Abstract
To maintain intracellular redox homeostasis, genes encoding many antioxidants and detoxification enzymes are transcriptionally upregulated upon deleterious oxidative stress through the cis antioxidant responsive elements (AREs) in their promoter regions. Nrf2 is the critical transcription factor responsible for ARE-dependent transcription. We and others have previously demonstrated that Nrf2 is targeted for ubiquitin-mediated degradation by Keap1 in a redox-sensitive manner through modifications of distinct cysteine residues of Keap1. Here, we report that p300/CBP directly acetylates Nrf2 in response to arsenite-induced stress. We have identified multiple acetylated lysine residues within the Nrf2 Neh1 DNA-binding domain. Combined lysine-to-arginine mutations on the acetylation sites, with no effects on Nrf2 protein stability, compromised the DNA-binding activity of Nrf2 in a promoter-specific manner. These findings demonstrated that acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 and established acetylation as a novel regulatory mechanism that functions in concert with Keap1-mediated ubiquitination in modulating the Nrf2-dependent antioxidant response.
Oxidative stress, known as adverse effects of oxidants on physiological functions, has long been shown to play important roles in both acute toxicity induced by many environmental insults and the pathogenesis of cancer, neurodegenerative disorders, and other aging-related diseases (27, 31, 48, 51, 64). The cause of oxidative stress is the production of reactive oxygen species exceeding the capacity of the body's natural antioxidant defense mechanisms. Understanding the molecular signaling events that govern these antioxidant defense mechanisms is critical for antioxidant therapeutic intervention (9).
Nrf2 is a critical transcription factor that regulates endogenous antioxidants, phase II detoxification enzymes, and other cellular defensive proteins through the antioxidant responsive elements (AREs) in the promoter regions of these genes (42, 43, 63). Among the best-characterized Nrf2 downstream genes are those encoding NAD(P)H quinone oxidoreductase 1 (NQO1), heme oxygenase 1 (HO-1), thioredoxin reductase 1 (TXNRD1), glutathione S-transferase A1 (GSTA1, also known as GST Ya in mice), and glutamate-cysteine ligase (also known as γ-glutamylcysteine synthetase) modifier subunit (GCLM) and catalytic subunit (GCLC) (2, 37, 40, 60). A growing body of evidence has established the Nrf2-dependent antioxidant response as a pivotal protection system against the detrimental effects of many environmental insults. Nrf2 knockout mice display increased sensitivity to chemical toxicants and carcinogens and are refractory to the protective actions of chemopreventive compounds (5, 46).
The activity of Nrf2 is negatively regulated by Keap1 (24). We and others have shown that under redox homeostasis conditions, Keap1 constitutively targets Nrf2 for ubiquitin conjugation and subsequent proteasome degradation in the cytoplasm by acting as a substrate adaptor for the Cul3-based E3 ubiquitin ligase complex (8, 14, 32, 68). Upon exposure of cells to oxidative stress or chemopreventive compounds, multiple cysteine residues on Keap1 are thought to be alkylated with electrophilic groups present in many Nrf2 inducers, such as tert-butylhydroquinone and sulforophane (10, 12, 19, 61, 67). Such alkylations compromise the ability of Keap1 to efficiently ubiquitinate Nrf2, resulting in elevated Nrf2 protein levels and enhanced Nrf2-dependent gene expression (8, 14, 32, 57, 66, 68, 69).
Within the nucleus, Nrf2 is believed to exert its transcriptional function by forming heterodimers with small Maf (v-maf musculoaponeurotic fibrosarcoma oncogene family) proteins, binding to ARE-containing promoters, and recruiting transcription coactivators to help remodel chromatin structures and facilitate the formation of basal transcription machinery (22, 29, 30, 41, 50, 74). Along with Nrf2, histone acetyltransferase (HAT) p300/CBP (CREB-binding protein) were detected in the ARE-binding complex, as measured by microinjection of antibodies against p300 or CBP (74). CBP was shown to interact with Nrf2 and to enhance Nrf2-dependent reporter gene activities (29). However, it is not clear whether or how p300/CBP might actively contribute to the dynamic regulation of Nrf2-dependent transcription.
p300 was cloned based on its interaction with the adenovirus-transforming protein E1A, while CBP was identified by its association with the transcription factor CREB (6, 11). p300 and CBP share a high degree of homology and possess intrinsic HAT activity. p300/CBP are believed to serve as transcription coactivators by acetylating core histones to facilitate chromatin decondensation and recruiting basic RNA polymerase machinery (44, 47). Recent findings have shown that many nonhistone proteins, particularly transcription factors, are substrates for p300/CBP, which greatly expands the possible mechanisms of p300/CBP in transcriptional activation (16, 65). On the other hand, an increasing list of signal-related proteins have been discovered to have intrinsic acetyltransferase activity. For instance, acetylation of p53 by p300 and Tip60 at multiple lysine residues is indispensable for p53 activation (56). Acetylation of type I interferon (alpha interferon) receptor, IRF9, and STATs by p300/CBP is a critical regulatory event in alpha interferon signaling (54). Acetylation of BMAL1 by CLOCK is crucial for circadian control (18). EcoI-mediated acetylation of a cohesion subunit is required to establish the sister chromatid cohesion during S phase in the cell cycle (3, 58, 72). Clearly, acetylation as a general posttranslational modification plays important roles in diverse signal transduction pathways other than “histone code.”
In this report, we demonstrate that p300/CBP directly binds and acetylates Nrf2 in response to arsenite-induced oxidative stress. We have identified multiple lysine residues as major acetylation sites within the Nrf2 Neh1 DNA-binding domain. Combined lysine-to-arginine (K→R) mutations of the acetylation sites, with no effects on Nrf2 protein stability, compromise the DNA-binding activity of Nrf2 in a promoter-specific manner. These findings demonstrate that acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 and establish acetylation as a novel regulatory mechanism of the Nrf2-mediated antioxidant responses.
MATERIALS AND METHODS
Recombinant DNA molecules.
Plasmids expressing the p300 wild type (WT) and the acetylase-deficient (DY) mutant were generous gifts from T. P. Yao (21). The vector for Flag-tagged p300 was kindly provided by M. A. Ikeda (45). The Nrf2 K→R point mutants were generated by site-directed mutagenesis using the PCR- and DpnI-based method as previously described (52). The construction of other plasmids is described in the supplemental material.
Cell culture, transfection, and reporter gene assay.
HCT116, HEK293T, MDA-MB-231, and COS-1 cells were purchased from the American Type Culture Collection. Nrf2−/− mouse embryonic fibroblasts (MEFs) were a generous gift from J. Y. Chan. HCT116 p300−/− and p300+/+ cells were kindly provided by C. Caldas (25). CBP−/− and CBP+/+ MEFs were kind gifts from D. Fang. The cells were maintained in either Dulbecco's modified Eagle's medium or Eagle's minimal essential medium in the presence of 10% fetal bovine serum. Transfection and reporter gene assays are described in the supplemental material.
Antibodies and immunoblot, immunoprecipitation, and immunofluorescence analyses.
Antibodies against hemagglutinin (HA) epitope (Covance); chitin binding domain (CBD) epitope (New England Biolabs); acetylated lysines (Cell Signaling); Flag and ubiquitin (Sigma); and Nrf2, p300, and α-tubulin (Santa Cruz Biotechnology) were purchased from commercial sources. Immunoprecipitation and immunofluorescence analysis are described in the supplemental material.
In vivo acetylation assay, ubiquitination assay, and MS.
For detecting acetylation in vivo, cells were lysed directly in Tris-buffered saline (10 mM Tris-HCl, pH 8.0, 150 mM NaCl) containing 2% sodium dodecyl sulfate (SDS) and 1 mM dithiothreitol. The cell lysates were boiled, sonicated, and diluted five times with Tris-buffered saline without SDS. The solution was then subjected to immunoprecipitation with anti-HA antibody and immunoblotting with anti-acetylated lysine antibodies. For ubiquitination assays, cells were treated with 50 μM MG132 for 4 h to block proteasome degradation before lysis for immunoprecipitation was performed as described above. Ubiquitination was detected by antiubiquitin antibodies. For MS, immunoprecipitated proteins were visualized by Coomassie staining and recovered from the gel. Liquid chromatography-tandem MS (LC-MS/MS) analysis was performed by the Harvard Taplin MS core facility.
In vitro acetylation and GST pulldown.
Glutathione S-transferase (GST)-Nrf2 proteins were expressed in Escherichia coli Rosetta (DE3) LysS cells and purified with glutathione Sepharose 4B matrix (Amersham Biosciences). Flag-p300 proteins were purified from COS-1 cells 48 h after transfection by using immunoprecipitation with anti-Flag M2 matrix and eluted with 3× Flag peptide elution buffer (Sigma). GST-Nrf2 proteins were incubated with p300 proteins in the presence of 14C-labeled acetyl-coenzyme A (acetyl-CoA) (55 mci/mmol; Amersham) in reaction buffer at 30°C for 1 h. The reaction mixtures were then resolved on SDS-polyacrylamide gel electrophoresis (PAGE), followed by autoradiography.
For GST pulldown assays, WT and truncated forms of p300 were radiolabeled with [35S]methionine using the in vitro TNT transcription/translation system (Promega) and were incubated with Sepharose beads conjugated with GST-Nrf2 fusion proteins in binding buffer at room temperature for 1 h. The Sepharose beads were then washed four times. The proteins were eluted by boiling them in SDS sample buffer, followed by SDS-PAGE and autoradiography. Detailed information is provided in the supplemental material.
Biotin-DNA pulldown.
