Abstract
Characterizing mechanisms regulating mammary cell growth and differentiation is vital, as they may contribute to breast carcinogenesis. Here, we examine a cross talk mechanism(s) downstream of prolactin (PRL), a primary differentiation hormone, and epidermal growth factor (EGF), an important proliferative factor, in mammary epithelial cell growth and differentiation. Our data indicate that EGF exerts inhibitory effects on PRL-induced cellular differentiation by interfering with Stat5a-mediated gene expression independent of the PRL-proximal signaling cascade. Additionally, our data show that PRL is a potent inhibitor of EGF-induced cell proliferation. We identify tyrosine phosphorylation of the growth factor receptor-bound protein 2 (Grb2) as a critical mechanism by which PRL antagonizes EGF-induced cell proliferation by attenuating the activation of the Ras/mitogen-activated protein kinase (MAPK) pathway. Together, our results define a novel negative cross-regulation between PRL and EGF involving the Jak2/Stat5a and Ras/MAPK pathways through tyrosine phosphorylation of Grb2.
Prolactin (PRL) plays an important regulatory role in mammary gland development. PRL is a primary differentiation factor for mammary epithelial cells and is essential for normal alveolar development and morphogenesis (13). In mammary cells, a major pathway activated downstream of the PRL receptor (PRLR) is the Janus kinase 2 (Jak2)/signal transducer and activator of transcription 5a (Stat5a) pathway. Gene deletion studies of PRL, PRLR, Jak2, and Stat5a have indicated the critical role of PRL signaling in alveolar differentiation (14, 21, 30, 36).
While the role of PRL in the functional differentiation of mammary epithelial cells is well known, the contribution of PRL to breast cancer development and progression is yet to be fully elucidated. PRL acts through an autocrine/paracrine loop to promote cell viability and accelerate oncogene-induced mammary tumorigenesis (4, 29, 32, 35). However, the role of PRL in breast cancer is more complex, and recent evidence has highlighted its role as a potential suppressor of breast cancer progression. For example, Stat5a has been shown to promote cellular adhesion, and activated Stat5a in breast cancer tissues was correlated with a good prognosis and response to endocrine therapy (27, 34, 37). Furthermore, PRL signaling was shown to suppress the epithelial mesenchymal transformation process and the invasive potential of breast cancer cells (28). Importantly, the ability of PRL to suppress the mitogen-activated protein kinase pathway (MAPK [Erk1/Erk2]) was found to be required for these anti-invasive properties of PRL. Thus, it is necessary to further establish this effect of PRL and to characterize the mechanism(s) by which PRL negatively regulates the MAPK pathway.
The epidermal growth factor (EGF) family of ligands, which also regulates mammary gland growth and differentiation, signals through members of the EGF receptor (EGFR) family (38). The role of EGF in mammary gland development has been examined using a variety of transgenic models, highlighting the important role of EGF in ductal outgrowth (23). Even though EGF plays an important role in mammary gland development, it is also implicated in the development and progression of breast cancer (17). Although EGF signals through a variety of pathways, activation of the Ras/MAPK cascade has been associated with the mitogenic/oncogenic role of this growth factor (16).
The mechanism of PRL/EGF cross talk in the regulation of mammary epithelial cell growth and differentiation remains controversial and not well characterized. While a cooperative interplay between PRL and EGF in the morphogenesis and functional differentiation of mammary epithelial organoids has been documented (6), other studies have indicated an inhibitory role for EGF in PRL-induced mammary epithelial cellular differentiation (25). Furthermore, while PRL was shown to block EGF-induced mammary cell growth, PRL was also reported to cooperate with EGF in breast cancer cells (10, 15).
In this study, we examine PRL and EGF regulation of mammary epithelial cell growth and differentiation. Our results highlight the antagonistic properties exhibited by PRL and EGF in mammary cells. While EGF blocked PRL-induced expression of the β-casein gene, a Stat5a target gene and a marker of mammary epithelial cell differentiation, PRL inhibited EGF-induced cell proliferation. We show that EGF-mediated inhibition of PRL-induced β-casein gene expression is at the level of gene transcription, without affecting upstream PRL signaling events. Interestingly, PRL-mediated inhibition of EGF-induced cell proliferation was related to its ability to block the EGF-mediated MAPK (Erk1/Erk2) pathway at the level of Ras activation through induction of tyrosine phosphorylation of Grb2. Together, these results provide new insights into the mechanisms of cross-regulation of PRL and EGF signaling and their implications for mammary epithelial growth, differentiation, and carcinogenesis.
MATERIALS AND METHODS
Reagents.
Reagents used in this study were ovine PRL, insulin, hydrocortisone, and mouse EGF (Sigma-Aldrich). Human PRL was provided by Vincent Goffin (Institut National de la Santé et de la Recherche Médicale, Paris, France). Monoclonal antibodies used in the study were phosphotyrosine (4G10; Upstate); Stat5, Ras, Sos1, and SHP-2 (BD Transduction Laboratories); Myc and GST (Santa Cruz); phospho-Stat5a (Y694; Zymed Laboratories Invitrogen); and β-tubulin (Sigma). Polyclonal antibodies used were Jak2 and Myc (Upstate); phosphotyrosine (BD Transduction Laboratories); Grb2, ribosomal S6 kinase 1 (RSK-1), and phospho-RSK1/2 (T359/S363) (Santa Cruz); and phospho-p44/42 (Erk1/Erk2) and p44/42 (Erk1/Erk2) (Cell Signaling). Other reagents used were goat anti-mouse antibody-horseradish peroxidase and goat anti-rabbit antibody-horseradish peroxidase (Santa Cruz), Jak2 inhibitor II (catalog no. 420132; Calbiochem), G418 (Sigma-Aldrich), a SuperSignal kit (Pierce), nitrocellulose membranes (Whatman), and protein A-Sepharose beads (GE Healthcare).
Plasmid constructs.
Expression plasmids encoding the long form of the PRLR and Jak2 have been described previously (18). Grb2 wild-type (Grb2WT) and Grb2W36,193K (a mutant form of Grb2 in which tryptophans 36 and 193 were mutated to lysine) expression plasmids were obtained from B. Mayer (Connecticut Health Center, Farmington, CT). myc-Grb2WT, Grb2Y7/37/52/209F (Grb2YF), and myc-Grb2YF were kindly provided by R. Van Etten (Tufts-New England Medical Center, Boston, MA) and S. Li (The Jackson Laboratory, Bar Harbor, ME). The expression plasmid encoding the catalytically inactive form of SHP-2, SHP-2CA, was previously described (26). The Grb2 short hairpin RNA (shRNA) construct (pU6+27-ShGrb2) directed against Grb2 nucleotides 310 to 330 (GAT GTG CAG CAC TTC AAG GTT) used for knockdown studies is as described previously (8). The strategy to generate the Grb2YF form unrecognizable by the Grb2 shRNA, the Sil-Grb2YF construct, is as described previously (8). This was achieved by incorporating seven silent mutations in the myc-Grb2YF plasmid within the region targeted by Grb2 shRNA (GAC GTC CAA CAT TTT AAA GTA; the silent mutations are underlined) (GenScript Corporation, Piscataway, NJ).
Cell culture.
HC11 and HC11-Lux cells (transfected stably with the β-casein gene promoter luciferase reporter construct) obtained from N. Hynes (Friedrich Miescher Institute, Basel, Switzerland) and B. Groner (Institute for Biomedical Research, Frankfurt am Main, Germany) were cultured until confluence in RPMI 1640 (Wisent) (supplemented with 10% fetal bovine serum [FBS], EGF [10 ng/ml], and insulin [5 μg/ml]); cells were then differentiated for 3 days in RPMI 1640 (supplemented with 10% FBS, insulin [5 μg/ml], and hydrocortisone [1 μM]), starved overnight (o/n) in RPMI 1640 (supplemented with fetuin [0.5 mg/ml], transferrin [10 μg/ml], insulin [5 μg/ml], and hydrocortisone [1 μM]) and stimulated with PRL (1 μg/ml) or EGF (10 ng/ml or 100 ng/ml) as indicated in the figure legends. In some experiments, cells were pretreated with the tyrosine phosphatase inhibitor sodium vanadate (50 μg/ml) for 30 min before stimulation. HC11-vector and HC11-Grb2YF sublines were cultured in media (described above) supplemented with G418. NMuMG cells obtained from Peter Siegel (McGill University, Montreal, Quebec, Canada) were grown in Dulbecco's modified Eagle's medium (DMEM) (Wisent) supplemented with 10% FBS and insulin (5 μg/ml). Before ligand stimulation, cells were starved o/n in DMEM/F-12 medium (Invitrogen) and stimulated with PRL (1 μg/ml) or EGF (10 ng/ml or 100 ng/ml) as indicated in the figure legends. The human embryonic kidney 293 cells were cultured in DMEM (10% FBS) and starved in DMEM/Ham's F-12 medium. Human breast cancer cell lines MCF-7 and T47D were grown in DMEM (10% FBS) and starved in DMEM (0% FBS).
Cell lysis, immunoprecipitation, and Western blotting.
Total cell lysates were isolated as previously described (1). Equal amounts of proteins obtained by whole-cell lysis were loaded and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoprecipitations were preformed as previously described (1). Western blot analyses were performed with the indicated antibodies, and proteins were revealed with the use of chemiluminescence (Pierce) according to the manufacturer's instructions.
Transient transfections and generation of stable cell lines.
