Abstract
The short form (S1b) of the prolactin receptor (PRLR) silences prolactin-induced activation of gene transcription by the PRLR long form (LF). The functional and structural contributions of two intramolecular disulfide (S-S) bonds within the extracellular subdomain 1 (D1) of S1b to its inhibitory function on the LF were investigated. Mutagenesis of the paired cysteines eliminated the inhibitory action of S1b. The expression of the mutated S1b (S1bx) on the cell surface was not affected, indicating native-like folding of the receptor. The constitutive JAK2 phosphorylation observed in S1b was not present in cells expressing S1bx, and JAK2 association was disrupted. BRET50 (BRET50 represents the relative affinity as acceptor/donor ratio required to reach half-maximal BRET [bioluminescence resonance energy transfer] values) showed decreased LF/S1bx heterodimeric-association and increased affinity in S1bx homodimerization, thus favoring LF homodimerization and prolactin-induced signaling. Computer modeling based on the PRLR crystal structure showed that minor changes in the tertiary structure of D1 upon S-S bond disruption propagated to the quaternary structure of the homodimer, affecting the dimerization interface. These changes explain the higher homodimerization affinity of S1bx and provide a structural basis for its lack of inhibitory function. The PRLR conformation as stabilized by S-S bonds is required for the inhibitory action of S1b on prolactin-induced LF-mediated function and JAK2 association.
The prolactin receptor (PRLR) belongs to the class I cytokine receptor superfamily (4, 5). It binds the pituitary hormone prolactin (PRL) with high affinity and triggers intracellular responses that participate in diverse biological functions in target tissues, including the mammary gland, organs of the reproductive system, the central nervous system, pituitary, and adrenal. At least nine variants are generated by alternative splicing of the human PRLR (hPRLR) gene (16-18, 21, 35); they differ in the lengths and compositions of their cytoplasmic domains and/or extracellular (EC) domains (http://atlasgeneticsoncology.org/Genes/PRLRID42891ch5p14.html). The full-length receptor, or long form (LF), is composed of a ligand binding EC domain, a single transmembrane domain, and a cytoplasmic domain required for signal transduction (5, 20). PRL acts through the LF to stimulate cell proliferation and differentiation. The short forms (SFs) of the receptor, S1a and S1b, are derived from alternative splicing of exons 10 and 11 and contain unique cytoplasmic sequences (16). S1a is a 376-amino-acid (aa) receptor that contains partial exon 10 sequences and a unique 39-aa C terminus derived from exon 11. S1b is a 288-aa variant that lacks the entire exon 10 and contains 3 aa derived from exon 11 at the C terminus. Unlike the LF, neither of the SF receptors can mediate activation of the β-casein gene promoter induced by PRL, and both SFs are inhibitory of the transcriptional activation induced by PRL through the LF (16). S1b is more effective than S1a in inhibiting LF actions due to its higher stability (16). The reduced expression of S1a and S1b relative to the LF in breast cancer tissues and cell lines, compared to adjacent normal tissues/cells, suggests that the relatively reduced expression of SFs in cancer could lead to gradations of unopposed PRL-induced LF stimulatory function and contribute to breast tumor development and progression (28).
The initiation of signal transduction associated with members of the cytokine receptor family depends on the interaction of cognate ligand preformed receptor dimers (9, 13, 24, 32). In the case of the PRLR, immunoprecipitation studies (12, 31) and bioluminescence resonance energy transfer using coelenterazine (BRET1) analysis (31) demonstrated that hetero- and homodimerization of hPRLR can occur independently of ligand binding. This indicated that PRL is a conformational modifier that induces activation of the JAK2/STAT5 pathway through the LF and possibly through other JAK2-dependent pathways via the SFs. The dominant-negative effect of the SFs results from their heterodimerization with the LF, with the consequent functional inactivation due to the absence of cytoplasmic sequences of the dimerized LF partner (SF) required for downstream JAK2/STAT signaling (16, 31). Compared to the LF, S1b does not contain the conserved cytoplasmic structural motif beyond the Box-1 JAK2 docking site. In the homodimerized LF, the ligand-induced activation of JAK2 phosphorylates the receptor, preferentially at Y580 (rat) or Y587 (human) in its C terminus (2, 22). This induces phosphorylation, dimerization, and nuclear translocation of STAT5, which causes transcriptional activation of PRL-responsive genes, including the β-casein gene (8). Alternatively, Src has been demonstrated to participate in PRL-induced LF-mediated biological function by recruiting either Fak, Erk1/2, or P13K independently of the classic JAK2/STAT5 pathway (1, 5). These additional pathways have been proposed to operate in SF-mediated action in rodent species (10).
Our studies of the mutagenesis of unpaired conserved cysteines adjacent to and within the transmembrane region of the PRLR (S1b) showed no significant changes in receptor homo- or heterodimerization (31). In the present study, we evaluated the relevance of the four conserved extracellular Cys residues that form two intramolecular disulfide (S-S) bonds in subdomain 1 (D1) of the EC domain to the formation of homo- and/or heterodimers and their relevance to the inhibitory function of the SF on the LF. These residues are known to be crucial for ligand binding (33) and hence for PRL-induced activation of the JAK2/STAT5 pathway through the PRLR LF and also presumably in the hormonal activation of S1a and S1b. These intramolecular S-S bonds might provide strong spatial constraints for tertiary folding. Moreover, these bonds might control the quaternary arrangement of the receptor, affecting the formation of homo- and/or heterodimers and the inhibitory function of the SF on the LF. We observed that these residues are essential for maintaining the conformation that favors SF/LF heterodimerization and are also important for the constitutive activity of JAK2 bound to the SF. These studies further revealed that the intramolecular S-S bonds are relevant to the inhibitory action of the SF on the LF function. In addition, molecular modeling of the PRLR wild type and Cys mutants in the EC domain provides insights into the molecular mechanism by which the structure impacts homodimer/heterodimer formation, JAK2 association, and the PRL-induced PRLR action.
MATERIALS AND METHODS
Mutagenesis.
hPRLR SF S1b, constructed in the mammalian expression vector phRL-CMV-luciferase (RL) (Promega, Madison, WI) (referred to as S1b-RL) (RL, Renilla luciferase), or the yellow variant of the green fluorescent protein (GFP)-topaz vector (pEYFP-N1; BD BioSciences, Palo Alto, CA) (referred to as S1b-Y) (Y, Aequora enhanced yellow fluorescent protein [YFP]), described previously (31), were used as the templates to generate the mutants (x) in the study. Point mutation of cysteine (C) to serine (S) in the EC D1 subdomain of S1b-RL or S1b-Y was incorporated to generate S1b(C36,46S), S1b(C75,86S), and S1b(C36,46,75,86S), referred to as the S1b4x mutant construct. They were prepared using the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Briefly, PCR was performed using mutagenesis grade Pfu-Turbo DNA polymerase with oligonucleotide primers containing the required point mutation: C36S (forward, 5′-CCT GAG ATC TTT AAA TCT CGT TCT CCC AAT AAG G-3′; reverse, 5′-C CTT ATT GGG AGA ACG AGA TTT AAA GAT CTC AGG-3′), C46S (forward, 5′-GA AAC ATT CAC CAG CTG GTG GAG GCC-3′; reverse, 5′-GGC CTC CAC CAG CTG GTG AAT GTT TC-3′), C75S (forward, 5′-CA CTC ATG CAT GAA TCT CCA GAC TAC ATA ACC-3′; reverse, 5′-GGT TAT GTA GTC TGG AGA TTC ATG CAT GAG TG-3′), and C86S (forward, 5′-GT GGC CCC AAC TCC AGT CAC TTT GGC AAG CAG-3′; reverse, 5′-CTG CTT GCC AAA GTG ACT GGA GTT GGG GCC AC-3′). The PCR product was treated with DpnI to digest the methylated parental DNA template. Newly synthesized DNAs containing mutations were then selected for the desired mutation(s) after transformation into XL1-Blue cells. The mutated plasmid was isolated, sequenced, and checked for expression.
