Abstract
In mammals, AU-rich elements (AREs) are critical regulators of mRNA turnover. They recruit ARE-binding proteins that inhibit or stimulate rapid mRNA degradation in response to stress or developmental cues. Using a bioinformatics approach, we have identified AREs in Drosophila melanogaster 3′ untranslated regions and validated their cross-species conservation in distant Drosophila genomes. We have generated a Drosophila ARE database (D-ARED) and established that about 16% of D. melanogaster genes contain the mammalian ARE signature, an AUUUA pentamer in an A/U-rich context. Using candidate ARE genes, we show that Drosophila AREs stimulate reporter mRNA decay in cultured cells and in the physiological context of the immune response in D. melanogaster. In addition, we found that the conserved ARE-binding protein Tis11 regulates temporal gene expression through ARE-mediated decay (AMD) in D. melanogaster. Our work reveals that AREs are conserved and functional cis regulators of mRNA decay in Drosophila and highlights this organism as a novel model system to unravel in vivo the contribution of AMD to various processes.
In mammals, some major posttranscriptional regulators of mRNA levels are AU-rich elements (AREs) located in the 3′untranslated region (3′UTR) of mRNAs (5). Both pioneer and recent investigations have revealed a positive correlation between mammalian mRNA decay and the presence of AREs in 3′UTRs (12, 27, 32, 40). AREs have been classified into three subclasses according to the presence and distribution of the most frequently found motif, the AUUUA pentamer (13). Whereas class I AREs contain dispersed AUUUA motifs within U-rich regions, class II corresponds to tandem repeats of the UUAUUUA(U/A)(U/A) motif. Class III AREs display U-rich regions devoid of the AUUUA motif. All classes stimulate deadenylation as the first step of mRNA decay, either mediating a processive (classes I and III) or distributive (class II) poly(A) shortening (13). Since their discovery in immune and inflammatory genes (12, 32), the number of ARE genes has increased considerably. More recently, a computational-based approach has predicted a large repertoire of ARE genes in mammals (5 to 8% of total mRNAs) (3, 4). The quantitative assessment of ARE occurrence in human, mouse, and rat transcripts revealed that AREs are conserved in 75% of mammalian ARE genes (21). The significant overrepresentation of ARE genes in various cellular processes such as adhesion, growth, differentiation, and apoptosis strongly suggests that AREs regulate gene expression during normal mammalian development, in addition to their established connection with pathological situations (37). This is supported by the finding that several signal transduction pathways that are critically required throughout development control the turnover of many ARE mRNAs (17, 19; reviewed in reference 18). However, the relevance of ARE-mediated gene regulation remains largely underdocumented during normal development in mammals, largely due to the difficulty of manipulating higher eukaryotes and of assessing mRNA turnover in vivo.
ARE-mediated decay (AMD) is regulated in response to intra- or extracellular cues. Molecular mechanisms underlying this regulation involve changes in the subcellular localization or posttranslational modifications of ARE-binding proteins (ARE-BPs), leading to the alteration of their ARE-binding activity and/or interaction with the degradation machinery (14, 18). While most ARE-BPs, such as AUF1, TTP, or KSRP, attract the degradation machinery, others, like the Hu/ELAV family of proteins, protect ARE mRNAs from rapid destruction (5, 6). An interesting finding is that ARE-BPs are evolutionarily conserved from yeast to mammals, as exemplified by Cth2, the yeast orthologue of TTP, which regulates the global expression of ARE genes in response to nutritional cues (26). Notably, recent studies point to the conservation of ARE-mediated gene regulation in invertebrates. In the leech Helobdella robusta, notch mRNA stability might be regulated by conserved AUUUA pentamers in its 3′UTR (20). In addition, mammalian AREs, such as those from tumor necrosis factor α and interleukin-6, induce reporter mRNA decay in Drosophila melanogaster cells (22), in which orthologues to both mammalian ARE-BPs and components of AMD pathways (e.g., 5′ to 3′ and 3′ to 5′) are conserved (16, 22). However, the genome-wide occurrence of AREs has not yet been investigated in invertebrates, so the potential contribution of AMD to global gene expression is unknown.
Using a computation-based approach, we have searched for AREs in the 3′UTRs of D. melanogaster genes and generated the first repertoire of Drosophila ARE genes. Combining both ex vivo and in vivo approaches, we have validated experimentally that our analysis has uncovered genuine AMD targets. Our analysis shows that AMD regulates temporal gene expression in D. melanogaster and predicts the widespread contribution of AREs to posttranscriptional regulation in this organism.
MATERIALS AND METHODS
Computational analysis. (i) Generating D-ARED.
3′UTR sequences for all D. melanogaster transcripts were downloaded from the FlyBase (http://flybase.org) FTP repository of precomputed files (Flybase2008_07 [DMEL R5.10] release). We used the batch query feature to obtain annotations and gene ontology associations for genes. For every transcript, the following analysis was carried out. We first searched for the poly(A) signal (AWTAAA) in the last 50 bases of transcripts. This position demarcated the search boundary, and no motifs past that position were considered. We then progressively scanned the 3′UTR of each transcript with the five ARE group motifs (Table 1), allowing for a single mismatch anywhere for groups 1 to 3 and only outside the core AUUUA motif for groups 4 and 5. We then assigned each transcript to the lowest-numbered matching group (corresponding to the longest-sequence match to an ARE motif). Lastly, for each gene, all transcript variants were compared, and the one with the lowest group number was assigned as the ARE gene representative. If no recognizable ARE motifs were detected, the gene was declared non-ARE. The characteristics of the algorithm used for ARE scanning reduce the false-positive rate with respect to truly functional sequence elements, as we have used (i) previously validated functional and conserved patterns of AREs in mammals, and (ii) a larger-than-typical length of search motifs (13 to 21 bases).
TABLE 1.
