Abstract
E proteins are a family of helix-loop-helix transcription factors that play important roles in cell differentiation and homeostasis. They contain at least two activation domains, AD1 and AD2. ETO family proteins and the leukemogenic AML1-ETO fusion protein are corepressors of E proteins. It is thought that ETO represses E-protein activity by interacting with AD1, which competes away p300/CBP histone acetyltransferases. Here we report that E proteins contain another conserved ETO-interacting region, termed DES, and that differential associations with AD1 and DES allow ETO to repress transcription through both chromatin-dependent and chromatin-independent mechanisms. At the chromatin level, AD1 and AD2 cooperatively recruit p300. ETO interacts with AD1 to abolish p300 recruitment and to allow HDAC-dependent silencing. At the post-chromatin-remodeling level, binding to DES enables ETO to directly inhibit activation of the basal transcription machinery. This novel repression mechanism is conserved in ETO family proteins and in the AML1-ETO fusion protein. In addition, the repression capacity exerted by each mechanism is differentially modulated by cross talk among various ETO domains and the AML1 domain of AML1-ETO. In particular, the oligomerization domain of ETO plays a major role in targeting ETO to the DES region and independently potentiates the TAFH domain-mediated AD1 interaction. The ability to exert repression at different levels not only may allow these corepressors to impose robust inhibition of signal-independent transcription but may also allow a rapid response to signals. In addition, our newly defined domain interactions and their interplays have important implications in effectively targeting both E-protein fusion proteins and AML1-ETO found in cancers.
In eukaryotic cells, transcriptional coactivators and corepressors play important roles in facilitating signal-dependent gene activation (21). In the absence of signaling, transcription is repressed by corepressors through their physical association with transcription factors. Upon stimulus, corepressors are released from transcription factors, allowing coordinated recruitment of multiple coactivators to overcome various rate-limiting steps in the transcription process. At the chromatin level, coactivators, such as p300 and CBP histone acetyltransferases (HATs), modify histone lysines to facilitate chromatin remodeling. Subsequent recruitment of other coactivators that directly contact the basal transcription machinery facilitates the assembly of a functional preinitiation complex (PIC), which contains RNA polymerase II (Pol II) to perform the transcription reaction. Contrary to the case with activation, repression is often achieved by the recruitment of a single corepressor or corepressor complex. Effective repression of transcription is a prerequisite for achieving a physiologically correct level of signal-dependent gene activation (48, 53). Conceivably, binding of a single corepressor to transcription factors such as nuclear receptors, which use the common AF2 domain to communicate with different coactivators, is likely sufficient to block the recruitment of multiple coactivators. However, how a corepressor exerts its dominant inhibition with transcription factors such as E proteins, which contain multiple activation domains, is less well understood.
The E proteins, including HEB (HeLa E-box binding protein), E2A, and E2-2, belong to a family of basic helix-loop-helix (HLH) transcription factors that can activate transcription from E-box (CANNTG) sites (43). E proteins contain multiple activation domains, including the two conserved but distinct AD1 and AD2 domains (8). AD1 recruits p300 and CBP through a highly conserved LXXLL motif (7, 67). This motif overlaps an adjacent LDSF motif, which recruits the SAGA (Spt-Ada-Gcn5-acetyltransferase) coactivator complex in yeast (42). AD2 also has the ability to interact with p300 and CBP (7, 10). It has been shown that AD1 and AD2 cooperate to maximally activate E-protein-mediated transcription (7, 8).
Accumulating evidence indicates that E proteins not only are critical to lymphopoiesis (43, 51) but also are intimately involved in the development of other hematopoietic lineages, including myeloid, erythroid, megakaryocyte, and plasmacytoid dendritic cells (8, 13, 14, 16, 30, 32). In addition, E proteins have independent functions in regulating cell proliferation and survival and exert a tumor suppressor activity that is lost when E-protein inhibitors are overexpressed in cancer. Examples include the Id proteins (inhibition of differentiation/DNA-binding) (54), which are overexpressed in a variety of cancers, and SCL/TAL1, which is overexpressed in more than 60% of T-cell acute lymphoblastic leukemias (47). Further emphasizing the importance of E proteins in maintaining normal cellular activities, E2A and HEB are associated with oncogenic chromosomal translocations that cause the loss of one allele of E2A or HEB genes and the aberrant expression of fusion proteins containing their activation domains. Both events may contribute to malignancy (5, 38, 57).
The family of ETO proteins and the leukemogenic AML1-ETO fusion protein are newly identified E-protein inhibitors (67). The eight-twenty-one gene product ETO (also called MTG8) is a prototypical member of a small family of transcriptional corepressors that also includes ETO-2 and MTGR1 (MTG8-related protein 1) (29). ETO family proteins are expressed in a variety of cell types and tissues, including brain, intestine, and adipose tissues and the hematopoietic compartments (1, 11, 12, 22, 36, 44, 52, 56). Like E proteins, members of this family of proteins (namely, ETO and ETO-2) are also targeted by oncogenic chromosomal translocations (27, 29, 34, 46, 49). In particular, nearly 15% of acute myeloid leukemia (AML) is caused by the t(8;21) translocation, which generates the AML1-ETO fusion product by combining an N-terminal DNA-binding domain of AML1 and an almost full-length ETO. Unlike Id proteins, which interact with the HLH region of E proteins to block their DNA-binding activity, ETO family proteins and AML1-ETO associate with an E protein's activation domain and actively repress its transcriptional activity (22, 24, 56, 67). It has been shown that dynamic associations between wild-type ETO family proteins and E proteins facilitate a switch from cell proliferation to differentiation in erythroid-megakaryocyte development (22, 24, 56). In contrast, in leukemic cells, aberrant expression of AML1-ETO causes constitutive associations between AML1-ETO and E proteins, resulting in deregulation of E-protein function (67).
Like E proteins, the ETO family proteins exhibit modular domain structures that include four conserved regions, designated Nervy homology regions 1 to 4 (NHR1 to -4), which are homologous to the corresponding regions of the Drosophila melanogaster Nervy protein. The NHR1 domain is also referred to as the TAFH domain (TAF4 homology), given its similarity in both primary and tertiary structures (50, 61, 62) to a conserved region within the TAF4 (TBP-associated factor 4) proteins that are part of TFIID. The NHR2 domain mediates tetramerization of ETO family proteins through a conserved hydrophobic heptad repeat (HHR) (37). NHR2 also interacts with the Sin3A corepressor (2), which associates with histone deacetylase 1 (HDAC1). The NHR4 region contains two MYND-type zinc fingers that allow ETO to recruit the nuclear receptor corepressors N-CoR/SMRT and associated HDACs (20, 40, 60).
The conserved LXXLL motif within the AD1 domain of E proteins is also referred to as PCET (p300/CBP and ETO target) because it mediates mutually exclusive binding of p300/CBP coactivators and the TAFH domain of ETO family proteins and AML1-ETO (67) (see Fig. 1A, left). Through this interaction, ETO is recruited to the AD1 domain of E proteins and blocks its interaction with p300/CBP. Here, by introducing a point mutation into the TAFH domain of ETO, we found that interaction with AD1 is dispensable for ETO to repress E-protein activity. This is because ETO can be independently recruited to another conserved E-protein region through novel interactions involving the TAFH domain and the oligomerization domain of ETO. Furthermore, we show that ETO, through its differential binding to the two E-protein regions, can mediate dual repression mechanisms that target either chromatin or the basal transcription machinery and that the potency of the repression exerted by each mechanism is differentially modulated by cross talk among various ETO domains and the AML1 domain of the AML1-ETO fusion protein.
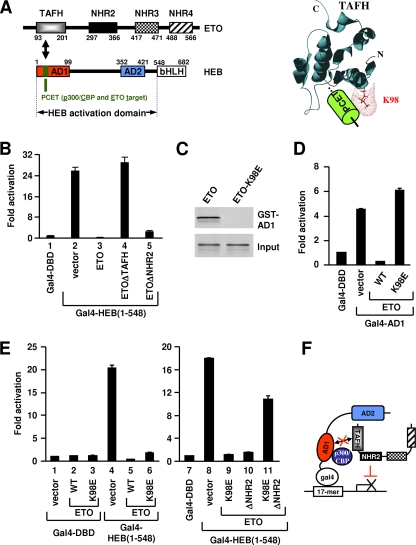
FIG. 1.
ETO does not need to interact with AD1 to repress HEB-dependent transcription. (A) Left, schematic representation of ETO and HEB domains showing the specific interaction between TAFH and PCET. Right, three-dimensional structure of the ETO TAFH domain in complex with PCET showing surface representation of K98. (B, D, and E) Luciferase assays of 293T cells transfected with expression plasmids for proteins indicated at the bottom, along with the Gal4 UAS-driven luciferase reporter. (C) GST pull-down analyses of in vitro-translated ETO and ETO-K98E with GST-AD1. (F) Schematic representation showing repression of HEB-dependent transcription by ETO in the absence of AD1 interaction. The p300/CBP coactivators are expected to bind AD1 in the absence of TAFH-AD1 interaction.
MATERIALS AND METHODS
Cell culture and luciferase assay.
