Abstract
While hypoxia-inducible factor (HIF) is a major actor in the cell survival response to hypoxia, HIF also is associated with cell death. Several studies implicate the HIF-induced putative BH3-only proapoptotic genes bnip3 and bnip3l in hypoxia-mediated cell death. We, like others, do not support this assertion. Here, we clearly demonstrate that the hypoxic microenvironment contributes to survival rather than cell death by inducing autophagy. The ablation of Beclin1, a major actor of autophagy, enhances cell death under hypoxic conditions. In addition, the ablation of BNIP3 and/or BNIP3L triggers cell death, and BNIP3 and BNIP3L are crucial for hypoxia-induced autophagy. First, while the small interfering RNA-mediated ablation of either BNIP3 or BNIP3L has little effect on autophagy, the combined silencing of these two HIF targets suppresses hypoxia-mediated autophagy. Second, the ectopic expression of both BNIP3 and BNIP3L in normoxia activates autophagy. Third, 20-mer BH3 peptides of BNIP3 or BNIP3L are sufficient in initiating autophagy in normoxia. Herein, we propose a model in which the atypical BH3 domains of hypoxia-induced BNIP3/BNIP3L have been designed to induce autophagy by disrupting the Bcl-2-Beclin1 complex without inducing cell death. Hypoxia-induced autophagy via BNIP3 and BNIP3L is clearly a survival mechanism that promotes tumor progression.
The evolutionarily conserved hypoxia-inducible factor (HIF) transcriptional complex is rapidly activated when the O2 tension decreases (26, 33). HIF orchestrates the expression of a myriad of genes, the function of which primarily is to ensure cell survival under a short- and long-term hypoxic stress, thereby attempting to restore O2 homeostasis (32). By exploring the functionality of HIF-1α and, in particular, the role of its two transactivation domains (N-TAD and C-TAD), we recently brought to light the bifunctional activity of HIF-1α (8). This duality of action is discriminated by FIH (factor inhibiting HIF-1), an inhibitor of the C-TAD. Among the genes expressed only by the N-TAD is the putative proapoptotic gene bnip3 (Bcl-2/adenovirus E1B 19-kDa interacting protein 3). This was an intriguing finding. Why would a death-promoting protein be induced under conditions of moderate hypoxia, where HIF-1 would be expected to promote cell survival? However, a closely related gene, bnip3l (Bcl-2/adenovirus E1B 19-kDa interacting protein 3 like, also known as BNIP3α and Nix), was classified as an FIH-inhibited gene, and it is induced by both the N-TAD and C-TAD domains. Its expression reached its maximum in severe hypoxia, which is encountered close to necrotic areas of tumors.
Therefore, we specifically focused our interest on understanding HIF-1-mediated cell death by studying the role of the HIF-dependent gene products BNIP3 and BNIP3L (7, 12) in both normal and cancer cells. BNIP3 and BNIP3L are members of the so-called BH3-only subfamily of Bcl-2 family proteins (40) that heterodimerize and antagonize the activity of the prosurvival proteins (Bcl-2 and Bcl-XL) (4). Both BNIP3 and BNIP3L have been reported to promote cell death (14, 39); however, the precise role of these two proapoptotic proteins remains unclear. We and others (24) failed to reproduce the proapoptotic or necrotic cell death features of ectopically expressed BNIP3 or BNIP3L in various cell types, including mouse embryonic fibroblasts (MEFs) and MCF7, PC3, and LS174 cells.
We questioned the cell death function of BNIP3/BNIP3L in a hypoxic environment and postulated a positive role in activating the autophagic cell survival process (26). Very recently, Zhang et al. demonstrated the hypoxia-induced mitochondrial autophagy via the expression of BNIP3. These results support the idea that hypoxia plays a protective role by decreasing reactive oxygen species production (41). In the meantime, Tracy et al. identified BNIP3 as a direct target of transcriptional repression acting through pRB/E2F, which is required for hypoxia-induced autophagy in numerous tumor cell lines and MEFs (37). However, the role of BNIP3L was not discussed, and the function of hypoxia-induced autophagy as a survival process was not clearly defined.
In this report, we demonstrate that hypoxia-induced autophagy is part of a general mechanism of cell survival that is controlled by HIF-1. In parallel, we show that the expression of both BNIP3 and BNIP3L is required for the optimal induction of autophagy in hypoxia, implicating their atypical BH3 domains as determinants for dissociating the Bcl-2-Beclin1 complexes. These results point to a new BH3-dependent process for hypoxia-induced autophagy.
MATERIALS AND METHODS
Cell culture.
DLD1, LS174, MCF7, PC3, 786-O, HIF+/+ and HIF−/− MEF (29), and CCL39 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL) supplemented with 5 or 10% inactivated fetal bovine serum (FBS) as appropriate. DLD1 and LS174 cells expressing the tetracycline (Tet) repressor were kindly provided by M. van de Wetering (38). The antibiotics penicillin G (50 U/ml) and streptomycin sulfate (50 μg/ml) were added, plus 10 μg/ml blasticidin (Gibco-BRL) in the case of the DLD1 and CCL39 cells.
Hypoxic conditions were induced by the incubation of cells in a sealed Bug-Box anaerobic workstation (Ruskinn Technology Biotrace International, Plc.). The oxygen level was maintained at 1 or 0.1%, with the residual gas mixture being 94.0 to 94.9% nitrogen and 5% carbon dioxide.
Treatment of cells with R8-BH3 peptides.
The R8-BH3 peptides were synthesized and purified by high-performance liquid chromatography (>95% purity) by NeoMPS (Strasbourg) and used as previously reported (11). BH3 peptide sequences of BNIP3/BNIP3L (as shown in Fig. 5B) were modified with a D-isomer homopolymer of eight Arg residues followed by a glycine linker to function as a peptide membrane-transducing domain. The disruption of the BH3 domain was ensured by two mutations of the well-conserved residues Leu>Ala and Asp>Ala of the BH3 core region (see Fig. 5B). Exponentially growing LS174 cells in 10% FBS were exposed to 10 μM of peptide at three intervals during 24 h. No cell death was measured, even after 3 days of incubation with the peptides.
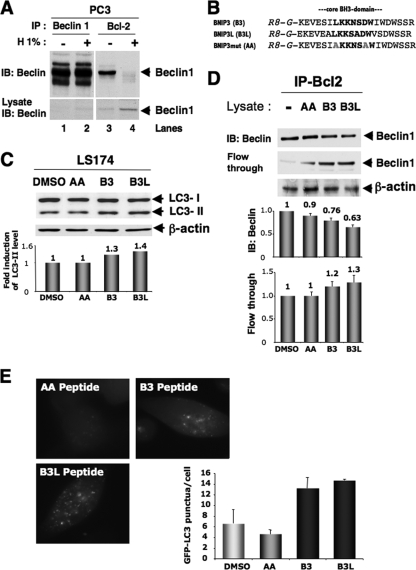
FIG. 5.
Hypoxia triggers autophagy in promoting survival. (A) Coimmunoprecipitation of endogenous Beclin1 with Bcl-2. PC3 cells were subjected to hypoxia (H 1%; 48 h), and cell lysates were immunoprecipitated with an anti-Beclin1 antibody resin. The presence of Beclin1 protein in the lysates and the immunoprecipitates is shown (arrow). IB, immunoblot; IP, immunoprecipitate. (B) Sequences of the three BH3 peptides used, BNIP3L, BNIP3, and a mutated form of BNIP3 (BNIP3 mut). The stretch of arginine residues (R8) and the glycine linker (G) are shown in italics, and the central core of the BH3 domain is in boldface. B3, R8 peptide of the BNIP3 BH3 domain; B3L, R8 peptide of the BNIP3L BH3 domain; AA, R8 peptide of the mutated BNIP3 BH3 domain. (C) R8-BH3 peptides of BNIP3 and BNIP3L trigger autophagy (i.e., an increase in the LC3-II/LC3-I ratio) in normoxia in LS174 cells. LS174 cells were exposed in complete medium three times during a 24-h period to 25 μg/ml of the R8-modified peptides in normoxia. Experiments were done twice in LS174 cells, giving identical results (results from one experiment are shown). In the two experiments, the addition of the mutated AA form gave results identical to those for dimethyl sulfoxide (DMSO) with no peptide. Total cell extracts were analyzed by immunoblotting with antibodies against LC3 and β-actin. (D) Disruption of the Bcl-2-Beclin1 complex by BH3 peptides derived from BNIP3 or BNIP3L. An acellular assay was performed with a normoxic lysate of PC3 cells incubated in the absence or presence of BNIP3 or BNIP3L peptide. The coimmunoprecipitation of endogenous Beclin1 was performed with an anti-Bcl-2 antibody. The presence of Beclin1 in the lysates and the immunoprecipitate is illustrated (arrow). (E) BNIP3 and BNIP3L peptides trigger autophagy in stable GFP-LC3-expressing PC3 cells. PC3 cells were infused five times with the AA (20 μM), BNIP3 (20 μM), or BNIP3L peptide (20 μM) for 24 h. An example of GFP-LC3 punctate structures observed in GFP-LC3 PC3 cells under normoxic and hypoxic conditions is given. GFP fluorescence was examined under a Zeiss fluorescence microscope. The quantification of the number of GFP aggregates per cell was performed using the analyze particles tool in the Image J software. *, P < 0.00001.
