Abstract
Epac1 is a guanine nucleotide exchange factor (GEF) for the small G protein Rap and is directly activated by cyclic AMP (cAMP). Upon cAMP binding, Epac1 undergoes a conformational change that allows the interaction of its GEF domain with Rap, resulting in Rap activation and subsequent downstream effects, including integrin-mediated cell adhesion and cell-cell junction formation. Here, we report that cAMP also induces the translocation of Epac1 toward the plasma membrane. Combining high-resolution confocal fluorescence microscopy with total internal reflection fluorescence and fluorescent resonance energy transfer assays, we observed that Epac1 translocation is a rapid and reversible process. This dynamic redistribution of Epac1 requires both the cAMP-induced conformational change as well as the DEP domain. In line with its translocation, Epac1 activation induces Rap activation predominantly at the plasma membrane. We further show that the translocation of Epac1 enhances its ability to induce Rap-mediated cell adhesion. Thus, the regulation of Epac1-Rap signaling by cAMP includes both the release of Epac1 from autoinhibition and its recruitment to the plasma membrane.
Cyclic AMP (cAMP) is an important second messenger that mediates many cellular hormone responses. It has become more and more appreciated that, along with the cAMP effector protein kinase A (PKA), Epac proteins also play pivotal roles in many cAMP-controlled processes, including insulin secretion (23, 39), cell adhesion (9, 17, 25, 49, 60), neurotransmitter release (22, 53, 63), heart function (13, 35, 54), and circadian rhythm (38). Epac1 and Epac2 are cAMP-dependent guanine nucleotide exchange factors (GEFs) for the small G proteins Rap1 and Rap2 (12, 24). They contain a regulatory region with one (Epac1) or two (Epac2) cAMP-binding domains, a Dishevelled, Egl-10, Pleckstrin (DEP) domain, and a catalytic region for GEF activity (11). The binding of cAMP is a prerequisite for catalytic activity in vitro and in vivo (11). Recently, the structures of both the inactive and active conformations of Epac2 were solved (51, 52). This revealed that in the inactive conformation, the regulatory region occludes the Rap binding site, which is relieved by a conformational change induced by cAMP binding.
Like all G proteins of the Ras superfamily, Rap cycles between an inactive GDP-bound and active GTP-bound state in an equilibrium that is tightly regulated by specific GEFs and GTPase-activating proteins (GAPs). The GEF-induced dissociation of GDP results in the binding of the cellularly abundant GTP, whereas GAPs enhance the intrinsic GTPase activity of the G protein, thereby inducing the inactive GDP-bound state. Besides Epac, several other GEFs for Rap have been identified, including C3G, PDZ-GEF, and RasGRP, and these act downstream of different signaling pathways (7). Since Rap localizes to several membrane compartments, including the Golgi network, vesicular membranes, and the plasma membrane (PM) (2-4, 37, 42, 48), the spatial regulation of its activity is expected to be established by the differential distributions of its upstream GEFs, each activating distinct pools of Rap on specific intracellular locations.
Similarly to Rap, Epac1 also is observed at many locations in the cell, including the cytosol, the nucleus, the nuclear envelope, endomembranes, and the PM (5, 11, 14, 21, 29, 47). These various locations may reflect the many different functions assigned to Epac1, such as the regulation of cell adhesion, cell junction formation, secretion, the regulation of DNA-dependent protein kinase by nuclear Epac1, and the regulation of the Na+/H+ exchanger NHE3 at the brush borders of kidney epithelium (19, 21, 26). Apparently, specific anchors are responsible for this spatial regulation of Epac1. Indeed, Epac1 was found to associate with phosphodiesterase 4 (PDE4) in a complex with mAKAP in cardiomyocytes (13), with MAP-LC bound to microtubules (62), and with Ezrin at the brush borders of polarized cells (M. Gloerich, J. Zhao, and J. L. Bos, unpublished data).
In this study, we report the unexpected observation that, in addition to the temporal control of Epac1 activity, cAMP also induces the translocation of Epac1 toward the plasma membrane. Using confocal fluorescence microscopy, total internal reflection fluorescence (TIRF) microscopy, and fluorescence resonance energy transfer (FRET)-based assays for high spatial and temporal resolution, we observed that the translocation of Epac1 is immediate and that Epac1 approaches the PM to within ∼7 nm. In line with this, Epac1-induced Rap activation was registered predominantly on this compartment. Epac1 translocation results directly from the cAMP-induced conformational change and depends on the integrity of its DEP domain. We further show that Epac1 translocation is a prerequisite for cAMP-induced Rap activation at the PM and enhances Rap-mediated cell adhesion. Thus, cAMP exerts dual regulation on Epac1 for the activation of Rap, controlling both its GEF activity and targeting to the PM.
MATERIALS AND METHODS
Reagents and antibodies.
Forskolin, IBMX, and H89 were from Calbiochem-Novabiochem Corp. (La Jolla, CA); isoproterenol, epidermal growth factor, cytochalasin D, latrunculin A, and nocodazol were from Sigma Chemical Co. (St. Louis, MO); 1-(2-nitrophenyl)ethyl adenosine-3′,-5′-cyclic monophosphate (NPE-caged cAMP) was from Jena Bioscience GmbH (Jena, Germany); 8-pCPT-2′-O-Me-cAMP (007) (15) and 8-pCPT-2′-O-Me-cAMP-AM (007-AM) (59) were from Biolog Life Sciences (Bremen, Germany); Fura-Red-AM and BAPTA-AM were from Invitrogen; the Rap1 antibody (SC-65) was from SantaCruz Biotechnology; and the Rap2 antibody (610216) was from BD Transduction Laboratories. The Epac1 antibody (5D3) has been described previously (46). Fibronectin was purified as described previously (45).
DNA constructs.
The following expression vectors were described elsewhere: pcDNA3 CFP-Epac1-YFP and pcDNA3 CFP-Epac1(ΔDEP,C.D.)-YFP (44), pcDNA3 CFP-Epac1(ΔDEP,C.D.)-Venus (55), pMT2-SM-HA Rap1A(G12V) and pMT2-SM-HA RapGAP1 (50), pMT2-SM-HA Epac1 (11), and pcDNA3 CFP-CAAX (56). Epac1 (RapGEF3, Homo sapiens) was cloned C terminally to yellow fluorescent protein (YFP) in a pCDNA3 vector. Mutations were introduced by site-directed mutagenesis. The separate regulatory (amino acids 1 to 328) and catalytic (amino acids 330 to 881) regions of Epac1 were cloned into pcDNA3 with an N-terminal cyan fluorescent protein (CFP) and YFP tag, respectively. GFP-RBD(RalGDS) was a kind gift from Mark Philips.
Cell culture.
HEK293 cells and A431 human carcinoma cells were cultured in Dulbecco's modified Eagle's medium (DMEM); OVCAR-3 and the Jurkat T-cell line JHM1 2.2 were grown in RPMI medium, each supplemented with 10% serum and antibiotics.
Live cell experiments.
Cells were seeded in 6-well plates on 25-mm glass coverslips and cultured in 3 ml medium. Constructs were transiently transfected using Fugene 6 transfection reagent (Roche Inc.). Experiments were performed in a culture chamber mounted on an inverted microscope in bicarbonate-buffered saline (containing 140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM glucose, 23 mM NaHCO3, 10 mM HEPES), pH 7.2, kept under 5% CO2 at 37°C. Agonists and inhibitors were added from concentrated stocks.
Dynamic monitoring of YFP/CFP FRET.
