBS or 3% bovine serum albumin-1% sucrose in PBS (kinetochore staining) for 1 h and stained with the indicated antibody at 4°C overnight. After a washing step, coverslips were stained with Alexa Fluor-conjugated secondary antibody for 1 h at room temperature. The coverslips were washed five times and then mounted onto slides with Fluoromount-G (Southern Biotechnologies, Birmingham, AL) or ProLong Gold antifade reagent (Invitrogen) containing DAPI (4′,6′-diamidino-2-phenylindole; Sigma) for DNA staining. Images were collected on an Axioplan2 microscope (Carl Zeiss, Oberkochen, Germany) and processed using OpenLab software (Improvision, Waltham, MA). Where indicated, images were collected on a Deltavision (Applied Precision, Issaquah, WA) and digitally deconvolved using softWoRx (Applied Precision).
Plk1 kinase assay.
Control EGFP or EGFP-Plk1 transfected cells were solubilized in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 1% Triton X-100, 150 mM NaCl, 2 mM EDTA (pH 8.0), 0.5% SDS, 50 mM NaF, 2 mM Na3VO4, 2 mM dithiothreitol, and protein inhibitors and then centrifuged at 14,000 × g for 10 min at 4°C. Anti-GFP immunoprecipitates were incubated in kinase buffer containing 50 mM Tris-HCl (pH 7.5), 200 μM Na3VO4, 5 mM dithiothreitol, 5 mM NaF, 50 μM ATP, 2 mM EGTA, and 10 mM MgCl2 at 30°C. Then, 20 μCi of [γ-32P]ATP and 1 μg of casein (Sigma) were added, the reaction was incubated for an additional 30 min at 30°C, and the products were analyzed by SDS-PAGE and autoradiography.
Live imaging and FRAP analysis.
U2OS cells grown on 12-by-12-mm or 18-by-18-mm coverslips were used for transfection with EGFP-Plk1 constructs. At 24 to 36 h after transfection, the coverslips were immersed in 37°C prewarmed PBS containing 0.9 mM CaCl2 and 0.33 mM MgCl2, and FRAP images of the cells then obtained by using an Olympus (Tokyo, Japan) FV 300/IX70 inverted laser-scanning confocal microscope (objective lens[×60], NA 1.42) with excitation at 488 nm (or 568 nm for DsRed) and 6% laser output intensity. All FRAP experiments were performed on an ambient temperature stage within 15 min of removal of the coverslips from the 37°C incubator. Under these conditions, the temperature of the PBS medium slowly decreased with time but remained at or above 30°C at the end of the measurement period. For photobleaching of EGFP-Plk1 constructs, the apertures were adjusted to encompass the smallest possible area that contained the mitotic substructure of interest, and irradiated using the 488-nm laser line of an argon laser (385-mW actual output). The region of interest was scanned 20 times with 1-s lapse (mitotic centrosome and kinetochore) or 2-s lapse (interphase centrosome and midbody) intervals at the same wavelength, with 100% laser output intensity after it was scanned 20 times with the same intervals with 6% intensity to establish a baseline, as described previously (23). Thereafter, imaging was continued at 6% intensity with the same intervals. In each FRAP experiment a minimum of 30 cells were measured. The mean fluorescence intensity was analyzed with MetaMorph software (Molecular Devices, Sunnyvale, CA) and plotted. Recovery was fitted to a single exponential curve (Y = TOP*[1 − exp(−K*X)] using GraphPad Prism software (San Diego, CA).
Statistical analysis.
Data are generally expressed as means ± the standard errors of the mean (SEM) unless otherwise indicated. The data were analyzed by analysis of variance with the Bonferroni correction for multiple-comparison tests or t tests (two-tailed) for determination of significance of the differences. A P value of <0.05 was considered to be statistically significant.
RESULTS
The subcellular localization pattern of EGFP-tagged Plk1 in live cells mimics that of the endogenous Plk1 protein.
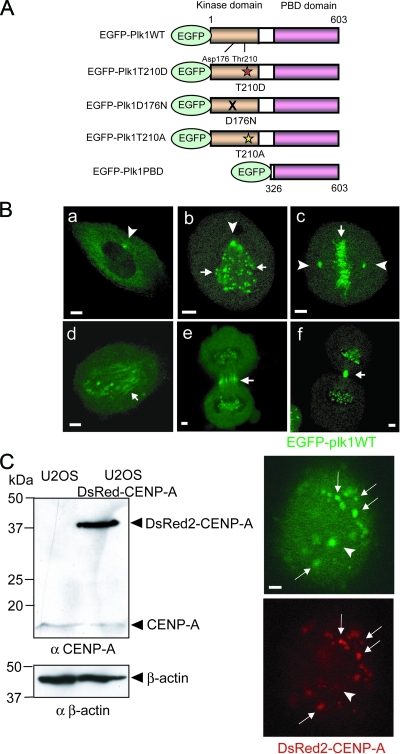
To analyze the dynamic exchange behavior of Plk1 at various mitotic structures by FRAP, we constructed an EGFP N-terminally tagged Plk1 (EGFP-Plk1) (Fig. 1A), and examined its subcellular localization after transient transfection in U2OS cells as they progressed through mitosis (Fig. 1B). When live cells were imaged, EGFP-Plk1 was observed at centrosomes in G2 (Fig. 1Ba) and persisted at mitotic centrosomes during prometaphase (Fig. 1Bb) and metaphase (Fig. 1Bc). In addition, during prometaphase and metaphase, EGFP-Plk1 became associated with both aligned and unaligned kinetochores, as revealed by the presence of punctate fluorescence in association with chromatin bodies (Fig. 1Bb and c, small arrows). Upon anaphase entry, EGFP-Plk1 transiently localized to spindle microtubules (Fig. 1Bd), followed by the appearance of strong Plk1 fluorescence at the spindle midzone during late anaphase (Fig. 1Be). Finally, during telophase and cytokinesis, EGFP-Plk1 accumulated at the midbody (Fig. 1Bf), although some residual kinetochore staining was also observed throughout these late mitotic stages.
FIG. 1.
Live cell images for FRAP during mitosis in U2OS cells expressing EGFP-Plk1. (A) Schematic representations of EGFP-tagged Plk1 constructs that were used for FRAP experiments. From top to bottom: full-length WT Plk (WT); a phosphorylation-mimicking T-loop mutation,Thr-210 to Asp (T210D) that renders Plk1 constitutively active; a kinase-dead Plk1 in which the catalytic base Asp-176, is mutated to Asn (D176N); a nonphosphorylatable T-loop form of Plk1 displaying only basal activity in which Thr-210 is mutated to Ala (T210A); an N-terminally truncated construct containing only the C-terminal PBD. (B) U2OS cells expressing were transfected with EGFP-Plk1 WT. Representative images of EGFP-Plk1 localization during mitosis are shown. Images represent G2 interphase (a), prometaphase (b), metaphase (c), anaphase (d), anaphase/telophase (e), and telophase (f), respectively. EGFP-Plk1 localized to the centrosomes in subpanels a, b, and c, as shown by arrowheads. Arrows indicate EGFP-Plk1 localized at kinetochores in subpanels b and c, at mitotic spindles in subpanel d, and at the midbody and/or spindle midzone in subpanel e and/or subpanel f. (C) U2OS cells stably expressing DsRed2-CENPA. A Western blot analysis of U2OS cell lysates using the indicated antibodies is shown in the left panel, with β-actin used as a loading control. The right panels show fluorescence images of EGFP-Plk1 (upper) and DsRed-CENPA (lower). Arrows indicate kinetochores, and arrowheads indicate centrosomes. Scale bars in all panels, 5 μm.
When examining U2OS cells in prometaphase, the centrosomal localization of EGFP-Plk1 often could not be unambiguously distinguished from kinetochore localization. This posed a potential problem for experiments measuring the dynamics of Plk1 exchange at centrosomes versus kinetochores by FRAP during this mitotic substage. To overcome this difficulty, we therefore established U2OS cells that stably expressed DsRed2-tagged CENP-A (DsRed-CENP-A [see Materials and Methods]), a known marker of kinetochores (29) (Fig. 1C). Expression of DsRed-CENP-A in these cells was confirmed by immunoblotting with anti-CENP-A antibodies (Fig. 1C, left panels), while localization of DsRed-CENP-A to kinetochores was verified by live cell fluorescence microscopy (Fig. 1C, lower right panel). When EGFP-Plk1 was coexpressed in the cells stably expressing DsRed-CENP-A, the kinetochore-localized Plk1 signal could be easily and unambiguously distinguished from the centrosome-localized Plk1 (Fig. 1C).
These imaging studies, performed in living cells, demonstrated that EGFP-Plk1 showed patterns of mitotic stage-specific localization identical to those reported previously in fixed cells stained for endogenous Plk1 (3, 20, 36) or for a similar GFP-tagged Plk1 construct (2). In addition, U2OS cells transiently overexpressing low to moderate amounts of EGFP-Plk1 showed normal cell cycle profiles and rates of mitotic progression compared to U2OS cells transfected using a spectrin-GFP control plasmid, although at the highest expression levels Plk1 induced some mitotic cell accumulation (data not shown).
