Abstract
HuR is an RNA-binding protein implicated in a diverse array of pathophysiological processes due to its effects on the posttranscriptional regulation of AU- and U-rich mRNAs. Here we reveal HuR's requirement in embryonic development through its genetic ablation. Obligatory HuR-null embryos exhibited a stage retardation phenotype and failed to survive beyond midgestation. By means of conditional transgenesis, we restricted HuR's mutation in either embryonic or endothelial compartments to demonstrate that embryonic lethality is consequent to defects in extraembryonic placenta. HuR's absence impaired the invagination of allantoic capillaries into the chorionic trophoblast layer and the differentiation of syncytiotrophoblast cells that control the morphogenesis and vascularization of the placental labyrinth and fetal support. HuR-null embryos rescued from these placental defects proceeded to subsequent developmental stages but displayed defects in skeletal ossification, fusions in limb elements, and asplenia. By coupling gene expression measurements, data meta-analysis, and HuR-RNA association assays, we identified transcription and growth factor mRNAs controlled by HuR, primarily at the posttranscriptional level, to guide morphogenesis, specification, and patterning. Collectively, our data demonstrate the dominant role of HuR in organizing gene expression programs guiding placental labyrinth morphogenesis, skeletal specification patterns, and splenic ontogeny.
The Elavl/Hu proteins are a family of RNA-binding proteins (RBPs) named after the lethal phenotypes of mutants in the Drosophila orthologue ELAV (embryonic lethal-abnormal vision) and their appearance as specific tumor antigens in humans with paraneoplastic neurological disorders (21, 38, 58, 61). In mammals, the family consists of four highly conserved members that include the ubiquitously expressed HuR/HuA/Elavl1 and the neuronal-specific Hel-N1/HuB/Elavl2, HuC/Elavl3, and HuD/Elavl4 (9, 29). All family members contain RNA recognition motifs with high affinity for U- and AU-rich sequences (AREs). The widespread distribution of AREs in 7 to 8% of eukaryotic mRNAs involved in a diversity of cellular responses (31) points to a critical role of ARE-binding proteins (4) in the regulation of many biological processes. However, the difference in the tissue distribution among Elavl/Hu proteins suggests that HuR may play a more diverse role in posttranscriptional mechanisms than its neuronal homologues.
HuR is predominantly nuclear but shuttles between the nucleus and the cytoplasm via a nucleocytoplasmic shuttling domain (16). This process is facilitated by the interaction of this domain with transport adaptors like pp32 and APRIL or directly with transport receptors such as CRM1 for nuclear export and transportins 1 and 2 (TRN1/2) for nuclear import (8, 18, 24, 48). Although not fully resolved, HuR's translocation and binding to target mRNAs appear to rely on specific stress signals resulting in HuR's phosphorylation or methylation (2, 32, 37). This has led to the hypothesis that HuR may initially bind mRNAs in the nucleus and accompany them into the cytoplasm while competing with RBPs that induce mRNA decay like members of the tristetraprolin and hnRNP families as well as exosomal/degrading enzymes (10, 42). HuR's molecular functions extend also to the level of translational processing but with differential outcomes (28, 34, 39). The molecular details underlying HuR's effects on mRNA turnover and translation remain poorly understood but include a complex interplay with degrading RNP structures and micro-RNAs (6, 30).
In the context of physiology and pathology, HuR is known to target several developmentally relevant mRNAs, including growth factors, cyclins and cyclin inhibitors, proto-oncogenes, and cytokines (1, 9, 29). This knowledge supported the notion of HuR's involvement in several pathological states from cancer to inflammation. Moreover, HuR is variably expressed throughout embryonic development (20). Additional studies using small interfering RNA silencing of HuR in unicellular cultures or transgenic HuR overexpression in mice indicated its putative role in the differentiation of specific cellular lineages, including spermatocytes, myocytes, and adipocytes (12, 17, 36, 56, 57). However, the in vivo requirement for HuR's biological functions in specific developmental and pathophysiological programs has not been elucidated. To address this issue, we induce obligatory and conditional deletions of the Elavl1 gene encoding HuR in mice and demonstrate its requirement in selective processes affecting extraembryonic and embryonic development.
MATERIALS AND METHODS
Targeting the murine Elavl1 locus.
The Elavl1 locus was isolated from a 129Ola mouse genomic PAC library (MRC Genesciences, United Kingdom). The gene contains six exons spanning a genomic region of 25 kb. A targeting vector was designed to flank a 0.77-kb XbaI/EcoRI fragment containing the second exon of the Elavl1 gene with loxP sequences and placed adjacent to a loxP-flanked PMC-neo-pA neomycin resistance gene. 5′ and 3′ homology regions of 4.4 kb (EcoRI/XbaI) and 5.7 kb (EcoRI), respectively, were inserted on either side of the modified exon 2 sequences. Finally, the complete modified locus was subcloned at the 5′ end of a herpes simplex virus thymidine kinase susceptibility gene for the negative selection of random integrants. Targeting procedures in 129Ola embryonic stem cells and subsequent injections in C57BL/6 blastocysts for chimera production and germ-line transmission from three independent clones were performed using standard procedures. Targeted clones, germ-line transmitters, and Cre recombinant progeny were identified using either Southern blot or PCR analyses of genomic DNA from cellular extracts or tail biopsies.
Mice.
Elavl1+/− and Elavl1fl/fl mice were maintained in a mixed C57BL/6J × 129Ola background or an inbred C57BL/6J background following backcrossing for 12 generations. Deleter Cre transgenic mice (51) were kindly provided by K. Rajewsky (Harvard University, MA); Sox2 Cre transgenic mice (25) were kindly provided by L. Robertson (University of Oxford, Oxford, United Kingdom); and Tie1 Cre transgenic mice (23) were kindly provided by R. Fassler (Max-Planck Institute, Martinsried, Germany). Mice were bred and maintained in the animal facilities of the Alexander Fleming Biomedical Sciences Research Center (BSRC) under specific-pathogen-free conditions. Experiments on live animals were approved by the Hellenic Ministry of Rural Development (Directorate of Veterinary Services) and by the Animal Research and Ethics Committee of the BSRC for compliance with FELASA regulations.
Cell culture and transfections.
Mouse embryonic fibroblasts (MEFs) were derived from embryonic day 12.5 (E12.5) embryos. After the removal of the heads and livers, the embryos were trypsinized for 30 min at 37°C, disaggregated, and cultured in complete Dulbecco's modified Eagle's medium plus 10% fetal bovine serum (Biochrom AG). Adherent fibroblasts were cultured for three passages and then used for most experiments. For the introduction of exogenous HuR, MEFs were transfected with 5 μg of a plasmid vector driving the expression of a hemagglutinin (HA)-tagged form of human HuR (pBBHuR) (28), empty plasmid (pBB), or a pCMV-GFP expression vector using the Amaxa/MEF1 nucleofection system according to the manufacturer's instructions. The transfected cells were left to rest for 24 h and then used for mRNA decay assays following incubation with actinomycin D (10 μg/ml; Sigma).
Histology.
For timed-mating embryos, mice were crossed overnight and the females were inspected for vaginal plugs the following morning. Noon on the day of plugging was defined as 0.5 day postcoitum. Embryos from the timed matings were isolated from pregnant females at the times indicated by standard dissection protocols in phosphate-buffered saline, and part of the yolk sac was kept for genotyping. The embryos, placentas, and yolk sacs were fixed in 10% buffered formalin overnight at 4°C. Fixation was followed by dehydration in an ethanol series prior to embedding in paraffin and sectioning. Sections of 4 to 7 μm were stained with hematoxylin and eosin for general histology according to standard techniques or used for immunohistochemistry and in situ hybridizations. Alternatively, specimens were directly embedded onto OCT cryopreservative (TissueTec) and mounted via freezing for cryostat sections.
RNA analysis and in situ hybridization.
Total RNA was derived from E12.5 MEFs using the Ribopure kit according to the manufacturer's instructions. For Northern blotting, equimolar amounts of RNA were resolved through electrophoresis in denaturing agarose gels and blotted onto nylon Hybond N+ membranes (Amersham). Blots were hybridized with 32P-labeled PCR-amplified probes for Elavl1 sequences corresponding to exons 2 and 5. For in situ hybridizations, linearized plasmids or M13 PCR-amplified cDNA inserts from Peg1, Prl3b1, Plf, and Tpbpa mRNAs were used to generate digoxigenin (DIG)-labeled riboprobes using the DIG RNA-labeling protocol according to the manufacturer's instructions (Roche). In situ hybridizations were carried out at 52°C overnight onto 7-μm-thick paraffin sections using standard procedures. Signals were detected by using an anti-DIG-alkaline phosphatase (AP)-conjugated antibody (Roche), and staining was performed overnight using Nitro Blue Tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate) (Promega). The sections were counterstained with nuclear fast red (Sigma). For the microarray analysis, please refer to Methods and Data in the supplemental material. For reverse transcriptase PCR, 5 to 10 μg of total RNA was used for cDNA synthesis with Moloney murine leukemia virus-RT (Promega). Quantitative real-time PCR (qRT-PCR) was performed using Platinum SYBR green qPCR SuperMix UDG (Invitrogen) on a RotorGene 6000 machine (Corbett Research). The primer sets are presented in Methods in the supplemental material. Expression was normalized to β2-microglobulin and Gapdh (for glyceraldehyde-3-phosphate dehydrogenase) mRNAs. The relative mRNA expression in the test samples was calculated as the difference from the control values that were assigned an arbitrary expression value of 1, using Bio-Rad RelQuant and REST 2005 (Corbett Research). The mRNA half-lives of actinomycin D-treated cells were calculated as previously described (22) and by setting the normalized amount of target mRNA (Nt = 2Ct[GAPDH] − Ct[Target], where Ct is the fractional cycle number at which fluorescence passes the fixed threshold) in unstimulated samples at 100 and extrapolating from the corresponding semilogarithmic plots of percentages versus time. HuR-RNA immunoprecipitation (IP) experiments were performed as previously described using agarose-conjugated anti-HuR (3A2; Santa-Cruz) or mouse immunoglobulin G1 (IgG1) antibodies (28). RNA was extracted from the IP material to perform the qRT-PCR. Target mRNAs were identified by the enrichment of a transcript in test IPs relative to control IPs, normalizing to the low-level contaminating transcript of Gapdh as previously described (46). For the analysis of polysome-coupled RNA, cytoplasmic fractions containing monosomes and polysomes were isolated from fibroblasts as previously described (28). The RNA in monosomal (1-12) and polysomal (13-24) fractions was pooled according to a UV 260/280 profile and used for qRT-PCR. Fibroblast nuclei run-on reactions were performed as previously described (28), using PCR-amplified products spotted onto nylon filters and analyzed using a STORM phosphorimager device. Steady-state, immunoprecipitated, transcribed, and polysomal RNAs were derived from, at least, three individual fibroblast cultures per genotype (one culture per embryo) and were different from the ones used in the microarray assays. Statistical analysis was performed using a two-tailed Student's t test to determine the P values.
