Abstract
p27kip1 (p27) is a cell cycle inhibitor and tumor suppressor whose expression is tightly regulated in the cell. Translational control of p27 mRNA has emerged as a prominent mechanism to regulate p27 expression during differentiation, quiescence, and cancer progression. The microRNAs miR-221 and miR-222 repress p27 expression in various cancer cells, and this repression promotes tumor cell proliferation. In addition, the presence of an internal ribosome entry site in the 5′ untranslated region (UTR) of p27 mRNA has been reported. Here, we show that p27 mRNA is translated via a cap-dependent mechanism in HeLa and HL60 cells and that the previously reported IRES activity can be attributed to cryptic promoters in the sequence corresponding to the p27 5′ UTR. Furthermore, cap-dependent translation of p27 mRNA is repressed by miR-181a in undifferentiated HL60 cells. Repression by miR-181a is relieved during differentiation of HL60 into monocyte-like cells, allowing the accumulation of p27, which is necessary to fully block cell cycle progression and reach terminal differentiation. These results identify miR-181a as a regulator of p27 mRNA translation during myeloid cell differentiation.
Translational control of gene expression plays an essential role in many biological processes, including early embryonic development, cell growth, differentiation, apoptosis, and metabolism. During the mitotic cell cycle, for example, timely translation of the mRNAs encoding the p58PITSLRE kinase or the cyclin-dependent kinase Cdk1 allows progression through the G2/M phase (40; reviewed in reference 50). These mRNAs are translated in a cap-independent manner, via structures in their 5′ untranslated regions (UTRs) named internal ribosome entry sites (IRES) that allow direct ribosome recruitment. Conversely, cap-dependent translation is reduced in the G2/M phase. Moreover, alterations in the levels of the cap binding protein eIF4E and other initiation factors, as well as in the signaling pathways that regulate their functions, promote uncontrolled cell proliferation and the development of tumors (49).
p27kip1 (hereafter referred to as p27) is another regulator of cell cycle progression whose expression is controlled at the translation level (1, 27, 41). p27 is a cyclin-dependent kinase inhibitor that blocks the cell cycle in G0/G1 upon differentiation signals or cellular insults. In addition to its roles in cell cycle progression, p27 has been shown to regulate cell motility and apoptosis (3, 5). p27 has been considered a tumor suppressor, although mutations or deletions of the p27 gene are rare in human cancers (51). Mice lacking p27 spontaneously develop adenomas of the pituitary gland and are more susceptible to tumorigenesis induced by chemical carcinogens or irradiation (16, 33, 45). Heterozygous p27+/− mice, containing half the p27 dose of wild-type animals, show an intermediate phenotype, indicating that the fine regulation of p27 levels is essential for appropriate cell proliferation (17). Furthermore, in many human cancers, low levels of p27 correlate with tumor aggressiveness and poor prognosis (9). Recently, p27 has been proposed to behave as an oncogene under certain circumstances, since point mutations that prevent its interaction with cyclins and cyclin-dependent kinases lead to hyperplasic lesions and tumors in multiple organs (2). Thus, given the complexity of p27 functions, an accurate knowledge of the mechanisms controlling p27 expression is likely to improve our capacity to develop successful therapies against cancer.
Expression of p27 is tightly regulated in a cell-type- and condition-specific manner. Although the best-characterized regulatory mechanisms are the modulation of p27 protein stability and subcellular localization, translational control has emerged as a prominent mode of p27 regulation during differentiation, quiescence, and cancer progression (reviewed in references 5, 9, and 37). In HeLa cells, the accumulation of p27 in early G1 is partially due to an increase in the translational efficiency of p27 mRNA (27). Accordingly, the 5′ UTR of p27 mRNA was shown to enhance the translation of a luciferase reporter mRNA and to display IRES activity (8, 30, 35, 42, 44). The proteins HuR and PTB (polypyrimidine tract-binding protein) recognize the p27 5′ UTR and inhibit and activate IRES-dependent translation, respectively (8, 35). In addition, hnRNPs C1/C2 interact with a U-rich sequence located within the IRES, although the exact roles of these proteins in p27 mRNA translation remain unexplored (42). The 3′ UTR of p27 transcript plays a negative regulatory role in translation (24, 42, 53). Recently, the related microRNAs (miRNAs) miR-221 and miR-222 have been shown to bind to the 3′ UTR and repress p27 mRNA expression in cell lines derived from glioblastoma, melanoma, hepatocarcinoma, and papillary thyroid and prostate carcinomas (15, 19, 20, 23, 38, 54). Importantly, inhibition of miR-221/miR-222 blocked the proliferation of glioblastoma cells, whereas no effect was observed when p27 was depleted, establishing a causal relationship between miR-221/miR-222, p27, and cell proliferation (38). These results suggest an important role for the translational regulation of p27 mRNA in preventing the development of certain types of tumor.
Here, we study the mechanisms that regulate p27 mRNA translation in HeLa and human promyelocytic leukemia (HL60) cells. We show that translation of p27 mRNA takes place by a cap-dependent mechanism and that the proposed IRES activity is due to the presence of cryptic promoters in the sequence corresponding to the p27 5′ UTR. In addition, we have identified the miRNA miR-181a as a repressor of p27 mRNA translation in undifferentiated HL60 cells. Phorbol-12-myristate-13-acetate (TPA)-induced differentiation of these cells into monocytes/macrophages results in the relief of miR-181a-mediated repression, allowing the accumulation of sufficient p27 to ensure proper differentiation. Consistent with these results, overexpression of miR-181a in the more differentiated myeloid cell line THP1 results in reduction of p27 levels. These results identify miR-181a as a novel regulator of p27 mRNA translation that functions during myeloid differentiation.
MATERIALS AND METHODS
Cell culture and transfections.
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. HL60 and THP1 cells were grown in RPMI 1640 medium supplemented with 0.3 mg/ml l-glutamine and 10% fetal bovine serum. HL60 cell differentiation into monocyte/macrophage-like cells was induced by treatment with 30 nM TPA (Calbiochem) during the indicated times.
Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For DNA transfections, 6 × 105 cells per well were grown on a six-well plate and transfected with 0.5 μg of reporter plasmid and 3.5 μg of pCR3-β-Gal (14). The cells were analyzed 36 h after transfection. For RNA transfection of HeLa cells, 2 μg of reporter mRNA and 2 μg of a non-translationally competent β-galactosidase mRNA were mixed with 10 μl of Lipofectamine, added to the cells, and incubated for 1 h at 37°C. The medium was changed and the cells were analyzed after incubation for 5 h at 37°C. For RNA transfection of HL60 cells, 10 × 106 to 15 × 106 cells were incubated in 9 ml of RPMI medium containing 12 μg of reporter mRNA mixed with 30 μl of Lipofectamine for 1 h at 37°C. The medium was changed and the cells were analyzed after incubation for 4 h at 37°C.
Plasmids.
The bicistronic reporter plasmid pRF (52) was a kind gift of A. E. Willis (University of Nottingham, United Kingdom). The p27 5′ UTR (positions −456 to +9) was amplified by PCR using specific oligonucleotides and fused in frame to the firefly luciferase open reading frame (ORF) in the pRF plasmid to yield pR-p27-F. The same PCR product was cloned in the inverted orientation at the same position to generate pR-p27inv-F. The promoterless pR-p27-F construct was derived from pR-p27-F by deletion of the simian virus 40 promoter.