Plk1 present in distinct mitotic structures shows different rates of exchange with the free Plk1 pool.
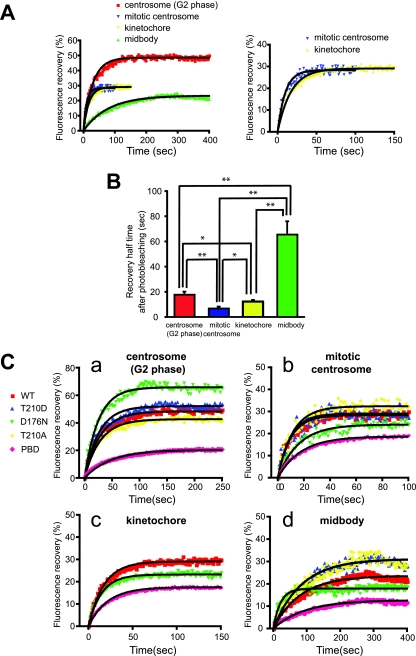
Next, we probed the dynamic properties of Plk1 at different mitotic substructures in living cells during G2- and M-phase using FRAP at 30 to 37°C. Individual centrosomes, kinetochores, or midbodies in the EGFP-Plk1-transfected cells showing low to moderate levels of expression were photobleached, and the recovery of the fluorescence signal in the bleached area was monitored by time-lapse imaging at 2-s intervals for G2 centrosomes and midbodies and at 1-s intervals for mitotic centrosomes and kinetochores (Fig. 2A and B; see Fig. S9 in the supplemental material). FRAP analysis revealed that the rate of Plk1 exchange was strongly dependent on its subcellular localization (Table 1). WT EGFP-Plk1 at G2 centrosomes, for example, was highly dynamic, with a mean time for 50% recovery of the total net fluorescence recovered (t1/2) of 17.9 s and a mean total net fluorescence recovery intensity of 48.3% (Fig. 2A and Table 1). At mitotic centrosomes, the rate of exchange of EGFP-Plk1 was even faster (50% recovery time t1/2 = 7.0 ± 1.3 s), although the mean net fluorescence recovery intensity was somewhat reduced (28.3%) (Fig. 2A and Table 1). Exchange rates very similar to those measured for mitotic centrosomes were also observed for mitotic kinetochores (t1/2 = 12.4 ± 1.3 s, mean net fluorescence recovery intensity = 29.1%) (Fig. 2A and Table 1). In marked contrast to the rapid rate of exchange observed at mitotic centrosomes and kinetochores, the exchange rate for WT EGFP-Plk1 localized to the midbody during the terminal stages of mitosis was significantly slower (t1/2 = 65.7 × 10.0 s; mean net fluorescence recovery intensity = 23.3%) (Fig. 2A and Table 1). Similar half-lives for recovery were observed over a range of bleaching conditions (cf. Fig. S1 and S2 in the supplemental material).
FIG. 2.
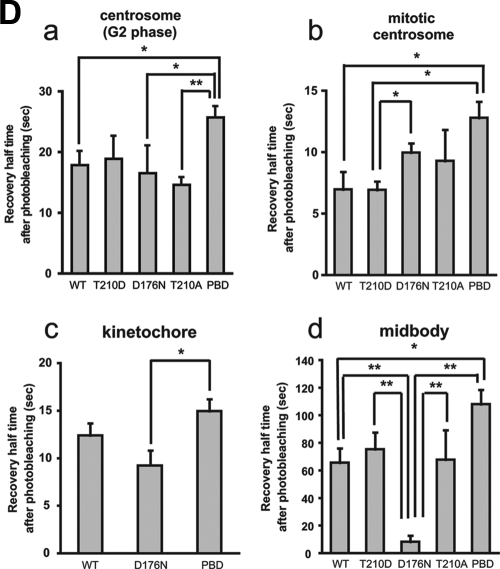
FRAP analysis of Plk1 at centrosomes, kinetochores, and midbodies. (A) Centrosomes during interphase and mitosis, kinetochores, and midbodies were targeted for laser photobleaching and monitored by fluorescence time-lapse microscopy. Photobleaching was performed at 100% laser transmission by scanning the bleached region of interest. Recovery intensity data were collected at 6% laser transmission at 1- or 2-s intervals. The data points represent the means from 7 to 17 experiments (see Table 1), with ≥30 cells measured in each experiment. Black curves were fitted to data by using GraphPad Prism software. The right panel shows the enlarged plotted intensities and fitted curves of mitotic centrosomes and kinetochores. (B) The 50% recovery times were calculated from each data curve from independent experiments. Mean values are indicated with bar graphs (G2 phase centrosomes [red], mitotic centrosomes [blue], kinetochores [yellow], or midbodies [green]). Error bars indicate the SEM. (C) FRAP for EGFP-Plk1 WT (WT) and mutants localized at G2 (a) and mitotic centrosomes (b), kinetochores (c), and midbodies (d) was performed as described in panel A. The fluorescence intensities after photobleaching are plotted (WT [red squares], T210D [blue triangles], D176N [green triangles], T210A [yellow diamonds], or PBD [pink diamonds]). Each data point represents the mean of 5 to 19 experiments, with ≥25 cells measured in each experiment. (D) The 50% recovery times for Plk1 localized at G2 centrosomes (a), mitotic centrosomes (b), kinetochores (c), and midbodies (d) were calculated from each data curve from independent experiments. Averages are indicated in the bar graphs with error bars showing the SEM. Statistically significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.01).
TABLE 1.
Dynamics of Plk1 at different subcellular locations
| Location | Plk1 | Mean 50% recovery time (s) ± SEM | Mean net fluorescence recovery intensity (%) ± SEM | No. of independent expts |
|---|---|---|---|---|
| G2 centrosomes | WT | 17.9 ± 2.2 | 48.3 ± 4.1 | 14 |
| T210D | 19.0 ± 3.5 | 52.1 ± 4.4 | 9 | |
| D176N | 16.6 ± 4.2 | 65.8 ± 8.9 | 6 | |
| T210A | 14.7 ± 2.0 | 42.6 ± 9.1 | 14 | |
| PBD | 25.8 ± 1.8 | 20.4 ± 5.8 | 15 | |
| WT-AKAP | 58.2 ± 6.8 | 31.7 ± 3.6 | 11 | |
| Mitotic centrosomes | WT | 7.0 ± 1.3 | 28.3 ± 2.0 | 7 |
| T210D | 6.9 ± 0.7 | 30.3 ± 4.6 | 6 | |
| D176N | 10.0 ± 0.8 | 21.8 ± 1.8 | 6 | |
| T210A | 9.3 ± 2.2 | 31.8 ± 4.4 | 8 | |
| PBD | 16.7 ± 1.0 | 17.4 ± 1.8 | 19 | |
| Kinetochores | WT | 12.4 ± 1.3 | 29.1 ± 2.5 | 17 |
| D176N | 9.4 ± 0.8 | 23.2 ± 1.5 | 14 | |
| PBD | 15.0 ± 1.5 | 17.5 ± 2.0 | 12 | |
| Midbodies | WT | 65.7 ± 10.0 | 23.3 ± 5.5 | 12 |
| T210D | 75.4 ± 10.9 | 31.0 ± 4.6 | 5 | |
| D176N | 8.3 ± 4.2 | 17.9 ± 3.1 | 8 | |
| T210A | 67.8 ± 19.5 | 31.2 ± 6.0 | 6 | |
| PBD | 108.1 ± 9.8 | 12.7 ± 1.3 | 9 |
Intrinsic protein kinase activity stimulates Plk1 release from mitotic centrosomes but delays Plk1 release from midbodies.
To determine the importance of kinase domain activity in determining the dynamic behavior of full-length Plk1, constructs of EGFP-Plk1 containing a series of mutations and/or deletions in the kinase domain were transfected into U2OS cells, and the kinetics of Plk1 exchange at the same G2 and mitotic substructures described above were analyzed by FRAP. Mutations that were studied included a constitutively active phosphorylation-mimicking mutation in the T-loop (T210D), a nonphosphorylatable T-loop mutant (T210A) that should retain basal kinase activity but cannot be further activated by mitotic phosphorylation (16, 45, 71), a kinase-dead mutant (D176N), and a truncation mutant solely containing the PBD (Fig. 1A; see Fig. S3 in the supplemental material) (15, 16, 44). After transient transfection, all of the constructs were expressed at similar levels and migrated at the expected sizes when examined by SDS-PAGE and Western blot analysis (see Fig. S3 and Table S1 in the supplemental material). The kinase activities of the WT and mutant EGFP-Plk1 constructs was directly measured by immunoprecipitating the proteins from asynchronous cells and assaying their ability to phosphorylate casein. As expected, roughly similar amounts of kinase activity were detected in the cells expressing Plk1 WT or T210A, increased by the expression of T210D and absent in cells expressing D176N (see Fig. S3 in the supplemental material). FRAP analysis revealed that the exchange kinetics (t1/2) for the WT, T210D, and T210A forms of EGFP-Plk1 were roughly similar at both G2 and mitotic centrosomes (Fig. 2C and D and Table 1). Interestingly, for mitotic centrosomes, but not G2 centrosomes, the kinase-dead D176N form of Plk1 showed a significantly slower recovery time and a reduced percentage of net fluorescence recovery compared to the T210D form, suggesting that after photobleaching the kinase-dead Plk1 has a reduced ability to dissociate from mitotic centrosomes and exchange with the free pool of cytoplasmic Plk1 than the constitutively active form. We interpret this as evidence that the kinase activity of full-length Plk1 may be important for stimulating the release of Plk1 from centrosomes as cells progress through mitosis.