The ARE motifs
| Groupa | Pattern |
|---|---|
| I | AUUUAUUUAUUUAUUUAUUUA |
| II | AUUUAUUUAUUUAUUUA |
| III | AUUUAUUUAUUUA |
| IV | WW[AUUUAUUUA]WWb |
| V | WWWU[AUUUA]UWWW |
Group I genes contain five or more AUUUA tandem repeats, and group IV is for a minimum of two repeats. For each group, a single mismatch is allowed anywhere in the motif used, except for groups IV and V, where the mismatch is tolerated only in the bases flanking the bracketed core.
W = A or U.
(ii) Conserved motif scanning.
The full set of conserved and potentially regulatory motif instances in the 3′UTRs of D. melanogaster published previously (36) was obtained from Manolis Kellis (personal communication). This set was compared to the list of ARE motifs identified in the D. melanogaster 3′UTRs by means of positional overlaps. ARE motifs were retained only if they overlapped at least partially with any conserved motif instance. Since both the conserved regulatory motifs and AREs were identified in the same genome, conservation automatically applies to the overlapped AREs. The original ARED set contains 4,940 distinct instances in 3,305 transcripts of 1,753 genes. The set of conserved AREs contains 2,662 sites, 1,877 transcripts, and 922 genes, respectively. The two most commonly overlapping motifs were MO37 (ATTTATT) and MO52 (TATTTAT), corresponding to a partial canonical ARE, as expected.
(iii) ARE conservation between D. melanogaster and Drosophila pseudoobscura.
D. pseudoobscura 3′UTR computationally predicted sequences assembled previously (34) were obtained from Alexander Stark (personal communication). Since the data set contains a complete set of one-to-one matched and aligned 3′UTR sequences for both D. melanogaster and D. pseudoobscura, we ran both 3′UTR sets through the ARED pipeline described above, producing an ARE transcript list for each species. The D. melanogaster list of ARE transcripts (data not shown) and the D-ARED (see Table S1 in the supplemental material) do not exactly match due to their assembling from different releases of Flybase. To assess the conservation of ARE motifs between the two species, we tabulated the joint distribution of non-ARE, class I, and class II ARE transcripts in D. melanogaster and D. pseudoobscura in a three-by-three contingency table (Table 2). The reported P value for a chi-squared test of independence (−log P > 15; all logs to base 10) indicates that D. melanogaster and D. pseudoobscura orthologues are extremely unlikely to contain an ARE by random chance alone. ARE conservation in orthologous branchless-RC (bnl-RC) and cecropinA1 (cecA1) 3′UTRs was assessed by performing sequence alignments using the UCSC Genome Bioinformatics Browser (http://genome.ucsc.edu/).
TABLE 2.
D. melanogaster versus D. pseudoobscura cross-species ARE conservation analysisa
| Drosophila melanogaster |
Drosophila pseudoobscura
|
|||
|---|---|---|---|---|
| Non-ARE | Class I | Class II | Total | |
| Non-ARE | 6,383 | 1,450 | 281 | 8,114 |
| Class I | 999 | 927 | 161 | 2,087 |
| Class II | 201 | 208 | 197 | 606 |
| Total | 7,583 | 2,585 | 639 | 10,807 |
Counts are in terms of transcripts with respect to the ARE class conservation. Out of 10,807 pairs of transcripts analyzed, 29.8% contain AREs (2,585 class I AREs and 639 class II AREs) in D. pseudoobscura, while 24.9% contain AREs (2,087 class I AREs and 606 class II AREs) in D. melanogaster. In addition, the table indicates that out of a total of 606 class II ARE transcripts in D. melanogaster, 66.8% (208 of class I and 197 of class II) contain a conserved ARE (either class I or II) in D. pseudoobscura.
Plasmids and cell culture.
3′UTRs of bnl-RA, bnl-RC, and cecA1 plus additional 3′ flanking sequences were amplified from genomic DNA using the primer pairs RA-1/RA-2, RB-1/RB-2, and cec-1/cec-2, respectively (see Table S4 in the supplemental material), and cloned downstream of the green fluorescent protein (GFP) coding sequence in the vector pRmHa, which contains the metallothionein promoter (11). Deletions in bnl transcripts were made by inverse PCR as described on the website of the Berkeley Drosophila Genome Project (www.fruitfly.org) (see Fig. S1 in the supplemental material for primers). The AUUUA-to-ACCCA alterations in the 3′UTR of cecA1 were introduced using oligonucleotide-mediated site-directed mutagenesis and inverse PCR (with oligonucleotides cec-3/cec-4). Schneider 2 (S2) and mbn-2 cells were cultured in fetal calf serum-supplemented Schneider's medium (Invitrogen) and transfected using Cellfectin reagent (Invitrogen) in 6-well plates. When required, mbn-2 cells were exposed to lipopolysaccharide (LPS) (Escherichia coli strain O111:B4; Sigma). The Tis11 double-stranded RNA (dsRNA) template (about 700 bp long) was generated as indicated previously (15) and is distributed by Open Biosystems. This dsRNA was transcribed in vitro using a T7 RiboMAX system (Promega) and targets the second half of the Tis11 coding sequence. The dsRNA was denatured at 75°C for 15 min and reannealed by being slowly cooled to room temperature. The extent of annealing was assessed by agarose gel electrophoresis. Six milligrams of dsRNA/106 cells was added directly to the culture medium and replaced three times during 6 days before reporter mRNA analysis.
RNA analysis.