Generation of a HEK293 stable cell line expressing the LF or wild-type S1b hPRLR.
Full-length cDNA for the LF or the SF S1b of hPRLR in the pcDNA vector (LF-pcDNA or S1b-pcDNA; 1 μg) (16), which contains the neomycin resistance gene, was transiently transfected into HEK293 cells growing in the presence of 10% fetal bovine serum (FBS) using Lipofectamine TM2000 reagent (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. Cells were selected by treatment with Geneticin (600 μg/ml; Invitrogen) for 2 weeks to obtain stable transfectants. Clones were expanded and tested for LF expression by Western blot analysis using anti-PRLR (H300) antibody (an epitope corresponding to aa 323 to 622 at the C terminus of the hPRLR LF, but not in S1b) (Santa Cruz Biotechnology, Santa Cruz, CA). Because S1b lacks the C terminus, it is not recognized by H300 antibody. The mouse monoclonal antibody (U5) (Affinity BioReagents, Golden, CO), which is only known to react with the EC of the LF, weakly reacts with S1b. For this reason, S1b expression was assessed by its functional activity (JAK2 phosphorylation) in the presence (activation) or absence (basal) of human PRL (hPRL) (150 ng/ml; NIDDK S1AFP-B2; AFP-1969A) after the cells were transfected with JAK2 cDNA (0.2 μg) (Homo sapiens Janus kinase 2; Origene, Rockville, MD).
Cell culture and transfection.
HEK 293 cells (American Type Culture Collection, Manassas, VA) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) containing 10% FBS (Invitrogen) at 37°C in a CO2 incubator and cultured in six-well dishes for the various experiments. Lipofectamine 2000 reagent (Invitrogen) was used according to the manufacturer's instructions in all the transient-transfection studies. In the functional characterization of the SF dominant-negative effect, wild-type or mutated (x) (see below) hPRLR variant constructs in frame with YFP (LF-Y S1b-Y or S1bx-Y alone or in different combinations [see the specific experimental designs]) were transiently cotransfected with β-casein-luciferase reporter plasmid (0.1 μg), which contains three repeats of the GAS motif from the β-casein promoter and the cytomegalovirus minimal promoter subcloned into the pGL-2 basic vector (Promega) (kindly provided by Warren Leonard, NHLBI, National Institutes of Health, Bethesda, MD) (3) for luciferase assays or JAK2 cDNA (0.2 μg) (Homo sapiens Janus Kinase 2; Origene, Rockville, MD) for phosphorylation analysis. The β-galactosidase reporter gene (0.05 μg) was cotransfected for normalization purposes. pEYFP-N1 (empty vector) was used for equalization of DNA transfection. Six hours after transfection, HEK293 cells were incubated in serum-free, phenol red-free medium with or without hPRL (150 ng/ml) for 16 h. Luciferase activity was measured using a luciferase reagent kit (Promega) and a luminometer (Autolumat Plus LB953; Berthold Technologies, Bad Wildbad, Germany). JAK2 phosphorylation following treatment with and without hPRL for 30 min was determined by Western blotting.
Cell surface biotinylation.
HEK293 cells (with or without a stably expressed LF) were transiently transfected with S1b-Y or S1b4x-Y plasmids, followed by washing with ice-cold phosphate-buffered saline (PBS), and labeled with the biotinylation reagent Ez-Link Sulfo-NHS-LC-Biotin (Pierce Chemical, Rockford, IL) for 30 min at room temperature. The biotinylated cells were washed with PBS/100 mM glycine to quench and remove excess biotin reagent and by-products. In a parallel group, trypsin was used to eliminate cell surface protein by incubating cells with 0.05% trypsin for 10 min at 37°C. “Mock-treated” cells were incubated with PBS alone. The cells were detached, washed with PBS, and lysed in PBS containing 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS) for 30 min at 4°C. After preclearing steps, the supernatants were incubated with 50 μl of ImmunoPure immobilized streptavidin gel (Pierce Chemical) with rotation for 2 h at 4°C. The streptavidin-agarose was washed three times with lysis buffer and once with PBS, resuspended in 50 μl SDS protein gel loading solution, and boiled for 10 min. Samples were subjected to Western blot analysis using anti-GFP antibody (BD Biosciences), antibody to the membrane protein marker N-cadherin (Santa Cruz Biotechnology), or anti-PRLR (H300) antibody (Santa Cruz Biotechnology) for endogenous LF expression in stably expressed cells.
Coimmunoprecipitation (co-IP) analysis.
HEK293 cells were plated in a 75-cm2 flask and cotransfected with JAK2 (2 μg) and S1b-Y or S1b4x-Y construct (4 μg) (see Fig. 3A) or a combination of LF-RL, S1b-RL, S1b4x-RL, LF-Y, S1b-Y, and S1b4x-Y constructs (3 μg) (see Fig. 2E) for 24 h in the presence of DMEM/10% FBS. The cells were lysed using RIPA buffer (for JAK2 studies) or 50 mM HEPES buffer, 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA, and 0.1 mM Na3VO3 for association studies of the PRLR forms LF and S1b wild type and mutant S1b4x in the presence of protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). After the preclearing steps, the recovered supernatants were incubated in the presence of protease inhibitors with anti-GFP antibody (see Fig. 3A) (BD Biosciences, Palo Alto, CA), luciferase antibody (Invitrogen; 3 μg), or normal mouse immunoglobulin (immunoglobulin G) overnight at 4°C on a rotary shaker. Protein G-Sepharose beads (Invitrogen) were added and further incubated for 1 h at 4°C with rotation. The protein G-protein complex recovered by brief centrifugation was washed five times (three times with lysis buffer and two times with 1× PBS). The beads were resuspended in 30 μl of SDS-polyacrylamide gel electrophoresis loading buffer and denatured by being heated at 95°C for 10 min in the presence of 5% β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO), followed by Western blot analysis.
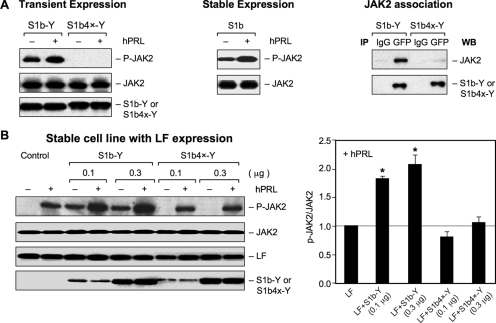
FIG. 3.
Effect of the Cys mutation on JAK2 phosphorylation. (A) (Left) HEK293 cells were cotransfected with wild-type S1b-Y or S1b4x-Y (0.2 μg) with JAK2 (0.2 μg) and incubated in the presence (+) or absence (−) of hPRL (150 ng/ml) for 0.5 h. (Middle) JAK2 phosphorylation in HEK293 cells stably expressing S1b transfected with JAK-2 and incubated in the presence or absence of hPRL. (Right) Co-IP analysis of JAK2 association with S1b wild type and the Cys mutant. (B) (Left) HEK293 cells stably expressing LF were transiently cotransfected with JAK2 (0.2 μg) with different doses of S1b wild-type or S1b4x-Y constructs and incubated in the presence or absence of hPRL (150 ng/ml) for 0.5 h. (Right) Levels of JAK2 phosphorylation stimulated by hPRL from three independent experiments were quantified and normalized by transiently expressed JAK2. The values (mean plus standard error) are presented relative to LFs (1; horizontal line). *, P < 0.05. S1b4x-Y, Cys-to-Ser mutation at aa 36, 46, 75, and 86 of the wild-type SF S1b (S1b-Y). Cell extracts were analyzed by Western blot analysis using anti-phospho-JAK2 (p-JAK2) for JAK2 phosphorylation and anti-JAK2 for transfected JAK2 expression. Anti-PRLR (H300) antibody was used for endogenous LF expression and anti-GFP for expression of transfected S1b-Y and S1b4x-Y.