Cells on coverslips were placed on an inverted Nikon microscope equipped with a ×63 magnification lens (numeric aperture [N.A.], 1.30) and excited at 425 nm. The emission of CFP and YFP was detected simultaneously by two photon multiplier tubes through bandpass filters (470 ± 20 and 530 ± 25 nm, respectively). Data were digitized by Picolog acquisition software (Picotech), and FRET was expressed as the normalized ratio of YFP/CFP signals. The ratio was adjusted to 1 at the onset of the experiment, and changes are expressed as the percent deviation from this initial value. For some of these experiments, a CFP-tagged version of K-Ras-CAAX was used as a PM marker. Whereas in hippocampal neurons K-Ras-CAAX may translocate to endomembranes under certain conditions (16), under our conditions CFP-CAAX localizes to the PM, as we have extensively documented (56, 57). For the experiment described in Fig. 3C, the YFP/CFP FRET ratio was determined in imaging mode by detecting CFP and YFP images simultaneously on a Leica fluorescence SP2 microscope equipped with a dual-view attachment and a Coolsnap-HQ charge-coupled device camera (Roper Scientific) using ASMDW acquisition software.
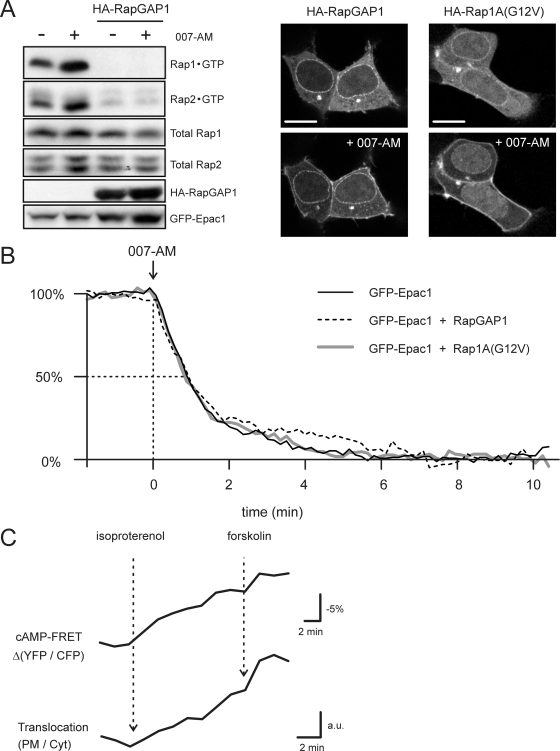
FIG. 3.
Rap activity is not involved in Epac1 translocation. (A) Western blotting of the Rap activation assay (see Materials and Methods) confirming that RapGAP1 overexpression effectively inhibits the 007-AM-mediated activation of Rap1 and Rap2. Images are HEK293 cells expressing GFP-Epac1 plus overexpressed RapGAP1 (left) or constitutively active Rap1A(G12V) (right). Neither RapGAP1 nor Rap1A(G12V) overexpression had any effect on the distribution of unstimulated GFP-Epac1 (upper) or on the 007-AM-induced translocation (lower). Scale bars, 10 μm. (B) Kinetic analyses were performed on cells transfected as described for panel A. Kinetics of the 007-AM-induced translocation of GFP-Epac1 were not affected by the coexpression of RapGAP1 or Rap1A(G12V). Translocation was quantified from the depletion of cytosolic fluorescence. Traces of >10 experiments per condition were averaged after normalization to the basal level (set to 100%) and the end level (set to 0%). (C) A431 cells expressing CFP-Epac1-YFP were imaged by the simultaneous detection of CFP and YFP emission (see Materials and Methods), allowing the analysis of both the Epac1 activation state (FRET) (YFP/CFP ratio) and its translocation to the PM (PM/Cyt ratio). Cyt, cytosol. The submaximal stimulation of A431 cells with 0.5 nM isoproterenol evokes slow cAMP accumulation and thereby induces the gradual activation of Epac1; subsequently, forskolin (25 μM) was added to saturate CFP-Epac1-YFP. The experiment illustrates that the kinetics of the construct's PM translocation strongly resemble its gradual activation (representative for n = 6).
Confocal microscopy.
Coverslips with cells expressing various constructs were mounted in a culture chamber and imaged at 37°C using an inverted TCS-SP5 confocal microscope equipped with a ×63 magnification oil immersion lens (N.A., 1.4; Leica, Mannheim, Germany). Imaging settings were the following: for CFP, excitation at 442 nm and emission at 465 to 500 nm; for GFP, excitation at 488 nm and emission at 510 to 560 nm; for YFP, excitation at 514 nm and emission at 522 to 570 nm.
For the detection of endogenous Epac1 in OVCAR-3 cells, cells were grown on 12-mm glass coverslips for 72 h, and after 10 min of stimulation with 25 μM forskolin they were fixed with 3.8% formaldehyde, permeabilized using 0.1% Triton X-100, and blocked in 2% bovine serum albumin (BSA). Cells were incubated with the Epac1 antibody (5D3) and subsequently with Alexa-conjugated secondary antibodies (Invitrogen). Mounted slides were examined using an Axioskop2 CLSM microscope (Zeiss) (×63 magnification lenses; N.A., 1.4).
Digital image analysis.
For translocation studies, series of confocal images were taken from a medial plane at 5- or 10-s intervals. To quantitate the translocation of constructs, the ratio of cytosolic to PM fluorescence was calculated by the postacquisition automated assignment of regions of interest using Leica Qwin software (56). Note that in the individual traces, the gain of fluorescence at the PM appears small compared to the loss in the cytosol because of the underrepresentation of the membrane area in medial sections; the majority of the membrane is present in the basal membrane and in the strongly curved apical parts of the cell. Postacquisition brightness and contrast adjustments were performed with ImageJ software (NIH).
TIRF microscopy.
Cells expressing GFP-Epac1 or YFP-RBD(RalGDS) were mounted in a culture chamber and imaged on a Leica TIRF setup equipped with a 488 argon excitation laser and a Hamamatsu EM charged-couple display detector. A ×63 magnification, 1.45 N.A. objective was used, and the evanescent field penetration depth was set to 90 nm. TIRF imaging was at ambient temperature, and analysis was performed with LAS-AF software.
Loading and flash photolysis of NPE-caged cAMP.
Cells were loaded by incubation with 100 μM NPE-caged cAMP for 15 min. Uncaging was done with brief pulses of UV light (340 to 410 nm) from a 100-W xenon mercury are lamp using a shutter. To define exposure times and UV light intensities for the desired cAMP release, cAMP was monitored ratiometrically (CFP/YFP) using the Epac-based sensor CFP-Epac(ΔDEP-C.D.)-Venus (55). The translocation of GFP-Epac1 was monitored in parallel experiments.
Rap activation assay.
Rap activity was assayed as described previously (58). Briefly, HEK293 cells grown in 9-cm plates were lysed in buffer containing 1% NP-40, 150 mM NaCl, 50 mM Tris-HCl (pH 7.4), 10% glycerol, 2 mM MgCl2, and protease and phosphatase inhibitors. Lysates were cleared by centrifugation, and active Rap was precipitated with a glutathione S-transferase fusion protein of the Ras-binding domain of RalGDS precoupled to glutathione-Sepharose beads.
Adhesion assay.
The adhesion of Jurkat T cells to fibronectin was measured as described previously (10). In brief, 96-well Nunc Maxisorp plates were coated with 5 μg/ml fibronectin and blocked with 1% bovine serum albumin. Jurkat cells (1.2 × 107) were transiently transfected by electroporation (950 μF, 250 V) with 5 μg cytomegalovirus-luciferase plasmid and pcDNA3 YFP-Epac1 plasmid adjusted to get equal expression levels and supplemented with pcDNA3 empty vector to a total of 40 μg plasmid DNA using a Gene Pulser II (Bio-Rad). Cells were harvested 2 days after transfection and resuspended in TSM buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM CaCl2, 2 mM MgCl2). Cells (2.5 × 104) for each well were allowed to adhere for 45 min, and nonadherent cells were removed with 0.5% BSA in TSM buffer. Adherent cells were lysed and subjected to a luciferase assay as described previously (33). For Rap1 activity measurements in Jurkat T cells, transfected cells were subjected to the Rap activation assay after resuspension in TSM buffer and stimulation with 007 (100 μM, 10 min).
RESULTS
cAMP induces translocation of Epac1 toward the PM.