The expression of the GFP reporter mRNAs was induced in S2 or mbn-2 cells for 2 h with 0.7 mM of CuSO4. For the determination of mRNA half-life (t1/2), cells were incubated with actinomycin D (5 μg/ml) for the times indicated in the figures. Total RNA then was prepared using an RNeasy RNA isolation kit (Qiagen). For Northern blot analysis, ∼8 μg of total RNA was heat denatured and separated by electrophoresis on a nondenaturing 1% agarose gel in Tris-acetate-EDTA and electrotransferred to a nylon membrane (Hybond-N+; Amersham Pharmacia Biotech). Hybridization was carried out using in vitro-transcribed probes directed against GFP and rp49 mRNAs synthesized using 32P-labeled CTP. Total RNAs from whole flies or fat bodies were prepared with TRIzol reagent (Invitrogen). For quantitative reverse transcription-PCR (RT-PCR) analysis, 1 μg of total DNase-free RNA was retrotranscribed using Superscript II RT (Invitrogen). Real-time PCR was performed on cDNA obtained from 0.1 μg of total RNA using platinum SYBR green (Invitrogen) according to the manufacturer's instructions using an iCycler IQ real-time detection system (Bio-Rad). All samples were analyzed in triplicate. The levels of mRNAs were determined by cycling threshold analysis and were normalized using rp49 as a control. Normalized data then were used to quantify the relative levels of mRNA using the ΔΔCT method (see Table S4 in the supplemental material for primers used for PCRs).
Transgenic flies.
The cecA1 promoter region from −760 to +30 bp was amplified from the pA10 plasmid (28) and was used to drive the firefly luciferase coding sequence fused to the wild-type (WT) or mutant cecA1 3′UTR. The cecA1-luc-3′UTRcecA1 constructs were cloned into a pattB-white+ derivative plasmid (V. Daburon, unpublished data) to allow for phiC31-targeted site-specific recombination into the attP landing site 3R-86F on the Drosophila melanogaster genome (7). luciferase (luc) reporter plasmids (250 ng/μl) were injected into embryos, and transgenic lines were established. The UAS-Tis11 RNA interference (RNAi) stock is from the Vienna Drosophila RNAi Center.
Septic injury and in vivo luciferase monitoring.
Five-day-old females were fed for 24 h with beetle luciferin (1 mM final concentration; Promega). Flies were collected under CO2 and individually infected by being pricked with a concentrated culture of E. coli, as described previously (8). Infected flies were placed in 96-well microtiter assay plates prefilled with luciferin-containing food as described previously (9). Plates were placed in a LucyI luminometer (Anthos) programmed for the automatic monitoring of relative luminescence units emanating from individual wells for 17 s each for 15 min.
RESULTS AND DISCUSSION
In silico screening identifies conserved AREs in Drosophila.
To identify ARE genes in D. melanogaster, we computationally searched for the consensus ARE motif WWWU[AUUUA]UWWW (W = A/U), which was previously used to establish the mammalian ARE database (ARED) (3), in the 3′UTRs of a collection of 10,844 nonredundant D. melanogaster genes. We found that 1,753 genes (∼16%) contain this motif in at least one transcript, and we compiled the results in a Drosophila ARE gene database (D-ARED) (see Table S1 in the supplemental material). The occurrence of this motif within 3′UTRs is significant, as 3′UTRs are 4- and 20-fold enriched in this motif compared to 5′UTRs and coding sequences, respectively. Interestingly, in analyzing the transcripts of 1,104 genes whose 3′UTR ends contained a poly(A) signal motif, we found that the mean length of ARE 3′UTRs is 2.5-fold longer (813 nucleotide [nt]; n = 1,104) than that of non-ARE 3′UTRs (323 nt; n = 5,745) (−log P > 6 by t test). These data suggest that shorter 3′UTRs could have been selected to escape ARE-mediated control and support the hypothesis that 3′UTR-mediated gene regulation impacts 3′UTR length (35). A closer analysis of the distribution of the AUUUA pentamer in 3′UTRs reveals that 352 genes (∼20% of all ARE genes) contain at least two contiguous repeats of the AUUUA pentamer, a feature of mammalian class II ARE genes (13). Those 352 genes were further categorized into four groups (I to IV) (Table 1) depending on their number of AUUUA repeats (see Table S1 in the supplemental material).
A number of conserved sequence motifs have previously been characterized by the alignment of 12 Drosophila genomes from species up to 45 million years distant (36). To test the functional significance of the mammalian consensus ARE sequence used to establish the D-ARED, we first examined whether the ARE sites overlap with positions of the evolutionarily conserved motifs in 3′UTRs. We found that 63.4% of the class II AREs in the D-ARED (or 53.9% of both class I and II AREs) overlap with conserved motifs, some of them containing the AUUUA core sequence (see Table S2 in the supplemental material). This result most probably is underestimated, since only AREs with strictly conserved positions are retained in this analysis, whereas no positional constraint in 3′UTRs has been reported for ARE activity in mammals or in yeast.
We then performed a direct cross-species analysis of ARE conservation between D. melanogaster and D. pseudoobscura using a previously assembled data set of computationally aligned 3′UTRs (10). We first found that AREs are similarly enriched in 3′UTRs of these two distantly related species (∼25 million years) (29.8 and 24.9% of transcripts in D. pseudoobscura and D. melanogaster, respectively) (Table 2). In addition, as found in D. melanogaster, the mean length of non-ARE 3′UTRs in D. pseudoobscura (1,191 nt; n = 7,583) is significantly shorter than that of ARE-containing 3′UTRs (1,549 nt; n = 3,224). Finally and most remarkably, 52.1 and 66.8% of D. melanogaster class I and II ARE transcripts, respectively, exhibit a conserved ARE (class I or II) in orthologous 3′UTRs in D. pseudoobscura (Table 2). This result is highly significant (P ≈ 0 by a binomial test) compared to the expected random values of 30%, assuming factor independence (Table 2). Taken together, our data establish that AREs are widespread in D. melanogaster 3′UTRs and evolutionarily conserved across Drosophila species.
