Sp1 is a ubiquitously expressed, prototypic C2H2-type zinc finger-containing DNA binding protein that can activate or repress transcription in response to physiologic and pathological stimuli. It was originally found to selectively transactivate the early and late simian virus 40 promoters without influencing numerous other promoters (16) and has since been shown to regulate the expression of thousands of genes implicated in the control of a diverse array of cellular processes, such as cell growth (26, 57), differentiation (48), apoptosis (26), angiogenesis (43), and immune response (25), to name just a few. Sp1 is a 785-amino-acid, 100- to 110-kDa nuclear transcription factor which regulates gene expression via multiple mechanisms. It binds GC-rich motifs (such as 5′-G/T-GGGCGG-G/A-G/A-C/T-3′ or 5′-G/T-G/A-GGCG-G/T-G/A-G/A-C/T-3′) with high affinity (4, 27, 28) and can regulate the expression of TATA-containing and TATA-less genes via protein-protein interactions or interplay with other transcription factors (47), such as Ets-1 (56), c-myc (51), c-Jun (44), Stat1 (6), and Egr-1 (31), and/or components of the basal transcriptional machinery. Sp1 has been linked to chromatin remodeling through interactions with chromatin-modifying factors such as p300 (62) and histone deacetylases (HDACs) (71).
Sp1 was once thought to serve mainly as a constitutive activator of housekeeping genes. However, growing evidence indicates that phosphorylation, acetylation, sumoylation, ubiquitylation, and glycosylation are among the posttranslational modifications that can influence the transcriptional activity and stability of Sp1. Here we will discuss recent developments in our understanding of the role of posttranslational modifications influencing Sp1-dependent transcription, focusing mainly on phosphorylation.
PHOSPHORYLATION OF Sp1
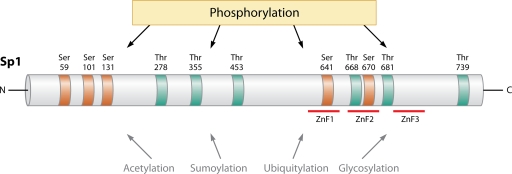
It is now clear that the phosphorylation state of Sp1 influences its transcriptional activity (Table 1). For example, Sp1 phosphorylation is linked with shear stress induction of the tissue factor promoter (39), hepatocyte growth factor-inducible vascular endothelial growth factor (VEGF)/vascular permeability factor gene transcription (46), epidermal growth factor (EGF)-inducible apolipoprotein A-I expression (73), and T-cell receptor-inducible interleukin-21 receptor gene expression (68). Sp1 is phosphorylated by various kinases at different sites within the protein (Table 2), and the influence of these modifications on the function of the transcription factor is only beginning to be understood. Kinases regulating Sp1 binding or transactivity include cyclin-dependent kinase (CDK) (17), atypical protein kinase C-ζ (PKC-ζ) (30, 52, 63), extracellular signal-regulated kinase (ERK) (3, 46), casein kinase II (CKII) (1), and DNA-dependent protein kinase (11). Human Sp1 (NCBI accession number P08047) has 61 putative phosphorylation sites, with 48 of these residues being Ser, 10 Thr, and 3 Tyr (from NetPhos 2.0; data not shown). Sp1 phosphorylation can both positively and negatively influence DNA binding and transcriptional activity. A survey of phosphorylated residues in Sp1 regulating its activity is illustrated in Fig. 1 and discussed below.
TABLE 1.
Agonists, kinases, and Sp1 phosphorylationa
| Protein | Cell type | Stimulus | Kinase(s) | DNA bindingb | Transcriptional changeb | Reference |
|---|---|---|---|---|---|---|
| PDGFR-α | SMCs | FGF-2 | ERK | ↑ | ↓ | 3 |
| PDGF-B | SMCs | NOG | PKC-ζ | ↑ | ↑ | 52 |
| PDGF-D | SMCs | Ang II | PKC-ζ | ↑ | ↑ | 63 |
| FasL | SMCs | CAM | PKC-ζ | ↑ | ↑ | 30 |
| MMP1 | Rat endothelial cells | Shear stress | PKC-ζ | ↑ | ↓ | 33 |
| Nitric oxide synthase | HUVEC | Lysophosphatidylcholine | PP2A | ↑ | ↑ | 13 |
| VEGF | Fibroblasts | Neu differentiation factor | p42/p44 MAPK | ↑ | ↑ | 46 |
| Tyrosine kinase | Fibroblasts | Cyclin A | CDK-2 | ↑ | ↑ | 18 |
| DHFR | Fibroblasts | Cyclin A | CDK-2 | ↑ | ↑ | 17 |
| Gastrin | Gastric carcinoma cells | EGF | ERK2 | ↑ | ↑ | 12 |
| VEGF | Keratinocytes | HGF | PI3K, MEK1/2, PKC-ζ | ↑ | 54 | |
| Cyclin D3 | Megakaryocytes | Thrombopoietin | PP1 | ↓ | ↓ | 67 |
| HIV-1 LTR | T lymphocytes | CD2/CD28 | PP2A | ↓ | ↓ | 35 |
| Aldolase | Hepatoma cells | Glucose | PP1 | ↓ | ↓ | 58 |
| Pyruvate kinase | Hepatoma cells | Glucose | PP1 | ↓ | ↓ | 58 |
| Acetyl-coenzyme A carboxylase | Proadipocytes | Glucose | PP1 | ↓ | ↓ | 14 |
| MMP2 | Human lung cancer | NSAID | ERK | ↑ | ↑ | 50 |
| α-ENaC2 | Lung epithelial cells | Thrombopoietin | PP1 | ↑ | ↑ | 9 |
| LHR | Choriocarcinoma and breast cancer cells | TSA | PKC-ζ | ↑ | ↑ | 69 |
| HO-1 | Human embryonic kidney | NGF | PI3K, PKC-ζ, MEK, ERK | ↑ | ↑ | 55 |
Abbreviations: CAM, calmodulin; NSAID, nonsteroidal anti-inflammatory drug; NOG, nogalamycin; NGF, nerve growth factor; LTR, long terminal repeat; HUVEC, human umbilical vein endothelial cells; DHFR, dihydrofolate reductase; α-ENaC2, α Na+ channel 2; MMP1, matrix metalloproteinase 1; HGF, hepatocyte growth factor.