Plasmids, small interfering RNA (siRNA), and lentivirus.
Conventional cloning procedures were used to insert the human BNIP3 cDNA obtained first by PCR using the primers (forward) 5′-GAAAGCTTAATGTCCGAGAACGGAGCGCCCGGGAT-3′ and (reverse) 5′-ATGAATTCTTCATCAAAAGGTGCTGGTGGAG-3′ into pCRII-TOPO (Invitrogen, Carlsbad, CA). The pBNIP3 plasmid was obtained by subcloning the EcoRI-HindIII fragment from pCRII-TOPO BNIP3 into the EcoRI-HindIII sites of the pTREX-C vector (also named pcDNA4/TO/myc-His N; Invitrogen). pBNIP3L was kindly provided by N. Denko (24). The partial hamster BNIP3 cDNA, obtained first by PCR using the primers (forward) 5′-GGGCTCCTGGGTAGAACTGC-3′ and (reverse) 5′-GTTGTCAGACGCCTTCCAAT-3′, was directly sequenced.
The 21-nucleotide RNAs were chemically synthesized (Eurogentec, Seraing, Belgium). The set of siRNAs targeting hamster BNIP3 (two sequences), human BNIP3 (two sequences), human BNIP3L (two sequences), and human ATG5 (one sequence) were the following: (forward) 5′-GGACGAAGUAGCUCCAAGATT-3′ and (reverse) 5′-UCUUGGAGCUACUUCGUCCTT-3′, (forward) 5′-UCUGUCUGAGGAAGAUUAUTT-3′ and (reverse) 5′-AUAAUCUUCCUCAGACAGATT-3′, (forward) 5′-ACACGAGCGUCAUGAAGAATT-3′ and (reverse) 5′-UUCUUCAUGACGCUCGUGUTT-3′, (forward) 5′-CAGCCUCGGUUUCUAUUUATT-3′ and (reverse) 5′-UAAAUAGAAACCGAGGCUGTT-3′, (forward) 5′-AACACGUACCAUCCUCAUCCUTT-3′ and (reverse) 5′-AGGAUGAGGAUGGUACGUGUUTT-3′, (forward) 5′-AACAGUUCCUGGGUGGAGCUATT-3′ and (reverse) 5′-UAGCUCCACCCAGGAACUGUUTT-3′, and (forward) 5′-GCAACUCUGGAUGGGAUUGTT-3′ and (reverse) 5′-CAAUCCCAUCCAGAGUUGCTT-3′. siRNA sequences targeting SIMA (also known as Drosophila HIF-α) have been described previously (2). The stable ablation of Beclin1 in PC3 cells was obtained using NM_003766 Beclin MISSION short hairpin RNA lentiviral transduction particles (Sigma, St. Louis, MO) as described in the manufacturer's protocol. Sequences TRCN0000033549 (5′-CCGGCCCGTGGAATGGAATGAGATTCTCGAGAATCTCATTCCATTCCACGGGTTTTTG-3′) and TRCN0000033552 (5′-CCGGCTCAAGTTCATGCTGACGAATCTCGAGATTCGTCAGCATGAACTTGAGTTTTTG-3′) were used in our experiments.
Stable clones.
CCL39, DLD1, and LS174 cells were transfected with pBNIP3. Zeocin-resistant clones were tested for their ability to overexpress human BNIP3. Clones incubated in the absence or presence of 10 μg/ml Tet to upregulate BNIP3 were screened by immunoblotting. CCL39-pBNIP3 and LS174- pBNIP3 clones then were transfected with pBNIP3L and pBabepuro. Zeocin- and puromycin-resistant clones were tested for their ability to stably overexpress BNIP3L and to overexpress BNIP3 in the presence of Tet. CCL39 were transfected with pca9. Zeocin-resistant clones were tested for their ability to overexpress human CAIX. Clones incubated in the absence or presence of 10 μg/ml Tet to upregulate CAIX were screened by immunoblotting. PC3 cells were transfected with pGFP-LC3. Neomycin-resistant clones were tested for their ability to increase the frequency of puncta under hypoxic conditions (for 48 h at 1% O2).
Cell death assay.
The mortality rate of all cell types used was assessed after vital staining using trypan blue exclusion (Sigma, St. Louis, MO).
Immunoblotting.
The BNIP3 and Beclin1 antibodies were purchased from Abcam and Novus Biochemicals, respectively. Antiserum against amino acids 98 to 116 of BNIP3L was described previously (24). Anti-LC3 was raised in rabbits immunized against the N-terminal 14 amino acids of human LC3 (PSEKTFKQRRSFEQC for coupling to keyhole limpet hemocyanin). Anti-mouse immunoglobulin G and anti-rabbit immunoglobulin G secondary antibodies were obtained from Promega. Cells were lysed in 1.5× Laemmli buffer, and the protein concentration was determined using the bicinchoninic acid assay. Forty micrograms of each whole-cell extract was resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane (Millipore). Membranes were blocked in 5% nonfat milk in TN buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl) and incubated in the presence of the primary and then secondary antibodies in 5% nonfat milk in TN buffer. After being washed in TN buffer containing 1% Triton X-100 and then in TN buffer, immunoreactive bands were visualized with the ECL system (Amersham Biosciences).
Immunoprecipitation.
Immunoprecipitations were performed using the Catch and Release v2.0 reversible immunoprecipitation system (Upstate) according to the manufacturer's instructions. Briefly, cells were lysed in cold radioimmunoprecipitation assay (RIPA) buffer without SDS, and then 1 mg of cell lysate was incubated overnight with 4 μg of anti-Beclin1 (Novus Biological) or 5 μg of anti-Bcl-2 (BD Pharmingen). The immunoprecipitates then were analyzed by SDS-polyacrylamide gel electrophoresis using a 10% acrylamide-bisacrylamide running gel and immunoblotting using a secondary antibody from ExactaCruz F (Santa Cruz Biotechnology), which strongly limits the detection of the heavy and light chains of the immunoprecipitated antibody.
An acellular assay of the disruption of the Bcl-2-Beclin1 complex by BH3 peptides derived from BNIP3 or BNIP3L was performed. Briefly, cells were washed three times in phosphate-buffered saline (PBS) and then incubated for 30 min on ice before being centrifuged at 15,000 × g for 15 min at 4°C. The supernatant then was collected. The assay was performed with 500 μg of a whole-cell lysate, in RIPA buffer without SDS, of cells incubated with 40 μM of BH3 peptides derived from BNIP3, BNIP3L, or a mutated BH3 of BNIP3, called BNIP3AA, at 30°C for 1 h. After incubation, Bcl-2 was immunoprecipitated according the experimental procedure described earlier in order to monitor the remaining pool of Beclin1 associated with Bcl-2.
Immunofluorescence microscopy.
Cells were grown on glass coverslips and then fixed in 3.3% paraformaldehyde for 30 min at room temperature (RT) and then permeabilized with 0.2% Triton X-100 for 5 min. For the detection of BNIP3, cells were blocked with phosphate-buffered saline containing 0.2% gelatin, 2% bovine serum albumin (PGB) for 30 min at RT and then incubated with the polyclonal anti-mouse BNIP3 antibody (1:250) and anti-rabbit BNIP3L (1:200) in PGB for 3 h at RT. After being washed, cells were incubated in the presence of a biotinylated anti-mouse secondary antibody conjugated to Alexa 594 (1:400) and anti-rabbit secondary antibody conjugated to Alexa 488 (1:400) for 1 h at RT. After being washed, coverslips were mounted in Cytifluor (Amersham Biosciences), the detection of the fluorescence was performed with a Leica DMR fluorescence microscope, and images were recorded using RSImage software.
MDC staining.
PC3 cells were seeded on coverslips overnight and then incubated for 48 h in hypoxia (1% O2), and during the last 1 h 0.1 μM monodansylcadaverine (MDC) was added. Cells then were washed with three rinses with PBS and fixed with a solution of 3.3% paraformaldehyde for 30 min. After being washed, coverslips were mounted in Cytifluor (Amersham Biosciences), the detection of the fluorescence was performed with a Leica DMR fluorescence microscope, and images were recorded using RSImage software.
Electron microscopy.
