Abstract
Many transcription factors are controlled through SUMO modification, and in the majority of cases this modification results in enhancements in their repressive properties. In some instances, SUMO modification and its associated repressive activities can be reversed by the action of intracellular signaling pathways, leading to enhanced transcriptional capacities of transcription factors. Here we have investigated sumoylation of the ETS domain transcription factor PEA3 and its interplay with the extracellular signal-regulated kinase (ERK) mitogen-activated protein (MAP) kinase signaling pathway. PEA3 is modified by SUMO in vitro and in vivo on multiple sites in its N-terminal region. Activation of the ERK MAP kinase pathway promotes sumoylation of PEA3. Importantly, sumoylation of PEA3 is required for maximal activation of target gene promoters, including MMP-1 and COX-2. Molecularly, sumoylation is selectively required for synergistic activation of target gene expression with the coactivator CBP. Moreover, sumoylation of PEA3 is required for ubiquitination of PEA3 and promotes its degradation, suggesting that SUMO-mediated recycling of PEA3 plays a role in PEA3-mediated promoter activation. Thus, in contrast to the majority of other transcription factors studied, sumoylation of PEA3 plays a positive role in PEA3-mediated transcriptional activation and the ERK MAP kinase pathway cooperates with rather than antagonizes this process.
SUMO is a small polypeptide which is structurally related to ubiquitin. Indeed, many components of the pathway that conjugate ubiquitin to substrate proteins also show structural and functional homology with the SUMO pathway (reviewed in references 17 and 22). However, unlike ubiquitin, which predominantly targets proteins for degradation, SUMO modification has been shown to exhibit a number of disparate functions depending on the protein substrates (reviewed in references 17, 22, and 55). Indeed, one of the initial roles demonstrated for SUMO was to oppose ubiquitin-mediated degradation in IκB (7). However, more recently, one of the major roles emerging for sumoylation is in imparting repressive properties on transcriptional regulatory proteins (reviewed in references 10 and 11). Conversely, in a limited number of cases, sumoylation has also been associated with transcriptional activation, such as with GRIP1 (25), myocardin (54), p45/NF-E2 (42), GATA-4 (53), NFAT1 (49), Smad4 (29), glucocorticoid receptor (27), p53 (38), and HSF-1/-2 (13, 20).
Protein sumoylation is not a static process. Indeed, sumoylation appears to be quite dynamic, with typically only a small proportion of any protein being modified. The latter observation has been suggested to equate to a role of SUMO in establishing a particular state which is then maintained through the action of other proteins and/or modifications (reviewed in reference 17). One way in which sumoylation can be controlled is through the action of protein kinase cascades in response to extracellular signals (reviewed in reference 16). An example of this is in the case of HSF-1, whose sumoylation status is enhanced following heat shock-mediated phosphorylation (18). Molecularly, this occurs through an extended SUMO recognition motif known as the PDSM, where a proline-directed serine phosphorylation site is located three amino acids downstream from the core consensus motif (ψKXEXXSP) (19). This type of regulation has also been demonstrated for a number of other proteins, including MEF2 family members (14, 23, 41; reviewed in reference 59) and Hic1 (44). In contrast, extracellular signal-regulated kinase (ERK) mitogen-activated protein (MAP) kinase pathway signaling reduces sumoylation of the ETS domain transcription factor Elk-1 (58) and the progesterone receptor (PR) (4). In the latter case, Elk-1 desumoylation is a component of the transcriptional activation process that is orchestrated by this transcription factor in response to ERK pathway signaling.
PEA3 (also known as E1AF and ETV4) is a member of the ETS domain transcription factor family, is in a distinct subclass from Elk-1, and shares a domain structure and extensive sequence similarities with ER81 and ERM (reviewed in reference 6). In common with Elk-1, PEA3 is an activator protein whose transactivation capacity is enhanced following activation of the ERK and JNK MAP kinase signaling pathways (37). However, PEA3 has a distinct biological role and is associated with neuronal pathfinding (31, 52) and in a disease context with tumor metastasis (reviewed in reference 6). In common with the latter observation, numerous PEA3 target genes have been identified, including MMP-1 and COX-2 (40, 51). Recently, a proteomic study identified ER81/ETV1 as a potential SUMO target (12). Furthermore, ERM was recently also shown to be modified by SUMO (5). We therefore asked whether PEA3 is also sumoylated and what the consequences of SUMO modification on PEA3 function might be.
We demonstrate that PEA3 is modified by SUMO in vitro and in vivo on multiple sites in its N-terminal region. Importantly, we show that activation of the ERK MAP kinase pathway promotes PEA3 sumoylation. Sumoylation subsequently triggers PEA3 ubiquitination and destabilization. Furthermore, we show that sumoylation of PEA3 is required for maximal activation of target gene promoters, including MMP-1 and COX-2, thereby implicating a dynamic series of regulatory events in the activation cycle of PEA3. Thus, in contrast to the majority of other transcription factors, sumoylation destabilizes PEA3 and plays a positive role in PEA3-mediated gene expression, and the ERK MAP kinase pathway cooperates with, rather than antagonizes, this process.
MATERIALS AND METHODS
Plasmid constructs.
The following plasmids were used in mammalian cell transfections. pCH110 (Pharmacia), pColI-Luc (containing the MMP1 promoter [−517/+63]; kindly provided by Olivier Kassel) (40), and phPES2 (nucleotides −327 to +59 of the human COX-2 promoter) (21) have been described previously. pAS1801 (encoding full-length mouse PEA3) (24), pCW7 (encoding His-tagged ubiquitin; kindly provided by Jean-Luc Baert) (1), pCDNA3-dnUbc9, pCDNA3-Ubc9, and pCDNA3-His-SUMO-2 (kindly provided by Ron Hay), pCMV-Flag-SENP1 (kindly provided by Edward Yeh), and cytomegalovirus (CMV)-driven plasmids encoding wild-type and catalytically inactive Ubp41 (kindly provided by Kyungjin Kim) (28) have been described previously. pCMV-MEK1 encodes constitutively active MEK-1(ΔNS218E-S222D). The vector encoding CBP (pPA8) was provided by Marleen Petit.
The following plasmids encode full-length PEA3 with mutations in combinations of the lysine residues K96 (K1), K222 (K2), K256 (K3), K330 (K4), and K437 (K5) or glutamate residues E98 (E1), E224 (E2), E258 (E3), E332 (E4), and E439 (E5) and were constructed by QuikChange mutagenesis (Stratagene). pAS1029, pAS1030, pAS1031, pAS1032, and pAS1033 [encoding PEA3(K1R), PEA3(K2R), PEA3(K3R), PEA3(K4R), and PEA3(K5R)] were constructed using the template pAS1801 and the primer pairs ADS1936/1937, ADS1938/1939, ADS1940/1941, ADS1942/1943, and ADS1944/1945, respectively. Additional compound mutations were then created by using the appropriate primer pairs on templates already containing individual or multiple mutations to create pAS1034, pAS1036, pAS1040, pAS1041, pAS1042, pAS2651, and pAS2652 [encoding PEA3(K12R), PEA3(K123R), PEA3(K1234R), PEA3(K12345R), PEA3(K2345R), PEA3(K1345R), and PEA3(K1245R)]. Similarly, the mutants pAS1043, pAS1044, p1045, pAS1046, and pAS1047 [encoding PEA3(E1A), PEA3(E2A), PEA3(E3A), PEA3(E4A), and PEA3(E5A)] and the compound mutants pAS1048, pAS1049, and pAS1050 [encoding PEA3(E23A), PEA3(E123A), and PEA3(E12345A)] were created by sequential insertion of mutations at the indicated glutamate residues using the primer pairs ADS1946/1947 (E1), ADS1948/1949 (E2), ADS1950/1951 (E3), ADS1952/1953 (E4), and ADS1954/1955 (E5). pAS2653, pAS2654, and pAS2655 [encoding PEA3(S101A), PEA3(S101E), and PEA3(P102A)] were constructed using the template pAS1801 and the primer pairs ADS1956/1957, ADS1958/1959, and ADS1960/1961, respectively.
For bacterial expression, pGEX2T-Ubc9 and pGEX2T-SUMO-1 have been described previously (8).
For in vitro transcription/translation, pAS1801 [encoding full-length mouse PEA3(1-480)] and its mutant derivatives were used.
Tissue culture, cell transfections, reporter gene assays, and reverse transcription-PCR (RT-PCR).
HEK293 and HCT116 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. SW480 cells were grown in L15 Leibovitz culture medium supplemented with 10% fetal bovine serum. Where indicated, cells were treated with phorbol myristate acetate (PMA; 10 nM), cerulenin (5 μg/ml), etoposide (10 μM; Sigma), the MEK inhibitor U0126 (10 μM), or the proteosome inhibitor MG132 (5 μM). Plasmid transfections for HEK293T cells were performed using Polyfect (Qiagen) according to the manufacturer's instructions. For transfected cells, the inhibitors were added 20 h after transfection and proteins were analyzed by Western blotting or luciferase assays typically 6 h later. To analyze protein stability, the protein synthesis inhibitor cycloheximide (50 μg/ml) was added to cells for the times indicated in the figure legends. Cells were then washed and samples analyzed by Western analysis over a 9-h period. Small interfering RNA (siRNA) transfections were performed with 5 μl Lipofectamine 2000 per well in a six-well plate and 40 nM SMARTpool siRNA duplexes against glyceraldehyde-3-phosphate dehydrogenase (GAPDH), RNF4, and CBP or a scrambled duplex (Dharmacon) for 24 h before treatment of cells with PMA.
For reporter gene assays, typically 0.25 μg of reporter plasmid and 50 ng of pCH110 were cotransfected with 0.005 to 2 μg of expression plasmids. Cell extracts were prepared and equal amounts of protein were used in luciferase and β-galactosidase assays as described previously (15).
Standard and real-time RT-PCR of MMP1 and COX-2 expression was carried out using the primer pairs ADS1962 (5′-CGTCTTACGAATTTGCCGACAGA-3′) with ADS1963 (5′-GTTCTAGGGAAGCCAAAGGAGCTG-3′) and ADS2058 (5′-TCAGCCATACAGCAAATCCT-3′) with ADS2059 (5′-CTTGAAGTGGGTAAGTATGTAGT-3′) as described previously (36).