In marked contrast to the behavior of kinase-dead Plk1 at mitotic centrosomes, the same kinase-dead D176N mutant of EGFP-Plk1 showed a dramatically enhanced ability to dissociate from the midbody, exchanging with the free Plk1 pool at a rate that was nearly eightfold faster than that observed with WT Plk1 (t1/2 = 8.3 ± 4.2 s versus 65.7 × 10.0 s, respectively) although, curiously, the mobile fraction of the D176N protein remains low (17.9%) and comparable to WT Plk1. We interpret this as evidence that intrinsic Plk1 kinase activity is required to maintain association of this small pool of highly dynamic Plk1 with the midbody as cells progress through telophase and cytokinesis.
The PBD, located C-terminal to the kinase domain, acts as a localization domain that recognizes short phosphorylated sequence motifs on a significant number of Plk1 substrates (15, 16, 44, 70) and has been shown to be critical for Plk1 localization to centrosomes, kinetochores, and midbodies (15, 25, 28, 33, 49, 50, 55, 61, 63). An EGFP-PBD construct completely lacking the kinase domain was found to exhibit a significantly slower rate of exchange at G2 and mitotic centrosomes and midbodies than that observed with any of the full-length Plk1 constructs, either WT or point mutants (Table 1). In addition, the mean net fluorescence recovery intensities of the PBD at these structures were considerably reduced, suggesting that the photobleached EGFP-PBD remains tightly associated with these structures. These data suggest that the mere presence of the Plk1 kinase domain, regardless of its catalytic activity, negatively modulates the ability of the PBD to engage its ligands at these mitotic structures in live cells.
Construction and analysis of a centrosome-localized Plk1 molecule.
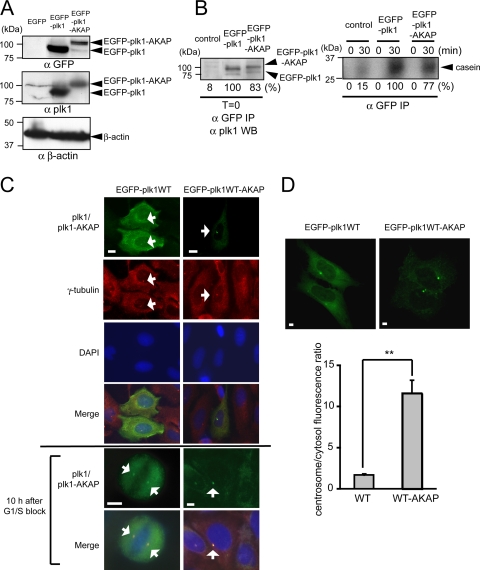
Given the relatively rapid exchange kinetics of Plk1 at centrosomes and kinetochores (Table 1) and the observation that kinase activity facilitates the dissociation of Plk1 from centrosomes as cells move from metaphase to anaphase (Fig. 2B to D), we next investigated whether cells expressing a much less mobile form of centrosomal Plk1 would be impaired in their ability to transit through mitosis. To accomplish this, we created a Plk1 construct that preferentially associates with the centrosome by fusing the C terminus of full-length Plk1 to the pericentrin-AKAP450 centrosomal targeting domain of AKAP450 (19). The resulting construct, EGFP-tagged Plk1-AKAP (EGFP-Plk1-AKAP), was expressed at moderate levels after transfection of both U2OS and 293T cells, as confirmed by immunoblotting with anti-GFP and anti-Plk1 antibodies (Fig. 3A and B). Although EGFP-Plk1-AKAP appeared at the expected molecular weight, its expression level was about 5- to 10-fold lower per μg of transfected DNA than that of EGFP-Plk1 in both cell types (Fig. 3A and B). EGFP-Plk1 and EGFP-Plk1-AKAP, however, displayed similar levels of kinase activity when comparable amounts were assayed (Fig. 3B and see Fig. S4 in the supplemental material). Next, we examined the subcellular localization of Plk1-AKAP by immunostaining for EGFP in fixed U2OS cells and by fluorescence microscopy in live U2OS cells (Fig. 3C and D). Both assays showed that the Plk1-AKAP strong localized at the centrosomes of interphase cells, as confirmed by complete overlap with γ-tubulin staining (Fig. 3C). Importantly, the ratio of centrosomal to cytoplasmic fluorescence was clearly higher in live U2OS cells expressing Plk1-AKAP than in those expressing Plk1 WT (Fig. 3D), confirming that Plk1-AKAP predominantly localizes to centrosomes.
FIG. 3.
Plk1-AKAP expression, kinase activity, and localization. (A) EGFP-Plk1 WT or Plk1-AKAP protein expression in transfected U2OS cells. Western blotting with the indicated antibodies is shown. (B) Plk1 kinase activities in transfected 293T cells. The amounts of immunoprecipitated EGFP-Plk1/EGFP-Plk1-AKAP proteins using Western blot analysis (left) and Plk1 kinase activities against β-casein (right) are shown. The extent of casein phosphorylation was quantitated by phosphorimager analysis and are indicated as the percentage observed compared to EGFP-Plk1, which was set at 100%, and are not normalized for the protein recoveries shown in the left panel. Empty vector was used for control transfections. (C) EGFP-Plk1/EGFP-Plk1-AKAP localization in transfected U2OS cells. Transfected U2OS cells were fixed and immunostained with anti-γ-tubulin antibody (red). EGFP-Plk1-AKAP is localized almost exclusively at the centrosome (arrows), compared to EGFP-Plk1, which displays both centrosomal and cytoplasmic localization. Chromosomal DNA is stained with DAPI (blue). In last two rows, transfected U2OS cells were released at 10 h after thymidine block, fixed, and stained. (D) Quantification of centrosome-localized EGFP-Plk1/EGFP-Plk1-AKAP in live cells. EGFP fluorescence of a fixed area (5 μm by 5 μm) at the centrosome or in the cytosol was analyzed (≥25 cells), and the ratio of centrosome/cytosolic fluorescence was calculated. Representative pictures (upper panels) and the average ratios (lower panel) are shown. Error bars represent the standard deviations. Statistically significant differences are indicated by asterisks (**, P < 0.01). All scale bars represent 5 μm.
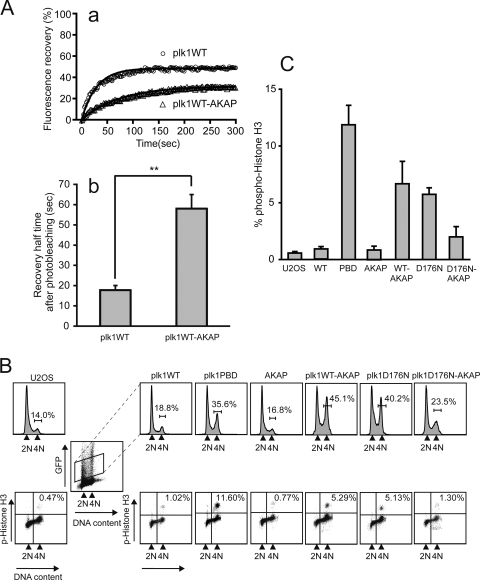
The mobility of EGFP-Plk1-AKAP was examined using FRAP (Fig. 4A). At centrosomes, the EGFP-Plk1-AKAP protein showed a >3-fold prolongation of the recovery half time after photobleaching compared to WT EGFP-Plk1 (t1/2 = 58.2 ± 6.8 s versus t1/2 = 17.9 ± 2.2 s, respectively). Likewise, the mean net fluorescence intensity recovered at the photobleached sites was reduced for EGFP-Plk1-AKAP (31.7%) compared to EGFP-Plk1 (48.3%), a finding consistent with more stable tethering of the Plk1-AKAP construct to centrosomes.
FIG. 4.
Expression of Plk1-AKAP induces mitotic arrest. (A) The dynamics of EGFP-Plk1 or EGFP-Plk1-AKAP at the centrosome in transfected U2OS cells were analyzed by FRAP. Average fluorescence recovery curves (a) and 50% recovery times after photobleaching (b) are shown. Each data point represents the mean of 14 and 11 experiments, respectively, in which >30 cells were imaged in each experiment (see Table 1). The black curves in subpanel a show the best single exponential fits to the data. Error bars in subpanel b indicate the SEM with statistically significant differences are indicated by asterisks (**, P < 0.01). (B) FACS profiles of DNA content and phospho-histone H3 staining in Plk1 and Plk1-AKAP cells. U2OS cells were transfected with the indicated EGFP-fused constructs for 24 h and then arrested with a thymidine block for an additional 24 h. At 24 h after release of the block, progression through the cell cycle was analyzed by FACS. Upper panels show the DNA profiles (PI) from control (U2OS) cells or from the transfected cells when gated for low to moderate EGFP expression (boxed). Lower panels show the phospho-histone H3 levels in the GFP-positive cells. The results from a typical experiment are shown. (C) Expression of either the PBD, a WT form of Plk1 that is tethered to the centrosome, or a freely diffusible kinase-dead Plk1 causes mitotic arrest. The average percentage of phosphohistone H3-positive cells in the GFP-expressing population from three independent experiments was determined. Error bars represent the standard deviations.