↑, increase; ↓, decrease.
TABLE 2.
Residues phosphorylated in Sp1
| Region in Sp1 | Residue | Reference(s) |
|---|---|---|
| N-terminal activation domain | Ser59 | 17,65 |
| N-terminal activation domain | Ser101 | 22,47a |
| N-terminal activation domain | Ser131 | 11 |
| N-terminal activation domain | Thr278 | 10 |
| N-terminal activation domain | Thr355 | 73 |
| N-terminal activation domain | Thr453 | 3,46 |
| High-charge-density region/zinc finger 1 | Ser641 | 69 |
| Zinc finger 2 | Thr668 | 1,63 |
| Zinc finger 2 | Ser670 | 63 |
| Zinc finger 2 (just outside) | Thr681 | 63,65 |
| C-terminal D domain | Thr739 | 3,10,46 |
FIG. 1.
Published Ser and Thr phosphorylation sites in Sp1. Ser and Thr residues are indicated in red and blue, respectively. ZnF, zinc finger domain. Acetylation, sumoylation, ubiquitylation, and glycosylation are among other posttranslational modifications that influence the transcriptional activity and stability of Sp1. See text for details.
The influence of Sp1 phosphorylation on altered gene expression was first observed in the context of human immunodeficiency virus (HIV). HIV type 1 (HIV-1) Tat and Sp1 form a tight protein-protein complex compared with other transcription factors such as Oct and NF-κB (24). This protein-protein interaction was found in a subsequent study by the same group to be mediated via Tat phosphorylation of Sp1 at Ser131 in Sp1 and amino acids 30 to 55 in Tat (11). Phosphorylation of Sp1 is dependent on DNA-dependent protein kinase binding Tat. HIV-1 long terminal repeat expression was limited by Sp1 phosphorylation and not by Sp1 total amount, indicating the importance of Sp1 modification as a trigger of inducible gene expression.
KINASES PHOSPHORYLATING Sp1 INFLUENCE ITS DNA BINDING AND TRANSCRIPTIONAL ACTIVITY
Fojas de Borja et al. showed that, in cyclin A-overexpressing NIH 3T3 cells, Sp1 is phosphorylated by cyclin A-dependent kinase at (murine) Ser61 (corresponding to Ser59 in human Sp1) (17). Moreover, these investigators found that cyclin A forms a complex with CDK, which phosphorylates Sp1 at its N terminus and increases its interaction with the dihydrofolate reductase promoter. Increased DNA binding by Sp1 was independent of changes in total levels of Sp1 (17).
Studies by Milanini-Mongiat and colleagues showed that p42/p44 mitogen-activated protein kinase (MAPK) phosphorylates Sp1 at two Thr residues: Thr453 and Thr739 (46). When these sites were mutated to Ala, the MAPK-dependent transcriptional activity of Sp1 in the context of the VEGF promoter, in Sp1-deficient Drosophila SL2 cells, decreased by 50%. Both sites are required for Sp1 activity after ERK activation (46). Building on these findings, Legros et al. demonstrated that the inhibitor of Bcr/Abl protein tyrosine kinase imatinib mesylate (STI571) perturbs VEGF transcription in K562 cells by inhibiting ERK and reducing Sp1's and Sp3's affinity for a proximal binding element in the VEGF promoter (38). However, this study did not demonstrate the specific involvement of Thr453 and Thr739 phosphorylation in imatinib's influence on Sp1 binding.
We showed that, in rat pup smooth muscle cells (SMCs), fibroblast growth factor 2 (FGF-2) negatively regulates platelet-derived growth factor (PDGF) receptor alpha (PDGFR-α) expression. Transcription of all known PDGF ligands (15, 31, 32, 40, 45, 53) and at least PDGFR-α (2, 3) is under the control of Sp1 and related zinc finger proteins. Mutation of both Thr453 and Thr739 in Sp1 perturbed FGF-2 repression of PDGFR-α transcription (3). FGF-2 stimulated Sp1 phosphorylation in an ERK- but not p38-dependent manner and increased Sp1 interaction with the PDGFR-α promoter (3). Recent studies by Chuang et al. indicate that Jun N-terminal kinase 1 phosphorylates Sp1 at Thr278 and Thr739 (10), protecting the transcription factor from ubiquitin-dependent degradation and increasing its stability during mitosis in tumor cell lines. Overexpression of green fluorescent protein-Sp1 in HeLa cells increased HeLa cell proliferation, whereas overexpression of green fluorescent protein-Sp1 mutants (T278A and T739A, alone or together) did not. Accumulation of Sp1 in tumors may therefore be a consequence of Jun N-terminal kinase 1-dependent Sp1 phosphorylation (10).