Cells were fixed in situ with 1.6% glutaraldehyde in 0.1 M phosphate buffer at RT and then held overnight at 4°C. Samples were rinsed in the same buffer and then postfixed with 1% osmium tetroxide and 1% potassium ferrocyanide in 0.1 M cacodylate buffer for 1 h at RT to enhance the staining of cytoplasmic membranes (10). Cells were rinsed with distilled water and embedded in epoxy resin. Embedded samples then were conventionally processed for thin sectioning and observed with a Philips CM12 transmission electron microscope equipped with an Olympus SIS charge-coupled display camera.
RESULTS
Hypoxia induces autophagy in an HIF-dependent manner in normal and cancer cells.
We explored the hypoxia-induced autophagic process in normal cells and in a variety of cancer cell lines. When autophagy is activated, for example, after amino acid starvation, the LC3-I protein localized in the cytoplasm is cleaved, lipidated, and inserted as LC3-II into autophagosome membranes (36). Thus, an increase in the amount of the smaller-molecular-weight LC3-II protein and an increase in the LC3-II/LC3-I ratio are a hallmark of autophagy and correlate with an increased number of autophagosomes. We first assessed hypoxia-induced autophagy in normal cells (CCL39 hamster fibroblasts) and cancer cells (MCF7 breast carcinoma cells, used as a control, since these cells do not express detectable levels of endogenous Beclin1 [17]), LS174 colon carcinoma cells, and PC3 prostate carcinoma cells by immunoblotting for LC3-I/LC3-II. In CCL39 cells, the conversion of LC3-I (18 kDa) to LC3-II (16 kDa) and an increase in the amount of LC3-II already were apparent after 24 h of hypoxia (1% O2) and significantly increased after 48 h (Fig. 1A). In MCF7, LS174, and PC3 cells, autophagy (i.e., an increase in the amount of LC3-II protein) was significantly induced in the first 24 h of hypoxia in both 1% (moderate hypoxia) and 0.1% O2 (severe hypoxia; data not shown) and was further increased after 48 h of hypoxia (Fig. 1B). It is essential to note that after 48 h, the time period of maximally induced levels of BNIP3 and BNIP3L, cell death did not exceed 5% for any cell type (as measured by trypan blue exclusion; data not shown) of the cell lines subjected to hypoxic treatment. To confirm that hypoxia is a strong stimulus of autophagy, four independent approaches for measuring autophagy were developed. First, PC3 cells were stably transfected with a green fluorescent protein-LC3 (GFP-LC3) plasmid and analyzed by fluorescence microscopy. GFP-LC3 PC3 cells showed a diffuse distribution of green fluorescence and a few dots (18 dots/cell) in normoxia (see Fig. S1 in the supplemental material). Following 48 h of hypoxia, a large number of cells displayed the GFP-LC3 marker in association with an increase in the punctate pattern and an increase in the number of cytoplasmic autophagic vacuoles (122 dots/cell) (see Fig. S1 in the supplemental material). Autophagosome structures also were observed (see Fig. S1 in the supplemental material). Second, we examined the ultrastructure of PC3 and LS174 cells subjected to hypoxia (1% O2) for 24 and 48 h. After 24 h, hypoxic PC3 and LS174 cells showed newly formed autophagosomes (see Fig. S2 and S3 in the supplemental material), which was not detectable in normoxia (data not shown). Note that autophagosomes were detected in both PC3 and LS174 cells, and in some cells a substantial increase in the number of multiple double membranes was observed in PC3 cells, but only under hypoxic conditions (see Fig. S2 in the supplemental material). After 48 h, cells also showed extensive multiple vesicular bodies and heterolysosomes (see Fig. S2 in the supplemental material). Most double-membrane-bound vacuoles contained sequestrated cell organelles. These results demonstrate that the entire autophagic process clearly is engaged after 24 h of hypoxia and that the presence of a large number of heterolysosomes and lysosomes reflects the completion of the process after 48 h. Third, we used bafilomycin A1, a specific inhibitor of vacuolar H+-ATPases and a blocker of autophagosome-lysosome fusion (20, 31). CCL39 cells were incubated in hypoxia (1% O2) for 24 h in the presence of bafilomycin A1 (1 and 4 h) prior to the end of the hypoxic treatment (Fig. 1C). As expected, bafilomycin A1 treatment leads to an increase in the amount of LC3-II. This increase was substantiated in hypoxia (H 1%), which argues strongly in favor of a hypoxia-induced autophagic flux. Fourth, we used MDC (100 μM for 1 h), a lysomotropic fluorescent compound that accumulates in acidic autophagic vacuoles (3, 21). After 48 h of hypoxia, MDC accumulated in vacuoles (Fig. 1D), which is in agreement with the observations from electron microscopy. As expected, in the presence of bafilomycin A1, an increase in the autofluorescent substance MDC was observed in hypoxia, thus confirming the hypoxia-dependent enhancement of the autophagic flux. Finally, the knockdown of Beclin1 (shBCN; a more than 90% reduction) (Fig. 1E, inset) by lentivirus short hairpin RNA interference totally abolished hypoxia-induced autophagy (Fig. 1E) compared to that of the control (empty lentivirus), as represented by the reduced detection of LC3-II.
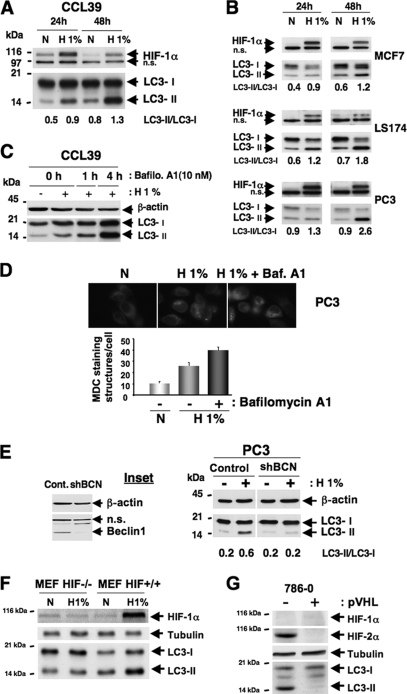
FIG. 1.
Hypoxia induces autophagy in an HIF-dependent manner in normal and cancer cell lines. (A) Conversion of LC3-I to LC3-II in CCL39 cells subjected to 24 and 48 h of hypoxia (H 1%) compared to that of cells in normoxia (N). Total cellular extracts were analyzed by immunoblotting with antibodies against HIF-1α and LC3. The LC3-I and LC3-II bands were quantified, and autophagy was reported as a variation in the ratio of LC3-II/LC3-I for each condition. n.s., nonspecific. (B) Conversion of LC3-I to LC3-II in MCF7, LS174, and PC3 cells subjected to 24 and 48 h of hypoxia for the indicated times. Total cell extracts were analyzed by immunoblotting with antibodies against HIF-1α and LC3. (C) Accumulation of LC3-II in the presence of bafilomycin A1 (Bafilo. A1) in CCL39 cells. CCL39 cells were incubated in hypoxia (24 h) and treated with bafilomycin A1 (10 nM for 1 or 4 h) prior to the end of hypoxic treatment. Total cellular extracts were analyzed by immunoblotting with antibodies against LC3. β-Actin was used as a control. (D) MDC staining of PC3 cells. PC3 cells were treated for 48 h in normoxia or hypoxia in the absence (− bafilomycin A1) or presence (+ bafilomycin A1) of bafilomycin A1 (1 nM for 48 h), and during the last hour 0.1 μM MDC was added. The quantification of MDC-stained structures was performed using the analyze particles tool of the Image J software on an average of 100 cells per treatment. Baf. A1, bafilomycin A1. (E) Ablation of Beclin1 totally suppresses hypoxia-induced autophagy. The inset shows the ablation of Beclin1 with a shBeclin1 lentivirus. Cont., control. PC3 cells were infected with shBCN. Total cell extracts were analyzed by immunoblotting with antibodies against Beclin1. β-Actin was used as a control. For immunoblotting, PC3 control and shBCN cells were subjected to either normoxia or hypoxia (48 h). Total cell extracts were analyzed by immunoblotting with antibodies against LC3. β-Actin was used as a control. (F) Knockout of HIF-1α suppresses hypoxia-induced autophagy. MEFs derived from wild-type (HIF+/+) and HIF-1α knockout embryos (HIF−/−) were subjected to either normoxia or hypoxia (48 h). Total cell extracts were analyzed by immunoblotting with antibodies against HIF-1α and LC3. Tubulin was used as a control. (G) Suppression of HIF-2α totally represses hypoxia-induced autophagy. Total cell extracts of renal clear cell carcinoma 786-O cells not expressing pVHL (−) or expressing pVHL (+) in normoxia were analyzed by immunoblotting with antibodies against HIF-1α, HIF-2α, and LC3. Tubulin was used as a control.