In vitro and in vivo SUMO and ubiquitination assays.
In vitro SUMO assays were performed essentially as described previously (58) using in vitro-translated PEA3 proteins with 0.08 μg SAE1/2 (Alexis Biochemicals), 0.5 μg Ubc9, and 0.5 μg SUMO-1 or glutathione S-transferase (GST)-SUMO-1 (see Fig. 1, below) according to the manufacturer's instructions (Biomol) (see Fig. 2, below). Sumoylated species were monitored on 12% (see Fig. 1) or 8% (see Fig. 2) sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels.
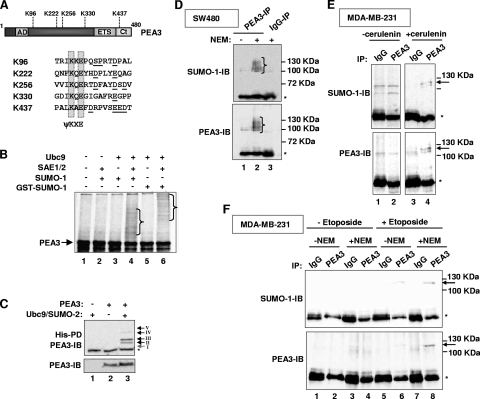
FIG. 1.
PEA3 is SUMO modified. (A) Schematic of the PEA3 structure, illustrating the location of the SUMO consensus sites with respect to the known domains (AD and Ct are activation domains and ETS is the DNA binding domain). The local context of the core ψKXE SUMO motifs is indicated, with downstream acidic residues or SP motifs underlined. (B) In vitro sumoylation of PEA3. In vitro-translated PEA3 was incubated with the indicated components of the SUMO pathway and either SUMO-1 or GST-SUMO-1. The positions of the high-molecular-weight SUMO-conjugated species are indicated by brackets. (C) In vivo sumoylation of PEA3. 293 cells were cotransfected with the indicated combinations of PEA3, Ubc9, and His-tagged SUMO-2. SUMO-conjugated proteins were isolated by nickel affinity pull down (PD) and PEA3-derived species were identified by immunoblotting (IB). Total input levels of PEA3 were determined by IB (bottom panel). The positions of discrete major bands corresponding to SUMO-conjugated species are indicated by arrows and roman numerals. (D, E, and F) Sumoylation of endogenous PEA3. PEA3 was immunoprecipitated (IP) from SW480 cells in the presence and absence of NEM (D and F) or from MDA-MB-231 cells treated with NEM (E) and the precipitated proteins identified by IB with SUMO-1 or PEA3 antibodies. Cells were also treated with cerulenin (E) or etoposide (F) where indicated. IgG precipitates were used as a control. The positions of the high-molecular-weight SUMO-conjugated species are indicated by brackets (B and D) or an arrow (E and F), or specific bands are indicated by numbered arrows (C). Asterisks represent a nonspecific band resulting from the co-IP (IgG heavy chain).
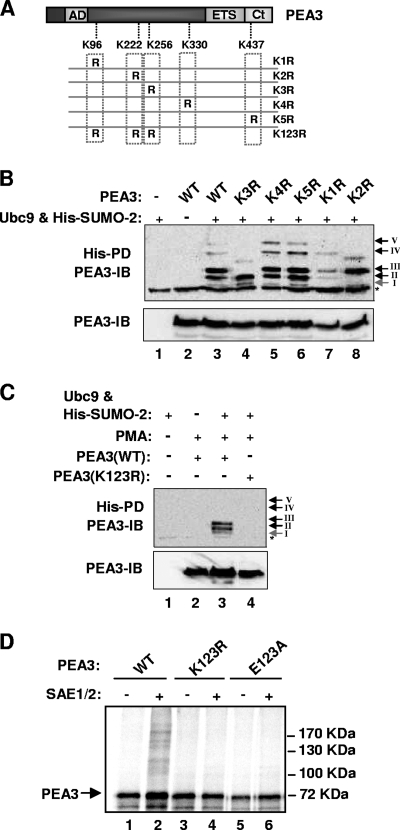
FIG. 2.
Mapping the SUMO modification sites in PEA3. (A) Schematic of the PEA3 structure, illustrating the locations of the sumoylation sites that were mutated. (B and C) In vivo sumoylation of the indicated PEA3 derivatives was probed as described for Fig. 1C. Arrows and associated roman numerals indicate the positions of the major SUMO-dependent high-molecular-weight PEA3 species. Cells were either untreated (B) or treated with PMA for 6 h before harvesting where indicated (C). (D) In vitro sumoylation of the indicated PEA3 derivatives was performed as described for Fig. 1B.
In vivo sumoylation of overexpressed proteins was detected by cotransfection of His-tagged SUMO-2 and Ubc9 and wild-type or mutant PEA3 derivatives into 293T cells. After transfection, the cells were left overnight and then treated with different reagents as detailed in the figure legends, followed by Ni-nitrilotriacetic acid purification of the conjugates under denaturing conditions in the presence of 8 M guanidine-HCl and 10 mM imidazole as described previously (58). In vivo ubiquitination of PEA3 was examined by cotransfection of His-ubiquitin, wild-type, or mutant derivatives of PEA3 into HEK293T cells, followed by similar procedures as used for purification of His-tagged SUMO conjugates. For analyzing sumoylation of endogenous proteins, cells were lysed in buffer containing the SUMO protease inhibitor N-ethylmaleimide (NEM) (150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 10% [vol/vol] glycerol, 1% [vol/vol] Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 10 mM aprotinin, 10 mM NEM), and subsequent immunoprecipitations were also performed in the presence of this inhibitor.
Western blot analysis.
Western blotting was carried out with the primary antibodies PEA3 (sc-113; Santa Cruz), SUMO-1 (sc-5308; Santa Cruz), and RNF4 (kindly provide by Jorma Palvimo) essentially as described previously (15). Data were quantified using Quantity One software (Bio-Rad).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays using control immunoglobulin G (IgG; Upstate) or acetyl histone H4 (Upstate 06-866) were performed as described previously (43). Bound promoters were detected by PCR using the following primers: for the MMP-1 promoter, ADS1968, 5′-GGGGACTCCAAGGCTCTATT-3′, and ADS1969, 5′-TCAGGAAAGCAGCATGTGAC-3′; for the COX-2 promoter, ADS2052, 5′-AAAGACGTACAGACCAGACACG-3′, and ADS2053, 5′-GCTTCCGAGAGCCAGTTC-3′, or SRF intron 3 (24).
RESULTS
PEA3 is modified by SUMO.
PEA3 contains five sites which fit the core SUMO consensus sequence ψKXE (Fig. 1A). These sites are conserved in PEA3 family proteins from humans to zebrafish in otherwise poorly conserved regions of the proteins, suggesting functional importance. Three of the sites, K222, K256, and K437, conform to the longer NDSM consensus sequence, where clusters of acidic residues are located downstream from the core motif (57). A fourth site, K96, conforms to the PDSM consensus sequence (19), as it contains a potential proline-directed serine phosphorylation site three residues downstream from the end of the core motif.
To establish whether PEA3 is a potential substrate of the SUMO pathway, we first carried out in vitro sumoylation assays. Incubation of PEA3 with components of the SUMO pathway caused the appearance of multiple higher-molecular-weight species indicative of several sumoylation events (Fig. 1B, lane 4). These bands were confirmed as sumoylated species as their mobility decreased upon substitution of SUMO-1 with a GST-SUMO-1 fusion protein (Fig. 1B, lane 6).
To confirm that PEA3 could be sumoylated in vivo, we cotransfected 293 cells with a PEA3 expression vector along with plasmids encoding Ubc9 (the E2 ligase) and His-tagged SUMO-2. Subsequent isolation of His-tagged protein conjugates demonstrated that PEA3 can be sumoylated to give rise to several high-molecular-weight bands in vivo (Fig. 1C; see also Fig. 2B, below). The number of bands detected varied, but there were two predominant ones (bands II and III). Furthermore, we determined whether endogenous PEA3 could be sumoylated. Little PEA3 protein can be detected in the majority of cultured cell lines; however, we were able to detect PEA3 in several high-molecular-weight forms in SW480 cells (Fig. 1D, bottom panel) and HCT116 cells (data not shown). These high-molecular-weight forms were also identified using an anti-SUMO-1 antibody (Fig. 1D, top panel) and their appearance was dependent on the inclusion of the SUMO protease inhibitor NEM, demonstrating that they represent SUMO-conjugated species of PEA3. Importantly, the molecular weights of these SUMO-modified PEA3 forms closely matched the mobilities of the sumoylated species seen with exogenous PEA3 (see Fig. 3A, below). We also examined whether we could detect SUMO-modified PEA3 in MDA-MB-231 cells that had been treated with the fatty acid synthase inhibitor cerulenin. This agent has previously been shown to enhance the levels of PEA3 protein in cancer cells (33). A high-molecular-weight form of PEA3 could be specifically precipitated from these cells upon cerulenin treatment, which corresponded to a SUMO-modified form (Fig. 1E, lanes 3 and 4). This was not apparent in cells where PEA3 levels had not been enhanced due to cerulenin treatment (Fig. 1E, lanes 1 and 2). Similarly, we treated MDA-MB-231 cells with etoposide, which was previously shown to increase PEA3 levels (56). Again, a higher-molecular-weight conjugated form of PEA3 could be detected with anti-PEA3 and -SUMO antibodies but only in the presence of etoposide and NEM (Fig. 1F, lane 8).