The transfected cells were lysed in RIPA buffer (10 mM sodium phosphate, pH 7.2, 150 mM NaCl, 1% sodium deoxycholate, 2 mM EDTA, 0.1% SDS, 1% NP-40). The cell lysates were precleared with protein A beads and incubated with 2 μg biotinylated 25- to 41-bp double-stranded DNA probes that contained the ARE sequences in the promoter regions of Nrf2 target genes. The DNA-protein complexes were captured by streptavidin beads. After the beads were washed three times, DNA-bound proteins were eluted from the beads by heating them in SDS sample buffer. The proteins were resolved on SDS-PAGE and subjected to immunoblotting with anti-HA antibodies to detect Nrf2. The sequences of the probes are given in the supplemental material.
Mobility shift assay.
Mobility shift assays were conducted as previously described (52). MafG was synthesized using the in vitro TNT Transcription/Translation system (Promega) with nonradiolabeled methionine. GST-tagged Nrf2 proteins were purified and acetylated in vitro as described above. Two microliters MafG and 4 μl (20 ng) Nrf2 proteins were preincubated with poly(dI-dC) in binding buffer (50 mM HEPES, pH 7.5, 60 mM KCl, 2 mM MgCl2, 0.004% NP-40, 5 mM EDTA, 10% glycerol, 100 μg/ml bovine serum albumin) for 10 min at room temperature. 32P-end-labeled DNA probes were added and further incubated for 20 min before being loaded on a 4% native gel. The gel was dried and analyzed by autoradiography. For cold-probe competition, a 150-fold excess of cold probes was used. For supershift, 1 μg antibody was added to the preincubation mixture. The sequences of the probes were identical to those used in the biotin-DNA pull-down assay.
ChIP, qPCR, and qRT-PCR.
Chromatin immunoprecipitation (ChIP) was previously described (52). Quantitative PCR (qPCR) and quantitative reverse transcription (qRT)-PCR were performed with the LightCycler 480 system (Roche) for ChIP and mRNA profiling, respectively. Detailed information and primer sequences are provided in the supplemental material.
Statistical test.
Student's t test was used to determine the significant difference between two samples from three independent assays in reporter gene analysis and qPCR analysis.
RESULTS
Nrf2 is acetylated by p300/CBP in vivo and in vitro.
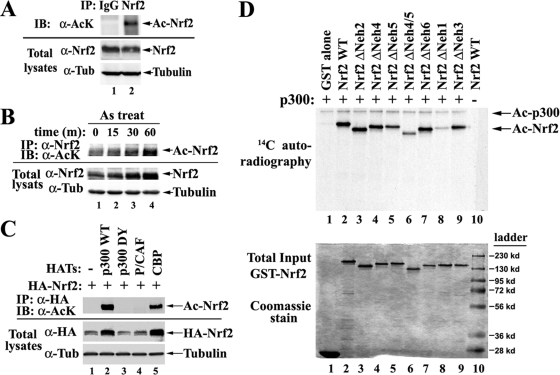
To determine if endogenous Nrf2 is acetylated, HCT116 cells cultured in 150-mm dishes were directly lysed under denaturing conditions to inactivate deacetylases and to disrupt protein-protein interactions. Diluted cell lysates were subjected to immunoprecipitation by normal immunoglobulin G (IgG) or anti-Nrf2 antibodies, followed by immunoblotting with antibodies specific for acetylated lysines. Acetylated Nrf2 was detectable under basal conditions (Fig. 1A, lane 2). To assess whether acetylation is upregulated in response to oxidative stress, cells were treated for different times with 20 μM sodium arsenite [As(III)], an environmental carcinogen and a strong Nrf2 inducer. There was a steady increase of Nrf2 acetylation, along with an accumulation of Nrf2 proteins, after As(III) treatment (Fig. 1B). Since the accumulated Nrf2 protein level is always coupled with increased Nrf2 nuclear entry, it is conceivable that the observed enhancement in acetylation is due to prompt acetylation of Nrf2 by acetyltransferase(s) within the nucleus, which contribute to the quick activation of Nrf2 in response to oxidative stress.
FIG. 1.
Nrf2 is acetylated by p300/CBP in vivo and in vitro. (A) Endogenous Nrf2 is acetylated. HCT116 cells were lysed under denaturing conditions. The cell lysates were diluted and subjected to immunoprecipitation (IP) by normal IgG or anti-Nrf2 (α-Nrf2) antibodies, followed by immunoblotting (IB) with antibodies specific for acetylated lysine (AcK). Tub, tubulin. (B) Sodium arsenite enhances acetylation of Nrf2. HCT116 cells were treated with 20 μm As(III) for the indicated times. Nrf2 acetylation levels were measured as described for panel A. (C) Nrf2 is acetylated by p300/CBP, not P/CAF. HEK293T cells were cotransfected with vectors expressing HA-tagged Nrf2 and the indicated HATs. The cell lysates were immunoprecipitated with anti-HA antibodies, followed by immunoblotting with antibodies specific for acetylated lysine. (D) Nrf2 is a bona fide substrate of p300. (Top) Purified GST-tagged Nrf2 proteins were incubated in the presence of 14C-labeled acetyl-CoA with immunoprecipitated p300 proteins from COS-1 cells overexpressing Flag-p300. The reaction mixtures were resolved on SDS-PAGE, followed by autoradiography. (Bottom) The same amounts of GST-Nrf2 proteins as used in the in vitro acetylation assay described above were subjected to Coomassie staining.
To identify which acetyltransferase(s) acetylates Nrf2, HEK293T cells were cotransfected with vectors expressing HA-tagged Nrf2 and different HATs, including p300, CBP, and P/CAF. The cells were lysed under denaturing conditions to preserve the modification. The diluted cell lysates were subjected to immunoprecipitation with anti-HA antibodies, followed by immunoblotting with antibodies specific for acetylated lysine. Nrf2 was acetylated only by p300 and CBP, but not P/CAF (Fig. 1C, lanes 2, 4, and 5). A DY point mutant of p300 failed to acetylate Nrf2 (Fig. 1C, lane 3) (21). p300 was the most potent in acetylating Nrf2 and therefore was chosen for the subsequent studies. Enhanced Nrf2 expression in the presence of p300/CBP was observed (Fig. 1C, lanes 2 and 5). This was likely due to indirect effects of p300/CBP on the transcription of the Nrf2 transgene, since similar observations were made with other transgenes carried in the same expression vector. This notion is further supported by the fact that Nrf2 protein half-lives were not changed when p300 was overexpressed (see Fig. S4 in the supplemental material).
To determine if p300 is self-sufficient in acetylating Nrf2, an in vitro approach was utilized. Purified GST-tagged Nrf2 proteins were incubated with purified p300 proteins in the presence of 14C-labeled acetyl-CoA. Acetylation of Nrf2 was detected by autoradiography (Fig. 1D, top, lane 2). The sample with GST alone (lane 1) did not give any positive signals, indicating that the acetylation reactions were specific for Nrf2. These results indicate that Nrf2 is a bona fide substrate of p300.
In the in vitro acetylation reaction, a series of Nrf2 deletion mutants were constructed in an effort to identify the major acetylation regions. The boundary of each domain is defined in Fig. 2D, bottom. Deletion of the Neh1 DNA-binding domain almost completely abolished acetylation of Nrf2, suggesting that the Neh1 domain contains the major acetylation sites (Fig. 1D, lane 8). Deletion of Neh4 and Neh5 also significantly decreased the acetylation levels of Nrf2 (Fig. 1D, lanes 5 and 6), which is consistent with the finding that Neh5, in coordination with Neh4, mediates the binding of Nrf2 to p300 (Fig. 2E, lanes 4 and 5). Deletion of other domains did not significantly alter the overall acetylation levels of Nrf2. Taken together, these results demonstrate that Nrf2 is acetylated by p300/CBP both in vivo and in vitro. The acetylation of Nrf2 is enhanced in response to arsenite exposure, and the Neh1 DNA-binding domain contains the majority of acetylated lysine residues.
FIG. 2.