Immunoblotting, immunohistochemistry, and histochemistry.
For the Western blots, whole-embryo, tissue, or MEF lysates were prepared in Laemmli or IP assay buffers. Equimolar amounts of protein were analyzed on sodium dodecyl sulfate-containing polyacrylamide gels (10 to 12%) and blotted onto nitrocellulose membranes (Schleicher & Schuell). The primary antibodies used were 3A2 and T17 for HuR, C-11 for actin, H-40 for Hoxd13, C-20 for Ets-2, H-121 for FGF10, and N-19 for TBX4 and were purchased from SantaCruz Biotech. For the detection of Tbx4, nuclear extracts were isolated from MEFs using NE-PER reagent (Pierce) according to the manufacturer's instructions. For the detection of FGF10, protein secretion was blocked by culturing MEFs for 4 h with 1 μg/ml of protein transport inhibitor (GolgiPlug; BD) according to the manufacturer's instructions. Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (Southern Biotechnologies) by enhanced chemiluminescence (ECL+; Amersham). For immunohistochemistry, 4- to 6-μm paraffin sections were deparaffinized and used directly or treated with microwaves in citrate buffer for epitope unmasking. Cryostat sections were used directly. Following blocking, sections were incubated with the following antibodies: peroxidase-conjugated lectin from Bandeiraea simplicifolia BS-I B4 (40 ng/μl; Sigma, St. Louis, MO) and 3A2 for HuR (1:150; Santa Cruz Biotech). The detection was performed using streptavidin-conjugated secondary antibodies (Southern Biotech). Development was performed using DAB (3,3′-diaminobenzidine) substrate (Sigma). Nuclear counterstaining was carried out with hematoxylin (Sigma). For AP stains, placental cryostat sections were washed in NT solution (0.15 NaCl, 0.1 M Tris [pH 7.5]) and then in NTMT solution (0.1 M NaCl, 0.1 M Tris [pH 9.5], 0.05 MgCl2, 0.1% Tween-20). Enzymatic activity was detected using fast blue substrate (Sigma) and counterstained with nuclear fast red (Vector).
Assays for cellular proliferation and apoptosis.
For cell death determination, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed on paraffin-embedded sections using an in situ cell death detection kit (cat. no. 11 684 795 910; Roche). The sections were permeabilized with proteinase K (Roche; 10 mg/ml for 20 min) or 0.1% Triton, 0.1% tri-sodium citrate (4°C for 2 min) for the paraffin and frozen sections, respectively, and the in situ labeling assay was carried out according to the manufacturer's instructions. For detection, the AP converter kit (Roche) was used, and the signal was detected with fast blue substrate (Sigma); sections were counterstained using nuclear fast red dye for the visualization of the nuclei. For proliferation, pulse BrdU experiments were performed by the intraperitoneal injection of the pregnant females 3 h prior to sacrifice. The placentas were collected, formalin fixed, and paraffin embedded. For the BrdU immunodetection, 7-μm sections were deparaffinized, rehydrated through an ethanol series, and permeabilized with proteinase K. Subsequently, sections were treated with 2 N HCl for 1 h following incubation for 10 min in borate buffer (pH 7.6) before blocking and overnight incubation with anti-BrdU antibody (M0744; Dako- Cytomation). On the following day, sections were incubated with anti-mouse IgG1 secondary antibody (Southern Biotech), visualized using DAB, and counterstained with hematoxylin.
Skeletal preparations.
E18.5 embryos were fixed in 10% formalin for 2 days, followed by a 3-day wash in H2O, skin removal, and evisceration. For the cartilage stains, embryos were stained with alcian blue 8GS (Applichem) in ethanol/acetic acid for 2 days, followed by washing in ethanol and rehydration. Maceration, clearing, and bone staining were performed in 0.5% KOH saturated with alizarin red S (Applichem) followed by several washes in 0.5% KOH-glycerol. Specimens were stored in pure glycerol for stereophotography.
RESULTS
HuR is required for midgestational embryonic development.
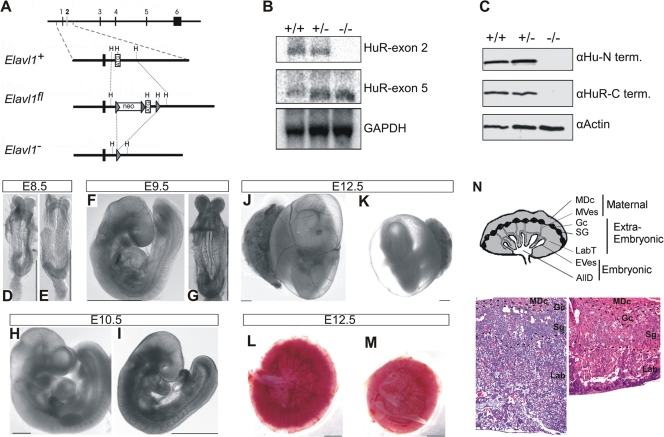
To investigate the in vivo functions of HuR, we generated a conditional allele for the murine Elavl1 gene by flanking its ATG-containing second exon with two loxP sites (Fig. 1A). Mutant Elavlflox/+ mice were produced via standard gene targeting procedures in embryonic stem cells and contained a fully functional Elavl1 allele. To generate an Elavl1 null allele, Elavl1flox/+ mice were crossed with transgenic mice expressing the Cre recombinase in the germ line (51) to yield Elavl1+/− mice. Heterozygous Elavl1+/− mice appeared phenotypically normal and were subsequently intercrossed for the generation of homozygous Elavl1−/− mice. An examination of the F2 progenies in either mixed (B6, 129Ola; see Table S1 in the supplemental material) or inbred (B6; data not shown) genetic backgrounds revealed the total absence of Elavl1−/− newborns, suggesting that the mutation resulted in embryonic lethality. A comparative macroscopic evaluation of staged embryos indicated that Elavl1−/− embryos were present at a correct Mendelian frequency till E9.5, suggesting that death was not consequent to preimplantation or early postimplantation defects. However, the percentage of live Elavl1−/− embryos gradually diminished from E10.5 to E12.5, correlating with an increased number of resorptions and empty deciduas, while no viable Elavl1−/− embryos were identified beyond E14.5 (see Table S1 in the supplemental material). A Northern analysis of RNA from E12.5 MEFs revealed the presence of a mutant mRNA lacking the second exon (Fig. 1B), whereas immunoblotting of the protein extracts from E11.5 embryos and MEFs with different anti-HuR antisera verified the complete absence of HuR protein (Fig. 1C and data not shown).
FIG. 1.
Midgestational phenotype of HuR-deficient embryos. (A) Schematic of the complete exon/intron orientation of the Elavl1 locus on mouse chromosome 8 and magnifications of the region containing the second exon (white box) as a wild-type (Elavl1+), targeted (Elavl1fl), and inactive, Cre-recombined locus (Elavl1−). The neo selection gene and loxP genes (arrowheads) are indicated. Restriction sites for HindIII (H) are also shown for alignment. (B) Northern analysis of RNA extracts from Elavl1+/+ (+/+), Elavl1+/− (+/−), and Elavl1−/− (−/−) MEFs with specific probes for exons 2 and 5, indicating the presence of shorter mHuR transcripts lacking exon 2. The Gapdh mRNA is shown for quantitation. (C) Western blotting of protein extracts from E11.5 Elavl1+/+ (+/+), Elavl1+/− (+/−), and Elavl1−/− (−/−) embryos with antibodies against the amino (3A2) and carboxy (T17) termini of mHuR depicting its complete absence. Actin is shown as a loading control. (D to K) Representative stereophotographs of staged Elavl1+/+ (D, F, H, and J) and Elavl1−/− (E, G, I, and K) embryos and conceptuses, indicating the stage retardation phenotype of the latter and the diminished blood flow in the corresponding yolk sacs. The mutant phenotype correlates with smaller placentas (M) as opposed to control placentas (L). Size bars correspond to 1 mm. (N) Diagram of placental compartments and representative hematoxylin/eosin histology of E12.5 placentas from control (+/+) and mutant (−/−) conceptuses. Dotted lines indicate the maternal decidua, giant cell, spongiotrophoblast, and labyrinth layers. Magnification, ×100. MDc, maternal decidua; MVes, maternal vessels; Gc, giant cells; Sg, spongiotrophoblasts; Lab, labyrinth; LabT, labyrinthine trophoblasts; EVes, embryonic vessels; AllD, nonendothelial allantoic derivatives.
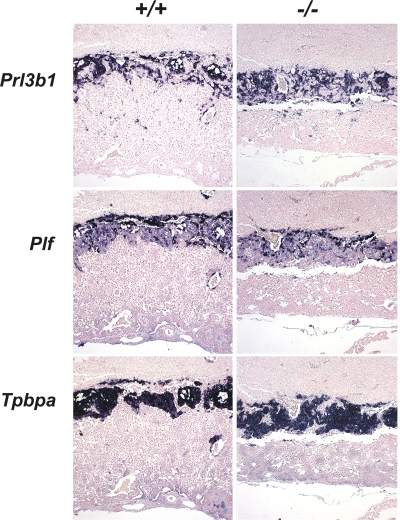
Phenotypically, Elavl1−/− embryos displayed no apparent morphological abnormalities till E8.0 and possessed proper allantoic structures; thus, HuR is not required for the progression from the primitive streak to head fold stages. However, a stage retardation phenotype became apparent between E8.5 and E9.0; at this age, control Elavl1+/+ and Elavl1+/− embryos possessed 8 to 10 pairs of somites, whereas >40% of the Elavl1−/− embryos remained at the presomite stage (Fig. 1D and E). This difference was maximized at E9.5 when control embryos possessed <17 pairs of somites and “turned,” whereas the Elavl1−/− embryos (Fig. 1F and G) possessed an E8.5 phenotype despite the presence of chorioallantoic fusion. Between E10.5 and E12.5 (Fig. 1H to K), surviving Elavl1−/− embryos displayed a 1.0- to 1.5-day-postcoitum lag; interestingly, blood vessels, although rudimentary, could be detected, and the visceral yolk sacs of the mutant embryos had a pale appearance, indicating problems with either vascularization or blood circulation (Fig. 1J and K and data not shown). Most strikingly however, the placentas from the Elavl1−/− embryos at these stages appeared smaller and less vascularized than the control placentas (Fig. 1L to M). Histologically, the maternally derived decidua, the giant cell zone and the spongiotrophoblast layers appeared to be present but smaller in E12.5 Elavl1−/− placentas (Fig. 1N). Similarly, the in situ distribution of placental lactogen/Prl3b1, Proliferin/Plf, and Tpbpa/4311 mRNAs was invariable between the mutant and control E12.5 placentas (Fig. 2), indicating that the secondary trophoblastic, trophoblast giant, and spongiotrophoblast cell layers were properly organized in Elavl1−/− placentas. However, the corresponding labyrinth was disproportionately decreased, showing reduced branching complexity and a loss of structural organization (Fig. 1N). Thus, the midgestational lethality of HuR-deficient embryos could result from placental failure.