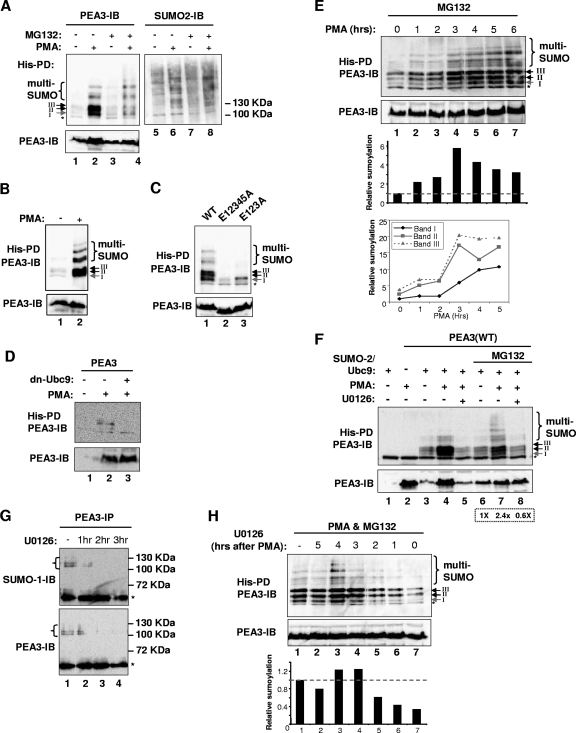
FIG. 3.
ERK MAP kinase signaling enhances PEA3 sumoylation. (A to F) In vivo sumoylation of wild-type PEA3 or the indicated mutant forms of PEA3 in transfected 293 cells was probed as described for Fig. 1C. (A) PMA and/or MG132 was added 6 h before harvesting, as indicated. Ubc9 and His-tagged SUMO-2 were cotransfected with PEA3. SUMO-conjugated PEA3 was detected by immunoblotting (IB) with anti-PEA3 antibodies (left panel) and total precipitated SUMO conjugates were detected by anti-SUMO-2 antibodies (right panel). (B) The levels of input PEA3 protein in panel A were normalized and the amounts of precipitated samples run on the gel were normalized, due to PMA affecting overall PEA3 levels. This enabled comparisons to be made based on equal amounts of PEA3 protein. (C) Cells were cotransfected with Ubc9, His-tagged SUMO-2, and the indicated PEA3 derivatives and sumoylation was detected following 6 h of treatment with PMA. (D) PEA3 was contransfected with His-tagged SUMO-2 and dnUbc9, and cells were treated with PMA where indicated. (E) Cells transfected with PEA3, SUMO-2, or Ubc9 were treated with MG132 and for the indicated times with PMA. The amount of sumoylation at each time point relative to total PEA3 level is shown in the bar graph beneath the figure. The line graph represents quantification of the appearance of the three major sumoylated PEA3 species (bands I to III) over the first 5 h after PMA stimulation relative to the basal levels of band I (taken as 1). (F) Ubc9 and His-tagged SUMO-2 were cotransfected with PEA3 as indicated, and PMA and the MEK inhibitor U0126 were added 6 h before harvesting. MG132 was added (lanes 6 to 8) 1 h prior to stimulation of cells with PMA. The amount of sumoylation relative to total PEA3 levels in lanes 6 to 8 is shown in the box beneath the figure. (G) Sumoylation of endogenous PEA3 in SW480 cells was probed as described for Fig. 1D. The MEK inhibitor U0126 was added at the indicated times prior to harvesting. (H) 293 cells were transfected with PEA3, SUMO-2, or Ubc9 and were treated with PMA for 6 h in the presence of MG132. U0126 was added at the indicated times after PMA addition. The amount of sumoylation at each time point relative to total PEA3 levels is shown in the bar graph beneath the figure. Arrows and associated roman numerals indicate the positions of the major SUMO-dependent high-molecular-weight PEA3 species. Multisumoylated species are indicated by brackets.
Together these results demonstrate that PEA3 can be sumoylated in vitro and in vivo at several sites.
Mapping the SUMO sites in PEA3.
To identify which of the potential SUMO sites are functional in PEA3, we mutated each one individually and examined PEA3 sumoylation status in vivo. Mutation of either K222 (K2) or K256 (K3) resulted in the loss of one of the two major sumoylated species (Fig. 2B, lanes 4 and 8; bands II and III, respectively). In contrast mutation of either K330 (K4) or K437 (K5) had little effect on the high-molecular-weight sumoylated species (Fig. 2B, lanes 5 and 6; bands IV and V, respectively). The minor sumoylated band (band I) was lost upon mutation of K96 (K1) along with reductions in bands II and III, but only band I loss was reproducibly observed (Fig. 2B, lanes 7). Thus, in vivo, the major sites of sumoylation are K222 and K256, and to a lesser extent K96. Moreover, as each band could be specifically attributed to a particular sumoylation event at a defined lysine residue, the different mobility bands most likely arise due to the abnormal mobility commonly observed for branched polypeptides formed when SUMO is conjugated at different positions in the protein sequence. To establish whether these three sites were the only ones used for SUMO conjugation, we mutated all three in the mutant PEA3(K123R) and tested its sumoylation status in vivo. Sumoylation was not detectable in this mutant (Fig. 2C). Finally, we confirmed the in vivo results in an in vitro sumoylation assay. Consistent with the in vivo results, in vitro analysis demonstrated that the PEA3 mutant K123R was devoid of sumoylation (Fig. 2D). Similarly, the mutant PEA3(E123A), which contains single amino acids substitutions at the conserved glutamic acid residues (to alanine) within the three core sumoylation motifs, also causes abolition of PEA3 sumoylation in vitro (Fig. 2D) and multisite sumoylation in vivo (data not shown). Thus, K96 (K1), K222 (K2), and K256 (K3) are also the major sites of sumoylation in PEA3 in vitro.
Collectively these results therefore demonstrate that PEA3 can be sumoylated at multiple sites, chiefly involving lysines K96, K222, or K256.
ERK MAP kinase pathway activation enhances PEA3 sumoylation.
The sumoylation levels of the ETS domain transcription factor Elk-1 and the PR are reduced upon activation of the ERK MAP kinase pathway (4, 58). As PEA3 is also regulated by ERK MAP kinase signaling, we therefore investigated whether this pathway affected PEA3 sumoylation levels. In contrast to the effects on Elk-1 and PR, upon treatment of cells with PMA, a potent activator of the ERK pathway, we observed an overall increase in PEA3 sumoylation levels. In particular, band I, corresponding to sumoylation at K96, was clearly visible (Fig. 3A, lanes 1 and 2). PMA treatment also caused an increase in PEA3 levels, but importantly, when PEA3 protein levels were normalized through either stabilization through proteosomal inhibition with MG132 (Fig. 3A, lanes 3 and 4) or through rerunning volumes of precipitated samples that corresponded to equal amounts of input PEA3 protein (Fig, 3B), increased sumoylation levels were still observed after PMA treatment. Equal precipitation of SUMO-modified proteins was confirmed under all treatment conditions (Fig. 3A, lanes 5 to 8). To confirm that the enhanced levels of sumoylation occurred at the same sites as those observed under steady-state conditions, we tested the triple E123A and multiple E12345A mutants which are mutated in the first three or all five of the consensus SUMO modification sites. In both cases, only residual low-level SUMO modification was observed following PMA stimulation, indicating that K96, K222, and K256 are the major sites of conjugation under both basal and activating signaling conditions (Fig. 3C). Importantly, inhibition of the endogenous SUMO pathway with a dominant-negative version of Ubc9 (dn-Ubc9) blocked the PMA-induced appearance of these sumoylated forms of PEA3 (Fig. 3D). To monitor the kinetics of appearance of the sumoylated species, we treated cells with PMA in the presence of the proteosome inhibitor MG132. This ensured that we had equal amounts of PEA3 present, as PMA potently stimulates the accumulation of PEA3 protein and confounds analysis of increases in sumoylation levels (Fig. 3A and D). Under these conditions, the increase in abundance of the bands corresponding to the three major sumoylation sites (I, II, and III) increased significantly between 2 and 3 h after PMA stimulation (Fig. 3E, lanes 1 to 4). Further increases in the intensity of band I were then observed, peaking between 5 and 6 h, whereas the others had already reached a plateau (Fig. 3E, lanes 5 to 7). Thus, while sumoylation of all three sites was enhanced in response to PMA stimulation, the kinetics of K96 sumoylation clearly differs, suggesting that its regulation differs from the other sites.
To demonstrate that the effect of PMA on PEA3 sumoylation was through the ERK pathway rather than some other signaling route, we treated cells with PMA in the presence and absence of the MEK inhibitor U0126. In the presence of inhibitor, the enhancement of PEA3 sumoylation was blocked, although PEA3 levels also decreased (Fig. 3F, lanes 4 and 5). We therefore stabilized PEA3 levels by cotreatment with MG132, and under these conditions, a clear reduction in PEA3 sumoylation levels could be observed upon inhibition of ERK pathway signaling, despite the equal levels of total PEA3 protein (Fig. 3F, lanes 7 and 8). Furthermore, treatment of SW480 cells with the MEK inhibitor also caused a decrease in the levels of sumoylated endogenous PEA3 (Fig. 3G).
Due to the relatively slow kinetics of ERK pathway-dependent enhancement of PEA3 sumoylation, we tested when ERK pathway activity was required by adding the U0126 inhibitor at different times following PMA stimulation. Addition of the inhibitor between 0 and 2 h after PMA addition substantially blocked the enhancement of PEA3 sumoylation (Fig. 3H, lanes 5 to 7). However, addition of the inhibitor after 3 h had little effect on PEA3 sumoylation levels (Fig. 3H, lanes 1 to 4). These results demonstrate that ERK kinase pathway activity must therefore be sustained for more than 2 h to trigger PEA3 sumoylation and is consistent with the kinetics of PEA3 sumoylation, which begin to peak between 3 and 4 h after ERK pathway activation.
Together these data therefore indicate that sustained ERK pathway signaling increases the levels of PEA3 sumoylation.
