Abstract
We previously observed that nitric oxide (NO) exposure increases the stability of mRNAs encoding heme oxygenase 1 (HO-1) and TIEG-1 in human and mouse fibroblasts. Here, we have used microarrays to look broadly for changes in mRNA stability in response to NO treatment. Using human IMR-90 and mouse NIH 3T3 fibroblasts treated with actinomycin D to block de novo transcription, microarray analysis suggested that the stability of the majority of mRNAs was unaffected. Among the mRNAs that were stabilized by NO treatment, seven transcripts were found in both IMR-90 and NIH 3T3 cells (CHIC2, GADD45B, HO-1, PTGS2, RGS2, TIEG, and ID3) and were chosen for further analysis. All seven mRNAs showed at least one hit of a signature motif for the stabilizing RNA-binding protein (RBP) HuR; accordingly, ribonucleoprotein immunoprecipitation analysis revealed that all seven mRNAs associated with HuR. In keeping with a functional role of HuR in the response to NO, a measurable fraction of HuR increased in the cytoplasm following NO treatment. However, among the seven transcripts, only HO-1 mRNA showed a robust increase in the level of its association with HuR following NO treatment. In turn, HO-1 mRNA and protein levels were significantly reduced when HuR levels were silenced in IMR-90 cells, and they were elevated when HuR was overexpressed. In sum, our results indicate that NO stabilizes mRNA subsets in fibroblasts, identify HuR as an RBP implicated in the NO response, reveal that HuR alone is insufficient for stabilizing several mRNAs by NO, and show that HO-1 induction by NO is regulated by HuR.
Nitric oxide (NO) is a diatomic free-radical gas generated by NO synthases with the conversion of the substrate l-arginine to citrulline. Exposure to NO from exogenous sources such as air pollution and tobacco smoke can also occur (4, 21, 44). NO participates in a wide variety of pathological and physiological processes (6, 19, 39). At high concentrations, NO can react with other oxidants to form reactive secondary products such as peroxynitrite and dinitrogen trioxide (12). The ability of these species to oxidize cysteine and tyrosine residues, lipids, and DNA may play a role in the etiology of diseases such as Parkinson's disease, Alzheimer's disease, and cancer (7, 48, 57, 58). At lower concentrations, NO reacts with cellular targets such as the iron present in both heme- and non-heme-containing proteins, zinc finger proteins, and sulfhydryl groups and participates in a large number of normal physiological processes including vasodilation, respiration, and synaptic transmission (20, 36, 43). NO can also affect signaling molecules such as mitogen-activated protein kinases (MAPKs) by directly activating upstream kinases such as p21Ras and regulates gene expression through the action of MAPKs on downstream transcription factors (30, 31, 52).
We and others previously reported that NO induces the expression of heme oxygenase 1 (HO-1), encoded by HMOX1 mRNA (hereafter HO-1 mRNA) (8, 10, 34), a protein involved in the cellular response to numerous damaging agents (55). Our studies showed that NO exposure dramatically increases the half-life of HO-1 mRNA in certain cell types (8, 34, 38). Although the process of mRNA turnover is complex and is not yet fully understood, several critical mechanisms have been described (3, 5, 15, 18, 37, 42, 56). mRNA is usually degraded by the action of several exoribonucleases and endoribonucleases (49). Several mRNA sequences, generally residing in the 3′-untranslated regions (3′UTRs), have been implicated in controlling mRNA stability. Such turnover regulatory sequences include adenosine/uracil-rich elements, GU-rich elements, and various other sequences that modulate mRNA degradation (11, 46, 47, 51, 59).
Two main classes of trans-binding factors interact with RNA turnover elements and influence mRNA half-life: microRNAs (miRNAs) and RNA-binding proteins (RBPs). While information about the role of miRNAs in mRNA decay is still emerging (50), the role of RBPs in this process is understood in significant detail (1, 23, 56). Decay-promoting RBPs include AUF1, KSRP, BRF1, CUG-BP1, and tristetraprolin (1). Stability-promoting RBPs include NF90, αCP1, nucleolin, RNPC1, CUG-BP2, PAIP2, and members of the Hu/elav family (comprising the ubiquitous HuR and the primarily neuronal HuB, HuC, and HuD) (1, 9).
In the present study, we used Affymetrix arrays to examine the regulation of mRNA stability in primary human lung IMR-90 fibroblasts and in mouse NIH 3T3 fibroblasts. This analysis revealed that a small fraction of mRNAs was significantly stabilized following NO treatment; in addition, seven mRNAs shared in both study groups were found to be HuR targets. HuR cytoplasmic abundance increased following NO treatment, but only the HuR interaction with HO-1 mRNA was robustly increased in NO-treated cells. Accordingly, HuR was required for elevating HO-1 mRNA half-life and steady-state levels; in addition, HuR contributed to elevating HO-1 translation and HO-1 protein levels after NO treatment.
MATERIALS AND METHODS
Cell culture and treatment.
IMR-90 and NIH 3T3 cells were acquired from the American Type Culture Collection (Manassas, VA) and were maintained in Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% (vol/vol) bovine calf serum (HyClone, Logan, UT), 50 μg/ml streptomycin and penicillin, and 40 μM glutamine in a 10% CO2 incubator. Both cell lines were routinely passaged every 3 days, and IMR-90 cells were used between passages 7 and 9 for all experiments. All experiments were carried out at 80 to 90% confluence.
IMR-90 and NIH 3T3 cells were seeded at a density of 3 × 105 and 3 × 106 cells, respectively, and allowed to reach 80 to 90% confluence prior to the start of the experiments. IMR-90 and NIH 3T3 cells were treated with 0.5 mM of the NO donor (Z)-1-{N-[3-aminopropyl]-N-[4-(3-aminopropylammonio)butyl]- amino}-diazen-1-ium-1,2-diolate] (SperNO) (Alexis Biochemicals, Carlsbad, CA) for 1 h. The media were then removed and replaced with conditioned media containing the transcriptional inhibitor actinomycin D (Act D) (Sigma, St. Louis, MO). At the times indicated following the addition of Act D, cells were lysed, and RNA was extracted from cells using RNeasy minikits for microarray analysis (Qiagen Inc., Valencia, CA) or Trizol (Invitrogen) for the analysis of individual mRNAs by reverse transcription (RT) followed by real-time, quantitative PCR (qPCR).
For silencing of HuR expression, Oligofectamine (Invitrogen) was used to transfect IMR-90 cells with small interfering RNAs (siRNAs). The control siRNA used was TTCTCCGAACGTGT; siRNAs (20 nM each) targeting HuR consisted of a mixture of AATCTTAAGTTTCGTAAGTTA (HuR U1), TTCGTAAGTTATTTCCTTTAA (HuR U3), and AAGTGCAAAGGGTTTGGCTTT (HuR H4). A plasmid vector used to overexpress HuR (pHuR-TAP) as well as the corresponding control vector (pTAP) were previously reported (28). Plasmids were transfected using Lipofectamine 2000 (Invitrogen).
Microarray analysis.
At the indicated time points, cells were harvested, and total RNA was extracted from either IMR-90 or NIH 3T3 cells. The mRNA expression patterns were studied using human U133 (for IMR-90 cells) and mouse 430A 2.0 (for NIH 3T3 cells) Affymetrix cDNA arrays, according to Affymetrix (Santa Clara, CA) protocols (see Data Sets S1 and S2 in the supplemental material). Samples were processed at the Bauer Center for Genomic Research at Harvard University. Each experiment was repeated in its entirety three times.
DNA-chip analyzer (dChip) software was used to perform the microarray analysis. Microarray data were first normalized using the invariant-set method to allow for comparisons between different arrays. The data were then filtered using the following criteria: (i) variation across samples after pooling replicates 0.5 < standard error of the mean < 10 and (ii) a Wilcoxon signed-rank test for hybridization specificity for all the arrays used.
Analysis of individual mRNAs.
To measure HO-1 mRNA levels by Northern blot analysis, RNA (10-μg aliquots) was separated on 1% agarose gels. RNA was transferred onto nylon membranes by vertical capillary transfer, UV cross-linked, and hybridized with 32P-labeled oligonucleotide probes for HO-1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA using standard methodologies (38). Northern blots were imaged using either film or a PhosphorImager (Storm 840; Amersham Biosciences Inc., Piscataway, NJ). HO-1 mRNA levels were quantified by densitometry and normalized to GAPDH mRNA band intensities to control for loading differences.