Sp1 phosphorylation at Thr355 (Thr266 in the originally reported shorter form of Sp1) activates the ApoA1 promoter in HepG2 cells exposed to EGF and insulin (72). This modification of Sp1 by EGF and insulin was mediated via the MAPK/ERK pathway. However with insulin, PKC also influenced Sp1 phosphorylation. Deletion and mutational analysis revealed that the insulin-responsive core element within the ApoA1 promoter was responsible for the actions of EGF. This motif bound Sp1 specifically but not Sp2 or Sp3 (72). When Thr355 in Sp1 was mutated to Ala in cells stimulated with EGF, ApoA1 was no longer induced compared with the wild type, indicating the functional importance of this residue (72).
Studies performed a decade ago with rat hepatoma cells revealed that Sp1 is phosphorylated by CKII at a consensus site located within the second zinc finger, Thr579 (corresponding to Thr668 in human Sp1) (1). Phosphorylation of Sp1 at this site reduced Sp1's capacity to bind DNA without affecting total levels of Sp1. Mutation of Thr579 in the consensus CKII site did not eliminate CKII phosphorylation of Sp1, though it did perturb CKII's ability to inhibit Sp1 binding to DNA in vitro. Treatment of K562 cells with okadaic acid to inhibit endogenous phosphatase increased Sp1 phosphorylation and reduced DNA binding activity (1).
We recently demonstrated that Thr668, along with Ser670 (and Thr681), is a target of phosphorylation by PKC-ζ using a combination of approaches including in vitro peptide and protein phosphorylation analysis with Ala mutant counterparts, mass spectrometry phosphopeptide analysis, and coimmunoprecipitation analysis (63). This follows investigations by Pal et al. demonstrating that PKC-ζ binds to and phosphorylates the zinc finger region of Sp1 (49). Phospho-specific antibodies targeting pThr668/pSer670 or pThr681 were used in a variety of applications to demonstrate that angiotensin II (Ang II) stimulates PKC-ζ (pThr410) and Sp1 phosphorylation via the AT1 receptor in SMCs without influencing total levels of Sp1. Extending our previous findings that Ang II activates the PDGF-D promoter and that Sp1 binds the PDGF-D promoter (40), we found that Ang II-inducible PDGF-D expression involves PKC-ζ-dependent Sp1 phosphorylation at Thr668, Ser670, and Thr681. Triple mutation, but not single and double mutations, of these amino acids blocked Ang II activation of the PDGF promoter despite the fact that the triple mutation did not perturb Sp1's ability to bind DNA.
We found phosphorylated Sp1 (p681) in SMCs of human atherosclerotic plaques, as well as in SMCs of the mechanically injured rat carotid artery wall. This is the first demonstration of phosphorylated Sp1 in diseased animal and human tissue (63). We previously showed that activated PKC-ζ (pThr410) is also expressed in atherosclerotic plaques in the context of the Fas ligand (29). Numerous other studies have linked aberrant phosphorylation of Tyr, Ser, or Thr residues to cancer (36), diabetes (41), hypertension (21), and cardiac hypertrophy (20). Earlier studies by Vicart et al. found that Thr681 and Ser59 are targets for protein phosphatase 2A (PP2A) (65). Interestingly these investigators found that Ser59 and Thr681 can function independently or together for PP2A regulation of Sp1 activity during interphase (65).
CHROMATIN RECRUITMENT AND DNA DAMAGE
More recently, Vicart and colleagues found that the phosphorylation state of Ser59 influences Sp1's association with chromatin and Sp1 global O glycosylation (65). Dephosphorylated Sp1 is preferentially recruited to chromatin, and Ser59 is a target of PP2A. Although a detailed mechanism is yet to be established, the increased association of Sp1 with chromatin via PP2A's effects on Ser59 may serve as a permissive step in transcription. They further showed that Thr681, which resides in the second zinc finger linker, is another target of PP2A.
Sp1 is hyperphosphorylated during DNA damage. Iwahori et al. recently demonstrated that Ser101 is phosphorylated in response to ionizing radiation, UV irradiation, or hydroxyurea-induced replicative stress (22). Ser101 is phosphorylated by ataxia telangiectasia mutated (ATM) kinase (22, 47a). The proportion of chromatin-bound Sp1 phosphorylated at Ser101 increases rapidly after ionizing radiation (22), and Sp1 colocalizes in the nuclei with ATM, which is itself phosphorylated at Ser1981, at sites of double-strand breaks.
Sp1 phosphorylation plays a role in the derecruitment of repressor proteins from the promoter. Zhang and colleagues used coimmunoprecipitation studies to demonstrate that treatment with the HDAC inhibitor trichostatin A (TSA) stimulated Sp1 and PKC-ζ interaction in a TSA dose-dependent manner (69). TSA activated PKC-ζ (Thr410 phosphorylation [8]) and Sp1 phosphorylation at Ser, which was blocked by PKC-ζ small interfering RNA or a dominant negative, whereas Sp3 was not phosphorylated in response to TSA. Mutational analysis revealed that PKC-ζ phosphorylation of Sp1 at Ser641 is required for a critical role in TSA-inducible luteinizing hormone receptor (LHR) gene activation (69). Further studies using wortmannin or LY294002 demonstrated that phosphatidylinositol 3-kinase (PI3K) acts upstream of PKC-ζ and that the activity of this kinase is needed for PKC-ζ-mediated phosphorylation of Sp1. Sp1 phosphorylation (at Ser641) is required for the release of the pRB homologue p107 inhibitor protein from the LHR gene promoter, where it acts as a repressor (70). Interestingly, cell-type-specific differences were apparent despite the involvement of Ser641. TSA stimulated LHR gene promoter-localized histone hyperacetylation in choriocarcinoma (JAR) cells without influencing DNA methylation status, whereas in mammary carcinoma cells (MCF-7) cells, TSA induced a 160-fold derepression of LHR gene expression through histone hyperacetylation and DNA demethylation at its promoter (69).