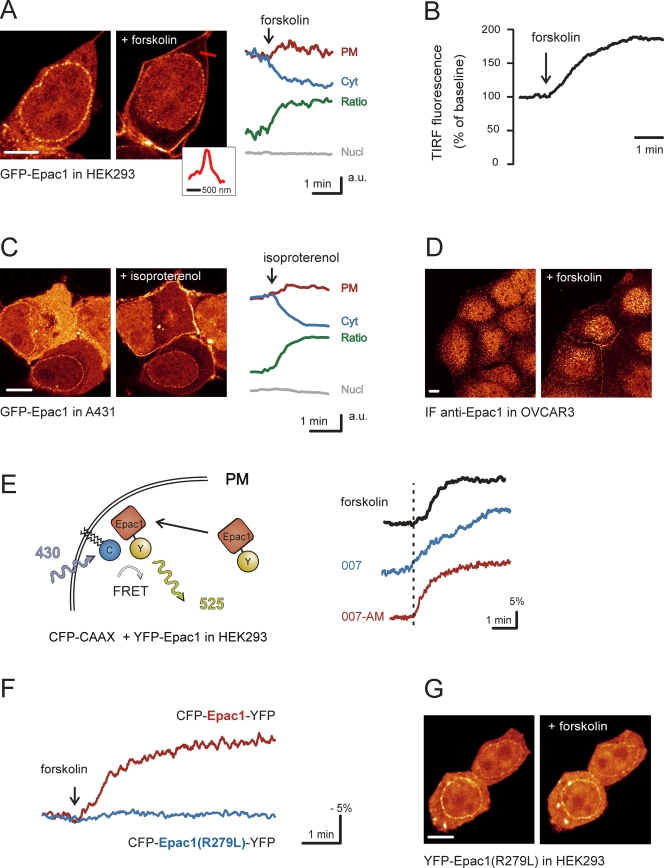
To study the subcellular localization of Epac1 during activation by cAMP, we monitored GFP-Epac1 using time-lapse confocal imaging in HEK293 cells. In accordance with previous reports (5, 11, 14, 29, 47), GFP-Epac1 is observed at the nuclear envelope, in the nucleus, at the PM, at endomembranes, and in the cytosol. Upon the addition of forskolin, which activates adenylate cyclase to produce cAMP, we observed a pronounced redistribution of GFP-Epac1 toward the periphery of the cell (Fig. 1A; also see the movie in the supplemental material). Automated image analysis (56) showed that the GFP-Epac1 redistribution is manifested both as a decrease in cytosolic fluorescence and an increase in PM-localized fluorescence, occurring within 2 min after stimulation (half time, ∼40 s) (Fig. 1A). In contrast, fluorescence in the nucleus was constant throughout the experiment (Fig. 1A). TIRF microscopy was employed to monitor GFP-Epac1 selectively at the basal membrane. The forskolin-induced accumulation of GFP-Epac1 was observed as an increase of 60% ± 4% (means ± standard errors of the means [SEM]; n = 17) relative to prestimulus levels (Fig. 1B), suggesting that Epac1 translocation represents a large-scale recruitment to the PM.
FIG. 1.
Epac1 translocates toward the PM upon elevation of cAMP levels. (A) In HEK293 cells, stimulation with forskolin (25 μM) induces the translocation of GFP-Epac1 toward the cell periphery. The inset shows fluorescence intensity along the red line, showing a sharp demarcation of the plasma membrane (width at half maximum, ∼300 nm). On the right, fluorescence intensity at the PM (red) and in the cytosol (Cyt) (blue) as well as the PM/Cyt ratio (green) during the response to forskolin is shown; fluorescence levels in the nucleus (Nucl) (gray) were constant. Traces are representative for n > 15. (B) Representative TIRF experiment showing the forskolin-induced accumulation of GFP-Epac1 at the basal membrane of HEK293 cells. The mean increase relative to prestimulus levels was 60% ± 4% (means ± SEM; n = 17). (C) A431 cells expressing GFP-Epac1 were imaged during isoproterenol (1 nM) stimulation. The translocation of GFP-Epac1 was observed within 1 min (n = 8). (D) Immunofluorescence of OVCAR-3 cells stained for endogenous Epac1. In resting cells, little PM localization of Epac1 is observed. After forskolin stimulation (25 μM, 10 min), the amount of PM-localized Epac1 is markedly increased. Note that the nuclear immunofluorescence is background staining rather than that of Epac1, as it is insensitive to the small interfering RNA-mediated silencing of Epac1 (data not shown). (E) Measurement of FRET between the PM marker CFP-CAAX (see Materials and Methods) and translocating YFP-tagged Epac1 (traces are representative for n > 5). FRET, expressed as a YFP/CFP ratio, increases upon the addition of forskolin (25 μM) (black). FRET increases also were induced by the Epac-specific cAMP analogue 007 (100 μM) (blue) or the more membrane-permeable analogue 007-AM (1 μM) (red). (F) Forskolin (25 μM) treatment of HEK293 cells transfected with the FRET sensor CFP-Epac1-YFP. The sensor reports the cAMP-induced conformational change as a loss of intramolecular FRET. In contrast to the wild-type sensor (red), the mutant FRET construct CFP-Epac1(R279L)-YFP (blue) lacks the ability to change conformation upon changing cAMP concentrations. (G) Mutagenesis of arginine 279 to leucine eliminated the ability of YFP-Epac1(R279L) to translocate upon forskolin stimulation (25 μM). Scale bars, 10 μm.
Similarly, isoproterenol stimulation, which induces physiological cAMP increases via the activation of β-adrenergic receptors, induced rapid Epac1 translocation in A431 cells (Fig. 1C). Epac1 translocation was observed in all cell types tested (HEK293, A431, OVCAR-3, ACHN, RCC10, MDCK, N1E-115, HeLa, Rat-1, GE11, and H1299) and across the full range of expression levels (data not shown). Importantly, the staining of OVCAR-3 cells with a monoclonal Epac1 antibody showed an increased presence of endogenous Epac1 at the PM after treatment with forskolin, reflecting the PM translocation of endogenous Epac1 (Fig. 1D). Similar results were obtained with a polyclonal Epac1 antibody (data not shown).
To further confirm the translocation of Epac to the PM, FRET was monitored between YFP-Epac1 and CFP-CAAX, which is membrane anchored by its prenylated K-Ras CAAX motif and, in these cells, constitutes a marker of the PM (see Materials and Methods). Forskolin induced immediate increases in the YFP/CFP ratio (10 to 15%) (Fig. 1E), indicating that translocated GFP-Epac1 approaches the PM to within approximately 7 nm. Similar results were obtained using a C-terminally tagged Epac1 (data not shown).
The translocation of Epac1 also can be induced by the Epac-specific cAMP analogue 8-pCPT-2′-O-Me-cAMP (007) and the more cell-permeable 8-pCPT-2′-O-Me-cAMP-AM (007-AM) (Fig. 1E), suggesting that cAMP induces Epac1 translocation through the activation of Epac1 itself rather than via parallel pathways such as that of PKA. Indeed, Epac1(R279L), which is mutated in the cAMP-binding domain and thereby locked in the autoinhibited, inactive conformation (47) (Fig. 1F), lacked the ability to translocate (Fig. 1G), indicating that Epac1 must be in its open conformation to translocate. In addition, the forskolin-induced translocation of GFP-Epac1 was not affected by the inhibition of PKA with H89 (data not shown). Thus, we show that Epac1 translocates toward the PM when it is bound by cAMP.
Epac1 translocation dynamically follows cAMP levels.
High-resolution confocal imaging shows that translocating GFP-Epac1 is recruited from a homogeneous cytosolic pool. Indeed, fast fluorescence recovery after photobleaching experiments (56) confirmed that the mobility of GFP-Epac1 approaches that of free GFP (data not shown). Conversely, active transport appears not to be involved, since Epac1 translocation is insensitive to the disruption of the actin cytoskeleton (with cytochalasin D [1 μg/ml] or latrunculin A [1 μM]) or the microtubule network (with nocodazole [25 ng/ml]) (data not shown). Thus, upon cAMP binding, Epac1 finds the PM by passive diffusion.