To measure the levels of CHIC2, GADD45B, HO-1, PTGS2, RGS2, TIEG, ID3, MKP-1, ATF3, BCL-2, GAPDH, and inducible NO synthase mRNAs as well as those of 18S rRNA, RT reactions were followed by qPCR amplification using specific primer pairs: TCCGATGGGTCCTTACACTC and TAAGGAAGCCAGCCAAGAGA for HO-1, ACTTTGTGGCTGCCTTTGTT and TCAGTCTCCAATGCAAGCAC for CHIC2, TCGGATTTTGCAATTTCTCC and GACTCGTACACCCCCACTGT for GADD45B, TGAGCATCTACGGTTTGCTG and TGCTTGTCTGGAACAACTGC for PTGS2, CAACTGCCCAGAAAAGGGTA and ATGGCAGGTCACAGTCCTTC for RGS2, AAAGTTCCCATCTGAAGGCCCA and GGTTGGAGGTAGAGCAATGTCA for TIEG, GGAGCTTTTGCCACTGACTC and TTCAGGCCACAAGTTCACAG for ID3, CAAGTGCATCTTTGCCTCAA and CCACCCGAGGTACAGACACT for ATF3, ACATCAAGAAGGTGGTGAAGCAGG and CCAGCAAGGATACTGAGAGCAAGAG for GAPDH, ATTTGGGTCGCGGTTCTTG and TGCCTTGACATTCTCGATGGT for UBC, and CCCTATCAACTTTCGATGGTAGTCG and CCAATGGATCCTCGTTAAAG GATTT for 18S. The primer pairs used to amplify MKP-1 and BCL-2 were described previously (24, 26).
To measure the relative mRNA stabilities in either untreated or NO-treated IMR-90 cells, cultures were treated with Act D (2 μg/ml) for the times shown. At subsequent time points after the addition of Act D, mRNA levels were measured by RT-qPCR, normalized to 18S rRNA levels, and plotted on a semilogarithmic scale to calculate the time required for each mRNA to reach one-half of its initial abundance (50%).
Immunofluorescence.
After IMR-90 cells were treated with NO (0.5 mM for 4 h) or CGP74514A (2 μM for 2 h as a positive control for HuR translocation to the cytoplasm) (24), cells were washed with ice-cold phosphate-buffered saline (PBS) and fixed for 10 min using 2% formaldehyde in PBS. Cells were permeabilized using 0.1% Triton X-100 in PBS for 5 min, washed with ice-cold PBS, and blocked with 5% bovine serum albumin in PBS for 1 h at 25°C. Following incubation with anti-HuR antibody (in 5% bovine serum albumin for 16 h at 4°C) and additional washes with ice-cold PBS, the cell preparations were incubated with a secondary antibody (Alexa 488), washed with PBS, and incubated with a solution of DAPI (4′,6′-diamidino-2-phenylindole) for 10 min. After washing thoroughly, the preparations were embedded using Prolong Gold antifade reagent (Invitrogen); 24 h later, photographs were taken using a fluorescence microscope (Zeiss Axiovert 35).
Western blot analysis.
Whole-cell, cytoplasmic, and nuclear extracts were prepared as described previously (26). Proteins were resolved by 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. HO-1 was detected using a polyclonal antibody from Santa Cruz Biotechnology. Monoclonal antibodies recognizing HuR and α-tubulin as well as polyclonal antibodies recognizing hnRNPC1/C2, AUF1, TIA-1, and TIAR were obtained from Santa Cruz Biotechnology; a monoclonal antibody against NF90 was obtained from BD Biosciences; a β-actin antibody was obtained from Abcam. After secondary-antibody incubations, signals were detected by enhanced chemiluminescence (Amersham Biosciences). HuR was also tested by Western blot analysis after immunoprecipitation (IP) with anti-HuR and immunoglobulin G (IgG) antibodies.
Ribonucleoprotein (RNP) IP assays.
For IP of endogenous RNA-protein complexes from whole-cell (1 mg) extracts, reactions were carried out for 2 h at 4°C with protein A-Sepharose beads (Sigma) that had been precoated with 30 μg of either mouse IgG1 (BD Biosciences), goat IgG (Santa Cruz Biotechnology), or antibodies recognizing HuR, TIA-1, TIAR, AUF1, or NF90. Beads were washed with NT2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl2, 1 mM MgCl2, and 0.05% Nonidet P-40), incubated with 20 U of RNase-free DNase I (15 min at 30°C), and further incubated in 100 μl NT2 buffer containing 0.1% SDS and 0.5 mg/ml proteinase K (30 min at 55°C).
The RNA isolated from the IP material was reverse transcribed using random hexamers and SSII reverse transcriptase (Invitrogen). Transcript abundance was assayed by amplification of the cDNA using gene-specific primer pairs and either conventional PCR (22 to 29 cycles, as indicated) or real-time qPCR employing SYBR green PCR master mix (Applied Biosystems). PCR primers for the detection of specific mRNAs are described above.
Biotin pull-down analysis.
For in vitro synthesis of biotinylated transcripts, cDNA from IMR-90 was used as a template for PCRs whereby the T7 RNA polymerase promoter sequence [CCAAGCTTCTAATACGACTCACTATAGGGAGA(T7)] was added to the 5′ end of all fragments. Primers used for the amplification of sequences of the GAPDH 3′UTR were previously described (26). Primers used for the preparation of biotinylated transcripts spanning the HO-1 mRNA (GenBank accession number NM_002133) were as follows: (T7)ATGGAGCGTCCGCAACCCGACA and TCACATGGCATAAAGCCCTACA for the coding region of HO-1 and (T7)ATGCAGGCATGCTGGCTCCCAG and CAGACAATGTTGTTTATTATTTCACAC for the 3′UTR of HO-1. PCR-amplified products were used as templates for the synthesis of the corresponding biotinylated RNAs using T7 RNA polymerase and biotin-CTP. Whole-cell lysates (40 μg per sample) were incubated with 3 μg of purified biotinylated transcripts for 1 h at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal), and bound proteins in the pull-down material were assayed by Western blotting using antibodies recognizing AUF1, HuR, NF90, TIA-1, or TIAR, as described above.
Analysis of translation: polysome gradients and nascent translation assay.
For polysome analysis, 48 h after transfection of control siRNA or HuR siRNA, untreated or NO-treated IMR-90 cells were incubated with 0.1 mg/ml cycloheximide for 10 min. Cytoplasmic extracts (1 mg each) were prepared and fractionated through a linear sucrose gradient (10 to 50% [wt/vol]), as previously reported (26). Ten fractions were collected using a fraction collector (Brandel) and monitored by optical density measurements (A254). The RNA in each fraction was isolated with Trizol LS (Invitrogen). Following RT, qPCR analysis was performed using primer pairs for HO-1 or for GAPDH (above).
The levels of nascent (de novo-translated) HO-1 and GAPDH were measured by incubating IMR-90 cells briefly (20 min) with 1 mCi l-[35S]methionine and l-[35S]cysteine (Easy Tag Express; NEN, Perkin-Elmer, Boston, MA) per 60-mm plate, as described previously (26). Cells were lysed in radioimmunoprecipitation buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS, and 1 mM dithiothreitol), and the IP reactions were carried out using 1 ml TNN buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 5 mM EDTA, 0.5% NP-40) for 16 h at 4°C using anti-HO-1 (Santa Cruz Biotechnology), IgG1 (BD Pharmingen), or anti-GAPDH (Santa Cruz Biotechnology) antibody. Following extensive washes in TNN buffer, the IP samples were resolved by SDS-polyacrylamide gel electrophoresis, transferred onto polyvinylidene difluoride filters, and visualized and quantified using a PhosphorImager apparatus (Molecular Dynamics).
RESULTS
Exposure to NO broadly affects mRNA stability.
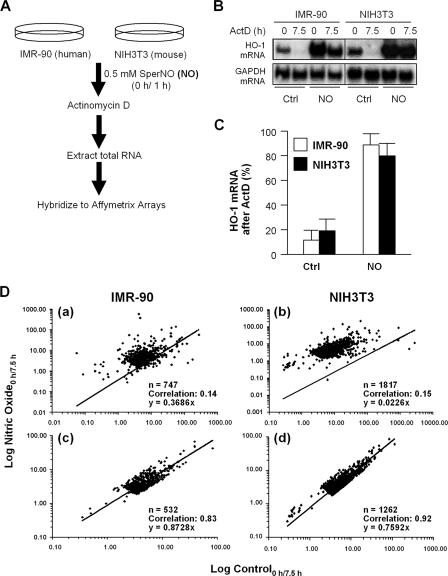
Our laboratory previously reported that exposure to pure NO gas (34) or to NO generated from SperNO (as described in Materials and Methods) increases the half-life of HO-1 mRNA from 2 to ≥10 h in both IMR-90 and NIH 3T3 fibroblasts (8, 32, 34). Using the protocol outlined in Fig. 1A, we monitored the posttranscriptional stabilization of collections of mRNAs using Affymetrix arrays (see Data Sets S1 and S2 in the supplemental material). As shown in Fig. 1B, treatment with NO significantly increased HO-1 mRNA levels in both IMR-90 and NIH 3T3 cells (n = 4; P < 0.01), as expected. That this induction was mediated, at least in part, by increased HO-1 mRNA stability was evidenced by the fact that treatment with Act D (an inhibitor of RNA polymerase II) only modestly reduced HO-1 mRNA levels in NO-treated cells (80 to 90%) compared to untreated cells (10 to 20%) (Fig. 1C).