Plasmids used to synthesize reporter mRNAs in vitro were generated using pBSK′As as the vector (21). The fragment containing Renilla and firefly luciferase ORFs from either pRF or pR-p27-F was inserted into pBSK′As to produce R-F and R-p27-F, respectively. The firefly luciferase ORF sequence flanked by the p27 5′ UTR and the full-length 3′ UTR in either the sense or inverted orientation was cloned into pBSK′As to generate p27-F-p27 and p27-F-p27inv, respectively. A small palindrome with the sequence 5′-CCCGGAGCGCCCAGATCTGGGCGCTCCGGGGTAC-3′ (28) was cloned 13 nucleotides (nt) downstream from the 5′ end to produce Hp27-F-p27 and Hp27-F-p27inv. Plasmids with the 5′ UTR of p27 in the inverted orientation were derived from p27-F-p27 and p27-F-p27inv by inverting the NaeI-SmaI fragment of the p27 5′ UTR.
Fragments of the p27 3′ UTR were amplified by PCR and cloned downstream of the firefly luciferase ORF, in either the sense or inverted orientation, to produce the following plasmids: p27-F-[1-551], p27-F-[551-1], p27-F-[552-1341], p27-F-[1341-552], p27-F-[1-234], p27-F-[234-1], p27-F-[235-551], and p27-F-[551-235]. The numbers in brackets indicate the 3′ UTR positions included in each fragment.
To generate the plasmids p27-F-p27-181mut, -221mut, -181mut1, and -181mut2, the binding sites of miR-181a or miR-221 in the p27 3′ UTR were mutated by overlap extension PCR mutagenesis.
The lentivirus-derived plasmid pLVTHM-931 was used to knock down p27 from HL60 cells. This plasmid generated a short hairpin RNA (shRNA) against positions 466 to 487 of the p27 ORF and was constructed by cloning the preannealed primers 931s (5′-CGCGTCCCCGCAACCGACGATTCTTCTACTCGAAAGTAGAAGAATCGTCGGTTGCTTTTTGGAAAT-3′) and 931a (5′-CGATTTCCAAAAAGCAACCGACGATTCTTCTACTTTCGAGTAGAAGAATCGTCGGTTGCGGGGA-3′) into the MluI and ClaI sites of pLVTHM (55).
To overexpress miR-181a in THP1 cells, the primary miR181a sequence was amplified by PCR from genomic DNA using the primers gen181a-se (5′-AGCTAAGCTTCTGTAAAGTGAGTAGAATTCTG-3′) and gen181a-an (5′-GCTATCTAGACTTCAGCGAATTCTGAGCACC-3′) and cloned into pLNHX (Clontech).
In vitro transcription.
mRNAs were synthesized as described previously (22). All mRNAs contained a poly(A) tail of 72 residues and either a 5′ m7GpppG cap or a non-translationally competent ApppG cap, as indicated. mRNAs used in the same experiment were synthesized and quantified in parallel.
Luciferase reporter assays.
Cells were lysed with Pasive lysis buffer (Promega), and the enzymatic luciferase activity was measured using the Promega luciferase kits.
Northern blot analysis and quantitative reverse transcription (qRT)-PCR.
Total RNA was extracted from cells using Trizol (Invitrogen) according to the manufacturer's instructions. Ten micrograms of HeLa or 50 μg of HL60 total RNA was resolved in a 1% agarose denaturing gel, transferred, and hybridized as described previously (10). Random-primed probes were used against the full-length firefly luciferase ORF, the β-actin ORF, and nt 590 to 2198 of p27 mRNA. To determine miRNA expression, 30 μg of total RNA were resolved in a 15% acrylamide-7.5 M urea gel and transferred to Zeta-Probe GT membranes (Bio-Rad) in 0.5× Tris-buffered EDTA at 200 mA overnight at 4°C. The membranes were UV cross-linked and heated at 80°C for 1 h. Hybridization was performed at 50°C as described above, using 5′-32P-labeled anti-miR-181a and anti-miR-221 oligoribonucleotide probes (Ambion) or an oligonucleotide complementary to U6 (a gift from J. Valcárcel, Centre de Regulació Genòmica, Spain).
qRT-PCR was carried out from 4 μg of total RNA using avian myeloblastosis virus retrotranscriptase (Promega), followed by PCR with a LightCycler FastStart DNA MasterPlus SYBR green I kit (Roche). The oligonucleotides were complementary to the ORF positions 1033 to 1052 and 1158 to 1178 of firefly luciferase, 2179 to 2200 and 2259 to 2280 of β-actin, and −16 to +5 and 146 to 165 of p27.
Western blotting.
Total-protein extracts were obtained by lysis with RIPA buffer (150 mM NaCl, 10 mM Tris, pH 7.2, 5 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 1% sodium deoxycholate, 1% Triton X-100, 1× protease inhibitor cocktail [Roche]). Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride membranes. Anti-p27kip1 (c-19; Santa Cruz Biotechnology), anti-eIF4A (a gift from W. Merrick, Case Western Reserve University, Cleveland, OH), anti-eIF4E (c-20; Santa Cruz Biotechnology), anti-caspase 3 (5A1; Cell Signaling Technology), and anti-β-actin (Sigma) antibodies were used, and proteins were visualized by chemiluminescence (enhanced chemiluminescence; GE Healthcare).
Metabolic labeling and pulse-chase experiments.
Untreated or TPA-treated HL60 cells were incubated in medium without methionine and cysteine for 1 h at 37°C. One hundred microcuries per milliliter of [35S]methionine/[35S]cysteine (Amersham Biosciences) was added, and the incubation was extended for 1 h. The cells were washed, and the medium was replaced by complete medium. The cells were subsequently lysed with RIPA buffer at 0, 15, 45, and 120 min. p27 was precipitated from equivalent amounts of total-protein extracts, resolved on a 15% SDS-polyacrylamide gel, and transferred to polyvinylidene difluoride membranes. Labeled p27 was visualized by autoradiography and by immunoblotting with α-p27 antibodies.
Virus production, infection, and cell clone selection.
Recombinant lentiviruses were produced by transient transfection of 293T cells as previously described (55). Briefly, cells were cotransfected with 20 μg of either pLVTHM-931 plasmid or pLVTHM, 15 μg of pLHVAR, and 5 μg of pVSV-G by calcium phosphate precipitation. After 24 h, the medium was changed, and recombinant lentiviruses were collected 24 h and 48 h after medium replacement, filtered through 0.45-μm sieves, and concentrated by centrifugation at 26,000 rpm for 90 min at 4°C. Viruses were resuspended in culture medium.
Recombinant retroviruses were produced by transient transfection of GP2-293 cells (Clontech) with 15 μg of pLNHX-pri-miR-181a or pLNHX and 10 μg of pVSV-G plasmids. Recombinant viruses were collected as described above.
Infection of HL60 or THP1 cells was performed by adding increasing amounts of virus to the cell culture medium, followed by centrifugation of the culture plate at 1,200 rpm for 2 h at room temperature and incubation at 37°C for 24 h. Selection of HL60 cells expressing the shRNA against p27 was performed by detection of the green fluorescent protein marker (included in the vector) using fluorescence-activated cell sorting (FACS). To select THP1 cells expressing pri-miR-181a, infected cells were grown in RPMI medium containing 400 μg/ml Geneticin (Gibco-BRL).
FACS analysis.