FIG. 2.
HuR is not required for the differentiation of trophoblasts into spongiotrophoblasts and giant cells. Prl3b1, Plf, and Tpbpa/4311 mRNAs were detected in situ (AP [blue]) in E12.5 placentas from Elavl1+/+ and Elavl1−/− conceptuses, marking the presence of secondary trophoblast giant, trophoblast giant, and spongiotrophoblast cells among genotypes. The counterstain was nuclear fast red. Magnification, ×40.
Extraembryonic defects cause the placental failure and lethality of HuR-null embryos.
A previous report demonstrated that in murine placenta, the expression of HuR peaks between E12.5 and E13.5 (20). An immunocytochemical analysis of the E12.5 placentas revealed that HuR is expressed in both embryonic and extraembryonic layers; however, its nucleocytoplasmic localization varied from the maternal to the fetal side. HuR appeared predominantly in the nucleus of maternal decidua cells, as well as in embryo-derived trophoblast giant cells and spongiotrophoblasts (Fig. 3E). In contrast, labyrinthine trophoblasts showed both nuclear and strong cytoplasmic HuR protein staining, suggesting that HuR's cytoplasmic functions are increased in these cells (Fig. 3F and G). As expected, Elavl1−/− placentas did not display any HuR immunostaining in the embryonic layers but only in the maternal decidua which was derived from the Elavl1+/− uterus (Fig. 3H to J).
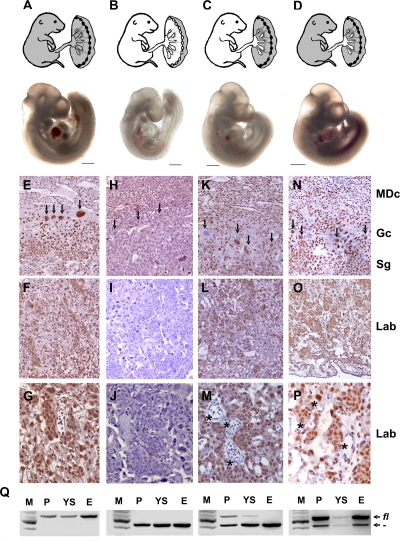
FIG. 3.
The midgestational lethality of HuR-deficient embryos results from extraembryonic defects in the corresponding placentas. Shown are diagrams of Cre-induced HuR deletions (white areas) in presumptive E14.5 embryos and representative stereophotographs of Elavl1fl/fl (A), Elavl1−/− (B), Sox2 Cre Elavl1fl/− (C), and Tie1 Cre Elavl1−/− (D) embryos at E10.5. Size bars = 1 mm. (D to P) HuR protein detection (brown) on sections from Elavl1fl/fl (E to G), Elavl1−/− (H to J), Sox2 Cre Elavl1fl/− (K-M), and Tie1 Cre Elavl1−/− (N to P) E12.5 placentas counterstained with hematoxylin. MDc, maternal decidua; Gc, giant cell (arrows); Sg, spongiotrophoblast; Lab, labyrinthine. Asterisks indicate HuR− embryonic endothelia. Magnification, ×100 for panels E, F, H, I, K, L, N, and O and ×400 for panels G, J, M, and P. (Q) PCR detection of Elavl1fl (fl) and recombined Elavl1− (−) alleles in DNA extracts from placentas (P), yolk sacs (YS), and embryos (E) from Elavl1fl/fl, Elavl1−/−, Sox2 Cre Elavl1fl/−, and Tie1 Cre Elavl1−/− conceptuses corresponding to the embryos depicted in panels A to D. M, DNA ladder.
In light of the histological appearance of the Elavl1−/− placentas and the distribution of HuR expression in placental layers, the developmental retardation and midgestational death of the Elavl1−/− embryos could result from defects in either embryonic or extraembryonic layers comprising the murine placenta. To dissect the effects of HuR in either of these layers, we utilized the conditional Elavl1flox allele to induce selective HuR deletions. To test whether an extraembryonic defect drives the phenotype of HuR-deficient embryos, we crossed Elavl1fl/fl mice with transgenic mice expressing Cre recombinase under the control of the Sox2 promoter. The Sox2 Cre transgene permits the efficient loss of gene function within only epiblast cells that give rise to the entire embryo proper as well as to the extraembryonic mesoderm that forms the fetal vasculature in the placental labyrinth but not in the trophectoderm-derived trophoblast cell types of the placenta (25, 54). Sox2 Cre+ Elavl1−/− embryos were recovered alive past E14.5 at the expected Mendelian frequencies (see Table S2 in the supplemental material). In addition, the number of resorptions or empty deciduas was almost negligible as opposed to those detected in the Elavl1+/− intercrosses (data not shown), whereas E9.5 to E12.5 Sox2 Cre+ Elavl1−/− embryos appeared indistinguishable from the control embryos (Fig. 3A to C). DNA analyses verified the presence of unrecombined cells in the Sox2 Cre+ Elavl1−/− placentas and visceral yolk sacs, whereas the corresponding embryos possessed an Elavl1−/− genotype (Fig. 3Q). The immunohistochemical detection of HuR in E12.5 Sox2 Cre+ Elavl1−/− placentas demonstrated the predominant presence of HuR-containing cells in maternal and trophoblastic layers, although some trophoblastic cells were HuR deficient probably due to a marginal ectopic expression of Cre recombinase. In contrast, all of the embryonic endothelial cells in the labyrinth were devoid of HuR (Fig. 3K to M).
To address whether the embryonic lethality of mutant embryos was solely due to a trophoblastic defect or an additional defect in the development of embryonic blood vessels, Elavl1flox/flox mice were crossed with transgenic mice expressing Cre recombinase under the control of the endothelial Tie-1 promoter (23). In this setting, only the fetal endothelia of the embryo and placenta were HuR deficient, as confirmed by immunochemistry and DNA recombination approaches (Fig. 3N to Q and data not shown), while all other tissues retained physiological HuR levels. The Tie1 Cre+ Elavl1flox/flox embryos generated from these crossings did not appear to have any retardation defect (Fig. 3D) but developed to term, maintaining a normal Mendelian ratio (data not shown). In addition, Tie1 Cre+ Elavl1flox/flox mice were fertile and did not exhibit any overt phenotype up to the end of the examination (6 months). Similar results were obtained when both maternal and embryonic endothelia were made devoid of HuR via the crossing of female tie-1 Cre+ Elavl1flox/flox mice with male Elavl1flox/flox mice (data not shown). Thus, the midgestational embryonic lethality of the Elavl1−/− mice results from placental failure due to extraembryonic trophoblast defects.
HuR is required for labyrinth branching morphogenesis.
The placenta comprises trophoblastic layers that have distinct ontogeny and function. These layers arise from either (a) the ectoplacental cone (EPC) that gives rise to the spongiotrophoblast and trophoblast giant cells located in the proximity of the maternal decidua to control the promotion of maternal blood flow or (b) trophoblast stem cells in the chorion that give rise to syncytiotrophoblast cells in the labyrinth. Labyrinth development is initiated by the fusion of the chorion with the allantois followed by the invagination of allantoic capillaries into the chorionic trophoblast layer that concomitantly starts to differentiate into syncytiotrophoblast cells. An extensive process of branching morphogenesis then gives rise to an intricate network of syncytiotrophoblast-lined maternal blood spaces and allantoic mesoderm-derived fetal blood vessels. The close proximity of the maternal and fetal blood circulations that is achieved by the labyrinthine architecture ensures sufficient nutrient transport to the embryo.
The aberrant appearance of the labyrinth layer in the E12.5 Elavl1−/− placentas and the normal presence of EPC-derived trophoblasts prompted us to investigate the earlier stages of chorioallantoic placental development. As indicated above, chorioallantoic attachment was normal in E8.0 to E8.5 Elavl1−/− conceptuses. In contrast, the morphogenesis of the chorioallantoic interface and the intrusion of underlying fetal blood vessels were not as apparent as in the case of the control conceptuses, and the chorionic plate appeared flat (Fig. 4A). Two days later, the chorioallantoic interface showed signs of interdigitation with the allantoic mesoderm and with a few blood vessels but did not progress to the complete formation of branched structures (Fig. 4A). The number, organization, and density of allantoic blood vessels beneath the chorionic plate of Elavl1−/− conceptuses seemed normal; however, these vessels appeared partially occluded and filled with enucleated blood cells, suggesting that the blood flow was obstructed (data not shown). Thus, in the absence of HuR, the initiation of branching is delayed and progresses poorly toward labyrinth morphogenesis.
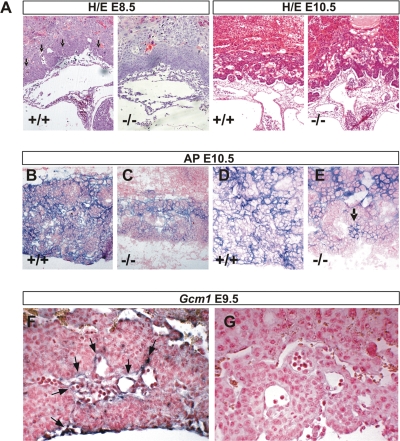
FIG. 4.