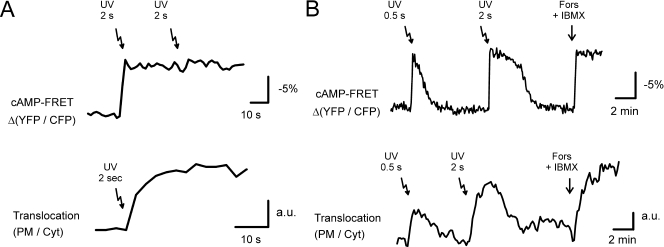
To examine the dynamics of Epac1 translocation, we loaded HEK293 cells with NPE-caged cAMP and transiently released cAMP by the UV-induced photolysis of the NPE cage. Using CFP-Epac(ΔDEP-CD)-Venus (55), an improved variant of the previously published cAMP sensor (44), we first established experimental conditions to instantly saturate Epac1 with cAMP (Fig. 2A). When applying identical UV pulses to cells expressing comparable levels of GFP-Epac1, we observed very rapid translocation that was halfway complete within 5 s and nearly complete in approximately 20 s (Fig. 2A).
FIG. 2.
Epac1 translocation is a highly dynamic and reversible event. (A) cAMP-uncaging experiments in HEK293 cells. The upper trace shows that the release of NPE-caged cAMP (arrows; see Materials and Methods for details) saturates the FRET-based cAMP sensor CFP-Epac(ΔDEP,C.D.)-Venus, in that a subsequent UV flash did not induce further FRET changes. The lower trace shows the ratio between PM and cytosolic fluorescence of GFP-Epac1 [expressed at levels comparable to those of CFP-Epac1(ΔDEP,C.D.)-Venus] showing the immediate translocation upon the identical photolysis of caged-cAMP (τ1/2 < 5 s). (B) cAMP uncaging in GE11 cells. The upper trace shows that due to the high speed of cAMP clearing in these cells, the dosed release of NPE-caged cAMP evokes transient cAMP rises. The amounts of released cAMP are approximately proportional to the duration of UV flashes. The lower trace shows that the release of identical amounts of caged cAMP in GE11 cells induces transient translocations of GFP-Epac1. The PM/cytosol ratio shows that the degree of translocation correlates with the dose of released cAMP.
To investigate whether the PM recruitment of Epac1 is a reversible event, we photoreleased NPE-caged cAMP in GE11 cells, which rapidly clear cAMP, likely due to high levels of PDE activity (43, 44). This allowed cAMP transients to be evoked repetitively, as detected by the FRET-based cAMP sensor (Fig. 2B). When similar amounts of cAMP were released in GE11 cells expressing GFP-Epac1, we observed rapid translocations to the PM followed by relocation to the cytosol (Fig. 2B). Analyses of the PM/cytosol ratio show that the translocation kinetics closely resemble the dynamic course of the cAMP levels. These experiments indicate that Epac1 translocation is a rapidly reversible event and that the momentary cAMP levels dictate the degree of PM localization.
Epac1 conformational change, rather than downstream signaling, is required for its translocation.
As the opening up of Epac1, which is essential for cAMP-induced translocation (Fig. 1G), also releases its catalytic activity, we examined whether downstream signals are required for its recruitment to the PM. As shown in Fig. 3A, the overexpression of RapGAP1 inhibits cAMP-induced Rap activation by keeping both Rap1 and Rap2 in the GDP-bound, inactive state. RapGAP1 overexpression did not affect the translocation of GFP-Epac1 (Fig. 3A, B), suggesting that Rap activity is not required. In line with this, the coexpression of constitutively active Rap1A(G12V) did not affect GFP-Epac1 localization in unstimulated cells (Fig. 3A), nor did it affect the magnitude or kinetics of the 007-AM-induced translocation (Fig. 3A, B). Furthermore, in OVCAR-3 cells, the cAMP-binding mutant Epac1(R279L) does not translocate when Rap is transiently activated through the activation of endogenous Epac1 (data not shown). Taken together, these data indicate that Epac1 translocation results from its conformational change rather than downstream signaling via Rap. This was further supported by using the CFP-Epac1-YFP probe, which allows the simultaneous visualization of localization as well as the conformational state via intramolecular FRET. These experiments showed that the kinetics of Epac1 translocation closely follow those of its conformational state (Fig. 3C).
The DEP domain is essential but not sufficient for Epac1 translocation.
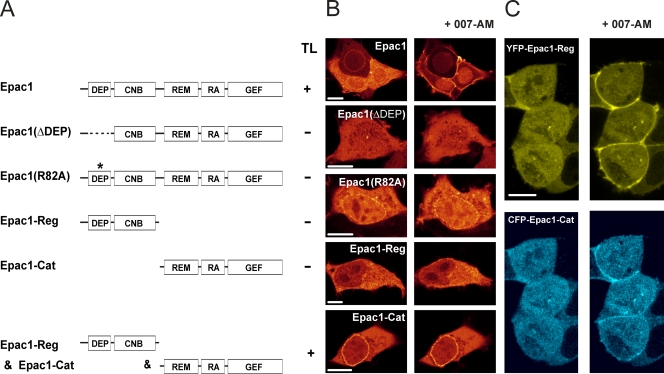
To determine which domains of Epac1 are involved in the translocation, a series of deletion mutants were analyzed (Fig. 4A). The removal of the DEP domain, which is essential for the proper intracellular targeting and functioning of DEP domain-containing proteins such as Dishevelled (40) and numerous RGS proteins (8, 20, 27, 31, 32), completely abolished Epac1 translocation (Fig. 4B). The mutation of arginine 82 within the DEP domain of Epac1, which localizes within a proposed interaction surface that is crucial for the DEP-mediated targeting of Dishevelled-1 and Ste2 (1, 40, 41, 61), also abolished the cAMP-induced translocation, further illustrating the critical role of the DEP domain in mediating the cAMP-induced translocation. Nonetheless, GFP-DEP (comprising amino acids 50 to 148) was not observed at the PM (data not shown). Similarly, the YFP-tagged regulatory region of Epac1 (YFP-Epac1-Reg), comprising the DEP domain and the cAMP-binding domain, did not localize at the PM, nor did it translocate to the PM upon cAMP elevation (Fig. 4A, B). However, the application of 007-AM to cells transfected with both YFP-Epac1-Reg and the CFP-tagged complementary catalytic region of Epac1 (CFP-Epac1-Cat), which are able to reconstitute the structure of the full-length protein (data not shown), induced the combined translocation of both fragments (Fig. 4C). This demonstrates that the DEP domain is required for the translocation of Epac1 to the PM, but that it can function only in conjunction with the catalytic region.
FIG. 4.
DEP domain is essential but not sufficient for Epac1 translocation. (A and B) Overview of Epac1 mutants and their abilities to translocate (TL) upon the addition of 007-AM (1 μM). Full-length Epac1 consists of a DEP domain (amino acids 50 to 148), the cyclic nucleotide-binding domain (CNB), the Ras exchange motive (REM), a putative RA domain, and the catalytic CDC25 homology GEF domain. The removal of the DEP domain (amino acids 50 to 148) or the disruption of the interaction surface within the DEP domain (R82A) abolishes the translocation of Epac1 in response to 007-AM. The separate regulatory region (YFP-Epac1-Reg; amino acids 1 to 328), containing both the DEP domain and the cAMP-binding domain (CNB), did not translocate after 007-AM stimulation, indicating that the DEP domain cannot mediate the localization of Epac1 at the PM in the absence of the catalytic region. In accordance with the crucial role of the DEP domain, the complete catalytic region (CFP-Epac1-Cat; amino acids 330 to 881) was not present at the PM either. (C) In contrast to the separately expressed regulatory and catalytic region, the coexpression of YFP-Epac-Reg (upper images) and CFP-Epac-Cat (lower images) restored the 007-AM-induced translocation, resulting in the colocalization of both constructs at the PM. Scale bars, 10 μm.