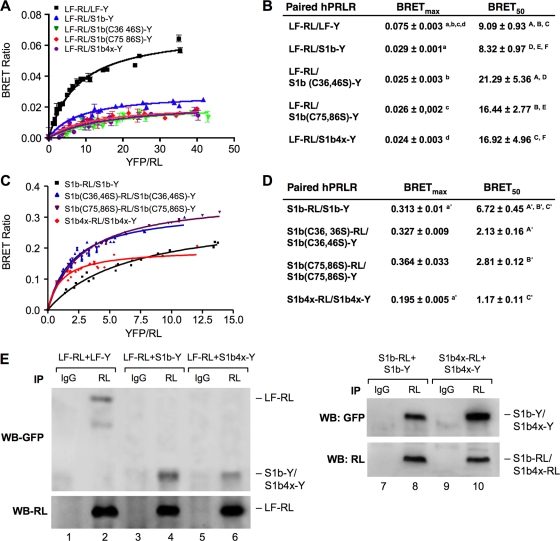
FIG. 2.
BRET saturation curve of hetero- and homodimerization of wild-type and mutated hPRLR variants. HEK293 cells were cotransfected with a constant LF-RL (0.2 μg) (A) or wild-type or mutated S1b-RL (0.2 μg) (C) with increasing DNA concentrations of YFP fusion construct (LF-Y, wild-type S1b-Y, or mutant S1b-Y). In all cases, the amounts of the YFP constructs (determined by fluorescence levels) were similar at each designated dose. The BRET ratio, total luminescence, and total fluorescence were measured 16 h after transfection. BRET levels were plotted as a function of the ratio of the expression level of the YFP construct (quantitated by the total fluorescence of the cells) over the RL construct (quantitated by the luminescence of the cells) (YFP/RL). This ratio was reflected as the change at the corresponding receptor expression level. The results are representative of three independent experiments carried out in triplicate. S1b(Cn, nS), mutation of the cysteine pair to serine occurred at the designated amino acid location; S1b4x, Cys-to-Ser mutation at aa 36, 46, 75, and 86 of the SF S1b. (B and D) Parameters derived from a BRET saturation curve of hetero- and homodimerization of wild-type and mutated hPRLR variants (A and C). BRETmax is the maximal BRET ratio obtained for a given pair. The results are representative of three independent experiments carried out in triplicate. Identical superscripts indicate statistical significance between experimental groups (P < 0.01). (E) Effect of the S1b4x mutant on the formation of homodimers and heterodimerization with LF. Shown is co-IP analysis of transfected wild-type (LF-RL, LF-Y, S1b-RL, and S1b-Y) and mutant (S1b4x-RL and S1b4x-Y) receptors into HEK293 cells, IP with RL antibody and immunoglobulin G (IgG) (negative control), and Western blotting (WB) with GFP or RL antibody. WB with RL antibody was used as a loading control (IP).
Western blot analysis.
Twenty-four hours after transient transfection, cells were washed twice with PBS and lysed using immunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% Na deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg/μl aprotinin, 1 μg/μl leupeptin, 1 μg/μl pepstatin, 1 mM Na3VO3, 1 mM NaF; Upstate, Charlottesville, VA). The lysates were incubated on ice for 20 min and centrifuged at 13,000 × g for 10 min at 4°C. The supernatants (30 μg protein) were resolved in 7.5% SDS-polyacrylamide gel electrophoresis gels and transferred to polyvinylidene difluoride membranes. After being blocked in 5% nonfat milk in PBS with 0.1% Tween 20, pH 7.6, the membranes were incubated with the appropriate primary antibodies (1:1,000): anti-rabbit phospho-JAK2 (Tyr 1007/1008 activation loop; Cell Signaling) for assessment of JAK2 activation or anti-rabbit JAK2 (Santa Cruz Biotechnology) for determination of total JAK2 expression; anti-GFP (monoclonal antibody; BD Biosciences) for assessment of total LF-Y, S1b-Y, and S1b-Y mutant expression; or anti-luciferase antibody for assessment of LF-RL, S1b-RL, and S1b4x-RL in CoIP studies (at 4°C overnight) and exposed to either goat anti-rabbit or anti-mouse secondary antibodies (1:5,000) for 2 h at room temperature. Immunoreactive proteins were visualized using the enhanced-chemiluminescence system (Pierce Chemical).
BRET analysis.
HEK293 cells were plated in six-well plates and cotransfected with wild-type and mutated PRLR variants (constructed earlier) in frame with either phRL-CMV-luciferase (RL) (Promega) or pEYFP-N1 (Y) vector (BD Biosciences) for BRET1 analysis (30); 0.2 μg of all the RL constructs, along with increasing concentrations of the respective Y constructs in excess, were transfected into the cells. The empty vector pEYFP-N1 for BRET1 was used for equalization of DNA transfection (total DNA, 4 μg). Six hours after transfection, the media were replaced with fresh DMEM containing 10% FBS. Sixteen hours later, the cells were detached and washed with BRET buffer (Dulbecco's PBS with 0.9 mM CaCl2, 0.5 mM MgCl2, and 5.5 mM d-glucose). After centrifugation, the cells were suspended in 300 μl BRET buffer, and 50 μl of cells was taken for protein estimation using a bicinchoninic acid protein assay kit (Pierce Chemical). Cells containing 50 μg of protein were distributed in a 96-well microplate. Coelenterazine (BRET1 analysis) (Nanoligh Technology, Pinetop, AZ) was added at a final concentration of 5 μM, and a reading was taken immediately for the light emitted at between 400 and 600 nm using a Mithras LB940 (Berthold Technologies). The BRET ratio was calculated using the following formula: [(emission at 530 nm) − (emission at 485 nm) × Cf]/(emission at 485 nm), where Cf corresponds to (emission at 530 nm)/(emission at 485 nm) for the LF-RL, wild-type, or mutated S1b-RL construct transfected individually in parallel experiments. The total fluorescence and luminescence were used as relative measures of the expression level of the acceptor (YFP) and donor (RL) proteins, respectively. After the cells were distributed in 96-well microplates, the total fluorescence of the cells was first measured using an excitation filter of 485 nm and an emission filter of 530 nm. The same cells were kept in the dark for 18 min with coelenterazine H incubation for the determination of RL expression as assessed by the luminescence at 400 nm. Saturation curves were plotted with GraphPad Prism4 software using a nonlinear regression curve assuming one-site binding.
Molecular modeling.
Langevin dynamics simulations of the EC wild-type hPRLR and the double-pair mutant (C36/46,75/86S) monomer and their corresponding homodimers were carried out. The hPRLR monomer was obtained from the hGH-hPRLR complex (Protein Data Bank [PDB] code 1bp3); the template for the dimers was the rat PRLR (rPRLR) dimer in the ovine prolactin-rPRLR complex (PDB code 1f6f). The simulations were performed under ambient conditions at zero ionic strength, the protonation states of titratable groups were fixed at standard values compatible with physiological pH, the N-terminal domain absent in the crystal structure was not modeled, and both termini were capped with acetylated and amidated groups. The screened Coulomb potential continuum model was used to represent the electrostatic and hydrophobic effects of the solvent (14, 15). The potential energy of the system was represented by the CHARMM force field (version c33b1) (6), with the CMAP correction (23, 25) of the all-atom representation (26). The simulations were extended up to 50 ns, preceded by an equilibration phase of 1 ns. The computational setup used here can be configured automatically (29) through the Web interface at http://www.charmming.org. Residues 230 to 234 and 283 to 287 of hPRLR were not resolved in the X-ray structure. These missing fragments are parts of two interacting loops in the D1 subdomain and were modeled prior to the simulations. The three-dimensional structure of these loops was calculated using a scaled-collective-variables technique in Monte Carlo simulations (27). High populations of two distinct conformations were obtained (data are available upon request), and the conformation with the lowest effective energy was used as the initial coordinates in the simulations. The dynamic trajectories were analyzed with the CHARMM program and with in-house scripts.