293T cells were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum. For reporter assays, subconfluent 293T cells grown in 24-well plates were transfected with equal total amounts of plasmids using the FuGENE 6 reagent (Roche). Luciferase assays were performed 36 to 48 h later. The amounts of plasmids used for transfection are as follows unless otherwise specified: ETO and ETO mutants, 75 ng; AML1-ETO and its derivatives, 150 ng; Gal4-HEB(1-548) and its derivatives, 10 ng or 50 ng; Gal4-E2A(1-493) and its derivatives, 5 ng or 10 ng; HEB or HEBΔAD1, 40 ng; E47 and E12, 5 ng. The 293T-E-box cell line was generated by stable transfection of the E-box-driven luciferase reporter (see below) with a pBabe-puro vector (20:1 ratio). Stable clones were screened by selection with 2 μg/ml puromycin and maintained with 1 μg/ml puromycin. Luciferase reporters contained either five copies of a Gal4-binding site (Gal4 UAS) or four copies of an E-box (CAGATG) site. Luciferase units (Promega) were normalized to β-galactosidase activity, which served as an internal control for transfection efficiency. Activation or repression values were relative to values for Gal4-DBD (DNA-binding domain) or other relevant empty vector controls unless otherwise indicated. Each result represents the average for duplicate samples in representative experiments.
Plasmids and protein expression.
Mammalian expression vectors for HEB, E2A, AML1-ETO, and ETO have been described previously (67) and contain a CMV promoter (human cytomegalovirus immediate-early promoter) to drive expression. Mammalian expression vectors for ETO derivatives either have been described previously (67) or were generated following standard molecular cloning/PCR procedures and verified by DNA sequencing. Plasmids used for in vitro translation with the TNT kit (Promega) were derived from the pCMX vector, which contains a T7 promoter. Plasmids for bacterial expression of glutathione S-transferase (GST) fusion proteins have been described previously (67). Baculovirus expression vectors were derived from pFastBac (Invitrogen).
Recombinant proteins (ACF-1, ISWI, p300, Gal4-HEB derivatives, and Gal4-VP16) were expressed and affinity purified from baculovirus-infected Sf9 cells via an in-frame FLAG tag by anti-FLAG M2-agarose beads (Sigma). ETO and ETO derivatives were affinity purified either from 293T cells following transient transfection or from baculovirus-infected Sf9 cells via an in-frame FLAG. The same results were obtained by using either source, and proteins used in a given experiment were from the same source. AML1-ETO, MTGR1, and ETO-2 were affinity purified from transfected 293T cells via an in-frame FLAG.
Coimmunoprecipitation assay.
293T cells were transfected using the FuGENE 6 reagent or Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions. Cells were lysed in lysis buffer (20 mM HEPES, pH 7.9, 1 mM EDTA, 20% glycerol, and protease inhibitors) containing 300 mM NaCl and 0.5% NP-40. Cell extracts were incubated with anti-FLAG M2-agarose beads (Sigma) at 4°C for 6 h. Following extensive washing with lysis buffer, bound proteins (immunoprecipitates) were subjected to Western blot analysis. Input lanes show 2% of the total.
GST pull-down assay.
In vitro-translated 35S-labeled proteins were incubated with bacterially expressed GST fusion proteins immobilized on glutathione-Sepharose beads (Amersham Pharmacia). Following extensive washing with buffer BC300-0.1% NP-40, bound polypeptides were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by autoradiography. Input lanes show a fraction of total input.
In vitro chromatin assembly and nucleosomal array HAT assay.
Chromatin assembly was performed as described previously (17) using supercoiled template, purified Drosophila ACF-1, ISWI, mouse NAP-1, and HeLa-derived core histone octamers. Micrococcal nuclease partial digestion and nucleosomal array HAT assays were performed as described previously (3). Typically, 20 ng template, 20 ng activators, and 200 ng corepressors were used.
In vitro transcription on naked DNA and chromatin templates.
Transcription reactions on supercoiled naked DNA templates using nuclear extracts were carried out essentially as described previously (58) following a stepwise protocol including the following: (i) template binding with activator (20 min), (ii) preinitiation complex formation by the addition of nuclear extract (30 min), and (iii) transcription (30 min) by the addition of nucleoside triphosphates. The final 50-μl reaction mixture contained 40 ng of supercoiled plasmid DNA template, 20 ng of purified DNA-bound activators, 200 ng of ETO or ETO derivatives, 25 mM HEPES (pH 7.8), 6% glycerol, 6 mM MgCl2, 70 mM KCl, 5 mM dithiothreitol, 0.4 mM (each) ATP, UTP, CTP, and GTP, 10 U of recombinant RNasin, and various amounts of different cofactors. RNA transcripts were measured by a primer extension assay with a primer located 110 nucleotides downstream of the transcription start site. For quantitation, the relative activity of transcription was based on a phosphorimager scan normalized to the basal level. In vitro transcription using purified recombinant factors was performed with supercoiled DNA templates as described previously (19). The final complete reaction contained TFIIA (10 ng), TFIIB (10 ng), TFIID (equivalent of 1 ng of TBP), TFIIEα (20 ng), TFIIEβ (20 ng), TFIIF (25 ng), TFIIH (20 ng), Pol II (50 ng), and PC4 (150 ng).
Chromatin-templated transcription was carried out by using a previously described protocol (4), which includes an additional p300-facilitated chromatin-remodeling step, as illustrated in Fig. 5G. Assembled chromatin templates (containing 40 ng of DNA) were incubated with cofactors, followed by an additional p300-dependent chromatin-remodeling step. Nuclear extract was then added, and thereafter the protocol was the same as that for in vitro transcription on naked DNA. The final 50-μl reaction mixture contained 25 mM HEPES (pH 7.8), 6.8 mM MgCl2, 65 mM KCl, 5 mM dithiothreitol, 6% glycerol, 0.4 mM (each) ATP, UTP, CTP, and GTP, 10 U of recombinant RNasin, 2 mM sodium butyrate, and various amounts of different cofactors. Typically, the final reaction in both naked DNA template-based and chromatin template-based transcription assays contained 40 ng of naked or chromatinized template; 20 ng of purified DNA-bound activators; 200 ng of ETO or ETO derivatives; and for chromatin transcription, 50 ng of p300 and 3 μM acetyl-coenzyme A unless otherwise indicated.
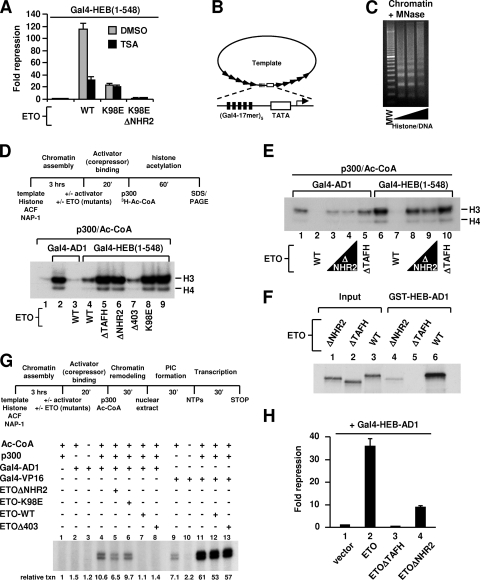
FIG. 5.
AD1 interaction is required for ETO repression at the chromatin level and is regulated by the NHR2 domain. (A) Luciferase reporter analyses of 293T cells transfected with expression plasmids indicated at the bottom, along with a Gal4 UAS-driven luciferase reporter. Repression is relative to results with Gal4-HEB(1-548) alone. TSA (150 nM) or dimethyl sulfoxide was added 24 h posttransfection, and the luciferase assays were performed 16 h later. WT, wild type. (B) Schematic representation of the DNA template used for chromatin assembly. The template contains five copies of the Gal4 UAS element upstream of a minimal TATA promoter derived from adenovirus E4 flanked by five copies of the 5S ribosomal DNA nucleosome-positioning sequence. (C) Micrococcal nuclease partial digestion assay. The histone/DNA ratios are 1.2:1, 1.3:1, and 1.4:1. The 1.3:1 ratio was used in the following experiments. MW, 100-bp DNA ladder. (D and E) Effects of ETO and ETO mutants on Gal4-AD1- or Gal4-HEB(1-548)-mediated p300-dependent histone acetylation of nucleosomal arrays. Top of panel D shows the stepwise experimental scheme. In panel D, 40 ng of Gal4-AD1 or Gal4-HEB(1-548) was used instead of the typical 20 ng. In panel E, a higher dose (300 ng) of ETOΔNHR2 was also used, in addition to the typical 200 ng for ETO and its derivatives. (F) GST pull-down analyses of in vitro-translated ETO and ETO mutants with GST-HEB-AD1. (G) Effects of ETO and ETO mutants on Gal4-AD1- or Gal4-VP16-mediated p300-dependent transcription from chromatin templates. Top panel, the stepwise experimental scheme. (H) Luciferase reporter analyses of 293T cells transfected with expression plasmids indicated at the bottom, Gal4-HEB-AD1, and a Gal4 UAS-driven luciferase reporter. Repression is relative to results with Gal4-HEB-AD1 alone.
RESULTS
ETO does not need to interact with AD1 to repress HEB-dependent transcription.
Deletion of TAFH abolishes ETO's ability to inhibit HEB-dependent transcription from either the E-box (67) or the Gal4 template (Fig. 1B). The latter experiment used a Gal4-DBD fusion protein containing the complete HEB activation domain (amino acids 1 to 548) [Gal4-HEB(1-548)]. To test whether the failure to repress transcription was due to a loss of TAFH-mediated interaction with AD1 (amino acids 1 to 99), a conserved lysine within the TAFH domain was mutated to glutamic acid (K98E). K98 is located on the surface of TAFH and may directly contact PCET (Fig. 1A, right) (50). As expected, the K98E mutation abolished the abilities of ETO to interact with AD1 (Fig. 1C) and to repress Gal4-AD1-dependent transcription (Fig. 1D).