Persistent activity of centrosome-tethered Plk1 causes G2 delay and mitotic arrest.
To assess whether altering the subcellular dynamics of Plk1 influences its mitotic function, we compared the cell cycle profiles of U2OS cells expressing low to moderate levels of EGFP-Plk1-AKAP, EGFP-Plk1, and EGFP-PBD (Fig. 4B) at 20 h after release from a thymidine block. As shown in the upper panel of Fig. 4B, expression of Plk1 WT-AKAP, but not of Plk1 WT or the AKAP protein alone, led to an accumulation of 4N DNA-containing cells, indicating that Plk1-AKAP cells were delayed in G2 and/or M phase. Similar results were also obtained after transfection of a constitutively active centrosomally tethered form of Plk1, Plk1T210D-AKAP (data not shown). Further examination revealed that a fraction of the Plk1-AKAP-expressing cells was blocked in mitosis, since they stained positively for histone H3 phosphorylation (Fig. 4B, lower panel, and 4C) with levels approximately half of those observed after expression of the isolated Plk1PBD, a treatment previously established to cause a preanaphase mitotic arrest (61). The difference in phosphohistone H3 levels between Plk1-AKAP and Plk1-PBD-expressing cells suggests that Plk1-AKAP-expressing cells may have also undergone a G2 delay. This was further supported by examination of cells at 10 h after thymidine release. As shown in the bottom two rows of Fig. 3C, at this time point nearly all of the Plk1-AKAP-expressing cells remained in G2, whereas a fraction of the Plk1-expressing cells had already entered into mitosis (phospho-histone H3 positivity of Plk1 WT-expressing cells, 10.8%; Plk1 WT-AKAP, 1.1% based on fluorescence-activated cell sorting [FACS] analysis). In addition, Plk1-AKAP-expressing cells were found to have defects in centrosome separation, γ-tubulin recruitment, and functional maturation (data not shown), all of which occur during G2 or shortly after the G2/M transition (46).
In the experiments described above, cells contained endogenous Plk1 in addition to the transiently expressed EGFP-Plk1-AKAP. To exclude the possibility of artifacts contributing to the observed phenotype, such as dominant inhibitory interactions between endogenous Plk1 and the Plk1-AKAP constructs, we studied the effects of anchoring Plk1 to centrosomes on cell cycle progression in cells depleted of endogenous Plk1. To accomplish this, we used vector-driven shRNA against Plk1 (pSuper-Plk1, pS-Plk1) (4, 74) and reconstituted these cells with WT or mutant Plk1. Two silent mutations within the shRNA-target region of the Plk1 constructs were introduced to allow expression of the exogenous Plk1, as described previously (74). We could demonstrate by Western blotting that this approach was successful, since endogenous Plk1 expression was eliminated, while simultaneously the expression of EGFP-Plk1 or EGFP-Plk1-AKAP was observed (see Fig. S5A in the supplemental material). Combining this reconstitution assay with flow cytometry analysis, we found that expression of EGFP-Plk1-AKAP in Plk1-depleted cells resulted in a mitotic arrest similar to that observed after Plk1-AKAP expression in Plk1-containing cells, whereas reconstitution with low to moderate levels of WT EGFP-Plk1 did not significantly alter the cell cycle (see Fig. S5B and C in the supplemental material). These results are identical to those obtained with U2OS cells that were not depleted of endogenous Plk1, indicating that tethering of Plk1 at centrosomes with reduced dynamic exchange dominantly interferes with cell cycle progression, regardless of the presence or absence of endogenous Plk1.
Expression of a kinase-dead form of Plk1, D176N, also induced a mitotic arrest (Fig. 4B and C), a finding similar to what has been reported previously for the K82M hypomorphic allele of the kinase (61). Intriguingly, when the D176N kinase-dead Plk1 was tethered to the centrosome by fusion to the pericentrin-AKAP450 centrosomal targeting domain of AKAP450, the population of cells undergoing mitotic arrest was dramatically reduced compared to cells expressing the cytosolic, fully dynamic form of kinase-dead Plk1 (Plk1D176N), as evidenced by both a decrease in the population of cells containing 4N DNA, and the percentage of cells staining positively for phosphohistone H3 (Fig. 4B and C). These data indicate that cytosolic Plk1 kinase activity is important for mitotic progression and that tethering Plk1 to centrosomes causes a dominant cell cycle arrest phenotype only if kinase activity is present. Taken together, these data strongly suggest that Plk1 activity at the centrosome must be extinguished and Plk1 activity in the cytoplasm activated in a coordinated manner in order for cells to progress properly through the various stages of mitosis.
Centrosomally tethered Plk1 induces mitotic spindle defects and activates the spindle assembly checkpoint to trigger prometaphase arrest.
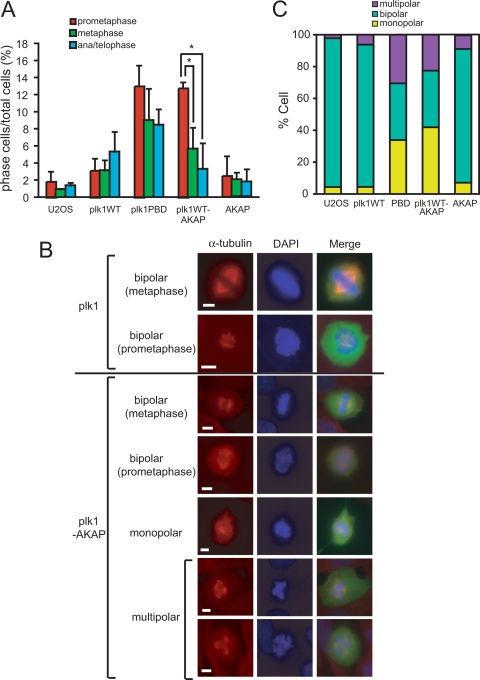
To explore the mitotic arrest phenotype of Plk1-AKAP cells in detail, the distribution of cells in each mitotic substage was evaluated by scoring the morphology of the DAPI-stained DNA masses in GFP-positive cells at 20 h after release from a thymidine block. More than 95% of the control EGFP-transfected cells had completed mitosis at this time and resided in interphase, with only 1.9% of the total cells population in prometaphase, and smaller percentages in metaphase and anaphase/telophase. At this time point cells expressing WT Plk1 showed only a slightly higher percentage of total cells in mitosis cells (∼10%), with most of the cells in anaphase or telophase. In contrast, expression of the isolated PBD resulted in a significant accumulation of preanaphase mitotic cells (Fig. 4B and C) (61), with 13.0% of the cells in prometaphase and 9.0% in metaphase (Fig. 5A). Similarly, expression of Plk1-AKAP resulted in the marked accumulation of prometaphase arrested cells compared to the increase in metaphase and anaphase/telophase cell populations (Fig. 5A). It therefore appears that the mitotic arrest induced by the expression of Plk1-AKAP predominantly consists of prometaphase cells.
FIG. 5.
Expression of Plk1-AKAP induces prometaphase arrest and aberrant spindle morphologies. (A) Transfected U2OS cells prepared as in Fig. 4 were released at 20 h after a thymidine block and subsequently fixed and stained with DAPI. GFP-positive cells were scored for mitotic subphases using confocal microscopy. The percentages of prometaphase (red), metaphase (green), and ana/telophase (cyan) cells are shown. The data are averages of three independent experiments. Error bars represent the standard deviations. (B) Transfected U2OS cells were released at 20 h after a thymidine block and subsequently fixed and stained with anti-α-tubulin antibody and DAPI. Representative images are shown. All scale bars represent 5 μm. (C) Cells were stained as in panel B. The percentages of GFP-positive cells displaying normal bipolar or mutant (monopolar or multipolar) spindle morphologies are shown.
To probe the underlying cause of the prometaphase arrest induced by Plk1-AKAP expression, mitotic spindle formation was analyzed by immunostaining the cells using an anti-α-tubulin antibody at 20 h after release from a thymidine block. As shown in Fig. 5B and C, 93% of the control EGFP-transfected U2OS cells and 87% of the WT Plk1-expressing cells exhibited bipolar spindle structures, with monopolar or multipolar spindles only occasionally observed (Fig. 5C). Expression of the PBD has been previously reported to induce a defect in spindle stability (61), presumably by interfering with the recruitment of WT Plk1 to centrosomes, microtubules, and/or kinetochores. In agreement with this, we observed that 70% of Plk1-PBD-expressing cells lacked bipolar spindles (Fig. 5C). Importantly, Plk1 WT-AKAP-expressing cells also displayed both monopolar spindles and multipolar spindles with high frequency (42 and 26%, respectively), indicating that persistent centrosomal Plk1 activity results in a set of centrosomal abnormalities with defective assembly of bipolar spindles similar to that observed when Plk1 recruitment is blocked.