To determine the HL60 cell cycle distribution, cells were fixed in 70% cold ethanol and stained with 50 μg/ml propidium iodide (Sigma) in the presence of 10 μg/ml RNase A (Roche) for 1 h at 37°C. The cells were analyzed for DNA content on a FACSCanto (BD Bioscience). To evaluate cell differentiation, HL60 cells were incubated with α-CD11b-allophycocyanin antibody (BD Bioscience) in phosphate-buffered saline for 30 min on ice, washed, and analyzed for CD11b expression on a FACSCanto.
RESULTS
p27 mRNA is translated by a cap-dependent mechanism in HeLa cells.
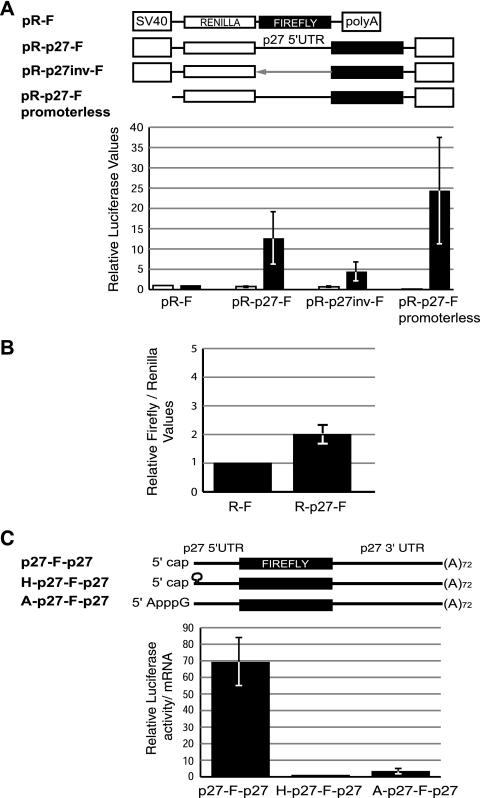
In order to study the mechanisms of p27 mRNA translational regulation, we first tested whether the 5′ UTR displayed IRES activity in HeLa cells. We generated a bicistronic reporter construct containing the 5′ UTR of the human p27 transcript (nt −456 to +9) between the two cistrons of the pR-F vector (Fig. 1A, top). Because the efficiency of translation may be influenced by the length of the UTRs, we used the same 5′-UTR fragment cloned in an inverted orientation as a negative control. In addition, to eliminate the possibility that the observed activity was derived from promoter activity of the cloned intercistronic sequence, we deleted the promoter (simian virus 40) from the vector. The empty vector was also included as a control. Asynchronous HeLa cells were transfected with these constructs, and the activities from the two cistrons were measured and corrected for cotransfected β-galactosidase. As shown in Fig. 1A (bottom), the p27 5′ UTR enhanced the translation of the second cistron (firefly luciferase) ∼12-fold compared to the empty pR-F vector, whereas the inverted segment increased translation by only 4-fold. As expected, the activity of the first cistron (Renilla luciferase) remained constant for these constructs. Surprisingly, however, the promoterless control showed elevated firefly luciferase activity while the Renilla luciferase activity was almost undetectable. These results suggest that the intercistronic sequence contains cryptic promoters that produce artifactual IRES activity.
FIG. 1.
p27 mRNA is translated via a cap-dependent mechanism in HeLa cells. (A) The 5′-UTR sequence of p27 contains cryptic promoters. HeLa cells were transiently transfected with bicistronic plasmids containing the ORFs of Renilla and firefly luciferases (open and filled boxes, respectively) and the 5′ UTR of p27 (nt −456 to +9) inserted in the intercistronic region in either sense (pR-p27-F) or inverted (pR-p27inv-F) orientation. The empty vector (pR-F) and a promoterless plasmid were used as controls. Luciferase values were normalized by cotransfected β-galactosidase and plotted relative to pR-F. The average of three independent experiments is shown. SV40, simian virus 40. (B) HeLa cells were transfected with bicistronic capped and polyadenylated mRNAs that either contained (R-p27-F) or lacked (R-F) the 5′ UTR of p27 in the intercistronic region. Firefly luciferase values were corrected for Renilla luciferase activity and plotted relative to R-F. The average of three independent experiments is shown. (C) Translation of p27 mRNA occurs by a cap-dependent mechanism. HeLa cells were transfected with polyadenylated reporter mRNAs containing either a canonical m7GpppG cap (cap) (p27-F-p27), a cap followed by a stable hairpin (H-p27-F-p27), or the non-translationally competent cap analog ApppG (A-p27-F-p27) at the 5′ end. mRNAs contained the 5′ (456-nt) and 3′ (1,341-nt) UTRs of the p27 transcript. Five hours after transfection, protein and RNA extracts were obtained. The luciferase activity was measured, corrected for mRNA levels, and plotted relative to H-p27-F-p27. The average of three independent experiments is shown.
To assess this possibility directly, we transfected bicistronic RNA constructs synthesized in vitro, either containing or lacking the 5′ UTR of p27 in the intercistronic region (Fig. 1B). The second cistron showed negligible expression under these conditions, challenging the presence of an IRES element in the p27 5′ UTR. To further confirm this hypothesis, we transfected monocistronic mRNAs containing the firefly luciferase ORF flanked by the 5′ (456-nt) and 3′ (1,341-nt) UTRs of the p27 transcript and a poly(A) tail of 72 residues (Fig. 1C, top). To evaluate the contribution of cap-dependent versus cap-independent (i.e., IRES) translation, the mRNAs contained either a canonical 7mGpppG cap or a non-translationally competent ApppG cap analog. In addition, a stable hairpin downstream of the canonical cap structure was used as an alternative mode to inhibit cap-dependent translation (28). Luciferase values were corrected for the levels of the reporter mRNAs after transfection, allowing the exclusion of effects due to changes in the stability of the different mRNA constructs. The presence of either the noncanonical cap analog or the hairpin decreased luciferase activity by ∼70-fold, indicating that efficient translation of p27 mRNA requires a functional cap structure (Fig. 1C, bottom). Furthermore, RNase protection assays from HeLa cells transfected with a monocistronic plasmid containing the full-length 5′ UTR of p27 showed at least four transcripts with reduced 5′-UTR length, confirming the existence of promoter activity in the 5′-UTR sequence (data not shown). Taken together, these data show that the translation of p27 mRNA takes place by a cap-dependent mechanism in HeLa cells.
The p27 3′ UTR represses cap-dependent translation.
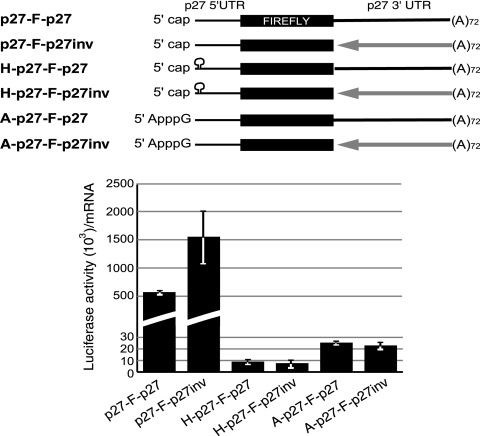
To evaluate the influence of the 3′ UTR on cap-dependent translation of p27 mRNA, we transfected HeLa cells with monocistronic transcripts containing the 3′ UTR in either the sense or inverted orientation (Fig. 2, top). An inverted 3′ UTR should lack regulatory sequences while preserving the same length as the wild-type 3′ UTR. The results showed that inversion of the 3′ UTR increased luciferase activity by ∼3-fold (Fig. 2, bottom). This increase was not observed when cap-dependent translation was hampered by the use of a hairpin or an ApppG cap analog, indicating that sequence elements in the 3′ UTR regulate cap-dependent translation of p27 mRNA.