Statistical analysis.
The significance of the differences in the BRET saturation analyses of homo- and heterodimer formation between wild-type and mutated hPRLR variants (LF and S1b) was determined by Tukey's multiple-comparison test (one-way analysis of variance).
RESULTS
Functional characterization of the S1b mutant with disrupted S-S bonds.
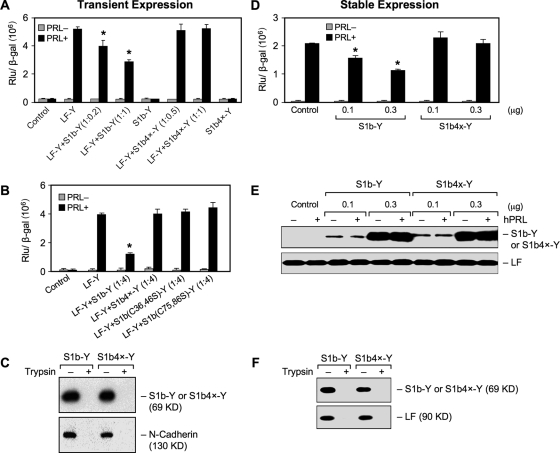
To determine whether disruption of the S-S bonds in the EC domains of the SF S1b affects its dominant-negative effect on the PRL-stimulated JAK2/STAT5-dependent transcription mediated by LF, we assessed the function of the PRLR S1b form mutated at either the first (aa 35/46) or second (aa 75/86) Cys-Cys pair, or both Cys-Cys pairs (S1b4x-Y), in the D1 subdomain. The dose-dependent inhibitory action of wild-type S1b on the β-casein promoter activation induced by hPRL through the LF of hPRLR was not observed upon cotransfection of LF with S1b with both Cys pairs mutated (S1b4x) (Fig. 1A). Similarly, the dominant-negative effect of S1b was not present upon cotransfection with either single-pair Cys-Cys S1b mutant (Fig. 1B). In all cases where Cys mutants were cotransfected with LF-Y, hPRL-stimulated β- casein promoter activities were comparable to the values observed in the LF-Y control group. No significant differences in the degree of loss of inhibitory function was found among the Cys mutant constructs examined [S1b4x-Y (Fig. 1A) and S1b (C36,45S)-Y or (C65,86S)-Y (Fig. 1B)]. Comparable results were observed in HEK293 cells stably expressing the LF cotransfected with increasing amounts of the S1b or S1b4x-YFP construct (Fig. 1D), where the inhibitory effect observed for wild-type S1b was negated by the Cys mutation (S1b4x-Y). Western blotting showed similar LF levels in the different experimental groups and increasing expression levels of S1b-Y or S1b4x-Y, depending on the amounts of the respective transfected constructs observed (Fig. 1E). Using a biotin-avidin labeling procedure, we demonstrated that S1b and its mutant (S1b4x) were similarly expressed at the surfaces of HEK 293 cells (Fig. 1C and F).
FIG. 1.
(A to C) Functional characterization of a S1b mutant lacking S-S bonds in transient-cotransfection studies. Reporter gene analyses of β-casein-luciferase reporter gene activity in HEK293 cells transfected with a constant hPRLR LF in frame with YFP (Y) (LF-Y; 0.2 μg) with increasing doses of DNA of either the wild-type S1b-Y or S1b4x-Y construct (at the ratio of 1:0.2- to 1-fold [A] of the LF-Y/S1b-Y or S1b4x-Y) or a single-pair cysteine mutant [S1b (C36, 46S)-Y or S1b (C75, 86S)-Y] (1:4-fold; LF-Y:S1b mutant) (B), along with the β-casein-luciferase reporter gene (0.1 μg). The cells were treated with 150 ng/ml hPRL for 16 h before termination. A β-galactosidase (β-Gal) plasmid (0.1 μg) was also included in the transfection, and its activity was measured for normalization of the reporter activity. The highest DNA concentration for the SF (either wild type or mutant) cotransfected with the LF was used in the control S1b-Y- or S1b4x-Y-only group. Empty YFP vector was used as the control and for equalization of DNA transfection. The results were representative of at least three independent experiments (mean plus standard error). The asterisks indicate changes stimulated by hPRL in the group compared to LF-Y with statistical significance (P < 0.01). S1b(Cn, nS), mutation of the cysteine pair to serine occurred at the designated amino acid location in the D1 subdomain; S1b4x, Cys-to-Ser mutation at aa 36, 46, 75, and 86 of the SF S1b. (C) Cell surface expression of wild-type S1b and S1b4x. Biotin-avidin labeling products of transiently expressed PRLR SF (S1b and S1b4x) in HEK293 cells were analyzed by Western blotting using anti-GFP antibody to assess SF expression. The membrane protein marker N-cadherin was used as the positive control to normalize cell surface expression. Normalized cell surface expression of S1b4x-Y relative to S1b was 1.0 ± 0.1 versus 1.14 ± 0.09. Trypsin treatment was used as the negative control. (D to F) Functional characterization of the S1b mutant lacking S-S bonds in HEK293 stably expressing hPRLR LF. HEK293 cells stably expressing the hPRLR LF were transiently transfected with increasing doses of DNA of either wild-type S1b-Y or S1b4x-Y constructs, along with the β-casein-luciferase reporter gene (0.1 μg). The cells were treated with 150 ng/ml hPRL for 16 h before termination. A β-galactosidase plasmid (0.1 μg) was also included in the transfection, and its activity was measured for normalization of the reporter activity (D). Empty YFP vector was used as the control and for equalization of DNA transfection. The results were representative of at least three independent experiments (mean plus standard error). The asterisks indicate changes stimulated by hPRL in the group compared to LF-Y with statistical significance (P < 0.01). (E) Western analysis of endogenous LF and transfected SF (S1b-Y or S1b4x-Y) expression. (F) Cell surface expression of wild-type S1b and S1b4x. Biotin-avidin-labeled products of transiently expressed PRLR SF (S1b and S1b4x) in HEK293 cells stably expressing LF were analyzed by Western blotting using anti-GFP antibody to assess SF expression. Cells stably expressing LF were used as the positive control to normalize cell surface expression. The normalized cell surface expression of S1b4x-Y relative to S1b was 1.0 ± 0.1 versus 0.82 ± 0.09. Trypsin treatment was used as a negative control.
Homo- and heterodimerization of hPRLR variants.