Surprisingly, unlike Gal4-AD1, Gal4-HEB(1-548) remained strongly inhibited by ETO-K98E (Fig. 1E, lane 6). This result shows that although TAFH is needed, its interaction with AD1 is dispensable for repression of HEB-mediated transcription (Fig. 1F). Nevertheless, ETO-K98E was unable to further repress the transcription to a below-basal level as ETO did (Fig. 1E, lanes 1, 5, and 6). Thus, the ability of ETO to silence basal transcription, presumably through ETO-associated HDACs, still requires its interaction with AD1.
ETO-K98E-mediated repression requires NHR2.
We next asked which other NHR domain(s) account for ETO-K98E-mediated repression. Like K98E mutation, deletion of NHR2 (ΔNHR2) or a C-terminal region containing NHR3 and NHR4 (Δ403) partially relieved repression to slightly above (ΔNHR2) or near (Δ403) basal levels (Fig. 1B; see Fig. S1 in the supplemental material). These results are consistent with their reported roles in HDAC recruitment and ETO-mediated repression (28, 29, 39, 66). Remarkably, simultaneous K98E mutation and NHR2 deletion essentially abolished the repression (Fig. 1E; see also Fig. S1 in the supplemental material). In contrast, the Δ403 deletion slightly increased the repression by ETO-K98E (see Fig. S1 in the supplemental material). Thus, NHR2, in conjunction with an AD1-interaction-independent function of TAFH, mediates a novel repression pathway that does not involve direct TAFH-AD1 interaction.
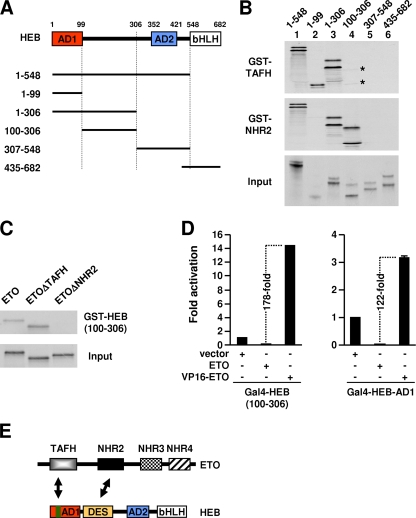
NHR2 physically interacts with an AD1 downstream region of HEB.
NHR2 may directly interact with HEB to facilitate ETO-K98E recruitment. This hypothesis was tested by GST pull-down assays using the various in vitro-translated HEB fragments depicted in Fig. 2A. Indeed, like TAFH, NHR2 interacted strongly with HEB(1-548) and its N-terminal 306 amino acids but not with the AD2 or HLH regions (Fig. 2B). Interestingly, within HEB(1-306), TAFH and NHR2 showed distinct binding specificities. As expected, AD1 interacted only with TAFH (lane 2). In contrast, HEB(100-306) (amino acids 100 to 306) interacted strongly with NHR2, although a weak but specific interaction with TAFH was also observed (lane 4; see also Fig. 4A).
FIG. 2.
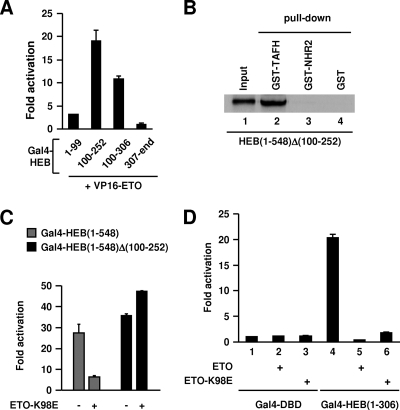
NHR2 physically interacts with an AD1 downstream region of HEB. (A) Schematic representation of HEB and truncated HEB derivatives. (B) GST pull-down analyses of in vitro-translated HEB and HEB derivatives with GST-TAFH and GST-NHR2. Weak interaction of HEB(100-306) with TAFH is marked by asterisks. (C) GST pull-down analyses of in vitro-translated ETO and ETO derivatives with GST-HEB(100-306). (D) Luciferase assays of 293T cells transfected with ETO and VP16-ETO along with Gal4-HEB-AD1 or Gal4-HEB(100-306) and the Gal4 UAS-driven reporter. (E) Schematic representation of two independent interaction interfaces (TAFH-AD1 and NHR2-DES) between ETO and HEB.
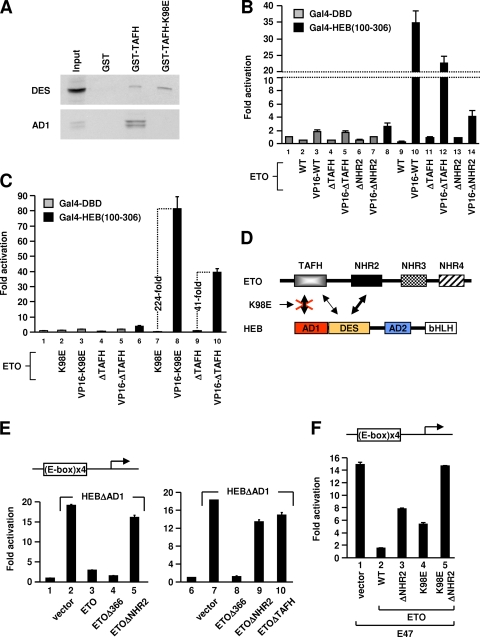
FIG. 4.
TAFH and NHR2 cooperatively target DES to repress transcription in vivo. (A) GST pull-down analyses of in vitro-translated Gal4-HEB-DES (i.e., amino acids 100 to 306) and Gal4-HEB-AD1 with GST, GST-TAFH, or GST-TAFH-K98E. (B) Luciferase assays of 293T cells transfected with ETO, ETO mutants, or their VP16 fusions along with Gal4-DBD or Gal4-HEB(100-306), as well as the Gal4 UAS-driven reporter. WT, wild type. (C) Luciferase assays of 293T cells transfected with ETOΔTAFH (ΔTAFH), ETO-K98E (K98E), or their VP16 fusion proteins, along with Gal4-DBD or Gal4-HEB(100-306) and the Gal4 UAS-driven reporter. (D) Schematic representation showing that the K98E mutation selectively abolishes AD1 interaction. (E and F) Luciferase assays of 293T cells transfected with an E-box-driven reporter and expression vector for HEBΔAD1(E) or E47(F), with ETO and ETO mutants indicated at the bottom.
Reciprocal GST pull-down assays confirmed the in vitro interaction between full-length ETO and HEB(100-306). They also showed that NHR2 and its conserved hydrophobic repeat region HHR are required for this interaction (Fig. 2C; see also Fig. S2 in the supplemental material). In contrast, deletion of TAFH did not reduce but rather slightly increased the interaction between ETO and HEB(100-306) (Fig. 2C). These results indicate that the TAFH domain within ETOΔNHR2 or ETO does not readily interact with HEB(100-306) in vitro.
The intracellular interaction between ETO and HEB(100-306) was then confirmed by mammalian two-hybrid assays (Fig. 2D). Whereas ETO repressed the transcriptional activity of Gal4-HEB(100-306), fusion of ETO with the VP16 activation domain (VP16-ETO) overcame this repression and further potentiated transcription. These results confirmed a strong interaction between ETO and HEB(100-306) in vivo (178-fold increase due to VP16 activity), which is comparable in magnitude to the interaction between ETO and AD1 assayed in the same experiment.
Taken together, these results indicate that ETO can be recruited independently to AD1 or its downstream ETO-interacting sequence (DES), namely, HEB(100-306), through TAFH-AD1 interaction or NHR2-DES interaction, respectively (Fig. 2E).
DES region is required for repression by ETO-K98E.
Given the findings described above, disrupting DES-NHR2 interaction should abolish repression by ETO-K98E. This was tested by deleting a DES subregion (Δ100-252), which is sufficient for ETO interaction (Fig. 3A) and is required for HEB(1-548) to interact with NHR2 (Fig. 3B). Using luciferase assays, we showed that ETO-K98E completely lost the ability to repress Gal4-HEB(1-548)Δ(100-252)-mediated transcription (Fig. 3C). Furthermore, the presence of a DES region allowed ETO-K98E to strongly repress the transcription mediated by Gal4-HEB(1-306) (Fig. 3D), indicating that DES is sufficient to recruit ETO to repress transcription.
FIG. 3.
NHR2 interaction with DES is required for AD1 interaction-independent repression by ETO. (A) Mammalian two-hybrid analysis showing HEB(100-252) is sufficient to interact with ETO. Activation is normalized to the luciferase activity observed with the individual Gal4 fusion protein alone. (B) GST pull-down analyses of in vitro-translated HEB(1-548)Δ(100-252) with GST-TAFH, GST-NHR2, or GST. (C and D) Luciferase assays of 293T cells transfected with a Gal4 UAS-driven luciferase reporter along with indicated expression plasmids.