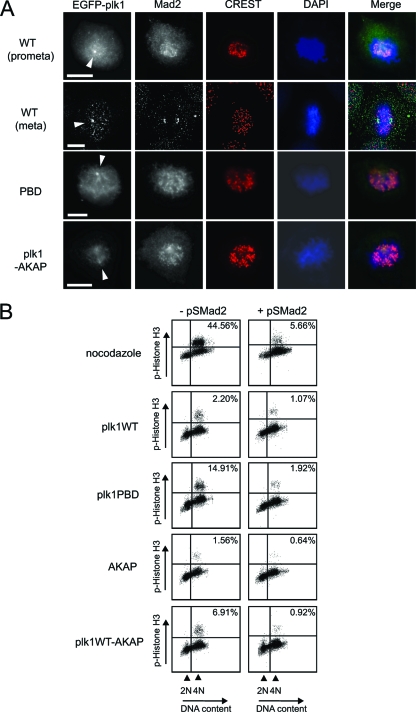
Abnormal mitotic spindles can lead to defects in stable microtubule-kinetochore attachments that persistently activate the spindle assembly checkpoint. To examine whether the accumulation of 4N DNA-containing cells after expression of Plk1-AKAP was the result of activation of the spindle checkpoint, we analyzed cells for accumulation of Mad2 at kinetochores (Fig. 6). Mad2, a spindle assembly checkpoint protein, localizes to kinetochores that lack proper microtubule attachment and is released from kinetochores once they form stable bivalent attachments to microtubules from opposing spindle poles and equal tension is exerted upon the chromosome arms (75). U2OS cells expressing WT Plk1 displayed enrichment of Mad2 on kinetochores during prometaphase but not at metaphase, as expected (Fig, 6A, rows 1 and 2). Plk1-AKAP-expressing cells, as well as PBD-expressing cells, retained Mad2 on most of their kinetochores (Fig. 6A, rows 3 and 4), suggesting that the prometaphase arrest induced by Plk1-AKAP is spindle checkpoint dependent. To further examine this, we performed RNA interference-mediated depletion of Mad2 (pS-Mad2) to inactivate the spindle checkpoint (38). As a control for the efficiency of spindle checkpoint bypass by this approach, U2OS cells were transfected with the control vector (pS) or with pS-Mad2, released from a thymidine block into medium containing the microtubule depolymerizing agent nocodazole to induce a spindle assembly-dependent mitotic arrest, and analyzed for phosphohistone H3 staining by flow cytometry. As shown in the upper panel in Fig. 6B, the Mad2-depleted cells no longer arrested in mitosis in the presence of nocodazole, indicating the functional inactivation of the spindle assembly checkpoint. U2OS cells were then transfected with the various Plk1 constructs in combination with control vector (pS) or pS-Mad2 and analyzed for spontaneous mitotic arrest 20 h after release from a thymidine block in the absence of nocodazole. As shown in the lower panels of Fig. 6B, the increase in phosphohistone H3 positivity observed after expression of the isolated Plk1 PBD was significantly suppressed by pS-Mad2 coexpression, showing that the PBD induces a spindle checkpoint-dependent mitotic arrest, a finding consistent with the findings of Seong et al. (61). Importantly, depletion of Mad2 also efficiently reversed the accumulation of phosphohistone H3-positive cells induced by expression of Plk1-AKAP. Thus, the mitotic arrest, caused by tethering a less dynamic form of Plk1 to centrosomes is mediated through activation of the Mad2-dependent spindle assembly checkpoint.
FIG. 6.
Spindle checkpoint activation in Plk1-AKAP-expressing cells. (A) Mad2 remains associated with kinetochores in Plk1-AKAP expressing cells, U2OS cells were transfected with the indicated constructs as in Fig. 4, and released at 20 h after thymidine block. Cells were fixed and stained with anti-Mad2 antibody, CREST, and DAPI. Images were collected and processed using OpenLab (Improvision) or a Deltavision microscope system (Applied Precision). Plk1 at mitotic centrosomes is indicated by arrowheads. Merged images show Mad2 in green, CREST staining in red, and DAPI in blue. All scale bars represent 5 μm. (B) U2OS cells were transfected with the indicated EGFP-tagged Plk1 plasmids with or without an shRNA-expressing vector targeting Mad2 (pS-Mad2). At 20 h after release from a thymidine block, cells were harvested and fixed for FACS analysis. In the control experiments examining spindle checkpoint activation by nocodazole, cells were transfected with pS vector control or pS-Mad2, along with spectrin-GFP, and nocodazole was added for 20 h after release of the thymidine block (top panels). Phospho-histone H3 positivity and DNA content (PI) of GFP-positive cells are shown.
Dynamic Plk1 facilitates mitotic reentry after DNA damage.
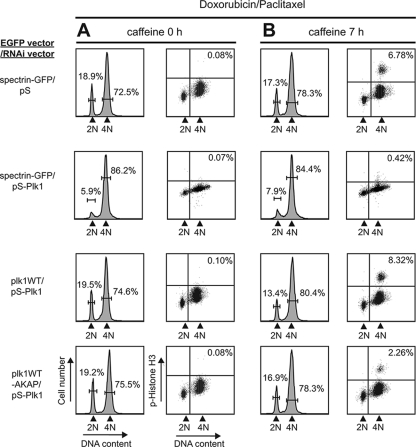
There is growing evidence that centrosomes are involved in the cellular response to DNA damage (43). Plk1 is implicated as a target of DNA damage checkpoint and is also crucial for mitotic entry after recovery after DNA damage (62, 72). We therefore tested whether a less dynamic, centrosomally tethered form of Plk1, which delays progression through the later stages of mitosis, was still sufficient for release of the DNA damage checkpoint upon recovery from DNA damage. In these experiments U2OS cells in which endogenous Plk1 was silenced by pS-Plk1 were used to examine the effect of exogenous centrosomally tethered Plk1 as the sole source of Plk1. When control U2OS cells released from a thymidine block are treated with 1 μM doxorubicin, a highly synchronous population arrests at G2 from a DNA damage checkpoint (Fig. 7A, top row). The addition of 5 mM caffeine, an ATM/ATR inhibitor, for 7 h after DNA damage allows recovery from the checkpoint-induced arrest and a fraction of the cells progress into mitosis, where they can be trapped by paclitaxel and assayed for phosphohistone H3 by flow cytometry (Fig. 7B, top row) (72). Cells depleted of endogenous Plk1 by shRNA showed an identical G2 arrest as control U2OS cells after treatment with doxorubicin (Fig. 7A, rows 2 to 4), regardless of whether they they also coexpressed Plk1 WT or Plk1 WT-AKAP. However, the Plk1-depleted cells were unable to exit from the DNA damage-induced G2 arrest (Fig. 7B, row 2), in agreement with the findings of van Vugt et al. (72). Coexpression of an RNA interference-resistant form of either Plk1 WT or Plk1 WT-AKAP in these Plk1-depleted cells allowed them to override the G2 arrest and enter mitosis (Fig. 7B, rows 3 and 4), although the relative percentage of pH3 positivity in cells that contain the centrosomally tethered Plk1 WT-AKAP was only ∼30% of that seen in cells expressing Plk1 WT protein. Similar results were also obtained using the constitutively active T210D forms of Plk1 and Plk1-AKAP (62; data not shown). Importantly, doxorubicin treatment had no effect on the ability of the Plk1-AKAP fusion protein to localize predominantly to centrosomes (see Fig. S6 in the supplemental material).
FIG. 7.
Centrosomally tethered less dynamic Plk1 impairs mitotic reentry after a DNA damage G2 arrest. (A) Normal activation of the G2 DNA damage checkpoint in the absence or presence of WT or centrosomally tethered Plk1. U2OS cells were cotransfected with the EGFP fusion protein and shRNA expression vector pair spectrin-GFP/pS, spectrin-GFP/pS-Plk1, EGFP-Plk1 WT/pS-Plk1, or EGFP-Plk1 WT-AKAP/pS-Plk1 at 24 h from the thymidine block for 6 h, and the samples were then treated with 1 μM doxorubicin for 1 h, washed, and grown in the presence of paclitaxel for 18 h. (B) Recovery from G2 DNA damage arrest in Plk1-AKAP-expressing cells. After the treatments shown in panel A, caffeine was added for 7 h. In panels A and B, DNA content (PI) and phospho-histone H3 staining of the GFP-positive cells was examined by FACS as shown.
These results indicate that signals emanating from a less dynamic from of Plk1 tethered at the centrosome reduces the ability of DNA-damaged cells to progress from G2 into M when upstream signals from the phosphatidylinositol 3-kinase-like kinases ATM, ATR and DNA-PK are silenced by caffeine. Thus, it appears that while the critical substrates for Plk1-directed mitotic reentry likely reside at the centrosome, a fully dynamic form of Plk1 is required for the process to occur in a maximally effective manner.