To assess the functional diversity of the ARE genes in our database, we determined gene ontology categories and analyzed the overrepresented molecular functions, biological processes, and pathways using the Panther database tool (http://www.pantherdb.org) (39). We found that D. melanogaster ARE genes are involved in a large variety of cellular and developmental processes, ranging from cell homeostasis to neurogenesis (see Table S3 in the supplemental material). We observed a marked enrichment of genes encoding transcription factors, as previously reported for ARE genes in human cells (4), where transcription factor-coding mRNAs are largely short lived (40). We also observed a significant enrichment of ARE genes in several receptor tyrosine kinase pathways (platelet-derived growth factor, epidermal growth factor, and fibroblast growth factor [FGF]) sharing the common RAS/mitogen-activated protein kinase signaling cascade and in the Wnt signaling pathway. Thus, ARE-mediated gene regulation could modulate the magnitude and/or duration of signaling pathway activation in Drosophila through the concerted regulation of several transduction components. It is noteworthy that a major effector in the Wnt/β-catenin signaling cascade is encoded by the β-catenin mRNA, whose stability is regulated by AMD in mammals (19). Our finding that D. melanogaster β-catenin mRNA contains a conserved ARE in its 3′UTR (see Tables S1 and S2 in the supplemental material) suggests that the investigation of the ARE-mediated regulation of signaling pathways in D. melanogaster will be relevant to related signaling pathways in mammals.
Drosophila AREs mediate mRNA decay in cultured cells.
As contiguous repeats of the AUUUA pentamer are efficient destabilizing cis elements in mammals (17, 41), we have focused on class II ARE genes as promising AMD targets in Drosophila. RNA expression data were scarce and available for only 6 out of the 16 group 1 and 2 genes of this class. We selected branchless (bnl) for further analysis, as it exhibits a remarkable dynamic mRNA expression pattern in vivo that could reflect a well-orchestrated combination of transcription and stability controls (38). bnl codes for an FGF homologue playing a crucial role in the morphogenesis of the tracheal (respiratory) system (38). bnl produces two transcripts with distinct 3′UTRs, bnl-RA and bnl-RC (see Fig. S1 in the supplemental material). bnl-RC contains eight AUUUA pentamers in its 3′UTR, with four in tandem (see Table S1 in the supplemental material; also see Fig. 2A). bnl-RA is absent from the D-ARED even though it contains four dispersed AUUUA (see Fig. 2A), a pattern more related to the one present in class I ARE genes in mammals (13). D. melanogaster class I ARE genes (see Table S1 in the supplemental material) thus are likely to be underestimated in our database due to the high stringency of the ARE motif used for screening. In addition to bnl, we also selected cecA1 from group III for the further examination of ARE activity in living flies, since cecA1 mRNA decay is likely to be rapid (23) and can easily be assessed in vivo (see below). cecA1 codes for a small antimicrobial peptide (AMP) produced in response to bacterial and/or fungal infection (31). Its 3′UTR contains five AUUUA motifs, with two in tandem (see Table S1 in the supplemental material).
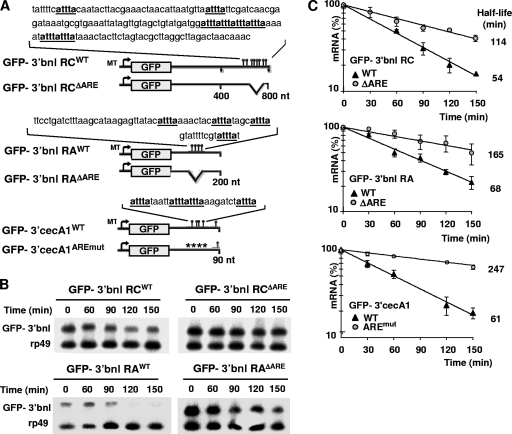
FIG. 2.
Drosophila AREs accelerate mRNA decay in cultured cells. (A) Schemes of the GFP reporters bearing the WT or the ARE mutant 3′UTRs of bnl-RC and bnl-RA and cecA1. AUUUA pentamers are indicated in boldface. AUUUA-containing regions (indicated above the constructs) have been entirely deleted (ΔARE) or mutagenized at four AUUUA repeats into ACCCA (indicated by asterisks in AREmut). MT, metallothionein promoter. (B and C) Stability of the GFP reporters was assessed by Northern blotting (B), using probes specific for GFP and rp49 mRNAs, and by quantitative RT-PCR (qRT-PCR) (C). Reporters harboring bnl or cecA1 3′UTRs were transiently transfected and expressed in S2 or mbn-2 cells, respectively. Total RNA was isolated at different points following the addition of actinomycin D (5 μg/ml). Levels of reporter mRNA shown in panel C were normalized to those of rp49 mRNA, and averages and standard deviations from three independent experiments are plotted.
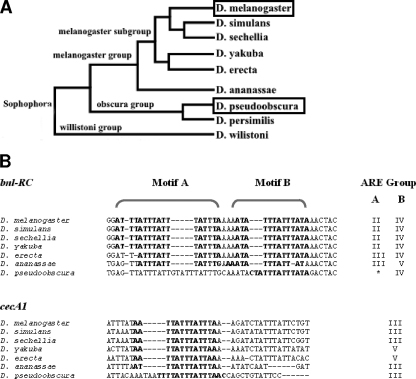
To assess the evolutionary conservation of class II AREs in these candidate genes, we collected the orthologues of both bnl-RC and cecA1 3′UTRs in seven Drosophila species. Figure 1B shows that bnl-RC and cecA1 class II AREs are detectably conserved between D. melanogaster and D. pseudoobscura, supporting that they are functional cis-regulatory elements.
FIG. 1.
bnl-RC and cecA1 class II ARE conservation across Drosophila species. (A) Phylogenic tree relating the seven Drosophila species used for the evaluation of ARE conservation. (B) Sequences (in boldface) and groups of AREs (as defined in Table 1) are shown. The asterisk indicates the loss of class II ARE not converted into a class I ARE (group V).