FIG. 1.
(A) Experimental design. (B) IMR-90 and NIH 3T3 cells were treated with DMEM or DMEM supplemented with 0.5 mM NO for 1 h. HO-1 mRNA levels were determined at 0 and 7.5 h following the addition of Act D using Northern blot analysis. (C) HO-1 mRNA levels were normalized to GAPDH mRNA levels to control for differences in loading and expressed as the percent mRNA remaining at 7.5 h versus 0 h after Act D treatment (n = 4; P < 0.01). Ctrl, control. (D) Population-wide changes in mRNA stability following NO treatment in IMR-90 (a and c) and NIH 3T3 (b and d) cells. The change in mRNA levels between 0 and 7.5 h after Act D treatment for each gene in the filtered gene set was calculated in both untreated and NO-treated cells.
RNA extracted from similarly treated cells was used to hybridize human U133A and mouse 430A 2.0 Affymetrix arrays containing 22,283 and 22,690 probe sets, respectively (representing approximately 14,500 well-characterized genes). The array data were processed as described in Materials and Methods. Because some transcripts (such as HO-1 mRNA) may also be transcriptionally induced by NO exposure, we examined whether treatment with NO caused broad changes in mRNA stability by comparing the change in mRNA levels for each gene in Act D-treated cells using a parameter which we called the “mRNA decay ratio”: log(expression level0 h after Act D/expression level7.5 h after Act D) = remaining mRNA ratio. We then compared the slopes for this value when plotted between control and NO-treated cells for each gene. A slope of 1 would represent no change in stability, as the values in both the numerator and denominator of the equation above would be the same. As shown in Fig. 1Da and b, the slopes of fitted trend lines were 0.37 and 0.02 in IMR-90 and NIH 3T3 cells, respectively. When we removed genes in which we observed at least a threefold increase in the mRNA decay ratio between NO treatment and controls, the slopes increased to 1.00 and 0.74 for IMR-90 and NIH 3T3 cells, respectively (Fig. 1Dc and d). Interestingly, this marked shift was achieved by removing only less than 10% of the total transcripts according to the above-described criterion. These results are consistent with the notion that NO treatment does not cause broad changes in mRNA stability by generally disrupting the cell's mRNA degradation machinery. Moreover, in both IMR-90 and NIH 3T3 cells, at least 70% of the mRNAs in the filtered set had mRNA decay ratios smaller then 2, which indicated minimal changes in mRNA stability between control and NO-treated cells (Fig. 2). This approach therefore provided a useful method to identify a subset of mRNAs that were strongly stabilized in NO-treated cells.
FIG. 2.
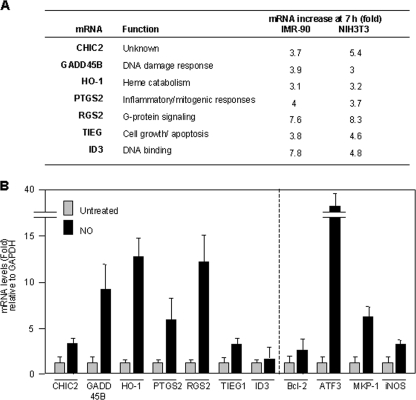
(A) List of mRNAs upregulated in the presence of Act D when IMR-90 and NIH 3T3 cells were treated with NO (0.5 mM for 1 h). Indicated are the functions of the encoded proteins as well as the relative mRNA abundances on microarrays by 7.5 h after Act D treatment in the NO-treated relative to the untreated cell populations. (B) Relative mRNA levels in IMR-90 cells following NO treatment (0.5 mM for 4 h), as calculated by RT followed by qPCR analysis. Data are shown as the means ± standard deviations (SD) from three independent experiments.
As noted above, we previously observed that NO treatment caused both the transcriptional induction and the stabilization of HO-1 mRNA (8). In the present experiments, we found that levels of most mRNAs were not dramatically elevated after NO treatment; instead, their levels were increased 1.1- to 2-fold above the levels in control cells (see Data Sets S1 and S2 in the supplemental material). However, 63 transcripts in IMR-90 fibroblasts (∼8% of array signals) and 191 transcripts in NIH 3T3 fibroblasts (∼9% of array signals) showed >3-fold-higher mRNA levels after the addition of Act D in the NO group than in the untreated group. These findings suggest that a distinct group of mRNAs was stabilized by NO treatment. Through this analysis, we further identified a subset of seven mRNAs that were NO stabilized in both cell types (Fig. 2A). This group includes mRNAs that we previously observed to be stabilized by NO, such as HO-1 and TIEG (8, 38), as well as mRNAs encoding GADD45B and PTGS2 (also known as cyclooxygenase-2), whose stabilities are known to be regulated by genotoxic and inflammatory agents (13, 16, 22).
Validation of microarray results.
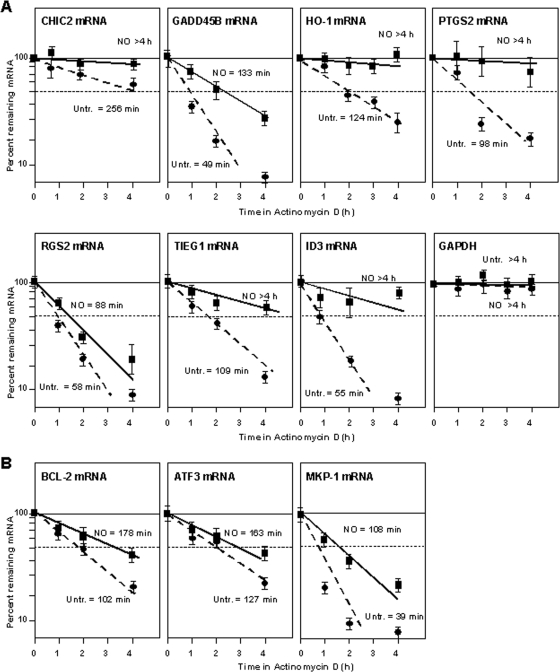
To validate the microarray results, we first quantified differences in the steady-state levels of the above-mentioned seven mRNAs as a function of NO exposure. As shown in Fig. 2B (left), the levels of these mRNAs (except ID3 mRNA) increased after NO treatment, as measured by RT followed by real-time qPCR analysis; additional NO-inducible mRNAs were also included in this analysis (Fig. 2B, right). To study if the NO-triggered elevations were due to increases in mRNA stability, we measured the half-lives of the mRNAs listed in Fig. 2A directly. Following NO treatment, IMR-90 cells were treated with Act D, and the time necessary to reduce a given mRNA to one-half of its initial abundance (its half-life) was measured by RT-qPCR analysis. As shown in Fig. 3A, the half-lives of all seven transcripts (CHIC2, GADD45B, HO-1, PTGS2, RGS2, TIEG1, and ID3 mRNAs) were higher in the NO-treated groups. The half-life of the housekeeping GAPDH mRNA (a highly stable transcript that was included as a negative control in this analysis) showed no difference in stability within the time period studied. Several mRNAs (BCL-2, ATF3, and MKP-1) that showed increased stability in IMR-90 cells were also analyzed, revealing similarly elevated half-lives after NO treatment (Fig. 3B). Collectively, these findings indicate that microarray analysis using Act D is a suitable method to identify mRNA subsets that are stabilized in response to a stimulus such as NO.
FIG. 3.
(A) Stabilities of the mRNAs shown in Fig. 2A. Following NO treatment (0.5 mM for 1 h), IMR-90 cells were incubated with Act D (2 μg/μl) for the times indicated. After RNA extraction and RT, the levels of the indicated mRNAs were measured by RT-qPCR, normalized to 18S rRNA values, and plotted as a function of the levels of that mRNA at time zero. The half-lives of each mRNA were calculated by regression analysis. The data reflect the means ± SD from three independent experiments. (B) mRNA stabilities were calculated as described for A for transcripts that were predicted to be specifically stabilized in IMR-90 cells.