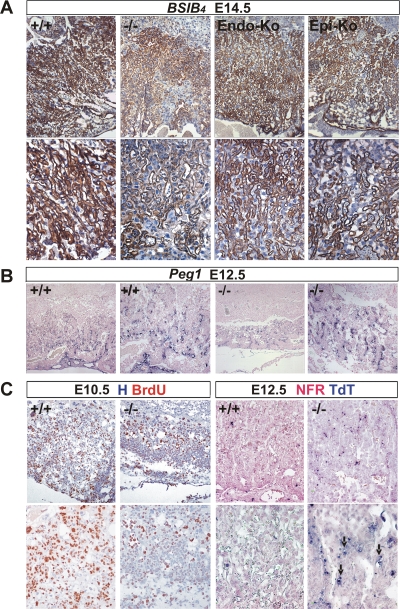
Defective labyrinth branching initiation and syncytiotrophoblast differentiation in HuR-deficient chorioallantoic placentas. (A) Histology (hematoxylin/eosin; magnification, ×100) of E8.5 and E10.5 chorioallantoic placentas from Elavl1+/+ and Elavl1−/− conceptuses. Arrows mark points of chorioallantoic branching which are missing in mutant placentas. (B to E) Histochemical detection of placental AP activity (blue) marking trophoblasts in E10.5 placentas from Elavl1+/+ and Elavl1−/− conceptuses. The counterstain was nuclear fast red. Magnification, ×100 for panels B and D and ×200 for panels C and E. Arrow marks the focal presence an AP-positive cluster in a mutant labyrinth. (F to G) In situ detection (magnification, ×200) of the syncytiotrophoblast-specific marker Gcm1 (blue [arrows]) in E9.5 Elavl1+/+ (F) and Elavl−/− (G) placentas. The counterstain was nuclear fast red.
To examine whether the deletion of HuR affected the differentiation of chorionic trophoblasts to labyrinth syncytiotrophoblasts, we first assessed the distribution of AP staining which marks differentiating trophoblast cells and becomes later restricted to syncytiotrophoblasts lining maternal blood sinuses (60). As can be seen in Fig. 4B and D, control placentas at E10.5 showed high levels of AP staining in all layers; in contrast, E10.5 Elavl1−/− placentas show signals in the spongiotrophoblastic layers, whereas the chorionic plate/labyrinth staining was diminished and restricted in a few clustered cells (Fig. 4C and E). Next, we examined the presence of Gcm1-expressing cells via in situ hybridization, since this transcription factor is essential for the initiation of chorioallantoic branching as well as for the differentiation of chorionic trophoblasts to syncytiotrophoblasts (3). As previously described, Gcm1-expressing cells were identified in the distal tips of the elongated branches in E9.5 control placentas, indicating active morphogenesis (Fig. 4F). In sharp contrast, the number of Gcm1-expressing cells in E9.5 Elavl1−/− placentas was almost nominal—as also verified by reverse transcriptase PCR analyses (data not shown)—and the in situ staining was barely detectable only in very few chorionic trophoblast cells (Fig. 4G and data not shown). To examine whether labyrinth morphogenesis could progress in subsequent stages, we examined the distribution of the isolectin B4 glycoprotein that marks the syncytiotrophoblasts lining the maternal lacunas in E12.5 to E14.5 control and Elavl1−/− placentas derived from surviving embryos. As can be seen in Fig. 5A, the vessel density and the surface area between the maternal and fetal blood spaces were extensively reduced in Elavl1−/− placentas relative to those of the control and the epiblast- and endothelial-specific HuR mutants. Thus, branching morphogenesis initiates at later stages but is distorted and unable to support placental vascularization. This was verified further by the limited distribution of Peg1-expressing cells in the mutant labyrinths that mark placental vessels (Fig. 5B). Thus, it appears that in the absence of HuR, branching morphogenesis is extensively delayed and proceeds poorly toward the generation of a functional labyrinth.
FIG. 5.
Abnormal labyrinth branching extension, vascularization, proliferation, and apoptosis in mature HuR-deficient placentas. (A) Immunohistochemical detection of isolectin B4 (brown staining) in E14.5 placentas from Elavl1+/+ (+/+), Elavl1−/− (−/−), Tie1 Cre Elavl1fl/fl (Endo-Ko), and Sox2 Cre Elavl1−/− (Epi-Ko) conceptuses revealing the matrix between the endothelium of the fetal vessels and the labyrinth trophoblast of the maternal lacunas. The counterstain was hematoxylin. Magnification, ×100 (top panels) and ×200 (bottom panels). (B) In situ detection of Peg1 mRNA (blue) in E12.5 placentas from Elavl1+/+ (+/+) and Elavl1−/− (−/−) conceptuses, marking placental endothelia. Magnification, from the left, ×100 for the first and third panels and ×200 for the second and fourth panels. Note the reduction in the area of the labyrinth endothelia in the mutant placentas. (C) Left panels, immunohistochemical detection of BrdU incorporation (brown) in E10.5 chorioallantoic placentas from Elavl1+/+ (+/+) and Elavl1−/− (−/−) conceptuses showing reduced proliferation in the mutant labyrinth layers. The counterstain was hematoxylin (H). Magnification, ×100 (top panels) and ×200 (bottom panels). Right panels, TUNEL detection of apoptotic cells (TdT; blue) in E12.5 placentas from Elavl1+/+ (+/+) and Elavl1−/− (−/−) conceptuses. Arrows indicate apoptotic trophoblastic cells. The counterstain was nuclear fast red (NFR). Magnification, ×100 (top panels) and ×200 (bottom panels).
To identify cellular defects that could relate to the poor morphogenesis of the Elavl1−/− placental labyrinth, we examined cellular proliferation following BrdU administration and immunohistochemical detection of proliferating cells in chorioallantoic placentas from E10.5 conceptuses. In the control placentas, proliferating BrdU-positive cells were present in giant cells and spongiotrophoblast and labyrinth layers as well as in allantoic vessels; in sharp contrast, the “poorly” branched labyrinth from the Elavl1−/− placentas was almost devoid of proliferating cells, whereas the remaining layers contained a normal distribution of BrdU staining (Fig. 5C). The reduction of proliferating cells in the mutant placentas from E10.5 conceptuses appeared to reflect a lack of proliferating/differentiating cells and not an increase in apoptotic cells as indicated by TUNEL assays (data not shown). However, numerous apoptotic cells were detected in E12.5 mutant placentas from surviving embryos and particularly in remnant labyrinthine trophoblast clusters (Fig. 5C), suggesting that the HuR-related defects in syncytiotrophoblast differentiation and labyrinthine vascularization result in labyrinthine apoptosis, obstructing further the placental/fetal interaction.
Collectively, our data demonstrate that in the absence of HuR, labyrinth branching morphogenesis and syncytiotrophoblast differentiation are impaired, leading to insufficient vascularization, labyrinthine apoptosis, and insufficient nutrient transport that do not sustain fetal development and survival.
HuR-null embryos show defects in skeletal and splenic development.
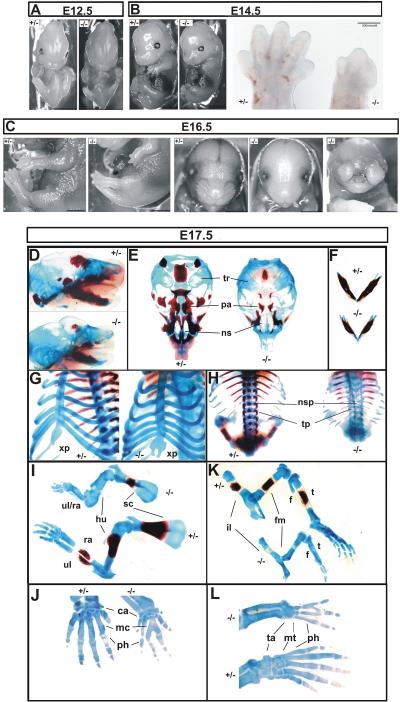
The epiblast-specific deletion of HuR in Sox2 Cre+ Elavl1−/− embryos promoted their survival through midgestation. However, most of these embryos appeared smaller past E14.5 and died between E17.5 and E19.5; a few HuR-null dead pups were identified, indicating that mutant mice could be delivered as stillborns but most probably were cannibalized by their parents. A macroscopic examination of Sox2 Cre+ Elavl1−/− embryos at different stages revealed prominent defects in skeletal development. Although both posterior and anterior limb buds were present in mutant embryos at E14.5, all HuR-null embryos possessed limbs of reduced size, which in many cases lacked definable digits and resembled E12.5 limb buds (Fig. 6A and B). These deformities were more extensive at late embryonic stages (Fig. 6C). Bone (alizarin red S) and cartilage (alcian blue) staining of embryos between E17 and 19.5 indicated that the skeletal structures in HuR-null embryos remained primarily cartilaginous, demonstrating a delay in endochondral ossification (Fig. 6D to L) while revealing additional skeletal anomalies. The sternums in E17.5 to E18.5 mutant embryos possessed a physiological number of ribs but the xiphoid process was open and bifid, demonstrating an incomplete fusion of the thoracic cage (Fig. 6G). Similarly, spines contained equal numbers of vertebrae between control and HuR-null embryos but showed a complete absence of ossified neural spines and zygapophyses in the latter (Fig. 6H). The long bones were shorter in null embryos and showed minimal ossification zones in scapulae, femurs, and tibia (Fig. 6I and K). Besides the defects in ossification, mutant limbs showed signs of synostoses underlying the macroscopic observations. In mutant forelimbs, humeri were present but radial and ulnar bones were fused in more than 40% of the HuR-null embryos (Fig. 6I). Most strikingly, mutant embryos displayed syndactyly in both forelimbs (carpal to metacarpal) and hind limbs (tarsal to metatarsal) and variable fusions among digits (Fig. 6J and L). In 80% of the mutant embryos analyzed, craniofacial osteogenic ossification was also delayed in most parts, with the exception of the mandibles that appeared properly ossified (Fig. 6D to F). However, cartilaginous structures were present, and the palate was fusing properly in the midline, as was also indicated in the few mutant newborns (data not shown). Interestingly, a less penetrant phenotype (<15% of embryos) was also observed in which mutant mice developed severe nasal clefting (Fig. 6C).
FIG. 6.