DISCUSSION
In this study we examined the mobility and exchange of Plk1 at different mitotic substructures using FRAP and explored the relative roles of the kinase domain and the PBD in controlling Plk1 dynamics. We found that Plk1 displayed a range of turnover rates that were strongly dependent on its subcellular localization and the mitotic state. At mitotic centrosomes, for example, Plk1 was highly dynamic with a t1/2 of ∼7 s, whereas at the midbody Plk1 was considerably less mobile, with a t1/2 of ∼66 s. The rapid centrosomal kinetics seen on both G2 and mitotic centrosomes are in good agreement with data for other centrosomal proteins, including the kinases Aurora-A and Nek2, which were reported to be in rapid flux with the cytoplasmic population (22, 27, 65, 66).
All FRAP experiments were performed between 30 and 37°C, although the microscope stage was not temperature controlled. In a number of our FRAP experiments, the mean net fluorescence recoveries after photobleaching were low. One possible explanation for this is that a significant fraction of Plk1 may be stably associated with each of the mitotic substructures we imaged. In agreement with this, the dynamic fraction of Plk1 that we measured gave essentially identical recoveries after repeated photobleaching trials (see Fig. S7 in the supplemental material). Furthermore, under the conditions that we could measure, the fluorescence of a similar unbleached mitotic structure within the bleached cell did not appear to change (see Fig. S8 in the supplemental material), arguing against an apparent reduction in recovery due to nonspecific bleaching effects. Finally, the extent of fluorescence for that we observed for Plk1 at centrosomes is similar to that reported for another centrosomal mitotic kinase, Nek2 (22).
We observed that complete removal of the kinase domain, leaving only the phospho-binding PBD, resulted in stable association of the protein with centrosomes (16, 61, 70), and the least dynamic form of centrosomal Plk1. These data fit well with previous in vitro observations of a mutually inhibitory intramolecular interaction between the PBD and the kinase domain (15, 25), resulting in enhanced PBD binding when the kinase domain is absent in our in vivo experiments (16, 25). Intriguingly, as cells progressed from G2 into mitosis, the exchange rate of full-length WT Plk1 at the centrosome increased >2-fold. However, the rate at which kinase-defective Plk1 mutants at mitotic centrosomes exchanged with the free cytosolic pool was slower, although the exchange rate of both WT and kinase-dead Plk1 at G2 centrosomes was unchanged. We interpret these data as evidence that much of the centrosomal localization of Plk1 during early mitosis results from strong direct interactions between the PBD of Plk1 and phosphorylated centrosomal proteins, likely through the generation of PBD-binding sites through Cdk1 (49, 50) or Plk1 itself (37, 58), and that these centrosome-localizing interactions are disrupted during subsequent stages of mitosis through Plk1 kinase activity, facilitating the release and replacement of the photobleached protein. Both cyclin B/Cdk1 and Plk1 kinase activities are high throughout early to mid mitosis, and Plk1 is thought to phosphorylate cyclin B and may promote its nuclear translocation (67, 76). Thus, during prometaphase or metaphase, when much of the cyclin B/cdk1 dissociates from centrosomes, a Plk1 kinase-dependent activity that drives the dissociation of centrosomal Plk1 may become dominant. Our findings are in excellent agreement with prior reports of PBD-engaging centrosomal ligands such as hCenexin1 (64). In addition, the kinase-dependent exchange of centrosomal Plk1 that we observed, probably mediated through phosphorylation of substrates at or near the centrosome, may constitute a physiological trigger for Plk1 release from the centrosome and retargeting to kinetochores and spindle structures as cells progress beyond prophase/prometaphase (Fig. 8A).
FIG. 8.
Model for Plk1 relocalization during mitosis and effect of centrosomally tethered Plk1 on mitotic progression. (A) Model for Plk1 localization and dynamics during normal mitosis. In prophase and prometaphase, Plk1 kinase activity facilitates its release from the centrosome and promotes its localization to kinetochores, the spindle midzone and the midbody through self-priming. Some residual Plk1 localization on kinetochores remains during telophase. (B) Model for the effect of centrosomally tethered Plk1 (Plk1-AKAP) during mitosis. Plk1 tethering to centrosomes induces spindle checkpoint activation and mitotic arrest, indicating that Plk1 dissociation from centrosome is important for further mitotic progression. (C) Centrosomally tethered Plk1 (Plk1-AKAP) (bottom, thin arrow) is less effective than fully dynamic WT Plk1 (top, thick arrow) at stimulating mitotic reentry after DNA damage.
An alternative possibility is that the rapid exchange behavior observed for Plk1 at mitotic centrosomes may be a consequence of cyclin B/Cdk1 activation and substrate phosphorylation. Plk1 and cyclin B/Cdk1 influence each other heavily during mitotic entry, as well as during mitotic progression. Plk1 phosphorylates cyclin B and is involved in the robust activation of cyclin B/Cdk1 at centrosomes (24). Consistent with this, depletion of Plk1 in both normal cells and cancer cells causes a delay in progression from G2 to M (31, 41, 57). Later in mitosis, Plk1 is required for correct spindle formation (52, 74), as well as in the downregulation of cyclin B/Cdk1 through the direct activation of the APC/C leading to Cyclin B degradation (39, 47). Thus, since Plk1 and Cdk1 activities are so interdependent, the protein dynamics we observed for Plk1 may depend on cyclin B/Cdk1 as well. Addition of the Cdk1 inhibitor roscovitine to cells already in mitosis, however, did not affect the centrosomal dynamics of Plk1 (data not shown), suggesting that the role of Cdk1 as a regulator of Plk1 dynamics is limited. Finally, our observation that the Plk1 kinase domain facilitates the release, rather than the binding of Plk1 to centrosomes contrasts with the opposite conclusions reached by Montoya and coworkers (18), who reported that the isolated Plk1 kinase domain could localize to centrosomes in asynchronous cells; however, that study that did not directly examine the effect of kinase activity on mitotic centrosomal localization of Plk1 in vivo.
Surprisingly, although kinase activity seems to promote release of Plk1 from mitotic centrosomes, kinase activity appeared to dramatically prolong the retention of Plk1 at the midbody by nearly an order of magnitude. At the midbody, as at the centrosome, the isolated PBD showed the least dynamic behavior of all of the constructs examined. Together, these data suggest that Plk1 kinase activity at the midbody likely creates its own PBD-binding sites via a self-priming process. Similar self-priming mechanisms for Plk1 localization have been elegantly described in a series of studies by Neef et al. and others, who showed that during anaphase Plk1 generates its own PBD-binding sites on the microtubule-associated protein PRC1 and on the mitotic kinesin MKlp2 to localize Plk1 to the central spindle and drive cytokinesis (3, 49, 50). Plk1 self-priming for PBD binding has also been recently implicated in recruiting Plk1 to centromeres/kinetochores via binding to PBIP1 (28, 34) and, indeed, we observed a statistically significant increase in Plk1 mobility for the kinase-dead D176N mutant of Plk1, but not for WT Plk1, compared to the isolated PBD alone (Fig. 3C). As Neef et al. point out, this process of self-priming is likely to be especially important in the later stages of mitosis when Cdk1 activity has declined (49). Furthermore, our observation that Plk1 kinase activity seems to promote Plk1 release from early mitotic structures and enhance relocalization to late mitotic structures helps explain the prior conundrum of why Plk1 kinase activity, which could self-prime for PBD binding, does not result in cell cycle arrest by creating a form of Plk1 that becomes stuck on centrosomes at early stages in mitosis (49).
The central importance of dynamic release of Plk1 from the centrosome as cells progress through mitosis was revealed by the studies shown in Fig. 3 to 6, in which we observed that centrosome-anchored Plk1 induced both a G2 delay and an M-phase arrest, with significant accumulation of prometaphase cells (Fig. 8A and B). Why does mild overexpression of centrosome-associated Plk1 prevent mitotic progression, while overexpression of cytosolic Plk1 does not do the same thing? One possibility is that there are both stimulatory and inhibitory pathways for mitotic progression mediated by Plk1. In addition to stimulating centrosome maturation, centrosome-localized kinase-active (but not kinase-dead) Plk1 might also trigger an inhibitory pathway that activates the spindle checkpoint through phosphorylation of proteins such as Cdc20. Alternatively, inappropriate and excessive phosphorylation of centrosomal substrates by Plk1-AKAP could result in disruption of normal Plk1-dependent events at the pericentriolar material, leading to abnormal numbers of spindle poles. For example, Plk1 activity directly stimulates the β-TrCP-dependent degradation of Bora, an adaptor protein that regulates Aurora-A activation and/or localization (6, 59). Knockdown of hBora using small interfering RNA, mimicking excessive Plk1-mediated degradation, has been shown to induce a variety of spindle abnormalities, including multipolar spindles, resulting in activation of the spindle checkpoint (6). Similarly, excessive and inappropriate kinase activity could interfere with the normal regulation of the centrosomal Plk1 substrate Kizuna, the knockdown of which has been shown to result in dissociation of the pericentriolar material, multipolar spindle formation, and prometaphase/metaphase arrest (53). Additional detailed studies are necessary to further elucidate the exact mechanism responsible for Plk1-AKAP inhibition of mitotic progression.