293 cells (5 × 105) were cotransfected with 1 μg of expression plasmids encoding the rat long PRLR form with either Jak2 or Src and the indicated forms of Grb2, as described for each experiment, by using the calcium-phosphate method. Twenty-four hours posttransfection, cells were serum starved and stimulated with PRL (1 μg/ml) for 10 min before cell lysis. For transient transfections in HC11, cells were grown to confluence and differentiated for 24 h and then transfected with either a vector (mock transfection) or an expression plasmid encoding myc-Grb2YF, with the use of a Lipofectamine transfection kit (Invitrogen) according to the manufacturer's protocol. Cells were left in differentiation media for 24 h before o/n starvation. Cells were then stimulated as indicated in the figure legends. To establish stable cell lines, HC11 cells were transfected with a vector or with a plasmid expressing myc-Grb2YF using a Lipofectamine 2000 transfection kit (Invitrogen) as described in the manufacturer's protocol. Stably expressing clones were isolated using 250 mM G418 (Sigma-Aldrich). For Grb2 knockdown and rescue studies, HC11 cells were grown to 80% confluence before transfection with a vector, a vector plus pU6+27-ShGrb2, or pU6+27-ShGrb2 plus Sil-Grb2YF plasmids using a Lipofectamine 2000 transfection kit (Invitrogen) according to the manufacturer's protocol. At 24 h following transfection, cells were selected for 48 h with G418.
RNA isolation and qRT-PCR measurement.
Differentiated HC11 cells were left untreated or were treated with PRL or the combination of PRL and EGF (PRL/EGF) for 16 h. Cells were lysed in 500 μl of TRIzol. Total RNA was isolated as described by the manufacturer (Invitrogen Life Technologies, Burlington, Ontario, Canada). Samples were quantified by absorbance at 260 nm. Aliquots of 300 to 400 ng of total RNA were used for reverse transcription and PCR amplification in one step using Brilliant II SYBR green quantitative real-time PCR (qRT-PCR) Master Mix kit, 1-Step (Stratagene Amsterdam, Zuidoost, The Netherlands) according to the manufacturer's recommendation. The q-PCR amplification was under the following conditions: 50°C for 30 min; 95°C for 10 min; and 40 cycles at 95°C for 30 s, 55°C for 1 min, and 72°C for 30 s. The specificity of the primers was then tested by a dissociation program at 95°C for 1 min, with a ramp-down to 55°C and then a ramp-up to 95°C (at the instrument default rate of 0.2°C/s). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was amplified as an internal control. GAPDH reverse (CTCAGTGTAGCCCAGGATGC) and GAPDH forward (ACCACCATGGAGAAGGCTGG) primers and β-casein forward (CTATTGCTCAACCCCCTGTG) and β-casein reverse (AGAGTTTATGAGGCGGAGCA) primers were used for qRT-PCR measurements.
Dissociation curve analysis was performed after the completed q-PCR. Data were obtained by slowly ramping up the temperatures of reaction solutions from 55 to 95°C. The four identical peaks confirm that the primers for β-casein gene are specific, resulting in one amplified product.
Luciferase assays.
HC11-Lux cells were left untreated or were treated for 16 h with PRL (1 μg/ml) or PRL (1 μg/ml)/EGF (10 ng/ml). Luciferase activity was determined as described previously (18). Luciferase activity was normalized to protein levels for each sample.
MTT and cell counting assays.
HC11, HC11-vector, and HC11-Grb2YF were plated for 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays or cell counting assays as described in the figure legends. Cells were grown o/n in the absence or presence of PRL (1 μg/ml) in RPMI 1640 (supplemented with 2% FBS, insulin [5 μg/ml], and hydrocortisone [1 μM]). Cells were then left untreated or treated with EGF for 72 h (MTT assay) or 24 h and 48 h (cell counting assay). In Jak2 inhibitor experiments, HC11 cells (2.5 × 103) were treated with either dimethyl sulfoxide (DMSO) or Jak2 inhibitor (1 μM) and grown o/n in the absence or presence of PRL in media containing 2% FBS and HI. Cells were then left untreated or were treated with EGF for 48 h. MTT assays were preformed as previously described (5). For Grb2 knockdown and rescue studies, HC11 cells were transfected with a vector, a vector and pU6+27-ShGrb2, or Sil-Grb2YF and pU6+27-ShGrb2 for 24 h. Cells were then selected in G418 for 48 h and processed for MTT assays as described above. NMuMG cells were grown o/n in DMEM containing (2% FBS) in the presence or absence of PRL. Cells were then left untreated or treated with EGF (10 ng/ml or 100 ng/ml) for 72 h and processed for the MTT assay. MCF-7 and T47D cells were grown o/n in 2% serum and then left untreated or treated with 10 ng/ml or 100 ng/ml human PRL for 72 h. Results are presented as means ± standard errors of the means (SEM) for triplicates of three separate experiments. Statistical significance was assessed through one-way analysis of variance.
Ras pulldown assay.
The Ras binding domain of human c-raf-1 (amino acids 1 to 149) was expressed as a glutathione S-transferase (GST) fusion protein in bacteria (provided by J. Woodgett, Samuel Lunenfeld Research Institute, Toronto, Canada) and then bound to glutathione-Sepharose beads (40 μg of protein for each 15 μl of packed beads). Cell lysates were incubated with GST fusion protein-coupled beads at 4°C for 30 min. Collected cell precipitates were processed for Western analysis.
Nuclear extracts and EMSA.
Nuclear extracts were prepared as previously described (9). Briefly, cells were lysed with a hypotonic buffer [10 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 1 mM Na3VO4, 20 mM NaF, 4 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF; Pefabloc), 5 μg/ml aprotonin, and 2 μg/ml leupeptin] and three freeze-thaw cycles. The cytoplasmic fraction was discarded. The nuclear fraction was lysed with a high-salt buffer (20 mM HEPES-KOH [pH 7.9], 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM Na3VO4, 20 mM NaF, 4 mM AEBSF, 5 μg/ml aprotonin, and 2 μg/ml leupeptin). Nuclear extracts were processed for electrophoretic mobility shift assays (EMSA) using the Stat5a binding site from bovine β-casein promoter as previously described (5).
RESULTS
PRL blocks EGF-mediated mammary epithelial cell proliferation.
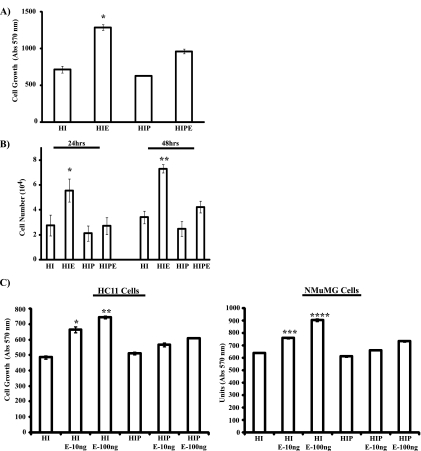
Since PRL and EGF represent two key regulators of mammary epithelial cell growth and differentiation and both have an impact on breast carcinogenesis, it is vital to better define the coregulation of signaling events downstream of these two ligands. The HC11 mouse mammary epithelial cell line, isolated from mice in mid-pregnancy, has been a widely used model system to study mammary epithelial cell proliferation and differentiation. While EGF has a strong mitogenic effect in these cells, a combinatorial treatment of PRL, insulin, and glucocorticoids has been shown to induce terminal differentiation as measured by the induction of milk protein synthesis, a marker of mammary epithelial cell differentiation (2). Using this cell model system, we first investigated PRL and EGF cross talk in regulating mammary epithelial cell proliferation. For this experiment, HC11 cells were grown in media containing 2% serum and hydrocortisone and insulin in the absence of PRL (HI) or in the presence of PRL (HIP) o/n before stimulation with EGF (HIF and HIPE) for 72 h (Fig. 1A). As shown, in the absence of PRL, EGF treatment led to a significant increase in cell growth (Fig. 1A, compare HI and HIE). Interestingly, the presence of PRL led to a significant inhibition in EGF-induced cell growth (Fig. 1A, compare HIE and HIPE). These data indicate that in HC11 cells, whereas EGF is a potent mitogen, PRL exhibits properties antagonistic to EGF-induced cell growth. The inhibitory effects of PRL on EGF-mediated cell growth were further confirmed by a direct cell counting assay (Fig. 1B). HC11 cells were maintained in medium containing either HI or HIP o/n. Cells were then stimulated with EGF for 24 h and 48 h. As can be seen, PRL significantly suppressed EGF-induced cell growth at both time points examined (Fig. 1B, compare HIE and HIPE). Furthermore, we observed that PRL itself did not induce growth of HC11 cells, in contrast to a trend toward cell growth arrest (20 to 30%), which was observed most evidently at the 48-h time point (Fig. 1B, compare HI and HIP). To further confirm the effects of PRL, we evaluated the inhibitory role of PRL on EGF-mediated cell growth using the NMuMG mouse mammary epithelial cell model system. As can be seen in Fig. 1C, PRL blocked EGF-induced NMuMG cell growth at two different concentrations of EGF, results similar to those for HC11 cells. Together, our data indicate that PRL itself does not induce proliferation of mammary epithelial cells. Moreover, PRL was found to significantly suppress EGF-induced cell growth.
FIG. 1.
PRL attenuates EGF-induced cell proliferation of mammary epithelial cells. (A) HC11 cells (1.5 × 103 cells) were plated in assay media (2% FBS, HI) in the absence or presence of PRL (1 μg/ml) o/n. Cells were then left untreated or treated with EGF (10 ng/ml) for 72 h. MTT assays were performed as described in Materials and Methods. Results are the means ± SEM for triplicates of three experiments (*, P = 0.043). (B) HC11 cells (2.5 × 104 cells) were plated as described for panel A, and cells were then left untreated or treated with EGF for 24 h and 48 h. Cell numbers were determined following trypan blue exclusion assays. Results are the means ± SEM for triplicates of three experiments (*, P = 0.07; **, P = 0.049). (C) HC11 cells (1.5 × 103 cells) (left panel) and NMuMG cells (3 × 103 cells) (right panel) were plated as described for panel A, and cells were then left untreated or treated with either 10 ng/ml or 100 ng/ml EGF (E-10ng and E-100ng, respectively) for 72 h. MTT assays were performed, and results are the means ± SEM for triplicates of three experiments (*, P = 0.0189; **, P = 0.0165; ***, P = 0.002; ****, P = 0.0004). Abs, absorbance.