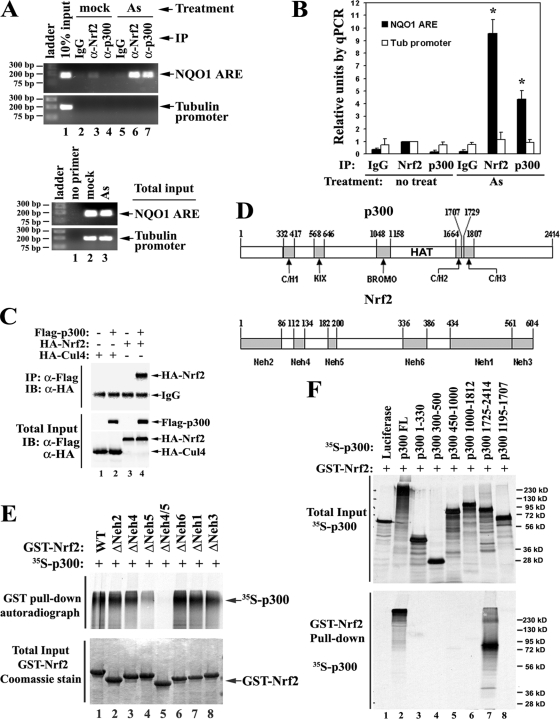
Nrf2 associates with p300. (A) Endogenous Nrf2 and p300 are coordinately recruited to the ARE in response to As(III)-induced stress. ChIP analysis was performed in HCT116 cells with the indicated antibodies after 4 h of treatment of 20 μM As(III). (Top) The genomic DNA fragments bound to either Nrf2 or p300 protein were recovered and quantified by qPCR using primer pairs specific for the NQO1-ARE region or the tubulin promoter region as a negative control. IP, immunoprecipitation. (Bottom) The total DNAs were amplified and visualized as described above to ensure equal input between mock and As treatment groups. (B) The precipitated DNA fragments in panel A were quantified by qPCR. The error bars indicate standard deviations. Asterisks indicate significant differences from IgG controls and the untreated group. IP, immunoprecipitation; Tub, tubulin. (C) Nrf2 interacts with p300 in vivo. HEK293T cells expressing Flag-tagged p300 and HA-tagged Nrf2 were lysed. The cell lysates were immunoprecipitated with anti-Flag (α-Flag) M2 matrix, followed by immunoblotting (IB) with anti-HA antibodies. An unrelated protein, Cul4, was included as a negative control. (D) Schematic of conserved domains in p300 and Nrf2 proteins. p300 contains five conserved domains: C/H1, KIX, BROMO, C/H2, and C/H3. The catalytic region, also known as the “HAT domain,” is between amino acids 1195 and 1673. Nrf2 contains six conserved domains: Neh2, Neh4, Neh5, Neh6, Neh1, and Neh3. (E) Nrf2 transactivation domains Neh4 and Neh5, especially Neh5, directly bind p300. (Top) p300 proteins were labeled with [35S]methionine and pulled down by GST-Nrf2 with different deletion mutants. The Nrf2-bound p300 was visualized by autoradiography. (Bottom) The same amounts of GST-Nrf2 proteins as used above were visualized by Coomassie blue staining. (F) The p300 C terminus directly interacts with Nrf2. (Top) Total inputs of different truncated forms of 35S-labeled p300 proteins were visualized by autoradiography. (Bottom) 35S-labeled p300 proteins were pulled down by GST-Nrf2 and visualized by autoradiography. Luciferase served as a negative control. FL, full length.
Nrf2 associates with p300.
ChIP analysis was performed to assess specific and coordinated recruitment of p300 and Nrf2 to the ARE in response to oxidative stress. HCT116 cells were either left untreated or treated with 20 μM As(III) for 4 h before being cross-linked and harvested. The cell lysates were subjected to immunoprecipitation with either anti-Nrf2 or anti-p300 antibody (with normal serum IgG as a negative control). The genomic DNA fragments bound to either Nrf2 or p300 proteins were recovered and quantified by qPCR using primer pairs specific for the NQO1-ARE region or the tubulin promoter region as negative controls. The amounts of NQO1-ARE bound to p300 or Nrf2 were increased nearly 10-fold compared to the mock-treated samples in response to As(III) treatment, while the amounts of tubulin promoter DNA bound to p300 or Nrf2 remained unchanged (Fig. 2A and B). This demonstrated that endogenous p300, along with Nrf2, was specifically recruited to the ARE-containing promoters in response to arsenite.
Interaction between Nrf2 and p300 was tested in HEK293T cells cotransfected with expression vectors for Flag-tagged p300 and HA-tagged Nrf2. Cell lysates were immunoprecipitated with anti-Flag M2 matrix. Both the immunoprecipitates and the total cell lysates were analyzed by immunoblotting with anti-HA antibodies. Nrf2 was detected in the Flag-p300 immunoprecipitates, while an unrelated protein, Cul4, was not detected (Fig. 2C), consistent with the previous finding for interactions between Nrf2 and CBP (29).
Direct interactions between Nrf2 and p300 and interaction domains were assessed. p300 contains five major conserved domains, C/H1, KIX, BROMO, C/H2, and C/H3 (Fig. 2D, top), whereas Nrf2 contains six major conserved domains, Neh2, Neh4, Neh5, Neh6, Neh1, and Neh3 (Fig. 2D, bottom). The Neh2 domain is the “degron” that is bound and ubiquitinated by Keap1 (24, 68). The Neh4 and Neh5 domains are transactivation domains (29, 50). Neh1 contains a CNC (“cap'n'collar”)-type basic leucine zipper structure responsible for dimerization with Maf proteins and DNA binding (22, 23).
To identify the p300 binding domain(s) in Nrf2, a series of domain-specific deletion mutants of Nrf2 were used. GST-tagged Nrf2 proteins containing the deletions were purified. Equal amounts of GST-Nrf2 proteins were used for GST pull-down analysis with p300 proteins that were radiolabeled with [35S]methionine. Consistent with a previous report (29), the Neh4 and Neh5 domains of Nrf2, but mainly the Neh5 domain, are required for interaction with p300 (Fig. 2E, lanes 4 and 5). In another set of experiments, a series of truncated forms of p300 were constructed and radiolabeled with [35S]methionine (Fig. 2F, top). These 35S-labeled p300 proteins were subjected to GST pulled down with GST-tagged Nrf2 WT proteins, followed by SDS-PAGE and autoradiography (Fig. 2F, bottom). Interactions with Nrf2 were mediated by p300 C-terminal positions 1725 to 2414 (Fig. 2F, lanes 7). Radiolabeled luciferase served as a negative control to show the specificity of the in vitro interaction (Fig. 2F, lanes 1). Collectively, these data suggest that the Neh4 and Neh5 domains of Nrf2 directly interact with the C/H3-containing C terminus of p300.
Identification of multiple acetylated lysines within the Neh1 DNA-binding domain of Nrf2.
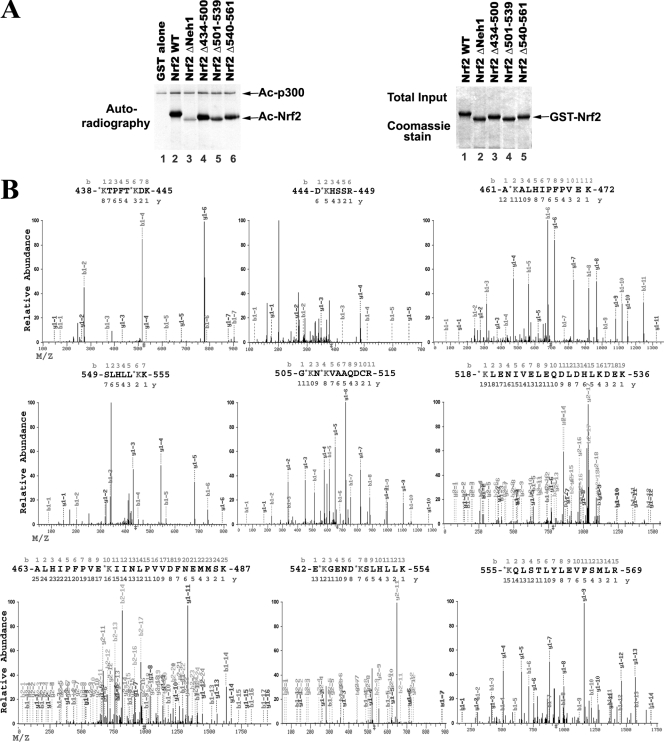
Figure 1D shows that the Neh1 domain (amino acids 434 to 561) of Nrf2 contains major acetylation sites. There are 18 lysines within this domain in human Nrf2 (see Fig. 5A). To further determine which lysine residues are major acetylation sites, GST-tagged Nrf2 with subdomain deletions within the Neh1 domain was used for the in vitro acetylation assay. None of subdomain deletion mutants were able to abolish Nrf2 acetylation to the same degree as the Neh1 deletion mutant, demonstrating that multiple lysine residues in Neh1 are acetylated (Fig. 3A, compare lanes 4, 5, and 6 to lane 3).
FIG. 5.
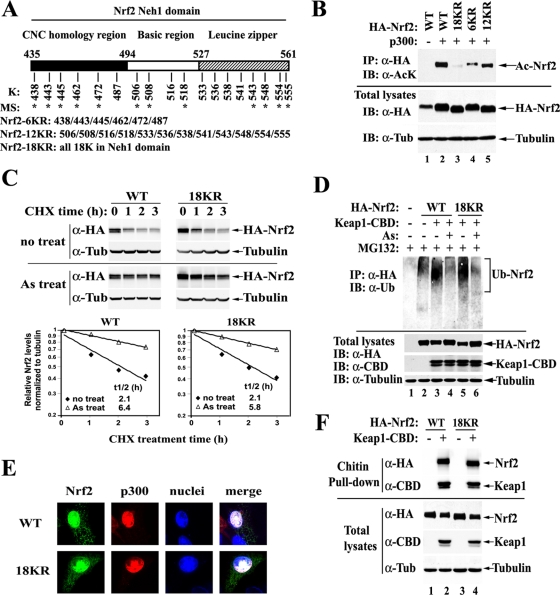
Acetylation on the Neh1 domain does not regulate the stability of Nrf2 proteins. (A) Distribution of lysines within the Neh1 domain of human Nrf2 protein. The asterisks indicate acetylated lysines as identified by MS. (B) Arginine substitution for all 18 lysines in the Neh1 domain abolishes acetylation of Nrf2 by p300. Acetylations on Nrf2 were analyzed as described for HEK293T cells expressing the indicated HA-Nrf2 and p300. IP, immunoprecipitation; IB, immunoblotting; α, anti; Tub, tubulin. (C) Mutation of 18KR in Nrf2 does not affect the half-lives of Nrf2 proteins under both basal and As-induced conditions. MDA-MB-231 cells expressing HA-Nrf2, Keap1, and p300, as indicated, were either left untreated or treated with 20 μM As(III) for 3 h, followed by cotreatment with 50 μM cycloheximide (CHX) for the indicated times. The cell lysates were analyzed by immunoblotting with anti-HA antibodies. The relative intensities of the Nrf2 bands were quantified, normalized to tubulin, and plotted on a semilog scale. The calculated half-lives (t1/2) of Nrf2 in each group are shown. (D) Mutation of 18KR in Nrf2 does not affect overall Nrf2 ubiquitination levels. MDA-MB-231 cells expressing HA-Nrf2, CBD-tagged Keap1, and p300, as indicated, were cotreated with 20 μM As(III) and 10 μM proteasome inhibitor MG132 as indicated for 4 h and then lysed under denaturing conditions. The cell lysates were diluted and subjected to immunoprecipitation with anti-HA antibodies, followed by immunoblotting with antiubiqutin (α-Ub) antibodies. (E) Mutation of 18KR in Nrf2 does not affect colocalization of Nrf2 with p300 in the nucleus. MDA-MB-231 cells expressing the indicated HA-Nrf2 and Flag-tagged p300 were subjected to indirect immunofluorescence analysis using anti-HA and anti-Flag antibodies. (F) Mutation of 18KR in Nrf2 does not affect the interaction between Nrf2 and Keap1. HEK293T cells expressing the indicated HA-Nrf2, Keap1-CBD, and p300 were lysed. Keap1-containing protein complexes were pulled down with chitin beads and immunoblotted with anti-HA antibodies.