Further studies addressed the molecular nature of the loss of inhibitory function of the Cys mutants. BRET saturation curves were generated to study the formation of either hetero- or homodimerization of LF-RL to S1b-Y or S1b-RL to S1b-Y compared to the respective S1b mutants. The maximal BRET (BRETmax) signals observed for LF homodimers were about threefold higher than for LF heterodimers with S1b or S1bx (Fig. 2A and B). BRET50 (BRET50 represents the relative affinity as an acceptor/donor ratio required to reach half-maximal BRET values) values were similar for LF homodimers and LF heterodimers with wild-type S1b (9.09 versus 8.32). In contrast a significant increase in BRET50 was found in LF heterodimers with S1b mutants (Fig. 2A and B). BRETmax values were similar among S1b wild-type and single-pair Cys mutants (C36,46S or C75,86S), with values of 0.31 to 0.36 (Fig. 2C and D). In contrast, a lower BRETmax signal of 0.195 was observed for S1b4x, and this was reached at a lower acceptor/donor ratio. However, BRET50 values were similar among the mutants (1.17 ± 0.11, 2.13 ± 0.16, and 2.81 ± 0.12) compared to the much higher value observed for the wild-type S1b (6.72 ± 0.45) (Fig. 2D). The S1b Cys mutants displayed higher affinity for forming homodimers than the wild type (S1b mutant BRET50, 1.17 to 2.81; S1b wild-type BRET50, 6.72) and lower affinity for forming heterodimers with the LF than the S1b wild type (S1b mutant BRET50, 16 to 21; S1b wild-type BRET50, 8.32). This would favor S1bx homodimerization, impairment of heterodimer formation with the LF, and, consequently, an increase in the propensity of the LF to form homodimers with competence to mediate PRL action through JAK2/STAT5 downstream signaling, which in turn causes β-casein promoter activation.
Co-IP studies comparing LF and SF (wild-type) forms revealed the formation of homodimers (LF-RL/LF-Y and S1b-RL/S1b-Y) (Fig. 2E, left, lane 2, and right, lane 8) and heterodimers (LF-RL/S1b-Y) (Fig. 2E, left, lane 4), as previously described (31). In contrast, heterodimer formation in cells transfected with LF-RL and S1b4x-Y was markedly reduced (LF-RL/S1b4x-Y) (Fig. 2E, left, lane 6). On the other hand, the band intensities of homodimers of S1b4x-RL/S1b4x-Y were significantly increased compared to the wild type (S1b-RL/S1b-Y) (Fig. 2E, right, lane 10 versus lane 8). These results are consistent with the affinity derived from BRET analysis (Fig. 2A to D).
Effects of the Cys mutations on JAK2 phosphorylation.
To learn about the impact of the Cys mutation on JAK2 phosphorylation, we initially performed analysis of its phosphorylation status in HEK293 cells transiently transfected with the S1b wild type and mutant (S1b4X) (Fig. 3A, left). In the absence of hPRL, a basal level of JAK2 phosphorylation was observed in cells transfected only with wild-type S1b-Y but not with the S1b4x-Y mutant. Following hPRL treatment, significant increases in JAK2 phosphorylation over control were observed in cells transfected with wild-type S1b but not with S1b4x (Fig. 3A, left). Additional studies showed that cells stably transfected with S1b displayed JAK2 basal phosphorylation that was stimulated (two- to threefold) upon exposure to PRL (Fig. 3A, middle). Similarly, a significant amount of basal JAK2 phosphorylation was observed in cells stably expressing the LF transfected with S1b (Fig. 3B, lanes 3 and 5). In contrast, JAK phosphorylation was not present either in control cells stably expressing LF (Fig. 3B, lane 1) or upon transfection of S1b4x-Y (Fig. 3B, lanes 7 and 9). PRL-stimulated JAK2 phosphorylation was observed in the control group and in cells transfected with S1b or S1b4x (Fig. 3B, left, upper gel, lanes 2, 4, 6, 8, and 10 from left). In the case of cells transfected with S1b-Y (0.1 to 0.3 μg DNA), JAK2 phosphorylation induced by PRL was significantly increased compared to that in untransfected cells expressing LF (Fig. 3B, left, upper gel, lanes 4 and 6 versus lane 2, and graph). However, in cells transfected with S1b4x-Y, the increase on JAK2 phosphorylation induced by PRL was not different from that of the control (Fig. 3B, left, upper gel, lanes 8 and 10 versus lane 2, and graph). Western blots revealed a constant JAK2 and LF expression level in all groups (Fig. 3B, left, upper gel, lanes 2 and 3, and graph). Similar expression levels of S1b-Y and S1b4x-Y were observed at each transfected dose (0.1 or 0.3 μg) (Fig. 3B, bottom). Co-IP studies of cells transiently cotransfected with either S1b-Y or S1b4x-Y with JAK2 using anti-GFP antibody followed by Western analysis showed loss of JAK2 association with S1b4x compared to wild-type S1b (Fig. 3A, right). Overall, these findings indicate an effect of the Cys mutations (S1b4x) on the conformation of the SF homo- and heterodimer with LF that compromise the JAK2 basal phosphorylation activity and JAK2 association observed in wild-type S1b.
Molecular modeling of wild-type and Cys-mutated EC domains of PRLR.
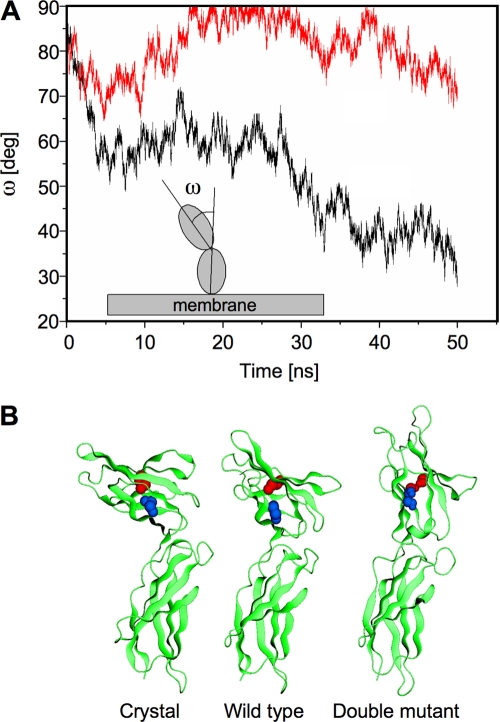
Computer simulations were carried out to investigate the structural and dynamic properties of hPRLR monomers and homodimers in aqueous solution and the effects of the Cys mutations at the atomic level (see Materials and Methods). The molecular dynamics of the EC wild-type monomer showed that the elements of secondary and tertiary structure in the crystal structure were preserved during the course of the simulation. The D1 and D2 domains behaved as semirigid bodies whose relative orientations fluctuated over time. To characterize these movements, the moments of inertia of each spheroid subdomain were calculated as a function of time. The angle ω formed by the main axes of inertia of each domain was monitored, and the results are shown in Fig. 4A. The relative orientation between D1 and D2 changed little with respect to that in the crystal, fluctuating around an ω of ∼70° to 90° (Fig. 4A). Next, four mutations were introduced simultaneously at positions 36, 46, 75, and 86 to study the effects of the two S-S bonds on the structure and dynamics of the PRLR monomer. Since comparable levels of expression at the cell surface were observed in the wild-type and mutant PRLR S1b, misfolding of the mutant receptor is unlikely. Thus, the tertiary fold of the D1 subdomain is expected to be native-like. To create an initial structure that mimicked the mutagenesis experiments, all four Cys residues were replaced by serine residues. These residues have similar physicochemical characteristics (polarity, electronic polarization, geometry, and volume), so the overall folding of the mutant PRLR construct was assumed to be the same as in the wild-type PRLR (sequence identity above ∼30% usually yields reliable structures in homology modeling; in this case, the identity is ∼98%). As in the dynamics of the wild-type PRLR, the elements of secondary structure were preserved during the simulation, although changes in the tertiary structure were apparent. Such changes propagated throughout the EC domain, affecting the movements and relative arrangement of D1 and D2. During the course of the simulation, both domains showed a tendency to align to each other, with the angle ω decreasing substantially with respect to its value in the wild-type monomer, stabilizing at an ω of ∼30° to 40° (Fig. 4A). Figure 4B shows the crystal structure of the wild-type receptor and the conformations of the wild-type and mutant monomers at the end of the 50 ns of dynamics. These final conformations are representative of the monomers in an aqueous environment as opposed to the crystal environment (11, 34). The conformational changes observed suggest that the absence of S-S bonds in the mutant leads to a structural relaxation of D1 with respect to its native fold, akin to a “breathing” movement, that is forbidden in the wild-type monomer due to the constrains of the bonds. Although this relaxation has no effect on the protein structural integrity, the perturbations propagate to the D1-D2 interface, leading to repositioning and more favorable relative orientation of these subdomains. This rearrangement modifies the secondary structure of the short sequence that connects D1 and D2 from an extended to α-helix turn, as shown in Fig. 4B.