OTHER POSTTRANSLATIONAL MODIFICATIONS INFLUENCING Sp1 ACTIVITY
Acetylation, sumoylation, ubiquitylation, and glycosylation are among other posttranslational modifications that influence the transcriptional activity and stability of Sp1. Glycosylation occurs at O-GlcNAc linkages at Ser and Thr residues in Sp1 (23) and is reversible by the action of O-GlcNAc-selective N-acetyl-d-glucosaminidase (34). Sp1 glycosylation can either stimulate or repress DNA binding and transcription (23). Studies with rat hepatoma cells showed that Sp1 glycosylation is critical for nuclear localization; however, once in the nucleus, Sp1 may be phosphorylated, thereby activating calmodulin gene expression (42). When Sp1 is hypoglycosylated under conditions of nutrition insufficiency, it is quickly proteolytically degraded (19), indicating that O-GlcNAc modification of Sp1 may play a role as a nutritional checkpoint (19).
Acetylation of Sp1 occurs in the DNA binding domain (62), and numerous studies investigating this modification have used HDAC inhibitors such as TSA. Recent studies by Chen et al. suggested that TSA treatment inhibits EGF induction of 12(S)-lipoxygenase by increasing Sp1 acetylation, which reduced Sp1 binding to the 12(S)-lipoxygenase promoter (7). Acetylation of Sp1 in this region along with the recruitment of c-Jun transcription factor and p300 increases the activity of the 12(S)-lipoxygenase gene promoter (7). The acetylation of Sp1 appears to play a protective role in neuronal cells undergoing oxidative stress via elevated cyclooxygenase 2 (COX-2) expression (37). COX-2 also plays a protective role in the context of vascular injury. COX-2-deficient mice undergo increased ischemic injury in the heart (5).
More recently it has been discovered that Sp1 is sumoylated at the N terminus under basal conditions, which negatively regulates Sp1 transcriptional activity (59). Sumoylation controls transcription by initiating chromatin structure changes that render DNA inaccessible to the transcriptional machinery. Sp3 sumoylation can suppress transcription by compacting repressive chromatin and provoking local heterochromatic gene silencing (61). SUMO-dependent transcriptional repression appears to be independent of HDACs (64). Horowitz and colleagues found that Sp3 isoforms (Sp3, M1, and M2) are sumoylated at Lys551 (60). Recent studies indicate that levels of sumoylated Sp1 are attenuated during tumorigenesis (66). Sumoylated Sp1 is found in the cytosol interacting with rpt6, and this interaction leads to increased Sp1 proteolysis and degradation (66). Decreased levels of cytosolic Sp1 as a consequence of sumoylation reduce the pool of Sp1 that would otherwise migrate to the nucleus (60). Modifications of Sp1 can occur simultaneously and synergistically. For example, glycosylation of Sp1 can lead to its translocation from the cytoplasm to the nucleus, where it is modified via phosphorylation to activate calmodulin gene expression (42). Sp1 phosphorylation and desumoylation can also occur simultaneously (59).
The influence of phosphorylation on the transcriptional activity of Sp1, particularly how it changes Sp1's affinity for DNA and/or other proteins, its roles in pathobiology, and the impact of other posttranslational modifications, is only beginning to become apparent, and further work is needed. For example, although we understand that certain kinases and residues in Sp1 mediate Sp1-dependent transcription, as in the case of FasL (30), PDGF B-chain (52), PDGF-D (63), PDGFR-α (3), and VEGF (49), it is unclear whether or how Sp1 phosphorylation influences chromatin remodeling, conformational changes, the recruitment of coactivators or repressors, and other posttranslational modifications. The availability of phospho-specific antibodies, such as those targeting pSer59 (65), pThr453 (46), p668/670 (63), p681 (63), and pThr739 (46), will serve as important tools in future investigations to more precisely define the roles of phosphorylation in the functions of this important transcription factor in health and disease.
Acknowledgments
We regret that the excellent studies of many other investigators could not be included due to space constraints.
This work was supported by grants from NHMRC and ARC.
Biography
 Nicole Y. Tan has research interests in the activation of genes in response to vascular smooth muscle cell stress. She has won Young Investigator Awards from the Australia Vascular Biology Society and International Vascular Biology Meeting. She received her B.Sc. with first class honors in pathology from UNSW under the guidance of Levon Khachigian and the support of an Australian Postgraduate Award.
Nicole Y. Tan has research interests in the activation of genes in response to vascular smooth muscle cell stress. She has won Young Investigator Awards from the Australia Vascular Biology Society and International Vascular Biology Meeting. She received her B.Sc. with first class honors in pathology from UNSW under the guidance of Levon Khachigian and the support of an Australian Postgraduate Award.
 Levon M. Khachigian is a vascular biologist whose research has centered on greater understanding of fundamental transcriptional mechanisms that lead to the inappropriate expression of harmful genes in our blood vessels. He has also pioneered the development of novel strategies targeting key regulatory genes in a variety of vascular disorders such as postangioplasty restenosis, arthritis, ocular neovascularization, tumor angiogenesis, and growth. His innovative research has been recognized by the Commonwealth Health Minister's Award for Excellence in Health and Medical Research, a GlaxoSmithKline Australia Award for Research Excellence, a Gottschalk Award from the Australian Academy of Science, and a Eureka Prize for Scientific Research and a Eureka Prize for Medical Research from the Australian Museum. He received his B.Sc. with first class honors in biochemistry and Ph.D. in cell and molecular biology from UNSW and then studied transcriptional control at Harvard Medical School, Boston, MA. He was later awarded a D.Sc. He is an NHMRC Australia Fellow and director of the Centre for Vascular Research at UNSW, Sydney, Australia.