FIG. 3.
Identification of multiple acetylated lysine residues within the Neh1 DNA-binding domain of Nrf2. (A) The Neh1 domain contains multiple acetylation sites. GST-tagged Nrf2 proteins with the indicated deletions within the Neh1 domain were subjected to in vitro acetylation analysis as described in the legend to Fig. 1D. Ac, acetylated. (B) Multiple lysines in the Neh1 domain were identified as acetylation sites by LC-MS/MS. Immunoprecipitation was performed in HEK-293T cells expressing HA-Nrf2 and p300 with anti-HA antibodies. Immunoprecipitated Nrf2 proteins were visualized by Coomassie staining, isolated, and analyzed by LC-MS/MS.
Next, MS was used to identify exact acetylated lysine residues on Nrf2. HEK293T cells were transfected with expression vectors for HA-Nrf2 and p300. Cell lysates were subjected to immunoprecipitation with anti-HA antibodies, followed by SDS-PAGE and Coomassie staining (see Fig. S1 in the supplemental material). The bands containing Nrf2 were isolated and analyzed by LC-MS/MS. Multiple acetylated lysines were detected (Fig. 3B; see Fig. 5A). Almost all acetylated lysines were within the Neh1 domain, which is consistent with the observation that deletion of Neh1 almost completely abolished acetylation of Nrf2 in the in vitro acetylation assay (Fig. 1D and 3A).
Functional redundancy among different acetylation sites.
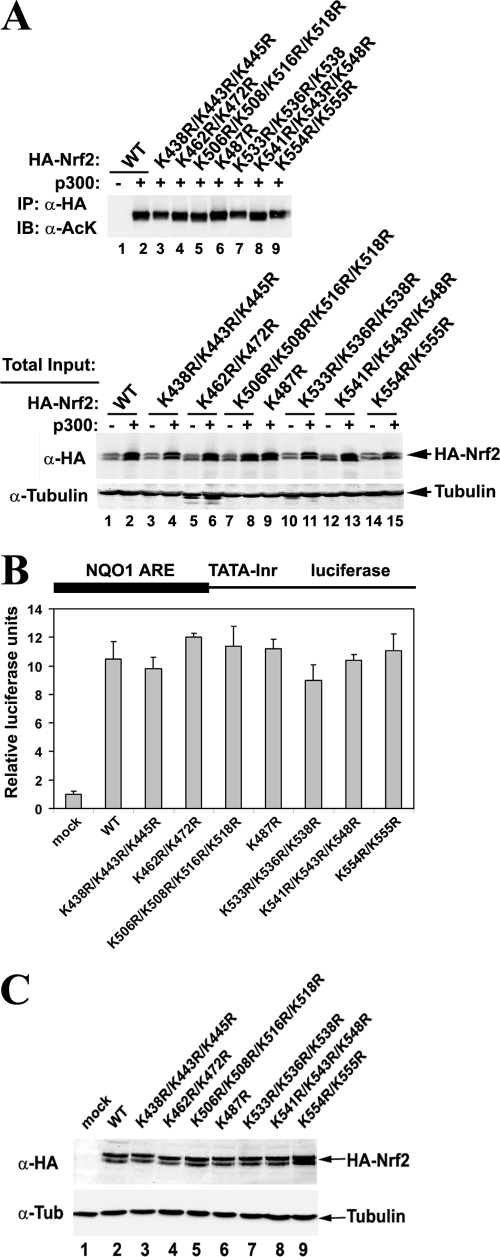
To elucidate the function of acetylation, lysine-to-arginine (K→R) substitutions were constructed in all lysine clusters within the Neh1 domain, and their effects on the overall Nrf2 acetylation levels (Fig. 4A) and Nrf2 transcriptional activity (Fig. 4B and C) were tested. As expected, none of the substitution constructs completely abolished acetylation of Nrf2, although a decrease in acetylation levels was visible for the K438R/K443R/K445R and K533R/K536R/K538R clusters (Fig. 4A, top, lane 2 and lane 7). None of the cluster mutations had significant impacts on Nrf2 transcriptional activities, as measured with the luciferase reporter gene (Fig. 4B). This observation was confirmed by qRT-PCR analysis of NQO1 and HO1 mRNA levels from the Nrf2−/− MEFs expressing the cluster mutants of Nrf2 (see Fig. S2 in the supplemental material for one example). Since none of the K→R mutations within each lysine cluster alter the transcriptional activity of Nrf2, it is likely that there is intrinsic functional redundancy among acetylations on different sites. In other words, Nrf2 partial acetylation is sufficient for Nrf2 to reach its maximum transcriptional activity.
FIG. 4.
Functional redundancy among different acetylation sites. (A) Arginine substitution for single or several adjacent lysine residue(s) does not change overall Nrf2 acetylation levels. Acetylation on Nrf2 was analyzed as described for HEK293T cells expressing the indicated HA-Nrf2 and p300. IP, immunoprecipitation; IB, immuno-blotting; α, anti. (B) HEK293T cells were cotransfected with vectors for the NQO1-ARE-dependent firefly luciferase reporter gene, TK Renilla luciferase gene, the indicated HA-Nrf2, and p300. Luciferase reporter gene activities were analyzed using the Promega dual-luciferase reporter gene assay system. Relative luciferase activities and standard deviations were calculated from three independent experiments. Inr, initiator. (C) Total cell lysates from the luciferase assay in panel B were subjected to immunoblot analysis with anti-HA antibodies. Tub, tubulin.
Acetylation of the Neh1 domain does not regulate the stability of Nrf2 proteins.
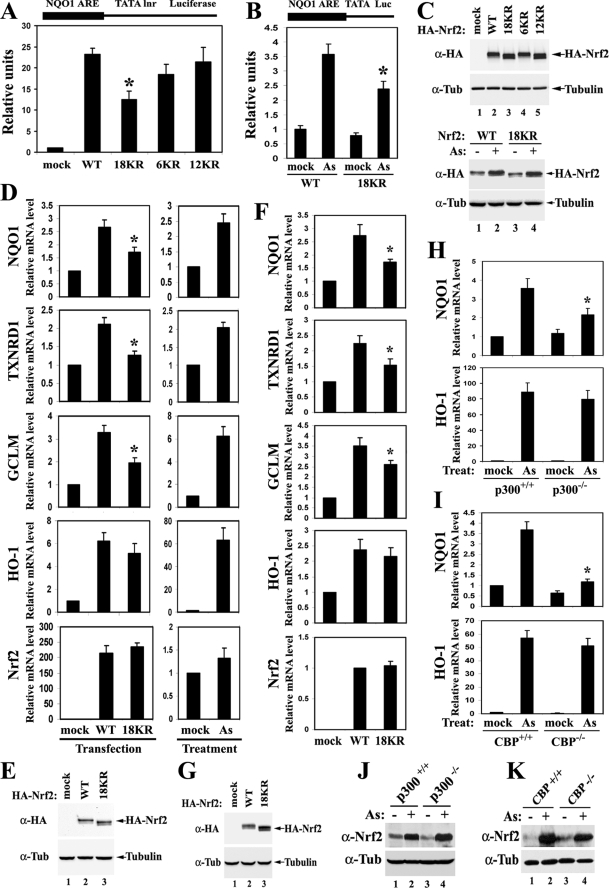
To resolve the redundancy issue, a mutant with combined arginine replacement of all 18 lysines within the Neh1 domain was constructed and named 18KR. In addition, mutants with two other combined mutations of the six lysines within the CNC homology region and the 12 lysines within the bZIP (basic leucine zipper) region were also constructed and were named 6KR and 12KR, respectively (Fig. 5A).
The effects of the combined mutations on overall Nrf2 acetylation levels were tested. The 18KR mutation almost completely abolished acetylation of Nrf2 by p300 (Fig. 5B, lane 3). The 6KR mutation seemed to decrease acetylation levels more than 12KR (Fig. 5B, lanes 4 and 5), implying that the CNC homology region may be more heavily acetylated than the bZIP region. However, a conclusion about the relative acetylation levels of each site is difficult to draw because the acetylation-specific antibody may have a certain bias toward specific amino acid contexts of the acetylated residues when different acetylated lysine-specific antibodies are compared (see Fig. S3 in the supplemental material). Nevertheless, the 18 lysines in the Neh1 domain were established as the major acetylation sites by both in vivo and in vitro acetylation assays. Therefore, the combined 18KR mutant was chosen for the functional study of Nrf2 acetylation.