FIG. 2.
Sequence elements in the 3′ UTR of p27 inhibit cap-dependent translation in HeLa cells. A scheme of the reporter mRNAs used in the experiment is shown at the top. mRNAs were as described in the legend to Fig. 1C and contained the 3′ UTR in either sense (solid line) or inverted (arrow) orientation. Luciferase values were normalized for mRNA levels after transfection. The average of at least three independent experiments is shown.
Expression of p27 is regulated at the translation level during HL60 cell differentiation.
To identify the regulatory elements, we first searched for a cellular model with robust translational control of p27 mRNA between two distinct biological situations, which would allow us later to evaluate the biological significance of p27 translational regulation. We tested HL60 cells, since previous reports had shown that p27 mRNA displays increased association with polysomes when these cells are induced to differentiate to monocyte/macrophage-like cells by the addition of TPA, suggesting increased translation during this process (41). To confirm these observations, we performed pulse-chase experiments and measured both the de novo synthesis and stability of p27, followed by Western and Northern blotting to evaluate the steady-state amounts of p27 protein and transcript. Western blot analysis showed that the amount of p27 increased dramatically during TPA-induced differentiation, especially during the first 24 h of treatment (Fig. 3A, top). This protein accumulation could not be explained by increased p27 mRNA levels (Fig. 3A, bottom). Metabolic labeling with [35S]methionine followed by immunoprecipitation showed that the synthesis of p27 increased 3.5-fold at 24 h (Fig. 3A, middle). In addition, pulse-chase experiments showed that the stability of p27 increased only marginally (∼1.5-fold) in TPA-treated compared to untreated cells (Fig. 3B). These results indicate that translational regulation accounts for most of the increase of endogenous p27 protein levels observed during HL60 cell differentiation.
FIG. 3.
Translation of p27 mRNA increases during HL60 cell differentiation. (A) HL60 cells were treated with 30 nM TPA for the indicated times. The levels of p27 were assessed by Western blotting from 50 μg of total protein extracts using eIF4A as a loading control (top). The amounts of p27 were corrected for the loading control, and the values are shown relative to the amount present in undifferentiated cells. p27 de novo synthesis was determined by metabolic labeling with [35S]methionine for 1 h, followed by immunoprecipitation (middle). After SDS-PAGE, the signal was quantified using a phosphorimager. p27 mRNA levels were evaluated by Northern blotting and corrected for the 18S rRNA loading control, and the values are shown relative to the amount present in undifferentiated cells (bottom). (B) The stability of p27 increases marginally after TPA treatment. The half-life of p27 was determined by pulse-chase experiments. After a labeling period of 1 h, cells were washed and samples were taken at 0, 15, 45, and 120 min. p27 was immunoprecipitated from either 700 μg (untreated) or 200 μg (24 h TPA-treated) of total-protein extract and separated by SDS-PAGE. A representative gel is shown on top. Data from three independent experiments were quantified and plotted as a regression curve (graph).
Roles of p27 during HL60 cell differentiation.
To evaluate the biological significance of p27 translational increase, we depleted p27 by RNA interference and tested the effects of this depletion on the cell cycle, differentiation, and survival of HL60 cells. A homogeneous population of cells expressing an shRNA against the coding sequence of p27 mRNA was obtained by lentiviral infection and FACS selection. As a control, cells infected with viruses containing the empty vector were used. The efficiency of p27 depletion reached ∼80%, as estimated by Western blotting (Fig. 4A). After TPA treatment, HL60 cells exit the cell cycle and accumulate in G0, a step prior to terminal differentiation (48). Analysis of the cell cycle distribution showed that p27-depleted cells exhibited a delay in G0/G1 accumulation (Fig. 4B). This delay was first detected at 24 h of TPA treatment, when the greatest changes in p27 levels were observed (Fig. 3). Accordingly, p27-depleted cells also showed delayed differentiation starting at this time point, as measured by the presence of the monocyte surface marker CD11b (Fig. 4C). In addition, p27-depleted cells showed accumulation of activated caspase 3, indicative of increased apoptosis (Fig. 4D). These results indicate that appropriate p27 levels, which are mostly obtained by translational control, are necessary for full differentiation of HL60 cells, in addition to protecting differentiating cells from apoptosis.
FIG. 4.
p27 is required for terminal differentiation of HL60 cells. (A) Depletion of p27 was achieved by infection of HL60 cells with a lentivirus expressing a specific shRNA against the p27 ORF. Control cells were infected with viruses containing the empty vector. p27 levels were analyzed by Western blotting in p27 knockdown (kd) and control cells at different times of TPA addition. The amounts of p27 relative to control cells are indicated. eIF4A was used as a loading control. (B) Cell cycle withdrawal induced by TPA treatment is delayed in p27 kd cells. Control and p27 kd cells were treated with TPA for the indicated times and stained with propidium iodide. The cell cycle distribution was determined by FACS analysis and is represented as the percentage of cells in G0/G1, S, and G2/M. The average of three independent experiments is shown. (C) Low levels of p27 delay differentiation. Cells were treated with TPA, stained with anti-CD11b antibody (a specific marker for monocytes), and analyzed by FACS. The percentage of CD11b-positive cells at each time point is plotted. The graph shows the average of three independent experiments. (D) p27 promotes cell survival during TPA-induced differentiation. Cell death was determined by detection of active caspase 3 (a cleaved product of 17 kDa) by Western blotting. The signals of two independent experiments were quantified, corrected for the eIF4A loading control, and plotted.
Determinants of p27 mRNA translational control.
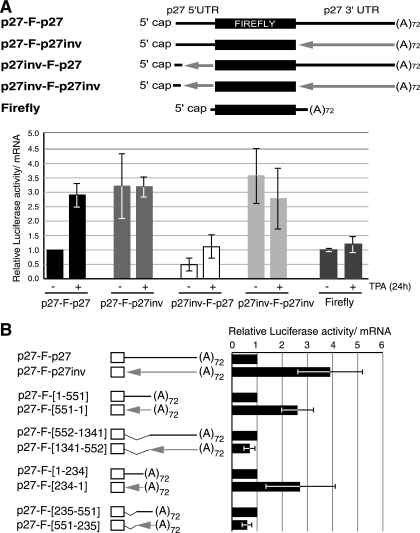
To identify which UTR elements influence p27 mRNA translation in HL60 cells, untreated or 24-h TPA-treated cells were transfected with reporter p27 transcripts containing the 5′ and/or 3′ UTR in wild-type or inverted orientation (Fig. 5A, top). Similar to the endogenous p27 transcript, translation of the wild-type reporter p27 mRNA increased ∼3-fold in differentiated cells, while translation of a reporter lacking p27 sequences remained constant (Fig. 5A, bottom, compare p27-F-p27 with Firefly). Inversion of the 3′ UTR resulted in increased translation in undifferentiated cells, with no further increase during differentiation (Fig. 5A, bottom, p27-F-p27inv). These results indicate that sequence elements in the 3′ UTR repress translation of p27 in undifferentiated cells and that this repression is relieved during differentiation. Inversion of the 5′ UTR reduced translation ∼2-fold in undifferentiated cells and hampered translational activation during differentiation (Fig. 5A, bottom, p27inv-F-p27), indicating that elements in the 5′ UTR are required for translational activation. The fact that inversion of the 5′ UTR has no effect in the context of a reporter containing the inverted 3′ UTR suggests that translational regulation by the 5′ and 3′ UTRs is functionally connected, especially during differentiation (Fig. 5A, bottom, p27inv-F-p27inv).