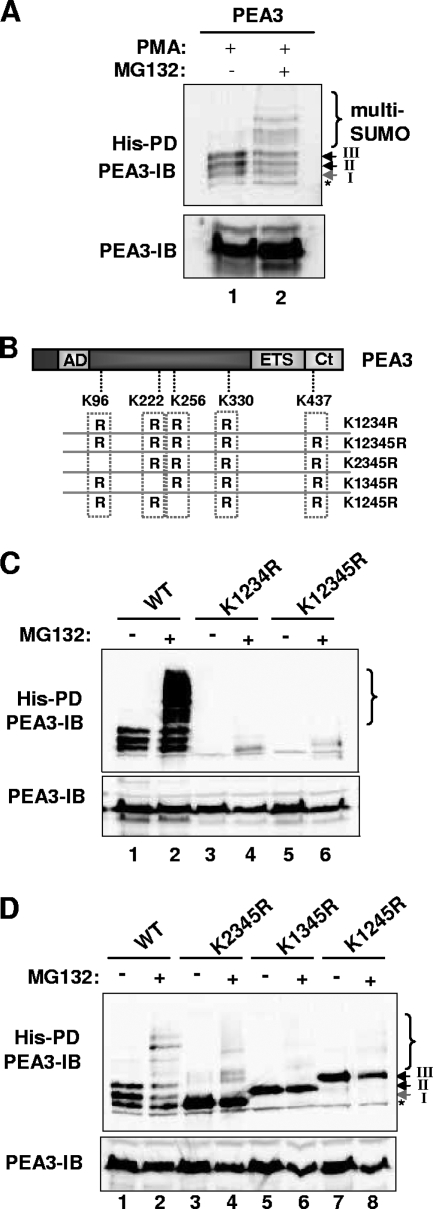
PEA3 is transiently multisumoylated.
In several experiments where we used the proteosomal inhibitor MG132, we noticed an accumulation of higher-molecular-weight conjugates (Fig. 3A, B, C, and E). To establish unequivocally that MG132 treatment was preserving these high-molecular-weight conjugates, we compared samples from PMA-treated cells in the presence and absence of MG132 cotreatment, following normalization of total PEA3 levels. Proteosomal inhibition led to the clear accumulation of high-molecular-weight lower-mobility species (Fig. 4A). The appearance of these species was dependent on the sumoylation sites, as removal of four or five of the consensus sumoylation sites abolished the induction of the high-molecular-weight conjugates (Fig. 4C). A similar effect was observed in the K123R and E123A mutants, which lacked the major SUMO modification events (data not shown). To establish whether any of the individual sumoylation sites were more important for the generation of these high-molecular-weight conjugates, we tested the effect of MG132 treatment on the sumoylation status of PEA3 proteins containing only one of the three sumoylation sites following PMA treatment. While high-molecular-weight conjugates could be observed on wild-type PEA3, mutant proteins containing the single sumoylation sites K96, K222, or K256 exhibited reduced levels of these lower-mobility species (Fig. 4D, lanes 4, 6, and 8). Moreover, the residual banding patterns did not resemble that observed with the wild-type protein (Fig. 4D, compare lanes 4 and 2).
FIG. 4.
PEA3 is multisumoylated. (A) Ubc9 and His-tagged SUMO-2 were cotransfected with PEA3. PMA was added 6 h before harvesting in the presence or absence of MG132 as indicated. The levels of input proteins and precipitated samples were normalized prior to loading. (B) Schematic of the PEA3 structure, illustrating the locations of the sumoylation sites that were mutated. (C and D) Ubc9 and His-tagged SUMO-2 were cotransfected with the indicated PEA3 derivatives. PMA was added 6 h before harvesting in the presence or absence of MG132 as indicated. IB, immunoblotting results; PD, pull-down assay results.
Together these results demonstrate that proteosomal inhibition traps a transient population of multisumoylated PEA3 and suggest that multiple SUMO sites are required for the production of the high-molecular-weight sumoylated species.
K96 sumoylation is controlled through a PDSM.
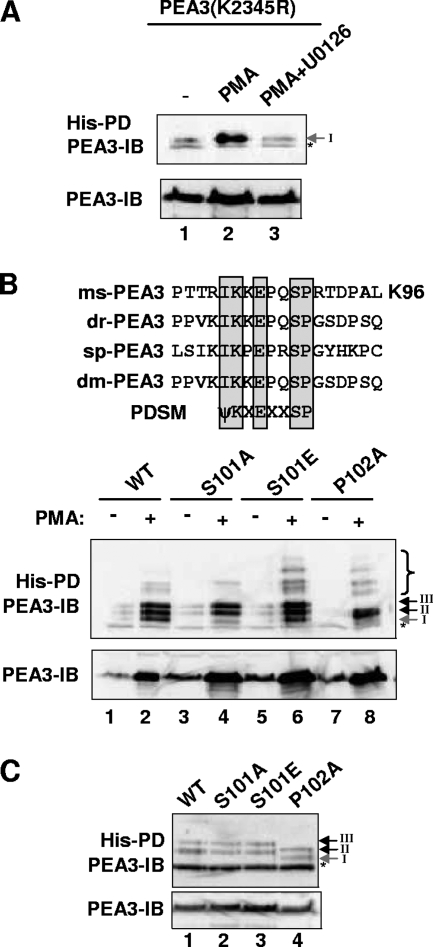
The induction kinetics of K96 sumoylation in response to MAP kinase signaling differs from that of K222 and K256. Indeed, the sequence of this site conforms to the PDSM consensus site (Fig. 5B) (19), suggesting that sumoylation at this site might be controlled by phosphorylation. Moreover, the key features of this site (i.e., the core and SP motifs) are evolutionarily conserved with PEA3 homologues in species as distant as Drosophila melanogaster and the sea urchin Strongylocentrotus pupuratus (Fig. 5B). To establish whether sumoylation of this site is responsive to MAP kinase-mediated signaling, we examined the sumoylation status of PEA3(K2345R), which lacks all potential sumoylation sites other than K96. Importantly, sumoylation of K96 was enhanced in this mutant following PMA treatment in an ERK pathway-dependent manner (Fig. 5A), demonstrating that prior sumoylation at other sites is not a prerequisite for K96 sumoylation.
FIG. 5.
Role of the PDSM in ERK MAP kinase-mediated PEA3 sumoylation. (A to C) In vivo sumoylation of wild-type PEA3 in transfected 293 cells was probed as described for Fig. 1C. (A) Ubc9 and His-tagged SUMO-2 were cotransfected with PEA3(K2345R). PMA and U0126 were added 6 h before harvesting as indicated. The levels of input proteins and precipitated samples were normalized prior to loading. (B and C) The indicated PEA3 mutants were cotransfected with Ubc9 and His-tagged SUMO-2. Cells were left untreated (C) or treated with PMA (6 h) as indicated (B). A longer exposure of the samples derived in the absence of PMA is shown in panel C to visualize the basal levels of sumoylation. Asterisks represent a nonspecific band resulting from the pull-down assay (PD). IB, immunoblotting results.
To probe the potential role for phosphorylation of the SP motif located immediately downstream from the core ψKXE SUMO site at K96, we mutated the SP motif and examined the consequences for MAP kinase-mediated sumoylation of PEA3. Mutation of the serine residue in PEA3(S101A) caused the virtual abolition of PMA-induced sumoylation of K96 (band I) but did not affect the enhancement of sumoylation at other sites (Fig. 5B, lane 4). Similarly, mutation of the adjacent proline residue in PEA3(P102A), also caused a loss of K96 sumoylation, although a change in mobility of the bands corresponding to K222 and K256 sumoylation was observed, most likely due to conformational changes induced by this mutation (Fig. 5B, lane 8). In contrast, replacement of serine 101 with a phospho-mimetic residue in PEA3(S101E) restored the PMA-inducible sumoylation of K96 (Fig. 5B, lane 6). However, enhanced sumoylation of K96 by introduction of this phospho-mimetic residue was not observed when the ERK pathway was not activated (Fig. 5C), demonstrating that negative charge at this position (and presumably phosphorylation) is only permissive and not causative in the enhancement of PEA3 sumoylation by ERK signaling.
Thus, the PDSM motif in PEA3 behaves in a similar manner to the PDSMs found in other substrates (59). The putative proline-directed serine phosphorylation site within the PDSM surrounding K96 in PEA3 is functionally important and is required for ERK pathway-dependent enhancement of K96 sumoylation.
Sumoylated PEA3 is unstable.
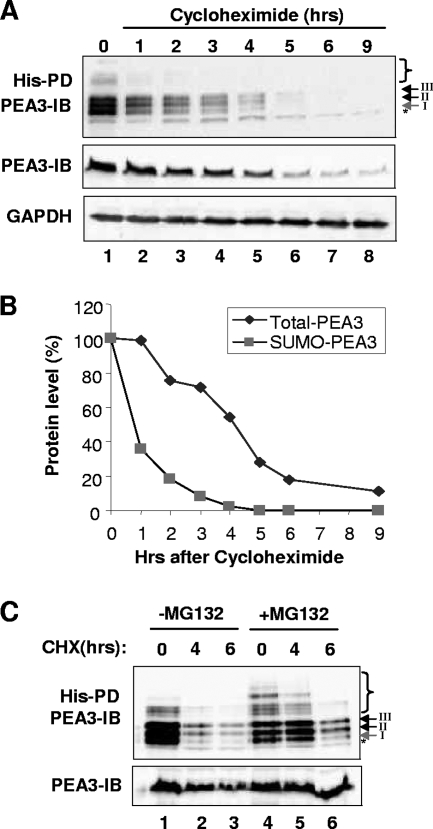
The trapping of high-molecular-weight sumoylated PEA3 species by proteosomal inhibition suggested a general role for sumoylation in enhancing PEA3 instability. To address this issue, we determined the stability of the sumoylated PEA3 species in comparison to the bulk PEA3 levels following treatment of cells with the protein synthesis inhibitor cycloheximide. Sumoylated PEA3 exhibited a much shorter half-life in comparison to the bulk PEA3 levels (less than 1 h compared to more than 3 h) (Fig. 6A and B). Among the different sites, sumoylation of K96 exhibited the most rapid turnover (Fig. 6A, band I, and data not shown). To confirm that the changes in sumoylation levels reflected a change in stability, rather than an indirect consequence of inhibiting a component of the sumoylation machinery and hence promoting sumoylation turnover, we compared the stability of PEA3 in the presence and absence of the proteosome inhibitor MG132. MG132 treatment did not cause dramatic increases or decreases in the overall levels of PEA3 sumoylation (Fig. 3A, lanes 3 and 4, and F, lanes 4 and 7), demonstrating that it does not have a gross effect on the SUMO pathway. Importantly, after 4 h of cycloheximide treatment, cotreatment with MG132 blocked the loss of sumoylated species, demonstrating that turnover was due to proteosomal degradation (Fig. 6C, lane 5). However, at a later time point, MG132 was unable to block the loss of sumoylation, demonstrating that this loss was an indirect consequence of cycloheximide treatment. Importantly, the majority of loss of sumoylated PEA3 was already achieved after 4 h cycloheximide treatment in the absence of MG132 (Fig. 6C, lane 2). Thus, sumoylated forms of PEA3 are more unstable, and higher-molecular-weight multi-SUMO-conjugated species appear particularly sensitive, as they are not readily detectable unless the proteosome inhibitor is inhibited.