PRL suppresses EGF-mediated activation of the MAPK (Erk1/Erk2) pathway.
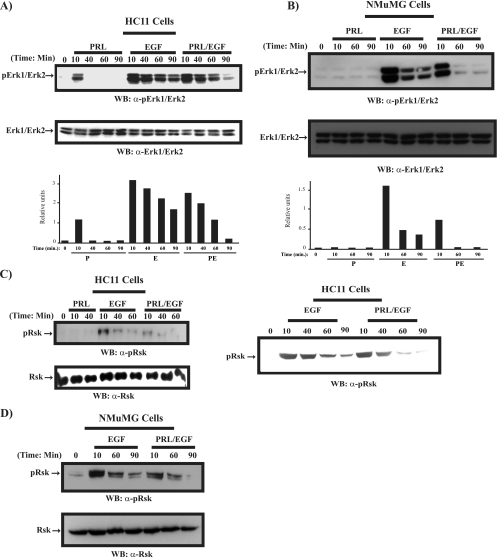
To examine the underlying mechanism of PRL inhibition of the EGF-induced proliferation of mammary epithelial cells, we next examined the effects of PRL on the EGF-induced MAPK (Erk1/Erk2) pathway, a major mitogenic pathway downstream of EGF. Since PRL itself is also known to activate the MAPK pathway (12), we therefore examined the activation pattern of the MAPK pathway by PRL, EGF, and the combination of PRL and EGF in HC11 cells (Fig. 2A). Treatment of cells with PRL induced a mild and transient activation of Erk1/Erk2. Indeed, MAPK activation was observed only at the 10-min time point, and it was already turned off by 40 min of stimulation. In contrast, EGF stimulation of HC11 cells led to more-pronounced and -prolonged activation than PRL stimulation did. As shown in Fig. 2A, MAPK activation was still maintained even at the 90-min time point of EGF stimulation. Interestingly, in cells cotreated with both ligands, there was a substantial decrease in MAPK activation starting at the 40-min time point and most noticeable following 60 min and 90 min of stimulation. Similar results were obtained with NMuMG mammary epithelial cells. As shown in Fig. 2B, PRL itself did not induce any significant activation of the MAPK pathway, and furthermore, PRL was able to block EGF-induced MAPK activation.
FIG. 2.
PRL suppresses EGF-induced MAPK (Erk1/Erk2)/RSK activation. Serum-starved differentiated HC11 cells (A) and NMuMG cells (B) were treated with PRL (P), EGF (E), or PRL/EGF (PE), as indicated. Cell lysates were immunoblotted with a polyclonal antibody to phospho-Erk1/Erk2 (upper panel) and Erk1/Erk2 (lower panel). (C) HC11 cell lysates prepared as in panels A and D. NMuMG cell lysates prepared as in panel B were immunoblotted with a polyclonal antibody to phospho-RSK (upper panel) and RSK (lower panel). WB, Western blot; α-, anti-.
To further characterize the activation of the MAPK pathway downstream of PRL and EGF in mammary cells, we examined the ability of the ligands to induce activation of p90 RSK kinase, a substrate of Erk1/Erk2 (3). To do so, we examined p90 RSK activation in both HC11 and NMuMG cells treated with PRL, EGF, or PRL/EGF. As shown in Fig. 2C (left and right panels) and Fig. 2D, in contrast to PRL, EGF induced a significant activation of p90 RSK. Interestingly, PRL was able to suppress EGF-induced RSK activation (Fig. 2C and D). These data indicate that in mammary cells, there is a significant difference in the abilities of PRL and EGF to induce MAPK activation. While EGF induces a robust and sustained activation, PRL weakly activates the MAPK pathway. More importantly, our data also illustrate the possibility that PRL suppresses growth factor-induced activation of the MAPK/RSK pathway. These data together further underscore the negative role of PRL in EGF signaling in mammary epithelial cells.
PRL-mediated activation of Jak2 is required for PRL inhibitory effects on EGF-induced cell proliferation.
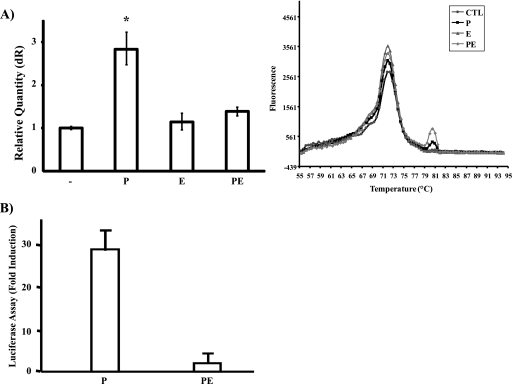
To examine the mechanism by which PRL negatively regulates EGF-induced MAPK activation and mammary cell proliferation, we next analyzed the regulatory role of EGF on PRL-induced signaling in mammary cells. First, we examined EGF regulation of PRL-induced differentiation signals as measured by induction of gene expression of the milk protein β-casein. Serum-starved differentiated HC11 cells were either left untreated or treated o/n with PRL, EGF, or PRL/EGF. Expression of the β-casein gene was assessed by qRT-PCR. As shown in Fig. 3A, PRL treatment led to a significant increase in β-casein gene expression. In contrast, EGF treatment of HC11 cells did not lead to induction of β-casein gene expression (Fig. 3A). Importantly, the presence of EGF led to a significant inhibition in PRL-induced β-casein gene expression (Fig. 3A, compare P and PE). We next investigated whether EGF can block PRL-mediated β-casein gene expression at the level of gene transcription. Serum-starved differentiated HC11 cells stably transfected with the β-casein gene promoter/luciferase construct (HC11-Lux) were treated with either PRL or PRL/EGF for 16 h. In agreement with a previous study (25), PRL induced a robust activation of the β-casein gene promoter (>25-fold), whereas cotreatment with EGF abolished PRL-induced effects (Fig. 3B). Together, these data indicate that EGF-mediated signals are inhibitory to PRL signaling to milk protein gene expression at the level of gene transcription.
FIG. 3.
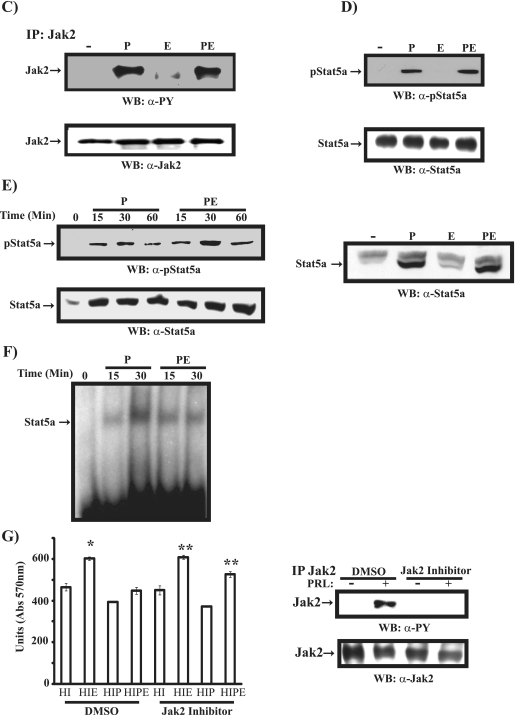
EGF regulation of PRL-induced Jak2/Stat5a pathway and β-casein gene expression. (A, left) Differentiated HC11 cells were left untreated or treated with PRL (P), EGF (E), or PRL/EGF (PE) for 16 h. qRT-PCR of the β-casein gene was performed, and results are the means from three experiments (*, P = 0.053). (Right) Dissociation curve analysis of the q-PCR. The four identical peaks confirm that the primers for the β-casein gene are specific, resulting in one amplified field. CTL, cytotoxic T lymphocytes. (B) Differentiated HC11-Lux cells were left untreated or treated with either PRL or PRL/EGF for 16 h. Results are the mean luciferase activity levels from quadruplicates of two experiments. (C) Serum-starved differentiated HC11 cells were untreated or treated with PRL, EGF, or PRL/EGF for 15 min. Cell lysates were immunoprecipitated (IP) with a polyclonal antibody to Jak2, immunoblotted with a monoclonal antibody to phosphotyrosine (upper panel), and reprobed with a polyclonal antibody to Jak2. (D) Cell lysates described for panel C were immunoblotted with a monoclonal antibody to phospho-Stat5a (upper panel) and reblotted with a monoclonal antibody to Stat5a (lower panel). (E, left) HC11 nuclear extracts were immunoblotted with a monoclonal antibody to phospho-Stat5a (upper panel) and Stat5a (lower panel). (Right) Nuclear extracts of HC11 cells treated with PRL, EGF, or PRL/EGF for 30 min were immunoblotted with a monoclonal antibody to Stat5a. (F) EMSA was performed using the Stat5a binding site of the β-casein gene promoter with nuclear extracts prepared as in panel E. (G, left) HC11 cells were treated with either DMSO or Jak2 inhibitor (1 μM) and grown o/n in 2% serum containing HI or HIP. Cells were then left untreated or treated with EGF for 48 h. An MTT assay was performed, and results are the means ± SEM for triplicates of three experiments (*, P = 0.017; **, P = 0.023). (Right) HC11 cells were pretreated with either DMSO or Jak2 inhibitor (25 μM) for 2 h. Cells were then stimulated with PRL for 15 min. Cell lysates were immunoprecipitated with a polyclonal antibody to Jak2 and immunoblotted with a monoclonal antibody to phosphotyrosine (PY) (upper panel) and reprobed with a polyclonal antibody to Jak2 (lower panel). Abs, absorbance; WB, Western blot; α-, anti-.