Skeletal defects in HuR-deficient embryos. Comparison of Sox2 Cre+ Elavl1+/− (+/−) and Sox2 Cre+ Elavl1−/− (−/−) embryos at E12.5 (A) and E14.5 (B) (size bars, 0.5 cm), indicating the aberrant limbs in the latter which are also presented in the inset in panel B. (C) Comparison of Sox2 Cre+ Elavl1+/− (+/−) and Sox2 Cre+ Elavl1−/− (−/−) embryos at E16.5, indicating the dominant-limb phenotype in the presence of formed facial skeletal structures and the less-penetrant nasal cleft phenotype. Size bars, 0.5 cm. (D to L) Stereophotographs of Sox2 Cre Elavl1+/− (+/−) and Sox2 Cre Elavl1−/− (−/−) skeletons at E17.5 following staining for cartilage (alcian blue) and bone (alizarin red). Presented are comparative lateral views of crania with mandibles (D), ventral views of vaults (E) and mandibles (F), tilted ventral views of the sternum (G), dorsal view of the thoracic cage (H), lateral views of forelimbs (I) and hind limbs (K), and dorsal magnifications of forepaws (J) and hind paws (L). tr, tympanic rings; pa, palate; ns, nasal capsule; xp, xiphoid process; nsp, neural spines; tp, transverse processes; ul, ulna; ra, radius; hu, humerus; sc, scapula; il, ilium; fm, femur; t, tibia; f, fibula; ca, carpal; mc, metacarpal; ta, tarsal; mt, metatarsal; ph, phalanges.
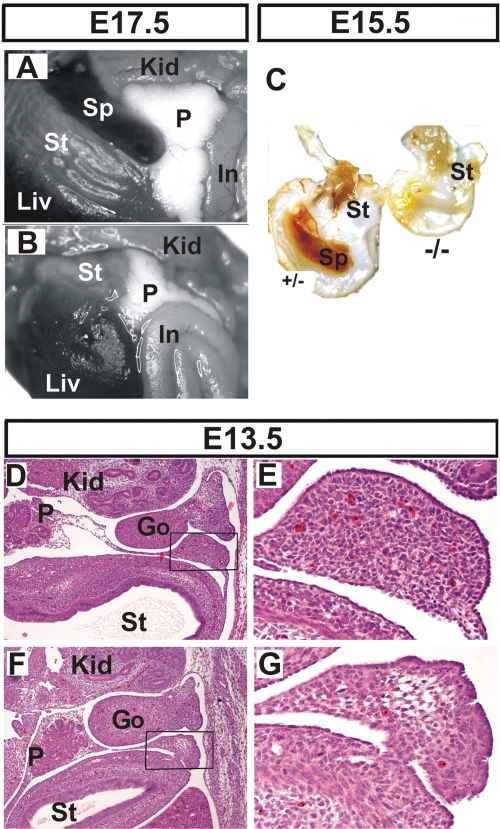
The gross anatomical analysis of internal organs below the thoracic cage in E17.5 to E18.5 mutant embryos revealed additional dysmorphologies in their lungs that will be described in more detail elsewhere; however, these embryos were also devoid of spleens (Fig. 7A and B). In contrast, the stomach and pancreas, which are derived from a region of splanchnic mesoderm adjacent to the splenic anlage, were properly located in the mutant. Similarly, the lack of spleen was not due to gross hematopoietic defects since the mutant embryos possessed hematopoietic centers in their fetal livers as well as fully developed thymuses (data not shown). Asplenia was also evident in E15.5 mutants (Fig. 7C). However, histological examination at E13.5 indicated the presence of a spleen anlage in both control and mutant embryos (Fig. 7D to G). This suggested that the splenic primordium was initially developing normally in earlier stages. However the mutant splenic anlage showed decreased cellularity, having an almost “empty” appearance as opposed to the control tissue. In addition, remnant cells possessed a condensed morphological appearance suggestive of increased apoptosis that might account for the lack of mature spleen in subsequent stages.
FIG. 7.
Asplenia in HuR-deficient embryos. Comparison of abdominal internal organs between Sox2 Cre Elavl1+/− (A) and Sox2 Cre Elavl1−/− (B) embryos at E17.5, indicating the complete absence of a spleen in the latter. (C) Stomachs from control (+/−) and mutant embryos (−/−) at E15.5, indicating asplenia in the latter. (D to G) Representative transverse sections of the abdominal region from control (D and E) and mutant (F and G) embryos at E13.5 marking the presence of a disorganized splenic anlage in the latter. Hematoxylin/eosin staining was used. Magnification, ×40 for panels D and F and ×200 for E and G. Liv, liver; St, stomach; Sp, spleen; Kid, kidney; P, pancreas; In, intestines; Go, gonads.
Collectively, our data demonstrate that HuR is involved in cellular processes affecting (i) endochondral and osteogenic ossification, (ii) skeletal patterning of long bones and digits, and (iii) splenic development and outgrowth.
Identification of HuR target mRNAs that determine extraembryonic and embryonic development.
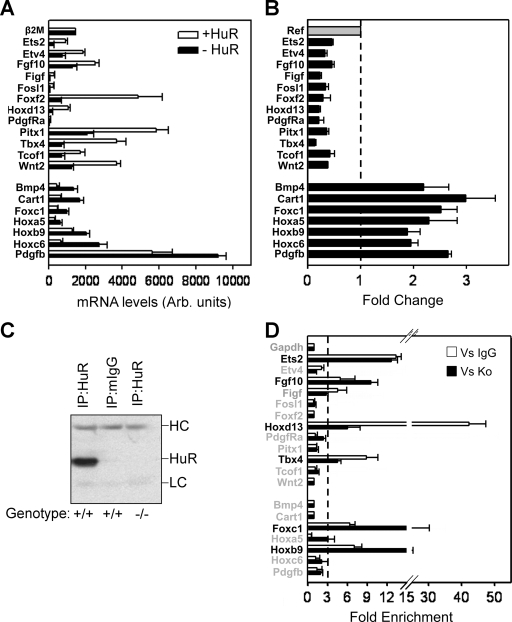
The complexity of the phenotypes of HuR− embryos are indicative of HuR's involvement in several developmental gene expression programs. To identify such programs, we hybridized total RNA from MEFs derived from Sox2 Cre+ Elavl1−/− (i.e., HuR−) and Sox2 Cre+ Elavl1+/− embryos (i.e., HuR+) onto Affymetrix microarrays (see Methods in the supplemental material). A collective of 395 differentially expressed genes (DEGs) were identified as up (240) or down (155) in HuR− MEFs (see Table S3a and S3b in the supplemental material). The frequency of Gene Ontology terms associated with these genes was suggestive of aberrations in cellular proliferation, adhesion, extracellular matrix modeling, anatomical structure specification, and developmental morphogenesis (see Table S4 in the supplemental material). To link gene expression changes to the specific phenotypes of the HuR− embryos, we used the software application Endeavor (55) and prioritized DEGs based on their relation to genetic mutations that induce (i) placental insufficiencies (46 genes) (50), (ii) limb deformities (167 genes relating to MGI's phenotypic ontology ID MP:0003937, abnormal limbs/digits/tail development [www.informatics.jax.org]), and (iii) asplenia (7) in mice (see Table S5 and S6 in the supplemental material). The cross comparison of the three individual priority lists revealed the common presence of 23 DEGs (see Table S7 in the supplemental material). Subsequently, qRT-PCRs validated consistent differences in the expression of 19/23 genes (12 up, 7 down) in HuR− MEFs (Fig. 8A and B) that were highly relevant to the developmental defects of HuR− embryos. The placentation defects correlated to the deregulated expression of genes guiding chorioallantoic fusion, branching morphogenesis, labyrinthine vascularization, and trophoblast differentiation (Tbx4; Fosl1/Fra1, Wnt2, Pdgfra/Pdgfb; and Ets2) (50). Similarly, embryonic defects correlated with the aberrant expression of genes controlling outgrowth, axis specification, and the patterning of skeletal segments (Fgf10, Wnt2, Bmp4, Tbx4, Pitx1, Hox paralogues, and Cart1) (15, 19, 33, 49, 52, 62).
FIG. 8.
Identification of HuR-interacting mRNAs of relevance to the phenotypes of HuR-deficient embryos. (A and B) Bar graphs depicting the actual and differential (change) levels of the 19 mRNAs relating to the phenotypes of Elavl1−/− embryos as predicted by the expression profiling of control (+HuR) and mutant (−HuR) MEFs. Data (arbitrary units and change ± standard error of the mean) were derived from qRT-PCR experiments performed with RNAs derived from 3 to 5 individual cultures per genotype. The dotted line in panel B represents the control value of 1 from which presented values deviate. With the exception of β2Μ, the values corresponding to the mutant MEFs in both graphs were significantly different from the control values (P < 0.01). (C) Representative anti-HuR immunoblot of the anti-HuR or mIgG1 IP material derived from HuR-proficient and -deficient MEFs. The heavy (HC) and light (LC) chains of the antibodies are indicated. (D) qRT-PCR detection of selected mRNAs immunoprecipitating with HuR in HuR+ MEFs. Data (means of biological triplicates ± standard errors of the means) are represented as the enrichment of each mRNA in HuR IP samples compared with its abundance in IgG1 IPs (Vs IgG) or HuR IPs from HuR− MEFs (Vs Ko). Enrichment levels were adjusted to the levels of the Gapdh mRNA. The dotted line indicates the cutoff value, above which enrichments were considered significant.
The abovementioned data reveal gene-to-phenotype relationships which can be under direct or indirect control by HuR. Interestingly, only three of the prioritized mRNAs (Fgf10, Pdgfb, and Hoxb6) have been reported to contain AU-rich elements according to the ARED Organism Database (see Table S8 in the supplemental material), whereas none has been reported to interact with HuR. Thus, we used HuR IP assays from HuR+ MEF extracts to detect whether any of the 19 mRNAs associated with HuR in the corresponding RNP complexes. The specificity of associations was controlled by (a) HuR IP assays from HuR− MEF extracts and (b) isotype mIgG1 IP assays from HuR+ MEF extracts, whereas the quality of the IP reactions was monitored by immunoblotting (Fig. 8C). In each IP sample, the presence of each mRNA was tested by the extraction of the IP-bound RNA and qRT-PCR. The association of an mRNA with HuR was considered positive only if its presence was more than threefold enriched compared to that of both control IP reactions. As shown, 6 out of the 19 mRNAs tested were found to associate distinctly with HuR (Fig. 8D). On the one end, four of these targets showed reduced mRNA abundance in HuR− cells: (i) the pleiotropic Ets2 transcription factor, which is implicated in the differentiation, polarity, and maintenance of the EPC-derived trophoblasts in the placenta (59); (ii) the homeotic Hoxd13 transcription factor, which is a key controller of limb patterning (19); (iii) the Tbx4 transcription factor, which controls chorioallantoic fusion and limb identity (43, 44); and (iv) FGF10, which is a powerful soluble growth factor driving the specification and outgrowth of many organs, including limbs, and is also under the transcriptional control of Tbx4 during limb development (44). On the other end, 2 HuR targets showed increased expression in HuR− MEFs: (i) the mRNA of the Hoxb9 transcription factor, which controls the specification of thoracic skeletal elements (11), and (ii) the mRNA of forkhead Foxc1 transcription factor implicated in ocular, meningeal, cardiac, and skeletal development (35). Thus, the extraembryonic and embryonic functions of HuR relate to its associations with mRNAs involved in these processes.