As HIF-1 is the key protein responsible for cellular adaptation to low oxygen tension, we checked if HIF-1 was involved in the hypoxia-induced autophagy. Using MEFs derived from wild-type (HIF+/+) and HIF-1α knockout embryos (HIF−/−), we showed that in the absence of HIF-1α, cells subjected to 48 h of hypoxia did not present an increase in the amount of LC3-II, whereas this occurred in HIF+/+ MEF cells (Fig. 1F). To reinforce this result, autophagy was measured in renal clear-cell carcinoma 786-O cells expressing only HIF-2α. As expected, autophagy was observed in the absence of pVHL (i.e., the stabilization of HIF-2α), whereas no autophagy was observed in cells in which pVHL was reestablished (i.e., no stabilization of HIF-2α) (Fig. 1G).
Taken together, these results demonstrate that hypoxic stress, mediated through HIF-1α or HIF-2α, is a strong signal that initiates the autophagic process. In addition, this hypoxia-induced autophagy is a permanent process, as cells treated for 1 week under hypoxic conditions at 1% O2 maintained a large amount of the LC3-II protein (data not shown).
Ablation of both BNIP3 and BNIP3L is required to suppress hypoxia-induced autophagy.
We next asked whether silencing BNIP3 would prevent hypoxia-induced autophagy. The ablation of 90% of the hypoxia-inducible expression of BNIP3 in CCL39 cells slightly decreased the level of LC3-II (Fig. 2A). Since the ablation of BNIP3 alone in CCL39 cells did not suppress hypoxia-induced autophagy, we examined the impact of the coablation of BNIP3 and BNIP3L in CCL39 and PC3 cells. The ablation of BNIP3L in a dose-dependent manner in CCL39 cells slightly diminished the level of LC3-II compared to that of hypoxic cells transfected with the siRNA control (data not shown). However, in the same cells, the coablation of BNIP3 and BNIP3L with 40 and 20 nM siRNA (data not shown) and 40 and 200 nM siRNA, respectively, reversed hypoxia-induced autophagy (Fig. 2B), and the amount of LC3-II clearly decreased. We next explored PC3 tumor cells in which the autophagic process in hypoxia is very pronounced, showing a robust increase in the amount of LC3-II (Fig. 2C). In these cells, the ablation of BNIP3L (20 nM) or BNIP3 (40 nM) alone with siRNA partially decreased to 50% the level of LC3-II seen in hypoxia (Fig. 2C). However, the most striking effect was obtained by the dual ablation of BNIP3/BNIP3L, for which hypoxia-induced autophagy was fully suppressed (Fig. 2C). The LC3-II level was reversed, close to the level found in normoxia. These results were confirmed using stable GFP-LC3 cells transfected under the same conditions, as described previously. The ablation of BNIP3, BNIP3L, or both strongly decreased the punctate GFP pattern (Fig. 2D). The GFP-LC3 PC3 assay appears to be more sensitive than the determination of the LC3-II/LC3-I ratio in estimating autophagy, since the ablation of either BNIP3 or BNIP3L impacts hypoxia-induced autophagy in the GFP assay, but this is not reflected in the LC3-II/LC3-I ratio. However, it is clear that the ablation of both proteins restores a normoxic level of autophagy in the different experiments (Fig. 2B, C, and D). These findings establish for the first time that the induction of both BNIP3 and BNIP3L was required to initiate the autophagic process in response to a hypoxic stress in both normal and tumor cells.
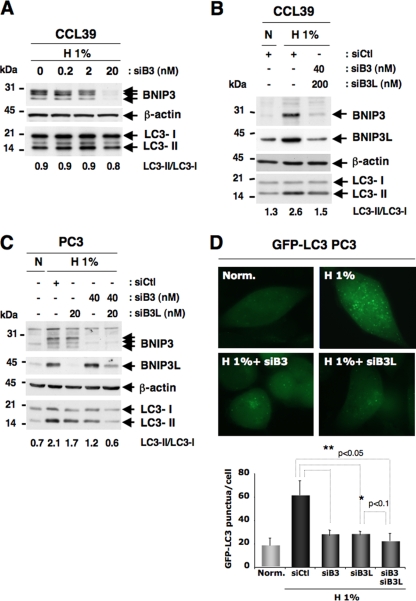
FIG. 2.
Silencing of BNIP3 and BNIP3L strongly affects the autophagic status of CCL39 and PC3 cells in hypoxia. (A) Specific silencing of BNIP3 in a dose-dependent manner using siRNA did not impact the conversion of LC3-I to LC3-II in CCL39 cells. Cell extracts were prepared from cells incubated in hypoxia (H 1%) 48 h after transfection with siRNA to BNIP3 (from 0.2 to 20 nM). Total cellular extracts were analyzed by immunoblotting with antibodies against BNIP3 and LC3. β-Actin was used as a control. siB3, siRNA to BNIP3. (B) Silencing of both BNIP3 and BNIP3L alters hypoxia-induced autophagy in CCL39 cells. Cells were transfected once with siRNA to BNIP3L (200 nM) and BNIP3 (40 nM), each alone or in combination, and maintained in normoxia or hypoxia for the last 48 h. Total cell extracts were analyzed by immunoblotting with antibodies against BNIP3, BNIP3L, and LC3. sictl, siRNA to SIMA; siB3L, siRNA to BNIP3L. (C) Silencing of both BNIP3 and BNIP3L alters hypoxia-induced autophagy in PC3 cells. PC3 cells were transfected once with siRNA to BNIP3L (20 nmol/liter) and BNIP3 (40 nmol/liter), each alone or in combination, and maintained in normoxia or hypoxia for the last 48 h. Total cellular extracts were analyzed by immunoblotting with antibodies against BNIP3, BNIP3L, and LC3. Two independent sets of siRNA to BNIP3 and BNIP3L were used. Experiments were done at least twice. Results are from one representative experiment. The band intensities of endogenous LC3-I and LC3-II were quantified, and the LC3-II/LC3-I ratio is indicated. (D) Silencing of BNIP3 and BNIP3L alters hypoxia-induced autophagy in stable GFP-LC3 PC3 cells. PC3 cells were transfected once with siRNA to BNIP3L (20 nmol/liter) and BNIP3 (40 nmol/liter), each alone or in combination, and maintained in normoxia or hypoxia for the last 48 h. An example of GFP-LC3 punctate structures observed in PC3 cells under normoxic and hypoxic conditions is shown. GFP fluorescence was examined under a Zeiss fluorescence microscope. The quantification of the number of GFP aggregates per cell was performed using the analyze particles command in the Image J software. *, P < 0.00001.
Ectopic expression of both BNIP3 and BNIP3L triggers autophagy in normoxia.
We then questioned whether the ectopic expression of BNIP3 and BNIP3L was sufficient to initiate the autophagic process. We attacked this question in three ways. We first asked whether it is possible to artificially promote autophagy in normoxia and in the presence of rich medium by the simple coexpression of BNIP3 and BNIP3L. Among the myriad of gene products expressed in response to hypoxia, are these two HIF-dependent gene products sufficient to induce autophagy? The coexpression of BNIP3 and BNIP3L in normoxia proved to be difficult. In many instances, we observed that the expression of BNIP3 slightly represses the expression of BNIP3L, suggesting an interplay between these two homologues. When BNIP3 expression was induced in response to Tet (Fig. 3A, lanes 3 and 4), a concomitant decrease in BNIP3L was observed. Under these conditions (Fig. 3A, lanes 1 to 4), no change in the level of LC3-II was observed in the presence of Tet. We hypothesized that BNIP3 reduces the half-life of the BNIP3L protein. We therefore conducted experiments in the presence of the proteasome inhibitor MG132. Under these conditions, BNIP3L expression persists with the strong coinduction of BNIP3. We thus observed an increase in the level of LC3-II and a decrease in the expression of LC3-I (Fig. 3A, lanes 5 to 8). Because MG132 inhibits the degradation of many cellular proteins, as a control we conducted the same experiment with a CCL39 clone expressing Tet-inducible carbonic anhydrase IX, but no change in the LC3-II level was detected in the presence of MG132 (see Fig. S4 in the supplemental material).
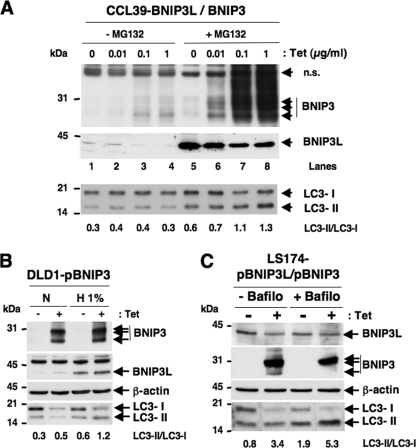
FIG. 3.