Because Nrf2 is mainly regulated by Keap1 at the levels of protein ubiquitination and degradation, it is important to know whether acetylation affects Nrf2 protein stability. To this end, the half-lives of Nrf2 proteins were measured under both basal and As(III)-treated conditions. MDA-MB-231 cells were chosen for this analysis because it is well established that the ubiquitination and stability of Nrf2 proteins are very sensitive to oxidative stress in this cell line (62, 67, 68). MDA-MB-231 cells expressing Nrf2 WT or Nrf2 18KR, along with Keap1 and p300, were either left untreated or treated with 20 μM As(III) for 3 h, followed by the addition of 50 μM cycloheximide to block protein synthesis. Cells were then lysed at different time points, and the Nrf2 protein levels were determined by immunoblotting. There was no significant difference in Nrf2 half-lives between WT and 18KR under both basal and As(III)-treated conditions (Fig. 5C). Consistent with this result, overexpression of p300 did not cause an increase in the Nrf2 protein half-life (see Fig. S4 in the supplemental material). To assess if 18KR affects Nrf2 ubiquitination, MDA-MB-231 cells expressing the HA-Nrf2 WT or 18KR mutant were cotreated with 20 μM As(III) and 10 μM MG132 for 4 h and were then lysed under denaturing conditions. The diluted cell lysates were subjected to immunoprecipitation with anti-HA antibodies, followed by immunoblotting with antiubiquitin antibodies. Ubiquitin conjugation on Nrf2 WT or Nrf2 18KR was reduced to similar levels by As(III) (Fig. 5D). These results demonstrated that acetylation has no effect on Nrf2 ubiquitination or degradation.
Considering the importance of both Nrf2 nuclear import and Keap1-mediated Nrf2 nuclear export in turning the Nrf2 signaling pathway on and off (26, 52, 59), the effects of 18KR on the subcellular localization of Nrf2 and on Keap1-Nrf2 interactions were tested. MDA-MB-231 cells expressing Nrf2 WT or 18KR and p300 were subjected to indirect immunofluorescent staining. Both Nrf2 WT and 18KR localized mainly in the nucleus with p300 (Fig. 5E). Furthermore, Nrf2 WT and 18KR bound to Keap1 equally well (Fig. 5F). Collectively, these results indicate that 18KR almost completely abolishes acetylation on Nrf2. Acetylation on the Nrf2 Neh1 domain does not regulate Nrf2 ubiquitination or protein stability, nor does it seem to contribute to the regulation of Nrf2 subcellular localization.
Acetylation plays a positive role in the transcriptional activity of Nrf2.
The transcriptional activity of the Nrf2 18KR mutant was measured in HEK293T cells cotransfected with expression vectors for either an NQO1-ARE-dependent (Fig. 6A) or GSTA1-ARE-dependent (see Fig. S5 in the supplemental material) firefly luciferase reporter gene, Nrf2 WT or 18KR, and p300. Luciferase reporter gene activities were measured. Nrf2 18KR showed a substantial decrease in its ability to drive the expression of both NQO1-ARE- and GSTA1-ARE-dependent luciferase, indicating that loss of acetylation at these 18 lysine residues impaired the transcriptional activity of Nrf2 (Fig. 6A). Although not as significant as the 18KR mutation, the 6KR mutation and the 12KR mutation also marginally decreased Nrf2-dependent transcription (Fig. 6A). An aliquot of cell lysates from the reporter gene assay was subjected to immunoblotting to ensure that the Nrf2 protein levels were equivalent among the different Nrf2 mutants (Fig. 6C, top). Similar results were obtained in MDA-MB-231 cells, suggesting that the observation is not specific to HEK293T cells (see Fig. S6 in the supplemental material).
FIG. 6.
Acetylation plays a positive role in the transcriptional activity of Nrf2. (A) Nrf2 18KR has decreased transcriptional activity compared to Nrf2 WT. Luciferase reporter gene analysis was performed with NQO1 ARE reporters, as described in the legend to Fig. 4B, in HEK293T cells expressing HA-Nrf2 and p300, as indicated. The asterisk indicates a significant difference from Nrf2 WT. The error bars indicate standard deviations. Inr, initiator. (B) Arsenite has decreased induction effects on Nrf2 18KR compared to Nrf2 WT. HEK293T cells expressing HA-Nrf2, Keap1, and p300, as indicated, were treated with 10 μM As(III) for 12 h. NQO1-ARE luciferase (Luc) reporter gene expression was analyzed as for panel A. The asterisk indicates a significant difference from Nrf2 WT. (C) Total cell lysates from the two luciferase assays in panels A and B were subjected to immunoblot analysis with anti-HA (α-HA) antibodies. Tub, tubulin. (D) Nrf2 18KR has decreased activity in driving the transcription of NQO1, TXNRD1, and GCLM, but not HO-1. qRT-PCR was performed in HEK293T cells either expressing the indicated HA-Nrf2 and p300 or treated with 20 μM As(III) for 12 h. The error bars indicate the standard deviations from three experiments. The asterisks indicate significant differences from Nrf2 WT. (E) Total cell lysates from HEK293T cells prepared in parallel with cells for the qRT-PCR analysis in panel D were subjected to immunoblot analysis with anti-HA antibodies. (F and G) MEF Nrf2−/− cells overexpressing HA-Nrf2 and p300, as indicated, were analyzed by qRT-PCR and immunoblotting. (H and J) HCT116 p300+/+ and p300−/− cells were treated with 20 μM As(III) for 12 h, followed by qRT-PCR and immunoblot analysis. The asterisk indicates a significant difference between p300+/+ and p300−/− cells within the same treatment group. (I and K) MEF CBP+/+ and CBP−/− cells were treated with 10 μM As(III) for 12 h, followed by qRT-PCR and immunoblot analysis. The asterisk indicates a significant difference between CBP+/+ and CBP−/− cells within the same treatment group.
Next, the differential effects of oxidative stress on cells expressing Nrf2 WT versus Nrf2 18KR were analyzed in HEK293T cells overexpressing either Nrf2 WT or Nrf2 18KR, along with Keap1 and p300. Cells were treated with 20 μM As(III) for 12 h, and the NQO1-ARE-dependent luciferase reporter gene expression levels were measured (Fig. 6B). Compared to cells expressing Nrf2 WT, cells expressing Nrf2 18KR had an obviously dampened ARE-dependent transcription induction in response to arsenite challenge, although Keap1-mediated control of the total Nrf2 protein level was not affected (Fig. 6C, bottom).
qRT-PCR was performed to confirm that acetylation positively regulates the transcriptional activity of Nrf2. mRNA was extracted from HEK293T cells expressing Nrf2 WT or Nrf2 18KR (Fig. 6D, left). mRNA was also analyzed in cells that were either left untreated or treated with 20 μM As(III) for 12 h (Fig. 6D, right). Overexpression of Nrf2 WT caused a two- to threefold increase in the mRNA levels of NQO1 and TXNRD1 compared to the mock-transfected control; the same increase in the mRNA levels of the two genes were observed upon As(III) treatment compared to the mock-treated control (Fig. 6D, top two rows). This indicated that overexpression of Nrf2 mimicked the induction of endogenous Nrf2 by As(III) very well. However, As(III) caused a more dramatic induction of GCLM and HO-1, especially HO-1, compared to overexpression of Nrf2 WT (Fig. 6D). This is likely due to the fact that As(III) may also induce Nrf2-independent signal pathways that function synergistically with Nrf2 to transactivate these genes (1). Consistent with the previous findings, As(III) did not change the mRNA levels of Nrf2 (Fig. 6D) (62). In line with the results from the luciferase reporter gene assay, the 18KR mutant significantly compromised the induction of NQO1, TXNRD1, and GCLM (Fig. 6D, left), demonstrating that acetylation plays a positive role in Nrf2-dependent transcription. Interestingly, the transcriptional activity of Nrf2 18KR on HO-1 was comparable to that of Nrf2 WT (Fig. 6D, left), suggesting that acetylation of Nrf2 may preferentially regulate certain Nrf2 downstream genes. A parallel set of samples were subjected to immunoblot analysis to ensure that the cells were expressing equal amounts of Nrf2 WT and Nrf2 18KR (Fig. 6E).
To ensure that the observations made in HEK293T cells were not specific to the cell type, the mRNA levels of NQO1, TXNRD1, HO-1, GCLM, and Nrf2 were measured by qRT-PCR in Nrf2−/− MEFs expressing HA-Nrf2 WT or 18KR and p300. Consistent with the results obtained in HEK293T cells, acetylation of Nrf2 was found to be important in transactivating NQO1, TXNRD1, and GCLM, but not HO-1 (Fig. 6F and G).
The gene-specific effects of Nrf2 acetylation were further confirmed by comparing HCT116 p300−/− to p300+/+ cells and MEF CBP−/− to CBP+/+ cells (Fig. 6H to K). Loss of either p300 or CBP HAT activity led to a significant decrease in NQO1 induction in response to arsenite treatment, while induction of HO-1 remained unchanged (Fig. 6H and I). Nrf2 protein levels were elevated to similar extents upon arsenite exposure regardless of the p300/CBP status (Fig. 6J and K), which is consistent with the observation that p300/CBP does not regulate the protein stability of Nrf2. These results provide independent support for the notion that acetylation of Nrf2 regulates its transcriptional activity in a gene-specific manner.
Acetylation augments promoter-specific DNA binding of Nrf2.