The loss of HuR elicits transcriptional and posttranscriptional alterations in the expression of its target mRNAs.
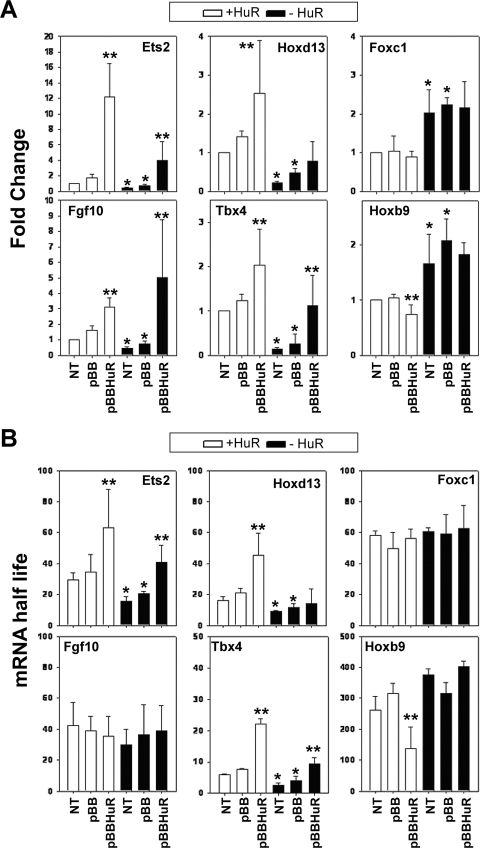
To gain further insight into the effects of HuR on its target mRNAs, we analyzed several stages of their biogenesis and utilization. Nucleus-run-on experiments revealed that a 50% reduction in the transcription of the Fgf10 gene was contributing to its reduced abundance in HuR− MEFs (see Fig. 10A; see also Fig. S1 in the supplemental material). In contrast, the transcription of the other HuR target RNAs was not altered in these cells, suggesting that their reduced expression was consequent to defects in posttranscriptional mechanisms, like the regulation of their stability. To assess this, we analyzed the decay of HuR target mRNAs following transcriptional inhibition by actinomycin D pulsing of MEF cultures (see Fig. S3 in the supplemental material) and calculated the mRNA half-lives as a measure of their stability (Fig. 9). The decay of Fgf10 and Foxc1 mRNAs appeared invariable between control and mutant MEFs. In contrast, the half-lives of Ets2, Hoxd13, and Tbx4 mRNAs were significantly reduced, whereas that of the Hoxb9 mRNA was increased in the same cells. To confirm that these changes were not due to the excessive alterations in mRNA abundance, we extended our analysis of control and mutant MEFs transfected with an HA-tagged human HuR expression vector (pBBHuR) (28) and examined for changes in mRNA accumulation and decay relative to transfectants bearing empty vector (pBB) or untransfected cells. Since the transfectability of the primary MEF varied from 5 to 43%, we only used those with an efficiency of 30 to 40% (see Fig. S2 in the supplemental material). The introduction of exogenous HuR in HuR+ and HuR− MEFs increased the levels of Fgf10 mRNA but not the half-lives (Fig. 9). In addition, the same cultures did not show any change in the abundance or stability of the Foxc1 mRNA, suggesting that the amount of exogenous HuR was not sufficient to control the utilization of this mRNA. In sharp contrast and in all MEF cultures, the abundance and the half-lives of the Ets2 and Tbx4 mRNAs were increased (Fig. 9). A similar effect was observed for the Hoxd13 mRNA in transfected HuR+ MEFs, whereas small but significant reductions were detected in the abundance and half-life of the Hoxb9 mRNA; however, the transfected HuR was not sufficient to affect the expression of the Hox mRNAs in HuR− MEFs. Still, our data demonstrate that changes in the abundance of the Ets2, Tbx4, Hoxd13, and Hoxb9 mRNAs in HuR MEFs were due to the loss of positive/negative stability control.
FIG. 10.
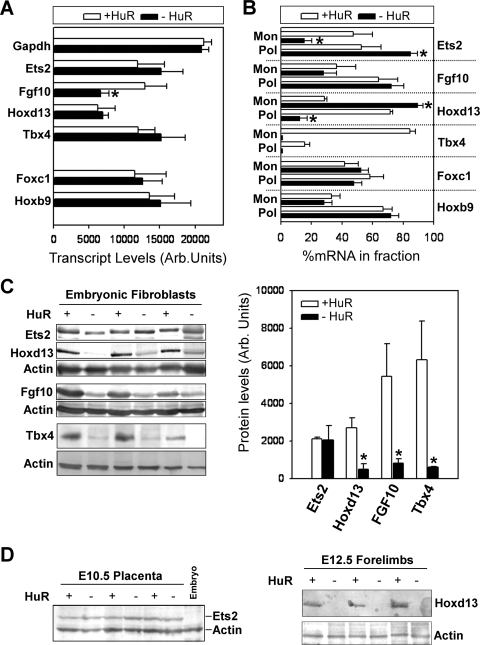
Differential effects of HuR's loss on the transcription and translation of its target mRNAs. (A) Densitometric quantitation of nuclei run-on experiments with radiolabeled RNAs from control and mutant MEFs hybridized to cold probes spotted onto nylon filters. Data (phosphorimager units ± standard deviations) were derived from three individual experiments. The asterisk denotes a significant difference with a P value of <0.01. (B) qRT-PCR detection of HuR target RNAs in monosomal (Mon)/polysomal (Pol) fractions from control and mutant MEFs. Data are derived from measurements in pooled fractions normalized to β2Μ mRNA and presented as percentages (±standard errors of the means) of total cytoplasmic mRNA. An asterisk denotes a significant difference with a P value of <0.05. (C) Detection and quantitation of immunoblotted proteins in total (for Ets2, FGF10, and Hoxd13) or nuclear (for Tbx4) extracts derived from control and mutant MEFs. Actin is shown as a loading control. Protein levels (phosphorimager units ± standard deviations from three independent samples) were normalized to actin. An asterisk denotes a significant difference with a P value of <0.05. (D) Detection of Ets2 (left) or Hoxd13 (right) in extracts from E10.5 placentas or E12.5 limbs, respectively. For Ets2, E10.5 embryonic extracts are shown as negative controls. Actin is shown as a loading control.
FIG. 9.
Differential effects of HuR's loss on the stability of its target mRNAs. (A) qRT-PCR detection of selected mRNAs in control and mutant MEFs in the absence (NT) or presence of empty vector (pBB) or vector expressing HA-hHuR (pBBHuR). Data are presented as the change (± standard deviation) from untransfected control values and derived from qRT-PCR measurements from three independent experiments. (B) Changes in the stability of selected mRNAs in actinomycin D-treated control and mutant, untransfected and transfected MEFs. Data are half-lives in minutes (± standard deviations) derived from decay plots from three independent experiments. For both panels A and B, a single asterisk denotes differences between the control and mutant MEFs; double asterisks denote differences between pBB- and pBBHuR-transfected cells.
To assess whether HuR could also affect the translation of its targets, we analyzed their presence in polysomal fractions from HuR− MEFs (Fig. 10B). The monosomal/polysomal fractions in control and HuR− MEFs were comparable (see Fig. S4 in the supplemental material), indicating that the mutation did not affect the central translational process. The polysomal representation of the Fgf10, Foxc1, and Hoxb9 mRNAs remained unaltered in HuR− MEFs, suggesting that HuR does not control their translation. In addition, the cytoplasmic levels of the Tbx4 mRNA in HuR− MEFs were below the limits of qRT-PCR detection, and thus, we cannot draw conclusions about its translatability. However, the polysomal representation of the Ets2 mRNA was increased in HuR− MEFs, whereas that of the Hoxd13 mRNA was reduced, suggesting that HuR can control both their turnover and translation but in a differential manner.
To evaluate the outcome of these alterations in gene expression, we analyzed the presence of selected proteins in HuR− MEFs. The availability of reagents for the corresponding mouse proteins was limited to immunoblot-compatible antibodies for Ets2, FGF10, Hoxd13, and Tbx4. The levels of Ets2 protein in HuR− MEFs and E10.5 Elavl1−/− placentas appeared unaffected (Fig. 10C and D), suggesting that the increased translation of its mRNA compensates for its reduced abundance. This finding provides an explanation for the presence of EPC-derived trophoblasts (e.g., spongiotrophoblasts, giant cells) in the absence of HuR, despite the collapse of the placental labyrinth since these cells require Ets2 for their maintenance. In contrast to Ets2, and despite our inability to detect it in tissue extracts, the Tbx4 protein was dramatically reduced in HuR− MEFs (Fig. 10C); thus, the lack of HuR's control over the posttranscriptional utilization of the Tbx4 mRNA may be the cause of the delay of the chorioallantoic interface and the limb aberrations. Similarly, the loss of the FGF10 and Hoxd13 proteins in both HuR− MEFs and E12.5 forelimbs (Fig. 10C and D) supports HuR's control over limb outgrowth and patterning. Together, our data demonstrate that HuR controls the expression of specific developmental mRNAs in a differential fashion for the prudent control of developmental morphogenesis, patterning, and specification.
DISCUSSION
In this study, we aimed to identify the functions of HuR in vivo by means of its genetic ablation from mice. In doing so, we revealed an array of embryonic phenotypes induced by HuR's permutation that point toward its role in the control of gene expression programs governing the development of specific organs and at different developmental stages.