The centrosome is increasingly recognized as a critical subcellular structure that regulates mitotic entry during an unpertubed cell cycle (13). During normal mitotic entry, cyclin B/Cdk1 complexes are preferentially activated on centrosomes, likely through the combined actions of Plk1 and CDC25B (9, 14, 24, 40), while inhibition of mitotic entry at the G2/M transition seems to involve the specific localization of checkpoint kinases, including ATM, ATR, Chk1, and/or Chk2, to the centrosome, where they may inhibit cyclin B/Cdk1 activation (30, 43, 68, 77). Whether the centrosome also plays a central role in coordinating the mitotic arrest and reentry functions in mammalian cells in response to DNA damage is unclear. In response to DNA damage, Plk1 kinase activity and phosphorylation at Thr210 has been shown to be inhibited in an ATM/ATR-Chk2/Chk1-dependent manner (62, 69, 73), while reactivation of Plk1 activity and T210 phosphorylation occurs upon recovery through an hBora:Aurora-A complex (45). Our finding that a centrosomally tethered form of Plk1 with reduced dynamic behavior impairs the ability of DNA damaged cells to reenter mitosis from G2 arrest (Fig. 8C) is consistent with both positive and negative regulators of the DNA damage checkpoint residing at the centrosome and indicates that fine-tuning of mitotic reentry from a G2 checkpoint requires communication between the centrosome and surrounding factors.
Supplementary Material
Acknowledgments
We thank D. Lim, J. Alexander, M. Stewart, E. Vasile, and M. Hayashi for reagents, assistance with techniques, and data analysis; S. Munro (MRC Laboratory of Molecular Biology, Cambridge, United Kingdom) for providing the AKAP 450 cDNA; and Rene Medema for helpful discussions.
K.K. was supported by the Manpei Suzuki Diabetes Foundation. M.V. was supported by an EMBO fellowship. This study was supported by NIH grants GM60594 and CA112967 to M.B.Y.
Footnotes
Published ahead of print on 23 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahonen, L. J., M. J. Kallio, J. R. Daum, M. Bolton, I. A. Manke, M. B. Yaffe, P. T. Stukenberg, and G. J. Gorbsky. 2005. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr. Biol. 151078-1089. [DOI] [PubMed] [Google Scholar]
- 2.Arnaud, L., J. Pines, and E. A. Nigg. 1998. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma 107424-429. [DOI] [PubMed] [Google Scholar]
- 3.Barr, F. A., H. H. Sillje, and E. A. Nigg. 2004. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell. Biol. 5429-440. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296550-553. [DOI] [PubMed] [Google Scholar]
- 5.Casenghi, M., P. Meraldi, U. Weinhart, P. I. Duncan, R. Korner, and E. A. Nigg. 2003. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 5113-125. [DOI] [PubMed] [Google Scholar]
- 6.Chan, E. H., A. Santamaria, H. H. Sillje, and E. A. Nigg. 2008. Plk1 regulates mitotic Aurora A function through βTrCP-dependent degradation of hBora. Chromosoma 117457-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, K. Y., E. D. Lowe, J. Sinclair, E. A. Nigg, and L. N. Johnson. 2003. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 225757-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Luca, M., P. Lavia, and G. Guarguaglini. 2006. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle 5296-303. [DOI] [PubMed] [Google Scholar]
- 9.De Souza, C. P., K. A. Ellem, and B. G. Gabrielli. 2000. Centrosomal and cytoplasmic Cdc2/cyclin B1 activation precedes nuclear mitotic events. Exp. Cell Res. 25711-21. [DOI] [PubMed] [Google Scholar]
- 10.Donaldson, M. M., A. A. Tavares, I. M. Hagan, E. A. Nigg, and D. M. Glover. 2001. The mitotic roles of Polo-like kinase. J. Cell Sci. 1142357-2358. [DOI] [PubMed] [Google Scholar]
- 11.Donaldson, M. M., A. A. Tavares, H. Ohkura, P. Deak, and D. M. Glover. 2001. Metaphase arrest with centromere separation in polo mutants of Drosophila. J. Cell Biol. 153663-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doxsey, S. 2001. Re-evaluating centrosome function. Nat. Rev. Mol. Cell. Biol. 2688-698. [DOI] [PubMed] [Google Scholar]
- 13.Doxsey, S., W. Zimmerman, and K. Mikule. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15303-311. [DOI] [PubMed] [Google Scholar]
- 14.Dutertre, S., M. Cazales, M. Quaranta, C. Froment, V. Trabut, C. Dozier, G. Mirey, J. P. Bouche, N. Theis-Febvre, E. Schmitt, B. Monsarrat, C. Prigent, and B. Ducommun. 2004. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J. Cell Sci. 1172523-2531. [DOI] [PubMed] [Google Scholar]
- 15.Elia, A. E., L. C. Cantley, and M. B. Yaffe. 2003. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 2991228-1231. [DOI] [PubMed] [Google Scholar]
- 16.Elia, A. E., P. Rellos, L. F. Haire, J. W. Chao, F. J. Ivins, K. Hoepker, D. Mohammad, L. C. Cantley, S. J. Smerdon, and M. B. Yaffe. 2003. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 11583-95. [DOI] [PubMed] [Google Scholar]
- 17.Gabrielli, B. G., C. P. De Souza, I. D. Tonks, J. M. Clark, N. K. Hayward, and K. A. Ellem. 1996. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J. Cell Sci. 109(Pt. 5)1081-1093. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Alvarez, B., G. de Carcer, S. Ibanez, E. Bragado-Nilsson, and G. Montoya. 2007. Molecular and structural basis of polo-like kinase 1 substrate recognition: implications in centrosomal localization. Proc. Natl. Acad. Sci. USA 1043107-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillingham, A. K., and S. Munro. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golsteyn, R. M., K. E. Mundt, A. M. Fry, and E. A. Nigg. 1995. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol. 1291617-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goto, H., T. Kiyono, Y. Tomono, A. Kawajiri, T. Urano, K. Furukawa, E. A. Nigg, and M. Inagaki. 2006. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 8180-187. [DOI] [PubMed] [Google Scholar]
- 22.Hames, R. S., R. E. Crookes, K. R. Straatman, A. Merdes, M. J. Hayes, A. J. Faragher, and A. M. Fry. 2005. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol. Biol. Cell 161711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi, M. K., H. M. Ames, and Y. Hayashi. 2006. Tetrameric hub structure of postsynaptic scaffolding protein homer. J. Neurosci. 268492-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackman, M., C. Lindon, E. A. Nigg, and J. Pines. 2003. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 5143-148. [DOI] [PubMed] [Google Scholar]
- 25.Jang, Y. J., C. Y. Lin, S. Ma, and R. L. Erikson. 2002. Functional studies on the role of the C-terminal domain of mammalian polo-like kinase. Proc. Natl. Acad. Sci. USA 991984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang, Y. J., S. Ma, Y. Terada, and R. L. Erikson. 2002. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J. Biol. Chem. 27744115-44120. [DOI] [PubMed] [Google Scholar]
- 27.Kallio, M. J., V. A. Beardmore, J. Weinstein, and G. J. Gorbsky. 2002. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 158841-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang, Y. H., J. E. Park, L. R. Yu, N. K. Soung, S. M. Yun, J. K. Bang, Y. S. Seong, H. Yu, S. Garfield, T. D. Veenstra, and K. S. Lee. 2006. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol. Cell 24409-422. [DOI] [PubMed] [Google Scholar]
- 29.Kingwell, B., and J. B. Rattner. 1987. Mammalian kinetochore/centromere composition: a 50-kDa antigen is present in the mammalian kinetochore/centromere. Chromosoma 95403-407. [DOI] [PubMed] [Google Scholar]
- 30.Kramer, A., N. Mailand, C. Lukas, R. G. Syljuasen, C. J. Wilkinson, E. A. Nigg, J. Bartek, and J. Lukas. 2004. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat. Cell Biol. 6884-891. [DOI] [PubMed] [Google Scholar]
- 31.Lane, H. A., and E. A. Nigg. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 1351701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, K. S., and R. L. Erikson. 1997. Plk is a functional homolog of Saccharomyces cerevisiae Cdc5, and elevated Plk activity induces multiple septation structures. Mol. Cell. Biol. 173408-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, K. S., T. Z. Grenfell, F. R. Yarm, and R. L. Erikson. 1998. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc. Natl. Acad. Sci. USA 959301-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, K. S., J. E. Park, Y. H. Kang, W. Zimmerman, N. K. Soung, Y. S. Seong, S. J. Kwak, and R. L. Erikson. 2008. Mechanisms of mammalian polo-like kinase 1 (Plk1) localization: self- versus non-self-priming. Cell Cycle 7141-145. [DOI] [PubMed] [Google Scholar]