FIG. 5.
Determinants of p27 translational control in HL60 cells. (A) The 3′ UTR of p27 mRNA represses translation in undifferentiated cells, and full derepression during differentiation requires an intact 5′ UTR. mRNA reporters containing the UTRs of the p27 transcript in either sense or inverted orientation, as depicted, were transfected into untreated (−) or 24-h TPA-treated (+) cells and processed as described in the legend to Fig. 1. The values are represented relative to that obtained for p27-F-p27 in untreated cells. A transcript lacking p27 sequences (firefly luciferase) was used as a control. The average of at least three independent experiments is shown. (B) Mapping the regulatory elements in the p27 3′ UTR. Reporter mRNAs containing the p27 5′ UTR and either the full-length p27 3′ UTR or the indicated fragments were transfected into HL60 cells. In each case, the mRNA with the corresponding 3′-UTR fragment in inverted orientation was used as a control (arrows). The luciferase values were corrected for mRNA levels and compared to the corresponding control. The average of three independent experiments is shown.
Because the 3′ UTR makes the strongest contribution to translational repression of p27 mRNA in undifferentiated cells, we next analyzed which segments of the 3′ UTR carried the repressor function. A set of mRNAs containing different fragments of the 3′ UTR was generated, as well as the corresponding mRNAs with inverted 3′ UTR sequences as controls (Fig. 5B). Undifferentiated HL60 cells were transfected with these transcripts, and their efficiency of translation was analyzed. The results showed that the regulatory sequences required for translational repression in undifferentiated cells are located within the first 234 nt of the 3′ UTR.
miR-181a inhibits cap-dependent translation of p27 mRNA.
Once the regulatory region was mapped, we tried to identify factors that could bind to this region and potentially participate in the translational control of p27 mRNA by UV cross-linking experiments. Because no specific protein cross-links were detected (data not shown), we considered the possibility that an miRNA could be responsible for regulation. In silico searches using the miRanda algorithm (http://microrna.sanger.ac.uk; Wellcome Trust Sanger Institute) yielded target sites for several miRNAs along the p27 3′ UTR. We focused on miR-181a because (i) one of its target sites fell within the mapped regulatory region and (ii) miR-181a is highly expressed in undifferentiated HL60 cells while low levels are observed in primary monocytes (47). Based on these data, we hypothesized that increased translation of p27 during HL60 cell differentiation is accompanied by reduced expression of miR-181a. Northern blot analysis showed that, indeed, miR-181a levels decreased about 40% during TPA treatment; in contrast, the expression of miR-221, known to regulate p27 expression in various cell lines, increased by 60% (data not shown) (31). We therefore set out to investigate whether miR-181a repressed p27 mRNA translation in our cell system.
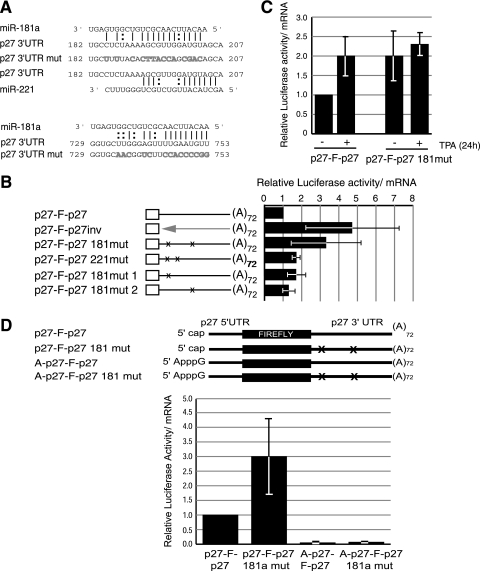
Two miR-181a target sites were predicted in the 3′ UTR of p27 mRNA, located at positions 182 to 204 and 729 to 753 (Fig. 6A). Reporter transcripts were obtained in which both sites were mutated either individually or in combination (the natures of the mutations are indicated in Fig. 6A). Curiously, the first miR-181a binding site overlaps with one of the sites predicted for miR-221, of which there are two in the p27 3′ UTR. To assess the relevance of miR-221 versus miR-181a in our cell system, we also carried transcripts with both miR-221 binding sites mutated. Wild-type or mutated reporter mRNAs were transfected into undifferentiated HL60 cells. As shown in Fig. 6B, translation of the miR-181a mutant mRNA increased ∼3-fold and was comparable to translation of the transcript with the inverted 3′ UTR. However, mutation of the miR-221 sites had a modest effect on translation. These results suggest that translational repression of p27 mRNA in undifferentiated cells is caused by miR-181a. Mutation of a single miR-181a binding site only slightly enhanced translation, suggesting that one site is enough to efficiently repress p27 synthesis, irrespective of its location (Fig. 6B). Upon differentiation, the translation of the reporter containing miR-181a mutated binding sites was not significantly increased, suggesting that regulation by miR-181a accounts for most of the observed changes in translational efficiency of p27 mRNA during differentiation (Fig. 6C).
FIG. 6.
miR-181a regulates p27 mRNA translation. (A) Schematic representation of the two miR-181a target sites in the p27 3′ UTR and the mutations (mut) introduced to evaluate their relevance (gray letters). The pairing of the first site with miR-221 is also shown. Numbers indicate the positions of miR-181a binding sites in the 3′ UTR of p27 mRNA. |, Watson-Crick pairing, :, G:U pairing. (B) miR-181a, but not miR-221, inhibits the translation of p27 mRNA in HL60 cells. The indicated mRNA reporters were transfected into HL60 cells, and their translational efficiencies were measured as usual. The average of at least three independent experiments is shown. The values are plotted relative to that obtained for the wild-type mRNA. ×, mutation of miR-181a binding sites. (C) miR-181a modulates the translation of p27 mRNA during differentiation. TPA-treated or untreated HL60 cells were transfected with wild-type or miR-181a mutated reporter mRNAs and processed as usual. The average of three independent experiments is shown. (D) miR-181a represses cap-dependent translation in HL60 cells. Reporter p27 mRNAs containing either a canonical 7mGpppG or an ApppG cap and wild-type or mutated miR-181a sites were transfected into undifferentiated HL60 cells. Samples were processed as usual, and the values are represented relative to that obtained for the wild-type 7mGpppG-containing RNA. The values represent the average of three independent experiments.
miRNAs have been proposed to inhibit translation initiation at the level of the cap structure, although some authors have reported an effect in subsequent steps of translation (reviewed in reference 18). To determine whether the cap structure was required for repression by miR-181a, we tested the efficiencies of translation of reporter mRNAs containing wild-type or mutated miR-181a binding sites and a canonical or an ApppG cap (Fig. 6D). Substitution of the canonical cap structure for an ApppG cap resulted in a dramatic decrease in the translational efficiencies of all reporters, indicating that, as for HeLa cells, p27 mRNA is translated via a cap-dependent mechanism in HL60 cells. In addition, no significant differences were observed between wild-type and miR-181a mutated mRNAs containing an ApppG cap, while the expected differences were exhibited by 7mGpppG-containing transcripts (Fig. 6D). We conclude that miR-181a inhibits cap-dependent translation of p27 mRNA.
miR-181a inhibits endogenous p27 expression.