Analysis of HuR in NO-treated cells.
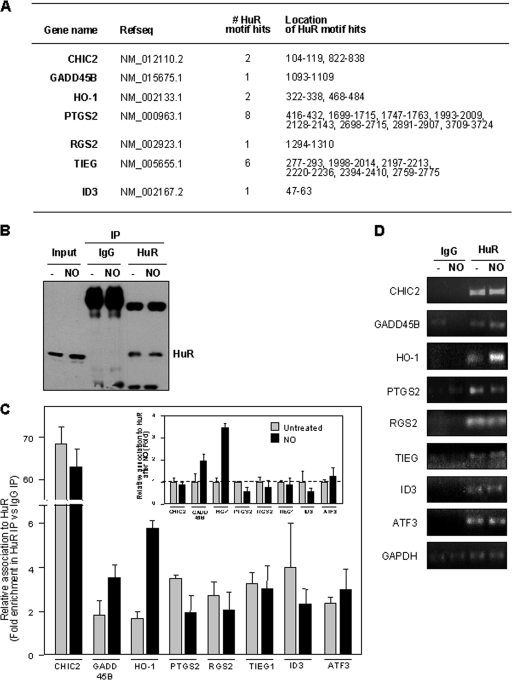
Given the stability-promoting influence of the RNA-binding protein HuR, we postulated that HuR might contribute to the NO-triggered stabilization of at least some mRNAs. A 17- to 20-base-long U-rich signature motif in HuR target mRNAs was previously described (33); this signature motif was used to successfully identify additional HuR target mRNAs in silico. As shown in Fig. 4A, all seven transcripts listed in Fig. 2A contained at least one hit of the HuR motif in their 3′UTRs; in the case of PTGS2 and TIEG, we identified a striking eight and six putative HuR motif hits, respectively.
FIG. 4.
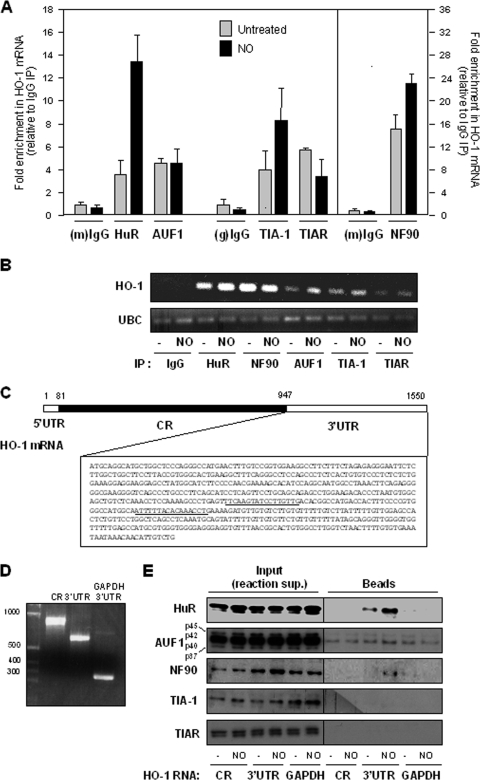
(A) Predicted hits for a signature HuR motif. The numbers and locations within of HuR motif hits within the 3′UTR for each transcripts are indicated. (B) Western blot analysis of HuR levels in lysates from untreated and NO-treated cells before IP (Input) (10 μg per lane) and after IP (from 100-μg aliquots) with IgG and with an anti-HuR antibody (25% of IP material loaded). (C) Relative association of HuR with the transcripts shown in IMR-90 cells that were either left untreated or treated with NO (0.5 mM for 2.5 h). Following HuR IP (and parallel control IgG IP), RNA isolation, RT-qPCR analysis, and normalization to GAPDH mRNA levels in each IP sample, the abundance of each mRNA in HuR IP compared with that in IgG IP was calculated. Data (means ± SD from three independent experiments) are plotted as relative enrichment in HuR IP versus IgG IP (main graph) and also as the levels of enrichment in NO relative to untreated populations (inset graph). (D) The enrichment of each mRNA in each IP sample was also visualized by RT followed by PCR amplification for 22 cycles for all mRNAs except for GAPDH, which was amplified for 28 cycles; PCR products were visualized in ethidium bromide-stained agarose gels.
To test if HuR associated with these mRNAs, we performed RNP IP assays. Following HuR IP and parallel control IgG IP reactions (which showed efficient IP of HuR, a ∼37-kDa RBP) (Fig. 4B), RNA was extracted from the IP materials, and the abundance of each mRNA of interest was measured by RT-qPCR analysis (Fig. 4C). The level of each mRNA in HuR IPs was compared to that in IgG IPs and was represented as the relative association in both IP groups; it was not represented as an absolute “number of transcripts” since RNP IP assays minimize mRNA degradation and rearrangement of mRNAs bound to HuR by reducing the time (2 h) and temperature (4°C) of the IP reaction. Accordingly, only a small fraction of HuR and associated mRNA is immunoprecipitated; for this reason, all of the steps are performed using parallel IgG IP samples, and the final results are shown as enrichment in HuR IP relative to IgG IP. The levels of GAPDH mRNA, a housekeeping mRNA which does not associate with HuR and is a nonspecific background contaminant in all IP samples, were also measured and used to account for any differences in sample input (i.e., it served as a normalization control). As shown, all of the mRNAs were found to be enriched by twofold or higher in the HuR IP, indicating that HuR did associate with this subset of mRNAs (Fig. 4C, main graph).
HuR has been shown to increase binding to some target mRNAs following exposure to various stimuli (54). We thus hypothesized that the abundance of HuR-mRNA complexes would increase following NO treatment. However, against our expectation, NO treatment did not generally lead to an increase in the level of the HuR association with these mRNAs, as shown in the inset graph of Fig. 4C, wherein the same data from the main graph are plotted as the difference in the association of HuR with each mRNA after NO treatment relative to that of untreated cells (the latter being defined as 1). In fact, only the GADD45B and HO-1 mRNAs associated more extensively with HuR following NO treatment, and this effect was most pronounced for HO-1 mRNA (Fig. 4C). The enrichment of each mRNA in each IP sample was also visualized by RT followed by conventional PCR (Fig. 4D; see Fig. S1 in the supplemental material).
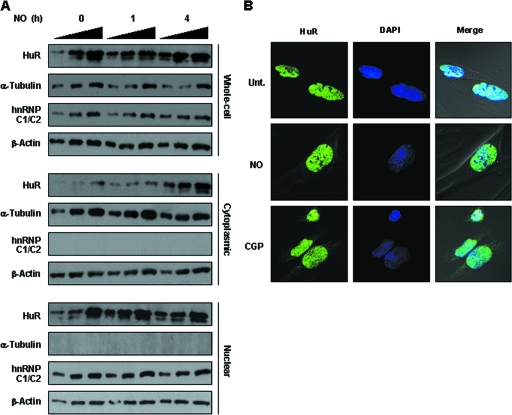
HuR function has been extensively linked to its translocation from the nucleus to the cytoplasm, where HuR is thought to stabilize target mRNAs; in some instances, HuR also modulates their translation (1). As reported previously for other stimuli (stress causing, mitogenic, and inflammatory), NO treatment of IMR-90 led to an increase in the cytoplasmic HuR pool. This increase was first studied by Western blot analysis of HuR abundance in fractionated nuclear and cytoplasmic lysates (Fig. 5A). The levels of α-tubulin and hnRNP C1/C2 were examined to monitor the purity of the sample preparation, while β-actin was included as a loading control. It should be noted that HuR levels in whole-cell lysates were also slightly elevated. Although whole-cell HuR levels are not generally found to increase in response to most treatments, they did increase in response to NO, together with the elevated HuR concentration in the cytoplasm. Nuclear HuR levels were not detectably changed following NO treatment (Fig. 5A and data not shown), as previously reported for a variety of stresses (54). The lack of a concomitant decrease in nuclear HuR levels when the level of cytoplasmic HuR increases is likely a consequence of the far-greater abundance of HuR in the nucleus than in the cytoplasm. Given this difference in relative levels, cytoplasmic HuR levels can increase severalfold without visibly depleting the levels of nuclear HuR. In addition, we cannot exclude the formal possibility that HuR de novo translation, and, hence, cytoplasmic accumulation, increases after NO treatment. Immunofluorescence analysis verified the cytoplasmic increase in HuR levels in IMR-90 cells (Fig. 5B) and NIH 3T3 cells (see Fig. S2 in the supplemental material).