First, HuR is required for the survival of midgestational embryos. The comparison of obligatory, epiblast, and endothelial deletions of HuR revealed that lethality at this stage was due to placental insufficiency underlined by an extraembryonic defect in the development of the placental labyrinth. To date, numerous mouse mutants have been described to possess a “small labyrinth” phenotype (14, 26) and have defects either in the initiation of chorioallantoic branching or in secondary branching and vascularization. It appears that HuR affects both of these processes but to a different extent. In mutant chorioallantoic placentas, branching initiation was delayed for 1 to 2 days; then branch extension progressed poorly, showing a block in the timely differentiation of syncytiotrophoblast cells that are required for normal labyrinth formation. Concomitantly, the vascularization of the mutant labyrinths was also compromised. This process appears to rely on the interdependence of three different cell types: the allantoic mesoderm, the endothelia forming the blood vessels, and the trophoblastic compartments. The absence of HuR in the allantoic mesoderm and embryonic endothelia (i.e., in epiblast mutants) or in maternal/fetal endothelia (i.e., in endothelial mutants) did not affect labyrinthine vascularization. Thus, and in light of HuR's cytoplasmic abundance in labyrinthine trophoblasts, we postulate that its functions in these cells are indirectly modulating labyrinthine vascularization, highlighting a necessary cross talk between both cell populations for the establishment of the normal placental architecture.
The molecular details governing chorioallantoic branching initiation, extension, and vascularization are very complex. However, a wealth of genetic data has pinpointed the requirement of chorionic signaling cascades induced by the allantoic mesoderm to drive labyrinthine morphogenesis and vascularization (26, 50). Although we did not analyze expression patterns in trophoblasts, coupling prior data to expression profiling in MEFs allowed us to identify HuR-interacting mRNAs whose dysfunction relates to placental insufficiency. The interaction of HuR with the Tbx4 mRNA and its reduced expression in HuR− cells suggested that HuR controls the initiation of branching following chorioallantoic fusion. In Tbx4null mouse mutants, the vascularization of allantois and its fusion to the chorion fails, whereas in heterozygote mutants fusion is delayed, phenocopying a HuR-deficient response (43). However, Tbx4 heterozygotes eventually form functional placentas, indicating that HuR's functions extend to additional processes such as chorioallantoic branching and labyrinthine vascularization. This is exemplified by the reduced expression of allantois-secreted factors (e.g., Wnt2), chorionic receptors (e.g., Frizzled Pdgfra), and downstream transcription factors (e.g., the fos-related transcription factor Fra-1) in HuR− cells, all of which are required for labyrinth formation and vascularization (27, 41). Most intriguingly, HuR can control mRNAs involved in trophoblast differentiation and function as exemplified by its association with Ets2 mRNA which induces the polarity of the EPC and controls the renewal of stem cells that give rise to spongiotrophoblasts and giant cells. The differential effect of HuR's loss on the abundance of Ets2 mRNA versus its translation suggests that HuR acts to modulate the thresholds of EPC-derived trophoblast differentiation for the proper development of non-EPC-derived placental layers. In addition, HuR could also be implicated in syncytiotrophoblast differentiation in the labyrinth via direct interference to fusion-induced signaling pathways affecting Gcm1 gene expression. However, the limited information on the temporal activation of this gene (3) and the difficult manipulation of primary trophoblastic cells hinder the assessment of a possible HuR-Gcm1 interaction.
The application of conditional genetic manipulation in the epiblast allowed us to dissect further the role of HuR in the development of the embryo proper. The most readily observable phenotypic distortions in HuR-null embryos were observed in skeletal segments. Mutant embryos showed delays in skeletal ossification that could contribute to the reduced size of HuR-deficient skeletons and the delay in the closing of the sternum. The process of ossification relies on condensations of mesenchymal tissue which can be either osteogenic or chondrogenic. Both types of condensations are modulated by BMP signals, including BMP-4 that shows an altered expression pattern in HuR-deficient cells (13). However, we failed to detect an interaction between HuR and Bmp4 mRNA, suggesting that yet-unidentified upstream interactions may be involved in the temporal modulation of osteoblast/chondrocyte differentiation and ossification.
Our data demonstrate a clear role for HuR in limb development. A previous study indicated that the expression of HuR peaks between E10.5 and E12.5 (20) which corresponds to the time of limb outgrowth and specification of its axes. In the mutant embryos, initial specification programs are active since limb buds are present and give rise to forelimbs and hind limbs. However, the poor outgrowth of these limb buds, the abnormal appearance of digital interzones between E10.5 and E12.5, the fusions among long and short limb elements, and the lack of definable anteroposterior specification in digits suggest that HuR controls a series of distinct processes in limb patterning. Such processes rely on FGF/Wnt and hedgehog signals and involve distinct transcription factors like Tbx4, Tbx5, Pitx1, and Hox paralogue genes that control the mitotic expansion of mesenchymal cells during limb outgrowth and the axial specification of the mesodermal tissue in the posterior of each limb bud (45). Our studies on MEFs are of direct relevance to these responses since they are prototypical mesenchymal representatives. The reduced expression of HuR-interacting Tbx4, FGF10, and HoxD13 mRNAs in mutant cells and the transcriptional control of the Fgf10 gene by TBX4 (44) point toward HuR's role as a spatiotemporal organizer of gene networks controlling distal limb outgrowth and patterning. Furthermore, the inverse correlation between Bmp4 and Wnt2 mRNAs in HuR− MEFs, which are not HuR targets, may reflect compensatory responses to defective outgrowth or defects in interior or interdigital apoptotic programs sculpting the bones which are dependent on counteracting BMP antagonist and WNT signals (45). This latter supposition is further supported by demonstrations of HuR's involvement in pro- and antiapoptotic signals (1, 18, 40).
The role of mesenchymal HuR in organogenesis is also demonstrated by the asplenic phenotype of the mutant embryos. The spleen arises from mesenchymal condensation that occurs in the dorsal pancreatic mesenchyme surrounding the pancreatic epithelium around E10.5. In mutant embryos, the pancreas develops and a splenic anlage is present, indicating that the development of the spleen from the splenopancreatic mesenchyme can initiate. However, the splenic primordium fails to expand, relating to the phenotypes of mutants lacking the transcription factors Hox11, Pbx1, Tcf21, and Sox11 (7). It is thus likely that HuR acts at a similar level as these transcription factors to influence the response of the splenic mesenchyme toward splenic outgrowth.
In molecular terms, our data demonstrate the complexity of HuR's functions over developmental gene expression programs. The involvement of ARE-binding proteins in such processes has been previously exemplified by mutations in genes encoding the tristetraprolin family member BRF1 (Zfp36L1/TIS11b). BRF1-deficient mutants die in midgestation due to placental insufficiency consequent to failures in chorioallantoic fusion or placental vascularization (5, 53). An antagonism between BRF1 and HuR toward the modulation of mRNA stability has been proposed (47) which could be extrapolated to their biological roles in placental morphogenesis. However, our data confer a wider role for HuR that extends beyond its prescribed role as a stabilizing ARE-binding protein. On the one end, most of the HuR target mRNAs identified in this study do not appear to possess a prototypical ARE signature, whereas other ARE-containing transcripts (e.g., Pdgfb and Hoxc6) did not associate with HuR in MEFs. Furthermore, we did not detect a posttranscriptional effect of HuR on the ARE-containing Fgf10 mRNA which interacts with HuR, suggesting that this interaction controls signal-restricted responses that have not been revealed in our experimental setting. On the other end, the posttranscriptional effects of HuR were dominant for the majority of the identified mRNA targets but were also variable and differential. As demonstrated herein, HuR can control the stability of some of its target mRNAs either in a positive (Ets2, Hoxd13, and Tbx4) or negative (Hoxb9) fashion. In addition, HuR's effect on translation was also either positive (Hoxd13) or negative (Ets2) and could differ from its effect on mRNA stability. Our data point toward a differential role of HuR in the posttranscriptional regulation of mRNAs which can be dictated by differences in the cis-element composition between HuR target mRNAs as well as their RBP associations in different RNP configurations.
Irrespective of the posttranscriptional process, our protein data demonstrate the importance of HuR's control over the biosynthesis of key developmental determinants. Our analysis is by no means exhaustive but is strongly indicative of the complexity of cellular interplays and signaling cascades that are modulated by HuR. In that context, the data presented in this study provide a framework for the future analysis of signals converging onto HuR and its RNP associations by means of its tissue-restricted modification and rigorous molecular analyses.
Supplementary Material
Acknowledgments
We thank Liz Robertson, Reinhard Fassler, and George Kollias for the provision of the Cre lines; Alexia Giannakopoulou for technical assistance in gene targeting; and Eumorphia Remboutsika for critical discussions.
This work was supported in part by European Commission grant MUGEN LSHG-CT-2005-005203, Hellenic Secretariat for Research and Technology grants PENED-KA-70-3-6705 and PENED2003-264, and the AICR (United Kingdom) grant 07-0548.