Levon M. Khachigian is a vascular biologist whose research has centered on greater understanding of fundamental transcriptional mechanisms that lead to the inappropriate expression of harmful genes in our blood vessels. He has also pioneered the development of novel strategies targeting key regulatory genes in a variety of vascular disorders such as postangioplasty restenosis, arthritis, ocular neovascularization, tumor angiogenesis, and growth. His innovative research has been recognized by the Commonwealth Health Minister's Award for Excellence in Health and Medical Research, a GlaxoSmithKline Australia Award for Research Excellence, a Gottschalk Award from the Australian Academy of Science, and a Eureka Prize for Scientific Research and a Eureka Prize for Medical Research from the Australian Museum. He received his B.Sc. with first class honors in biochemistry and Ph.D. in cell and molecular biology from UNSW and then studied transcriptional control at Harvard Medical School, Boston, MA. He was later awarded a D.Sc. He is an NHMRC Australia Fellow and director of the Centre for Vascular Research at UNSW, Sydney, Australia.
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 27213489-13495. [DOI] [PubMed] [Google Scholar]
- 2.Bonello, M. R., Y. V. Bobryshev, and L. M. Khachigian. 2005. Peroxide-inducible Ets-1 mediates platelet-derived growth factor receptor-α gene transcription in vascular smooth muscle cells. Am. J. Pathol. 1671149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonello, M. R., and L. M. Khachigian. 2004. Fibroblast growth factor-2 represses platelet-derived growth factor receptor-alpha (PDGFR-alpha) transcription via ERK1/2-dependent Sp1 phosphorylation and an atypical cis-acting element in the proximal PDGFR-alpha promoter. J. Biol. Chem. 2792377-2382. [DOI] [PubMed] [Google Scholar]
- 4.Briggs, M. R., J. T. Kadonaga, S. P. Bell, and R. Tjian. 1986. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 23447-52. [DOI] [PubMed] [Google Scholar]
- 5.Camitta, M. G., S. A. Gabel, P. Chulada, J. A. Bradbury, R. Langenbach, D. C. Zeldin, and E. Murphy. 2001. Cyclooxygenase-1 and -2 knockout mice demonstrate increased cardiac ischemia/reperfusion injury but are protected by acute preconditioning. Circulation 1042453-2458. [DOI] [PubMed] [Google Scholar]
- 6.Canaff, L., X. Zhou, and G. N. Hendy. 2008. The proinflammatory cytokine, interleukin-6, up-regulates calcium-sensing receptor gene transcription via Stat1/3 and Sp1/3. J. Biol. Chem. 28313586-13600. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. J., W. C. Chang, and B. K. Chen. 2008. Attenuation of c-Jun and Sp1 expression and p300 recruitment to gene promoter confers the trichostatin A-induced inhibition of 12(S)-lipoxygenase expression in EGF-treated A431 cells. Eur. J. Pharmacol. 59136-42. [DOI] [PubMed] [Google Scholar]
- 8.Chou, M. M., W. Hou, J. Johnson, L. K. Graham, M. H. Lee, C. S. Chen, A. C. Newton, B. S. Schaffhausen, and A. Toker. 1998. Regulation of protein kinase C zeta by PI 3-kinase and PDK-1. Curr. Biol. 81069-1077. [DOI] [PubMed] [Google Scholar]
- 9.Chu, S., C. A. Cockrell, and T. J. Ferro. 2003. Expression of alpha-ENaC2 is dependent on an upstream Sp1 binding motif and is modulated by protein phosphatase 1 in lung epithelial cells. Biochem. Biophys. Res. Commun. 3031159-1168. [DOI] [PubMed] [Google Scholar]
- 10.Chuang, J. Y., Y. T. Wang, S. H. Yeh, Y. W. Liu, W. C. Chang, and J. J. Hung. 2008. Phosphorylation by c-Jun NH2-terminal kinase 1 regulates the stability of transcription factor Sp1 during mitosis. Mol. Biol. Cell 191139-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun, R. F., O. J. Semmes, C. Neuveut, and K. T. Jeang. 1998. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol. 722615-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chupreta, S., M. Du, A. Todisco, and J. L. Merchant. 2000. EGF stimulates gastrin promoter through activation of Sp1 kinase activity. Am. J. Physiol. Cell. Physiol. 278C697-C708. [DOI] [PubMed] [Google Scholar]
- 13.Cieslik, K., C. M. Lee, J. L. Tang, and K. K. Wu. 1999. Transcriptional regulation of endothelial nitric-oxide synthase by an interaction between casein kinase 2 and protein phosphatase 2A. J. Biol. Chem. 27434669-34675. [DOI] [PubMed] [Google Scholar]
- 14.Daniel, S., S. Zhang, A. A. DePaoli-Roach, and K. H. Kim. 1996. Dephosphorylation of Sp1 by protein phosphatase 1 is involved in the glucose-mediated activation of the acetyl-CoA carboxylase gene. J. Biol. Chem. 27114692-14697. [DOI] [PubMed] [Google Scholar]
- 15.Delbridge, G. J., and L. M. Khachigian. 1997. FGF-1-induced PDGF A-chain gene expression in vascular endothelial cells involves transcriptional activation by Egr-1. Circ. Res. 81282-288. [DOI] [PubMed] [Google Scholar]