FIG. 5.
(A) IMR-90 cells were treated with NO for the times shown, whereupon whole-cell, cytoplasmic, and nuclear lysates (2.5, 5.0, and 7.5 h) were prepared and the levels of HuR, the cytoplasmic marker α-tubulin, the nuclear marker hnRNP C1/C2, and the loading control β-actin were determined. (B) Immunofluorescence analysis of HuR. HuR signals (green) in either untreated (Unt.) or NO-treated (0.5 mM for 4 h) IMR-90 cells were studied. Green, HuR fluorescence; blue, DAPI staining to visualize nuclei; merge, overlap of the two signals. Treatment of IMR-90 cells with CGP74514A (2 μM for 2 h), which induces HuR translocation to the cytoplasm, was included as a positive control.
Selective influence of HuR on HO-1 expression after NO treatment.
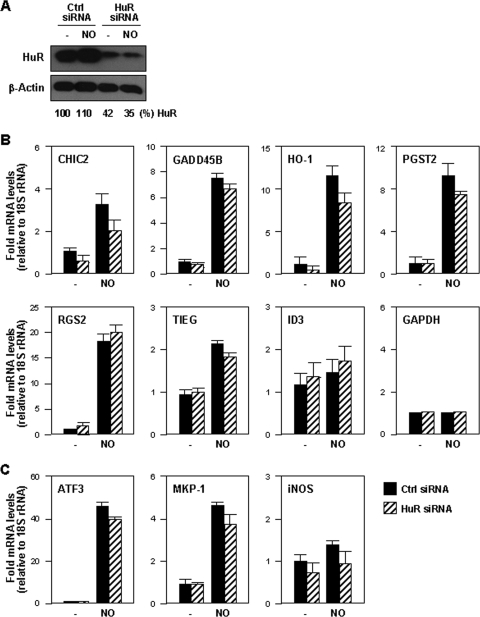
To test if HuR broadly modulated target mRNA expression levels, HuR was silenced in IMR-90 cells using siRNA (Fig. 6A). At 48 h after siRNA transfection, the levels of HuR target mRNAs were measured and normalized to the levels of 18S rRNA. Although HuR silencing lowered the levels of most mRNAs in NO-treated cells (CHIC2, GADD45B, HO-1, PGST2, and TIEG), these reductions were generally small; for ID3 mRNA levels, there was actually a slight increase in HuR-silenced cells. Only levels of CHIC2 and HO-1 mRNAs were substantially reduced (>25%) in the NO-treated group when HuR was silenced (Fig. 6B). These observations are in agreement with the strong association of HuR to CHIC2 and HO-1 mRNAs seen after NO treatment (Fig. 4C). The levels of housekeeping GAPDH mRNA were unchanged by these interventions. Testing of other HuR target mRNAs (ATF3, MKP-1, and inducible NO synthase) revealed various degrees of NO effects and HuR-dependent changes in mRNA levels (Fig. 6C).
FIG. 6.
(A) Western blot analysis of levels of HuR and loading control β-actin by 48 h after transfecting IMR-90 cells with either control siRNA or HuR siRNA. (B and C) The levels of the mRNAs, including the mRNAs listed in Fig. 4A (B), the GAPDH mRNA (B), and HuR target mRNAs (C), were calculated for untreated (−) or NO-treated (0.5 mM for 4 h) IMR-90 cells expressing either normal (control [Ctrl] siRNA) or reduced (HuR siRNA) levels. mRNA data were quantified by RT-qPCR normalized to the levels of 18S rRNA (also measured by RT-qPCR and shown as the means ± SD from three independent experiments).
Of the array-identified, HuR-regulated transcripts (CHIC2 and HO-1), very little is known about CHIC2; therefore, we sought to study the interaction of HuR with HO-1 mRNA in greater detail. Among several RBPs that stabilize target mRNAs, HuR and NF90 showed increased levels of association with HO-1 mRNA following NO treatment, while AUF1 did not show marked differences in binding. These associations were studied by RNP IP analysis followed by measurements of the levels of HO-1 mRNA (as well as housekeeping GAPDH mRNA for sample normalization) using RT-qPCR (Fig. 7A); they were also visualized using RT followed by conventional PCR analysis (Fig. 7B). In response to NO treatment, TIAR and TIA-1 (which repress the translation of some target mRNAs) showed different patterns of association with the HO-1 mRNA (Fig. 7A).
FIG. 7.
(A) Association of HO-1 mRNA with the RBPs shown (HuR, AUF1, TIAR, TIA-1, and NF90) was monitored by RNP IP (Materials and Methods) of IMR-90 cells that were either left untreated or treated with NO (0.5 mM for 2.5 h). Following IP of the RBPs indicated (alongside control mouse [m] or goat [g] IgG IPs), the levels of HO-1 mRNA were normalized to those of housekeeping control GAPDH mRNAs. The data were then plotted as enrichment in RBP IP relative to IgG IP. (B) RNP interactions were studied as described above (A) except that RNA in RNP IP samples was subjected to RT followed by conventional PCR analysis, whereupon PCR products were visualized using ethidium bromide-stained agarose gels. (C) Schematic of HO-1 mRNA depicting the short 5′UTR, the coding region (CR), and the complete 3′UTR sequence; the predicted HuR signature motif hits are underlined. (D) Image of biotinylated transcripts spanning the entire coding region, the entire 3′UTR, and (as a negative control) the GAPDH 3′UTR. (E) Equimolar amounts of the biotinylated transcripts shown in D were incubated with whole-cell lysates prepared from IMR-90 cells that were either left untreated or treated with NO (0.5 mM for 2.5 h). The presence of HuR, AUF1, NF90, TIA-1, and TIAR was then tested by Western blot analysis following pull-down using streptavidin-coated beads (Beads); control aliquots were tested to monitor the presence of other RBPs in the reaction supernatant (Input).
To gain further support for the putative interaction between HuR and HO-1 mRNA (Fig. 4C) and to find out if HuR associated with the HO-1 3′UTR, we analyzed these complexes in vitro using biotinylated transcripts that spanned the HO-1 coding region and 3′UTR (with the latter containing the two predicted HuR motif hits) (Fig. 7C). Incubation of equimolar quantities of the biotinylated transcripts (Fig. 7D) with whole-cell lysates from untreated and NO-treated cells revealed extensive binding by HuR and NF90 but little or no specific binding by the other RBPs; a biotinylated GAPDH 3′UTR transcript was included as a negative control (Fig. 7D and E). While NF90, TIAR, and TIA-1 associated with the endogenous HO-1 mRNA (Fig. 7A), little or no association was seen in the biotin pull-down assay (Fig. 7E). It should be noted that each of these two assays provides distinct information on the interaction on an RBP and a target mRNA and do not always yield identical results. As observed here, TIAR and TIA-1 show associations with HO-1 mRNA by the RNP IP assay but not by the pull-down assay, while NF90 showed a strong interaction by RNP IP analysis but not by the biotin pull-down test. While these discrepancies were not analyzed in depth, TIAR and TIA-1 are not easily studied by the biotin pull-down assay, which typically requires a larger amount of lysate than that used here (our unpublished observations). For NF90, the antibody appears to work efficiently in RNP IP assays (Fig. 7A), while NF90 appears to bind biotinylated RNA weakly in vitro, may actually bind the 5′UTR (which was not included in these experiments), and/or is relatively less efficiently detected by Western blot analysis (Fig. 7E), requiring larger amounts of lysate. Additionally, NF90 is a larger protein (90 kDa) and, thus, is better visualized after Western transfers optimized for larger proteins; here, the Western transfer was optimized for 30- to 50-kDa proteins. In conclusion, both by RNP IP and by biotin pull-down analyses, HuR was found to associate with HO-1 mRNA, and this interaction increased in response to NO treatment.
HuR promotes HO-1 mRNA stabilization and enhances HO-1 translation.