FIG. 6.
Sumoylated PEA3 is unstable. (A) 293 cells were cotransfected with PEA3, Ubc9, or His-tagged SUMO-2. Cells were treated with PMA for 3 h, and then cycloheximide (CHX) was added and cells harvested at the indicated times after cycloheximide addition. Sumoylation of PEA3 was probed as described for Fig. 1C. Bulk PEA3 levels were determined by harvesting in the absence of NEM (middle panel). GAPDH levels were probed as a loading control (bottom panel). (B) Graphical representation of the data from panel A. (C) 293 cells were transfected as described for panel A, but MG132 was added at the same time as PMA in lanes 4 to 6. PD, results from pulldown assay; IB, immunoblotting results.
ERK pathway signaling and sumoylation promote PEA3 ubiquitination.
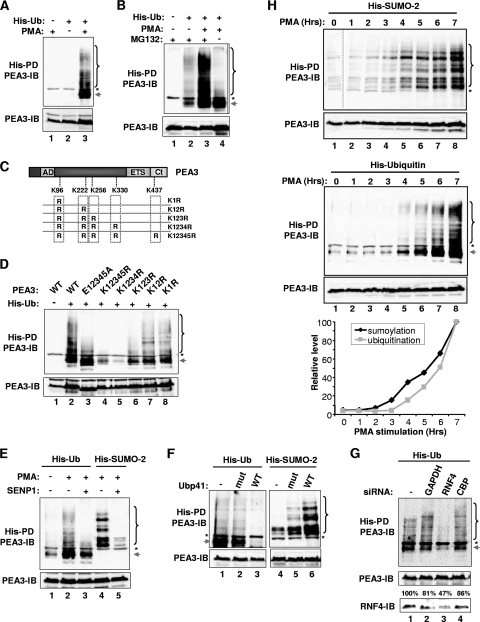
Ubiquitination is often associated with protein instability. We therefore tested whether PEA3 is ubiquitinated by cotransfecting a His-tagged ubiquitin construct and examining whether conjugated PEA3 could be detected in nickel affinity precipitates. Substantial amounts of PEA3 ubiquitination could only be observed following stimulation of cells with PMA (Fig. 7A, lane 3). Importantly, both multiubiquitinated high-molecular-weight species and a prominent lower-molecular-weight band, most likely representing mono-ubiquitination, were observed. These results are consistent with previous studies which demonstrated polyubiquitination of the human form of PEA3, E1AF, although the role of ERK signaling in this process was not addressed (47). To establish whether this polyubiquitination was associated with proteosome-mediated degradation, we repeated the experiment but omitted the proteosomal inhibitor MG132. Importantly PMA-inducible polyubiquitination was lost in the absence of MG132, although monoubiquitination was retained (Fig. 7B, lane 4), demonstrating that polyubiquitination of PEA3 is likely targeting it for degradation.
FIG. 7.
Sumoylation promotes PEA3 ubiquitination. 293 cells were cotransfected with PEA3, His-SUMO-2, or His-tagged ubiquitin (His-Ub) as indicated. SENP1 was cotransfected as indicated (E), and wild-type and catalytically inactive mutant Ubp41 was transfected as indicated (F). siRNA constructs against GAPDH, RNF4, and CBP were cotransfected as indicated (G). MG132 was applied in all cases apart from where indicated in panel B and lanes 4 to 6 of panel F. Cells were also treated with PMA for 6 h (D, F, and G), for 6 h as indicated (A and E), or for 0 to 7 h (H). Ubiquitin- or SUMO-conjugated proteins were isolated by nickel affinity pull-down assay (PD), and PEA3-derived species were identified by immunoblotting (IB). Asterisks represent a nonspecific band resulting from the co-IP. The arrow likely represents monoubiquitinated PEA3, and the bracket represents polyubiquitinated or multisumoylated PEA3. Total levels of PEA3 in the input lysates are shown in the panels below each PD. (C) Schematic of PEA3 structure, illustrating the locations of the sumoylation sites that were mutated. The graph in panel H shows the levels of multisumoylation or polyubiquitination at each time point. The numbers above the bottom portion of panel G represent the percentages of RNF4 protein remaining relative to the no-siRNA treatment.
Next we investigated whether any of the SUMO consensus sites were needed for ubiquitination of PEA3. We examined the ubiquitination status of a series of mutant PEA3 proteins in which additional lysine residues were sequentially mutated (Fig. 7C). Removal of the three N-terminal lysine residues, corresponding to the major sumoylation sites substantially reduced the levels of PEA3 ubiquitination, especially the multiubiquitinated species (Fig. 7D, lanes 6 and 7). Further loss of the lysine residues in the two C-terminal consensus sumoylation sites reduced the levels of ubiquitination even further, with only residual monoubiquitination remaining (Fig. 7D, lanes 4 and 5). As ubiquitination and sumoylation take place on lysine residues, these results suggested that these two modifications might occur on the same sites in PEA3. However, an alternative hypothesis is that sumoylation might promote ubiquitination. We therefore tested the ubiquitination levels of a mutant PEA3 protein (E12345A) in which the consensus sumoylation sites were disabled but the lysine residues left intact (Fig. 7C). This mutant exhibits substantially reduced levels of sumoylation (Fig. 3C). The pattern of ubiquitination of this mutant resembled that of the K123R mutant, with much reduced multiubiquitination, but retained high levels of monoubiquitination (Fig. 7D, lane 3), strongly implicating sumoylation as a requirement for subsequent ubiquitination.
To further explore the links between sumoylation and ubiquitination, we cotransfected the SUMO protease SENP1. As expected, SENP1 caused a dramatic loss in PEA3 sumoylation levels (Fig. 7E, lane 5). Importantly, SENP1 also caused a loss of polyubiquitination while again leaving monoubiquitination largely intact (Fig. 7E, lane 3). We also performed the reciprocal experiment and overexpressed the ubiquitin protease Ubp41. As expected a loss of ubiquitinated PEA3 was observed (Fig. 7F, lane 3); however, sumoylation of PEA3 was not reduced but instead was increased (Fig. 7F, lane 6). This increase in sumoylation is consistent with the notion that polyubiquitination leads to increased turnover of sumoylated PEA3, and reversing the polyubiquitination would be expected to preserve the sumoylation status of PEA3.
Recently, RNF4 was identified as a SUMO-dependent E3 ubiquitin ligase that provides a molecular link between SUMO-modified proteins and their subsequent degradation in a ubiquitin-dependent manner (26, 46, 48). We therefore depleted RNF4 by siRNA treatment and examined PMA-induced ubiquitination of PEA3. Treatment with control duplexes directed against GAPDH or CBP had little effect on polyubiquitination levels (Fig. 7G, lanes 2 and 4). However, loss of RNF4 was accompanied by a reduction in polyubiquitinated PEA3 (Fig. 7G, lane 3), demonstrating a further link between SUMO modification and subsequent polyubiquitination of PEA3. A further key prediction of these results is that sumoylation must kinetically precede ubiquitination. We therefore followed the appearance of multisumoylated and polyubiquitinated species following PMA treatment. Whereas increases in sumoylation could be detected after 3 to 4 h, ubiquitination was delayed, and increases were only detectable after 4 h (Fig. 7H).
Together these results therefore demonstrate that sumoylation precedes and is required for the subsequent efficient polyubiquitination of PEA3. However, monoubiquitination seems to occur largely independently of SUMO modification, most likely on one of the lysine residues within the consensus sumoylation sites.
PEA3 transactivation activity is dependent on sumoylation.
Transcription factor sumoylation generally causes a reduction in transactivation capacity or an increase in repressive activity associated with a transcription factor, although in a few cases, sumoylation has been associated with reciprocal effects resulting in activation (reviewed in reference 10). To establish how PEA3-mediated transcriptional activity is affected by sumoylation, we compared the activity of wild-type PEA3 with a mutant version in which the three major sumoylation sites had been disrupted [PEA3(E123A)]. We chose to mutate the glutamate residues within the core motif to avoid potential complications associated with other lysine modifications. However, similar effects were seen with the PEA3(K123R) mutant (data not shown).
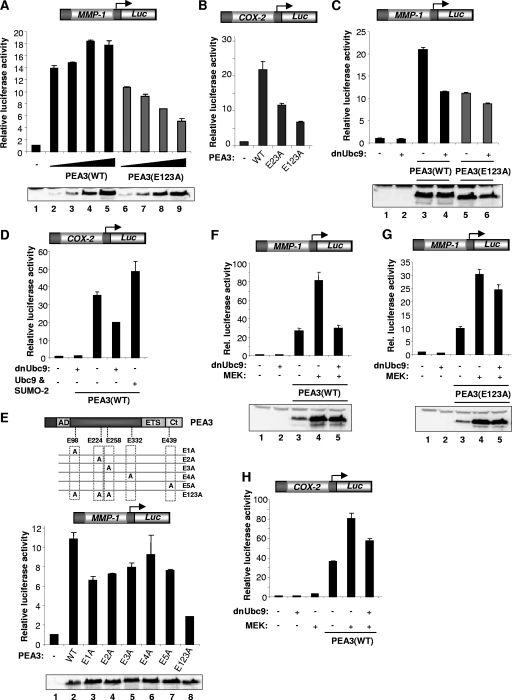
First we carried out a dose-response experiment using the MMP-1 promoter in which the levels of transfected wild-type and mutant PEA3 were varied over an eightfold range. In comparison to the wild-type protein, the activity of the nonsumoylatable mutant PEA3(E123A) was reduced at all expression levels (Fig. 8A). Similarly, the nonsumoylatable mutant PEA3(E123A) exhibited reduced transactivation properties compared to wild-type PEA3 on the COX-2 promoter (Fig. 8B). Furthermore, blocking the SUMO pathway by cotransfection with dnUbc9 reduced the activity of wild-type PEA3 on both the MMP-1 and COX-2 promoters to a similar extent as disruption of the SUMO modification sites (Fig. 8C and D). In contrast, dnUbc9 had little effect on the activity of the nonsumoylatable PEA3(E123A) mutant (Fig. 8C). Conversely, cotransfection of Ubc9 and SUMO-2 caused an increase in PEA3-mediated activation of the COX-2 promoter (Fig. 8D). To establish whether any single sumoylation site was important for the transactivation capacity of PEA3 we compared the activities of mutants disabled specifically at each individual consensus sumoylation site through mutation of glutamate residues in the SUMO consensus motif (Fig. 8E). Mutation of the first three sites which were mapped as the major sites of modification (Fig. 2) caused reductions in the transactivation capacity of PEA3, with the biggest effect being observed in the E1A mutant (corresponding to K96). However, the simultaneous mutation of all three sites caused an additive reduction in transactivation capacity. Similar results were seen from the analogous single-site lysine mutants, although the reduction seen upon disabling the K437 site was not as pronounced (data not shown). Thus, multiple sites contribute to the PEA3 transactivation capacity.