Since the Jak2/Stat5a pathway is critical for PRL signaling to β-casein gene promoter activation, we next examined the regulation of EGF on primarily PRL-mediated activation of the Jak2/Stat5a pathway. As shown in Fig. 3C and D, in contrast to results for EGF stimulation, PRL stimulation of HC11 cells led to tyrosine phosphorylation of Jak2 and Stat5a (on Y694), suggesting that PRL but not EGF induces activation of the Jak2/Stat5a pathway in mammary epithelial cells. Interestingly, Jak2 and Stat5a activation was still maintained in samples cotreated with these two ligands. These data suggest that the inhibitory effect of EGF on PRL-induced β-casein gene promoter activation is independent of the cytoplasmic activation of the Jak2/Stat5a pathway. Next, we investigated whether EGF may influence more downstream signaling events, such as Stat5a nuclear translocation and Stat5a DNA binding activity. Nuclear extracts were prepared from HC11 cells treated with either PRL or PRL/EGF for up to 1 h. As shown in Fig. 3E (left panel), Stat5a nuclear translocation was observed within 15 min of stimulation in samples treated with either PRL or PRL/EGF. Moreover, in accordance with the inability of EGF to induce tyrosine phosphorylation of Stat5a, EGF treatment of HC11 cells did not lead to the nuclear translocation of Stat5a (Fig. 3E, right panel). Furthermore, using an EMSA assay (Fig. 3F), we observed a rapid binding of Stat5a to its response element on the β-casein gene promoter in both PRL- and PRL/EGF-treated cells. Together, the above results show that EGF blocks PRL-induced β-casein gene promoter activation at the level of Stat5a-mediated gene transcription independently of activation of Jak2, Stat5a tyrosine phosphorylation, nuclear translocation, and DNA binding activity.
The above results indicate that Jak2 activation is maintained in cells cotreated with PRL and EGF. This result led us to hypothesize that the ability of PRL to inhibit EGF-induced cell proliferation observed in Fig. 1 is a Jak2-dependent event. Therefore, we next investigated whether PRL activation of Jak2 is required for PRL-mediated inhibitory effects on EGF-induced cell proliferation. HC11 cells were grown in media containing HI or HIP in the presence of DMSO (as the control) or the Jak2 inhibitor (1 μM) o/n before stimulation with EGF for 48 h or no stimulation. As shown in Fig. 3G, EGF treatment led to a significant increase in cell growth in the absence or the presence of the Jak2 inhibitor (Fig. 3G, compare HI and HIE results for DMSO with HI and HIE results for Jak2 inhibitor). As expected for the control DMSO-treated cells, the presence of PRL led to a complete inhibition of EGF-induced cell growth (Fig. 3G, DMSO, compare HIE and HIPE results). Interestingly, the presence of the Jak2 inhibitor reversed significantly the inhibitory effects of PRL on EGF-induced cell proliferation (Fig. 3G, compare the HIPE results for DMSO to those for Jak2 inhibitor). These data indicate the important role of Jak2 kinase in mediating the inhibitory effect of PRL on EGF-induced mammary cell growth.
PRL induces tyrosine phosphorylation of the adaptor protein Grb2.
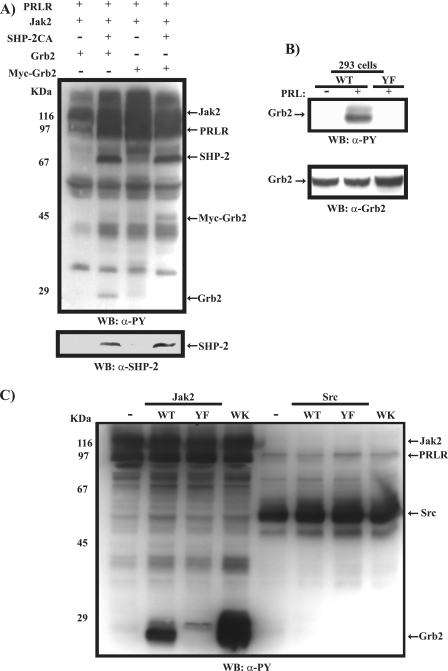
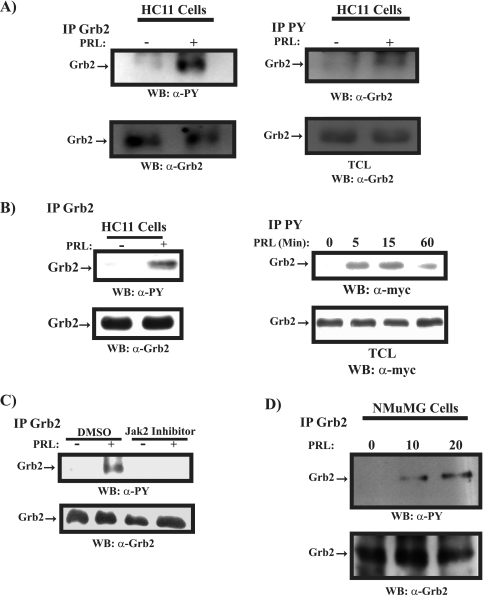
Our results thus far have indicated that in mammary epithelial cells, PRL attenuates EGF-induced cell proliferation. Our data also suggest that this is a Jak2-dependent early signaling event. To delineate a possible mechanism through which PRL/Jak2 blocks EGF signaling, we analyzed the contribution of the adaptor protein Grb2. We have previously shown that Grb2 or a 29-kDa protein associated with Grb2, a substrate of SHP-2, is tyrosine phosphorylated following PRLR/Jak2 activation (26). To examine whether this protein is a tyrosine phosphorylated form of Grb2, we overexpressed both an untagged and a myc-tagged form of Grb2 in 293 cells along with PRLR, Jak2, and the catalytically inactive form of SHP-2. As shown in Fig. 4A, we find that in the presence of catalytically inactive SHP-2, activation of the PRLR/Jak2 complex led to tyrosine phosphorylation of Grb2 (Fig. 4A, lanes 2 and 4). While it is generally believed that Grb2 itself does not undergo tyrosine phosphorylation in response to growth factor stimulation (22, 33), in Bcr/Abl-transformed cells, tyrosine phosphorylation of Grb2 on residues Y7, Y37, Y52, and Y209 in the SH3 domains has been reported and was shown to negatively regulate the Ras/MAPK pathway (20). However, the physiological relevance of this observation is yet to be evaluated. We hypothesized that Grb2 is a substrate of the PRLR/Jak2 complex and may contribute to PRL inhibition of EGF-induced MAPK activation and cell proliferation. To investigate whether PRL induces phosphorylation of Grb2 on tyrosine residues present within its SH3 domains, we overexpressed in 293 cells PRLR, Jak2, and either the Grb2WT or Grb2YF mutant form in which the four tyrosine residues, Y7, Y37, Y52, and Y209, were replaced by phenylalanine. As shown in Fig. 4B, PRLR activation led to tyrosine phosphorylation of the Grb2WT but not the Grb2YF mutant, suggesting that PRL induces tyrosine phosphorylation of Grb2 on the tyrosine residues within its SH3 domains. Moreover, tyrosine phosphorylation of Grb2 was detected only in cells overexpressing the PRLR along with the Jak2 kinase but not the Src kinase, indicating that Grb2 is a specific substrate of the PRLR/Jak2 complex (Fig. 4C). The importance of PRLR/Jak2-induced tyrosine phosphorylation of Grb2 is further emphasized with the Grb2W36,193K mutant form. This mutant has been shown previously to be incapable of activating Ras due to the loss of functionality of the SH3 domains and its interaction with Sos (11). As shown in Fig. 4C, Grb2W36,193K exhibited dramatic tyrosine phosphorylation upon activation of the PRLR/Jak2 complex, suggesting that tyrosine phosphorylation of Grb2 is a possible mechanism utilized by Grb2W36,193K to block Grb2/Sos complex formation and thereby MAPK activation. Together, these results prompted us to determine whether PRL stimulation of mammary epithelial cells would lead to tyrosine phosphorylation of Grb2. We thus immunoprecipitated Grb2 using a polyclonal antibody to Grb2 in differentiated HC11 following PRL stimulation. As shown in Fig. 5A (left panel), PRL was observed to induce the tyrosine phosphorylation of Grb2 within 15 min of stimulation. Furthermore, Grb2 was found to be present in phosphotyrosine immunoprecipitates of HC11 cells following stimulation by PRL, confirming the ability of PRL to induce tyrosine phosphorylation of Grb2 in mammary cells (Fig. 5A, right panel). Moreover, as shown in Fig. 5B (left and right panels), using HC11, we observed tyrosine phosphorylation of endogenous Grb2 and overexpressed myc-Grb2 in cells that were initially pretreated with a phosphatase inhibitor (sodium vanadate) before PRL stimulation, further confirming the ability of PRL to induce tyrosine phosphorylation of Grb2. As Jak2 kinase but not Src kinase was found to mediate PRL-induced tyrosine phosphorylation of Grb2 (Fig. 4C), we next used the Jak2 kinase inhibitor to determine whether the tyrosine phosphorylation of Grb2 requires Jak2 kinase activation in HC11 cells. As shown in Fig. 5C, PRL was unable to induce Grb2 tyrosine phosphorylation in the presence of the Jak2 inhibitor compared to results for the control DMSO-treated cells. Together, these data indicate that in mammary epithelial cells, PRL leads to tyrosine phosphorylation of Grb2 through Jak2 activation.
FIG. 4.
PRLR/Jak2-mediated tyrosine phosphorylation of Grb2. (A) Plasmids (1 μg) encoding the long form of the PRLR, Jak2, SHP2CA, Grb2, or myc-Grb2 were cotransfected in 293 cells as indicated. Serum-starved cells were treated with PRL for 10 min. Lysates were immunoblotted with a monoclonal antibody to phosphotyrosine (upper panel) and a monoclonal antibody to SHP2 (lower panel). (B) 293 cells were cotransfected with plasmids (1 μg) encoding PRLR, Jak2, and the indicated forms of Grb2. Serum-starved cells were left unstimulated or were stimulated with PRL for 15 min. Lysates were immunoblotted using a monoclonal antibody to phosphotyrosine (upper panel) or a polyclonal antibody to Grb2 (lower panel). (C) 293 cells were cotransfected with plasmids (1 μg) encoding the long form of the PRLR along with various forms of Grb2 with either Jak2 or Src, as indicated. Serum-starved cells were treated with PRL for 10 min. Lysates were immunoblotted with a monoclonal antibody to phosphotyrosine (PY). WB, Western blot; α-, anti-.