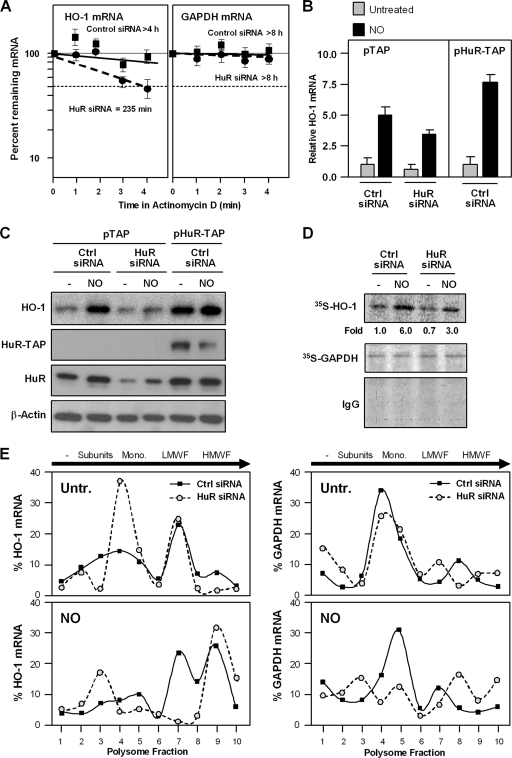
The influence of HuR upon HO-1 expression was further tested by modulating HuR levels and measuring HO-1 mRNA and protein levels. HO-1 mRNA stability was tested after NO treatment of IMR-90 cells that contained normal (control siRNA) or silenced (HuR siRNA) HuR levels. As shown in Fig. 8A, the NO-dependent stabilization of HO-1 mRNA was markedly lower in the HuR-silenced group. Conversely, when HuR was overexpressed as a tandem affinity purification (TAP)-tagged protein, the populations expressing higher HuR levels also expressed higher HO-1 mRNA levels, as measured by RT-qPCR (Fig. 8B). Importantly, the HuR-regulated changes in HO-1 mRNA levels led to changes in HO-1 protein levels, as determined by Western blot analysis (Fig. 8C). HuR silencing reduced HO-1 protein levels, while HuR overexpression enhanced HO-1 protein levels.
FIG. 8.
(A) Half-lives of HO-1 and GAPDH mRNAs were calculated for IMR-90 cells that were transfected with either control siRNA or HuR siRNA by using Act D as described in the legend to Fig. 3A. (B and C) IMR-90 cells were transfected with either control siRNA or HuR siRNA, together with either a control plasmid (pTAP) or a plasmid expressing the HuR-TAP chimeric protein; 48 h later, cells were left untreated or were treated with NO (0.5 mM for 4 h). The levels of HO-1 mRNA and those of normalization control 18S rRNA were measured by RT-qPCR (B), and the levels of HO-1 protein, endogenous and TAP-tagged HuR, and loading control β-actin were assessed by Western blot analysis (C). (D) Nascent HO-1 production was monitored following a brief (20-min-long) incubation of IMR-90 cells with l-[35S]methionine and l-[35S]cysteine after either no treatment (−) or treatment with NO (0.5 mM for 1 h). Following IP using either anti-HO-1 antibody, anti-GAPDH antibody, or control IgG, the incorporation of radiolabeled amino acids into the newly synthesized HO-1 and GAPDH proteins was assessed by electrophoresis through 12% SDS-containing polyacrylamide gels and visualization and quantitation using a PhosphorImager apparatus. (E) The relative association of HO-1 and GAPDH mRNAs with polysomes was tested by preparing cytoplasmic lysates from cells treated as described above (D), fractionating them through sucrose gradients, and collecting 10 fractions for analysis. RNA was extracted from each fraction, and the levels of HO-1 mRNA in each fraction from each population (untreated, top left graph [Untr.]; NO treated, bottom left graph) were measured by RT-qPCR. The levels of GAPDH mRNA were also measured in each fraction and plotted (right graphs). The data shown are representative of three independent experiments. The direction of sedimentation (arrows) as well as the components of the translational machinery in each fraction are indicated: no ribosome components (−), ribosome subunits (Subunits), monosome (Mono.), low-molecular-weight polysome fractions (LMWF), and high-molecular-weight polysome fractions (HMWF).
As the magnitude of NO-triggered changes in HO-1 protein levels appeared to exceed the changes in HO-1 mRNA abundance, we sought to test if NO additionally increased HO-1 translation. To this end, we performed two types of assays. First, we tested the rate of nascent (“de novo”) HO-1 translation by incubating cells briefly (for 20 min) with the 35S-labeled amino acids methionine and cysteine, whereupon cells were lysed and the newly synthesized HO-1 was visualized by IP using anti-HO-1 antibody. The short 35S-amino acid incorporation period ensured that any NO-triggered changes in HO-1 protein stability would be negligible, and hence, the radiolabeled signals would reflect the levels of newly translated HO-1. As shown in Fig. 8D, the level of HO-1 nascent translation increased dramatically after NO treatment, and this increase was dampened when HuR was silenced. The level of translation of a housekeeping protein, GAPDH, was unchanged among these treatment groups, underscoring the specificity of the NO- and HuR-dependent changes in HO-1 translation.
Second, we tested the level of HO-1 translation by comparing the relative association of HO-1 mRNA with actively translating polysomal fractions in untreated and NO-treated cells (Fig. 8E). In untreated cells, HuR silencing shifted the distribution of HO-1 mRNA from the polysomal fractions of the gradient (fractions 6 to 10) toward the left (fractions 1 to 5), where translation was less active. NO treatment moderately shifted the HO-1 mRNA toward the heavier polysomal fractions (fractions 6 to 10), supporting the view that NO promoted HO-1 translation. In HuR-silenced cells, a significant proportion of the HO-1 mRNA shifted toward fractions of lesser or no translation (fractions 1 to 5); the remaining pool of actively translating HO-1 mRNA in the HuR siRNA group (peaking at fraction 9) likely represents cells in which HuR was incompletely silenced, as we never achieved a full depletion of HuR (Fig. 6A). The relative distribution of a housekeeping transcript (GAPDH mRNA) on polysome gradients highlighted the specificity of the changes in HO-1 mRNA levels. The nascent translation and sucrose gradient analyses jointly revealed that the level of HO-1 translation was elevated in IMR-90 cells in an NO- and HuR-dependent manner. Together, our findings show that NO potently induces HO-1 expression through HuR-mediated stabilization of the HO-1 mRNA and HuR-enhanced translation of HO-1.
DISCUSSION
NO exposure stabilizes HO-1 mRNA in IMR-90 and NIH 3T3 cells (8, 10, 32). In the present study, we used Affymetrix gene arrays to investigate if other mRNA species were regulated in a similar way. By measuring mRNA levels in untreated and NO-treated cells in the presence of the transcriptional inhibitor Act D, we identified a fraction of mRNAs that appeared to be stabilized following treatment with NO in both IMR-90 and NIH 3T3 cells. Seven mRNAs that were identified as being stabilized by NO in both IMR-90 and NIH 3T3 cells were studied further; this analysis revealed that six of the seven mRNAs were NO inducible (ID3 mRNA was not inducible by NO), with increased half-lives in response to NO treatment. Moreover, all seven mRNAs were also discovered to bear one or several copies of a signature motif for the mRNA-stabilizing RBP HuR, and these mRNAs were shown to associate with HuR by RNP IP analysis. Against our prediction, however, HuR did not appear to enhance the abundance of all of the transcripts. Only CHIC2 mRNA (encoding a little-known cysteine-rich hydrophobic domain 2 protein) and HO-1 mRNA appeared to be stabilization targets of HuR, as the silencing of HuR decreased the stabilities of CHIC2 and HO-1 mRNAs after NO treatment. In addition, the HO-1 mRNA half-life and levels of HO-1 translation were markedly reduced when HuR was silenced.
It is unclear why the other five HuR-associated mRNAs (GADD45B, HO-1, PTGS2, RGS2, TIEG, and ID3) were unaffected by the silencing of HuR in IMR-90 cells. Two immediate reasons can be proposed to explain this result. First, although HuR silencing was strong, it was not complete (Fig. 6A), as it is notoriously difficult to transfect IMR-90 cells; thus, the possibility remained that the residual HuR still present was sufficient to stabilize these mRNAs. Second, the level of association of HuR with these five mRNAs was unchanged or was lower in the NO-treated groups (Fig. 4C), suggesting that the mRNAs could be the targets of other RBPs acting independently of HuR. This is a likely possibility, since many RBPs that modulate mRNA stability and translation tend to have affinities for similar sequences (U rich or AU rich), and in some instances, they have been shown to compete for binding shared targets (27) or to cooperate in their stabilizing influences (26). While the identification of alternative RBPs involved in NO-triggered mRNA stabilization is beyond the scope of this investigation, the influence of RBPs that promote or prevent mRNA stabilization warrants future analysis. In this regard, decay-promoting RBPs could function by dissociating from these mRNAs in response to NO treatment, as reported previously for GADD45A in cells treated with an alkylating agent (29). Whether miRNAs could mediate the stabilizing effects of NO must also be examined, as miRNAs can also promote mRNA decay (50).