Both AD1 and DES are highly conserved among E proteins. Alignment of DES regions shows a nearly 50% identity between HEB and E2A and an even greater conservation between HEB and E2-2 (see Fig. S3A in the supplemental material). Consistent with this homology, the results observed with HEB were recapitulated in E2A. Thus, the NHR2 domain of ETO interacted specifically with the corresponding DES region of E2A (see Fig. S3B to E in the supplemental material). Similarly, this interaction was both necessary and sufficient for ETO-K98E (which also failed to interact with E2A-AD1) to repress transcription mediated by Gal4-E2A(1-493), corresponding to Gal4-HEB(1-548), and Gal4-E2A(1-301), corresponding to Gal4-HEB(1-306) (see Fig. S4 in the supplemental material). Thus, like AD1-dependent repression (67), DES-dependent repression is likely conserved among E-protein family members.
TAFH and NHR2 cooperatively bind to DES to repress transcription.
Given that deleting TAFH completely abolished ETO-mediated repression of HEB-dependent transcription (Fig. 1B), the role of TAFH in DES-dependent repression of Gal4-HEB(1-548) was then investigated. Because this role is retained in ETO-K98E and the isolated TAFH domain elicits a direct interaction with DES (Fig. 2B), we first tested whether ETO-K98E can also interact with DES. Indeed, GST pull-down assays showed that whereas K98E mutation abolished TAFH interaction with AD1 as expected, it did not affect the DES interaction (Fig. 4A). Neither DES nor AD1 interacted with GST, demonstrating the specificity of the observed interactions. The finding that K98 is differentially recognized by AD1 and DES is not surprising. K98 is probably not required for the integrity of TAFH. Rather, it plays a specialized role in binding and stabilizing the LXXLL conformation of PCET (50). DES lacks the characteristic LXXLL motif, explaining why its interaction with TAFH is not affected by K98E mutation. These differences also suggest that DES and AD1 may bind to distinct regions of TAFH.
If the ability of TAFH to interact with DES underlies its role in repression, this interaction should also occur in vivo in the context of ETO. Such an interaction may be manifested as an ability of TAFH to enhance ETO's interaction with DES in vivo. Mammalian two-hybrid assays were performed to test this possibility. Deletion of TAFH significantly reduced ETO interaction with DES (Fig. 4B). Compared with results for VP16-ETO, both the absolute level of activation (lane 10 versus lane 12) and the increase relative to results for nonfused proteins were significantly reduced by deleting TAFH (109-fold, lanes 9 and 10, versus 25.5-fold, lanes 11 and 12). These results confirmed that in vivo, TAFH does contribute to maximal ETO interaction with DES. In further support of an intracellular TAFH-DES interaction, deletion of NHR2 did not completely abolish ETO interaction with DES (cf. lanes 13 and 14 and lanes 6 and 7). Finally, we confirmed that like wild-type ETO, ETO-K98E elicited a much stronger interaction with DES than ETOΔTAFH did (Fig. 4C), suggesting that K98E mutation does not affect TAFH-DES interaction in vivo as expected based on the in vitro data (Fig. 4A).
The in vivo association between TAFH and DES in the context of ETO contrasts with our earlier GST pull-down data (Fig. 2C; see also Fig. S2 in the supplemental material) showing that this interaction does not readily occur in vitro. One possible reason that may account for this difference is that the specific in vivo conditions, such as coexpression and the presence of chaperones or other cellular factors, may have assisted the cooperative binding of DES to both TAFH and NHR2.
Taken together, our data suggest that DES-dependent repression of HEB requires its simultaneous interaction with both TAFH and NHR2 (Fig. 4D). This explains why deletion of TAFH abolishes DES-dependent repression by ETO (Fig. 1B) and why K98E mutation preserves this repression capability (Fig. 1E). The data also indicate that the repression is not simply a matter of recruitment, because ETOΔTAFH does get recruited but is unable to inhibit Gal4-HEB(1-548). Rather, multiple contacts between ETO and HEB are needed.
DES-dependent repression can occur on E-box templates.
Binding to E-box elements may alter the protein-protein interaction capability of E proteins and thereby their consequent transcriptional activity (41). We therefore tested if DES-dependent repression can also occur on E-box-regulated promoters. In luciferase assays using an E-box-driven template, a HEB derivative (HEBΔAD1) lacking AD1 was still capable of strong activation of transcription (Fig. 4E). This is consistent with the independent function of multiple E-protein activation domains (8). Importantly, HEBΔAD1-mediated transcription was also repressed by ETO (Fig. 4E). Furthermore, the repression was dependent on both TAFH and NHR2 (Fig. 4E), consistent with the requirement of their interactions with DES for repression. In contrast, deletion of C-terminal NHR3 and NHR4 regions (Δ366) slightly enhanced the repression, similar to the effect of Δ403 deletion on ETO-K98E-mediated repression on Gal4 templates (see Fig. S1 in the supplemental material). Thus, DES-dependent repression and its underlying mechanism are probably independent of DNA binding elements.
We also confirmed DES-dependent repression of E47 on E-box templates (Fig. 4F). ETO strongly repressed E47, as expected (lane 2). Both K98E mutation and NHR2 deletion partially reduced but failed to abrogate the repression (lanes 3 and 4). Complete disruption of repression required simultaneous K98E mutation and NHR2 deletion to disrupt both AD1 and DES interactions (lane 5). These results indicate that both AD1-dependent and DES-dependent repression pathways can contribute to repression of E47 by ETO on E-box templates. We obtained similar results with HEB (C. Guo and J. Zhang, unpublished data).
DES-dependent repression does not target chromatin remodeling.
We next turned our attention to the mechanism that underlies DES-dependent repression. Earlier data showed that ETO-K98E was not able to repress Gal4-HEB(1-548)-mediated transcription to below basal levels (Fig. 1E). This would argue against a role for HDACs in ETO-K98E-mediated repression. To further test this possibility, we carried out reporter assays in the absence or presence of trichostatin A (TSA), a specific HDAC inhibitor. Treatment with TSA significantly reduced the ability of ETO to repress Gal4-HEB(1-548)-mediated transcription (Fig. 5A), confirming the involvement of HDACs in repression by wild-type ETO. In contrast, repression mediated by ETO-K98E was insensitive to TSA and was similar in magnitude to HDAC-independent repression by ETO, as manifested in the presence of TSA (Fig. 5A). These results exclude the possibility that the repression mediated by ETO-K98E involves an HDAC-associated activity, which is consistent with the data shown in Fig. 1E. As expected, repression mediated by ETO-K98E was dependent on NHR2 (Fig. 5A).
We then considered the possibility that binding of ETO to DES may interfere with p300 recruitment or its activity. To this end, an in vitro HAT assay system was set up using nucleosomal arrays assembled on Gal4 UAS-containing templates by purified ACF-1, ISWI, NAP-1, and HeLa histones (Fig. 5B and C; see also Fig. S5A in the supplemental material). This in vitro system confirmed that AD1 can mediate p300-dependent nucleosomal acetylation and that ETO inhibits this acetylation (Fig. 5D, lanes 1 to 3), presumably by blocking p300 interaction with AD1. The inhibition by ETO was specific, since ETO did not affect Gal4-VP16-dependent histone acetylation by p300 (see Fig. S5B in the supplemental material).
Gal4-HEB(1-548) was more potent than Gal4-AD1 in mediating histone acetylation (Fig. 5D, lanes 2 and 9). The effect was more dramatic when a lesser amount of activators was used (Fig. 5E, lanes 1 and 6). In both cases, Gal4-HEB(1-548)-dependent histone acetylation was essentially abolished by ETO (Fig. 5D, lane 4, and E, lane 7) but was unaffected by the ETO-K98E and ETOΔTAFH mutants, which cannot interact with AD1 (Fig. 5D, lanes 8 and 5). These results indicate that ETO interaction with AD1 is necessary and sufficient to block p300 recruitment and its HAT activity. In addition, the data also suggest that AD1 and another HEB region(s), likely AD2, can mediate cooperative recruitment of p300 and that this recruitment is dependent on AD1 interaction with p300.
NHR2 enhances TAFH-AD1 interaction and is required for optimal AD1-dependent repression on chromatin.
Although TAFH is sufficient to interact with AD1, deleting NHR2 dramatically reduced the ability of ETO to inhibit nucleosomal acetylation elicited by either Gal4-HEB(1-548) or Gal4-AD1 (Fig. 5D, lane 6, and E). In contrast, deletion of the C-terminal NHR3/4 regions (Δ403) had no effect (Fig. 5D, lane 7). Consistent with this result, the in vitro interaction between ETO and GST-AD1, which requires TAFH, was greatly reduced by deleting NHR2 (nearly eightfold as measured by densitometry) (Fig. 5F).
To document the functional significance of the interplay between TAFH and NHR2 in chromatin-dependent transcriptional repression by ETO, we performed in vitro transcription assays with chromatin templates. Consistent with the HAT assay results (Fig. 5D), ETO and ETOΔ403 abolished p300-dependent transcription by Gal4-AD1 (Fig. 5G, lanes 1 to 4, 7, and 8) while not affecting transcription elicited by Gal4-VP16 (lanes 11 to 13). Confirming the functional importance of both NHR2 and TAFH, the repression by ETO was dramatically reduced by deletion of NHR2 (lane 5) and was abolished by K98E mutation (lane 6). These in vitro results were essentially recapitulated by reporter assays with HEB-AD1 (Fig. 5H) or with E2A-AD1, each of which contains a highly conserved PCET motif (see Fig. S6 in the supplemental material). The reduced dependency of repression on NHR2 in vivo could reflect an independent function of HDAC and N-CoR/SMRT proteins, through binding to ETO, in stabilizing ETO occupancy on chromatin or promoting its repression.