To confirm the role of miR-181a in the regulation of p27 mRNA translation, we tried to inhibit the expression of the endogenous miRNA in HL60 cells using a specific 2′-O-methyl-oligoribonucleotide (Ambion Inc.) or custom-designed antagomirs (34). In addition, we obtained homogeneous populations of HL60 cells infected with lentiviruses expressing shRNAs against the miR-181a precursors or the mature miRNA. However, in no case could we reduce the expression or activity of endogenous miR-181a (data not shown). As an alternative approach, we overexpressed miR-181a in the cell line THP1 and tested the effect of this treatment on endogenous p27. THP1 cells represent a more differentiated stage of human monocytic leukemia cells than HL60 cells and express high levels of p27 and low levels of miR-181a (Fig. 7A and B). We expressed miR-181a at levels similar to those present in undifferentiated HL60 cells without affecting the levels of miR-221 (Fig. 7B). Overexpression of miR-181a caused a 5-fold reduction of p27 protein levels, while the amount of p27 mRNA was reduced only ∼1.5-fold (Fig. 7C and D). These results support the conclusion that miR-181a regulates the translation of p27 mRNA in myeloid cells.
FIG. 7.
Overexpression of miR-181a in THP1 cells inhibits the translation of endogenous p27 mRNA. (A) p27 protein levels in HL60 and THP1 cells measured by Western blotting. β-Actin was used as a loading control. (B) Analysis of miR-181a expression. THP1 cells were infected with either empty or primary miR-181a (pri-miR-181a)-expressing retroviruses and selected in neomycin-containing medium. Total RNA was isolated, and the amounts of miR-181a and miR-221 were analyzed by Northern blotting. Untreated HL60 cells were carried as a control. miRNA levels were normalized according to U6 RNA levels, and the resulting values are indicated relative to those found in control THP1 cells. (C) p27 expression decreases in THP1 cells that overexpress miR-181a. Protein levels were analyzed by Western blotting using eIF4E as a loading control. A nonspecific band is also shown as a reference. The p27 values are indicated after correction for eIF4E. (D) Analysis of endogenous p27 mRNA levels in control or miR-181a-overexpressing cells, as determined by qRT-PCR. The average of triplicate RT-PCRs for one biological sample is shown.
DISCUSSION
p27 regulates a variety of cellular processes, such as proliferation, differentiation, migration, and apoptosis, and its expression must be tightly controlled to prevent cell malfunction that could lead to tumor development. In this report, we have investigated the mechanisms that regulate the translation of p27 mRNA. Our results indicate that p27 mRNA is translated in a cap-dependent manner in HeLa and HL60 cells. In addition, we found that miR-181a inhibits cap-dependent translation of p27 mRNA in undifferentiated HL60 cells by virtue of two target sites in the 3′ UTR and that differentiation concurs with relief of miR-181a-mediated translational repression.
Several studies have reported the presence of an IRES in the 5′ UTR of p27 mRNA (8, 30, 35, 44). Our results, like those reported by Liu et al. (39), contradict these findings. First, the use of promoterless DNA bicistronic constructs revealed the presence of cryptic promoters in the sequence encoding the p27 5′ UTR (Fig. 1A). The presence of mRNA species transcribed from these promoters was confirmed by RNase protection assays (data not shown), and their sizes match those transcription initiation sites described in previous studies (11, 29, 43). Second, transfection of bicistronic mRNAs yielded no significant translation of the second cistron (Fig. 1B). Third, transfection of monocistronic mRNAs into HeLa or HL60 cells resulted in activity that was dramatically reduced by manipulation of the cap structure or insertion of a stable hairpin close to the cap, both treatments known to inhibit cap-dependent but not IRES-dependent translation (Fig. 1C and 6D). Although we did not detect IRES activity in the p27 5′ UTR, we did observe a stimulatory role of this UTR in the context of cap-dependent translation (see below).
Many regulatory factors that control the translation of specific mRNAs operate through the 3′ UTR (13). The importance of the 3′ UTR has been highlighted by the discovery of miRNAs and their essential role in the control of many cellular processes (reviewed in reference 6). miRNAs have been shown to act as either tumor suppressors or oncogenes and are better predictors of tumor type and prognosis than mRNAs (36). Recently, miRNAs have appeared as chief regulators of p27 mRNA translation. The first indications that miRNAs could play a role in the expression of p27 were obtained in Drosophila (26). Depletion of Dicer-1, the RNase III enzyme required for miRNA biogenesis, delays G1/S transition of germ line stem cells by increasing the amount of the p27 orthologue Dacapo. Subsequent studies have identified miR-221 and miR-222 as repressors of p27 synthesis in cell lines derived from glioblastoma, melanoma, hepatocarcinoma, papillary thyroid carcinomas, and prostate carcinoma (15, 19, 20, 23, 38, 54). Generally, inhibition of p27 expression by miR-221/miR-222 in these cells leads to increased proliferation and improved colony-forming potential. miR-221 is also expressed in HL60 cells, although it does not seem to control p27 expression in that cell line (Fig. 6). Rather, we have found that another miRNA, miR-181a, represses p27 mRNA translation in undifferentiated HL60 cells (Fig. 6). Furthermore, overexpression of miR-181a in THP1 cells leads to inhibition of endogenous p27 synthesis (Fig. 7). miR-181a is preferentially expressed in hematopoietic tissues and cell lines, where it plays a critical role in the development of B and T cells through regulation of the expression of the key factors Bcl-2, CD69, and TCRα (7, 46, 47). Regarding its function in myeloid differentiation, Debernardi et al. (12) found a strong correlation between the levels of miR-181a and the expression of putative target mRNAs associated with a specific acute myeloid leukemia subtype. These studies showed that in acute myeloid leukemia samples of the M2 subtype, from which HL60 cells are derived, the amount of miR-181a is elevated and some of its putative targets are downregulated, whereas in the more differentiated samples of the M5 subtype, from which THP1 cells were obtained, low levels of miR-181a correlate with upregulation of the same targets. We have found a similar inverse correlation between miR-181a and p27 expression in HL60 and THP1 cells (Fig. 7).
There are two target sites for miR-181a in the p27 3′ UTR that can function individually or synergistically to repress the translation of p27 reporters (Fig. 6B). The first site does not perfectly match the miR-181a seed region, but other features, including increased supplementary pairing, the presence of an adenine at position 1, and the AU-rich flanking regions, compensate for a suboptimal seed sequence (25). This site coincides with one of the two functional miR-221 binding sites in the p27 3′ UTR. However, mutation of the two miR-221 sites had no effect on p27 mRNA translation, suggesting that miR-221 does not regulate p27 expression in HL60 cells (Fig. 6). The distal site (positions 729 to 753) is located in a region for which we did not detect repressor activity in our initial 3′-UTR deletion analyses (Fig. 5B, p27-F-[552-1341] mRNA). This discrepancy could be explained by the sequence context in which these mutations were analyzed, because further analyses using smaller fragments containing this site (p27-F-[552-870] mRNA) indeed revealed translational repressor activity (data not shown).