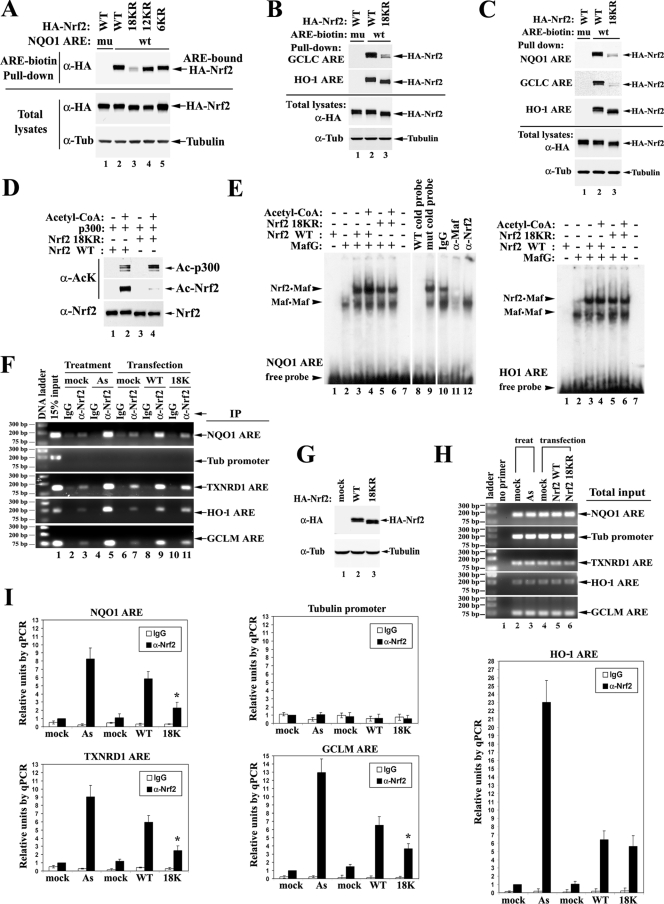
Given the fact that acetylation mainly occurs in the DNA-binding domain, the effects of Nrf2 acetylation on ARE DNA binding were analyzed in vitro. Biotinylated NQO1-ARE DNA was incubated with whole-cell lysates from HEK293T cells expressing Nrf2 WT or the indicated mutant. ARE-bound Nrf2 proteins were pulled down by streptavidin beads and detected by immunoblotting them with anti-HA antibodies. Nrf2 18KR, but not 6KR or 12KR, had a marked reduction in ARE binding compared to the WT (Fig. 7A, lanes 2 to 5). An ARE fragment with mutations in its core region was included as a control for binding specificity (Fig. 7A, lane 1). These results demonstrate that acetylation of Nrf2 in the Neh1 DNA-binding domain promotes interaction between Nrf2 and the ARE.
FIG. 7.
Acetylation augments promoter-specific DNA binding of Nrf2. (A) Nrf2 18KR has decreased binding affinity to NQO1 ARE compared to Nrf2 WT. HEK293T cells expressing the indicated HA-Nrf2 and p300 were lysed. The whole-cell lysates were incubated with either WT (wt) or mutated (mu) biotinylated ARE from the NQO1 promoter region. The protein-DNA-binding complexes were pulled down by streptavidin beads and analyzed by immunoblotting with anti-HA (α-HA) antibodies. Tub, tubulin. (B) Nrf2 18KR has decreased binding affinity to the GCLC ARE, but not to the HO-1 ARE, compared to Nrf2 WT. HEK293T cells expressing the indicated HA-Nrf2 and p300 were subjected to DNA-binding assays with the AREs from GCLC and HO-1 as described for panel A. Tub, tubulin. (C) The same DNA-binding assay was repeated in MDA-MB-231 cells. (D) Protein input for the mobility shift assay. Two hundred nanograms of purified GST-tagged Nrf2 WT or Nrf2 18KR protein was incubated with purified p300 protein in the absence or presence of acetyl-CoA in an in vitro 50-μl acetylation reaction; 1/10 of the reaction products were analyzed by immunoblotting with acetylation-specific antibody and Nrf2 antibody. Ac, acetylated. (E) A mobility shift assay was performed with 32P-labeled ARE probes in the presence of 2 μl of in vitro-translated MafG protein (Promega kit) and 4 μl of the indicated Nrf2 proteins from the in vitro acetylation reaction in panel D. (F) ChIP analysis was performed on HEK293T cells either treated with 20 μM As(III) or overexpressing the indicated HA-Nrf2 and p300. ChIP analysis was as described in the legend to Fig. 2A, with either IgG or anti-Nrf2 antibody. DNA fragments containing AREs of NQO1, TXNRD1, HO-1, and GCLM were amplified by PCR using specific primer sets and visualized on an agarose gel. The tubulin promoter region was also amplified to serve as a negative control. (G) Total cell lysates from HEK293T cells prepared in parallel with cells for the ChIP assay were subjected to immunoblot analysis with anti-HA antibodies. (H) Total DNA input was examined as for panel F. (I) The precipitated DNA fragments from the ChIP assay were quantified by qPCR. DNA precipitated by anti-Nrf2 antibodies in the mock-treatment group was set as 1. The error bars indicate the standard deviations from three experiments. The relative units of DNA amounts were plotted on the same scale for convenient comparison. The asterisks indicate significant differences from Nrf2 WT.
Since the function of acetylation seems to be promoter specific, the binding of Nrf2 18KR to the AREs from GCLC and HO-1 was analyzed similarly. Consistent with the results of the qRT-PCR analysis (Fig. 6D and F), acetylation of Nrf2 promoted the binding of Nrf2 to the GCLC ARE, but not the HO-1 ARE, in HEK293T cells (Fig. 7B). The same finding was obtained when MDA-MB-231 cells were used (Fig. 7C). Next, purified Nrf2 proteins were subjected to electrophoretic mobility shift assays (Fig. 7D and E). GST-tagged Nrf2 WT and Nrf2 18KR proteins were purified from bacteria and incubated with purified Flag-tagged p300 proteins in an in vitro acetylation reaction, as shown in Fig. 1D. Immunoblot analysis confirmed efficient conjugation of the acetyl group on Nrf2 only in the presence of acetyl-CoA, while the 18KR mutation almost completely abolished Nrf2 acetylation (Fig. 7D). In the mobility shift assay with the radiolabeled NQO1-ARE probe, the acetylated Nrf2 WT protein showed enhanced ability to form the ARE-binding heterodimer with in vitro-translated MafG, compared to unacetylated Nrf2 WT and Nrf2 18KR (Fig. 7E, left, lanes 3 to 6). On the other hand, Nrf2 18KR retained the ability to bind the ARE, suggesting that the K→R mutation itself does not affect DNA recognition or dimerization between Nrf2 and small Maf proteins. When HO1-ARE was used, acetylated Nrf2 WT did not show increased DNA binding compared to unacetlyated Nrf2 WT (Fig. 7E, right, lanes 3 and 4). These results demonstrate that acetylation of Nrf2 enhances its ARE binding in a promoter-specific manner.
To verify the above-mentioned findings obtained in vitro, the ARE-binding activity of 18KR was analyzed in vivo. ChIP analysis was performed in HEK293T cells either treated with 20 μM As(III) or overexpressing HA-Nrf2 WT or 18KR, as indicated, along with p300. Immunoprecipitation was performed with either IgG or anti-Nrf2 antibody. DNA fragments containing AREs from NQO1, TXNRD1, HO-1, and GCLM were amplified by PCR using specific primer sets and visualized on agarose gels (Fig. 7F). The tubulin promoter region was also amplified to serve as a negative control. Equal amounts of Nrf2 proteins and genomic DNA were used for each sample (Fig. 7G and H). The precipitated DNA from the ChIP assay was quantified by qPCR. The reading for the amount of DNA precipitated by anti-Nrf2 antibodies in the mock-treatment group was set as 1 (Fig. 7I). As(III) induced a seven- to ninefold increase in binding of endogenous Nrf2 to NQO1-ARE and TXNRD1-ARE; a comparable increase was observed in cells expressing exogenous WT Nrf2 (Fig. 7I, left). This suggests that overexpression of Nrf2 mimics As(III)-induced DNA binding of endogenous Nrf2 to some degree. Abolishing acetylation in the DNA-binding domain significantly decreased binding of Nrf2 to the AREs from NQO1, TXNRD1, and GCLM, but not the HO-1 ARE (Fig. 7I). These results demonstrate that acetylation augments promoter-specific DNA binding of Nrf2 in vivo.
DISCUSSION
In this report, we have demonstrated that acetylation of Nrf2 by p300/CBP constitutes a novel regulatory mechanism for Nrf2-dependent transcription. Our findings add Nrf2 to the increasing list of nonhistone proteins, especially transcription factors, that are substrates of p300/CBP (16). The Neh1 domain of Nrf2 is acetylated on multiple lysine residues. The relatively low substrate and site specificity of p300 can be explained by the Theorell-Chance catalysis model illuminated by the crystal structures of the p300 HAT domain (33). In this model, the substrate does not form tight and rigid complexes with p300; instead, it weakly associates with the surface of p300, and the target lysine residue(s) slides through the catalytic tunnel of p300 to react with acetyl-CoA (33). In keeping with this notion, multiple acetylation sites have been observed on many substrates of p300, such as p53, c-Myc, FoxOs, and STATs (4, 17, 54, 56, 73). In this study, the functional redundancy among the different acetylation sites was demonstrated by the observation that Nrf2-dependent transcription was not affected by mutations of any single lysine or combined adjacent lysines (Fig. 4; see Fig. S2 in the supplemental material). However, complete abolishment of acetylation by the 18KR mutation significantly compromised Nrf2-dependent transcription, as shown by luciferase reporter gene assays and qRT-PCR analysis performed in several cell lines (Fig. 6; see Fig. S5 and S6 in the supplemental material). The 18KR mutation does not affect Nrf2 ubiquitination or stability under either basal or arsenite-induced conditions, demonstrating that these lysines in the Neh1 domain are not targets for ubiquitination (Fig. 5C and D). This is consistent with the previous findings that the seven lysine residues in the N-terminal Neh2 domain are the sites for redox-sensitive ubiquitin conjugation and that the Neh6 domain is the region for redox-insensitive ubiquitin conjugation (28, 38, 39).