To test experimentally whether Drosophila AREs regulate mRNA turnover, we have either deleted or altered the AUUUA motifs in bnl (3′bnl RAΔARE and 3′bnl RCΔARE) and cecA1 3′UTRs (3′cecA1AREmut), respectively (Fig. 2A). WT (control) and mutant 3′UTRs were fused downstream of the GFP coding sequence, and the mRNA decay of resulting reporters was analyzed in D. melanogaster cultured cells. We found that all ARE mutant reporters are stabilized compared to controls (Fig. 2B and C). This result shows that Drosophila AREs are bona fide cis regulators of mRNA decay. Notably, bnl-RA and bnl-RC 3′UTRs induce similar rates of reporter decay (Fig. 2C), suggesting that tandem and dispersed AUUUA repeats are equivalent destabilizing sequences under these conditions. However, it remains possible that different spacing and/or numbers of AUUUA pentamers elicit a range of regulation in vivo, depending on the tissue-specific expression and regulation of ARE-BPs. An independent work recently has proposed that as little as a single AUUUA pentamer can regulate gcm mRNA decay in D. melanogaster (33). Our present work establishes D-ARED as a novel and valuable predictive tool to identify genuine AMD target genes in D. melanogaster, together with available data on gene expression and ARE sequence conservation.
Acute expression profile of cecA1 in cultured cells and in vivo relies on AMD.
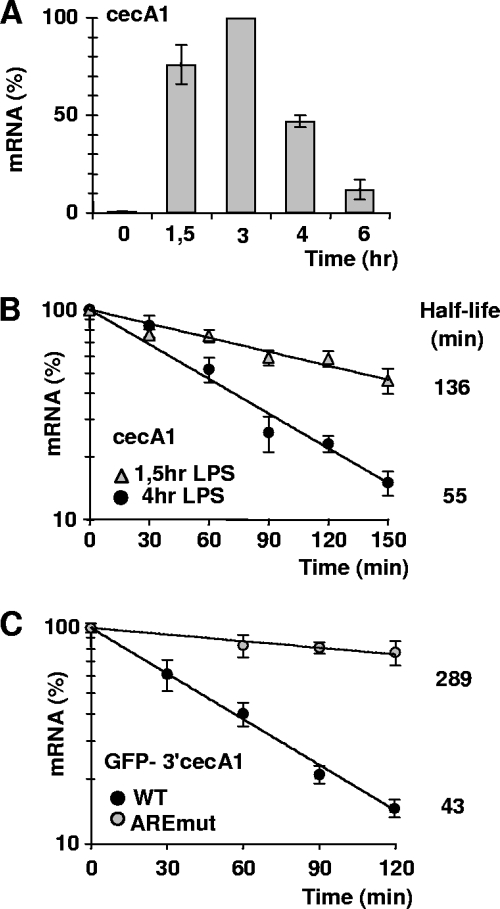
To validate that Drosophila AREs regulate gene expression in a physiological context, we examined the cecA1 ARE activity in LPS-stimulated cultured hemocytes (mbn-2), a cellular system allowing for the expression of immune-induced genes, including cecA1 (30). The cecA1 mRNA level increases and peaks 3 h after LPS treatment (inductive phase) and then rapidly declines (recovery phase) (Fig. 3A). We assessed cecA1 mRNA decay and found that its t1/2 drops from 136 to 54 min between the inductive and recovery phases, showing that transcription and mRNA turnover coordinately regulate cecA1 mRNA in response to LPS (Fig. 3B). To determine whether cecA1 AREs accelerates mRNA turnover during the recovery phase, the stability of both GFP-3′cecA1WT and GFP-3′cecA1AREmut reporters was analyzed 4 h following LPS treatment. The ARE mutant reporter is significantly stabilized (t1/2 = 289 min) compared to the control (t1/2 = 43 min) (Fig. 3C), strongly suggesting that AMD is responsible for the rapid decrease of cecA1 mRNA levels after an immune stimulation.
FIG. 3.
Rapid downregulation of endogenous cecA1 expression involves AMD. (A) Time course expression of cecA1 mRNA in mbn-2 cells exposed to LPS (10 μg/ml), as monitored by quantitative RT-PCR (qRT-PCR). The highest level of cecA1 mRNA was set as 100%. (B) Decay of cecA1 mRNA in mbn-2 cells following 1.5 or 4 h of LPS treatment. RNA was isolated at different points following the addition of actinomycin D and analyzed by qRT-PCR. (C) Decay of GFP-3′cecA1WT and GFP-3′cecA1AREmut mRNAs, as analyzed by qRT-PCR in mbn-2 cells following 4 h of LPS treatment. RNA was analyzed as described in the legend to Fig. 2B. In all panels, cecA1 mRNA levels were normalized to those of rp49 mRNA. Mean values ± standard deviations are given.