Overexpression of BNIP3 and BNIP3L strongly affects the autophagic status of CCL39, DLD1, and LS174 cells in normoxia. (A) Overexpression of BNIP3 and BNIP3L triggers autophagy in normoxia in CCL39-pBNIP3/BNIP3L cells. Cells were incubated in the absence (0 μg/ml) or presence (0.01, 0.1, or 1 μg/ml) of Tet for 4 days and maintained in the presence or absence of MG132 (5 μM) for the last 18 h. Total cell extracts were analyzed by immunoblotting with antibodies to BNIP3, BNIP3L, and LC3. (B) Combined BNIP3 and BNIP3L overexpression in DLD1-pBNIP3 cells did impact the conversion of LC3-I to LC3-II. DLD1-pBNIP3 cells were cultured in the absence (−) or presence (+) of Tet (10 μg/ml) for 4 days and maintained in either normoxia (N) or hypoxia (H 1%) for the last 24 h. Total cell extracts were analyzed by immunoblotting with antibodies against BNIP3, BNIP3L, and LC3. (C) BNIP3 and BNIP3L overexpression in combination in LS174-pBNIP3L/BNIP3 cells modified the conversion of LC3-I to LC3-II. LS174-pBNIP3L/BNIP3 clones were cultured in the absence (− Tet) or presence (+ Tet) of Tet (10 μg/ml) for 24 h. In the meantime, cells were cultured in the absence (− Bafilo) or presence (+ Bafilo) of bafilomycin A1 (1 nM) for the same 24 h. Total cell extracts were analyzed by immunoblotting with antibodies against BNIP3, BNIP3L, and LC3. The intensity of the bands of endogenous LC3-I and LC3-II was quantified, and the LC3-II/LC3-I ratio is indicated.
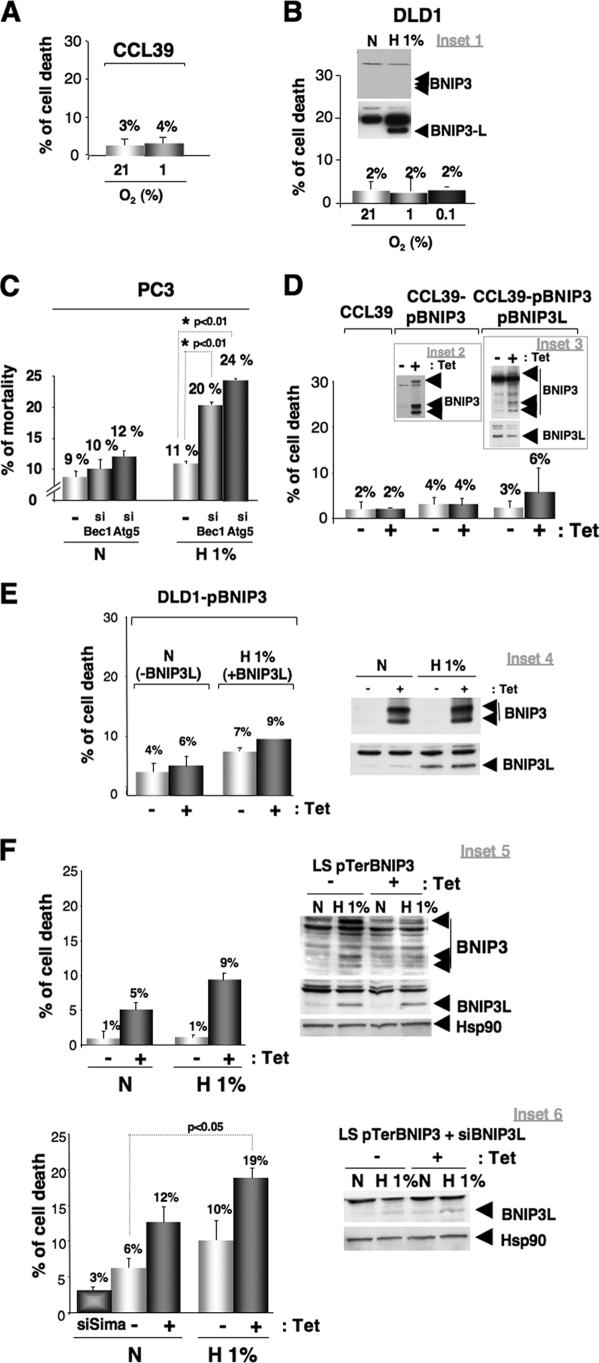
In the second approach, we hypothesized that restoring BNIP3 in DLD1 cells lacking BNIP3 expression (22 and data not shown) would restore the defective hypoxia-induced autophagic process. In the subclone DLD1-pBNIP3, which expresses only BNIP3L (in the absence of Tet), 48 h of hypoxia induced a low level of autophagy, as reflected by a slight variation in the level of LC3-II (Fig. 3B). In contrast, the restoration of BNIP3 via Tet addition slightly increased hypoxia-induced autophagy in this DLD1-defective cell line, since LC3-II and LC3-I almost totally disappeared (Fig. 3B).
Finally, a clone similar to that obtained in CCL39 cells (CCL39-pBNIP3L/BNIP3) was isolated from LS174 cells (LS174-pBNIP3/pBNIP3L). The basal expression of BNIP3L and the inducible expression of BNIP3 in response to Tet in normoxia clearly showed an increase in LC3-II and a clear decrease in LC3-I (Fig. 3C). In addition, the coexpression of BNIP3 and BNIP3L in the presence of bafilomycin A1 resulted in an enhanced accumulation of LC3-II compared to that of the other conditions (Fig. 3C). These data indicate that the ectopic expression of BNIP3 and BNIP3L is sufficient to induce an autophagic flux in normoxia, thus mimicking the hypoxia-dependent autophagic flux. The coexpression of BNIP3/BNIP3L had the capacity to promote the initiation of autophagy in the absence of any stress signals generated by O2 or by nutrient deprivation. In addition, these results confirm that the HIF-1-dependent expression of BNIP3/BNIP3L is a key mechanism in the activation of hypoxia-induced autophagy.
Hypoxia-induced autophagy is a survival process.
We first confirmed that autophagy, as a survival mechanism under hypoxic conditions of 1 or 0.1% O2 (for 24 or 48 h), does not lead to cell death in either nonneoplastic CCL39 or neoplastic DLD1 cells (Fig. 4A and B) or in any other cell line tested in our laboratory (LS174, PC3, HepG2, Hep3B, BE, A549, RCC4, 786-O, and HeLa; data not shown). In addition, we clearly demonstrate that the silencing of Beclin1 (Fig. 4C), a major actor and initiator of autophagy, and the ablation of ATG5 (Fig. 4C) or ATG7 (data not shown) enhances cell death in hypoxia, thereby suggesting that autophagy participates in cell survival rather than cell death in hypoxia.
FIG. 4.