FIG. 4.
Molecular dynamics simulation of wild-type and Cys-mutated EC domains of PRLR. (A) Evolution over time of the angle ω (inset) between the principal axes of inertia of the N-terminal (D1) and C-terminal (D2) EC subdomains of the wild-type (red line) and mutant (black line) monomers. (B) Ribbon representation of the crystal structure of the EC domain of hPRLR (PDB code 1bp3) and of the simulated structures of the wild-type and double-mutant monomers in water. The paired cysteines C36 and C46 are shown in blue, and the paired C75 and C86 are shown in red, in both the crystal and the wild-type monomers; the corresponding serine residues of the double mutant are shown in the same colors.
The conformations of the wild-type and mutant receptors at the end of the simulations were subsequently assembled in pairs to create initial models of the homodimers. The dimerization interfaces in both cases were assumed to be similar to that of the rPRLR dimer in the ovine prolactin-rPRLR complex (11, 34). Therefore, the D2 domains of the wild-type and mutant monomers were superimposed on the corresponding domains of the rPRLR monomers. Dynamic simulations of the dimers were carried out starting from these initial constructs to allow the monomers to evolve toward a refined complex. During the dynamics of the dimers, the secondary and tertiary structures of the monomer subdomains were preserved. The D2 subdomains approached each other as time passed, resulting in tight dimer interfaces with extensive hydrogen-bonding networks, as shown in Fig. 5A. In the initial construct based on the rPRLR complex, only one (wild type) or two (mutant) intermonomer H bonds were observed connecting D2s. However, during the simulations, these H bonds were not preserved. In particular, 13 new persistent H bonds developed during the simulation of the wild-type dimer, as detailed in Fig. 5A, bottom. Thus, the two monomers were able to recognize each other and adjusted their relative orientation and side chain conformations at the interface for favorable docking.
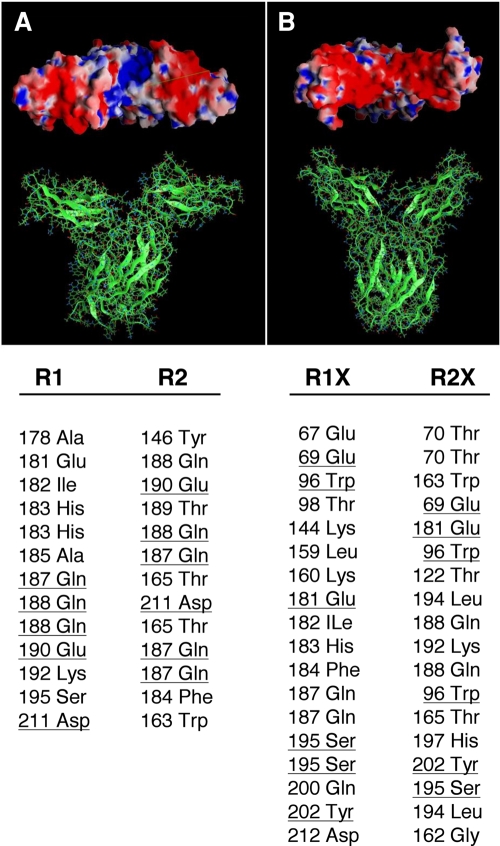
FIG. 5.
Molecular models of the wild-type (A) and double-mutant (B) EC domains of the hPRLR dimers. In the wild type, the intermolecular hydrogen bond network involves only the D2 subdomains, while both D1 and D2 subdomains are H bonded in the C36S, C46S, C75S, and C86S mutant. Electrostatic potentials on the molecular surfaces of the PRLR dimers are shown in the upper panels (viewed from the EC side toward the membrane plane). In the wild type, a groove of positive potential (blue) is flanked by two negative regions (red); this positive crevice is occluded in the mutant receptor by the D1 subdomains that are now H bonded. The lower panel lists the corresponding intermonomer H bonds of the EC domains as obtained in the molecular dynamics simulations of the receptors. The wild-type dimer contains 13 H-bonded pairs, involving 16 aa, with 4 aa common to both monomers (underlined); the mutant dimer contains 18 H-bonded pairs, involving 25 aa, with 5 aa common to both monomers (underlined). R1 and R2 denote the wild-type monomers; R1X and R2X denote the mutant monomers. Many of these intermonomer H bonds involve at least one backbone atom.
The differences observed in the relative orientation of the D1 and D2 subdomains in the monomers (Fig. 4B) are directly related to specific structural differences observed in the corresponding dimers (Fig. 5A and B). In the wild-type dimer, no intermonomer H bonds were observed involving D1s, and the dimers were stabilized only through interactions between D2s. The quaternary arrangement left a groove between D1s sufficiently accessible for ligand recognition and binding, as shown in Fig. 5A, top. In contrast, during the dynamics, the mutant dimer developed additional H bonds that bridged both D1s; Fig. 5B, bottom, lists the 18 persistent H bonds observed in the mutant dimers. These interactions further stabilize the mutant dimer with respect to the wild-type dimer but also close and lock the groove, making it inaccessible to the ligand (Fig. 5B, top). These observations may explain both the higher affinity suggested by the BRET analysis (Fig. 2) and the lack of function shown in Fig. 1 (see Discussion). It also provides a structural rationale for the lack of ligand binding to the mutant receptor, as previously reported (33).
DISCUSSION
In this study, we have provided evidence, derived from functional analysis, that the four conserved Cys residues in the D1 subdomain of the EC region of the hPRLR are essential to preserve the dominant-negative action of the S1b SF on ligand-induced LF-mediated STAT5-dependent transcriptional activation. These conclusions were supported by our computer modeling, which provided insights into the effects of the S-S bonds on the stabilization of the tertiary and quaternary structure of the receptor. Our studies have demonstrated that the inhibitory action of wild-type S1b on the LF function was abolished upon disruption of these S-S bonds. The lack of inhibitory action of mutated S1b forms may result from their higher affinity for forming homodimer association than the wild type (Fig. 2C and D) and the significant reduction in their affinity for forming heterodimers with the LF (Fig. 2A and B). The observed reduction in the affinity (Fig. 2B) between LF/S1b4x and LF/S1b was consistent with a marked reduction in heterodimer formation revealed by co-IP studies (Fig. 2E, lane 6 versus lane 4). Moreover, the marked increase in the affinity of the SF mutant (S1b4x-RL/S1b4x-Y) versus the wild type (S1b-RL/Sb-Y) was also reflected in the co-IP studies (Fig. 2E, lane 8 versus lane 10). The preference of the mutated S1b to form homodimers rather than heterodimers contrasted with the wild-type S1b propensity to form heterodimers with the LF. This in turn facilitated the formation of LF homodimers competent to mediate PRL-induced downstream signaling. Thus, the EC Cys mutant lacks the inhibitory action normally observed in the wild-type SF. Also, these studies demonstrated that either of the Cys pairs (36/46 and 75/86) was important in the heterodimerization process and for the inhibitory effect of the SF on LF-mediated PRL action.