FIG. 8.
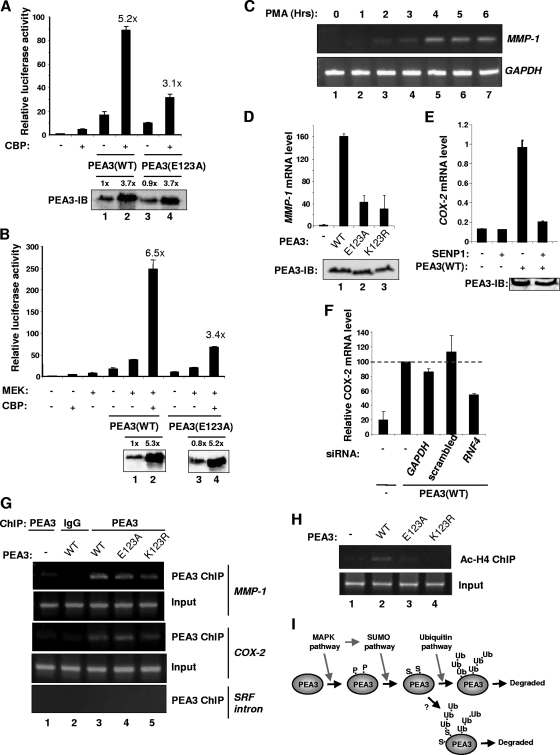
Sumoylation of PEA3 enhances its transactivation capacity. (A to H) Luciferase reporter gene assays using a MMP-1 (A, C, E, F, and G) or a COX-2 promoter-driven luciferase reporter (B, D, and H) in 293 cells. Data are presented relative to the activity of the reporter alone (taken as 1). Western blot results for the expression levels of PEA3 derivatives are shown below the graphs. (A) Increasing amounts of wild-type or mutant (E123A) PEA3 were transfected (50 ng, 100 ng, 200 ng, and 400 ng; increasing amounts are indicated schematically by a triangle). (B to E) Wild-type PEA3 or the indicated mutant version of PEA3 (500 ng) was transfected in the absence (B and E) or presence of cotransfected Ubc9 or SUMO-2 (D) or dnUbc9 (1 μg) (C and D). A schematic of the PEA3 structure, illustrating the locations of the glutamate residues mutated within the sumoylation sites, is shown above panel E. (F to H) Wild-type PEA3 (300 ng) (F and H) or mutant PEA3(E123A) (300 ng) (G) and constitutively active MEK (200 ng) and dnUbc9 (1 μg) were cotransfected as indicated.
Under conditions where the MAP kinase pathway was activated by cotransfection of constitutively active MEK, MMP-1 (Fig. 8F) and COX-2 (Fig. 8H) promoter activation by PEA3 was also inhibited by dnUbc9. PEA3 expression levels were unaffected by dnUbc9 (Fig. 8F and G). Again, under these conditions, the repressive effect of dnUbc9 was severely ablated in the presence of the nonsumoylatable PEA3(E123A) protein (Fig. 8G).
PEA3 cooperates with CBP/p300 to activate transcription (30; reviewed in reference 6). We therefore cotransfected CBP with wild-type PEA3 or PEA3(E123A) and monitored MMP-1 promoter activity. In the presence of CBP, both the magnitude and relative extent of induction of promoter activation were severely reduced in the presence of PEA3(E123A) under basal conditions (Fig. 9A) or where the ERK pathway was activated (Fig. 9B). Thus, sumoylation of PEA3 appears to be important for cooperativity with CBP.
FIG. 9.
Sumoylation of PEA3 is important for its responses to coactivators and activation of endogenous MMP-1. (A and B) Luciferase reporter gene assays using an MMP-1 promoter-driven luciferase reporter in 293 cells. Data are presented relative to the activity of the reporter alone (taken as 1). Western blots show the expression levels of PEA3 derivatives below the graphs. Numbers above each lane represent quantification of the PEA3 level relative to lane 1 in each panel. Wild-type PEA3 or PEA3(E123A) (300 ng) was contransfected with 2 μg CBP. Where indicated, constitutively active MEK (200 ng) was also cotransfected. (C) 293 cells were transfected with wild-type PEA3 expression vector (500 ng) and treated with PMA for the indicated times, and the expression of endogenous MMP-1 was detected by RT-PCR. (D, E, and F) 293 cells were transfected with the indicated PEA3 expression vectors (500 ng) and the expression of endogenous MMP-1 (D) or COX-2 (E and F) was detected by real-time RT-PCR. SENP1 (350 ng) was cotransfected as indicated (E) or siRNA constructs against GAPDH, RNF4, or a scrambled duplex was cotransfected (F). Data in panels D and E are the average of duplicate samples and representative of at least two independent experiments, and the data in panel F are the averages of two independent experiments, each with duplicate samples. (G and H) ChIP analysis was performed in 293 cells transfected with the indicated PEA3 expression vectors. Antibodies were used to detect PEA3 binding to SRF intron 3 or the MMP-1 or COX-2 promoter (H) or acetyl-histone H4 levels on the endogenous MMP-1 promoter (G). Cells were left untreated (G) or treated with PMA for 6 h (H) before ChIP analysis. (I) Model depicting the sequential action of different pathways on PEA3. MAP kinase signaling initiates this cascade, partly due to phosphorylation (P) of PEA3, and promotes subsequent sumoylation (S), which in turn is important for polyubiquitination (Ub) and eventual turnover of PEA3. The question mark represents an alternative potential route where polyubiquitination might occur on the SUMO moiety and/or PEA3 itself.
To probe whether the effects we observed could be detected at the endogenous MMP-1 promoter, we first determined the activation kinetics of MMP-1 in response to PMA stimulation in the presence of PEA3. Maximal levels of MMP-1 expression were detected after 4 h of PMA treatment (Fig. 9C). This time coincides with when we observed enhanced PEA3 sumoylation levels (Fig. 3). Next we compared the abilities of nonsumoylatable PEA3 derivatives and wild-type PEA3 to activate the endogenous MMP-1 gene. Consistent with the reporter gene analysis, the activating capacities of both PEA3(E123A) and PEA3(K123R) were significantly reduced (Fig. 9D). Furthermore, PEA3 can also enhance endogenous COX-2 expression, and this enhancement is ablated by cotransfection of the SUMO protease SENP1 (Fig. 9E). Consistent with these observations, dnUbc9 reduced the ability of wild-type PEA3 to activate MMP-1 expression, further emphasizing the link between PEA3 sumoylation and its transcriptional activation properties on endogenous target genes (data not shown). To probe the role of the interplay between sumoylation and ubiquitination in the transactivation properties of PEA3, we depleted RNF4, which functionally couples these two processes, and assessed the ability of PEA3 to activate endogenous COX-2 expression. Control siRNA duplexes had little effect on PEA3 activity. However depletion of RNF4 significantly reduced the level of PEA3-mediated activation of COX-2 expression (Fig. 9F).
Finally, ChIP analysis was performed to analyze PEA3 binding and histone acetylation levels at the COX-2 and MMP-1 promoters. First we demonstrated that binding could be detected for wild-type PEA3 and the SUMO site mutants PEA3(E123A) and PEA3(K123R) at both the COX-2 and MMP-1 promoters (Fig. 9G). Next we analyzed the levels of histone H4 acetylation at the endogenous MMP-1 promoter in the presence of wild-type and mutant PEA3 derivatives. While enhanced levels of histone acetylation could be detected in the presence of wild-type PEA3, acetylation levels were much reduced in the presence of either PEA3(E123A) and PEA3(K123R) (Fig. 9H). This observation is consistent with the reduced ability of the nonsumoylatable PEA3 derivatives to cooperate with CBP and to activate the MMP-1 promoter.
Together these results demonstrate that sumoylation of PEA3 is an important molecular determinant of its transactivation capacity at target genes like MMP-1 and COX-2.
DISCUSSION
Sumoylation appears increasingly important in controlling transcriptional events, and the prevailing view is that SUMO imparts repressive properties on transcription factors (reviewed in references 10, 17, and 32). Furthermore, links between MAP kinase pathway signaling and the sumoylation pathway have been made, most notably with the transcription factor Elk-1, for which MAP kinase pathway activation leads to desumoylation and hence contributes to transcriptional activation (16, 58). However, here we have demonstrated that sumoylation of PEA3 is an important event in promoting its transactivation properties and that activation of the ERK pathway promotes PEA3 sumoylation. Thus, different functional interactions between the ERK and SUMO pathways ultimately lead to the same event, transcriptional activation, but through different routes.