Epac1 translocation enhances Rap-dependent cell adhesion.
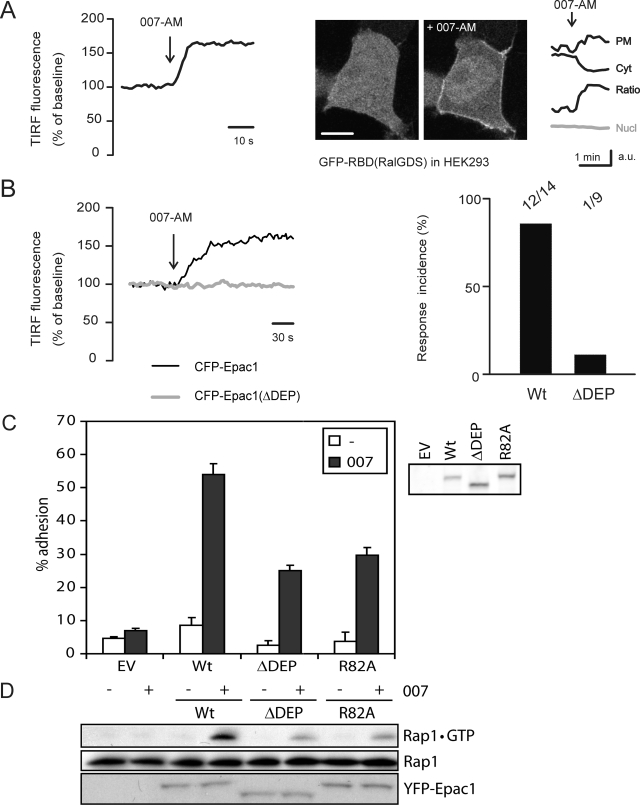
As the main pool of Epac1 redistributes to the PM after its activation, we explored whether the translocation of Epac1 is a prerequisite for the activation of Rap at this compartment. For this, HEK293 cells were transfected with the YFP-tagged Ras-binding domain of RalGDS [YFP-RBD(RalGDS)], which recognizes Rap1 specifically in its GTP-bound, activated state (4). When cells were cotransfected with hemagglutin-Epac1 (HA-Epac1), the addition of 007-AM resulted in the rapid accumulation of YFP-RBD(RalGDS) at the PM, as visualized both by TIRF (Fig. 5A, left) and confocal microscopy (Fig. 5A, right). Interestingly, such accumulation was not observed in other subcellular compartments, suggesting that in HEK293 cells cAMP signaling via Epac1 activates Rap predominantly at the PM (Fig. 5A). To test for the role of Epac1 translocation in Rap activation at the PM, we analyzed YFP-RBD(RalGDS) membrane recruitment in cells expressing either CFP-Epac1 or CFP-Epac1(ΔDEP) by TIRF microscopy. 007-AM induced the recruitment of the probe to the basal membrane in cells expressing CFP-Epac1 (12 of 14 cells), whereas this was almost absent in cells coexpressing CFP-Epac1(ΔDEP) (one of nine cells) (P ≪ 0.01) (Fig. 5B). Thus, the translocation of Epac1 is required for Rap activation at the PM.
FIG. 5.
Translocation of Epac1 enhances Rap activation and Rap-mediated adhesion of Jurkat T cells. (A) The trace on the left shows the in vivo TIRF imaging of Rap1 activation in HEK293 cells using YFP-RalGDS(RBD). When coexpressed with HA-Epac1, YFP-RalGDS(RBD) translocates to the basal membrane within seconds after 007-AM stimulation (1 μM). The 007-AM-induced increase was 46% ± 6% (means ± SEM) relative to prestimulus levels (n = 9). The images on the right are confocal pictures of YFP-RalGDS(RBD) translocation, showing accumulation at the PM as well as the simultaneous depletion of cytosol (Cyt); nuclear fluorescence was constant (Nucl) (gray). The accumulation of YFP-RalGDS(RBD) was not observed on intracellular membranes. Scale bar, 10 μm. (B) HEK293 cells were transfected with CFP-Epac1 or CFP-Epac1(ΔDEP), and the recruitment of cotransfected YFP-RBD(RalGDS) to the basal membrane was measured by TIRF microscopy. Cells expressing low levels of CFP- and YFP-tagged proteins were selected. Traces are from representative experiments. The bar graph shows the relative occurrence of the 007-AM-induced membrane accumulation of YFP-RBD(RalGDS): for CFP-Epac1, 12 out of 14 cells; for CFP-Epac1(ΔDEP), 1 out of 9 cells. (C) Jurkat T cells were transfected with either wild-type Epac1 or the nontranslocating Epac1 mutants [Epac1(ΔDEP) and Epac1(R82A)] together with a luciferase reporter. Transfected cells were allowed to adhere to a fibronectin-coated surface for 45 min, and adhesion subsequently was detected as luciferase emission. Wild-type Epac1, when activated by 007 (100 μM) (black bars), greatly enhanced the adhesion compared to that of unstimulated conditions (white bars). This effect was impaired when the translocation-deficient mutant Epac1(ΔDEP) or Epac1(R82A) was transfected, and it was absent in empty vector-transfected cells (EV). Shown are data from a representative experiment performed in triplicate (n = 4). Total luciferase levels were comparable in all transfections. On the right is a Western blot labeled with the Epac1 antibody (5D3) showing the expression levels of the transfected wild-type and mutant Epac1 used in the adhesion assay. (D) Jurkat T cells were transfected with similar amounts of either wild-type Epac1 or the translocation-deficient mutants Epac1(ΔDEP) and Epac1(R82A), and GTP-bound Rap1 was pulled down from lysates of cells after stimulation with 007 (100 μM, 10 min).
Epac1-Rap signaling is involved in integrin-mediated cell adhesion by regulating both the affinity and avidity of actin-associated integrin molecules (6). For Jurkat T cells, it has been shown that this requires the presence of active Rap at the PM (4). To study the role of Epac1 translocation in its ability to mediate integrin regulation, we measured the adhesion of Jurkat T cells in response to 007. For this, the cells were transfected with luciferase together with either wild-type Epac1 or the nontranslocating Epac1 variants mutated in their DEP domain, and adhesion was quantified by the detection of luciferase emission. Indeed, 007 induced strong adhesion to the fibronectin of wild-type Epac1-transfected cells (Fig. 5C). In contrast, this cAMP-induced effect on cell adhesion was impaired when the translocation-deficient mutant Epac1(ΔDEP) or Epac1(R82A) was expressed (Fig. 5C). Rap1GTP pulldown experiments in Jurkat T cells show that these mutants do indeed result in reduced Rap1 activation compared to that of wild-type Epac1 (Fig. 5D), implying that a significant fraction of Rap1 resides at the PM in these cells. These data indicate that the translocation of Epac1 significantly enhances the signaling cascade toward Rap-mediated cell adhesion.
DISCUSSION
In the current study, we have shown that cAMP induces the translocation of cytosolic Epac1 toward the PM. Epac1 translocation is a generally occurring, physiological event, as it was observed in a wide array of cell types, both with overexpressed and endogenous Epac1 (Fig. 1). The translocation depends solely on the cAMP-induced conformational state of Epac1, since it could be induced by the Epac-selective analogues 007 and 007-AM and was prevented by a mutation in the cAMP-binding pocket (R279L) (Fig. 1F, G). Furthermore, since neither the activation nor inhibition of Rap could affect Epac1 translocation, the involvement of Rap-mediated signaling could be excluded (Fig. 3). Finally, the degree of Epac1 translocation closely followed the levels of free cAMP within the cells and showed kinetics similar to those of the cAMP-induced conformational change (Fig. 2 and 3C). Thus, in addition to releasing Epac1 from autoinhibition, the direct binding of cAMP also regulates the translocation of Epac1 toward the PM.