- 16.Dynan, W. S., and R. Tjian. 1983. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell 32669-680. [DOI] [PubMed] [Google Scholar]
- 17.Fojas de Borja, P., N. K. Collins, P. Du, J. Azizkhan-Clifford, and M. Mudryj. 2001. Cyclin A-CDK phosphorylates Sp1 and enhances Sp1-mediated transcription. EMBO J. 205737-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haidweger, E., M. Novy, and H. Rotheneder. 2001. Modulation of Sp1 activity by a cyclin A/CDK complex. J. Mol. Biol. 306201-212. [DOI] [PubMed] [Google Scholar]
- 19.Han, I., and J. E. Kudlow. 1997. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol. Cell. Biol. 172550-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinrichsen, R., A. H. Hansen, S. Haunso, and P. K. Busk. 2008. Phosphorylation of pRb by cyclin D kinase is necessary for development of cardiac hypertrophy. Cell Prolif. 41813-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, C., A. Dandapat, L. Sun, M. R. Marwali, N. Inoue, F. Sugawara, K. Inoue, Y. Kawase, K. Jishage, H. Suzuki, P. L. Hermonat, T. Sawamura, and J. L. Mehta. 2008. Modulation of angiotensin II-mediated hypertension and cardiac remodeling by lectin-like oxidized low-density lipoprotein receptor-1 deletion. Hypertension 52556-562. [DOI] [PubMed] [Google Scholar]
- 22.Iwahori, S., Y. Yasui, A. Kudoh, Y. Sato, S. Nakayama, T. Murata, H. Isomura, and T. Tsurumi. 2008. Identification of phosphorylation sites on transcription factor Sp1 in response to DNA damage and its accumulation at damaged sites. Cell. Signal. 201795-1803. [DOI] [PubMed] [Google Scholar]
- 23.Jackson, S. P., and R. Tjian. 1988. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell 55125-133. [DOI] [PubMed] [Google Scholar]
- 24.Jeang, K. T., R. Chun, N. H. Lin, A. Gatignol, C. G. Glabe, and H. Fan. 1993. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J. Virol. 676224-6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, K. A., J. T. Kadonaga, P. A. Luciw, and R. Tjian. 1986. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science 232755-759. [DOI] [PubMed] [Google Scholar]
- 26.Kaczynski, J., T. Cook, and R. Urrutia. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadonaga, J. T., K. R. Carner, F. R. Masiarz, and R. Tjian. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 511079-1090. [DOI] [PubMed] [Google Scholar]
- 28.Kadonaga, J. T., and R. Tjian. 1986. Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. USA 835889-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kavurma, M. M., Y. Bobryshev, and L. M. Khachigian. 2002. Ets-1 positively regulates Fas ligand transcription via cooperative interactions with Sp1. J. Biol. Chem. 27736244-36252. [DOI] [PubMed] [Google Scholar]
- 30.Kavurma, M. M., F. S. Santiago, E. Bonfoco, and L. M. Khachigian. 2001. Sp1 phosphorylation regulates apoptosis via extracellular FasL-Fas engagement. J. Biol. Chem. 2764964-4971. [DOI] [PubMed] [Google Scholar]
- 31.Khachigian, L. M., V. Lindner, A. J. Williams, and T. Collins. 1996. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science 2711427-1431. [DOI] [PubMed] [Google Scholar]
- 32.Khachigian, L. M., A. J. Williams, and T. Collins. 1995. Interplay of Sp1 and Egr-1 in the proximal PDGF-A promoter in cultured vascular endothelial cells. J. Biol. Chem. 27027679-27686. [DOI] [PubMed] [Google Scholar]
- 33.Kim, J. I., A. C. Cordova, Y. Hirayama, J. A. Madri, and B. E. Sumpio. 2008. Differential effects of shear stress and cyclic strain on Sp1 phosphorylation by protein kinase Czeta modulates membrane type 1-matrix metalloproteinase in endothelial cells. Endothelium 1533-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudlow, J. E. 2006. Post-translational modification by O-GlcNAc: another way to change protein function. J. Cell. Biochem. 981062-1075. [DOI] [PubMed] [Google Scholar]
- 35.Lacroix, I., C. Lipcey, J. Imbert, and B. Kahn-Perles. 2002. Sp1 transcriptional activity is up-regulated by phosphatase 2A in dividing T lymphocytes. J. Biol. Chem. 2779598-9605. [DOI] [PubMed] [Google Scholar]
- 36.Larrea, M. D., J. Liang, T. Da Silva, F. Hong, S. H. Shao, K. Han, D. Dumont, and J. M. Slingerland. 2008. Phosphorylation of p27Kip1 regulates assembly and activation of cyclin D1-Cdk4. Mol. Cell. Biol. 286462-6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, J., B. Kosaras, H. Aleyasin, J. A. Han, D. S. Park, R. R. Ratan, N. W. Kowall, R. J. Ferrante, S. W. Lee, and H. Ryu. 2006. Role of cyclooxygenase-2 induction by transcription factor Sp1 and Sp3 in neuronal oxidative and DNA damage response. FASEB J. 202375-2377. [DOI] [PubMed] [Google Scholar]