We found that acetylation on Nrf2 directly modulates its DNA-binding function (Fig. 7). The role of acetylation in DNA binding was previously documented with several transcription factors (7, 13, 17, 20, 35, 36). In most cases, acetylation promoted DNA binding of the transcription factor and enhanced its transcription, which is in line with our findings and the overall positive function of p300/CBP in transcription. Acetylation on the bZIP region of MafG by CBP was shown to facilitate DNA binding of the NF-E2 heterodimer composed of MafG and p45 (20). Considering that MafG is also a common binding partner for Nrf2, it is conceivable that recruitment of p300/CBP by Nrf2 to the ARE may also facilitate the acetylation of small Mafs. Therefore, acetylation on both Nrf2 and MafG may have synergistic positive effects on their DNA-binding activities. Mechanistically, acetylation on the bZIP domains of those proteins may cause subtle allosteric or electrostatic changes that favor their heterodimerization and DNA binding.
A striking finding in this report is that transcription of the HO-1 gene by Nrf2 is not regulated by acetylation, while that of other Nrf2 target genes, such as the NQO1, TXNRD1, GCL, and GSTA1 genes, is (Fig. 6 and 7). This suggests that acetylation of Nrf2 contributes to its fine discrimination among different ARE sequences, considering that there is considerable flexibility and variability in ARE sequences among different genes and species (43, 63). Similar modes of gene-specific regulation by acetylation were also found on several trans-acting elements, particularly transcription factors. For example, acetylation of p53 by Tip60 on its DNA-binding domain modulates the expression of BAX and PUMA, but not p21 or MDM2 (53, 55). Acetylation of FoxOs within its forkhead box DNA-binding domain enhances the expression of proapoptotic genes while suppressing genes involved in stress resistance (15). Acetylation of p73 on multiple sites within a region between the DNA-binding domain and the oligomerization domain modulates the expression of a proapoptotic gene, p53AIP1, but has no effects on p21 (7). The precise molecular mechanism underlying the reported differential gene regulation by acetylation in the DNA-binding domains of transcription factors requires further investigation. It is possible that acetyl groups on lysine residues within the DNA-binding domain directly contribute to conformational change during DNA recognition and binding. It is also possible that the acetylation of Nrf2 might recruit another cofactor(s) to the ARE and that the cofactor(s) selectively modulates the binding of Nrf2 to specific AREs. Nevertheless, these findings suggest that acetylation within the DNA-binding domain of a transcription factor is more likely to serve as a fine-tuning mechanism rather than an on-and-off switch in modulating the transcription factors.
An intriguing question is why the HO-1 gene, unlike other Nrf2 target genes, is highly activated by Nrf2 independently of Nrf2 acetylation. Since acetylation of transcription factors is a reversible process that fine tunes transcription, it is conceivable that the ability of Nrf2 to tightly bind the HO-1 promoter, regardless of its acetylation status, is a strategy used by the cell to ensure efficient induction of HO-1 by Nrf2 at its full capacity in response to oxidative stress. HO-1 seems to play a specific role in stress responses. Compared to most other Nrf2 target genes, which are mainly involved in reactive oxygen species detoxification and redox cycling, the HO-1 gene has been implicated in a variety of physiological defense responses, including vasodilation, angiogenesis, anti-inflammation, antithrombosis, antiproliferation, and antiapoptosis. HO-1 degrades heme to produce biliverdin, carbon monoxide (CO), and ferrous iron. Biliverdin is reduced to bilirubin, accounting for the antioxidant activity of HO-1. CO, on the other hand, has long been recognized as a vasodilating reagent, like nitric oxide, and has been shown to play protective roles in many inflammatory diseases, pulmonary injuries, and cardiovascular diseases (34, 49). In accordance with its broad functions, HO-1 is rapidly induced at the transcriptional level in response to a plethora of stimuli and stress conditions. Compared to most other Nrf2 target genes, the HO-1 gene has a promoter region containing more cis-regulatory elements that ensure timely induction of HO-1 in response to diverse stress-associated stimuli by many transcription factors (1, 34). In support of this idea, we observed a huge (∼60-fold or more) induction of HO-1 gene transcription compared to that of other target genes (<8-fold) in response to arsenite-induced stress in HEK293T cells (Fig. 6D), HCT116 cells (Fig. 6H), MEFs (Fig. 6I), and UROtsa cells (62). The binding of Nrf2 to the promoter of the HO-1 gene was also increased to a greater extent than that to other Nrf2 target gene promoters in response to stress (∼23-fold for the HO-1 gene compared to ∼10-fold for other genes) (Fig. 7I). In line with our finding, selective regulation of HO-1 by Nrf2 was reported to be mediated by a SWI/SNF family chromatin-remodeling subunit, BRG1 (71). BRG1 interacts with Nrf2 and enhances HO-1 induction, without affecting NQO1, GCLC, or GCLM, by facilitating Z-DNA formation on the HO-1 promoter (70, 71).
We believe that p300/CBP-mediated Nrf2 acetylation functions in concert with, and downstream of, Keap1-mediated Nrf2 ubiquitination in modulating Nrf2 activity. In response to oxidative stress and environmental insults, distinct cysteine residues on Keap1 are modified, leading to compromised Nrf2 ubiquitination and enhanced Nrf2 protein levels. This allows Nrf2 accumulation in the nucleus, where it is acetylated by p300/CBP. Acetylation is essential for maximum binding of Nrf2 to specific ARE-containing promoters. As a dynamic and reversible process, acetylation of Nrf2 is determined by the relative activities of HATs and histone deacetylases, both of whose activities are tightly regulated by many signaling pathways and are subject to change under diverse pathophysiological conditions. Thus, identification of Nrf2 acetylation as a novel regulatory mechanism will shed light on how redox homeostasis is maintained and fine-tuned by diverse signal transduction pathways in different cellular environments.
Supplementary Material
Acknowledgments
We thank J. Y. Chan for Nrf2−/− MEFs, C. Caldas for HCT116 p300−/− and p300+/+ cells, D. Fang for CBP−/− and CBP+/+ MEFs, T. P. Yao and M. A. Ikeda for p300 DNA constructs, S. Sachdev for technical help with ChIP analysis, and N. F. Villeneuve and A. Lau for editing of the manuscript. We particularly thank C. Smith for critical reading.
This work was supported by research grants from NIEHS, ACS and SWEHSC to D. D. Zhang (1RO1ES015010, RSG-07-154, and ES006694).
