Abstract
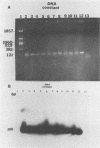
Serogrouping of Bacteroides nodosus is based on antigenic differences in fimbriae of the different New Zealand prototype strains. Because of the time needed to isolate and grow pure cultures of B. nodosus and the difficulty in distinguishing between different serogroups because of cross-agglutination, a new DNA-based diagnostic approach based on the fimbrial gene sequence of B. nodosus was developed. Published nucleotide sequences of the fimbrial genes for serogroups A, G, D, and H showed conservation at the 5' end, coding for the N terminus, and variability at the 3' end, coding for the C terminus. The polymerase chain reaction was used to amplify both the constant and variable regions of the fimbrial genes. Constant-region oligonucleotide primers were used to amplify a 100-base-pair fragment from the constant regions of the fimbrial genes of 10 New Zealand serogroups. Serogroup-specific oligonucleotide primers for serogroups A and H allowed amplification of a 282-base-pair fragment from serogroup A and a 363-base-pair fragment from serogroup H. Thus, amplification of the constant and variable regions of the fimbrial gene allows rapid detection and grouping of B. nodosus.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Claxton P. D., Ribeiro L. A., Egerton J. R. Classification of Bacteroides nodosus by agglutination tests. Aust Vet J. 1983 Nov;60(11):331–334. doi: 10.1111/j.1751-0813.1983.tb02834.x. [DOI] [PubMed] [Google Scholar]
- Egerton J. R., Roberts D. S., Parsonson I. M. The aetiology and pathogenesis of ovine foot-rot. I. A histological study of the bacterial invasion. J Comp Pathol. 1969 Apr;79(2):207–215. doi: 10.1016/0021-9975(69)90007-3. [DOI] [PubMed] [Google Scholar]
- Egerton J. R. Surface and somatic antigens of Fusiformis nodosus. J Comp Pathol. 1973 Jan;83(1):151–159. doi: 10.1016/0021-9975(73)90038-8. [DOI] [PubMed] [Google Scholar]
- Elleman T. C., Hoyne P. A., Emery D. L., Stewart D. J., Clark B. L. Isolation of the gene encoding pilin of Bacteroides nodosus (strain 198), the causal organism of ovine footrot. FEBS Lett. 1984 Jul 23;173(1):103–107. doi: 10.1016/0014-5793(84)81026-1. [DOI] [PubMed] [Google Scholar]
- Elleman T. C., Hoyne P. A., McKern N. M., Stewart D. J. Nucleotide sequence of the gene encoding the two-subunit pilin of Bacteroides nodosus 265. J Bacteriol. 1986 Jul;167(1):243–250. doi: 10.1128/jb.167.1.243-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman T. C., Hoyne P. A. Nucleotide sequence of the gene encoding pilin of Bacteroides nodosus, the causal organism of ovine footrot. J Bacteriol. 1984 Dec;160(3):1184–1187. doi: 10.1128/jb.160.3.1184-1187.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman T. C. Pilins of Bacteroides nodosus: molecular basis of serotypic variation and relationships to other bacterial pilins. Microbiol Rev. 1988 Jun;52(2):233–247. doi: 10.1128/mr.52.2.233-247.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman T. C., von Ahlefeldt D. A. Nucleotide sequence of the pilin gene from Bacteroides nodosus strain 238 (serogroup G). Nucleic Acids Res. 1987 Sep 11;15(17):7189–7189. doi: 10.1093/nar/15.17.7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Every D. Purification of pili from Bacteroides nodosus and an examination of their chemical, physical and serological properties. J Gen Microbiol. 1979 Dec;115(2):309–316. doi: 10.1099/00221287-115-2-309. [DOI] [PubMed] [Google Scholar]
- Every D., Skerman T. M. Protection of sheep against experimental footrot by vaccination with pili purified from Bacteroides nodosus. N Z Vet J. 1982 Oct;30(10):156–158. doi: 10.1080/00480169.1982.34921. [DOI] [PubMed] [Google Scholar]
- Fahey K. J., McWaters P. G., Stewart D. J., Peterson J. E., Clark B. L. Quantitation by ELISA of pili and sheep antibodies to the pili of Bacteroides nodosus. Aust Vet J. 1983 Apr;60(4):111–116. doi: 10.1111/j.1751-0813.1983.tb05907.x. [DOI] [PubMed] [Google Scholar]
- Hindmarsh F., Fraser J. Serogroups of Bacteroides nodosus isolated from ovine footrot in Britain. Vet Rec. 1985 Feb 16;116(7):187–188. doi: 10.1136/vr.116.7.187. [DOI] [PubMed] [Google Scholar]
- Lombard G. L., Dowell V. R., Jr Comparison of three reagents for detecting indole production by anaerobic bacteria in microtest systems. J Clin Microbiol. 1983 Sep;18(3):609–613. doi: 10.1128/jcm.18.3.609-613.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKern N. M., O'Donnell I. J., Inglis A. S., Stewart D. J., Clark B. L. Amino acid sequence of pilin from Bacteroides nodosus (strain 198), the causative organism of ovine footrot. FEBS Lett. 1983 Nov 28;164(1):149–153. doi: 10.1016/0014-5793(83)80039-8. [DOI] [PubMed] [Google Scholar]
- McKern N. M., O'Donnell I. J., Stewart D. J., Clark B. L. Primary structure of pilin protein from Bacteroides nodosus strain 216: comparison with the corresponding protein from strain 198. J Gen Microbiol. 1985 Jan;131(1):1–6. doi: 10.1099/00221287-131-1-1. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Schmitz J. A., Gradin J. L. Serotypic and biochemical characterization of Bacteroides nodosus isolates from Oregon. Can J Comp Med. 1980 Oct;44(4):440–446. [PMC free article] [PubMed] [Google Scholar]
- Schreckenberger P. C., Blazevic D. J. Rapid fermentation testing of anaerobic bacteria. J Clin Microbiol. 1976 Mar;3(3):313–317. doi: 10.1128/jcm.3.3.313-317.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D. J., Clark B. L., Peterson J. E., Griffiths D. A., Smith E. F. Importance of pilus-associated antigen in Bacteroides nodosus vaccines. Res Vet Sci. 1982 Mar;32(2):140–147. [PubMed] [Google Scholar]
- Stewart D. J. Studies on the antigenic structure of Bacteroides nodosus. Res Vet Sci. 1978 May;24(3):293–299. [PubMed] [Google Scholar]
- Whaley D. N., Dowell V. R., Jr, Wanderlinder L. M., Lombard G. L. Gelatin agar medium for detecting gelatinase production by anaerobic bacteria. J Clin Microbiol. 1982 Aug;16(2):224–229. doi: 10.1128/jcm.16.2.224-229.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wideman P. A., Citronbaum D. M., Sutter V. L. Simple disk technique for detection of nitrate reduction by anaerobic bacteria. J Clin Microbiol. 1977 Mar;5(3):315–319. doi: 10.1128/jcm.5.3.315-319.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D., Emery D. L., Stewart D. J. Monoclonal antibodies defining immunogenic regions of pili from Bacteroides nodosus strains 198 (A1), 265 (H1) and 336 (F1). Immunol Cell Biol. 1989 Feb;67(Pt 1):71–78. doi: 10.1038/icb.1989.9. [DOI] [PubMed] [Google Scholar]