- 38.Legros, L., C. Bourcier, A. Jacquel, F. X. Mahon, J. P. Cassuto, P. Auberger, and G. Pages. 2004. Imatinib mesylate (STI571) decreases the vascular endothelial growth factor plasma concentration in patients with chronic myeloid leukemia. Blood 104495-501. [DOI] [PubMed] [Google Scholar]
- 39.Lin, M.-C., F. Almus-Jacobs, H.-H. Chen, G. C. N. Parry, N. Mackmann, J. Y. Shyy, and S. Chen. 1997. Shear stress induction of the tissue factor gene. J. Clin. Investig. 99737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, M. Y., M. Eyries, C. Zhang, F. S. Santiago, and L. M. Khachigian. 2006. Inducible platelet-derived growth factor D-chain expression by angiotensin II and hydrogen peroxide involves transcriptional regulation by Ets-1 and Sp1. Blood 1072322-2329. [DOI] [PubMed] [Google Scholar]
- 41.Mack, E., E. Ziv, H. Reuveni, R. Kalman, M. Y. Niv, A. Jorns, S. Lenzen, and E. Shafrir. 2008. Prevention of insulin resistance and beta-cell loss by abrogating PKCepsilon-induced serine phosphorylation of muscle IRS-1 in Psammomys obesus. Diabetes Metab. Res. Rev. 24577-584. [DOI] [PubMed] [Google Scholar]
- 42.Majumdar, G., A. Harrington, J. Hungerford, A. Martinez-Hernandez, I. C. Gerling, R. Raghow, and S. Solomon. 2006. Insulin dynamically regulates calmodulin gene expression by sequential o-glycosylation and phosphorylation of sp1 and its subcellular compartmentalization in liver cells. J. Biol. Chem. 2813642-3650. [DOI] [PubMed] [Google Scholar]
- 43.Mazure, N. M., M. C. Brahimi-Horn, and J. Pouyssegur. 2003. Protein kinases and the hypoxia-inducible factor-1, two switches in angiogenesis. Curr. Pharm. Des. 9531-541. [DOI] [PubMed] [Google Scholar]
- 44.McDonough, P. M., D. S. Hanford, A. B. Sprenkle, N. R. Mellon, and C. C. Glembotski. 1997. Collaborative roles for c-Jun N-terminal kinase, c-Jun, serum response factor, and Sp1 in calcium-regulated myocardial gene expression. J. Biol. Chem. 27224046-24053. [DOI] [PubMed] [Google Scholar]
- 45.Midgley, V. C., and L. M. Khachigian. 2004. Fibroblast growth factor-2 induction of platelet-derived growth factor-C chain transcription in vascular smooth muscle cells is ERK-dependent but not JNK-dependent and mediated by Egr-1. J. Biol. Chem. 27940289-40295. [DOI] [PubMed] [Google Scholar]
- 46.Milanini-Mongiat, J., J. Pouyssegur, and G. Pages. 2002. Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases: their implication in vascular endothelial growth factor gene transcription. J. Biol. Chem. 27720631-20639. [DOI] [PubMed] [Google Scholar]
- 47.Naar, A. M., S. Ryu, and R. Tjian. 1998. Cofactor requirements for transcriptional activation by Sp1. Cold Spring Harbor Symp. Quant. Biol. 63189-199. [DOI] [PubMed] [Google Scholar]
- 47a.Olofsson, B. A., C. M. Kelly, J. Kim, S. M. Hornsby, and J. Azizkhan-Clifford. 2007. Phosphorylation of Sp1 in response to DNA damage by ataxia telangiectasia-mutated kinase. Mol. Cancer Res. 51319-1330. [DOI] [PubMed] [Google Scholar]
- 48.Opitz, O. G., and A. K. Rustgi. 2000. Interaction between Sp1 and cell cycle regulatory proteins is important in transactivation of a differentiation-related gene. Cancer Res. 602825-2830. [PubMed] [Google Scholar]
- 49.Pal, S., K. P. Claffey, H. T. Cohen, and D. Mukhopadhyay. 1998. Activation of Sp1-mediated vascular permeability factor/vascular endothelial growth factor transcription requires specific interaction with protein kinase C zeta. J. Biol. Chem. 27326277-26280. [DOI] [PubMed] [Google Scholar]
- 50.Pan, M. R., and W. C. Hung. 2002. Nonsteroidal anti-inflammatory drugs inhibit matrix metalloproteinase-2 via suppression of the ERK/Sp1-mediated transcription. J. Biol. Chem. 27732775-32780. [DOI] [PubMed] [Google Scholar]
- 51.Parisi, F., P. Wirapati, and F. Naef. 2007. Identifying synergistic regulation involving c-Myc and sp1 in human tissues. Nucleic Acids Res. 351098-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rafty, L. A., and L. M. Khachigian. 2001. Sp1 phosphorylation regulates inducible expression of platelet-derived growth factor B-chain gene via atypical protein kinase C-zeta. Nucleic Acids Res. 291027-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rafty, L. A., and L. M. Khachigian. 1998. Zinc finger transcription factors mediate high constitutive PDGF-B expression in smooth muscle cells derived from aortae of newborn rats. J. Biol. Chem. 2735758-5764. [DOI] [PubMed] [Google Scholar]
- 54.Reisinger, K., R. Kaufmann, and J. Gille. 2003. Increased Sp1 phosphorylation as a mechanism of hepatocyte growth factor (HGF/SF)-induced vascular endothelial growth factor (VEGF/VPF) transcription. J. Cell Sci. 116225-238. [DOI] [PubMed] [Google Scholar]
- 55.Rojo, A. I., M. Salina, M. Salazar, S. Takahashi, G. Suske, V. Calvo, M. R. de Sagarra, and A. Cuadrado. 2006. Regulation of heme oxygenase-1 gene expression through the phosphatidylinositol 3-kinase/PKC-zeta pathway and Sp1. Free Radic. Biol. Med. 41247-261. [DOI] [PubMed] [Google Scholar]