Blockade of autophagy in hypoxia triggers cell death through BNIP3. (A) Measurement of cell death in CCL39 cells. CCL39 cells were subjected to either normoxia (N) or hypoxia (H 1%) for 48 h before measuring cell death. (B) Measurement of cell death in DLD1 cells. Inset 1 shows the expression of BNIP3 and BNIP3L in DLD1 cells. The immunoblot shows DLD1 cells subjected to either normoxia (N) or hypoxia (H 1%) for 24 h. BNIP3 and BNIP3L expression was analyzed by immunoblotting. For the measurement of cell death, DLD1 cells were subjected to either normoxia (21% O2) or hypoxia (1 and 0.1% O2) for 48 h before measuring cell death. (C) Ablation of Beclin1 and ATG5 increase cell death in hypoxia. PC3 cells were transfected twice during a 24-h interval with siRNA to the control SIMA (40 nmol/liter), Beclin1 (40 nmol/liter), or ATG5 (40 nmol/liter), and cells were incubated for 48 h in hypoxia (H 1%) or normoxia (N). Beclin1 and ATG5 expression was analyzed by immunoblotting, and autophagy was examined. Two independent sets of siRNA to Beclin1 were used, but only one set of siRNA to ATG5 was used. Experiments were done at least twice. (D) Measurement of cell death in CCL39 clones overexpressing BNIP3/BNIP3L. CCL39 cells were incubated in the absence (−Tet) or presence (+Tet) of Tet (10 μg/ml) for 3 days before measuring cell death. Inset 2 shows the overexpression of BNIP3 in CCL39 cells. The immunoblot shows the overexpression of the BNIP3 protein in the presence of Tet. BNIP3 expression was analyzed by immunoblotting 3 days after Tet addition. The clone strongly expressed BNIP3 protein levels in normoxia. For the measurement of cell death, CCL39-pBNIP3 and CCL39-pBNIP3-BNIP3L clones were incubated in the absence (−Tet) or presence (+Tet) of Tet (10 μg/ml) for 3 days before measuring cell death. Inset 3 shows the overexpression of BNIP3 and BNIP3L in CCL39 cells. The immunoblot shows the stable overexpression of BNIP3L and the overexpression of the BNIP3 protein in the presence of Tet. BNIP3 expression was analyzed by immunoblotting 3 days after Tet addition. The clone strongly expressed BNIP3 and BNIP3L protein levels under normoxia. For cell death measurement, CCL39-pBNIP3-BNIP3L clones were incubated in the absence (−Tet) or presence (+Tet) of Tet (10 μg/ml) for 3 days before measuring cell death. (E) Measurement of cell death in Tet-induced BNIP3-expressing DLD1 cells. Inset 4 shows the expression of BNIP3 and BNIP3L in the DLD1 pBNIP3 clone. The immunoblot shows the DLD1 pBNIP3 clone incubated in the absence (−Tet) or presence (+Tet) of Tet (10 μg/ml) for 3 days before incubation for 24 h in hypoxia (1% O2). BNIP3 and BNIP3L expression was analyzed by immunoblotting. For the measurement of cell death, the DLD1 pBNIP3 clone was incubated in the absence (−Tet) or presence (+Tet) of Tet (10 μg/ml) for 3 days before incubation for 24 h in hypoxia (1% O2) before measuring cell death. (F) Ablation of BNIP3 increases cell death in normoxia and hypoxia. Inset 5 shows the effect of BNIP3 ablation. The immunoblot shows the ablation of BNIP3 in normoxia and hypoxia. LS pTerBNIP3 cells were incubated in the absence (−Tet) or presence (+Tet) of Tet (10 μg/ml) for 3 days before incubation for 48 h in hypoxia (H 1% O2). BNIP3 and BNIP3L expression was analyzed by immunoblotting. Hsp90 was used as a control. For the measurement of cell death, the LS pTerBNIP3 clone was incubated in the absence (−Tet) or presence (+Tet) of Tet (10 μg/ml) for 3 days before incubation in either normoxia (N) or hypoxia (H 1%) for 48 h. Inset 6 shows the effect of BNIP3 and BNIP3L ablation. The immunoblot shows the ablation of BNIP3L in normoxia and hypoxia in the absence of BNIP3 expression. The LS pTerBNIP3 clone was incubated in the absence (−Tet) or presence (+Tet) of Tet (10 μg/ml) for 3 days before transient transfection with siRNA to BNIP3L (40 nM). Cells then were subjected to normoxia (N) or hypoxia (H 1%) for 48 h. BNIP3 and BNIP3L expression was analyzed by immunoblotting. Hsp90 was used as a control. For the measurement of cell death, the LS pTerBNIP3 clone was incubated in the absence (−Tet) or presence (+Tet) of Tet (10 μg/ml) for 3 days before transient transfection with siRNA to BNIP3L (40 nM). An siRNA to SIMA (siSIMA; 40 nM) was used as a control. Cells then were subjected to normoxia (N) or hypoxia (H 1%) for 48 h before measuring cell death with trypan blue. Experiments were done in duplicate at least three times.
Thus, whereas HIF promotes cell survival, it was rather paradoxical that, in parallel, HIF could induce supposedly prodeath gene products like BNIP3 and BNIP3L. The forced induction of BNIP3 (CCL39-pBNIP3), the forced expression of BNIP3L (CCL39-pBNIP3L/pBNIP3 in the absence of Tet), or the forced coexpression of both BNIP3 and BNIP3L (CCL39-pBNIP3/pBNIP3L in the presence of Tet) in normoxia did not induce cell death (Fig. 4D), nor did the restoration of BNIP3 in the colorectal cell line DLD1, which does not express BNIP3 (Fig. 4E). Thus, these data confirm that BNIP3 and/or BNIP3L expression do not participate in cell death, as Papandreou et al. showed previously for MEFs and MCF7 cells (24). We finally resolved the paradox by establishing that BNIP3 and BNIP3L, contrary to previous belief, are not prodeath but prosurvival proteins.
Finally, the ablation of both BNIP3 and BNIP3L under normoxic conditions significantly increased cell death (19%), similarly to that for Beclin1 (siBec, 1 to 20%) or ATG5 (siATG, 5 to 24%) (Fig. 4F).
Taken together, these results strongly demonstrated that (i) hypoxia (1% O2) does not trigger cell death, (ii) hypoxia triggers autophagy, a survival process, and (iii) BNIP3 and BNIP3L are essential in the activation of hypoxia-induced autophagy.
BH3 domains of BNIP3 and BNIP3L trigger autophagy.
How do BNIP3 and BNIP3L trigger autophagy? A direct interaction between BNIP3 and BNIP3L using a two-hybrid system has been reported (28). BNIP3 is known to heterodimerize with Bcl-2 (27), but most importantly for this study, Bcl-2 also has been shown to interact directly with the evolutionarily conserved autophagic protein Atg6/Beclin1 (25). Therefore, autophagy is kept in check by the complex Bcl-2-Beclin1. In keeping with this notion, we anticipated that hypoxia leads to the dissociation of the Bcl-2-Beclin1 complex. Indeed, we found that the endogenous Bcl-2-Beclin1 complex, formed under normoxic conditions in PC3 cells, is fully dissociated following 48 h of hypoxia (Fig. 5A, compare immunoprecipitates of lanes 3 and 4). A recently identified BH3 domain of Beclin1 appears to mediate the interaction between Bcl-2, Bcl-XL, and Beclin1 (18, 23). This association is of low affinity (1 to 2 μM range) (23), and one could imagine that the BH3 domains of BNIP3 and BNIP3L, which have atypical features (6), easily could compete with Beclin1 and promote autophagy, whereas they fail to induce apoptosis because of their low affinity for Bcl-2, Bcl-XL, and other prosurvival members (6). We therefore tested whether BH3 peptides of BNIP3 and BNIP3L alone could promote autophagy in normoxia. These 20-mer peptides containing the BH3 core (Fig. 5B) were fused to a stretch of eight arginine residues for membrane transduction, as reported previously (11). BNIP3 and BNIP3L peptides were indeed capable of promoting a modest (1.3- to 1.4-fold induction) but reproducible autophagic response in LS174 cells, whereas the BH3 mutant peptide had no effect (Fig. 5C). The same peptides used in PC3 cells clearly increased the amount of LC3-II compared to that of the BH3 mutant peptide (data not shown). In contrast to the BH3 mimetic ABT737, which induces cell death in LS174 and PC3 cells (data not shown), the two BNIP BH3 peptides did not affect viability, as expected (data not shown).
To reinforce these results, an acellular assay was performed using whole-cell lysates from normoxic cells left untreated or treated with the BH3 mutant, BNIP3, or BNIP3L peptide (Fig. 5D). After incubation with the different peptides, the immunoprecipitation of Bcl2 was done to evaluate the amount of Beclin1 coimmunoprecipitated by immunoblot analysis, thus monitoring the ability of peptides to disrupt the interaction between Bcl-2 and Beclin1. As a result, we observed that only the BNIP3 or BNIP3L peptide decreased the amount of Beclin1 coprecipitated with Bcl-2 compared to that of the BH3 mutant peptide or the control. Consequently, we observed an increase in the amount of Beclin1 that no longer interacts with Bcl-2 (Fig. 5D, flow through). These results confirm the ability of the BH3 domains derived from BNIP3 and BNIP3L to trigger the disruption of the Bcl-2-Beclin1 complex.
Finally, BNIP3 or BNIP3L peptide was capable of increasing the intensity of the punctate pattern in stable GFP-LC3 PC3 cells under normoxic conditions (Fig. 5E). These data strengthen the role of the BNIP3/BNIP3L BH3 domain in the dissociation of the Bcl-2-Beclin1 complex.
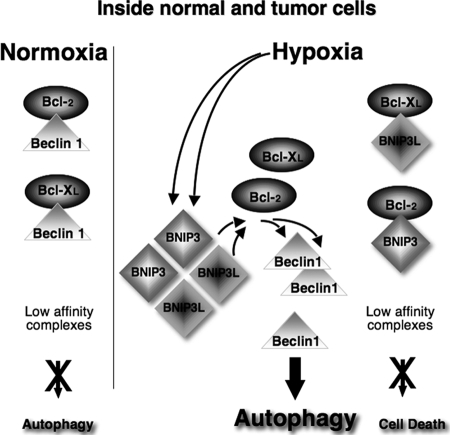
We thus propose a simple model (Fig. 6) in which the atypical BH3 domains of hypoxia-induced BNIP3/BNIP3L are central to the disruption of an interaction between Bcl-2 and Beclin1, thereby releasing the negative autophagic blockade.
FIG. 6.
Hypothetical model of HIF-induced autophagy. In normoxia (left), Beclin1 forms low-affinity complexes with Bcl-XL and Bcl-2 via its BH3 domain, thereby decreasing the rate of autophagy. In hypoxia (right), the rapid induction of the BH3-only proteins (BNIP3 and BNIP3L) displaces Beclin1 from Bcl-XL and Bcl-2, leading to autophagy. The affinity of the BH3 domains of the BNIP proteins is too low to form tight complexes with Bcl-XL and Bcl-2. Therefore, BNIP3 and BNIP3L fail to induce cell death.