It is not surprising that S1b mediated JAK2 phosphorylation activity upon PRL stimulation, since it contains a conserved proline-rich domain at the Box-1 docking site for JAK2 association that is known to engage constitutively with the PRLR LF (22). Janus kinases can trans phosphorylate themselves and/or undergo autophosphorylation (36). It is believed that the ligand, as the inducer of receptor aggregation or through conformational changes of existing dimers, causes tyrosine phosphorylation of the kinase at the activation loop and increases its catalytic activity. The significant increases in JAK2 phosphorylation induced by PRL in cells stably expressing the LF transfected with S1b (S1b versus control) reflects the additive effect of S1b and LF over the basal level present only in S1b (Fig. 3B). JAK2 activation associated with the SF with a short cytoplasmic sequence could have an impact on other potential signaling via mitogen-activated protein kinase, AKT, or JUNK, in contrast to STAT activation resulting from coupling to the additional cytoplasmic tail found in the LF. It was interesting to observe basal levels of JAK2 phosphorylation with S1b in the absence of PRL stimulation in cells both transiently and stably expressing S1b (Fig. 3A). Since no detectable basal JAK phosphorylation was associated with LF, it is possible that an impact of its extended cytoplasmic sequence on the overall conformation of the Box-1 region in the LF could prevent productive basal JAK2 phosphorylation. Thus, in the absence of hormone it appears that the shortened length of the cytoplasmic domain in S1b with only a breve sequence beyond Box-1 (8 aa identical to the LF and 3 unique aa) (16) would favor basal JAK2 phosphorylation (at Tyr 1007/1008 in the activation loop), presumably via auto- and/or trans phosphorylation. It is conceivable that the S1b association with JAK2 induces partial relaxation of the inhibition of the catalytic domain (JH1) by the pseudokinase (JH2) to cause basal activation. Additional disruption of the inhibition upon activation by a hormone would result in a further enhanced level (over the basal level) of JAK2 catalytic activity (Fig. 3A) (19). This could also apply to heterodimers, since basal JAK2 phosphorylation is not present with LF homodimers in the absence of hormone (Fig. 3B, control). Also, this may involve other associated kinase modules, as is the case for Src kinases in the activation of focal adhesion kinases (7). In contrast to S1b, complete loss of basal JAK2 phosphorylation of its Cys mutant (S1b4x-Y) was observed in transiently transfected cells. The apparent loss of JAK2 binding to S1b4x compared to the S1b wild type observed in the co-IP study revealed (Fig. 3A, right) a functional link between the EC domain conformation and the JAK2 association with its docking site at Box-1.
Wild-type S1b and the Cys mutant S1b4x were found to be similarly expressed at the cell surface by a biotin-avidin labeling procedure (Fig. 1C and F). This accounted for the effect of S1b on LF action at the cell membrane level and further validated the relevance of intramolecular Cys-Cys residues in the dominant-negative function of S1b on LF action. The loss of hormone activation of JAK2 in the Cys mutant of the S1b homodimer was expected, since these residues were found to be required for hormone binding by the LF homodimer in a point mutation study (33). In addition, impairment of JAK2 association with the Cys-mutated receptor demonstrated in this study (Fig. 3A) would render the receptor unresponsive to the hormonal stimulus. Thus, it is probable that the spatial constraints conveyed by the S-S bonds on tertiary folding of the PRLR, which is essential for PRL binding, would also participate in the association/dissociation properties of dimer/ heterodimer variants and/or their productive association with JAK2.
The molecular dynamics simulations of the EC domain of the wild-type and of the Cys mutant monomers showed substantial conformational changes in the latter, with less bending of D1 with respect to D2. These changes are triggered by the structural relaxation of the native-like D1 subdomain (Fig. 4B). In contrast, the wild-type monomer remained practically unchanged throughout the dynamics (Fig. 4B). The simulations were based on the X-ray structure of the EC domain of hPRLR and rPRLR (PDB codes 1bp3 [monomer] and 1f6f [dimer template]). The decrease in the angle between D1 and D2 observed during the simulations implies that both domains tend to align in the mutant monomer (ω, ∼30° to 40°), in contrast to the wild type, for which the relative orientation of the domains remained stable (ω, ∼70° to 90°) (Fig. 4A). The surface area buried in the hormone-receptor interface resides mainly in the subdomain D1 of the receptor, as suggested by the X-ray structure of the hGH-hPRLR complex (34). The conformational changes observed in the mutant reduce the accessible space for a ligand to bind the dimerized receptor. The simulations also showed that additional intermolecular H bonds can form in the mutant homodimers: in addition to the H bonds linking both D2 subdomains observed in the wild-type homodimer, several other residues form H bonds between the D1 subdomains. These additional H bonds in the mutant receptor are responsible for stabilizing the locked form of the dimer and may serve two functional roles: (i) prevent ligand binding to the receptor and (ii) increase the homodimer affinity of S1b4x. Estimation of the electrostatic potential on the molecular surface shows a positive region in the wild-type dimer interface (Fig. 5A, top) surrounded by a predominately negative potential. This electrostatic pattern could be a motif for ligand recognition and binding, which is absent in the mutant (Fig. 5B, top). The closure of the mutant binding site also explains structurally, based on simple steric arguments, why PRL may not bind to the PRLR mutant (33). Several of the H-bonded residues located in the D2 domains of the X-ray structure have been implicated in the formation of the receptor-receptor interface of dimerization (34). It is possible, then, that the additional H bonds observed in the mutant dimers could enhance the dimerization process and explain the comparatively significantly lower BRET50 signal.
Overall, the study reported here demonstrated the relevance of the S-S bonds of the PRLR for S1b inhibitory action on PRL-induced LF-mediated STAT5-dependent action and for cytoplasmic events related to JAK2 association/activity.
Acknowledgments
This work was supported by the NIH Intramural Research Program through the Eunice Kennedy Shriver National Institute of Child Health and Human Development and the Center for Information Technology.
The study utilized the high-performance computer capabilities of the Biowulf PC/Linux cluster at the NIH.