FIG. 5.
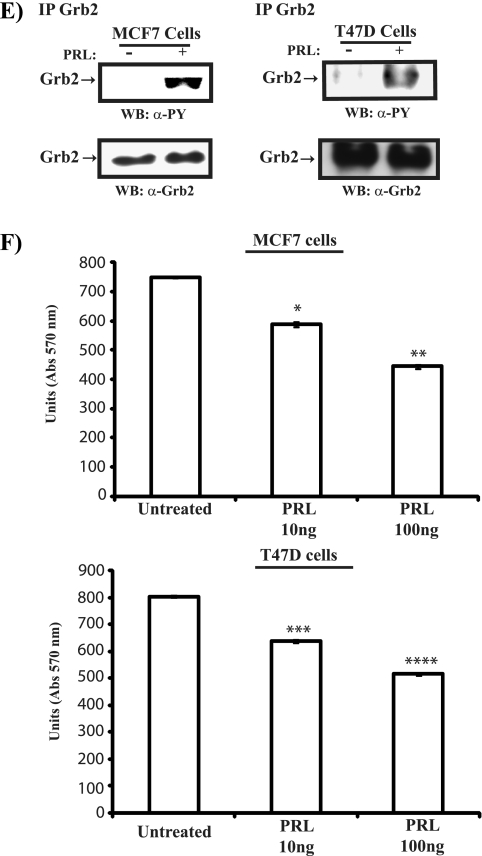
PRL induces tyrosine phosphorylation of Grb2 in mammary cells. (A, left) Serum-starved HC11 cells were untreated or treated with PRL for 15 min. Lysates were immunoprecipitated (IP) using a polyclonal antibody to Grb2 and immunoblotted with a monoclonal antibody to phosphotyrosine (upper panel) and a polyclonal antibody to Grb2 (lower panel). (Right) Lysates of HC11 cells prepared as described above were immunoprecipitated using a polyclonal antibody to phosphotyrosine and immunoblotted with a polyclonal antibody to Grb2 (upper panel). Cell lysates were immunoblotted with a polyclonal antibody to Grb2 (lower panel). (B, left) Serum-starved differentiated HC11 cells were pretreated with sodium vanadate (50 μg/ml) for 30 min and then stimulated with PRL for 15 min. Lysates were immunoprecipitated with a polyclonal antibody to Grb2, immunoblotted with a monoclonal antibody to phosphotyrosine (upper panel), and reblotted with a polyclonal antibody to Grb2 (lower panel). (Right) Differentiated HC11 cells were transfected with an expression vector encoding myc-Grb2. Serum-starved cells were pretreated with sodium vanadate (50 μg/ml) for 30 min, stimulated with PRL as indicated, and immunoprecipitated using a polyclonal antibody to phosphotyrosine, followed by immunoblotting with a monoclonal antibody to the myc tag (upper panel). Lysates from the same transfection were immunoblotted with a monoclonal antibody to the myc tag (lower panel). (C) Serum-starved differentiated HC11 cells were pretreated with DMSO or Jak2 inhibitor (25 μM) for 90 min. Cells were then left unstimulated or were stimulated with PRL for 15 min. Lysates were immunoprecipitated and immunoblotted as in panel A (left). (D) Serum-starved NMuMG cells were left untreated or were treated with PRL as indicated. Numbers indicate time (in minutes). Lysates were immunoprecipitated using a polyclonal antibody to Grb2 and immunoblotted with a monoclonal antibody to phosphotyrosine (upper panel) and a polyclonal antibody to Grb2 (lower panel). (E) Serum-starved MCF-7 (left panel) and T47D (right panel) cells were pretreated with sodium vanadate (50 μg/ml) for 30 min and then stimulated or not with human PRL (100 ng/ml). Cell lysates were immunoprecipitated with a polyclonal antibody to Grb2, immunoblotted with a monoclonal antibody to phosphotyrosine (upper panel), and reblotted with a polyclonal antibody to Grb2 (lower panel). (F) MCF-7 (upper panel) and T47D (lower panel) cells were plated o/n in 2% serum and left untreated or treated with human PRL for 72 h. An MTT assay was performed, and the results are the means ± SEM for triplicates of three experiments. Upper panel, *, P = 0.0216; **, P = 0.0075. Lower panel, *, P = 0.0153; **, P = 0.0024. Abs, absorbance; WB, Western blot; α-, anti-; PY, phosphotyrosine; TCL, total cell lysates.
We next examined the ability of PRL to induce tyrosine phosphorylation of Grb2 using NMuMG mammary epithelial cells. As shown in Fig. 5D, PRL was observed to induce the tyrosine phosphorylation of Grb2 within 10 min of stimulation. Furthermore, we assessed the ability of PRL to induce tyrosine phosphorylation of Grb2 in other PRL-responsive cell systems, such as human breast cancer MCF-7 and T47D cells. As shown in Fig. 5E, PRL was able to induce the tyrosine phosphorylation of Grb2 in both breast cancer cell lines, and this induction was correlated with the ability of PRL to suppress cell growth (Fig. 5F). Together, these data indicate that Grb2 is a downstream target of PRL/Jak2 signaling in mammary epithelial and breast cancer cells.
PRL-mediated suppression of EGF-induced MAPK activation is through tyrosine phosphorylation of Grb2.
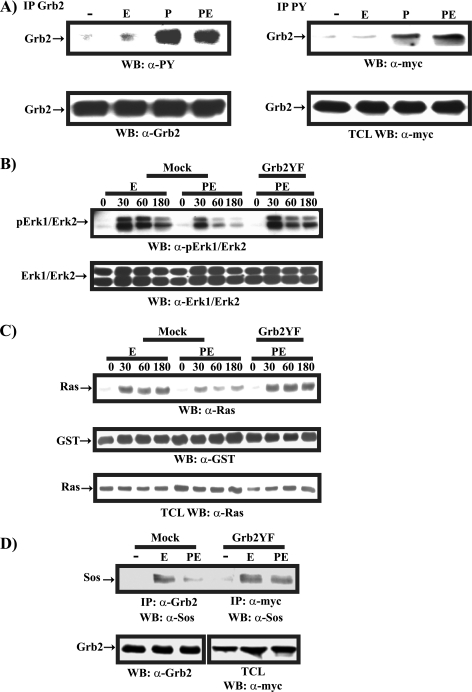
Next, we were interested in determining whether PRL utilizes tyrosine phosphorylation of Grb2 as a mechanism to inhibit EGF-induced MAPK activation in mammary cells. We first examined the pattern of tyrosine phosphorylation of Grb2 in response to treatment with PRL, EGF, and PRL/EGF. HC11 cells either were left untransfected (Fig. 6A, left panel) or were transfected with an expression vector encoding myc-Grb2 (Fig. 6A, right panel) and stimulated with PRL, EGF, or PRL/EGF for 15 min. Since Grb2 phosphorylation is tightly regulated (22, 33), we pretreated HC11 cells with the phosphatase inhibitor sodium vanadate to optimize ligand-induced Grb2 tyrosine phosphorylation. In comparison to EGF, PRL potently induced tyrosine phosphorylation of both endogenous (Fig. 6A, left panel, 41.6-fold) and overexpressed Grb2 (Fig. 6A, right panel, 22.3-fold). Furthermore, in cells cotreated with the combination of PRL and EGF, the level of Grb2 phosphorylation was comparable to that for PRL-treated cells (Fig. 6A, left panel, 38.8-fold, and right panel, 26-fold). Together, these data indicate that PRL is more effective in inducing tyrosine phosphorylation of Grb2 than EGF and further highlight the observation that Grb2 tyrosine phosphorylation is maintained in cells cotreated with both PRL and EGF.
FIG. 6.
PRL-induced tyrosine phosphorylation of Grb2 inhibits EGF-induced Ras/MAPK activation. (A, left) Serum-starved differentiated HC11 cells were pretreated with sodium vanadate for 30 min and were left unstimulated or stimulated with EGF (E), PRL (P), or PRL/EGF (PE) for 15 min. Cell lysates were immunoprecipitated using a polyclonal antibody to Grb2, immunodetected with a monoclonal antibody to phosphotyrosine, and reprobed with a polyclonal antibody to Grb2. (Right) Differentiated HC11 cells were transfected with an expression vector encoding myc-Grb2. Serum-starved cells were pretreated with sodium vanadate and stimulated as described above. Cell lysates were immunoprecipitated using a polyclonal antibody to phosphotyrosine and immunodetected using a monoclonal antibody to the myc epitope (upper panel). Cell lysates were immunoblotted with a monoclonal antibody to the myc epitope (lower panel). (B) Differentiated HC11 cells were transfected with a vector or a plasmid encoding myc-Grb2YF. Starved cells were stimulated with EGF or PRL/EGF as indicated. Numbers indicate time (in minutes). Lysates were immunoblotted using a polyclonal antibody to phospho-Erk1/Erk2 (upper panel) and Erk1/Erk2 (lower panel). (C) HC11 cells treated as described above were stimulated with EGF or PRL/EGF as indicated. A pulldown assay of Ras-GTP was performed, and immunodetection was done using a monoclonal antibody to Ras (upper panel) and a polyclonal antibody to GST (middle panel). Cell lysates were immunodetected with a monoclonal antibody to Ras (lower panel). (D) Differentiated HC11 cells were transfected as described above. Serum-starved cells were left unstimulated or were stimulated with EGF or PRL/EGF for 15 min. Lysates were immunoprecipitated with a polyclonal antibody to Grb2 or to myc, immunoblotted with a monoclonal antibody to Sos (upper panel), and reblotted with a polyclonal antibody to Grb2, or total cell lysates were immunodetected using a monoclonal antibody to myc (lower panels). α-, anti-; PY, phosphotyrosine; TCL, total cell lysates.