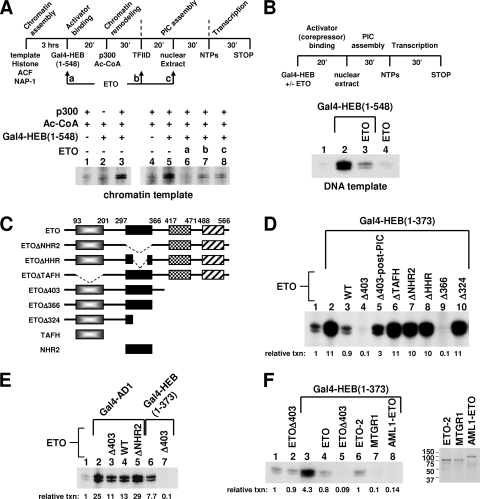
DES-dependent repression can occur on naked DNA templates and interferes with PIC assembly.
We continued to explore the mechanism underlying DES-dependent repression. Our data presented thus far indicate that DES-dependent repression by ETO can occur independently of AD1 but can dominantly inhibit AD1 activity. Furthermore, this repression does not involve HDAC activity, nor does it interfere with AD1-mediated p300 recruitment. We thus hypothesized that DES-dependent repression may allow ETO to block transcription at a step after chromatin remodeling. To test this, we varied the order of ETO addition during Gal4-HEB(1-548)-dependent transcription on chromatin templates (Fig. 6A). In these experiments, purified TFIID (see Fig. S7A in the supplemental material) was added to the reaction after the remodeling step. Although Gal4-HEB(1-548) can mediate a high level of nucleosomal acetylation, we found that its ability to facilitate Pol II transcription on chromatin templates was intrinsically weak but could be enhanced by additional TFIID added after the remodeling step. This observation agrees with the observed competitive binding of TFIID and p300 to chromatin (9) and also suggests that TFIID recruitment can be a rate-limiting step in HEB-mediated p300-dependent transcription.
FIG. 6.
Recapitulation of the DES-dependent chromatin-independent transcriptional repression by ETO in vitro. (A) Top panel, scheme for in vitro transcription from chromatin templates with ETO added at various time points. The experiment is essentially the same as that shown in Fig. 5G except that purified TFIID, equivalent to 5 ng of TBP, was included at the PIC assembly step to facilitate transcription by Gal4-HEB(1-548) from chromatin templates. Bottom panel, effects of ETO added at different indicated steps on transcriptional activation by Gal4-HEB(1-548). (B) Top panel, scheme for in vitro transcription from naked DNA templates. Bottom panel, effects of ETO on Gal4-HEB(1-548)-dependent activation and basal transcription. (C) Schematic representation of ETO and ETO derivatives. (D) Effects of ETO and ETO derivatives on Gal4-HEB(1-373)-dependent transcription from naked DNA templates. WT, wild-type ETO; Δ403-post-PIC, ETOΔ403 was added after PIC assembly (i.e., with nucleoside triphosphates). (E) Effects of ETO and ETO derivatives on Gal4-HEB-AD1- or Gal4-HEB(1-373)-dependent transcription from naked DNA templates. (F) Effects of ETO, ETOΔ403, AML1-ETO, ETO-2, or MTGR1 on Gal4-HEB(1-373)-dependent transcription from naked DNA templates. Right panel, Coomassie blue staining of purified ETO-2, MTGR1, and AML1-ETO proteins following sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Consistent with the HAT assay data, ETO strongly repressed transcription when added to the reaction before p300 (Fig. 6A, lane 6). Importantly, when ETO was added after the remodeling step, either with TFIID or with nuclear extracts, it was still capable of repressing transcription, close to the basal level (Fig. 6A, lanes 7 and 8). This post-chromatin-remodeling repression should reflect ETO's ability to directly interfere with the communication between HEB and the basal transcription machinery, since a similar inhibition of HEB-dependent transcription was also observed on naked DNA templates (Fig. 6B).
On naked DNA templates, Gal4-HEB(1-373), which contains only AD1 and DES, was also strongly repressed by ETO (Fig. 6D, lane 3). This finding allowed us to focus on HEB(1-373) in subsequent studies. We first asked whether the observed strong inhibition is dependent on DES by examining the effect of ETO on AD1's activity. Whereas ETO strongly repressed Gal4-HEB(1-373), it only slightly inhibited Gal4-AD1 (Fig. 6E, lanes 2 and 4). Additional evidence that strong repression requires DES is provided by the use of ETOΔ403. ETOΔ403 repressed Gal4-HEB(1-373)-mediated transcription to a greater extent than ETO did (Fig. 6D, lanes 3 and 4). In sharp contrast, like ETO, it manifested only a twofold inhibition of Gal4-AD1 (Fig. 6E, lane 3), which is consistent with their similar effects on AD1-mediated transcription from chromatin templates (Fig. 5G). It was not surprising that the twofold repression of Gal4-AD1 by ETO was NHR2 dependent (Fig. 6E, lane 5) given the importance of NHR2 in the association between ETO and AD1, as shown earlier.
To explore the transcription step that is blocked in DES-dependent repression, an “order-of-addition” experiment was performed. When ETOΔ403 was added to the reaction after the completion of PIC assembly, it lost the majority of its inhibitory function (Fig. 6D, lane 4 versus lane 5), although an approximately threefold inhibition was still evident (lane 2 versus lane 5). In another in vitro transcription study using purified general transcription factors, PC4 and Pol II (see Fig. S7B in the supplemental material), repression of Gal4-HEB(1-548)-dependent transcription by ETOΔ403 could largely be rescued by excess TFIID (see Fig. S7C in the supplemental material), suggesting that TFIID and ETOΔ403 may compete for binding to HEB. Together, these data indicate that DES-dependent repression occurs predominantly by interfering with PIC assembly and TFIID function.
DES-dependent repression in vitro also requires cooperative functions of TAFH and NHR2 and is enhanced in AML1-ETO and MTGR1.
Because DES interaction-dependent repression in vivo requires cooperative functions of NHR2 and TAFH, we tested whether such a requirement also holds in vitro by using the various ETO mutants shown in Fig. 6C (see also Fig. S8A in the supplemental material). Indeed, deletion of TAFH, NHR2, or the highly conserved HHR within NHR2 abolished the repression (Fig. 6D). Like ETOΔ403, the ETOΔ366 mutant lacking NHR3 and NHR4 also strongly repressed transcription (lane 9). Further confirming the importance of NHR2, the additional deletion of NHR2 to generate the ETOΔ324 mutant completely restored transcription (lane 10).
The effect of the K98E mutation was examined next to test if an interaction with AD1 is required for repression. We found that the K98E mutation largely abolished repression by ETOΔ403 (see Fig. S8B in the supplemental material). Also, ETOΔ403 failed to repress Gal4-HEB(100-306), which lacks the AD1 domain (see Fig. S8C in the supplemental material). Taken together, our domain requirement study indicates that the in vitro repression requires both TAFH-AD1 and NHR2-DES interactions. The failure of a combination of isolated TAFH and NHR2 fragments to elicit any repressive effect suggests that these interactions need to occur cooperatively in the context of ETO (see Fig. S8C in the supplemental material).
Our in vivo and in vitro results are both consistent with the view that DES-dependent repression requires cooperative binding of TAFH and NHR2 to HEB. However, we were somewhat surprised by the in vitro requirement for the TAFH-AD1 interaction because our earlier in vivo study has shown that this interaction is dispensable for DES-dependent repression. There are two possible explanations for this difference. The first reflects in vitro and in vivo variation in the ability of TAFH to interact with DES in the context of ETO. It is likely that in vitro, an initial TAFH interaction with AD1 may facilitate its subsequent association with DES, which is needed for DES-dependent repression in vivo. This is a likely possibility because our results suggest that AD1 and DES may bind distinct regions of TAFH. Second, TAFH interaction with AD1 may also help to suppress the constitutive activity of AD1 observed in vitro, whereas in cells such activity may depend on a later step (after chromatin remodeling) that may be blocked by ETO in its association with DES (see Discussion).
The TAFH and NHR2 domains are both retained in the AML1-ETO fusion protein and are conserved in the ETO-related proteins MTGR1 and ETO-2. It is thus not surprising that these proteins also repressed Gal4-HEB(1-373) on naked DNA templates (Fig. 6F). However, whereas the level of repression due to ETO-2 was comparable to that due to ETO, AML1-ETO and MTGR1 resembled the C-terminally truncated ETOΔ403 by causing a more potent repression that reduced transcription significantly below the basal level (Fig. 6F). These results suggest that the AML1 domain within the fusion protein can influence ETO's activity, possibly through cross talk with other ETO domains, such as the C-terminal region. The unique property of MTGR1 revealed here was surprising, because the three ETO family members are generally thought to be functionally equivalent. Interestingly, sequence analysis showed that MTGR1 has the lowest sequence conservation in all NHR domains among ETO family proteins, which may underlie its functional specificity elucidated here.
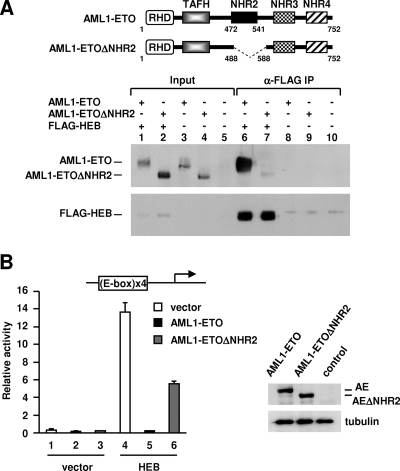
NHR2 is critical for AML1-ETO to associate with HEB and to repress HEB-dependent transcription.