DISCUSSION
Hypoxia-induced autophagy, a survival response.
Autophagy is an evolutionarily conserved mechanism involving the formation of autophagosomes that sequester cytoplasmic macromolecules and organelles before fusion with the endo/lysosomal compartment (13, 34, 35). Despite recent advances in understanding its molecular mechanisms and biological function, it remains controversial as to whether autophagy acts primarily as a cell survival or cell death pathway (9, 19). Autophagy has been described as a form of nonapoptotic or necrotic cell death based on morphological criteria observed for a variety of cell types during development. However, in the context of hypoxia, nutrient depletion, or growth factor deprivation, it is clear that autophagy is crucial in maintaining cellular ATP production and macromolecular synthesis and, therefore, represents an essential prosurvival pathway (19). In this study, we showed that hypoxia (1 or 0.1% O2 for 24 or 48 h) does not kill the numerous cells tested but rather induces autophagy, a survival process, as the ablation of Beclin1 or ATG5 increases cell death (Fig. 4C). In keeping with this notion, the recent work of Zhang et al. (41) revealed that hypoxia induces the autophagy of mitochondria to prevent an increase in the level of reactive oxygen species and cell death. Therefore, hypoxia appears to be an early prosurvival process, probably acting as a warning signal for cells to anticipate extreme nutritional stress.
BNIP3 and BNIP3L act together in autophagy.
In this study, we showed that the HIF-dependent coinduction of BNIP3 and BNIP3L is critical to activate hypoxia-induced autophagy. The foundation of this new finding is based on the following arguments: (i) hypoxia-induced autophagy is completely suppressed only when BNIP3 and BNIP3L are coinvalidated in normal and tumor cells; (ii) the dual ectopic expression of BNIP3 and BNIP3L suffices to trigger autophagy in normoxia; and finally, (iii) the ablation of BNIP3 or BNIP3L alone with siRNA increased cell death in hypoxia to 9 and 10%, respectively (Fig. 4F). However, a more striking effect was obtained by the dual ablation of BNIP3/BNIP3L (19% cell death). During the preparation of the manuscript, Tracy et al. reported that BNIP3 alone was essential for hypoxia-induced autophagy in Saos2 Rb-null cells (37). The authors showed that pRb attenuates the induction of BNIP3 by HIF, thereby acting as a transcriptional repressor. However, they did not study the possible impact of pRb on BNIP3L expression, suggesting that their results are a reflection of both BNIP3 and BNIP3L expression. Very recently, Sandoval et al. confirmed the critical role of BNIP3L in the autophagic maturation of erythroid cells by disturbing the mitochondrial membrane potential and by affecting the targeting of mitochondria into autophagosomes, but they did not study the impact of BNIP3 in Nix−/− mice (30). Taken together, our results bring additional support to the prosurvival feature of these two HIF target gene products. We clearly reveal that BNIP3 and BNIP3L can act separately, but the strength of the response is enhanced when BNIP3 and BNIP3L act together.
Mechanism of action of BNIP3 and BNIP3L.
By what mechanism do BNIP3 and BNIP3L trigger hypoxia-induced autophagy? The nutrient status of cells regulates the interaction between Bcl-2 and Beclin1, presumably via the mTOR kinase-dependent phosphorylation of Bcl-2. Although hypoxia reduces the activity of mTOR via the induction of Redd1 (5) and also via a direct link between BNIP3 and Rheb (16), this action could contribute to weakening the Bcl-2-Beclin1 complex but cannot account for the pronounced autophagy observed under hypoxic conditions. Indeed, rapamycin is only a weak activator of autophagy compared to hypoxia (data not shown), and the ectopic expression of BNIP3/BNIP3L in normoxia activates autophagy under conditions where mTOR is fully active. This observation suggests that BNIP3/BNIP3L act independently of mTOR to disrupt the Bcl-2-Beclin1 complex. How do they act? Recently, Beclin1, a key player in the initiation of the autophagy signaling platform, was defined as a novel BH3-only protein (18, 23), suggesting it has a dual role in apoptosis/autophagy or that it is an atypical BH3 domain. Levine et al. recently suggested that Beclin1 acts as an interactome in the regulation of apoptosis rather than autophagy (15). We hypothesize that BNIP3 and BNIP3L act as prosurvival proteins through their intrinsic atypical BH3 domain. Small peptides containing the BH3 core domains of BNIP3 or BNIP3L were capable of triggering cells into an autophagic response under normoxic conditions. Although this autophagic response was modest in intensity, it was reproducible in three independent experiments, as observed in the two cell lines tested (LS174 and PC3), and was specific for an active BH3 domain, since the double mutant LL/AA BH3 peptide had no effect. In addition, BNIP3 and BNIP3L peptides also were capable of triggering Bcl-2-Beclin1 complex dissociation, as shown in immunoprecipitation experiments (Fig. 5D). Stable GFP-LC3 PC3 cells, a more sensitive system, confirmed this conclusion, as the BNIP3 and BNIP3L peptides promoted autophagosome structures in normoxia, in contrast to the mutated peptide (Fig. 5E).
These results lead us to propose a model (Fig. 6) in which the BH3 domains of BNIP3 and BNIP3L expressed in hypoxia displace Beclin1 from the Bcl-2- or Bcl-XL-Beclin1 complex. Although we have not been able to measure the affinity of these BNIP BH3 peptides for Bcl-XL, we propose that their affinity is low, like that of the BH3 domain of Beclin1 (18, 23). This assumption is based on the fact that Beclin1, like BNIP3/BNIP3L, is not capable of triggering cell death, the association with the prosurvival proteins Bcl-2/Bcl-XL being too low. We hypothesize that within the large family of BH3-only proteins (1), BNIP proteins have been designed through evolution as low-affinity BH3 binders, a feature that is appropriate for triggering autophagy without inducing apoptosis (Fig. 6). This hypothesis, as well as the intimate mechanism by which BNIP dimers and/or heterodimer complexes disrupt the Beclin1-Bcl complexes, needs further investigation. To reinforce this model, we observed the expression levels of both Bcl-2 and Bcl-XL under hypoxic conditions (1% O2 for 48 h) in MCF7, PC3, and LS174 cells (Table 1). Interestingly, cells that show a high level of expression of Bcl-2 with a high level of expression of BNIP3 and BNIP3L, such as MCF7 cells, show only low levels of autophagy. However, in cells with levels of BNIP3 and BNIP3L similar to those of MCF7 cells but weakly expressing Bcl-2 or Bcl-XL, autophagy was strongly induced. These observations suggest that the ratio between the prosurvival proteins Bcl-2/Bcl-XL and the low-affinity BH3 binders BNIP3/BNIP3L/Beclin is crucial to understanding the mechanism by which autophagy is induced under hypoxic conditions.
TABLE 1.
Hypoxia-induced autophagy is dependent on Bcl-2/Bcl-XL expressiona
| Neoplastic cell line(s) | Expression of:
|
Autophagy | |||
|---|---|---|---|---|---|
| Bcl2 | Bcl-XL | BNIP3 | BNIP3L | ||
| DLD1 | − | − | − | + | +/− |
| MCF7 | +++ | +/− | + | + | +/− |
| PC3/LS174 | +/− | +/− | + | + | ++ |
MCF7, PC3, and LS174 cells were subjected to normoxia or hypoxia (48 h; H 1%) and analyzed by immunoblotting with antibodies against Bcl-2, Bcl-XL, BNIP3, BNIP3L, and LC3. Tubulin was used as the control.
In conclusion, our results clearly uncover one of the crucial functions of BNIP3 and BNIP3L in hypoxia-induced autophagy and show how hypoxia impacts cell survival. The identification of BNIP3 and BNIP3L as central mediators of autophagy definitively reveals their novel function as prosurvival proteins in opposition to pro-cell death proteins. These results give us good reason to think that the manipulation of HIF-induced autophagy via BNIP3 and BNIP3L is a good therapeutic option to investigate for the treatment of cancer.
Supplementary Material
Acknowledgments
This work was supported by the Ligue Nationale Contre le Cancer (Equipe labelisée), the Centre National de la Recherche Scientifique (CNRS), the Institut National du Cancer, and Canceropole PACA.
We thank John Hickman and Olivier Geneste (Servier) for helpful discussions and in vitro experiments with BH3 peptides and Patrice Codogno and Tamotsu Yoshimori for providing reagents. We thank M. Christiane Brahimi-Horn (INSERM) for the critical reading of the manuscript.