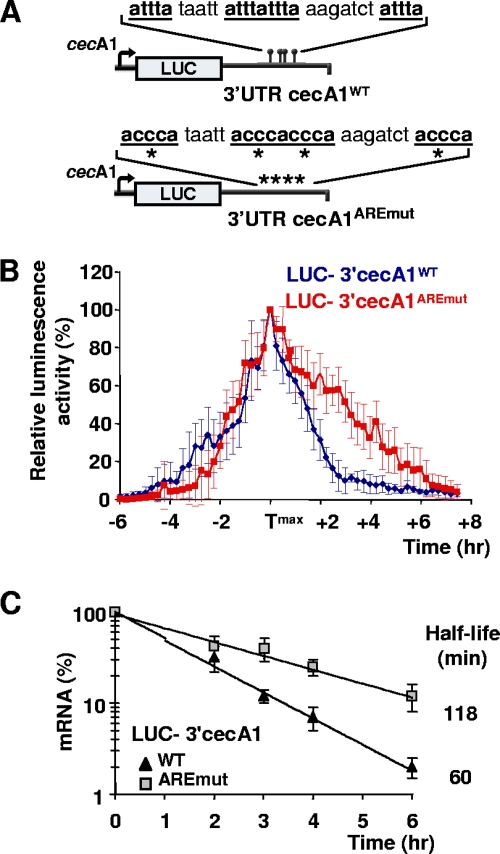
To validate that the ARE of cecA1 is functional in vivo, we designed a luc reporter assay and monitored the real-time mRNA turnover of luc in living flies. Luciferase, which turns over rapidly in many organisms, including flies, provides a powerful reporter system to continuously track mRNA decay (9). To assess mRNA decay independently of transcription, we took advantage of the acute-phase activation of the cecA1 promoter during the immune response (23) and expressed luc under the control of cecA1 5′ regulatory elements in the fat body and hemocytes. This cecA1-luc construct was fused to the WT (luc-3′cecA1WT) or the ARE mutant cecA1 3′UTR described above (luc-3′cecA1AREmut) (Fig. 4A). The resulting transgenes were inserted at the same chromosomal position in the genome of D. melanogaster, allowing for identical transcription contexts (see Materials and Methods). The bioluminescence captured from luciferin-fed adult flies bearing the luc-3′cecA1WT transgene shows that our luc reporter assay accurately reproduces the inducible and transient expression of cecA1 in vivo after septic injury with bacteria (Fig. 4B) (23). Remarkably, while both transgenes exhibit similar kinetics of induction, luc-3′cecA1AREmut transgenic flies produce substantially prolonged luminescence during the recovery phase, starting about 6 h postinfection. To determine whether this difference reflects different mRNA levels of expression, we quantified luc mRNA during the recovery phase (Fig. 4C). We observed an ∼2-fold slower mRNA clearance of luc-3′cecA1AREmut (t1/2 = 118 min) compared to that of luc-3′cecA1WT (t1/2 = 60 min). We conclude that the ARE of cecA1 accelerates mRNA turnover in vivo and likely contributes to the appropriate temporal cecA1 turnoff during the immune response in D. melanogaster. It is noteworthy that the four cec genes of D. melanogaster (cecA1, cecA2, cecB, and cecC) (31) are found in the D-ARED (see Table S1 in the supplemental material) and that cecB mRNA also exhibits short-lived expression (30). Interestingly, a recent report by Ryu et al. (29) revealed that constitutive and high levels of AMP expression, including that of CecA1, are detrimental for the commensal microbial homeostasis in the gut of adult flies, resulting in pathological consequences. Taken together, these data indicate that AMD is essential to regulate AMP expression temporally during the Drosophila immune response, thus preventing tissue damage.
FIG. 4.
cecA1 ARE accelerates gene expression turnoff in vivo. (A) Schemes of the transgenic reporters inserted in the genome of D. melanogaster. Transgenes contain the luc cDNA fused to either the WT or mutagenized cecA1 3′UTR (as indicated in the legend to Fig. 2A). Inducible expression of luc is driven by cecA1 regulatory sequences in the fat body and hemocytes upon septic injury. (B) Time course analysis of bioluminescence in living 5-day-old females expressing luc transgenes after septic injury with E. coli. Shown is the mean luminescence (± standard deviations) from 10 individuals plotted as a function of time. For a given fly, the highest level of luminescence was set as 100%, and other luminescence levels were expressed relative to the peak expression value. As the time required for reaching the maximal level of luminescence (Tmax) varies between flies (between 5 and 7 h after infection), curves were arbitrarily aligned on their Tmax. (C) Quantification of luc mRNA decay in whole adult flies by quantitative RT-PCR. Total RNA was prepared from pools of 10 infected flies starting 6 h after infection. luc mRNA levels were normalized to those of rp49 mRNA. Mean values ± standard deviations from 3 independent experiments are shown.
Tis11 regulates AMD ex vivo and in vivo.
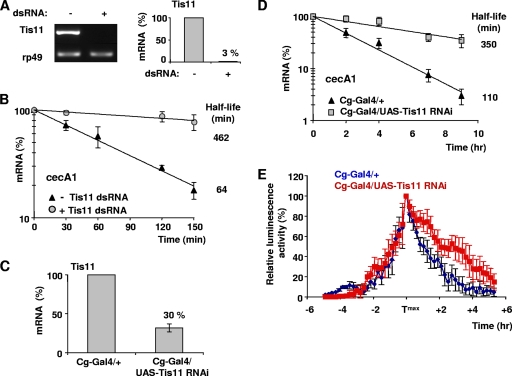
Tis11, the single characterized member of the mammalian TTP family in Drosophila (24), was shown to promote the mRNA decay of mammalian class II AREs in D. melanogaster cultured cells (22). To investigate its function in Drosophila ARE decay, we depleted Tis11 mRNA by RNAi in cultured hemocytes (to 3% of its WT level) (Fig. 5A). We found that cecA1 mRNA is stabilized up to sevenfold during the recovery phase after LPS treatment, showing that Tis11 triggers cecA1 mRNA decay (Fig. 5B). This effect is specific, as the decay of another immune-induced gene, reaper, containing dispersed AUUUA in its 3′UTR, remains unchanged in the same condition (see Fig. S2 in the supplemental material).
FIG. 5.