While sumoylation is frequently linked to transcriptional repression, it is important to emphasize that in a growing number of cases, it enhances transactivation properties. In some cases, this is through enhancing DNA binding (e.g., HSF-1/-2) (13, 20). However, functional interactions with coactivators could also be affected, as suggested here for PEA3, by the reduced ability of a nonsumoylatable PEA3 to cooperate with CBP (Fig. 9). Furthermore, sumoylation of PEA3 decreases its stability (Fig. 6), which differs from the commonly observed antagonism seen between sumoylation and ubiquitin-mediated proteolysis (reviewed in reference 50). Here we show that PEA3 ubiquitination is enhanced following ERK pathway activation. Ubiquitination is a conserved feature of the related PEA3 subfamily, as another member ERM has recently also been shown to be ubiquitinated and exhibits both poly- and monoubiquitination, as we have observed for PEA3 (1). However, for PEA3, sumoylation is also important for efficient PEA3 polyubiquitination. This is suggestive of a mechanism involving SUMO-dependent recruitment of a ubiquitin ligase, such as is observed with RNF4 (26, 46, 48) or between HIF-1α and VHL (3). Indeed, we have provided evidence for such a link in PEA3, as depletion of RNF4 reduces the amount of polyubiquitination of PEA3 (Fig. 7G). Moreover, depletion of RNF4 also reduces the transactivation effects of PEA3 (Fig. 9F), indicating that the coupling of sumoylation to ubiquitination has important consequences for the activity of PEA3. Interestingly, sumoylation has also been shown to decrease both the transcriptional repressing properties and the stability of a monomeric form of another ETS domain transcription factor, TEL, although direct links between the SUMO and ubiquitin pathways were not made (39). These findings raise the issue of how ubiquitination and destabilization might contribute to the enhanced activity of PEA3. Indeed, a recent study on BMAL1 demonstrated that sumoylation of this transcription factor also promoted polyubiquitination and degradation yet was important for its transactivation properties (28). One attractive hypothesis is that this mechanism might be important in recycling the transcription factor so that new productive interactions can be made with the basal transcriptional machinery. Such a role for transcription factor turnover has previously been hypothesized (reviewed in reference 34). In addition, ubiquitination might itself provide an activation signal independent from a role in degradation, as observed for other transcriptional regulators, such as yeast Gal4 (9). Indeed, in addition to polyubiquitination, there also appears to be a prominent monoubiquitinated species (Fig. 7), but the significance of this is not yet clear. This monoubiquitinated species is dependent on the lysine residues within the SUMO modification sites but not on sumoylation per se, as neither disruption of the sumoylation motif by mutating the glutamic acid residues (Fig. 7D) nor treatment with the SUMO-specific protease SENP1 (Fig. 7E) completely abolishes PEA3 monoubiquitination. Further studies are needed to probe how monoubiquitination is controlled.
It is important to note that the increase in PEA3 sumoylation is a relatively slow process and therefore allows for a temporal delay in its effects. Ubiquitination of PEA3 is delayed even further. First, MAP kinase signaling can activate (37) and stabilize (data not shown) PEA3 directly. Sumoylation and ubiquitination occur with slower kinetics, meaning that MAP kinase pathway activation can control the entire cascade of events with an inbuilt temporal delay (Fig. 9I). Importantly, we estimate that less than 5% of PEA3 is SUMO modified, meaning that only a subpopulation of PEA3 is sumoylated, and therefore that bulk PEA3 levels will be largely unaffected by sumoylation and subsequent ubiquitination. It is tempting to speculate that this subpopulation is the one that is actively engaged in controlling transcription.
It is currently unclear how MAP kinase pathway activation leads to increased sumoylation. However, while sumoylation at three sites is induced in response to MAP kinase activation, the kinetics of K96 sumoylation are delayed in comparison to K222 and K256, suggesting that a different regulatory mechanism is operative. This most likely is dictated by a functionally important potential proline-directed serine kinase site located downstream from K96. Indeed, a version of PEA3 with a phosphomimetic residue located in place of S101 is efficiently sumoylated in response to ERK pathway activation, despite being ineffectual in changing the basal levels of PEA3 sumoylation at K96. Importantly, this phosphomimetic residue enhances multisumoylation, consistent with the role of signaling in enhancing this molecular event. The putative phosphorylation event that occurs at S101 is initiated after the onset of the more generic enhancement of SUMO pathway activity toward PEA3, thereby accounting for the temporal delay in sumoylation kinetics at this site. It is unclear how this temporal delay is incurred and also what the identity of the S101 kinase is. Further studies are needed to establish whether MAP kinase-mediated inducible phosphorylation events on PEA3 play a direct role in enhancing its sumoylation status.
Another member of the PEA3 subfamily, ERM has previously been shown to be sumoylated (5). ERM is also modified at multiple sites, chiefly at the four most-N-terminal sites (corresponding to PEA3 K96, K222, K256, and K330). A key difference here appears to be that the fourth site in PEA3, K330, is not modified. However, recently, and in agreement with our results, both mouse PEA3 and its human homologue E1AF/ETV4 were also shown to be chiefly modified at the three most-N-terminally located SUMO consensus motifs (2, 35). In all cases, sumoylation at the individual sites appears to occur independently, and multisumoylated species are difficult to detect unless proteosomal inhibitors are added (Fig. 4). It is currently unclear whether each of the individual sites has a specific role, but single point mutations have only small effects on the activity of PEA3 in reporter gene assays (Fig. 8E), suggesting either that individual sites act in a functionally redundant manner or that multisite sumoylation might be the more relevant event. Knock-in studies with individual and composite PEA3 SUMO site mutants might be more revealing about their specific functions.
It is not clear whether sumoylation of ERM is controlled by MAP kinase signaling. However, in contrast to PEA3, sumoylation of ERM causes enhancement in its repressive properties rather than promoting transactivation. This is observed on both a natural promoter (ICAM-1) and a reporter driven by a reiterated PEA3 response element. Indeed, we also observed a repressive effect of sumoylation on a reiterated PEA3 response element (data not shown), but it is unclear whether this has any physiological relevance. This is in stark contrast to the role of sumoylation in promoting transcriptional activation in the context of natural promoters in reporter constructs or in their natural chromatin context (Fig. 8 to 9). The differences we observed might represent protein- or promoter-specific effects of sumoylation. Indeed, two recent studies concluded that sumoylation imparted repressive properties on mouse PEA3 (2) and human PEA3 (E1AF/ETV4) (35). However, these effects were modest on the MMP-7 promoter and were only revealed on reporter constructs containing reiterated binding elements. Indeed, a side-by-side comparison demonstrated that equivalent mouse and human PEA3 mutants behave similarly to the wild-type protein on the MMP-7 promoter but show similar defects in transcriptional activity on the MMP-1 and COX-2 promoters (data not shown). Thus, there are clearly promoter-specific effects involved. These effects are reminiscent of data reported for nuclear hormone receptors, for which sumoylation only causes repression on promoters containing multiple DNA binding sites, giving rise to the concept of synergy control elements (45). Whether PEA3-dependent promoters fall into two different categories containing SUMO-repressible multi-PEA3 binding sites and SUMO-activatable single PEA3 binding sites awaits the identification of a panel of PEA3-regulated promoters by global ChIP-based analysis.
It is likely that PEA3 sumoylation is important in cancer, as PEA3 has been linked to metastatic processes (reviewed in reference 6). Indeed, we have found that PEA3 is heavily modified by SUMO in several cancer cell lines, including SW480 colon cancer cells (Fig. 1). Importantly, this cell line contains oncogenic Ras, thus implicating MAP kinase signaling in this event. Thus, we have made an important connection between Ras-ERK pathway signaling and PEA3 activation through promoting its sumoylation, which is potentially an important molecular event in the metastatic process.
Acknowledgments
We thank Anne Clancy for excellent technical assistance; Paul Shore and members of our laboratory for comments on the manuscript and stimulating discussions; Jorma Palvimo, Kyungjin Kim, Ron Hay, Olivier Kassel, John Hassell, Jean-Luc Baert, and Marleen Petit for reagents; and Tamotsu Nishida for discussion of results on sumoylation of human E1AF.
This work was supported by the Wellcome Trust and a Royal Society-Wolfson award to A.D.S.
Footnotes
Published ahead of print on 23 March 2008.
