Abstract
Sir3, a component of the transcriptional silencing complex in the yeast Saccharomyces cerevisiae, has an N-terminal BAH domain that is crucial for the protein's silencing function. Previous work has shown that the N-terminal alanine residue of Sir3 (Ala2) and its acetylation play an important role in silencing. Here we show that the silencing defects of Sir3 Ala2 mutants can be suppressed by mutations in histones H3 and H4, specifically, by H3 D77N and H4 H75Y mutations. Additionally, a mutational analysis demonstrates that three separate regions of the Sir3 BAH domain are important for its role in silencing. Many of these BAH mutations also can be suppressed by the H3 D77N and H4 H75Y mutations. In agreement with the results of others, in vitro experiments show that the Sir3 BAH domain can interact with partially purified nucleosomes. The silencing-defective BAH mutants are defective for this interaction. These results, together with the previously characterized interaction between the C-terminal region of Sir3 and the histone H3/H4 tails, suggest that Sir3 utilizes multiple domains to interact with nucleosomes.
Transcriptional silencing in the yeast Saccharomyces cerevisiae occurs at the silent mating type loci, HML and HMR, at genes near telomeres, and at the ribosomal DNA. The silencer elements flanking the HM loci recruit the DNA binding proteins Rap1, Abf1, and Orc1, while telomeric sequences bind Rap1. These DNA-bound proteins in turn recruit the silent information regulator (SIR) proteins. Multiple protein-protein interactions lead to spreading of a Sir2, Sir3, and Sir4 complex to nearby nucleosomes (reviewed in references 11 and 35). Sir2 plays a crucial role in this spreading by deacetylating histone H4 K16, thus allowing Sir3 and Sir4 to bind to nucleosomes and allowing further spreading of the Sir complex (reviewed in references 11, 26, and 35). Sir3 protein levels seem to control the extent of silencing. Silencing normally spreads up to 4 kb from the telomeres but can go as far as 20 kb away from the telomeric ends when Sir3 is overexpressed (16, 34). While the exact mechanism by which transcription is silenced at these loci is not yet clear, it seems to involve a specialized chromatin structure that prevents transcription at either initiation or elongation (6, 36).
The N-terminal tails of histones H4 and H3 are important for silencing. Mutations in these tail regions show loss of silencing because they cause reduced binding of the Sir2, -3, and -4 complex (17, 19, 41). In addition to the N-terminal tails, a group of residues in the core domain of H3 and H4 are important for silencing (31, 42). These residues cluster around H3 K79, a site of methylation by Dot1, forming a patch on the surface of the nucleosome that could be a potential site for interaction with the silencing complex (23, 44, 49). Silencing is restricted to silent loci by many redundant mechanisms that prevent the spread of the silencing complex from the silent chromatin to active chromatin. Acetylation at H4 K16, methylation at H3 K4 and K79, and incorporation of the H2A.Z variant into nucleosomes are known hallmarks of active chromatin and, as such, antisilencing factors (25; reviewed in reference 45). Overexpression of Sas2, the acetyltransferase for H4 K16, or of Dot1, the methyltransferase for H3 K79, leads to loss of silencing as the silencing proteins are driven off the silent loci by the modified lysines (1, 44). On the other hand, loss of these active chromatin markers allows the silencing complex to spread from the silent loci to the active chromatin (20, 40, 44, 47). Such spreading dilutes the Sir proteins at the silent loci and, hence, weakens silencing. This explains why sas2, dot1, and set1 mutants have telomeric silencing defects. The delineation between active and silent chromatin by the modifications on histone proteins helps to localize the silencing complex to the silent loci.