Epac1 translocation is based on passive diffusion, since fluorescence distribution in the cytosol is homogeneous throughout the translocation without discernible discrete moving structures that would indicate active transport. In line with the diffusion model, Epac1 translocation shows rapid and reversible kinetics after cAMP uncaging (Fig. 2) and is not affected by the disruption of the actin cytoskeleton or the microtubule network (data not shown). These data imply that upon transition to its opened conformation, Epac1 acquires an affinity for an anchoring factor at the PM, to which it subsequently is targeted via passive diffusion.
Deletion and point mutations have indicated the DEP domain as an essential determinant of translocation. This is analogous to the targeting function of DEP domains in other proteins such as Dishevelled and RGS (1, 8, 27, 31, 40). However, crystal structure studies of Epac2 (52) suggest that the DEP domain is solvent exposed regardless of cAMP binding. Therefore, a model wherein the cAMP-induced conformational change renders the DEP domain accessible is unlikely. Indeed, the separate DEP domain or regulatory region of Epac1 did not localize to the PM (Fig. 4). Thus, the DEP domain can fulfill its function only in the context of the structure of the full-length protein. The interaction surface for PM anchoring thus is established by the combined structural features of the DEP domain and a determinant in the catalytic region, which are dependent on the cAMP-bound conformation. The identification of the membrane anchor likely would help to define the underlying structural mechanism.
Many downstream effects of Epac1 occur at the PM: cell adhesion, cell-cell junction formation, and the regulation of NHE3. These effects may require the localized activation of Epac1 and, thereby, the localized activation of Rap. Indeed, we showed that the translocation of Epac1 is a prerequisite for Rap activation (Fig. 5B). Furthermore, translocation strongly enhances Rap-mediated cell adhesion, as the translocation-deficient mutants Epac1(ΔDEP) and Epac1(R82A) were impaired in their ability to induce the Rap-dependent adhesion of Jurkat T cells to fibronectin. It is important to note here that purified Epac1(Δ1-148) mediates GDP dissociation from Rap1 in vitro equally as well as full-length Epac1 does (28), indicating that the DEP domain is not required for the catalytic activity. The residual effect of the Epac1 mutants on cell adhesion may be due to the relatively high expression of Epac1 in this system, allowing a fraction of the mutant Epac1 to locate near the PM regardless of the DEP-dependent translocation, thereby activating Rap. Indeed, TIRF experiments showed the membrane recruitment of YFP-RBD(RalGDS) in cells expressing CFP-Epac1(ΔDEP) at relatively high levels (data not shown) but not at lower levels (Fig. 5B).
Recently, the DEP domain of Epac1 has been shown to be essential for the ability of the regulatory region to disrupt TSH-mediated mitogenesis, further supporting the notion that the proper localization of Epac1 via its DEP domain is required for its function (18). Different anchors may target Epac1 to other cellular compartments and thereby regulate alternative functions of Epac1 that are not linked to (processes at) the PM. Indeed, in cardiomyocytes Epac1 was found in a complex with muscle-specific mAKAP, PDE4A, and PKA to regulate ERK5 activity (13). Epac1 also is present at the nuclear membrane (Fig. 1A), and this binding is maintained during the early time points of cAMP stimulation, indicating that only a subfraction of Epac1 translocates to the PM. Thus, in this respect Epac1 resembles PKA, which is targeted to distinct subcellular compartments through the binding to AKAPs.
PM localization also has been reported to be essential for signaling via Epac2, which is mediated by the binding of its Ras association (RA) domain to activated Ras (29, 30). Originally this was proposed to be regulated by cAMP (29). However, the binding of Epac2 to Ras does not require the open conformation of Epac2 and is not affected by cAMP (30 and our own unpublished data). In addition, the expression of an active Ras mutant suffices for targeting Epac2 to the PM (29, 30). Conversely, the deletion of the putative RA domain within Epac1 does not affect its cAMP-induced translocation, as demonstrated by increased FRET between CFP-CAAX and a YFP-Epac1 mutant lacking the RA domain (data not shown). These data exclude the possibility that the RA domain is the missing determinant within the catalytic region of Epac1. The different mechanisms of PM targeting distinguish the roles of Epac1 and Epac2, which may add to the understanding of their specific biological functions.
Based on our data, we propose a model in which the binding of cAMP regulates Epac1 in two manners: it targets Epac1 toward the PM and simultaneously releases the activity of its GEF domain. Such dual regulation imposes signal specificity by guaranteeing that cAMP predominantly affects PM-localized Rap molecules (Fig. 5A), whereas, for example, growth factors such as epidermal growth factor activate a perinuclear pool of Rap (34). Analogously, the negative regulation of Rap by GAPs also may be spatially confined, as it has been reported that a PM pool of RapGAP restricts Rap activation in COS1 cells (36). Thus, it appears that the localization of GEFs and GAPs rather than that of the G protein itself determines the intracellular location of G protein activity.
We can only speculate as to why the activation of Epac1 is dynamically regulated by the simultaneous cAMP-dependent translocation rather than by more static confinement via stable association to the PM. The translocation mechanism may serve to dynamically regulate the availability of Epac1 throughout the cell. In addition, the separation between Epac1 and Rap in resting conditions may be an ultimate guarantee against the stimulation of Rap by residual Epac1 activity. Adding a spatial component to the cAMP-mediated regulation of Epac1 undoubtedly renders the transition from unstimulated to stimulated Rap at the PM more pronounced.
Supplementary Material
Acknowledgments
We thank the members of our laboratories and J. de Rooij for support and stimulating discussions, W. Moolenaar, B. Burgering, and F. Zwartkruis for critical reading of the manuscript, M. Langeslag for help with TIRF experiments, and Y. Zhou for the generation of constructs.
This study was supported by Chemical Sciences (M.G.) and The Netherlands Genomics Initiative (J.L.B.) of The Netherlands Organization for Scientific Research (NWO) and by a material grant from ZonMW. H.R. is a recipient of the Hendrik Casimir-Karl Ziegler-Forschungspreis of the Nordrhein-Westfälischen Akademie der Wissenschaften and the Koninklijke Nederlandse Akademie van Wetenschappen.