REFERENCES
- 1.Baert, J. L., C. Beaudoin, D. Monte, C. Degerny, S. Mauen, and Y. de Launoit. 2007. The 26S proteasome system degrades the ERM transcription factor and regulates its transcription-enhancing activity. Oncogene 26415-424. [DOI] [PubMed] [Google Scholar]
- 2.Bojović, B. B., and J. A. Hassell. 2008. The transactivation function of the Pea3 subfamily Ets transcription factors is regulated by sumoylation. DNA Cell. Biol. 27289-305. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, J., X. Kang, S. Zhang, and E. T. Yeh. 2007. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 131584-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel, A. R., E. J. Faivre, and C. A. Lange. 2007. Phosphorylation-dependent antagonism of sumoylation derepresses progesterone receptor action in breast cancer cells. Mol. Endocrinol. 212890-2906. [DOI] [PubMed] [Google Scholar]
- 5.Degerny, C., D. Monte, C. Beaudoin, E. Jaffray, L. Portois, R. T. Hay, Y. deLaunoit, and J. L. Baert. 2005. SUMO modification of the Ets-related transcription factor ERM inhibits its transcriptional activity. J. Biol. Chem. 28024330-24338. [DOI] [PubMed] [Google Scholar]
- 6.de Launoit, Y., J. L. Baert, A. Chotteau-Lelievre, D. Monte, L. Coutte, S. Mauen, V. Firlej, C. Degerny, and K. Verreman. 2006. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim. Biophys. Acta 176679-87. [DOI] [PubMed] [Google Scholar]
- 7.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2233-239. [DOI] [PubMed] [Google Scholar]
- 8.Desterro, J. M., J. Thomson, and R. T. Hay. 1997. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417297-300. [DOI] [PubMed] [Google Scholar]
- 9.Ferdous, A., D. Sikder, T. Gillette, K. Nalley, T. Kodadek, and S. A. Johnston. 2007. The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding to promoters. Genes Dev. 21112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill, G. 2005. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15536-541. [DOI] [PubMed] [Google Scholar]
- 11.Girdwood, D. W., M. H. Tatham, and R. T. Hay. 2004. SUMO and transcriptional regulation. Semin. Cell. Dev. Biol. 15201-210. [DOI] [PubMed] [Google Scholar]
- 12.Gocke, C. B., H. Yu, and J. Kang. 2005. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J. Biol. Chem. 2805004-5012. [DOI] [PubMed] [Google Scholar]
- 13.Goodson, M. L., Y. Hong, R. Rogers, M. J. Matunis, O. K. Park-Sarge, and K. D. Sarge. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. 27618513-18518. [DOI] [PubMed]
- 14.Gregoire, S., A. M. Tremblay, L. Xiao, Q. Yang, K. Ma, J. Nie, Z. Mao, Z. Wu, V. Giguere, and X. J. Yang. 2006. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J. Biol. Chem. 2814423-4433. [DOI] [PubMed] [Google Scholar]
- 15.Guo, B., R. E. Sallis, A. Greenall, M. M. Petit, E. Jansen, L. Young, W. J. Van de Ven, and A. D. Sharrocks. 2006. The LIM domain protein LPP is a coactivator for the ETS domain transcription factor PEA3. Mol. Cell. Biol. 264529-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, B., S. H. Yang, J. Witty, and A. D. Sharrocks. 2007. Signalling pathways and the regulation of SUMO modification. Biochem. Soc. Trans. 351414-1418. [DOI] [PubMed] [Google Scholar]
- 17.Hay, R. T. 2005. SUMO: a history of modification. Mol. Cell 181-12. [DOI] [PubMed] [Google Scholar]
- 18.Hietakangas, V., J. K. Ahlskog, A. M. Jakobsson, M. Hellesuo, N. M. Sahlberg, C. I. Holmberg, A. Mikhailov, J. J. Palvimo, L. Pirkkala, and L. Sistonen. 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 232953-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hietakangas, V., J. Anckar, H. A. Blomster, M. Fujimoto, J. J. Palvimo, A. Nakai, and L. Sistonen. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA 10345-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong, Y., R. Rogers, M. J. Matunis, C. N. Mayhew, M. Goodson, O. K. Park-Sarge, and K. D. Sarge. 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 27640263-40267. [DOI] [PubMed] [Google Scholar]
- 21.Inoue, H., C. Yokoyama, S. Hara, Y. Tone, and T. Tanabe. 1995. Transcriptional regulation of human prostaglandin-endoperoxide synthase-2 gene by lipopolysaccharide and phorbol ester in vascular endothelial cells. Involvement of both nuclear factor for interleukin-6 expression site and cAMP response element. J. Biol. Chem. 27024965-24971. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73355-382. [DOI] [PubMed] [Google Scholar]
- 23.Kang, J., C. B. Gocke, and H. Yu. 2006. Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem. 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasza, A., A. O'Donnell, K. Gascoigne, L. A. Zeef, A. Hayes, and A. D. Sharrocks. 2005. The ETS domain transcription factor Elk-1 regulates the expression of its partner protein, SRF. J. Biol. Chem. 2801149-1155. [DOI] [PubMed] [Google Scholar]
- 25.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J. Biol. Chem. 27730283-30288. [DOI] [PubMed] [Google Scholar]
- 26.Lallemand-Breitenbach, V., M. Jeanne, S. Benhenda, R. Nasr, M. Lei, L. Peres, J. Zhou, J. Zhu, B. Raught, and H. de Thé. 2008. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10547-555. [DOI] [PubMed] [Google Scholar]
- 27.Le Drean, Y., N. Mincheneau, P. Le Goff, and D. Michel. 2002. Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology 1433482-3489. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J., Y. Lee, M. J. Lee, E. Park, S. H. Kang, C. H. Chung, K. H. Lee, and K. Kim. 2008. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol. Cell. Biol. 286056-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, M., F. Melchior, X. H. Feng, and X. Lin. 2004. Regulation of Smad4 sumoylation and transforming growth factor-beta signaling by protein inhibitor of activated STAT1. J. Biol. Chem. 27922857-22865. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y., G. L. Borchert, and J. M. Phang. 2004. Polyoma enhancer activator 3, an ets transcription factor, mediates the induction of cyclooxygenase-2 by nitric oxide in colorectal cancer cells. J. Biol. Chem. 27918694-18700. [DOI] [PubMed] [Google Scholar]
- 31.Livet, J., M. Sigrist, S. Stroebel, V. De Paola, S. R. Price, C. E. Henderson, T. M. Jessell, and S. Arber. 2002. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron 35877-892. [DOI] [PubMed] [Google Scholar]
- 32.Lyst, M. J., and I. Stancheva. 2007. A role for SUMO modification in transcriptional repression and activation. Biochem. Soc. Trans. 351389-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menendez, J. A., L. Vellon, I. Mehmi, B. P. Oza, S. Ropero, R. Colomer, and R. Lupu. 2004. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in cancer cells. Proc. Natl. Acad. Sci. USA 10110715-10720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4192-201. [DOI] [PubMed] [Google Scholar]
- 35.Nishida, T., M. Terashima, K. Fukami, and Y. Yamada. 2007. Repression of E1AF transcriptional activity by sumoylation and PIASy. Biochem. Biophys. Res. Commun. 360226-232. [DOI] [PubMed] [Google Scholar]
- 36.O'Donnell, A., S.-H. Yang, and A. D. Sharrocks. 2008. MAP kinase-mediated c-fos regulation relies on a histone acetylation relay switch. Mol. Cell 29780-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Hagan, R. C., R. G. Tozer, M. Symons, F. McCormick, and J. A. Hassell. 1996. The activity of the Ets transcription factor PEA3 is regulated by two distinct MAPK cascades. Oncogene 131323-1333. [PubMed] [Google Scholar]
- 38.Rodriguez, M. S., J. M. Desterro, S. Lain, C. A. Midgley, D. P. Lane, and R. T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 186455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roukens, M. G., M. Alloul-Ramdhani, A. C. Vertegaal, Z. Anvarian, C. I. Balog, A. M. Deelder, P. J. Hensbergen, and D. A. Baker. 2008. Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function. Mol. Cell. Biol. 282342-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneikert, J., H. Peterziel, P. A. Defossez, H. Klocker, Y. Launoit, and A. C. Cato. 1996. Androgen receptor-Ets protein interaction is a novel mechanism for steroid hormone-mediated down-modulation of matrix metalloproteinase expression. J. Biol. Chem. 27123907-23913. [DOI] [PubMed] [Google Scholar]
- 41.Shalizi, A., B. Gaudilliere, Z. Yuan, J. Stegmuller, T. Shirogane, Q. Ge, Y. Tan, B. Schulman, J. W. Harper, and A. Bonni. 2006. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 3111012-1017. [DOI] [PubMed] [Google Scholar]
- 42.Shyu, Y. C., T. L. Lee, C. Y. Ting, S. C. Wen, L. J. Hsieh, Y. C. Li, J. L. Hwang, C. C. Lin, and C. K. Shen. 2005. Sumoylation of p45/NF-E2: nuclear positioning and transcriptional activation of the mammalian beta-like globin gene locus. Mol. Cell. Biol. 2510365-10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spencer, V. A., J. M. Sun, L. Li, and J. R. Davie. 2003. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods 3167-75. [DOI] [PubMed] [Google Scholar]
- 44.Stankovic-Valentin, N., S. Deltour, J. Seeler, S. Pinte, G. Vergoten, C. Guerardel, A. Dejean, and D. Leprince. 2007. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol. Cell. Biol. 272661-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian, L., M. D. Benson, and J. A. Iñiguez-Lluhí. 2003. A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J. Biol. Chem. 2789134-9141. [DOI] [PubMed] [Google Scholar]
- 46.Sun, H., J. D. Leverson, and T. Hunter. 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 264102-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi, A., F. Higashino, M. Aoyagi, K. Yoshida, M. Itoh, M. Kobayashi, Y. Totsuka, T. Kohgo, and M. Shindoh. 2005. E1AF degradation by a ubiquitin-proteasome pathway. Biochem. Biophys. Res. Commun. 327575-580. [DOI] [PubMed] [Google Scholar]
- 48.Tatham, M. H., M. C. Geoffroy, L. Shen, A. Plechanovova, N. Hattersley, E. G. Jaffray, J. J. Palvimo, and R. T. Hay. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10538-546. [DOI] [PubMed] [Google Scholar]
- 49.Terui, Y., N. Saad, S. Jia, F. McKeon, and J. Yuan. 2004. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J. Biol. Chem. 27928257-28265. [DOI] [PubMed] [Google Scholar]
- 50.Ulrich, H. D. 2005. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 15525-532. [DOI] [PubMed] [Google Scholar]
- 51.Vincenti, M. P., D. J. Schroen, C. I. Coon, and C. E. Brinckerhoff. 1998. v-src activation of the collagenase-1 (matrix metalloproteinase-1) promoter through PEA3 and STAT: requirement of extracellular signal-regulated kinases and inhibition by retinoic acid receptors. Mol. Carcinog. 21194-204. [PubMed] [Google Scholar]
- 52.Vrieseling, E., and S. Arber. 2006. Target-induced transcriptional control of dendritic patterning and connectivity in motor neurons by the ETS gene Pea3. Cell 1271439-1452. [DOI] [PubMed] [Google Scholar]
- 53.Wang, J., X. H. Feng, and R. J. Schwartz. 2004. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J. Biol. Chem. 27949091-49098. [DOI] [PubMed] [Google Scholar]
- 54.Wang, J., A. Li, Z. Wang, X. Feng, E. N. Olson, and R. J. Schwartz. 2007. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol. Cell. Biol. 27622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watts, F. Z. 2004. SUMO modification of proteins other than transcription factors. Semin. Cell Dev. Biol. 15211-220. [DOI] [PubMed] [Google Scholar]
- 56.Wei, Y., D. Liu, Y. Ge, F. Zhou, J. Xu, H. Chen, J. Gu, and J. Jiang. 2008. Identification of E1AF as a target gene of E2F1-induced apoptosis in response to DNA damage. J. Biochem. 144539-546. [DOI] [PubMed] [Google Scholar]
- 57.Yang, S. H., A. Galanis, J. Witty, and A. D. Sharrocks. 2006. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 255083-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, S. H., E. Jaffray, R. T. Hay, and A. D. Sharrocks. 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 1263-74. [DOI] [PubMed] [Google Scholar]
- 59.Yang, X. J., and S. Gregoire. 2006. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol. Cell 23779-786. [DOI] [PubMed] [Google Scholar]