Footnotes
Published ahead of print on 9 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, J. M., and S. Cory. 2007. Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr. Opin. Immunol. 19488-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berra, E., E. Benizri, A. Ginouves, V. Volmat, D. Roux, and J. Pouyssegur. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 224082-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biederbick, A., H. F. Kern, and H. P. Elsasser. 1995. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 663-14. [PubMed] [Google Scholar]
- 4.Boyd, J. M., S. Malstrom, T. Subramanian, L. K. Venkatesh, U. Schaeper, B. Elangovan, C. D'Sa-Eipper, and G. Chinnadurai. 1994. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell 79341-351. [DOI] [PubMed] [Google Scholar]
- 5.Brugarolas, J., K. Lei, R. L. Hurley, B. D. Manning, J. H. Reiling, E. Hafen, L. A. Witters, L. W. Ellisen, and W. G. Kaelin, Jr. 2004. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 182893-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Certo, M., V. Del Gaizo Moore, M. Nishino, G. Wei, S. Korsmeyer, S. A. Armstrong, and A. Letai. 2006. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 9351-365. [DOI] [PubMed] [Google Scholar]
- 7.Chen, G., J. Cizeau, C. Vande Velde, J. H. Park, G. Bozek, J. Bolton, L. Shi, D. Dubik, and A. Greenberg. 1999. Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J. Biol. Chem. 2747-10. [DOI] [PubMed] [Google Scholar]
- 8.Dayan, F., D. Roux, M. C. Brahimi-Horn, J. Pouyssegur, and N. M. Mazure. 2006. The oxygen sensor factor-inhibiting hypoxia-inducible factor-1 controls expression of distinct genes through the bifunctional transcriptional character of hypoxia-inducible factor-1alpha. Cancer Res. 663688-3698. [DOI] [PubMed] [Google Scholar]
- 9.Debnath, J., E. H. Baehrecke, and G. Kroemer. 2005. Does autophagy contribute to cell death? Autophagy 166-74. [DOI] [PubMed] [Google Scholar]
- 10.de Bruijn, W. C. 1973. Glycogen, its chemistry and morphologic appearance in the electron microscope. I. A modified OsO 4 fixative which selectively contrasts glycogen. J. Ultrastruct. Res. 4229-50. [DOI] [PubMed] [Google Scholar]
- 11.Goldsmith, K. C., X. Liu, V. Dam, B. T. Morgan, M. Shabbout, A. Cnaan, A. Letai, S. J. Korsmeyer, and M. D. Hogarty. 2006. BH3 peptidomimetics potently activate apoptosis and demonstrate single agent efficacy in neuroblastoma. Oncogene 254525-4533. [DOI] [PubMed] [Google Scholar]
- 12.Guo, K., G. Searfoss, D. Krolikowski, M. Pagnoni, C. Franks, K. Clark, K. T. Yu, M. Jaye, and Y. Ivashchenko. 2001. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ. 8367-376. [DOI] [PubMed] [Google Scholar]
- 13.Kondo, Y., T. Kanzawa, R. Sawaya, and S. Kondo. 2005. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer 5726-734. [DOI] [PubMed] [Google Scholar]
- 14.Lee, H., and S. G. Paik. 2006. Regulation of BNIP3 in normal and cancer cells. Mol. Cells 211-6. [PubMed] [Google Scholar]
- 15.Levine, B., S. Sinha, and G. Kroemer. 2008. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 4600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y., Y. Wang, E. Kim, P. Beemiller, C. Y. Wang, J. Swanson, M. You, and K. L. Guan. 2007. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J. Biol. Chem. 28235803-35813. [DOI] [PubMed] [Google Scholar]
- 17.Liang, X. H., S. Jackson, M. Seaman, K. Brown, B. Kempkes, H. Hibshoosh, and B. Levine. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402672-676. [DOI] [PubMed] [Google Scholar]
- 18.Maiuri, M. C., G. Le Toumelin, A. Criollo, J. C. Rain, F. Gautier, P. Juin, E. Tasdemir, G. Pierron, K. Troulinaki, N. Tavernarakis, J. A. Hickman, O. Geneste, and G. Kroemer. 2007. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 262527-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathew, R., V. Karantza-Wadsworth, and E. White. 2007. Role of autophagy in cancer. Nat. Rev. Cancer 7961-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima, N., and T. Yoshimori. 2007. How to interpret LC3 immunoblotting. Autophagy 3542-545. [DOI] [PubMed] [Google Scholar]
- 21.Munafo, D. B., and M. I. Colombo. 2001. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J. Cell Sci. 1143619-3629. [DOI] [PubMed] [Google Scholar]
- 22.Murai, M., M. Toyota, H. Suzuki, A. Satoh, Y. Sasaki, K. Akino, M. Ueno, F. Takahashi, M. Kusano, H. Mita, K. Yanagihara, T. Endo, Y. Hinoda, T. Tokino, and K. Imai. 2005. Aberrant methylation and silencing of the BNIP3 gene in colorectal and gastric cancer. Clin. Cancer Res. 111021-1027. [PubMed] [Google Scholar]
- 23.Oberstein, A., P. D. Jeffrey, and Y. Shi. 2007. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 28213123-13132. [DOI] [PubMed] [Google Scholar]
- 24.Papandreou, I., C. Krishna, F. Kaper, D. Cai, A. J. Giaccia, and N. C. Denko. 2005. Anoxia is necessary for tumor cell toxicity caused by a low-oxygen environment. Cancer Res. 653171-3178. [DOI] [PubMed] [Google Scholar]
- 25.Pattingre, S., A. Tassa, X. Qu, R. Garuti, X. H. Liang, N. Mizushima, M. Packer, M. D. Schneider, and B. Levine. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122927-939. [DOI] [PubMed] [Google Scholar]
- 26.Pouyssegur, J., F. Dayan, and N. M. Mazure. 2006. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 441437-443. [DOI] [PubMed] [Google Scholar]
- 27.Ray, R., G. Chen, C. Vande Velde, J. Cizeau, J. H. Park, J. C. Reed, R. D. Gietz, and A. H. Greenberg. 2000. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J. Biol. Chem. 2751439-1448. [DOI] [PubMed] [Google Scholar]
- 28.Rual, J. F., K. Venkatesan, T. Hao, T. Hirozane-Kishikawa, A. Dricot, N. Li, G. F. Berriz, F. D. Gibbons, M. Dreze, N. Ayivi-Guedehoussou, N. Klitgord, C. Simon, M. Boxem, S. Milstein, J. Rosenberg, D. S. Goldberg, L. V. Zhang, S. L. Wong, G. Franklin, S. Li, J. S. Albala, J. Lim, C. Fraughton, E. Llamosas, S. Cevik, C. Bex, P. Lamesch, R. S. Sikorski, J. Vandenhaute, H. Y. Zoghbi, A. Smolyar, S. Bosak, R. Sequerra, L. Doucette-Stamm, M. E. Cusick, D. E. Hill, F. P. Roth, and M. Vidal. 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature 4371173-1178. [DOI] [PubMed] [Google Scholar]
- 29.Ryan, H. E., M. Poloni, W. McNulty, D. Elson, M. Gassmann, J. M. Arbeit, and R. S. Johnson. 2000. Hypoxia-inducible factor-1α is a positive factor in solid tumor growth. Cancer Res. 604010-4015. [PubMed] [Google Scholar]
- 30.Sandoval, H., P. Thiagarajan, S. K. Dasgupta, A. Schumacher, J. T. Prchal, M. Chen, and J. Wang. 2008. Essential role for Nix in autophagic maturation of erythroid cells. Nature 454232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkar, S., E. O. Perlstein, S. Imarisio, S. Pineau, A. Cordenier, R. L. Maglathlin, J. A. Webster, T. A. Lewis, C. J. O'Kane, S. L. Schreiber, and D. C. Rubinsztein. 2007. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 3331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schofield, C. J., and P. J. Ratcliffe. 2004. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5343-354. [DOI] [PubMed] [Google Scholar]
- 33.Semenza, G. L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 3721-732. [DOI] [PubMed] [Google Scholar]
- 34.Shintani, T., and D. J. Klionsky. 2004. Autophagy in health and disease: a double-edged sword. Science 306990-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shintani, T., and D. J. Klionsky. 2004. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 27929889-29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanida, I., T. Ueno, and E. Kominami. 2004. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 362503-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tracy, K., B. C. Dibling, B. T. Spike, J. R. Knabb, P. Schumacker, and K. F. Macleod. 2007. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell. Biol. 176229-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webster, K. A., R. M. Graham, and N. H. Bishopric. 2005. BNip3 and signal-specific programmed death in the heart. J. Mol. Cell Cardiol. 3835-45. [DOI] [PubMed] [Google Scholar]
- 40.Willis, S. N., and J. M. Adams. 2005. Life in the balance: how BH3-only proteins induce apoptosis. Curr. Opin. Cell Biol. 17617-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, H., M. Bosch-Marce, L. A. Shimoda, Y. S. Tan, J. H. Baek, J. B. Wesley, F. J. Gonzalez, and G. L. Semenza. 2008. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 28310892-10903. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.