Footnotes
Published ahead of print on 9 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ballon, D. R., P. L. Flanary, D. P. Gladue, J. B. Konopka, H. G. Dohlman, and J. Thorner. 2006. DEP-domain-mediated regulation of GPCR signaling responses. Cell 1261079-1093. [DOI] [PubMed] [Google Scholar]
- 2.Benetka, W., M. Koranda, S. Maurer-Stroh, F. Pittner, and F. Eisenhaber. 2006. Farnesylation or geranylgeranylation? Efficient assays for testing protein prenylation in vitro and in vivo. BMC Biochem. 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beranger, F., B. Goud, A. Tavitian, and J. de Gunzburg. 1991. Association of the Ras-antagonistic Rap1/Krev-1 proteins with the Golgi complex. Proc. Natl. Acad. Sci. USA 881606-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bivona, T. G., H. H. Wiener, I. M. Ahearn, J. Silletti, V. K. Chiu, and M. R. Philips. 2004. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 164461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borland, G., M. Gupta, M. M. Magiera, C. J. Rundell, S. Fuld, and S. J. Yarwood. 2006. Microtubule-associated protein 1B-light chain 1 enhances activation of Rap1 by exchange protein activated by cyclic AMP but not intracellular targeting. Mol. Pharmacol. 69374-384. [DOI] [PubMed] [Google Scholar]
- 6.Bos, J. L. 2005. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17123-128. [DOI] [PubMed] [Google Scholar]
- 7.Bos, J. L., H. Rehmann, and A. Wittinghofer. 2007. GEFs and GAPs: critical elements in the control of small G proteins. Cell 129865-877. [Erratum, 130:385.] [DOI] [PubMed] [Google Scholar]
- 8.Cabrera-Vera, T. M., S. Hernandez, L. R. Earls, M. Medkova, A. K. Sundgren-Andersson, D. J. Surmeier, and H. E. Hamm. 2004. RGS9-2 modulates D2 dopamine receptor-mediated Ca2+ channel inhibition in rat striatal cholinergic interneurons. Proc. Natl. Acad. Sci. USA 10116339-16344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullere, X., S. K. Shaw, L. Andersson, J. Hirahashi, F. W. Luscinskas, and T. N. Mayadas. 2005. Regulation of vascular endothelial barrier function by Epac, a cAMP-activated exchange factor for Rap GTPase. Blood 1051950-1955. [DOI] [PubMed] [Google Scholar]
- 10.de Bruyn, K. M., S. Rangarajan, K. A. Reedquist, C. G. Figdor, and J. L. Bos. 2002. The small GTPase Rap1 is required for Mn2+- and antibody-induced LFA-1- and VLA-4-mediated cell adhesion. J. Biol. Chem. 27729468-29476. [DOI] [PubMed] [Google Scholar]
- 11.de Rooij, J., H. Rehmann, M. van Triest, R. H. Cool, A. Wittinghofer, and J. L. Bos. 2000. Mechanism of regulation of the Epac family of cAMP-dependent RapGEFs. J. Biol. Chem. 27520829-20836. [DOI] [PubMed] [Google Scholar]
- 12.de Rooij, J., F. J. Zwartkruis, M. H. Verheijen, R. H. Cool, S. M. Nijman, A. Wittinghofer, and J. L. Bos. 1998. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396474-477. [DOI] [PubMed] [Google Scholar]
- 13.Dodge-Kafka, K. L., J. Soughayer, G. C. Pare, J. J. Carlisle Michel, L. K. Langeberg, M. S. Kapiloff, and J. D. Scott. 2005. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature 437574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dremier, S., M. Milenkovic, S. Blancquaert, J. E. Dumont, S. O. Doskeland, C. Maenhaut, and P. P. Roger. 2007. Cyclic adenosine 3′,5′-monophosphate (cAMP)-dependent protein kinases, but not exchange proteins directly activated by cAMP (Epac), mediate thyrotropin/cAMP-dependent regulation of thyroid cells. Endocrinology 1484612-4622. [DOI] [PubMed] [Google Scholar]
- 15.Enserink, J. M., A. E. Christensen, J. de Rooij, M. van Triest, F. Schwede, H. G. Genieser, S. O. Doskeland, J. L. Blank, and J. L. Bos. 2002. A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK. Nat. Cell Biol. 4901-906. [DOI] [PubMed] [Google Scholar]
- 16.Fivaz, M., and T. Meyer. 2005. Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J. Cell Biol. 170429-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuhara, S., A. Sakurai, H. Sano, A. Yamagishi, S. Somekawa, N. Takakura, Y. Saito, K. Kangawa, and N. Mochizuki. 2005. Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signaling pathway. Mol. Cell. Biol. 25136-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hochbaum, D., K. Hong, G. Barila, F. Ribeiro-Neto, and D. L. Altschuler. 2008. Epac, in synergy with cAMP-dependent protein kinase (PKA), is required for cAMP-mediated mitogenesis. J. Biol. Chem. 2834464-4468. [DOI] [PubMed] [Google Scholar]
- 19.Honegger, K. J., P. Capuano, C. Winter, D. Bacic, G. Stange, C. A. Wagner, J. Biber, H. Murer, and N. Hernando. 2006. Regulation of sodium-proton exchanger isoform 3 (NHE3) by PKA and exchange protein directly activated by cAMP (EPAC). Proc. Natl. Acad. Sci. USA 103803-808. [Erratum, 103:4328.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt, R. A., W. Edris, P. K. Chanda, B. Nieuwenhuijsen, and K. H. Young. 2003. Snapin interacts with the N-terminus of regulator of G protein signaling 7. Biochem. Biophys. Res. Commun. 303594-599. [DOI] [PubMed] [Google Scholar]
- 21.Huston, E., M. J. Lynch, A. Mohamed, D. M. Collins, E. V. Hill, R. MacLeod, E. Krause, G. S. Baillie, and M. D. Houslay. 2008. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc. Natl. Acad. Sci. USA 10512791-12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko, M., and T. Takahashi. 2004. Presynaptic mechanism underlying cAMP-dependent synaptic potentiation. J. Neurosci. 245202-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashima, Y., T. Miki, T. Shibasaki, N. Ozaki, M. Miyazaki, H. Yano, and S. Seino. 2001. Critical role of cAMP-GEFII-Rim2 complex in incretin-potentiated insulin secretion. J. Biol. Chem. 27646046-46053. [DOI] [PubMed] [Google Scholar]
- 24.Kawasaki, H., G. M. Springett, N. Mochizuki, S. Toki, M. Nakaya, M. Matsuda, D. E. Housman, and A. M. Graybiel. 1998. A family of cAMP-binding proteins that directly activate Rap1. Science 2822275-2279. [DOI] [PubMed] [Google Scholar]
- 25.Kooistra, M. R., M. Corada, E. Dejana, and J. L. Bos. 2005. Epac1 regulates integrity of endothelial cell junctions through VE-cadherin. FEBS Lett. 5794966-4972. [DOI] [PubMed] [Google Scholar]
- 26.Kooistra, M. R., N. Dube, and J. L. Bos. 2007. Rap1: a key regulator in cell-cell junction formation. J. Cell Science 12017-22. [DOI] [PubMed] [Google Scholar]
- 27.Kovoor, A., P. Seyffarth, J. Ebert, S. Barghshoon, C. K. Chen, S. Schwarz, J. D. Axelrod, B. N. Cheyette, M. I. Simon, H. A. Lester, and J. Schwarz. 2005. D2 dopamine receptors colocalize regulator of G-protein signaling 9-2 (RGS9-2) via the RGS9 DEP domain, and RGS9 knock-out mice develop dyskinesias associated with dopamine pathways. J. Neurosci. 252157-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraemer, A., H. R. Rehmann, R. H. Cool, C. Theiss, J. de Rooij, J. L. Bos, and A. Wittinghofer. 2001. Dynamic interaction of cAMP with the Rap guanine-nucleotide exchange factor Epac1. J. Mol. Biol. 3061167-1177. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., S. Asuri, J. F. Rebhun, A. F. Castro, N. C. Paranavitana, and L. A. Quilliam. 2006. The RAP1 guanine nucleotide exchange factor Epac2 couples cyclic AMP and Ras signals at the plasma membrane. J. Biol. Chem. 2812506-2514. [DOI] [PubMed] [Google Scholar]
- 30.Liu, C., M. Takahashi, Y. Li, S. Song, T. J. Dillon, U. Shinde, and P. J. Stork. 2008. Ras is required for the cyclic AMP-dependent activation of Rap1 via Epac2. Mol. Cell. Biol. 287109-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martemyanov, K. A., P. V. Lishko, N. Calero, G. Keresztes, M. Sokolov, K. J. Strissel, I. B. Leskov, J. A. Hopp, A. V. Kolesnikov, C. K. Chen, J. Lem, S. Heller, M. E. Burns, and V. Y. Arshavsky. 2003. The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. J. Neurosci. 2310175-10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martemyanov, K. A., P. J. Yoo, N. P. Skiba, and V. Y. Arshavsky. 2005. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J. Biol. Chem. 2805133-5136. [DOI] [PubMed] [Google Scholar]