- 35.Lee, K. S., S. Song, and R. L. Erikson. 1999. The polo-box-dependent induction of ectopic septal structures by a mammalian polo kinase, plk, in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 9614360-14365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, K. S., Y. L. Yuan, R. Kuriyama, and R. L. Erikson. 1995. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol. Cell. Biol. 157143-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenart, P., M. Petronczki, M. Steegmaier, B. Di Fiore, J. J. Lipp, M. Hoffmann, W. J. Rettig, N. Kraut, and J. M. Peters. 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17304-315. [DOI] [PubMed] [Google Scholar]
- 38.Lens, S. M., R. M. Wolthuis, R. Klompmaker, J. Kauw, R. Agami, T. Brummelkamp, G. Kops, and R. H. Medema. 2003. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 222934-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindon, C., and J. Pines. 2004. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J. Cell Biol. 164233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindqvist, A., H. Kallstrom, A. Lundgren, E. Barsoum, and C. K. Rosenthal. 2005. Cdc25B cooperates with Cdc25A to induce mitosis but has a unique role in activating cyclin B1-Cdk1 at the centrosome. J. Cell Biol. 17135-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, X., M. Lei, and R. L. Erikson. 2006. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol. Cell. Biol. 262093-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, X., T. Zhou, R. Kuriyama, and R. L. Erikson. 2004. Molecular interactions of Polo-like-kinase 1 with the mitotic kinesin-like protein CHO1/MKLP-1. J. Cell Sci. 1173233-3246. [DOI] [PubMed] [Google Scholar]
- 43.Loffler, H., J. Lukas, J. Bartek, and A. Kramer. 2006. Structure meets function: centrosomes, genome maintenance, and the DNA damage response. Exp. Cell Res. 3122633-2640. [DOI] [PubMed] [Google Scholar]
- 44.Lowery, D. M., K. R. Clauser, M. Hjerrild, D. Lim, J. Alexander, K. Kishi, S. E. Ong, S. Gammeltoft, S. A. Carr, and M. B. Yaffe. 2007. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 262262-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macurek, L., A. Lindqvist, D. Lim, M. A. Lampson, R. Klompmaker, R. Freire, C. Clouin, S. S. Taylor, M. B. Yaffe, and R. H. Medema. 2008. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455119-123. [DOI] [PubMed] [Google Scholar]
- 46.Meraldi, P., and E. A. Nigg. 2002. The centrosome cycle. FEBS Lett. 5219-13. [DOI] [PubMed] [Google Scholar]
- 47.Moshe, Y., J. Boulaire, M. Pagano, and A. Hershko. 2004. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc. Natl. Acad. Sci. USA 1017937-7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mundt, K. E., R. M. Golsteyn, H. A. Lane, and E. A. Nigg. 1997. On the regulation and function of human Polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem. Biophys. Res. Commun. 239377-385. [DOI] [PubMed] [Google Scholar]
- 49.Neef, R., U. Gruneberg, R. Kopajtich, X. Li, E. A. Nigg, H. Sillje, and F. A. Barr. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9436-444. [DOI] [PubMed] [Google Scholar]
- 50.Neef, R., C. Preisinger, J. Sutcliffe, R. Kopajtich, E. A. Nigg, T. U. Mayer, and F. A. Barr. 2003. Phosphorylation of mitotic kinesin-like protein 2 by Polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nigg, E. A. 1998. Polo-like kinases: positive regulators of cell division from start to finish. Curr. Opin. Cell Biol. 10776-783. [DOI] [PubMed] [Google Scholar]
- 52.Ohkura, H., I. M. Hagan, and D. M. Glover. 1995. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 91059-1073. [DOI] [PubMed] [Google Scholar]
- 53.Oshimori, N., M. Ohsugi, and T. Yamamoto. 2006. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat. Cell Biol. 81095-1101. [DOI] [PubMed] [Google Scholar]
- 54.Preisinger, C., R. Korner, M. Wind, W. D. Lehmann, R. Kopajtich, and F. A. Barr. 2005. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 24753-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi, W., Z. Tang, and H. Yu. 2006. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol. Biol. Cell 173705-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qian, Y. W., E. Erikson, and J. L. Maller. 1999. Mitotic effects of a constitutively active mutant of the Xenopus Polo-like kinase Plx1. Mol. Cell. Biol. 198625-8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian, Y. W., E. Erikson, F. E. Taieb, and J. L. Maller. 2001. The Polo-like kinase Plx1 is required for activation of the phosphatase Cdc25C and cyclin B-Cdc2 in Xenopus oocytes. Mol. Biol. Cell 121791-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santamaria, A., R. Neef, U. Eberspacher, K. Eis, M. Husemann, D. Mumberg, S. Prechtl, V. Schulze, G. Siemeister, L. Wortmann, F. A. Barr, and E. A. Nigg. 2007. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol. Biol. Cell 184024-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seki, A., J. A. Coppinger, H. Du, C. Y. Jang, J. R. Yates III, and G. Fang. 2008. Plk1- and β-TrCP-dependent degradation of Bora controls mitotic progression. J. Cell Biol. 18165-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seki, A., J. A. Coppinger, C. Y. Jang, J. R. Yates, and G. Fang. 2008. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 3201655-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seong, Y. S., K. Kamijo, J. S. Lee, E. Fernandez, R. Kuriyama, T. Miki, and K. S. Lee. 2002. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. J. Biol. Chem. 27732282-32293. [DOI] [PubMed] [Google Scholar]
- 62.Smits, V. A., R. Klompmaker, L. Arnaud, G. Rijksen, E. A. Nigg, and R. H. Medema. 2000. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2672-676. [DOI] [PubMed] [Google Scholar]
- 63.Song, S., T. Z. Grenfell, S. Garfield, R. L. Erikson, and K. S. Lee. 2000. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol. Cell. Biol. 20286-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soung, N. K., Y. H. Kang, K. Kim, K. Kamijo, H. Yoon, Y. S. Seong, Y. L. Kuo, T. Miki, S. R. Kim, R. Kuriyama, C. Z. Giam, C. H. Ahn, and K. S. Lee. 2006. Requirement of hCenexin for proper mitotic functions of polo-like kinase 1 at the centrosomes. Mol. Cell. Biol. 268316-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stenoien, D. L., S. Sen, M. A. Mancini, and B. R. Brinkley. 2003. Dynamic association of a tumor amplified kinase, Aurora-A, with the centrosome and mitotic spindle. Cell Motil. Cytoskel. 55134-146. [DOI] [PubMed] [Google Scholar]
- 66.Thompson, H. M., H. Cao, J. Chen, U. Euteneuer, and M. A. McNiven. 2004. Dynamin 2 binds gamma-tubulin and participates in centrosome cohesion. Nat. Cell Biol. 6335-342. [DOI] [PubMed] [Google Scholar]
- 67.Toyoshima-Morimoto, F., E. Taniguchi, N. Shinya, A. Iwamatsu, and E. Nishida. 2001. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature 410215-220. [DOI] [PubMed] [Google Scholar]
- 68.Tsvetkov, L. 2004. Polo-like kinases and Chk2 at the interface of DNA damage checkpoint pathways and mitotic regulation. IUBMB Life 56449-456. [DOI] [PubMed] [Google Scholar]
- 69.Tsvetkov, L., and D. F. Stern. 2005. Phosphorylation of Plk1 at S137 and T210 is inhibited in response to DNA damage. Cell Cycle 4166-171. [DOI] [PubMed] [Google Scholar]
- 70.van de Weerdt, B. C., D. R. Littler, R. Klompmaker, A. Huseinovic, A. Fish, A. Perrakis, and R. H. Medema. 2008. Polo-box domains confer target specificity to the Polo-like kinase family. Biochim. Biophys. Acta 17831015-1022. [DOI] [PubMed] [Google Scholar]
- 71.van de Weerdt, B. C., M. A. van Vugt, C. Lindon, J. J. Kauw, M. J. Rozendaal, R. Klompmaker, R. M. Wolthuis, and R. H. Medema. 2005. Uncoupling anaphase-promoting complex/cyclosome activity from spindle assembly checkpoint control by deregulating polo-like kinase 1. Mol. Cell. Biol. 252031-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Vugt, M. A., A. Bras, and R. H. Medema. 2004. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol. Cell 15799-811. [DOI] [PubMed] [Google Scholar]
- 73.van Vugt, M. A., V. A. Smits, R. Klompmaker, and R. H. Medema. 2001. Inhibition of Polo-like kinase-1 by DNA damage occurs in an ATM- or ATR-dependent fashion. J. Biol. Chem. 27641656-41660. [DOI] [PubMed] [Google Scholar]
- 74.van Vugt, M. A., B. C. van de Weerdt, G. Vader, H. Janssen, J. Calafat, R. Klompmaker, R. M. Wolthuis, and R. H. Medema. 2004. Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J. Biol. Chem. 27936841-36854. [DOI] [PubMed] [Google Scholar]
- 75.Waters, J. C., R. H. Chen, A. W. Murray, and E. D. Salmon. 1998. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1411181-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan, J., F. Eckerdt, J. Bereiter-Hahn, E. Kurunci-Csacsko, M. Kaufmann, and K. Strebhardt. 2002. Cooperative phosphorylation including the activity of polo-like kinase 1 regulates the subcellular localization of cyclin B1. Oncogene 218282-8292. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, S., P. Hemmerich, and F. Grosse. 2007. Centrosomal localization of DNA damage checkpoint proteins. J. Cell Biochem. 101451-465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.