To determine the role of tyrosine phosphorylation of Grb2 in PRL inhibitory effects on the EGF-induced MAPK pathway in mammary cells, we transiently overexpressed the myc-Grb2YF mutant in HC11 cells and examined the ability of PRL to inhibit EGF-induced MAPK activation. As shown in Fig. 6B, as expected for mock-transfected cells, PRL was able to block EGF-induced MAPK activation (Fig. 6B, compare mock samples PE and E). However, in cells overexpressing the myc-Grb2YF mutant form, PRL was unable to block EGF-induced activation of the MAPK pathway (Fig. 6B, compare myc-Grb2YF sample PE to mock sample E). These data indicate that tyrosine phosphorylation of Grb2 is an essential mechanism utilized by PRL to suppress EGF-induced MAPK activation.
To further map the negative regulatory effects of PRL on EGF-induced MAPK activation and to determine the contribution of tyrosine phosphorylation of Grb2 to this process, we examined the influence of PRL on EGF-induced Ras activation. Using a pulldown assay of activated Ras, we observed that in control mock-transfected cells, EGF induced the activation of Ras and, interestingly, PRL attenuated EGF-induced Ras activation (Fig. 6C, upper panel). Furthermore, the overexpression of myc-Grb2YF reversed the inhibitory effects of PRL on EGF-induced Ras activation (Fig. 6C, upper panel). Reprobing of the membrane with a monoclonal antibody to GST indicated a similar GST-Ras binding domain of raf (GST-RBD) fusion protein present in all samples (Fig. 6C, middle panel). Moreover, immunodetection of cell lysates using a monoclonal antibody to the Ras protein confirmed that equal cell lysates were used in all samples (Fig. 6C, lower panel). These data together indicate that PRL inhibits EGF-induced Ras activation through tyrosine phosphorylation of Grb2.
Since Grb2 tyrosine phosphorylation impairs SH3-dependent binding to Sos, thereby blocking Ras activation (20), we next determined whether PRL is able to regulate EGF-induced Grb2/Sos complex formation in mammary epithelial cells through tyrosine phosphorylation of Grb2. Grb2/Sos coimmunoprecipitation in HC11 cells that were either mock transfected or transfected with myc-Grb2YF and treated with EGF or PRL/EGF was examined (Fig. 6D). In control mock-transfected cells, PRL treatment led to a significant decrease in EGF-induced Grb2/Sos complex formation. Interestingly, in immunoprecipitations of myc-Grb2YF, we observed no significant difference in the amounts of Sos in complex with Grb2YF in samples treated with EGF and with PRL/EGF (Fig. 6D). These data suggest that PRL utilizes the tyrosine phosphorylation of Grb2 to inhibit EGF-induced Grb2/Sos complex formation. Together, these results indicate that PRL suppresses the EGF-induced MAPK pathway at the level of Ras activation by blocking Grb2/Sos interaction. Furthermore, our data emphasize the contribution of PRL-induced tyrosine phosphorylation of Grb2 as a mechanism by which PRL exerts its negative regulatory role in EGF-induced MAPK activation in mammary epithelial cells.
PRL/Jak2-induced tyrosine phosphorylation of Grb2 attenuates EGF-induced cell proliferation.
Since PRL-induced tyrosine phosphorylation of Grb2 is critical in PRL negative regulation of EGF-induced Ras/MAPK activation, we next examined the contribution of PRL-induced tyrosine phosphorylation of Grb2 in EGF-mediated cell proliferation. To do so, we established stable cell lines overexpressing the myc-Grb2YF mutant in HC11 cells (Fig. 7A). Similarly to parental HC11 cells, vector-transfected cells (HC11-vector) show that in contrast to EGF, PRL weakly activated the MAPK pathway. Furthermore, PRL inhibited EGF-induced MAPK activation (Fig. 7B, left panel). In Grb2YF-overexpressing cells (HC11-Grb2YF), PRL also showed mild activation of the MAPK pathway in comparison to EGF-treated cells (Fig. 7B, right panel). As expected, PRL did not block EGF-induced MAPK activation, further confirming the importance of tyrosine phosphorylation of Grb2 in PRL-mediated suppressive effects on EGF-induced MAPK activation (Fig. 7B, right panel).
FIG. 7.
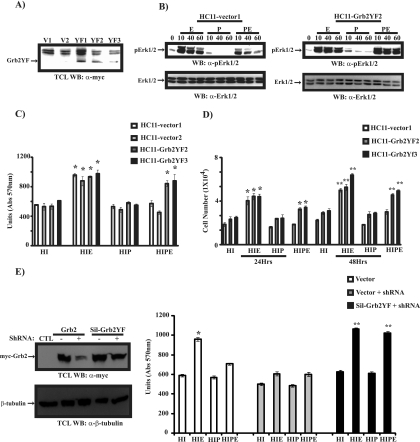
Tyrosine phosphorylation of Grb2 is essential for the inhibitory effects of PRL on EGF-induced cell proliferation. (A) HC11 cells were transfected with a vector (V) or a plasmid encoding myc-Grb2YF (YF). Cell lysates were immunoblotted with a monoclonal antibody to the myc epitope to detect stable expression of myc-Grb2YF. (B) Serum-starved differentiated HC11-vector1 (left) or HC11-Grb2YF2 (right) clones were treated with PRL (P), EGF (E), or PRL/EGF (PE), as indicated. Numbers indicate time (in minutes). Cell lysates were immunoblotted with a polyclonal antibody to phospho-Erk1/Erk2 (upper panels) and Erk1/Erk2 (lower panels). Blots shown are representative of HC11-vector and HC11-Grb2YF isolated clones. (C) Vector or Grb2YF-transfected HC11 cell lines were grown o/n in media containing 2% serum HI or HIP. Cells were then left untreated or were treated with EGF for 72 h. An MTT assay was performed, and the results are expressed as the means ± SEM for triplicates of three different experiments (*, P = 0.048). (D) Vector or Grb2YF HC11 clones were grown o/n in media containing 2% FBS HI or HIP. Cells were then left untreated or were treated with EGF for 24 h and 48 h. A cell counting assay was performed, and the results are expressed as the means ± SEM for triplicates of three experiments (*, P = 0.034; **, P = 0.041). (E, left) 293 cells were transfected with either myc-tagged Grb2 or Sil-Grb2YF in the absence or the presence of pU6+27-ShGrb2. At 48 h following transfections, cells were lysed and immunoblotted with a monoclonal antibody to myc or a monoclonal antibody to β-tubulin. (Right) HC11 cells were cotransfected with a vector, a vector and pU6+27-ShGrb2, or the Sil-Grb2YF mutant and pU6+27-ShGrb2. Transfected HC11 cells were selected in media containing G418 for 48 h. Cells were then grown o/n in media containing 2% FBS HI or HIP. Cells were then left untreated or were treated with EGF for 48 h. An MTT assay was performed, and the results are expressed as the means ± SEM for triplicates of three experiments (*, P = 0.002; **, P = 0.0017). Abs, absorbance; α-, anti-; TCL, total cell lysates.
To further study the importance of PRL-induced tyrosine phosphorylation of Grb2 in the inhibition of EGF-induced cell proliferation, we used MTT assays to examine the proliferation capacity of HC11-vector and HC11-Grb2YF cell lines in response to each ligand and the combination of both ligands (Fig. 7C). As expected, while EGF induced a significant increase in cell proliferation in vector-transfected HC11 cells (Fig. 7C, HC11-Vector1 and HC11-Vector2, compare HI and HIE samples), PRL suppressed EGF-mediated cell growth (Fig. 7C, HC11-Vector1 and HC11-Vector2, compare HIE and HIPE samples). In Grb2YF-overexpressing HC11 clones (Fig. 7C, HC11-Grb2YF2 and HC11-Grb2YF3), EGF induced cell proliferation to an extent similar to that seen for mock-transfected cells. Interestingly, PRL was unable to block EGF-induced cell proliferation (Fig. 7C, HC11-Grb2YF2 and HC11-Grb2YF3, compare HIE and HIPE samples). These data indicate that PRL-induced tyrosine phosphorylation of Grb2 is essential in mediating PRL-suppressive effects on EGF-induced cell growth. This finding was further confirmed using a cell-counting assay (Fig. 7D). As expected, in vector transfected cells, PRL was able to block EGF-induced cell growth (Fig. 7D, HC11-vector1, compare HIE and HIPE samples). Importantly, this antagonistic effect of PRL on EGF-induced cell growth was blocked in cells overexpressing Grb2YF at both the 24-h and 48-h time points (Fig. 7D, HC11-Grb2YF2 and HC11-Grb2YF3, compare HIE and HIPE samples), emphasizing the importance of Grb2 tyrosine phosphorylation in PRL inhibitory effects on EGF-induced cell proliferation. Furthermore, our data indicate that HC11-Grb2YF cells were unresponsive to the growth inhibitory effects of PRL (Fig. 7D, HC11-vector1 to HC11-Grb2YF2 and HC11-Grb2YF3, compare HIP samples). Interestingly, we observed that HC11-Grb2YF cells exhibit a higher proliferation rate than parental or vector-transfected cells, further suggesting Grb2 tyrosine phosphorylation as an inhibitory mechanism to cell proliferation.