Repression by miR-181a is relieved during TPA treatment of HL60 cells, contributing to the high p27 levels necessary for full differentiation (Fig. 4 to 6). Other factors, such as regulatory RNA-binding proteins, could collaborate in the translational control of p27 mRNA at this time. The RNA-binding protein Dead-end 1 (Dnd1) interacts with U-rich sequences in the vicinity of the miR-221 target sites and blocks miR-221-dependent repression (32). Therefore, it is conceivable that similar proteins could prevent the interaction of the residual miR-181a with its target sites during differentiation. In such cases, these proteins should bind to sequences different from those recognized by Dnd1, because mutation of these sequences does not affect translation of p27 in our model system (data not shown). The well-known regulators HuR and PTB could be good candidates, since both interact with p27 mRNA, albeit at the 5′ UTR, and have been shown to modulate p27 synthesis (8, 35). Interestingly, translational derepression during HL60 cell differentiation is hampered by the inversion of the p27 5′ UTR (Fig. 5A). Thus, although the effects of HuR and PTB were originally interpreted in the context of IRES-dependent translation, they could play roles in modulating the function of miR-181a. In support of this hypothesis, HuR performs a similar task in hepatocarcinoma cells under certain stress conditions, since it prevents the inhibition of the cationic amino acid transporter 1 (CAT-1) mRNA by miR-122 (4).
Our results, as well as those of other laboratories, highlight the relevance of miRNA-mediated translational control of p27 mRNA. Although miRNAs usually repress translation about twofold, the observation that p27 is haploinsufficient for tumor suppression implies that a reduction of this kind would be enough to promote tumor growth. Learning about the regulation of p27 mRNA translation and transcript stability, two largely unexplored aspects of p27 biology, will expand the therapeutic approaches to fight against cancer.
Acknowledgments
We thank Ramón Eritja for providing antagomirs against miR-181a and Juan Valcárcel, Albert Jordan, Gabriel Gil, Christian Thoma, Didier Trono, Anne E. Willis, Isabel Novoa, Bill Merrick, and Marisa de Andrés for cell lines, vectors, and reagents.
This work was supported by grants 02/028-00 from La Caixa Foundation and 2005SGR00669 from the Department of Universities, Information and Sciences of the Generalitat of Catalunya (DURSI) to F.G. and by grant PI05/2473 from the Spanish Ministry of Health to R.C. R.C. was supported by a Ramón y Cajal contract from the Spanish Ministry of Education and Science.
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Agrawal, D., P. Hauser, F. McPherson, F. Dong, A. Garcia, and W. J. Pledger. 1996. Repression of p27kip1 synthesis by platelet-derived growth factor in BALB/c 3T3 cells. Mol. Cell. Biol. 164327-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besson, A., H. C. Hwang, S. L. Donovan, S. Cicero, M. Gurian-West, D. Johnson, B. E. Clurman, M. A. Dyer, and J. M. Roberts. 2007. Discovery of an oncogenic activity in p27kip1 that causes stem cell expansion and a multiple tumor phenotype. Genes Dev. 211731-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besson, A., S. F. Dowdy, and J. M. Roberts. 2008. Cell cycle regulators and beyond. Dev. Cell 14159-169. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharyya, S. N., R. Habermacher, U. Martine, E. I. Closs, and W. Filipowicz. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 1251111-1124. [DOI] [PubMed] [Google Scholar]
- 5.Borriello, A., V. Cucciolla, A. Oliva, V. Zappia, and F. D. Ragione. 2007. p27kip1 metabolism. A fascinating labyrinth. Cell Cycle 61053-1061. [DOI] [PubMed] [Google Scholar]
- 6.Bushati, N., and S. M. Cohen. 2007. microRNA functions. Annu. Rev. Cell Dev. Biol. 23175-205. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, C. Z., L. Li, H. F. Lodish, and D. P. Bartel. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 30383-86. [DOI] [PubMed] [Google Scholar]
- 8.Cho, S., H. K. Kim, S. H. Back, and S. K. Jang. 2005. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 251283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, I. M., L. Hengst, and J. M. Slingerland. 2008. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat. Rev. 8253-267. [DOI] [PubMed] [Google Scholar]
- 10.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 811991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman, J., M. Hawkinson, R. Miskimins, and W. K. Miskimins. 2001. The major transcription initiation site of the p27kip1 gene is conserved in human and mouse and produces a long 5′ UTR. BMC Mol. Biol. 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debernardi, S., S. Skoulakis, G. Molloy, T. Chaplin, A. Dixon-Mclver, and B. D. Young. 2007. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia 21912-916. [DOI] [PubMed] [Google Scholar]
- 13.De Moor, C. H., H. Meijer, and S. Lissenden. 2005. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 1649-58. [DOI] [PubMed] [Google Scholar]
- 14.Feigenblum, D., and R. J. Schneider. 1996. Cap-binding protein (eukaryotic initiation factor 4E) and 4E-inactivating protein BP-1 independently regulate cap-dependent translation. Mol. Cell. Biol. 165450-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felicetti, F., M. C. Errico, L. Bottero, P. Segnalini, A. Stoppacciaro, M. Biffoni, N. Felli, G. Mattia, M. Petrini, M. P. Colombo, C. Peschle, and A. Care. 2008. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 152745-2754. [DOI] [PubMed] [Google Scholar]
- 16.Fero, M. L., M. Rivikin, M. Tasch, P. Porter, C. E. Carow, E. Firpo, K. Polyak, L. H. Tsai, V. Broudy, R. M. Perlmutter, K. Kaushansky, and J. M. Roberts. 1996. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(kip1)-deficient mice. Cell 85733-744. [DOI] [PubMed] [Google Scholar]
- 17.Fero, M. L., E. Randel, K. E. Gurley, J. M. Roberts, and C. J. Kemp. 1998. The murine gene p27kip1 is haplo-insufficient for tumour suppression. Nature 396177-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filipowicz, W., S. N. Bhattacharyya, and N. Sonenberg. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. 9102-114. [DOI] [PubMed] [Google Scholar]
- 19.Fornari, F., L. Gramantieri, M. Ferracin, A. Veronese, S. Sabbioni, G. A. Calin, G. L. Grazi, C. Giovannini, C. M. Croce, L. Bolondi, and M. Negrini. 2008. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 275651-5661. [DOI] [PubMed] [Google Scholar]
- 20.Gallardi, S., N. Mercatelli, E. Giorda, S. Massalini, G. V. Frajese, S. A. Ciafre, and M. G. Farace. 2007. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27 kip1. J. Biol. Chem. 28223716-23724. [DOI] [PubMed] [Google Scholar]
- 21.Gebauer, F., W. Xu, G. Cooper, and J. Ritcher. 1994. Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO J. 135712-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gebauer, F., D. Corona, T. Preiss, P. Becker, and M. Hentze. 1999. Translational control of dosage compensation in Drosophila by Sex-lethal: Cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A). EMBO J. 186146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillies, J. K., and I. A. J. Lorimer. 2007. Regulation of p27kip1 by miRNA 221/222 in glioblastoma. Cell Cycle 62005-2009. [DOI] [PubMed] [Google Scholar]