The importance of NHR2 in mediating E-protein interactions (both with AD1 and with DES) is reminiscent of its critical function in leukemogenic activities of AML1-ETO (37, 63, 66). Because NHR2 has important physiological implications in leukemia, we further examined its role in the formation of a complex between AML1-ETO and HEB, which occurs in both cultured and primary t(8;21) leukemic cells (67). An ETO region that includes NHR2 and a sequence immediately downstream of it was deleted to test the role of NHR2. This derivative, designated AML1-ETOΔNHR2, contains intact TAFH, NHR3, and NHR4 domains but is unable to inhibit myeloid differentiation (66). Coimmunoprecipitation revealed that wild-type AML1-ETO but not AML1-ETOΔNHR2 formed a stable complex with HEB in transfected cells (Fig. 7A). In functional studies, HEB-mediated transcription from a stably integrated E-box-driven luciferase template was abolished by AML1-ETO but was minimally affected by AML1-ETOΔNHR2 (Fig. 7B, lane 5 versus lane 6, 60-fold versus 2.4-fold repression), which was expressed at a level similar to that of AML1-ETO (Fig. 7B, right). A similar loss of inhibition by AML1-ETOΔNHR2 was also observed for E2A (E12 and E47; see Fig. S9 in the supplemental material). These results suggest that in the context of AML1-ETO, the NHR2 domain may have acquired more importance in E-protein targeting and silencing. Accordingly, the residual interaction derived from the TAFH domain of AML1-ETOΔNHR2 is not sufficient to support a significant inhibition of E-protein-mediated transcription.
FIG. 7.
NHR2 is critical for AML1-ETO to associate with HEB and to repress HEB-directed transcription. (A) Coimmunoprecipitation of AML1-ETO and AML1-ETOΔNHR2 with FLAG-HEB using anti-FLAG (α-FLAG) M2-agarose. AML1-ETO and AML1-ETOΔNHR2 were detected via an N-terminal hemagglutinin tag. FLAG-HEB was detected by anti-FLAG antibody. Top panel, schematic representation of domains of AML1-ETO and AML1-ETOΔNHR2. AML1-ETOΔNHR2 lacks amino acids 488 to 588, corresponding to 313 to 413 of the ETO moiety. IP, immunoprecipitation. (B) Luciferase assays of E-box-293T cells containing an integrated E-box luciferase reporter transfected with expression plasmids for FLAG-HEB, AML1-ETO, and AML1-ETOΔNHR2. Right panel, Western blot analyses of transfected AML1-ETO and AML1-ETOΔNHR2 via an N-terminal HA tag with α-tubulin serving as the control. AE, AML1-ETO.
DISCUSSION
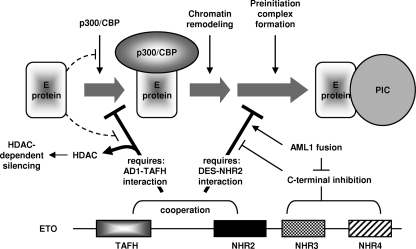
ETO family corepressors and AML1-ETO repress E-protein-mediated transcription through both chromatin-dependent and chromatin-independent mechanisms (Fig. 8).
FIG. 8.
Schematic model for E-protein targeting and inhibition by ETO. Solid arrows represent facilitation of activation or repression steps. T-shaped lines refer to inhibition. The major repression mechanisms are represented by thick lines underneath the activation steps. See the text for details.
ETO has served as a prototype for studying the mechanisms of its family members and AML1-ETO (20, 40, 60, 67). The current model proposes that these proteins repress transcription by reducing chromatin accessibility. For E proteins, this repression is thought to be mediated by a single interaction between the TAFH domain of ETO corepressors and the AD1 domain of E proteins, which competes with p300/CBP coactivators for binding to AD1 (67). We now show that ETO, in fact, interacts with two E-protein regions. The interaction with AD1, however, remains necessary and sufficient for repression of transcription from chromatin templates. In addition, we have identified a novel chromatin-independent repression mechanism that depends on the newly defined interaction. Mutation of K98E disrupts ETO interaction with AD1 and restores p300/CBP recruitment, but it did not abolish ETO-mediated repression of HEB or E2A as long as the second ETO binding site was functioning. This AD1-independent repression is not mediated by HDACs, since it is not affected by TSA. On the other hand, TSA reduces the repression by wild-type ETO to a level comparable to that observed with ETO-K98E. Furthermore, AD1, a region needed for p300/CBP recruitment, is dispensable for both HEB-mediated activation and repression of this activation by ETO. We find that HEBΔAD1 is expressed at a high level in transfected cells (C. Guo and J. Zhang, unpublished observation), which may increase its capacity to bind DNA and to stimulate PlC assembly. This effect is reminiscent of the direct stimulation of transcription by nuclear receptors expressed at high levels (33). Together, these in vivo studies suggest that productive transcription by E proteins involves overcoming a second rate-limiting step after chromatin remodeling and that ETO directly inhibits this process in addition to its inhibition of chromatin remodeling. Consistent with this idea, in vitro transcription confirms that ETO is capable of repressing transcription on chromatin templates when it is added after chromatin remodeling. Moreover, on naked DNA templates, ETO and AML1-ETO, as well as related family members, directly inhibit HEB-dependent transcription.
Our study has added ETO family proteins to the list of corepressors that can repress transcription by targeting both chromatin and the basal transcription machinery. These corepressors, including Tup1 and Groucho/TLE family proteins as well as N-CoR, have been studied mainly in association with repressors (15, 25). ETO is unique, however, in that it associates with a transcriptional activator that contains multiple activation domains, stressing the importance of inactivating coactivators in repression by ETO. Interestingly, like Tup1 and Groucho family corepressors, ETO is also able to interact with HDACs and to form oligomers. An HDAC-mediated mechanism should allow ETO to enforce long-term silencing, which may impose a slow response to activating stimuli. On the other hand, chromatin-independent repression may allow ETO to effectively maintain an inactive promoter region in an open chromatin state, thereby allowing rapid signal-dependent activation. The latter form of repression is particularly interesting given recent findings that many inactive promoters in eukaryotic cells are associated with active chromatin marks, but are defective in PIC recruitment or postrecruitment function (31).
Roles of E-protein domains in repression and activation.
E proteins contain multiple conserved domains. Among them, AD1 plays a dual role in both activation and ETO-mediated repression (Fig. 8). AD1 has been previously proposed to facilitate the recruitment of p300/CBP (7, 10, 67). This is consistent with our finding that it is sufficient to mediate p300-dependent nucleosomal acetylation and to activate transcription from chromatin templates. Interestingly, the ability of AD1 to recruit p300 to acetylate histone can be enhanced by additional HEB regions, likely by AD2 given its possible association with p300 (7, 10). The increased acetylation however, is abolished by ETO to a nearly basal level (Fig. 5D and E). These results indicate that cooperative recruitment of p300 and possibly CBP is dependent on initial binding by AD1. ETO abolishes this initial binding step, thereby blocking p300 recruitment. This model likely applies to other E proteins, because deletion of AD1 but not AD2 dramatically reduces CBP recruitment by E2A (7). It also suggests a strategy whereby corepressors target the first step of coactivator recruitment to effectively block the function of coactivators. Intriguingly, in addition to its important role in controlling p300/CBP recruitment, we show that AD1 is also critical to the HDAC-dependent repression function of ETO (Fig. 8). In this regard, future studies are needed to determine whether an interaction with AD1 is required for ETO to recruit HDACs or to facilitate their postrecruitment function.
Whereas AD1 plays a principal role in directing chromatin-dependent repression, our in vivo and in vitro results indicate that chromatin-independent repression by ETO requires its interaction with DES (Fig. 8). It should be mentioned that although an interaction with AD1 is dispensable for DES-dependent repression in cells, this interaction is required for repression in vitro on naked DNA templates. As noted earlier, the TAFH-AD1 interaction may promote a subsequent TAFH-DES interaction in vitro, which is required for DES-dependent repression based on the in vivo data. Furthermore, the TAFH-AD1 interaction may help suppress the in vitro AD1 activity. Such activity would not be expected to occur constitutively in cells, because AD1 needs to first recruit p300/CBP to remodel chromatin. At a later step, p300/CBP may need to be released from AD1 in order for such an activity to become effective. It has been shown that p300 is inhibitory to transcription (9, 23, 55) and that the release of p300 from a promoter is necessary for, and is coupled with, TFIID recruitment (9). Recruitment of TFIID, however, may be inhibited by ETO binding to DES (see below).
The hierarchical involvement of AD1 and DES in repression is consistent with our finding that their individual interactions with ETO are each sufficient for repression of E-protein-dependent transcription. Intriguingly, naturally occurring splice variants of HEB and E2-2, namely HEBAlt and ITF-2A, are lacking AD1 and most of DES (59). Similar variants of E2A have also been reported (26). Based on our data, replacing full-length E proteins at target promoters with these variants should bypass the repression imposed by ETO corepressors, leading to constitutive activation of target promoters during a time frame when these variants are expressed at high levels. Indeed, it has been shown that HEBAlt is transiently upregulated in early T-cell precursors and plays a critical role in facilitating early T-cell development from hematopoietic stem cells (59), where various ETO corepressors are present at high levels (22, 36, 44, 56).