Tis11-dependent AMD in vivo. (A) Semiquantitative (left) and quantitative (right) RT-PCR analysis of Tis11 mRNA silencing in mbn-2 cells treated with a specific Tis11 dsRNA. (B) Decay of cecA1 mRNA in untreated or Tis11-depleted mbn-2 cells analyzed by quantitative RT-PCR. Total RNA was prepared at the indicated time following 4 h of LPS exposure and the addition of actinomycin D. cecA1 mRNA levels were normalized to those of rp49 mRNA. Mean values ± standard deviations from three experiments are shown. (C) qRT-PCR analysis of Tis11 mRNA silencing in the dissected fat body of third-instar larvae carrying the Cg-Gal4 driver alone or together with the UAS-Tis11 RNAi transgene. Larvae were raised at 25°C 2 days before RNA isolation. Tis11 mRNA levels were normalized to those of rp49 mRNA and set arbitrarily as 100% in flies carrying the Gal4 driver alone. Mean values ± standard deviations are shown. (D) Prolonged cecA1 mRNA accumulation in Tis11 RNAi adult flies as analyzed by quantitative RT-PCR. Cg-Gal4 and Cg-Gal4/Tis11 RNAi transgenic females were raised for 5 days at 25°C before septic injury. Total RNA was prepared at the indicated time from pools of 10 infected flies starting 6 h after infection. cecA1 mRNA levels were normalized to those of rp49 mRNA. Mean values ± standard deviations are shown. (E) Time course analysis of bioluminescence in luc-3′cecA1WT reporter flies carrying only Cg-Gal4 drivers or together with UAS-Tis11 RNAi transgenes. Adults were raised for 5 days at 25°C before infection. The relative mean bioluminescence (± standard deviations) is represented as indicated in the legend to Fig. 4B.
We next tested whether Tis11 regulates cecA1 mRNA decay in vivo. We verified that Tis11 mRNA is detectable in the fat body of third-instar larvae (Fig. 5C) and adults (data not shown), which is the prominent site of cecA1 expression upon infection (28). As a Tis11 mutant was not available, we knocked down Tis11 in the fat body and hemocytes by RNAi. For this purpose, we generated transgenic flies expressing a UAS-Tis11 RNAi construct in these tissues using the Cg-Gal4 driver (2). We found that Tis11 mRNA is reduced to about 30% of its normal level in the fat body of Tis11 RNAi larva (Fig. 5C). In addition, Tis11 RNAi-infected adults exhibit an ∼3-fold slower decline of cecA1 mRNA after infection (Fig. 5D). To validate that Tis11 regulates cecA1 mRNA decay through its ARE-containing 3′UTR, the bioluminescence of WT and Tis11 RNAi flies expressing the luc-3′cecA1WT reporter was analyzed. We observed a significant prolonged bioluminescence of the reporter in Tis11 RNAi flies during the recovery phase (starting after the time to maximum level of bioluminescence [Tmax]) (Fig. 5E). Taken together, these data strongly suggest that Tis11 regulates cecA1 mRNA turnover in D. melanogaster through its ARE. This function is very reminiscent of the TTP-mediated downregulation of immune genes in mammals (1), and it will be of interest to further investigate whether Tis11 is a specific or general posttranscriptional regulator of immune genes in Drosophila.
AREs confer transient expression to early response genes during the inflammatory response in mammals (1). In D. melanogaster, many early immune-induced genes, including cec family genes, have a transient expression pattern (8). Remarkably, we found an ∼3-fold enrichment (−log P > 5 by a hypergeometric test) of genes containing the ARE motif WWWU[AUUUA]UWWW (W = A/U) among the Drosophila early immune-induced genes found previously (8). Thus, we propose that AMD contributes to terminate the expression of a wide variety of genes during D. melanogaster immune response, as described for mammals. Also tempting is the hypothesis that the conserved AUUUA pentamers in the 3′UTRs of D. melanogaster bnl and its mammalian orthologue FGF-10 (data not shown) contribute to the remarkable dynamic expression of those genes in vivo, which is essential to pattern the respiratory systems of both organisms (25). Thus, AMD might be conserved from Drosophila to humans to regulate physiological processes requiring the fine-tuning of gene expression. Due to its wide repertoire of genetic and molecular tools, D. melanogaster constitutes a powerful model system to investigate in vivo the relevance of ARE-mediated regulation.
Supplementary Material
Acknowledgments
We thank Ylva Engström for providing the pA10 plasmid and the mbn-2 cell line. We thank Alexander Stark and Manolis Kellis for providing sequence information. We thank M. Crozatier, A. J. Carpousis, and D. Cribbs for comments on the manuscript and members of the CBD for helpful discussions.
F.C. is supported by the Fundação para a Ciência e a Tecnologia (SFRH/BPD/21460/2005) and the Fundação Calouste Gulbenkian.