To further evaluate the role of tyrosine phosphorylation of Grb2 as a negative regulator of cell growth, we examined whether the Grb2YF mutant form may derive cell proliferation in the absence of endogenous Grb2. For this experiment, we generated a construct expressing a Grb2YF form that is unrecognizable to the Grb2 shRNA. Indeed, as shown in Fig. 7E (left panel), while co-overexpression of the shRNA against Grb2 suppressed expression of the myc-Grb2 wild type, expression of Sil-Grb2YF was resistant to the suppressive effects of the Grb2 shRNA. To test the ability of the Sil-Grb2YF form to rescue cell growth in the absence of endogenous Grb2, we transfected HC11 cells with either a vector plasmid or a plasmid expressing Grb2 shRNA in the absence or the presence of a plasmid expressing Sil-Grb2YF (Fig. 7E, right panel). As can be seen, EGF was not able to induce cell growth in samples overexpressing the Grb2 shRNA compared to samples overexpressing the vector alone (Fig. 7E, right panel, compare samples [HI and HIE] for vector and vector+shRNA). These data indicate that silencing of Grb2 impairs the ability of EGF to induce cell growth, emphasizing the critical role of Grb2 in EGF-induced mammary cell growth. Interestingly, this loss in cell growth was rescued in cells overexpressing the Sil-Grb2YF mutant form (Fig. 7E, compare samples [HI and HIE] for vector+shRNA and Sil-Grb2YF+shRNA). More importantly, PRL was also unable to block EGF-mediated cell growth in the presence of the Sil-Grb2YF form (Fig. 7E, compare samples HIE and HIPE for SilGrb2YF+shRNA). These data together indicate that the Grb2YF form is able to mediate proliferative signals and that PRL is ineffective in blocking EGF-mediated cell proliferation in the absence of tyrosine phosphorylation of Grb2. Together, these results point to the essential role of PRL-induced tyrosine phosphorylation of Grb2 in regulating growth factor-induced mammary epithelial cell proliferation.
DISCUSSION
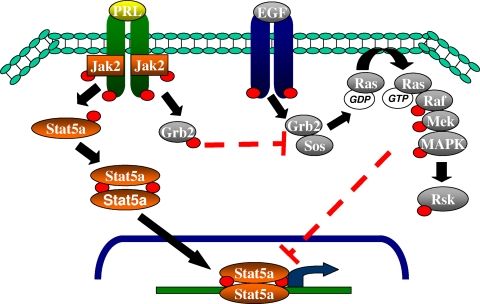
Multiple hormones and growth factors regulate mammary epithelial cell growth and differentiation. In the present study, we have examined the PRL and EGF cross talk mechanism in mammary epithelial cells. Our results indicate that PRL and EGF exhibit antagonistic properties that have an impact on mammary epithelial cell proliferation and differentiation. We have discovered an intricate signaling network mediating this negative cross talk. EGF was shown to block PRL-induced cellular differentiation by interfering with Stat5a-mediated gene transcription without inhibiting parts of the PRL-proximal signaling cascade, such as Jak2 activation, whereas PRL attenuated EGF-induced MAPK activation and cell proliferation through Jak2-dependent tyrosine phosphorylation of the adaptor protein Grb2. Together, these results define a novel signaling mechanism underlying PRL and EGF cross talk in mammary epithelial cells (Fig. 8).
FIG. 8.
PRL/EGF negative cross talk in mammary epithelial cells. Both PRL and EGF play important roles in mammary epithelial cell growth and differentiation. We propose multiple levels of cross-regulation in PRL/EGF cross talk in mammary cells. While EGF blocks PRL-induced cellular differentiation by interfering with Stat5a transcriptional activity without inhibiting the PRL-proximal signaling cascade, PRL suppressed EGF-induced MAPK activation and cell proliferation through tyrosine phosphorylation of Grb2.
The direct role of PRL in mammary epithelial cell proliferation is not fully elucidated and appears to be cell context dependent. While PRL has been described to play a proliferative and tumorigenic role in the mammary epithelium (4, 29, 32), the overexpression of PRL in the differentiated mammary epithelium resulted in altered alveologenesis, yet no carcinomas were detected (24), suggesting that the state of differentiation of mammary epithelial cells may be an important determinant in specifying the role of PRL as a proliferative agent. While these data indicate that the role of PRL in mammary cell proliferation is complex and requires further investigation, our data indicate that in differentiated mammary epithelial cells, PRL is nonproliferative, and moreover, PRL can attenuate growth factor-induced cell proliferation. Due to the well-known effect of EGF as a promoter of mammary epithelial cell proliferation and tumorigenesis and based on our data indicating the ability of PRL to significantly suppress EGF-induced cell proliferation, we were interested in defining the mechanism by which PRL exerts this effect on EGF-mediated signaling. While PRL has been reported to activate the MAPK (Erk1/Erk2) pathway (12, 15), we have previously reported that PRL, through Jak2, may suppress the MAPK pathway (28). Here, we observed that unlike EGF stimulation, PRL stimulation resulted in only minor and transient activation of Erk1/Erk2; in addition, we could not detect any PRL-induced activation of Rsk, a downstream target of the MAPK pathway. Moreover, we observed that PRL suppressed EGF-induced MAPK activation. Together, these data indicate that PRL is a poor activator of the MAPK pathway and that PRL may suppress growth factor-mediated MAPK activation in mammary epithelial cells. On the other hand, EGF was found to be a potent inhibitor of PRL-Stat5a-mediated gene transcription, which was found to be independent of the ability of PRL to activate Jak2 kinase. This result suggests that PRL may suppress EGF effects through a Jak2-dependent proximal signaling mechanism.
We have previously reported the presence of a 29-kDa protein, a substrate of SHP-2, in immunoprecipitates of Grb2 following PRLR/Jak2 activation (26). Moreover, in samples overexpressing myc-Grb2 (molecular size, 45 kDa) we observed a phosphorylated protein of 45 kDa and not 29 kDa, suggesting that p29 is indeed Grb2 (Fig. 4). Moreover, we previously found that the phosphorylation level of this protein was dependent on the catalytic activity of SHP-2 as well as SHP-2 C-terminal tyrosine residues (26). These data together suggest that Grb2 is recruited to the PRLR/Jak2 complex through the C-terminal tyrosine residues of SHP-2. Confirming previous observations (22, 33) these data suggest that tyrosine phosphorylation of Grb2 is a reversible process, tightly regulated by intracellular protein tyrosine phosphatases. Indeed, pretreatment of cells with sodium vanadate led to better detection of Grb2 phosphorylation, although the pattern of regulation by PRL or EGF was intact.
Tyrosine phosphorylation of Grb2 within its SH3 domains was previously observed for cells transformed by the Bcr/Abl oncogene as well as following EGFR activation in A431 cells (20). However, the physiological context of tyrosine phosphorylation of Grb2 had not been well established. Our data indicate that in mammary epithelial cells, PRL-induced tyrosine phosphorylation of Grb2 is essential for PRL inhibitory effects on EGF-induced signaling to MAPK activation and cell proliferation. Other mechanisms of PRL-induced negative regulation of EGF signaling have been proposed, such as through protein kinase C (10) and threonine phosphorylation of the EGFR (31). Although our study does not rule out the presence of other mechanisms, the observation that overexpression of the Grb2YF mutant was able to recover significant levels of EGF-induced Ras/MAPK activation and cell proliferation indicates the importance of Grb2 tyrosine phosphorylation in the suppression of EGF signaling by PRL in mammary epithelial cells.
Furthermore, while tyrosine phosphorylation of Grb2 was proposed as an auto-feedback mechanism to inhibit EGF activation of the MAPK pathway (20), our data, in accordance with previous studies (22, 33), indicate that EGF stimulation does not lead to significant tyrosine phosphorylation of Grb2. This result suggests that Grb2 phosphorylation might represent a cross-regulatory mechanism rather than an auto-regulatory mechanism for EGF, in that ligands that employ Grb2 might influence EGF signaling by increasing the pool of phosphorylated Grb2. Our results suggest a model in which the presence of PRL significantly increases the pool of phosphorylated Grb2, which impairs the ability of EGF to induce the activation of the Ras/MAPK pathway and hence cell proliferation.
The importance of the adaptor protein Grb2 in growth factor receptor signaling and in the development of cancers, such as breast cancer, has been well characterized. Therefore, multiple inhibitors of Grb2 have been developed as therapeutic agents (7, 19). The majority of the Grb2 inhibitors act by blocking the interaction between Grb2 and tyrosine kinase receptors or targeting SH3-ligand interaction. Our findings and those by Li et al. (20) suggest that interrupting the interaction of Grb2 and Sos via the induction of tyrosine phosphorylation of Grb2 may be a possible target of Grb2 inhibition. In this study, we have identified a novel cross-regulatory signaling mechanism operating downstream of PRL and EGF that influences mammary epithelial cell growth and differentiation.
Acknowledgments
We thank N. Hynes (Friedrich Miescher Institute, Basel, Switzerland) and B. Groner (Institute for Biomedical Research, Frankfurt am Main, Germany) for providing us with HC11 and HC11-Lux cell lines. We thank B. Mayer (Connecticut Health Center, Farmington, CT) for providing the expression plasmid encoding Grb2WT and Grb2W36,193K and R. Van Etten (Tufts-New England Medical Center, Boston, MA) and S. Li (The Jackson Laboratory, Bar Harbor, ME) for providing the expression plasmid encoding myc-Grb2YF. Also, we thank J. Woodgett (Samuel Lunenfeld Research Institute, Toronto, Ontario, Canada) for providing us with the GST-RBD construct. We thank N. Chughtai and N. Shahrzad for their excellent technical assistance.
A grant from the Canadian Institutes of Health Research (MOP-13681; S.A.) supported this study. E.H. is a recipient of the McGill University Health Center Studentship Award. S.A. is a recipient of the Fonds de la Recherche en Santé du Québec (F.R.S.Q.) (Senior Award) and the Girls for the Cure Breast Cancer Award, Montreal General Hospital Foundation. J.-J.L. is a recipient of the Scientist Award, National Cancer Institute of Canada.
We acknowledge the recent passing away of Strachan Hartley and we dedicate this study to his memory.