- 33.Medema, R. H., W. L. de Laat, G. A. Martin, F. McCormick, and J. L. Bos. 1992. GTPase-activating protein SH2-SH3 domains induce gene expression in a Ras-dependent fashion. Mol. Cell. Biol. 123425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mochizuki, N., S. Yamashita, K. Kurokawa, Y. Ohba, T. Nagai, A. Miyawaki, and M. Matsuda. 2001. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 4111065-1068. [DOI] [PubMed] [Google Scholar]
- 35.Morel, E., A. Marcantoni, M. Gastineau, R. Birkedal, F. Rochais, A. Garnier, A. M. Lompre, G. Vandecasteele, and F. Lezoualc'h. 2005. cAMP-binding protein Epac induces cardiomyocyte hypertrophy. Circ. Res. 971296-1304. [DOI] [PubMed] [Google Scholar]
- 36.Ohba, Y., K. Kurokawa, and M. Matsuda. 2003. Mechanism of the spatio-temporal regulation of Ras and Rap1. EMBO J. 22859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohba, Y., N. Mochizuki, K. Matsuo, S. Yamashita, M. Nakaya, Y. Hashimoto, M. Hamaguchi, T. Kurata, K. Nagashima, and M. Matsuda. 2000. Rap2 as a slowly responding molecular switch in the Rap1 signaling cascade. Mol. Cell. Biol. 206074-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Neill, J. S., E. S. Maywood, J. E. Chesham, J. S. Takahashi, and M. H. Hastings. 2008. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320949-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozaki, N., T. Shibasaki, Y. Kashima, T. Miki, K. Takahashi, H. Ueno, Y. Sunaga, H. Yano, Y. Matsuura, T. Iwanaga, Y. Takai, and S. Seino. 2000. cAMP-GEFII is a direct target of cAMP in regulated exocytosis. Nat. Cell Biol. 2805-811. [DOI] [PubMed] [Google Scholar]
- 40.Pan, W. J., S. Z. Pang, T. Huang, H. Y. Guo, D. Wu, and L. Li. 2004. Characterization of function of three domains in dishevelled-1: DEP domain is responsible for membrane translocation of dishevelled-1. Cell Res. 14324-330. [DOI] [PubMed] [Google Scholar]
- 41.Penton, A., A. Wodarz, and R. Nusse. 2002. A mutational analysis of dishevelled in Drosophila defines novel domains in the dishevelled protein as well as novel suppressing alleles of axin. Genetics 161747-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pizon, V., M. Desjardins, C. Bucci, R. G. Parton, and M. Zerial. 1994. Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J. Cell Sci. 1071661-1670. [DOI] [PubMed] [Google Scholar]
- 43.Ponsioen, B., L. van Zeijl, W. H. Moolenaar, and K. Jalink. 2007. Direct measurement of cyclic AMP diffusion and signaling through connexin43 gap junctional channels. Exp. Cell Res. 313415-423. [DOI] [PubMed] [Google Scholar]
- 44.Ponsioen, B., J. Zhao, J. Riedl, F. Zwartkruis, G. van der Krogt, M. Zaccolo, W. H. Moolenaar, J. L. Bos, and K. Jalink. 2004. Detecting cAMP-induced Epac activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO Rep. 51176-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poulouin, L., O. Gallet, M. Rouahi, and J. M. Imhoff. 1999. Plasma fibronectin: three steps to purification and stability. Protein Expr. Purif. 17146-152. [DOI] [PubMed] [Google Scholar]
- 46.Price, L. S., A. Hajdo-Milasinovic, J. Zhao, F. J. Zwartkruis, J. G. Collard, and J. L. Bos. 2004. Rap1 regulates E-cadherin-mediated cell-cell adhesion. J. Biol. Chem. 27935127-35132. [DOI] [PubMed] [Google Scholar]
- 47.Qiao, J., F. C. Mei, V. L. Popov, L. A. Vergara, and X. Cheng. 2002. Cell cycle-dependent subcellular localization of exchange factor directly activated by cAMP. J. Biol. Chem. 27726581-26586. [DOI] [PubMed] [Google Scholar]
- 48.Quinn, M. T., M. L. Mullen, A. J. Jesaitis, and J. G. Linner. 1992. Subcellular distribution of the Rap1A protein in human neutrophils: colocalization and cotranslocation with cytochrome b559. Blood 791563-1573. [PubMed] [Google Scholar]
- 49.Rangarajan, S., J. M. Enserink, H. B. Kuiperij, J. de Rooij, L. S. Price, F. Schwede, and J. L. Bos. 2003. Cyclic AMP induces integrin-mediated cell adhesion through Epac and Rap1 upon stimulation of the beta 2-adrenergic receptor. J. Cell Biol. 160487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reedquist, K. A., E. Ross, E. A. Koop, R. M. Wolthuis, F. J. Zwartkruis, Y. van Kooyk, M. Salmon, C. D. Buckley, and J. L. Bos. 2000. The small GTPase, Rap1, mediates CD31-induced integrin adhesion. J. Cell Biol. 1481151-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rehmann, H., E. Arias-Palomo, M. A. Hadders, F. Schwede, O. Llorca, and J. L. Bos. 2008. Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature 455124-127. [DOI] [PubMed] [Google Scholar]
- 52.Rehmann, H., J. Das, P. Knipscheer, A. Wittinghofer, and J. L. Bos. 2006. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature 439625-628. [DOI] [PubMed] [Google Scholar]
- 53.Sakaba, T., and E. Neher. 2003. Direct modulation of synaptic vesicle priming by GABA(B) receptor activation at a glutamatergic synapse. Nature 424775-778. [DOI] [PubMed] [Google Scholar]
- 54.Somekawa, S., S. Fukuhara, Y. Nakaoka, H. Fujita, Y. Saito, and N. Mochizuki. 2005. Enhanced functional gap junction neoformation by protein kinase A-dependent and Epac-dependent signals downstream of cAMP in cardiac myocytes. Circ. Res. 97655-662. [DOI] [PubMed] [Google Scholar]
- 55.van der Krogt, G. N., J. Ogink, B. Ponsioen, and K. Jalink. 2008. A comparison of donor-acceptor pairs for genetically encoded FRET sensors: application to the Epac cAMP sensor as an example. PLoS ONE 3e1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Wal, J., R. Habets, P. Varnai, T. Balla, and K. Jalink. 2001. Monitoring agonist-induced phospholipase C activation in live cells by fluorescence resonance energy transfer. J. Biol. Chem. 27615337-15344. [DOI] [PubMed] [Google Scholar]
- 57.van Rheenen, J., and K. Jalink. 2002. Agonist-induced PIP(2) hydrolysis inhibits cortical actin dynamics: regulation at a global but not at a micrometer scale. Mol. Biol. Cell 133257-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Triest, M., and J. L. Bos. 2004. Pull-down assays for guanoside 5′-triphosphate-bound Ras-like guanosine 5′-triphosphatases. Methods Mol. Biol. 25097-102. [DOI] [PubMed] [Google Scholar]
- 59.Vliem, M., B. Ponsioen, F. Schwede, W. Pannekoek, J. Riedl, M. Kooistra, K. Jalink, H. Genieser, J. Bos, and H. Rehmann. 2008. 8-pCPT-2′-O-Me-cAMP-AM: an improved Epac-selective cAMP analogue. Chembiochem 92052-2054. [DOI] [PubMed] [Google Scholar]
- 60.Wittchen, E. S., R. A. Worthylake, P. Kelly, P. J. Casey, L. A. Quilliam, and K. Burridge. 2005. Rap1 GTPase inhibits leukocyte transmigration by promoting endothelial barrier function. J. Biol. Chem. 28011675-11682. [DOI] [PubMed] [Google Scholar]
- 61.Wong, H. C., J. Mao, J. T. Nguyen, S. Srinivas, W. Zhang, B. Liu, L. Li, D. Wu, and J. Zheng. 2000. Structural basis of the recognition of the dishevelled DEP domain in the Wnt signaling pathway. Nat. Struct. Biol. 71178-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yarwood, S. J. 2005. Microtubule-associated proteins (MAPs) regulate cAMP signalling through exchange protein directly activated by cAMP (EPAC). Biochem. Soc. Trans. 331327-1329. [DOI] [PubMed] [Google Scholar]
- 63.Zhong, N., and R. S. Zucker. 2005. cAMP acts on exchange protein activated by cAMP/cAMP-regulated guanine nucleotide exchange protein to regulate transmitter release at the crayfish neuromuscular junction. J. Neurosci. 25208-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.