Cross talk between ETO and AML1-ETO domains governs repression specificity and potency.
Interactions with AD1 and DES allow ETO to inhibit transcription through separate mechanisms. We found that optimal repression exerted by each mechanism requires cooperation between TAFH and NHR2 (Fig. 8). Oligomerization by NHR2 likely accounts for its role in enhancing the TAFH-AD1 interaction and consequent repression at the chromatin level. Our assays have used heterologous domains (Gal4 and GST) that dimerize. Physiologically, oligomerization of ETO likely increases its affinity for the two PCET motifs in a given E-protein dimer (6, 43).
In addition to facilitating AD1 interaction, we show that NHR2 can directly interact with DES. TAFH cooperates with this interaction to mediate chromatin-independent repression (Fig. 8). Mechanistically, ETO, through its physical interaction with DES, may interfere with the recruitment of TFIID to inhibit PIC assembly, given previously demonstrated AD1-independent TFIID recruitment by HEB (67) and our finding that excess TFIID can rescue ETOΔ403-mediated repression in purified in vitro transcription assays. In addition to the block of activated transcription, we show that ETOΔ403, ETOΔ366, and AML1-ETO, as well as MTGR1, when complexing with HEB(1-373), can also repress basal transcription activity (Fig. 6D and F). This basal repression may conceivably involve physical associations between these proteins and components of the basal transcription machinery, which will be explored in future studies. The finding that fusion with AML1 (in AML1-ETO) and C-terminal deletion (in ETOΔ403 and ETOΔ366) similarly enhanced the repression activity of ETO suggests functional cross talk between the N-terminal and C-terminal regions of AML1-ETO (Fig. 8). One possibility is that the N-terminal AML1 region may override inhibition imposed by the C terminus on NHR2- and DES-dependent repression. This form of cross talk is reminiscent of that described for another leukemogenic fusion protein, PML-RAR. It has been shown that the N-terminal PML destabilizes the inhibitory conformation of the RAR C-terminal tail, thereby facilitating binding of RAR to N-CoR/SMRT (35, 45).
Tethering of AML1-ETO to E-protein binding sites should disrupt the delicate balance between E-protein coactivators and ETO family corepressors and thereby cause deregulation of E-protein function. Interestingly, a recent work showed that HEB can also form a complex with AML1-ETO in transduced U937 cells at AML1-ETO sites and at promoters that contain both HEB and AML1-ETO sites (18). Thus, cross talk between E proteins and AML1-ETO may be context dependent, resembling that between nuclear receptors and other transcription factors. Nevertheless, the transcriptional property of the AML1-ETO-E-protein complex should be largely governed by their interdomain interactions, which dictate cofactor associations. Our data support the involvement of multiple interdomain communications in regulating the repression function of ETO family corepressors and the AML1-ETO fusion protein. In particular, our data suggest that in the context of AML1-ETO, NHR2 plays a critical role in targeting AML1-ETO to E proteins to repress transcription. This suggests positive cross talk between the AML1 and NHR2 domains that may occur directly or indirectly through other regions, such as the C terminus, as described above. Physiologically, NHR2 is also critical for AML1-ETO's leukemogenic activities (37, 63, 66), whereas the ETO C terminus, containing NHR3/4 regions, is inhibitory (64, 65). These findings unexpectedly mirror our observations described here. We believe that further insight into the mechanisms underlying the various domain interactions and their interplay should not only increase our understanding of transcriptional mechanisms associated with E proteins, AML1-ETO, and ETO family corepressors but should also lead to novel therapeutic targets in cancers involving the expression of AML1-ETO and E-protein-derived fusion proteins.
Supplementary Material
Acknowledgments
This work is supported by awards to J. Zhang from the Leukemia and Lymphoma Society and from Ohio Cancer Research Associates and by start-up funds available to J. Zhang from the University of Cincinnati College of Medicine and the Department of Cancer and Cell Biology. C.Y. is partially supported by a scholarship from the Chinese Scholarship Council.
We thank Peter Stambrook and Sohaib Khan for stimulating discussions; Peter Stambrook for critical reading of the manuscript; Maryellen Daston for editorial assistance; Sohaib Khan, Jun Ma, Carolyn Price, and Nicholas Olshavsky for critical comments; and Robert G. Roeder for continuous support and encouragement.
Footnotes
Published ahead of print on 16 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amann, J. M., B. J. Chyla, T. C. Ellis, A. Martinez, A. C. Moore, J. L. Franklin, L. McGhee, S. Meyers, J. E. Ohm, K. S. Luce, A. J. Ouelette, M. K. Washington, M. A. Thompson, D. King, S. Gautam, R. J. Coffey, R. H. Whitehead, and S. W. Hiebert. 2005. Mtgr1 is a transcriptional corepressor that is required for maintenance of the secretory cell lineage in the small intestine. Mol. Cell. Biol. 259576-9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 216470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117735-748. [DOI] [PubMed] [Google Scholar]
- 4.An, W., and R. G. Roeder. 2004. Reconstitution and transcriptional analysis of chromatin in vitro. Methods Enzymol. 377460-474. [DOI] [PubMed] [Google Scholar]
- 5.Aspland, S. E., H. H. Bendall, and C. Murre. 2001. The role of E2A-PBX1 in leukemogenesis. Oncogene 205708-5717. [DOI] [PubMed] [Google Scholar]
- 6.Barndt, R. J., M. Dai, and Y. Zhuang. 2000. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant-negative mutation of HEB. Mol. Cell. Biol. 206677-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayly, R., L. Chuen, R. A. Currie, B. D. Hyndman, R. Casselman, G. A. Blobel, and D. P. LeBrun. 2004. E2A-PBX1 interacts directly with the KIX domain of CBP/p300 in the induction of proliferation in primary hematopoietic cells. J. Biol. Chem. 27955362-55371. [DOI] [PubMed] [Google Scholar]
- 8.Bhalla, S., C. Spaulding, R. L. Brumbaugh, D. E. Zagort, M. E. Massari, C. Murre, and B. L. Kee. 2008. differential roles for the E2A activation domains in B lymphocytes and macrophages. J. Immunol. 1801694-1703. [DOI] [PubMed] [Google Scholar]
- 9.Black, J. C., J. E. Choi, S. R. Lombardo, and M. Carey. 2006. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell 23809-818. [DOI] [PubMed] [Google Scholar]
- 10.Bradney, C., M. Hjelmeland, Y. Komatsu, M. Yoshida, T. P. Yao, and Y. Zhuang. 2003. Regulation of E2A activities by histone acetyltransferases in B lymphocyte development. J. Biol. Chem. 2782370-2376. [DOI] [PubMed] [Google Scholar]
- 11.Calabi, F., R. Pannell, and G. Pavloska. 2001. Gene targeting reveals a crucial role for MTG8 in the gut. Mol. Cell. Biol. 215658-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chyla, B. J., I. Moreno-Miralles, M. A. Steapleton, M. A. Thompson, S. Bhaskara, M. Engel, and S. W. Hiebert. 2008. Deletion of Mtg16, a target of t(16;21), alters hematopoietic progenitor cell proliferation and lineage allocation. Mol. Cell. Biol. 286234-6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cisse, B., M. L. Caton, M. Lehner, T. Maeda, S. Scheu, R. Locksley, D. Holmberg, C. Zweier, N. S. den Hollander, S. G. Kant, W. Holter, A. Rauch, Y. Zhuang, and B. Reizis. 2008. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 13537-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cochrane, S. W., Y. Zhao, R. S. Welner, and X. H. Sun. 2009. Balance between Id and E proteins regulates myeloid-versus-lymphoid lineage decisions. Blood 1131016-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 152786-2796. [DOI] [PubMed] [Google Scholar]
- 16.Dias, S., R. Mansson, S. Gurbuxani, M. Sigvardsson, and B. L. Kee. 2008. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity 29217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fyodorov, D. V., and J. T. Kadonaga. 2003. Chromatin assembly in vitro with purified recombinant ACF and NAP-1. Methods Enzymol. 371499-515. [DOI] [PubMed] [Google Scholar]
- 18.Gardini, A., M. Cesaroni, L. Luzi, A. J. Okumura, J. R. Biggs, S. P. Minardi, E. Venturini, D. E. Zhang, P. G. Pelicci, and M. Alcalay. 2008. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet. 4e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge, H., E. Martinez, C. M. Chiang, and R. G. Roeder. 1996. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 27457-71. [DOI] [PubMed] [Google Scholar]
- 20.Gelmetti, V., J. Zhang, M. Fanelli, S. Minucci, P. G. Pelicci, and M. A. Lazar. 1998. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 187185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14121-141. [PubMed] [Google Scholar]
- 22.Goardon, N., J. A. Lambert, P. Rodriguez, P. Nissaire, S. Herblot, P. Thibault, D. Dumenil, J. Strouboulis, P. H. Romeo, and T. Hoang. 2006. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 25357-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guermah, M., V. B. Palhan, A. J. Tackett, B. T. Chait, and R. G. Roeder. 2006. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125275-286. [DOI] [PubMed] [Google Scholar]