- 56.Rosmarin, A. G., M. Luo, D. G. Caprio, J. Shang, and C. P. Simkevich. 1998. Sp1 cooperates with the ets transcription factor, GABP, to activate the CD18 (beta2 leukocyte integrin) promoter. J. Biol. Chem. 27313097-13103. [DOI] [PubMed] [Google Scholar]
- 57.Santiago, F. S., H. Ishii, S. Shafi, R. Khurana, P. Kanellakis, R. Bhindi, M. Ramirez, A. Bobik, J. Martin, C. N. Chesterman, I. Zachary, and L. Khachigian. 2007. Yin Yang-1 inhibits intimal thickening by repressing p21WAF1/Cip1 transcription and p21WAF1/Cip1-Cdk4-Cyclin D1 assembly. Circ. Res. 101146-155. [DOI] [PubMed] [Google Scholar]
- 58.Schafer, D., B. Hamm-Kunzelmann, and K. Brand. 1997. Glucose regulates the promoter activity of aldolase A and pyruvate kinase M2 via dephosphorylation of Sp1. FEBS Lett. 417325-328. [DOI] [PubMed] [Google Scholar]
- 59.Spengler, M. L., and M. G. Brattain. 2006. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J. Biol. Chem. 2815567-5574. [DOI] [PubMed] [Google Scholar]
- 60.Spengler, M. L., S. B. Kennett, K. S. Moorefield, S. O. Simmons, M. G. Brattain, and J. M. Horowitz. 2005. Sumoylation of internally initiated Sp3 isoforms regulates transcriptional repression via a trichostatin A-insensitive mechanism. Cell. Signal. 17153-166. [DOI] [PubMed] [Google Scholar]
- 61.Stielow, B., A. Sapetschnig, C. Wink, I. Kruger, and G. Suske. 2008. SUMO-modified Sp3 represses transcription by provoking local heterochromatic gene silencing. EMBO Rep. 9899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki, T., A. Kimura, R. Nagai, and M. Horikoshi. 2000. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells 529-41. [DOI] [PubMed] [Google Scholar]
- 63.Tan, N., V. C. Midgley, M. M. Kavurma, F. S. Santiago, X. Luo, R. Peden, R. G. Fahmy, M. C. Berndt, M. P. Molloy, and L. M. Khachigian. 2008. Angiotensin II-inducible platelet-derived growth factor-D transcription requires specific Ser/Thr residues in the second zinc finger region of Sp1. Circ. Res. 102e38-e51. [DOI] [PubMed] [Google Scholar]
- 64.Valin, A., and G. Gill. 2007. Regulation of the dual-function transcription factor Sp3 by SUMO. Biochem. Soc. Trans. 351393-1396. [DOI] [PubMed] [Google Scholar]
- 65.Vicart, A., T. Lefebvre, J. Imbert, A. Fernandez, and B. Kahn-Perles. 2006. Increased chromatin association of Sp1 in interphase cells by PP2A-mediated dephosphorylations. J. Mol. Biol. 364897-908. [DOI] [PubMed] [Google Scholar]
- 66.Wang, Y. T., J. Y. Chuang, M. R. Shen, W. B. Yang, W. C. Chang, and J. J. Hung. 2008. Sumoylation of specificity protein 1 augments its degradation by changing the localization and increasing the specificity protein 1 proteolytic process. J. Mol. Biol. 380869-885. [DOI] [PubMed] [Google Scholar]
- 67.Wang, Z., Y. Zhang, J. Lu, S. Sun, and K. Ravid. 1999. Mpl ligand enhances the transcription of the cyclin D3 gene: a potential role for Sp1 transcription factor. Blood 934208-4221. [PubMed] [Google Scholar]
- 68.Wu, Z., H. P. Kim, H. H. Xue, H. Liu, K. Zhao, and W. J. Leonard. 2005. Interleukin-21 receptor gene induction in human T cells is mediated by T-cell receptor-induced Sp1 activity. Mol. Cell. Biol. 259741-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, Y., M. Liao, and M. L. Dufau. 2006. Phosphatidylinositol 3-kinase/protein kinase Cζ-induced phosphorylation of Sp1 and p107 repressor release have a critical role in histone deacetylase inhibitor-mediated depression of transcription of the luteinizing hormone receptor gene. Mol. Cell. Biol. 266748-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, Y., M. Liao, and M. L. Dufau. 2008. Unlocking repression of the human luteinizing hormone receptor gene by trichostatin A-induced cell-specific phosphatase release. J. Biol. Chem. 28324039-24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao, S., K. Venkatasubbarao, S. Li, and J. W. Freeman. 2003. Requirement of a specific Sp1 site for histone deacetylase-mediated repression of transforming growth factor beta type II receptor expression in human pancreatic cancer cells. Cancer Res. 632624-2630. [PubMed] [Google Scholar]
- 72.Zheng, X. L., S. Matsubara, C. Diao, M. D. Hollenberg, and N. C. Wong. 2000. Activation of apolipoprotein AI gene expression by protein kinase A and kinase C through transcription factor, Sp1. J. Biol. Chem. 27531747-31754. [DOI] [PubMed] [Google Scholar]
- 73.Zheng, X. L., S. Matsubara, C. Diao, M. D. Hollenberg, and N. C. Wong. 2001. Epidermal growth factor induction of apolipoprotein A-I is mediated by the Ras-MAP kinase cascade and Sp1. J. Biol. Chem. 27613822-13829. [DOI] [PubMed] [Google Scholar]