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Ali, S. 1998. Prolactin receptor regulates Stat5 tyrosine phosphorylation and nuclear translocation by two separate pathways. J. Biol. Chem. 2737709-7716. [DOI] [PubMed] [Google Scholar]
- 2.Ball, R. K., R. R. Friis, C. A. Schoenenberger, W. Doppler, and B. Groner. 1988. Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J. 72089-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blenis, J. 1993. Signal transduction via the MAP kinases: proceed at your own RSK. Proc. Natl. Acad. Sci. USA 905889-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clevenger, C. V. 2003. Role of prolactin/prolactin receptor signaling in human breast cancer. Breast Dis. 1875-86. [DOI] [PubMed] [Google Scholar]
- 5.Cocolakis, E., S. Lemay, S. Ali, and J. J. Lebrun. 2001. The p38 MAPK pathway is required for cell growth inhibition of human breast cancer cells in response to activin. J. Biol. Chem. 27618430-18436. [DOI] [PubMed] [Google Scholar]
- 6.Darcy, K. M., S. F. Shoemaker, P. P. Lee, M. M. Vaughan, J. D. Black, and M. M. Ip. 1995. Prolactin and epidermal growth factor regulation of the proliferation, morphogenesis, and functional differentiation of normal rat mammary epithelial cells in three dimensional primary culture. J. Cell. Physiol. 163346-364. [DOI] [PubMed] [Google Scholar]
- 7.Dharmawardana, P. G., B. Peruzzi, A. Giubellino, T. R. Burke, Jr., and D. P. Bottaro. 2006. Molecular targeting of growth factor receptor-bound 2 (Grb2) as an anti-cancer strategy. Anticancer Drugs 1713-20. [DOI] [PubMed] [Google Scholar]
- 8.Di Fulvio, M., K. M. Henkels, and J. Gomez-Cambronero. 2007. Short-hairpin RNA-mediated stable silencing of Grb2 impairs cell growth and DNA synthesis. Biochem. Biophys. Res. Commun. 357737-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenton, S. E., and L. G. Sheffield. 1997. Prolactin inhibits EGF-induced DNA synthesis in mammary epithelium via early signaling mechanisms: possible involvement of protein kinase C. Exp. Cell Res. 236285-293. [DOI] [PubMed] [Google Scholar]
- 11.Gupta, R. W., and B. J. Mayer. 1998. Dominant-negative mutants of the SH2/SH3 adapters Nck and Grb2 inhibit MAP kinase activation and mesoderm-specific gene induction by eFGF in Xenopus. Oncogene 172155-2165. [DOI] [PubMed] [Google Scholar]
- 12.Gutzman, J. H., D. E. Rugowski, M. D. Schroeder, J. J. Watters, and L. A. Schuler. 2004. Multiple kinase cascades mediate prolactin signals to activating protein-1 in breast cancer cells. Mol. Endocrinol. 183064-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennighausen, L., and G. W. Robinson. 2005. Information networks in the mammary gland. Nat. Rev. Mol. Cell Biol. 6715-725. [DOI] [PubMed] [Google Scholar]
- 14.Horseman, N. D., W. Zhao, E. Montecino-Rodriguez, M. Tanaka, K. Nakashima, S. J. Engle, F. Smith, E. Markoff, and K. Dorshkind. 1997. Defective mammopoiesis, but normal hematopoiesis, in mice with a targeted disruption of the prolactin gene. EMBO J. 166926-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, Y., X. Li, J. Jiang, and S. J. Frank. 2006. Prolactin modulates phosphorylation, signaling and trafficking of epidermal growth factor receptor in human T47D breast cancer cells. Oncogene 257565-7576. [DOI] [PubMed] [Google Scholar]
- 16.Hynes, N. E., and H. A. Lane. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5341-354. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H., and W. J. Muller. 1999. The role of the epidermal growth factor receptor family in mammary tumorigenesis and metastasis. Exp. Cell Res. 25378-87. [DOI] [PubMed] [Google Scholar]
- 18.Lebrun, J. J., S. Ali, V. Goffin, A. Ullrich, and P. A. Kelly. 1995. A single phosphotyrosine residue of the prolactin receptor is responsible for activation of gene transcription. Proc. Natl. Acad. Sci. USA 924031-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, P., M. Zhang, Y. Q. Long, M. L. Peach, H. Liu, D. Yang, M. Nicklaus, and P. P. Roller. 2003. Potent Grb2-SH2 domain antagonists not relying on phosphotyrosine mimics. Bioorg. Med. Chem. Lett. 132173-2177. [DOI] [PubMed] [Google Scholar]
- 20.Li, S., A. D. Couvillon, B. B. Brasher, and R. A. Van Etten. 2001. Tyrosine phosphorylation of Grb2 by Bcr/Abl and epidermal growth factor receptor: a novel regulatory mechanism for tyrosine kinase signaling. EMBO J. 206793-6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, X., G. W. Robinson, K. U. Wagner, L. Garrett, A. Wynshaw-Boris, and L. Hennighausen. 1997. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 11179-186. [DOI] [PubMed] [Google Scholar]
- 22.Lowenstein, E. J., R. J. Daly, A. G. Batzer, W. Li, B. Margolis, R. Lammers, A. Ullrich, E. Y. Skolnik, D. Bar-Sagi, and J. Schlessinger. 1992. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell 70431-442. [DOI] [PubMed] [Google Scholar]
- 23.Luetteke, N. C., T. H. Qiu, S. E. Fenton, K. L. Troyer, R. F. Riedel, A. Chang, and D. C. Lee. 1999. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development 1262739-2750. [DOI] [PubMed] [Google Scholar]
- 24.Manhes, C., C. Kayser, P. Bertheau, B. Kelder, J. J. Kopchick, P. A. Kelly, P. Touraine, and V. Goffin. 2006. Local over-expression of prolactin in differentiating mouse mammary gland induces functional defects and benign lesions, but no carcinoma. J. Endocrinol. 190271-285. [DOI] [PubMed] [Google Scholar]
- 25.Marte, B. M., M. Jeschke, D. Graus-Porta, D. Taverna, P. Hofer, B. Groner, Y. Yarden, and N. E. Hynes. 1995. Neu differentiation factor/heregulin modulates growth and differentiation of HC11 mammary epithelial cells. Mol. Endocrinol. 914-23. [DOI] [PubMed] [Google Scholar]
- 26.Minoo, P., N. Chughtai, M. Campiglio, M. Stein-Gerlach, J. J. Lebrun, A. Ullrich, and S. Ali. 2003. The adaptor function of SHP-2 downstream of the prolactin receptor is required for the recruitment of p29, a substrate of SHP-2. Cell. Signal. 15319-326. [DOI] [PubMed] [Google Scholar]
- 27.Nevalainen, M. T., J. Xie, J. Torhorst, L. Bubendorf, P. Haas, J. Kononen, G. Sauter, and H. Rui. 2004. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J. Clin. Oncol. 222053-2060. [DOI] [PubMed] [Google Scholar]
- 28.Nouhi, Z., N. Chughtai, S. Hartley, E. Cocolakis, J. J. Lebrun, and S. Ali. 2006. Defining the role of prolactin as an invasion suppressor hormone in breast cancer cells. Cancer Res. 661824-1832. [DOI] [PubMed] [Google Scholar]
- 29.Oakes, S. R., F. G. Robertson, J. G. Kench, M. Gardiner-Garden, M. P. Wand, J. E. Green, and C. J. Ormandy. 2007. Loss of mammary epithelial prolactin receptor delays tumor formation by reducing cell proliferation in low-grade preinvasive lesions. Oncogene 26543-553. [DOI] [PubMed] [Google Scholar]
- 30.Ormandy, C. J., A. Camus, J. Barra, D. Damotte, B. Lucas, H. Buteau, M. Edery, N. Brousse, C. Babinet, N. Binart, and P. A. Kelly. 1997. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev. 11167-178. [DOI] [PubMed] [Google Scholar]
- 31.Quijano, V. J., Jr., and L. G. Sheffield. 1998. Prolactin decreases epidermal growth factor receptor kinase activity via a phosphorylation-dependent mechanism. J. Biol. Chem. 2731200-1207. [DOI] [PubMed] [Google Scholar]
- 32.Rose-Hellekant, T. A., L. M. Arendt, M. D. Schroeder, K. Gilchrist, E. P. Sandgren, and L. A. Schuler. 2003. Prolactin induces ERalpha-positive and ERalpha-negative mammary cancer in transgenic mice. Oncogene 224664-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozakis-Adcock, M., J. McGlade, G. Mbamalu, G. Pelicci, R. Daly, W. Li, A. Batzer, S. Thomas, J. Brugge, P. G. Pelicci, et al. 1992. Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature 360689-692. [DOI] [PubMed] [Google Scholar]
- 34.Sultan, A. S., J. Xie, M. J. LeBaron, E. L. Ealley, M. T. Nevalainen, and H. Rui. 2005. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene 24746-760. [DOI] [PubMed] [Google Scholar]
- 35.Vomachka, A. J., S. L. Pratt, J. A. Lockefeer, and N. D. Horseman. 2000. Prolactin gene-disruption arrests mammary gland development and retards T-antigen-induced tumor growth. Oncogene 191077-1084. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, K.-U., A. Krempler, A. A. Triplett, Y. Qi, N. M. George, J. Zhu, and H. Rui. 2004. Impaired alveologenesis and maintenance of secretory mammary epithelial cells in Jak2 conditional knockout mice. Mol. Cell. Biol. 245510-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita, H., M. Nishio, Y. Ando, Z. Zhang, M. Hamaguchi, K. Mita, S. Kobayashi, Y. Fujii, and H. Iwase. 2006. Stat5 expression predicts response to endocrine therapy and improves survival in estrogen receptor-positive breast cancer. Endocr. Relat. Cancer 13885-893. [DOI] [PubMed] [Google Scholar]
- 38.Yarden, Y. 2001. The EGFR family and its ligands in human cancer: signalling mechanisms and therapeutic opportunities. Eur. J. Cancer 37(Suppl. 4)S3-S8. [DOI] [PubMed] [Google Scholar]