HuR also appeared to promote the translation of HO-1, as HuR silencing dramatically lowered basal and NO-induced HO-1 protein levels (Fig. 8B), while the change in HO-1 mRNA abundance was relatively less pronounced. HuR can enhance the translation of numerous other target mRNAs, including those that encode prothymosin α, MKP-1, hypoxia-inducible factor 1α, and p53 (17, 26, 28, 35). Further analysis of HO-1 translation showed that NO enhanced both the biosynthesis of HO-1 and the association of HO-1 mRNA with the translational apparatus (Fig. 8D and E). These studies further revealed that the level of HO-1 translation was diminished in HuR-depleted cells. Accordingly, HuR promoted HO-1 expression through two posttranscriptional events, mRNA stabilization and translational upregulation; recently, HuR was shown to have a similar dual influence on MKP-1 induction by H2O2 treatment, as it stabilized MKP-1 mRNA and increased MKP-1 translation in response to the oxidant (26). Together with its transcriptional control, HO-1 provides a prominent example of a gene product whose abundance is controlled at various stages. Such multileveled regulation is increasingly being recognized for proteins with important cellular functions, such as HO-1.
In addition to functioning as a signaling molecule, NO causes potent nitrosative and oxidative damage. Thus, following NO exposure, the cell activates a stress response program that includes well-documented changes in transcription (mediated, for example, by AP-1, Sp1, HIF-1, and NF-κB) (25, 40, 41, 45, 53, 60). In addition, there is growing appreciation that NO also triggers changes in mRNA stability. Using microarrays to study gene expression programs in lipopolysaccharide-stimulated human THP-1 monocytes, Wang and colleagues previously demonstrated that NO stabilized a large set of mRNAs through a mechanism that was partly dependent on the activation of the MAPK p38 (52). Those authors also proposed that the p38-regulated interaction of the RBPs HuR and hnRNP A0 with interleukin-8 mRNA contributed to the stabilization of interleukin-8 mRNA in NO-treated THP-1 mRNA. Our findings support the observations reported previously by Wang and coworkers but reveal that an interaction with an mRNA does not necessarily result in altered expression, as the levels of HuR target GADD45B, PTGS2, RGS2, TIEG, and ID3 mRNAs were essentially unaltered after silencing HuR. While the analysis of HuR was restricted to mRNAs stabilized in both human and mouse fibroblasts, numerous other mRNAs in each cell population are reported or predicted HuR targets. Future experiments must test these NO-inducible mRNAs systematically, since the two cell systems (IMR-90 and NIH 3T3) differ in many important respects.
As these studies progress, it will also be important to elucidate the NO-triggered events that elevate whole-cell and cytoplasmic HuR concentrations. In this regard, the cytoplasmic abundance of HuR has been linked to HuR phosphorylation by protein kinase C or Cdk1 in HuR's translocation domain (14, 24). Studies are also warranted to assess if NO influences protein kinase C and/or Cdk1 activities in fibroblasts and thereby modulates HuR cytoplasmic localization. Similarly, it will be important to determine if HuR function following NO exposure is regulated by the checkpoint kinase Chk2. Phosphorylation of HuR by Chk2 in response to another oxidant, hydrogen peroxide, modulated HuR binding to SIRT1 mRNA and other transcripts in human diploid fibroblasts (2). As Chk2 phosphorylation of HuR (at S88, S100, and T118) promoted HuR binding to some target mRNAs but reduced the binding to other target mRNAs, it will be interesting to examine if Chk2 action is also responsible for the differential effects of HuR upon the various NO-stabilized mRNAs. The answers to these questions will provide important insight into the posttranscriptional processes that regulate gene expression in cells confronting the challenge of toxic NO exposure. Such information would also allow the identification of targets for interventions aimed at modifying the cellular response to NO.
Supplementary Material
Acknowledgments
We acknowledge Jennifer Couget and Shufen Meng at the Bauer Center for Genomic Research at Harvard for assistance with the Affymetrix microarray analysis. We also thank Cheng Li and Xin Lu of the Department of Biostatistics (Harvard School of Public Health) for help with the statistical analysis of the microarray results. We thank Ming Zhan (NIA-IRP) for assistance with HuR motif identification.
Experimental work at Harvard was supported by NCI grant R01-CA082737 (to B.D.). A.R. was supported by radiation biology training grant NCI T32-CA009078. M.G., S.S., and Y.K. were supported by the National Institute on Aging-Intramural Research Program, National Institutes of Health.
Footnotes
Published ahead of print on 16 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdelmohsen, K., Y. Kuwano, H. H. Kim, and M. Gorospe. 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 389243-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdelmohsen, K., R. Pullmann, Jr., A. Lal, H. H. Kim, S. Galban, X. Yang, J. D. Blethrow, M. Walker, J. Shubert, D. A. Gillespie, H. Furneaux, and M. Gorospe. 2007. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25543-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abler, M. L., and P. J. Green. 1996. Control of mRNA stability in higher plants. Plant Mol. Biol. 3263-78. [DOI] [PubMed] [Google Scholar]
- 4.Arenillas, A., F. Rubiera, and J. J. Pis. 2002. Nitric oxide reduction in coal combustion: role of char surface complexes in heterogeneous reactions. Environ. Sci. Technol. 365498-5503. [DOI] [PubMed] [Google Scholar]
- 5.Beelman, C. A., and R. Parker. 1995. Degradation of mRNA in eukaryotes. Cell 81179-183. [DOI] [PubMed] [Google Scholar]
- 6.Bentz, B. G., R. L. Simmons, G. K. Haines III, and J. A. Radosevich. 2000. The yin and yang of nitric oxide: reflections on the physiology and pathophysiology of NO. Head Neck 2271-83. [DOI] [PubMed] [Google Scholar]
- 7.Boje, K. M. 2004. Nitric oxide neurotoxicity in neurodegenerative diseases. Front. Biosci. 9763-776. [DOI] [PubMed] [Google Scholar]
- 8.Bouton, C., and B. Demple. 2000. Nitric oxide-inducible expression of heme oxygenase-1 in human cells. Translation-independent stabilization of the mRNA and evidence for direct action of nitric oxide. J. Biol. Chem. 27532688-32693. [DOI] [PubMed] [Google Scholar]
- 9.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, K., and M. D. Maines. 2000. Nitric oxide induces heme oxygenase-1 via mitogen-activated protein kinases ERK and p38. Cell. Mol. Biol. (Noisy-le-Grand) 46609-617. [PubMed] [Google Scholar]
- 11.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20465-470. [DOI] [PubMed] [Google Scholar]
- 12.Davis, K. L., E. Martin, I. V. Turko, and F. Murad. 2001. Novel effects of nitric oxide. Annu. Rev. Pharmacol. Toxicol. 41203-236. [DOI] [PubMed] [Google Scholar]
- 13.Dixon, D. A., N. D. Tolley, P. H. King, L. B. Nabors, T. M. McIntyre, G. A. Zimmerman, and S. M. Prescott. 2001. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Investig. 1081657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doller, A., A. Huwiler, R. Müller, H. H. Radeke, J. Pfeilschifter, and W. Eberhardt. 2007. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell 182137-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehretsmann, C. P., A. J. Carpousis, and H. M. Krisch. 1992. mRNA degradation in procaryotes. FASEB J. 63186-3192. [DOI] [PubMed] [Google Scholar]
- 16.Faour, W. H., A. Mancini, Q. W. He, and J. A. Di Battista. 2003. T-cell-derived interleukin-17 regulates the level and stability of cyclooxygenase-2 (COX-2) mRNA through restricted activation of the p38 mitogen-activated protein kinase cascade: role of distal sequences in the 3′-untranslated region of COX-2 mRNA. J. Biol. Chem. 27826897-26907. [DOI] [PubMed] [Google Scholar]
- 17.Galbán, S., Y. Kuwano, R. Pullmann, Jr., J. L. Martindale, H. H. Kim, A. Lal, K. Abdelmohsen, X. Yang, Y. Dang, J. O. Liu, S. M. Lewis, M. Holcik, and M. Gorospe. 2008. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1α. Mol. Cell. Biol. 2893-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guhaniyogi, J., and G. Brewer. 2001. Regulation of mRNA stability in mammalian cells. Gene 26511-23. [DOI] [PubMed] [Google Scholar]