Footnotes
Published ahead of print on 9 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, P. 2008. Post-transcriptional control of cytokine production. Nat. Immunol. 9353-359. [DOI] [PubMed] [Google Scholar]
- 2.Asha, H., I. Nagy, G. Kovacs, D. Stetson, I. Ando, and C. R. Dearolf. 2003. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163203-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakheet, T., M. Frevel, B. Williams, W. Greer, and K. Khabar. 2001. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakheet, T., B. R. Williams, and K. S. Khabar. 2006. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 34D111-D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barreau, C., L. Paillard, and H. B. Osborne. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 337138-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bevilacqua, A., M. C. Ceriani, S. Capaccioli, and A. Nicolin. 2003. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell Physiol. 195356-372. [DOI] [PubMed] [Google Scholar]
- 7.Bischof, J., R. K. Maeda, M. Hediger, F. Karch, and K. Basler. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 1043312-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutros, M., H. Agaisse, and N. Perrimon. 2002. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 3711-722. [DOI] [PubMed] [Google Scholar]
- 9.Brandes, C., J. D. Plautz, R. Stanewsky, C. F. Jamison, M. Straume, K. V. Wood, S. A. Kay, and J. C. Hall. 1996. Novel features of drosophila period transcription revealed by real-time luciferase reporting. Neuron 16687-692. [DOI] [PubMed] [Google Scholar]
- 10.Brennecke, J., A. Stark, R. B. Russel, and S. M. Cohen. 2005. Principles of microRNA-target recognition. PLoS Biol. 3404-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunch, T. A., Y. Grinblat, and L. S. B. Goldstein. 1988. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 161043-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caput, D., B. Beutler, K. Hartog, R. Thayer, S. Brown-Shimer, and A. Cerami. 1986. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. USA 831670-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, C.-Y. A., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20465-470. [DOI] [PubMed] [Google Scholar]
- 14.Doller, A., J. Pfeilschifter, and W. Eberhardt. 2008. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. 202165-2173. [DOI] [PubMed] [Google Scholar]
- 15.Echard, A., G. R. Hickson, E. Foley, and P. H. O'Farrell. 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 141685-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 89-22. [DOI] [PubMed] [Google Scholar]
- 17.Frevel, M. A. E., T. Bakheet, A. M. Silva, J. G. Hissong, K. Khabar, and B. R. G. Williams. 2003. p38 mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 23425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garneau, N. L., J. Wilusz, and C. J. Wilusz. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8113-126. [DOI] [PubMed] [Google Scholar]
- 19.Gherzi, R., M. Trabucchi, M. Ponassi, T. Ruggiero, G. Corte, C. Moroni, C. Y. Chen, K. S. Khabar, J. S. Andersen, and P. Briata. 2006. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 5e5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Gonsalves, F. C., and D. A. Weisblat. 2007. MAPK regulation of maternal and zygotic Notch transcript stability in early development. Proc. Natl. Acad. Sci. USA 104531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halees, A. S., R. El-Badrawi, and K. S. Khabar. 2008. ARED organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. 36D137-D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing, Q., S. Huang, S. Guth, T. Zarubin, A. Motoyama, J. Chen, F. Di Padova, S. C. Lin, H. Gram, and J. Han. 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120623-634. [DOI] [PubMed] [Google Scholar]
- 23.Lemaitre, B., J. M. Reichhart, and J. A. Hoffmann. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 9414614-14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma, Q., and H. R. Herschman. 1994. The Drosophila TIS11 homologue encodes a developmentally controlled gene. Oncogene 93329-3334. [PubMed] [Google Scholar]
- 25.Metzger, R. J., and M. A. Krasnow. 1999. Genetic control of branching morphogenesis. Science 2841635-1639. [DOI] [PubMed] [Google Scholar]
- 26.Puig, S., E. Askeland, and D. J. Thiele. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 12099-110. [DOI] [PubMed] [Google Scholar]
- 27.Raghavan, A., R. L. Ogilvie, C. Reilly, M. L. Abelson, S. Raghavan, J. Vasdewani, M. Krathwohl, and P. R. Bohjanen. 2002. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 305529-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roos, E., G. Bjorklund, and Y. Engstrom. 1998. In vivo regulation of tissue-specific and LPS-inducible expression of the Drosophila cecropin genes. Insect Mol. Biol. 751-62. [DOI] [PubMed] [Google Scholar]
- 29.Ryu, J. H., S. H. Kim, H. Y. Lee, J. Y. Bai, Y. D. Nam, J. W. Bae, D. G. Lee, S. C. Shin, E. M. Ha, and W. J. Lee. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319777-782. [DOI] [PubMed] [Google Scholar]
- 30.Samakovlis, C., B. Asling, H. G. Boman, E. Gateff, and D. Hultmark. 1992. In vitro induction of cecropin genes—an immune response in a Drosophila blood cell line. Biochem. Biophys. Res. Commun. 1881169-1175. [DOI] [PubMed] [Google Scholar]
- 31.Samakovlis, C., D. A. Kimbrell, P. Kylsten, A. Engstrom, and D. Hultmark. 1990. The immune response in Drosophila: pattern of cecropin expression and biological activity. EMBO J. 92969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46659-667. [DOI] [PubMed] [Google Scholar]
- 33.Soustelle, L., N. Roy, G. Ragone, and A. Giangrande. 2008. Control of gcm RNA stability is necessary for proper glial cell fate acquisition. Mol. Cell Neurosci. 37657-662. [DOI] [PubMed] [Google Scholar]
- 34.Stark, A., J. Brennecke, R. B. Russel, and S. M. Cohen. 2003. Identification of Drosophila microRNA targets. PLoS Biol. 1397-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark, A., J. Brennecke, N. Bushati, R. B. Russell, and S. M. Cohen. 2005. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 1231133-1146. [DOI] [PubMed] [Google Scholar]
- 36.Stark, A., M. F. Lin, P. Kheradpour, J. S. Pedersen, L. Parts, J. W. Carlson, M. A. Crosby, M. D. Rasmussen, S. Roy, A. N. Deoras, J. G. Ruby, J. Brennecke, Harvard FlyBase curators, Berkeley Drosophila Genome Project, E. Hodges, A. S. Hinrichs, A. Caspi, B. Paten, S. W. Park, M. V. Han, M. L. Maeder, B. J. Polansky, B. E. Robson, S. Aerts, J. van Helden, B. Hassan, D. G. Gilbert, D. A. Eastman, M. Rice, M. Weir, M. W. Hahn, Y. Park, C. N. Dewey, L. Pachter, W. J. Kent, D. Haussler, E. C. Lai, D. P. Bartel, G. J. Hannon, T. C. Kaufman, M. B. Eisen, A. G. Clark, D. Smith, S. E. Celniker, W. M. Gelbart, and M. Kellis. 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450219-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinman, R. A. 2007. mRNA stability control: a clandestine force in normal and malignant hematopoiesis. Leukemia 211158-1171. [DOI] [PubMed] [Google Scholar]
- 38.Sutherland, D., C. Samakovlis, and M. A. Krasnow. 1996. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 871091-1101. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, P. D., A. Kejariwal, N. Guo, H. Mi, M. J. Campbell, A. Muruganujan, and B. Lazareva-Ulitsky. 2006. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 34W645-W650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, E., E. Nimwegen, M. Zavolan, N. Rajewsky, M. Schroeder, M. Magnasco, and J. E. Darnell. 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 131863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zubiaga, A. M., J. G. Belasco, and M. E. Greenber. 1995. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 152219-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.