Footnotes
Published ahead of print on 9 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Alam, J., and J. L. Cook. 2007. How many transcription factors does it take to turn on the heme oxygenase-1 gene? Am. J. Respir. Cell Mol. Biol. 36166-174. [DOI] [PubMed] [Google Scholar]
- 2.Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a Cap′n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 27426071-26078. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shahar, T. R., S. Heeger, C. Lehane, P. East, H. Flynn, M. Skehel, and F. Uhlmann. 2008. EcoI-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321563-566. [DOI] [PubMed] [Google Scholar]
- 4.Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 3032011-2015. [DOI] [PubMed] [Google Scholar]
- 5.Chan, K., X. D. Han, and Y. W. Kan. 2001. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. USA 984611-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365855-859. [DOI] [PubMed] [Google Scholar]
- 7.Costanzo, A., P. Merlo, N. Pediconi, M. Fulco, V. Sartorelli, P. A. Cole, G. Fontemaggi, M. Fanciulli, L. Schiltz, G. Blandino, C. Balsano, and M. Levrero. 2002. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell 9175-186. [DOI] [PubMed] [Google Scholar]
- 8.Cullinan, S. B., J. D. Gordan, J. Jin, J. W. Harper, and J. A. Diehl. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 248477-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton, T. P., H. G. Shertzer, and A. Puga. 1999. Regulation of gene expression by reactive oxygen. Annu. Rev. Pharmacol. Toxicol. 3967-101. [DOI] [PubMed] [Google Scholar]
- 10.Dinkova-Kostova, A. T., W. D. Holtzclaw, R. N. Cole, K. Itoh, N. Wakabayashi, Y. Katoh, M. Yamamoto, and P. Talalay. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 9911908-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8869-884. [DOI] [PubMed] [Google Scholar]
- 12.Eggler, A. L., G. Liu, J. M. Pezzuto, R. B. van Breemen, and A. D. Mesecar. 2005. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 10210070-10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuoka, M., H. Daitoku, M. Hatta, H. Matsuzaki, S. Umemura, and A. Fukamizu. 2003. Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int. J. Mol. Med. 12503-508. [PubMed] [Google Scholar]
- 14.Furukawa, M., and Y. Xiong. 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giannakou, M. E., and L. Partridge. 2004. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 14408-412. [DOI] [PubMed] [Google Scholar]
- 16.Glozak, M. A., N. Sengupta, X. Zhang, and E. Seto. 2005. Acetylation and deacetylation of non-histone proteins. Gene 36315-23. [DOI] [PubMed] [Google Scholar]
- 17.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90595-606. [DOI] [PubMed] [Google Scholar]
- 18.Hirayama, J., S. Sahar, B. Grimaldi, T. Tamaru, K. Takamatsu, Y. Nakahata, and P. Sassone-Corsi. 2007. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 4501086-1090. [DOI] [PubMed] [Google Scholar]
- 19.Hong, F., K. R. Sekhar, M. L. Freeman, and D. C. Liebler. 2005. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J. Biol. Chem. 28031768-31775. [DOI] [PubMed] [Google Scholar]
- 20.Hung, H. L., A. Y. Kim, W. Hong, C. Rakowski, and G. A. Blobel. 2001. Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J. Biol. Chem. 27610715-10721. [DOI] [PubMed] [Google Scholar]
- 21.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 201331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh, K., T. Chiba, S. Takahashi, T. Ishii, K. Igarashi, Y. Katoh, T. Oyake, N. Hayashi, K. Satoh, I. Hatayama, M. Yamamoto, and Y. Nabeshima. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236313-322. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, K., T. Ishii, N. Wakabayashi, and M. Yamamoto. 1999. Regulatory mechanisms of cellular response to oxidative stress. Free Radic Res. 31319-324. [DOI] [PubMed] [Google Scholar]
- 24.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1376-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer, N. G., J. Xian, S. F. Chin, A. J. Bannister, Y. Daigo, S. Aparicio, T. Kouzarides, and C. Caldas. 2007. p300 is required for orderly G1/S transition in human cancer cells. Oncogene 2621-29. [DOI] [PubMed] [Google Scholar]
- 26.Jain, A. K., D. A. Bloom, and A. K. Jaiswal. 2005. Nuclear import and export signals in control of Nrf2. J. Biol. Chem. 28029158-29168. [DOI] [PubMed] [Google Scholar]
- 27.Kanwar, J. R. 2005. Anti-inflammatory immunotherapy for multiple sclerosis/experimental autoimmune encephalomyelitis (EAE) disease. Curr. Med. Chem. 122947-2962. [DOI] [PubMed] [Google Scholar]
- 28.Katoh, Y., K. Iida, M. I. Kang, A. Kobayashi, M. Mizukami, K. I. Tong, M. McMahon, J. D. Hayes, K. Itoh, and M. Yamamoto. 2005. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch. Biochem. Biophys. 433342-350. [DOI] [PubMed] [Google Scholar]
- 29.Katoh, Y., K. Itoh, E. Yoshida, M. Miyagishi, A. Fukamizu, and M. Yamamoto. 2001. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6857-868. [DOI] [PubMed] [Google Scholar]
- 30.Katsuoka, F., H. Motohashi, T. Ishii, H. Aburatani, J. D. Engel, and M. Yamamoto. 2005. Genetic evidence that small Maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol. Cell. Biol. 258044-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klaunig, J. E., and L. M. Kamendulis. 2004. The role of oxidative stress in carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 44239-267. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 247130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, X., L. Wang, K. Zhao, P. R. Thompson, Y. Hwang, R. Marmorstein, and P. A. Cole. 2008. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451846-850. [DOI] [PubMed] [Google Scholar]
- 34.Loboda, A., A. Jazwa, A. Grochot-Przeczek, A. J. Rutkowski, J. Cisowski, A. Agarwal, A. Jozkowicz, and J. Dulak. 2008. Heme oxygenase-1 and the vascular bed: from molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 101767-1812. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuzaki, H., H. Daitoku, M. Hatta, H. Aoyama, K. Yoshimochi, and A. Fukamizu. 2005. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. USA 10211278-11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahon, M., K. Itoh, M. Yamamoto, S. A. Chanas, C. J. Henderson, L. I. McLellan, C. R. Wolf, C. Cavin, and J. D. Hayes. 2001. The Cap‘n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 613299-3307. [PubMed] [Google Scholar]
- 38.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 27821592-21600. [DOI] [PubMed] [Google Scholar]
- 39.McMahon, M., N. Thomas, K. Itoh, M. Yamamoto, and J. D. Hayes. 2004. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 27931556-31567. [DOI] [PubMed] [Google Scholar]
- 40.Moinova, H. R., and R. T. Mulcahy. 1999. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem. Biophys. Res. Commun. 261661-668. [DOI] [PubMed] [Google Scholar]
- 41.Motohashi, H., F. Katsuoka, J. D. Engel, and M. Yamamoto. 2004. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc. Natl. Acad. Sci. USA 1016379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen, T., P. J. Sherratt, and C. B. Pickett. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43233-260. [DOI] [PubMed] [Google Scholar]
- 43.Nioi, P., M. McMahon, K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 374337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87953-959. [DOI] [PubMed] [Google Scholar]
- 45.Ohshima, T., T. Suganuma, and M. Ikeda. 2001. A novel mutation lacking the bromodomain of the transcriptional coactivator p300 in the SiHa cervical carcinoma cell line. Biochem. Biophys. Res. Commun. 281569-575. [DOI] [PubMed] [Google Scholar]
- 46.Ramos-Gomez, M., M. K. Kwak, P. M. Dolan, K. Itoh, M. Yamamoto, P. Talalay, and T. W. Kensler. 2001. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA 983410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 7081-120. [DOI] [PubMed] [Google Scholar]
- 48.Ruef, J., K. Peter, T. K. Nordt, M. S. Runge, W. Kubler, and C. Bode. 1999. Oxidative stress and atherosclerosis: its relationship to growth factors, thrombus formation and therapeutic approaches. Thromb. Haemost. 82(Suppl. 1)32-37. [PubMed] [Google Scholar]
- 49.Ryter, S. W., J. Alam, and A. M. Choi. 2006. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol. Rev. 86583-650. [DOI] [PubMed] [Google Scholar]
- 50.Shen, G., V. Hebbar, S. Nair, C. Xu, W. Li, W. Lin, Y. S. Keum, J. Han, M. A. Gallo, and A. N. Kong. 2004. Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J. Biol. Chem. 27923052-23060. [DOI] [PubMed] [Google Scholar]
- 51.Simonian, N. A., and J. T. Coyle. 1996. Oxidative stress in neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 3683-106. [DOI] [PubMed] [Google Scholar]
- 52.Sun, Z., S. Zhang, J. Y. Chan, and D. D. Zhang. 2007. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell. Biol. 276334-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sykes, S. M., H. S. Mellert, M. A. Holbert, K. Li, R. Marmorstein, W. S. Lane, and S. B. McMahon. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24841-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang, X., J. S. Gao, Y. J. Guan, K. E. McLane, Z. L. Yuan, B. Ramratnam, and Y. E. Chin. 2007. Acetylation-dependent signal transduction for type I interferon receptor. Cell 13193-105. [DOI] [PubMed] [Google Scholar]
- 55.Tang, Y., J. Luo, W. Zhang, and W. Gu. 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24827-839. [DOI] [PubMed] [Google Scholar]
- 56.Tang, Y., W. Zhao, Y. Chen, Y. Zhao, and W. Gu. 2008. Acetylation is indispensable for p53 activation. Cell 133612-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong, K. I., B. Padmanabhan, A. Kobayashi, C. Shang, Y. Hirotsu, S. Yokoyama, and M. Yamamoto. 2007. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 277511-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unal, E., J. M. Heidinger-Pauli, W. Kim, V. Guacci, I. Onn, S. P. Gygi, and D. E. Koshland. 2008. A molecular determinant for the establishment of sister chromatid cohesion. Science 321566-569. [DOI] [PubMed] [Google Scholar]
- 59.Velichkova, M., and T. Hasson. 2005. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol. Cell. Biol. 254501-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venugopal, R., and A. K. Jaiswal. 1996. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA 9314960-14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wakabayashi, N., A. T. Dinkova-Kostova, W. D. Holtzclaw, M. I. Kang, A. Kobayashi, M. Yamamoto, T. W. Kensler, and P. Talalay. 2004. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 1012040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, X. J., Z. Sun, W. Chen, Y. Li, N. F. Villeneuve, and D. D. Zhang. 2008. Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: enhanced Keap1-Cul3 interaction. Toxicol. Appl. Pharmacol. 230383-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wasserman, W. W., and W. E. Fahl. 1997. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. USA 945361-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, C. S., J. M. Landau, M. T. Huang, and H. L. Newmark. 2001. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annu. Rev. Nutr. 21381-406. [DOI] [PubMed] [Google Scholar]
- 65.Yang, X. J., and E. Seto. 2008. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, D. D. 2006. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 38769-789. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, D. D., and M. Hannink. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 238137-8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, D. D., S. C. Lo, J. V. Cross, D. J. Templeton, and M. Hannink. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2410941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, D. D., S. C. Lo, Z. Sun, G. M. Habib, M. W. Lieberman, and M. Hannink. 2005. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem. 28030091-30099. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, J., T. Hosoya, A. Maruyama, K. Nishikawa, J. M. Maher, T. Ohta, H. Motohashi, A. Fukamizu, S. Shibahara, K. Itoh, and M. Yamamoto. 2007. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem. J. 404459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, J., T. Ohta, A. Maruyama, T. Hosoya, K. Nishikawa, J. M. Maher, S. Shibahara, K. Itoh, and M. Yamamoto. 2006. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol. Cell. Biol. 267942-7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, J., X. Shi, Y. Li, B. J. Kim, J. Jia, Z. Huang, T. Yang, X. Fu, S. Y. Jung, Y. Wang, P. Zhang, S. T. Kim, X. Pan, and J. Qin. 2008. Acetylation of Smc3 by EcoI is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31143-151. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, K., F. Faiola, and E. Martinez. 2005. Six lysine residues on c-Myc are direct substrates for acetylation by p300. Biochem. Biophys. Res. Commun. 336274-280. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, M., and W. E. Fahl. 2001. Functional characterization of transcription regulators that interact with the electrophile response element. Biochem. Biophys. Res. Commun. 289212-219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.