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Acosta, J. J., R. M. Munoz, L. Gonzalez, A. Subtil-Rodriguez, M. A. Dominguez-Caceres, J. M. Garcia-Martinez, A. Calcabrini, I. Lazaro-Trueba, and J. Martin-Perez. 2003. Src mediates prolactin-dependent proliferation of T47D and MCF7 cells via the activation of focal adhesion kinase/Erk1/2 and phosphatidylinositol 3-kinase pathways. Mol. Endocrinol. 172268-2282. [DOI] [PubMed] [Google Scholar]
- 2.Ali, S., and S. Ali. 2000. Recruitment of the protein-tyrosine phosphatase SHP-2 to the C-terminal tyrosine of the prolactin receptor and to the adaptor protein Gab2. J. Biol. Chem. 27539073-39080. [DOI] [PubMed] [Google Scholar]
- 3.Aman, M. J., T. S. Migone, A. Sasaki, D. P. Ascherman, M. Zhu, E. Soldaini, K. Imada, A. Miyajima, A. Yoshimura, and W. J. Leonard. 1999. CIS associates with the interleukin-2 receptor beta chain and inhibits interleukin-2-dependent signaling. J. Biol. Chem. 27430266-30272. [DOI] [PubMed] [Google Scholar]
- 4.Bazan, J. F. 1989. A novel family of growth factor receptors: a common binding domain in the growth hormone, prolactin, erythropoietin and IL-6 receptors, and the p75 IL-2 receptor beta-chain. Biochem. Biophys. Res. Commun. 164788-795. [DOI] [PubMed] [Google Scholar]
- 5.Bole-Feysot, C., V. Goffin, M. Edery, N. Binart, and P. A. Kelly. 1998. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr. Rev. 19225-268. [DOI] [PubMed] [Google Scholar]
- 6.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4182-217. [Google Scholar]
- 7.Calalb, M. B., T. R. Polte, and S. K. Hanks. 1995. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol. Cell. Biol. 15954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chughtai, N., S. Schimchowitsch, J. J. Lebrun, and S. Ali. 2002. Prolactin induces SHP-2 association with Stat5, nuclear translocation, and binding to the beta-casein gene promoter in mammary cells. J. Biol. Chem. 27731107-31114. [DOI] [PubMed] [Google Scholar]
- 9.Constantinescu, S. N., T. Keren, M. Socolovsky, H. Nam, Y. I. Henis, and H. F. Lodish. 2001. Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc. Natl. Acad. Sci. USA 984379-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das, R., and B. K. Vonderhaar. 1995. Transduction of prolactin's (PRL) growth signal through both long and short forms of the PRL receptor. Mol. Endocrinol. 91750-1759. [DOI] [PubMed] [Google Scholar]
- 11.Elkins, P. A., H. W. Christinger, Y. Sandowski, E. Sakal, A. Gertler, A. M. de Vos, and A. A. Kossiakoff. 2000. Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor. Nat. Struct. Biol. 7808-815. [DOI] [PubMed] [Google Scholar]
- 12.Gadd, S. L., and C. V. Clevenger. 2006. Ligand-independent dimerization of the human prolactin receptor isoforms: functional implications. Mol. Endocrinol. 202734-2746. [DOI] [PubMed] [Google Scholar]
- 13.Gent, J., P. van Kerkhof, M. Roza, G. Bu, and G. J. Strous. 2002. Ligand-independent growth hormone receptor dimerization occurs in the endoplasmic reticulum and is required for ubiquitin system-dependent endocytosis. Proc. Natl. Acad. Sci. USA 999858-9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan, S. A., F. Guarnieri, and E. L. Mehler. 2000. A general treatment of solvent effects based on screened Coulomb potentials J. Phys Chem. B 1046478-6489. [Google Scholar]
- 15.Hassan, S. A., E. L. Mehler, D. Zhang, and H. Weinstein. 2003. Molecular dynamics simulations of peptides and proteins with a continuum electrostatic model based on screened Coulomb potentials. Proteins 51109-125. [DOI] [PubMed] [Google Scholar]
- 16.Hu, Z. Z., J. Meng, and M. L. Dufau. 2001. Isolation and characterization of two novel forms of the human prolactin receptor generated by alternative splicing of a newly identified exon 11. J. Biol. Chem. 27641086-41094. [DOI] [PubMed] [Google Scholar]
- 17.Hu, Z. Z., L. Zhuang, J. Meng, M. Leondires, and M. L. Dufau. 1999. The human prolactin receptor gene structure and alternative promoter utilization: the generic promoter hPIII and a novel human promoter hP(N). J. Clin. Endocrinol. Metab. 841153-1156. [DOI] [PubMed] [Google Scholar]
- 18.Hu, Z. Z., L. Zhuang, J. Meng, C. H. Tsai-Morris, and M. L. Dufau. 2002. Complex 5′ genomic structure of the human prolactin receptor: multiple alternative exons 1 and promoter utilization. Endocrinology 1432139-2142. [DOI] [PubMed] [Google Scholar]
- 19.Ihle, J. N., and D. G. Gilliland. 2007. Jak2: normal function and role in hematopoietic disorders. Curr. Opin. Genet. Dev. 178-14. [DOI] [PubMed] [Google Scholar]
- 20.Ihle, J. N., and I. M. Kerr. 1995. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 1169-74. [DOI] [PubMed] [Google Scholar]
- 21.Kline, J. B., H. Roehrs, and C. V. Clevenger. 1999. Functional characterization of the intermediate isoform of the human prolactin receptor. J. Biol. Chem. 27435461-35468. [DOI] [PubMed] [Google Scholar]
- 22.Lebrun, J. J., S. Ali, A. Ullrich, and P. A. Kelly. 1995. Proline-rich sequence-mediated Jak2 association to the prolactin receptor is required but not sufficient for signal transduction. J. Biol. Chem. 27010664-10670. [DOI] [PubMed] [Google Scholar]
- 23.Li, X., S. A. Hassan, and E. L. Mehler. 2005. Long dynamics simulations of proteins using atomistic force fields and a continuum representation of solvent effects: calculation of structural and dynamic properties. Proteins 60464-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livnah, O., E. A. Stura, S. A. Middleton, D. L. Johnson, L. K. Jolliffe, and I. A. Wilson. 1999. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science 283987-990. [DOI] [PubMed] [Google Scholar]
- 25.MacKerell, A. D., Jr., M. Feig, and C. L. Brooks III. 2004. Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 126698-699. [DOI] [PubMed] [Google Scholar]
- 26.MacKerell, A. D., Jr., D. Bashford, M. Bellott, R. L. Dunbrack Jr., J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, I. W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1023586-3616. [DOI] [PubMed] [Google Scholar]
- 27.Mehler, E. L., S. A. Hassan, S. Kortagere, and H. Weinstein. 2006. Ab initio computational modeling of loops in G-protein-coupled receptors: lessons from the crystal structure of rhodopsin. Proteins 64673-690. [DOI] [PubMed] [Google Scholar]
- 28.Meng, J., C. H. Tsai-Morris, and M. L. Dufau. 2004. Human prolactin receptor variants in breast cancer: low ratio of short forms to the long-form human prolactin receptor associated with mammary carcinoma. Cancer Res. 645677-5682. [DOI] [PubMed] [Google Scholar]
- 29.Miller, B. T., R. P. Singh, J. B. Klauda, M. Hodoscek, B. R. Brooks, and H. L. Woodcock III. 2008. CHARMMing: a new, flexible web portal for CHARMM. J. Chem. Inf. Model 481920-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfleger, K. D., R. M. Seeber, and K. A. Eidne. 2006. Bioluminescence resonance energy transfer (BRET) for the real-time detection of protein-protein interactions. Nat. Protoc. 1337-345. [DOI] [PubMed] [Google Scholar]
- 31.Qazi, A. M., C. H. Tsai-Morris, and M. L. Dufau. 2006. Ligand-independent homo- and heterodimerization of human prolactin receptor variants: inhibitory action of the short forms by heterodimerization. Mol. Endocrinol. 201912-1923. [DOI] [PubMed] [Google Scholar]
- 32.Remy, I., I. A. Wilson, and S. W. Michnick. 1999. Erythropoietin receptor activation by a ligand-induced conformation change. Science 283990-993. [DOI] [PubMed] [Google Scholar]
- 33.Rozakis-Adcock, M., and P. A. Kelly. 1991. Mutational analysis of the ligand-binding domain of the prolactin receptor. J. Biol. Chem. 26616472-16477. [PubMed] [Google Scholar]
- 34.Somers, W., M. Ultsch, A. M. De Vos, and A. A. Kossiakoff. 1994. The X-ray structure of a growth hormone-prolactin receptor complex. Nature 372478-481. [DOI] [PubMed] [Google Scholar]
- 35.Trott, J. F., R. C. Hovey, S. Koduri, and B. K. Vonderhaar. 2004. Multiple new isoforms of the human prolactin receptor gene. Adv. Exp. Med. Biol. 554495-499. [DOI] [PubMed] [Google Scholar]
- 36.Yasukawa, H., H. Misawa, H. Sakamoto, M. Masuhara, A. Sasaki, T. Wakioka, S. Ohtsuka, T. Imaizumi, T. Matsuda, J. N. Ihle, and A. Yoshimura. 1999. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. 181309-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]