- 24.González, T., M. Seoane, P. Caamaño, J. Viñuelas, F. Domínguez, and J. Zalvide. 2003. Inhibition of Cdk4 activity enhances translation of p27kip1 in quiescent Rb-negative cells. J. Biol. Chem. 27812688-12695. [DOI] [PubMed] [Google Scholar]
- 25.Grimson, A., K. K.-H. Farh, W. K. Johnston, P. Garrett-Engele, L. P. Lim, and D. P. Bartel. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 2791-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatfield, S. D., H. R. Shcherbata, K. A. Fischer, K. Nakahara, R. W. Carthew, and H. Ruohola-Baker. 2005. Stem cell division is regulated by the microRNA pathway. Nature 435974-978. [DOI] [PubMed] [Google Scholar]
- 27.Hengst, L., and S. I. Reed. 1996. Translational control of p27kip1 accumulation during the cell cycle. Science 2711861-1864. [DOI] [PubMed] [Google Scholar]
- 28.Hundsdoerfer, P., C. Thoma, and M. W. Hentze. 2005. Eukaryotic initiation factor 4G1 and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. USA 10213421-13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito, E., Y. Iwahashi, Y. Yanagisawa, Y. Suzuki, S. Sugano, Y. Yuasa, and K. Maruyama. 1999. Two short sequences have positive effects on the human p27kip1 gene transcription. Gene 22893-100. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, H., J. Coleman, R. Miskimins, R. Srinivasan, and W. K. Miskimins. 2007. Cap-independent translation through the p27 5′ UTR. Nucleic Acids Res. 354767-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasashima, K., Y. Nakamura, and T. Kozu. 2004. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem. Biophys. Res. Commun. 322403-410. [DOI] [PubMed] [Google Scholar]
- 32.Kedde, M., M. J. Strasser, B. Boldajipour, J. A. F. Oude Vrielink, K. Slanchev, C. le Sage, R. Nagel, P. M. Voorhoeve, J. van Duijse, U. A. Orom, A. H. Lund, A. Perrakis, E. Raz, and R. Agami. 2007. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 1311273-1286. [DOI] [PubMed] [Google Scholar]
- 33.Kiyokawa, H., R. D. Kineman, K. O. Manova-Todorova, V. C. Soares, E. S. Hoffman, M. Ono, D. Khanam, A. C. Hayday, L. A. Frohman, and A. Koff. 1996. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27kip1. Cell 85721-732. [DOI] [PubMed] [Google Scholar]
- 34.Krutzfeld, J., N. Rajewsky, R. Braich, K. G. Rajeev, T. Tuschl, M. Manoharan, and M. Stoffel. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438685-689. [DOI] [PubMed] [Google Scholar]
- 35.Kullmann, M., U. Gopfert, B. Siewe, and L. Hengst. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′ UTR. Genes Dev. 163087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, Y. S., and A. Dutta. 25 September 2008. MicroRNAs in cancer. Annu. Rev. Pathol. doi. 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed]
- 37.Le Sage, C., R. Nagel, and R. Agami. 2007. Diverse ways to control p27kip1 function: miRNAs come into play. Cell Cycle 62742-2749. [DOI] [PubMed] [Google Scholar]
- 38.Le Sage, C., R. Nagel, D. A. Egan, M. Schrier, E. Mesman, A. Mangiola, C. Anile, G. Maira, N. Mercatelli, S. A. Ciafre, M. G. Farace, and R. Agami. 2007. Regulation of the p27kip1 tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 263699-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, Z., Z. Dong, B. Han, Y. Yang, Y. Liu, and J. T. Zhang. 2005. Regulation of expression by promoters versus internal ribosome entry site in the 5′-untranslated sequence of the human cyclin-dependent kinase inhibitor p27kip1. Nucleic Acids Res. 333763-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marash, L., N. Liberman, S. Henis-Korenblit, G. Sivan, E. Reem, O. Elroy-Stein, and A. Kimchi. 2008. DAP5 promotes cap-independent translation of Bcl-2 and Cdk1 to facilitate cell survival during mitosis. Mol. Cell 30447-459. [DOI] [PubMed] [Google Scholar]
- 41.Millard, S. S., J. S. Yan, H. Nguyen, M. Pagano, H. Kiyokawa, and A. Koff. 1997. Enhanced ribosomal association of p27kip1 mRNA is a mechanism contributing to accumulation during growth arrest. J. Biol. Chem. 2727093-7098. [DOI] [PubMed] [Google Scholar]
- 42.Millard, S. S., A. Vidal, M. Markus, and A. Koff. 2000. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell. Biol. 205947-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minami, S., N. Ohtani-Fujita, E. Igata, T. Tamaki, and T. Sakai. 1997. Molecular cloning and characterization of the human p27kip1 gene promoter. FEBS Lett. 4111-6. [DOI] [PubMed] [Google Scholar]
- 44.Miskimins, W. K., G. Wang, M. Hawkinson, and W. K. Miskimins. 2001. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol. Cell. Biol. 214960-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, D. Y. Loh, and K. Nakayama. 1996. Mice lacking p27kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85707-720. [DOI] [PubMed] [Google Scholar]
- 46.Neilson, J. R., G. X. Y. Zheng, C. B. Burge, and P. A. Sharp. 2007. Dynamic regulation of microRNAs expression in ordered stages of cellular development. Genes Dev. 21578-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramkissoon, S. H., L. A. Mainwaring, Y. Ogasawara, K. Keyvanfar, J. P. McCoy, E. M. Sloand, S. Kajigaya, and N. S. Young. 2006. Hematopoietic-specific microRNA expression in human cells. Leuk. Res. 30643-647. [DOI] [PubMed] [Google Scholar]
- 48.Rots, N. Y., A. Iavarone, V. Bromleigh, and L. P. Freedman. 1999. Induced differentiation of U937 cells by 1,25-dihydroxyvitamin D3 involves cell cycle arrest in G1 that is preceded by a transient proliferative burst and an increase in cyclin expression. Blood 932721-2729. [PubMed] [Google Scholar]
- 49.Schneider, R. J., and N. Sonenberg. 2007. Translational control in cancer development and progression, p. 401-431. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Sivan, G., and O. Elroy-Stein. 2008. Regulation of mRNA translation during cellular division. Cell Cycle 7741-744. [DOI] [PubMed] [Google Scholar]
- 51.Slingerland, J., and M. Pagano. 2000. Regulation of the cdk inhibitor p27 and its deregulation in cancer. J. Cell. Physiol. 18310-17. [DOI] [PubMed] [Google Scholar]
- 52.Stoneley, M., F. E. M. Paulin, J. P. C. Le Quesne, S. A. Chappell, and A. E. Willis. 1998. c-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16423-428. [DOI] [PubMed] [Google Scholar]
- 53.Vidal, A., S. S. Millard, J. P. Miller, and A. Koff. 2002. Rho activity can alter the translation of p27 mRNA and is important for Rasv12-induced transformation in a manner dependent on p27 status. J. Biol. Chem. 27716433-16440. [DOI] [PubMed] [Google Scholar]
- 54.Visone, R., L. Russo, P. Pallante, I. De Martino, A. Ferraro, V. Leone, E. Borbone, F. Petrocca, H. Alder, C. M. Croce, and A. Fusco. 2007. MicroRNAs miR-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27kip1 protein levels and cell cycle. Endocr. Relat. Cancer 14791-798. [DOI] [PubMed] [Google Scholar]
- 55.Wiznerowicz, M., and D. Trono. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 778957-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]