- 24.Hamlett, I., J. Draper, J. Strouboulis, F. Iborra, C. Porcher, and P. Vyas. 2008. Characterisation of megakaryocyte GATA1-interacting proteins: the corepressor ETO2 and GATA1 interact to regulate terminal megakaryocyte maturation. Blood 1122738-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 38743-48. [DOI] [PubMed] [Google Scholar]
- 26.Henthorn, P., M. Kiledijian, and T. Kadesch. 1990. Two distinct transcription factors that bind the immunoglobulin enhancer μE5/κE2 motif. Science 247467-470. [DOI] [PubMed] [Google Scholar]
- 27.Hiebert, S. W., J. R. Downing, N. Lenny, and S. Meyers. 1996. Transcriptional regulation by the t(8;21) fusion protein, AML-1/ETO. Curr. Top. Microbiol. Immunol. 211253-258. [DOI] [PubMed] [Google Scholar]
- 28.Hildebrand, D., J. Tiefenbach, T. Heinzel, M. Grez, and A. B. Maurer. 2001. Multiple regions of ETO cooperate in transcriptional repression. J. Biol. Chem. 2769889-9895. [DOI] [PubMed] [Google Scholar]
- 29.Hug, B. A., and M. A. Lazar. 2004. ETO interacting proteins. Oncogene 234270-4274. [DOI] [PubMed] [Google Scholar]
- 30.Jankovic, V., A. Ciarrocchi, P. Boccuni, T. DeBlasio, R. Benezra, and S. D. Nimer. 2007. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 1041260-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch, F., F. Jourquin, P. Ferrier, and J. C. Andrau. 2008. Genome-wide RNA polymerase II: not genes only! Trends Biochem. Sci. 33265-273. [DOI] [PubMed] [Google Scholar]
- 32.Kondo, M. 2008. Multitalented E2A: a new role in lymphoid-lineage priming. Immunity 29169-170. [DOI] [PubMed] [Google Scholar]
- 33.Lee, Y. H., S. S. Koh, X. Zhang, X. Cheng, and M. R. Stallcup. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 223621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Licht, J. D. 2001. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene 205660-5679. [DOI] [PubMed] [Google Scholar]
- 35.Lin, R. J., L. Nagy, S. Inoue, W. Shao, W. H. Miller, Jr., and R. M. Evans. 1998. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391811-814. [DOI] [PubMed] [Google Scholar]
- 36.Lindberg, S. R., A. Olsson, A. M. Persson, and I. Olsson. 2005. The leukemia-associated ETO homologues are differently expressed during hematopoietic differentiation. Exp. Hematol. 33189-198. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Y., M. D. Cheney, J. J. Gaudet, M. Chruszcz, S. M. Lukasik, D. Sugiyama, J. Lary, J. Cole, Z. Dauter, W. Minor, N. A. Speck, and J. H. Bushweller. 2006. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity. Cancer Cell 9249-260. [DOI] [PubMed] [Google Scholar]
- 38.Look, A. T. 1997. E2A-HLF chimeric transcription factors in pro-B cell acute lymphoblastic leukemia. Curr. Top. Microbiol. Immunol. 22045-53. [DOI] [PubMed] [Google Scholar]
- 39.Lutterbach, B., D. Sun, J. Schuetz, and S. W. Hiebert. 1998. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol. Cell. Biol. 183604-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lutterbach, B., J. J. Westendorf, B. Linggi, A. Patten, M. Moniwa, J. R. Davie, K. D. Huynh, V. J. Bardwell, R. M. Lavinsky, M. G. Rosenfeld, C. Glass, E. Seto, and S. W. Hiebert. 1998. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 187176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markus, M., Z. Du, and R. Benezra. 2002. Enhancer-specific modulation of E protein activity. J. Biol. Chem. 2776469-6477. [DOI] [PubMed] [Google Scholar]
- 42.Massari, M. E., P. A. Grant, M. G. Pray-Grant, S. L. Berger, J. L. Workman, and C. Murre. 1999. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell 463-73. [DOI] [PubMed] [Google Scholar]
- 43.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meier, N., S. Krpic, P. Rodriguez, J. Strouboulis, M. Monti, J. Krijgsveld, M. Gering, R. Patient, A. Hostert, and F. Grosveld. 2006. Novel binding partners of Ldb1 are required for haematopoietic development. Development 1334913-4923. [DOI] [PubMed] [Google Scholar]
- 45.Minucci, S., M. Maccarana, M. Cioce, P. De Luca, V. Gelmetti, S. Segalla, L. Di Croce, S. Giavara, C. Matteucci, A. Gobbi, A. Bianchini, E. Colombo, I. Schiavoni, G. Badaracco, X. Hu, M. A. Lazar, N. Landsberger, C. Nervi, and P. G. Pelicci. 2000. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol. Cell 5811-820. [DOI] [PubMed] [Google Scholar]
- 46.Nimer, S. D., and M. A. Moore. 2004. Effects of the leukemia-associated AML1-ETO protein on hematopoietic stem and progenitor cells. Oncogene 234249-4254. [DOI] [PubMed] [Google Scholar]
- 47.O'Neil, J., J. Shank, N. Cusson, C. Murre, and M. Kelliher. 2004. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell 5587-596. [DOI] [PubMed] [Google Scholar]
- 48.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116511-526. [DOI] [PubMed] [Google Scholar]
- 49.Peterson, L. F., A. Boyapati, E. Y. Ahn, J. R. Biggs, A. J. Okumura, M. C. Lo, M. Yan, and D. E. Zhang. 2007. Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood 110799-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plevin, M. J., J. Zhang, C. Guo, R. G. Roeder, and M. Ikura. 2006. The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain. Proc. Natl. Acad. Sci. USA 10310242-10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quong, M. W., W. J. Romanow, and C. Murre. 2002. E protein function in lymphocyte development. Annu. Rev. Immunol. 20301-322. [DOI] [PubMed] [Google Scholar]
- 52.Rochford, J. J., R. K. Semple, M. Laudes, K. B. Boyle, C. Christodoulides, C. Mulligan, C. J. Lelliott, S. Schinner, D. Hadaschik, M. Mahadevan, J. K. Sethi, A. Vidal-Puig, and S. O'Rahilly. 2004. ETO/MTG8 is an inhibitor of C/EBPβ activity and a regulator of early adipogenesis. Mol. Cell. Biol. 249863-9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 201405-1428. [DOI] [PubMed] [Google Scholar]
- 54.Ruzinova, M. B., and R. Benezra. 2003. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13410-418. [DOI] [PubMed] [Google Scholar]
- 55.Santoso, B., and J. T. Kadonaga. 2006. Reconstitution of chromatin transcription with purified components reveals a chromatin-specific repressive activity of p300. Nat. Struct. Mol. Biol. 13131-139. [DOI] [PubMed] [Google Scholar]
- 56.Schuh, A. H., A. J. Tipping, A. J. Clark, I. Hamlett, B. Guyot, F. J. Iborra, P. Rodriguez, J. Strouboulis, T. Enver, P. Vyas, and C. Porcher. 2005. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol. Cell. Biol. 2510235-10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sjogren, H., B. Wedell, J. M. Meis-Kindblom, L. G. Kindblom, G. Stenman, and J. M. Kindblom. 2000. Fusion of the NH2-terminal domain of the basic helix-loop-helix protein TCF12 to TEC in extraskeletal myxoid chondrosarcoma with translocation t(9;15)(q22;q21). Cancer Res. 606832-6835. [PubMed] [Google Scholar]
- 58.Wallberg, A. E., K. Pedersen, U. Lendahl, and R. G. Roeder. 2002. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell. Biol. 227812-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, D., C. L. Claus, G. Vaccarelli, M. Braunstein, T. M. Schmitt, J. C. Zuniga-Pflucker, E. V. Rothenberg, and M. K. Anderson. 2006. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J. Immunol. 177109-119. [DOI] [PubMed] [Google Scholar]
- 60.Wang, J., T. Hoshino, R. L. Redner, S. Kajigaya, and J. M. Liu. 1998. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. USA 9510860-10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, X., D. M. Truckses, S. Takada, T. Matsumura, N. Tanese, and R. H. Jacobson. 2007. Conserved region I of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators. Proc. Natl. Acad. Sci. USA 1047839-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei, Y., S. Liu, J. Lausen, C. Woodrell, S. Cho, N. Biris, N. Kobayashi, Y. Wei, S. Yokoyama, and M. H. Werner. 2007. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat. Struct. Mol. Biol. 14653-661. [DOI] [PubMed] [Google Scholar]
- 63.Wichmann, C., L. Chen, M. Heinrich, D. Baus, E. Pfitzner, M. Zornig, O. G. Ottmann, and M. Grez. 2007. Targeting the oligomerization domain of ETO interferes with RUNX1/ETO oncogenic activity in t(8;21)-positive leukemic cells. Cancer Res. 672280-2289. [DOI] [PubMed] [Google Scholar]
- 64.Yan, M., S. A. Burel, L. F. Peterson, E. Kanbe, H. Iwasaki, A. Boyapati, R. Hines, K. Akashi, and D. E. Zhang. 2004. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc. Natl. Acad. Sci. USA 10117186-17191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan, M., E. Kanbe, L. F. Peterson, A. Boyapati, Y. Miao, Y. Wang, I. M. Chen, Z. Chen, J. D. Rowley, C. L. Willman, and D. E. Zhang. 2006. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 12945-949. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, J., B. A. Hug, E. Y. Huang, C. W. Chen, V. Gelmetti, M. Maccarana, S. Minucci, P. G. Pelicci, and M. A. Lazar. 2001. Oligomerization of ETO is obligatory for corepressor interaction. Mol. Cell. Biol. 21156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, J., M. Kalkum, S. Yamamura, B. T. Chait, and R. G. Roeder. 2004. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science 3051286-1289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.