Footnotes
Published ahead of print on 23 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdelmohsen, K., A. Lal, H. H. Kim, and M. Gorospe. 2007. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle 61288-1292. [DOI] [PubMed] [Google Scholar]
- 2.Abdelmohsen, K., R. Pullmann, Jr., A. Lal, H. H. Kim, S. Galban, X. Yang, J. D. Blethrow, M. Walker, J. Shubert, D. A. Gillespie, H. Furneaux, and M. Gorospe. 2007. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25543-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anson-Cartwright, L., K. Dawson, D. Holmyard, S. J. Fisher, R. A. Lazzarini, and J. C. Cross. 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25311-314. [DOI] [PubMed] [Google Scholar]
- 4.Barreau, C., L. Paillard, and H. B. Osborne. 2006. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 337138-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, S. E., M. J. Sanchez, O. Spasic-Boskovic, T. Santalucia, L. Gambardella, G. J. Burton, J. J. Murphy, J. D. Norton, A. R. Clark, and M. Turner. 2006. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Dev. Dyn. 2353144-3155. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharyya, S. N., and W. Filipowicz. 2007. Argonautes and company: sailing against the wind. Cell 1281027-1028. [DOI] [PubMed] [Google Scholar]
- 7.Brendolan, A., M. M. Rosado, R. Carsetti, L. Selleri, and T. N. Dear. 2007. Development and function of the mammalian spleen. Bioessays 29166-177. [DOI] [PubMed] [Google Scholar]
- 8.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 1511-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20465-470. [DOI] [PubMed] [Google Scholar]
- 11.Chen, F., and M. R. Capecchi. 1997. Targeted mutations in hoxa-9 and hoxb-9 reveal synergistic interactions. Dev. Biol. 181186-196. [DOI] [PubMed] [Google Scholar]
- 12.Cherry, J., V. Karschner, H. Jones, and P. H. Pekala. 2006. HuR, an RNA-binding protein, involved in the control of cellular differentiation. In Vivo 2017-23. [PubMed] [Google Scholar]
- 13.Colnot, C. 2005. Cellular and molecular interactions regulating skeletogenesis. J. Cell Biochem. 95688-697. [DOI] [PubMed] [Google Scholar]
- 14.Cross, J. C., Z. Werb, and S. J. Fisher. 1994. Implantation and the placenta: key pieces of the development puzzle. Science 2661508-1518. [DOI] [PubMed] [Google Scholar]
- 15.DeLaurier, A., R. Schweitzer, and M. Logan. 2006. Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev. Biol. 29922-34. [DOI] [PubMed] [Google Scholar]
- 16.Fan, X. C., and J. A. Steitz. 1998. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 9515293-15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figueroa, A., A. Cuadrado, J. Fan, U. Atasoy, G. E. Muscat, P. Munoz-Canoves, M. Gorospe, and A. Munoz. 2003. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cell. Biol. 234991-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 2941895-1901. [DOI] [PubMed] [Google Scholar]
- 19.Goodman, F. R. 2002. Limb malformations and the human HOX genes. Am. J. Med. Genet. A 112256-265. [DOI] [PubMed] [Google Scholar]
- 20.Gouble, A., and D. Morello. 2000. Synchronous and regulated expression of two AU-binding proteins, AUF1 and HuR, throughout murine development. Oncogene 195377-5384. [DOI] [PubMed] [Google Scholar]
- 21.Graus, F., J. Dalmou, R. Rene, M. Tora, N. Malats, J. J. Verschuuren, F. Cardenal, N. Vinolas, J. Garcia del Muro, C. Vadell, W. P. Mason, R. Rosell, J. B. Posner, and F. X. Real. 1997. Anti-Hu antibodies in patients with small-cell lung cancer: association with complete response to therapy and improved survival. J. Clin. Oncol. 152866-2872. [DOI] [PubMed] [Google Scholar]
- 22.Gringhuis, S. I., J. J. Garcia-Vallejo, B. van het Hof, and W. van Dijk. 2005. Convergent actions of IκB kinase β and protein kinase Cδ modulate mRNA stability through phosphorylation of 14-3-3β complexed with tristetraprolin. Mol. Cell. Biol. 256454-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustafsson, E., C. Brakebusch, K. Hietanen, and R. Fassler. 2001. Tie-1-directed expression of Cre recombinase in endothelial cells of embryoid bodies and transgenic mice. J. Cell Sci. 114671-676. [DOI] [PubMed] [Google Scholar]
- 24.Guttinger, S., P. Muhlhausser, R. Koller-Eichhorn, J. Brennecke, and U. Kutay. 2004. Transportin2 functions as importin and mediates nuclear import of HuR. Proc. Natl. Acad. Sci. USA 1012918-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi, S., T. Tenzen, and A. P. McMahon. 2003. Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis 3751-53. [DOI] [PubMed] [Google Scholar]
- 26.Hemberger, M., and J. C. Cross. 2001. Genes governing placental development. Trends Endocrinol. Metab. 12162-168. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa, T., Y. Tamai, A. M. Zorn, H. Yoshida, M. F. Seldin, S. Nishikawa, and M. M. Taketo. 2001. Mouse Wnt receptor gene Fzd5 is essential for yolk sac and placental angiogenesis. Development 12825-33. [DOI] [PubMed] [Google Scholar]
- 28.Katsanou, V., O. Papadaki, S. Milatos, P. J. Blackshear, P. Anderson, G. Kollias, and D. L. Kontoyiannis. 2005. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell 19777-789. [DOI] [PubMed] [Google Scholar]
- 29.Keene, J. D. 1999. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. USA 965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keene, J. D., and S. A. Tenenbaum. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 91161-1167. [DOI] [PubMed] [Google Scholar]
- 31.Khabar, K. S. 2005. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J. Interferon Cytokine Res. 251-10. [DOI] [PubMed] [Google Scholar]
- 32.Kim, H. H., K. Abdelmohsen, A. Lal, R. Pullmann, Jr., X. Yang, S. Galban, S. Srikantan, J. L. Martindale, J. Blethrow, K. M. Shokat, and M. Gorospe. 2008. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 221804-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King, M., J. S. Arnold, A. Shanske, and B. E. Morrow. 2006. T-genes and limb bud development. Am. J. Med. Genet. A 1401407-1413. [DOI] [PubMed] [Google Scholar]
- 34.Kullmann, M., U. Gopfert, B. Siewe, and L. Hengst. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′ UTR. Genes Dev. 163087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehmann, O. J., J. C. Sowden, P. Carlsson, T. Jordan, and S. S. Bhattacharya. 2003. Fox's in development and disease. Trends Genet. 19339-344. [DOI] [PubMed] [Google Scholar]
- 36.Levadoux-Martin, M., A. Gouble, B. Jegou, V. Vallet-Erdtmann, J. Auriol, P. Mercier, and D. Morello. 2003. Impaired gametogenesis in mice that overexpress the RNA-binding protein HuR. EMBO Rep. 4394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, H., S. Park, B. Kilburn, M. A. Jelinek, A. Henschen-Edman, D. W. Aswad, M. R. Stallcup, and I. A. Laird-Offringa. 2002. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 27744623-44630. [DOI] [PubMed] [Google Scholar]
- 38.Manley, G. T., P. S. Smitt, J. Dalmau, and J. B. Posner. 1995. Hu antigens: reactivity with Hu antibodies, tumor expression, and major immunogenic sites. Ann. Neurol. 38102-110. [DOI] [PubMed] [Google Scholar]
- 39.Mazan-Mamczarz, K., S. Galban, I. Lopez de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 1008354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazroui, R., S. Di Marco, E. Clair, C. von Roretz, S. A. Tenenbaum, J. D. Keene, M. Saleh, and I. E. Gallouzi. 2008. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J. Cell Biol. 180113-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monkley, S. J., S. J. Delaney, D. J. Pennisi, J. H. Christiansen, and B. J. Wainwright. 1996. Targeted disruption of the Wnt2 gene results in placentation defects. Development 1223343-3353. [DOI] [PubMed] [Google Scholar]
- 42.Mukherjee, D., M. Gao, J. P. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naiche, L. A., and V. E. Papaioannou. 2003. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development 1302681-2693. [DOI] [PubMed] [Google Scholar]
- 44.Naiche, L. A., and V. E. Papaioannou. 2007. Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development 13493-103. [DOI] [PubMed] [Google Scholar]
- 45.Niswander, L. 2003. Pattern formation: old models out on a limb. Nat. Rev. Genet. 4133-143. [DOI] [PubMed] [Google Scholar]
- 46.Pullmann, R., Jr., H. H. Kim, K. Abdelmohsen, A. Lal, J. L. Martindale, X. Yang, and M. Gorospe. 2007. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 276265-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 321279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebane, A., A. Aab, and J. A. Steitz. 2004. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 10590-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert, B. 2007. Bone morphogenetic protein signaling in limb outgrowth and patterning. Dev. Growth Differ. 49455-468. [DOI] [PubMed] [Google Scholar]
- 50.Rossant, J., and J. C. Cross. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2538-548. [DOI] [PubMed] [Google Scholar]
- 51.Schwenk, F., U. Baron, and K. Rajewsky. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 235080-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sekine, K., H. Ohuchi, M. Fujiwara, M. Yamasaki, T. Yoshizawa, T. Sato, N. Yagishita, D. Matsui, Y. Koga, N. Itoh, and S. Kato. 1999. Fgf10 is essential for limb and lung formation. Nat. Genet. 21138-141. [DOI] [PubMed] [Google Scholar]
- 53.Stumpo, D. J., N. A. Byrd, R. S. Phillips, S. Ghosh, R. R. Maronpot, T. Castranio, E. N. Meyers, Y. Mishina, and P. J. Blackshear. 2004. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the tristetraprolin family. Mol. Cell. Biol. 246445-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka, M., M. Gertsenstein, J. Rossant, and A. Nagy. 1997. Mash2 acts cell autonomously in mouse spongiotrophoblast development. Dev. Biol. 19055-65. [DOI] [PubMed] [Google Scholar]
- 55.Tranchevent, L. C., R. Barriot, S. Yu, S. Van Vooren, P. Van Loo, B. Coessens, B. De Moor, S. Aerts, and Y. Moreau. 2008. ENDEAVOUR update: a web resource for gene prioritization in multiple species. Nucleic Acids Res. 36W377-W384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Giessen, K., S. Di-Marco, E. Clair, and I. E. Gallouzi. 2003. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J. Biol. Chem. 27847119-47128. [DOI] [PubMed] [Google Scholar]
- 57.van der Giessen, K., and I. E. Gallouzi. 2007. Involvement of transportin 2-mediated HuR import in muscle cell differentiation. Mol. Biol. Cell 182619-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verschuuren, J. J., J. Dalmau, R. Hoard, and J. B. Posner. 1997. Paraneoplastic anti-Hu serum: studies on human tumor cell lines. J. Neuroimmunol. 79202-210. [DOI] [PubMed] [Google Scholar]
- 59.Wen, F., J. A. Tynan, G. Cecena, R. Williams, J. Munera, G. Mavrothalassitis, and R. G. Oshima. 2007. Ets2 is required for trophoblast stem cell self-renewal. Dev. Biol. 312284-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, L., A. de Bruin, H. I. Saavedra, M. Starovic, A. Trimboli, Y. Yang, J. Opavska, P. Wilson, J. C. Thompson, M. C. Ostrowski, T. J. Rosol, L. A. Woollett, M. Weinstein, J. C. Cross, M. L. Robinson, and G. Leone. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421942-947. [DOI] [PubMed] [Google Scholar]
- 61.Yao, K. M., M. L. Samson, R. Reeves, and K. White. 1993. Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J. Neurobiol. 24723-739. [DOI] [PubMed] [Google Scholar]
- 62.Zhao, Q., R. R. Behringer, and B. de Crombrugghe. 1996. Prenatal folic acid treatment suppresses acrania and meroanencephaly in mice mutant for the Cart1 homeobox gene. Nat. Genet. 13275-283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.