- 19.Ignarro, L. J. 1996. Physiology and pathophysiology of nitric oxide. Kidney Int. Suppl. 55S2-S5. [PubMed] [Google Scholar]
- 20.Ignarro, L. J., G. Cirino, A. Casini, and C. Napoli. 1999. Nitric oxide as a signaling molecule in the vascular system: an overview. J. Cardiovasc. Pharmacol. 34879-886. [DOI] [PubMed] [Google Scholar]
- 21.Im, H., F. Rasouli, and M. Hajaligol. 2003. Formation of nitric oxide during tobacco oxidation. J. Agric. Food Chem. 517366-7372. [DOI] [PubMed] [Google Scholar]
- 22.Jackman, J., I. Alamo, Jr., and A. J. Fornace, Jr. 1994. Genotoxic stress confers preferential and coordinate messenger RNA stability on the five gadd genes. Cancer Res. 545656-5662. [PubMed] [Google Scholar]
- 23.Keene, J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8533-543. [DOI] [PubMed] [Google Scholar]
- 24.Kim, H. H., K. Abdelmohsen, A. Lal, R. Pullmann, Jr., X. Yang, S. Galban, S. Srikantan, J. L. Martindale, J. Blethrow, K. M. Shokat, and M. Gorospe. 2008. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 221804-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura, H., A. Weisz, Y. Kurashima, K. Hashimoto, T. Ogura, F. D'Acquisto, R. Addeo, M. Makuuchi, and H. Esumi. 2000. Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood 95189-197. [PubMed] [Google Scholar]
- 26.Kuwano, Y., H. H. Kim, K. Abdelmohsen, R. Pullmann, Jr., J. L. Martindale, X. Yang, and Gorospe, M. 2008. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell. Biol. 284562-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lal, A., K. Mazan-Mamczarz, T. Kawai, X. Yang, J. L. Martindale, and M. Gorospe. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 233092-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lal, A., T. Kawai, X. Yang, K. Mazan-Mamczarz, and M. Gorospe. 2005. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 241852-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lal, A., K. Abdelmohsen, R. Pullmann, Jr., T. Kawai, S. Galban, X. Yang, G. Brewer, and M. Gorospe. 2006. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol. Cell 22117-128. [DOI] [PubMed] [Google Scholar]
- 30.Lander, H. M., J. M. Tauras, J. S. Ogiste, O. Hori, R. A. Moss, and A. M. Schmidt. 1997. Activation of the receptor for advanced glycation end products triggers a p21(ras)-dependent mitogen-activated protein kinase pathway regulated by oxidant stress. J. Biol. Chem. 27217810-17814. [DOI] [PubMed] [Google Scholar]
- 31.Lander, H. M., D. P. Hajjar, B. L. Hempstead, U. A. Mirza, B. T. Chait, S. Campbell, and L. A. Quilliam. 1997. A molecular redox switch on p21(ras). Structural basis for the nitric oxide-p21(ras) interaction. J. Biol. Chem. 2724323-4326. [DOI] [PubMed] [Google Scholar]
- 32.Leautaud, V., and B. Demple. 2007. Regulation of heme oxygenase-1 mRNA deadenylation and turnover in NIH3T3 cells by nitrosative or alkylation stress. BMC Mol. Biol. 8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López de Silanes, I., M. Zhan, A. Lal, X. Yang, and M. Gorospe. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 1012987-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marquis, J. C., and B. Demple. 1998. Complex genetic response of human cells to sublethal levels of pure nitric oxide. Cancer Res. 583435-3440. [PubMed] [Google Scholar]
- 35.Mazan-Mamczarz, K., S. Galbán, I. López de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 1008354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMahon, T. J., R. E. Moon, B. P. Luschinger, M. S. Carraway, A. E. Stone, B. W. Stolp, A. J. Gow, J. R. Pawloski, P. Watke, D. J. Singel, C. A. Piantadosi, and J. S. Stamler. 2002. Nitric oxide in the human respiratory cycle. Nat. Med. 8711-717. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell, P., and D. Tollervey. 2000. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10193-198. [DOI] [PubMed] [Google Scholar]
- 38.Mitsumoto, M., A. Mitsumoto, and B. Demple. 2003. Nitric oxide-mediated upregulation of the TGF-beta-inducible early response gene-1 (TIEG1) in human fibroblasts by mRNA stabilization independent of TGF-beta. Free Radic. Biol. Med. 341607-1613. [DOI] [PubMed] [Google Scholar]
- 39.Moncada, S. 2000. Nitric oxide and cell respiration: physiology and pathology. Verh. K. Acad. Geneeskd. Belg. 62171-179, 179-181. [PubMed] [Google Scholar]
- 40.Pilz, R. B., M. Suhasini, S. Idriss, J. L. Meinkoth, and G. R. Boss. 1995. Nitric oxide and cGMP analogs activate transcription from AP-1-responsive promoters in mammalian cells. FASEB J. 9552-558. [DOI] [PubMed] [Google Scholar]
- 41.Ptasinska, A., S. Wang, J. Zhang, R. A. Wesley, and R. L. Danner. 2007. Nitric oxide activation of peroxisome proliferator-activated receptor through a p38 MAPK signaling pathway. FASEB J. 21950-961. [DOI] [PubMed] [Google Scholar]
- 42.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuman, E. M., and D. V. Madison. 1991. A requirement for the intercellular messenger nitric oxide in long-term potentiation. Science 2541503-1506. [DOI] [PubMed] [Google Scholar]
- 44.Schwarzbach, E. 1971. Formation of ozone and nitric oxide caused by welding. Zentralbl. Arbeitsmed. 21290-292. [PubMed] [Google Scholar]
- 45.Spiecker, M., H. B. Peng, and J. K. Liao. 1997. Inhibition of endothelial vascular cell adhesion molecule-1 expression by nitric oxide involves the induction and nuclear translocation of I B. J. Biol. Chem. 27230969-30974. [DOI] [PubMed] [Google Scholar]
- 46.Theil, E. C. 1993. The IRE (iron regulatory element) family: structures which regulate mRNA translation or stability. Biofactors 487-93. [PubMed] [Google Scholar]
- 47.Thomson, A. M., J. T. Rogers, and P. J. Leedman. 1999. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int. J. Biochem. Cell Biol. 311139-1152. [DOI] [PubMed] [Google Scholar]
- 48.Tieu, K., H. Ischiropoulos, and S. Przedborski. 2003. Nitric oxide and reactive oxygen species in Parkinson's disease. IUBMB Life 55329-335. [DOI] [PubMed] [Google Scholar]
- 49.Tourriere, H., K. Chebli, and J. Tazi. 2002. mRNA degradation machines in eukaryotic cells. Biochimie 84821-837. [DOI] [PubMed] [Google Scholar]
- 50.Valencia-Sanchez, M. A., J. Liu, G. J. Hannon, and R. Parker. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20515-524. [DOI] [PubMed] [Google Scholar]
- 51.Vlasova, I. A., N. M. Tahoe, D. Fan, O. Larsson, B. Rattenbacher, J. R. Sternjohn, J. Vasdewani, G. Karypis, C. S. Reilly, P. B. Bitterman, and P. R. Bohjanen. 2008. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell 29263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, S., J. Zhang, Y. Zhang, S. Kern, and R. L. Danner. 2008. AU-rich element-dependent and -independent mechanisms. Nitric oxide-p38 MAPK signaling stabilizes mRNA through AU-rich element-dependent and -independent mechanisms. J. Leukoc. Biol. 83982-990. [DOI] [PubMed] [Google Scholar]
- 53.Wang, S., W. Wang, R. A. Wesley, and R. L. Danner. 1999. A Sp1 binding site of the tumor necrosis factor promoter functions as a nitric oxide response element. J. Biol. Chem. 27433190-33193. [DOI] [PubMed] [Google Scholar]
- 54.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. J. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilks, A. 2002. Heme oxygenase: evolution, structure, and mechanism. Antioxid. Redox Signal. 4603-614. [DOI] [PubMed] [Google Scholar]
- 56.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2237-246. [DOI] [PubMed] [Google Scholar]
- 57.Wink, D. A., Y. Vodovotz, J. Laval, F. Laval, M. W. Dewhirst, and J. B. Mitchell. 1998. The multifaceted roles of nitric oxide in cancer. Carcinogenesis 19711-721. [DOI] [PubMed] [Google Scholar]
- 58.Xu, W., L. Z. Liu, M. Loizidou, M. Ahmed, and I. G. Charles. 2002. The role of nitric oxide in cancer. Cell Res. 12311-320. [DOI] [PubMed] [Google Scholar]
- 59.Yang, E., E. van Nimwegen, M. Zavolan, N. Rajewsky, M. Schroeder, M. Magnasco, and J. E. Darnell, Jr. 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 131863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, J., S. Wang, R. A. Wesley, R. L. Danner. 2003. Adjacent sequence controls the response polarity of nitric oxide-sensitive Sp factor binding sites. J. Biol. Chem. 27829192-29200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.