Sir3 interacts with Sir4, Rap1, Abf1, H3, and H4 N-terminal tails through the C-terminal two-thirds of its 978 amino acids (reviewed in references 9 and 35). However, mutants that lack the N-terminal domain are defective in silencing, suggesting that the N terminus plays an important role in silencing (14). Mutants with mutations in the N-terminal domain have also been isolated as enhancers of the minor sir1 silencing defect (38). The N-terminal region of Sir3 contains a bromo-adjacent homology (BAH) domain found in several other chromatin-associated proteins, such as Orc1 and Rsc1 and Rsc2, subunits of the RSC remodeling complex (5). Previous work from our laboratory has shown that the Sir3 BAH domain can establish some silencing in the absence of the rest of the Sir3 protein, especially if Sir1 is overexpressed (7). It was also shown that this short fragment of Sir3 can spread along the silent loci to establish silencing even though the Sir3 C-terminal region responsible for binding to the H3/H4 tails is missing (7). This suggests that the Sir3 BAH domain is able to mediate protein-protein interactions with Sir proteins or nucleosomes independent of the interactions mediated by the Sir3 C-terminal region. This has been directly demonstrated by Moazed's group, who showed that the Sir3 BAH domain can bind to nucleosomes and to H3/H4 tetramers (29). They also showed that the BAH domain binds with greater affinity to nucleosomes lacking acetylation on H4 Lys16 or methylation of H3 Lys79, results that agree well with the in vivo silencing results with sas2 or dot1 mutants (or overexpression plasmids) described above.
Previous work from our laboratory had also shown that the N-terminal alanine residue of Sir3, Ala2, plays an important role in silencing. Mutating this residue, or affecting its acetylation state by mutating the Nα-acetyltransferase, NatA, that acetylates it, weakened silencing (39, 48). Our initial goal in the work described here was to determine the protein to which the N-terminal residue of Sir3 bound. We used a genetic screen to look for mutants that suppressed the silencing defect of a sir3 mutant with Gly instead of Ala at its N terminus. Interestingly, we found that two mutations in the nucleosome core, H3 D77N and H4 H75Y, suppressed the silencing defect of the Sir3 A2G mutant. In the course of this work, we also identified several mutations in the specific regions of the Sir3 BAH domain that affect silencing. A recent report described similar mutants (3). The results described here, together with previously published results, provide genetic and biochemical evidence that the N-terminal BAH domain of Sir3 interacts with the nucleosome and thus explain why this domain is important for silencing.
MATERIALS AND METHODS
Strains and plasmids.
The strains used in this study are listed in Table 1. They were grown in yeast extract-peptone-dextrose (YPD) or synthetic complete (SC) medium (2). Plasmid transformations were performed according to standard protocols (2). Gene replacements were performed by replacing the open reading frame (ORF) with Streptomyces hygroscopicus hphMX, a hygromycin resistance cassette; Streptomyces noursei natMX, a nourseothricin resistance cassette; or other standard markers (13).
TABLE 1.
Yeast strains used in this study
| Strain | Genotypea |
|---|---|
| W303-1a | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
| JCY3 | W303-1a sir3Δ::kanMX6 |
| JCY4 | W303-1b sir3Δ::kanMX6 |
| JCY8 | W303-1a sir3Δ::kanMX6 sir1Δ::S.p.his5+ |
| JCY9 | W303-1b sir3Δ::kanMX6 sir1Δ::S.p.his5+ |
| JCY5 | W303-1a ard1Δ::kanMX6 |
| DC16 | MATahis1 |
| DC17 | MATα his1 |
| XRY16 | W303-1b TEL-VIIL-ADH4::URA3 sir3Δ::kanMX6 |
| PYY4 | MATahis3Δ200 trp1-901 leu2-3,112 ade2 LYS2::(4lexAop-HIS3) URA3::(8lexAop-lacZ) TEL-VIIL-ADH4::URA3 ppr1Δ::natMX4 sir3Δ::kanMX6 |
| XRY36 | MATα HML::URA TEL-VR::ADE2 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ura3-52 sir3Δ::kanMX6 sir1Δ::natMX hht1Δ-hhf1Δ::S.p.his5+ |
| VSY29 | MATα HML::URA3 TEL-VR::ADE2 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ura3-52 sir3Δ::kanMX6 sir1Δ::natMX hht1Δ-hhf1Δ::S.p.his5+hht2Δ-hhf2Δ::hphMX4; pHHT2-HHF2 LEU2 CEN |
| VSY30 | MATα HML::URA3 TEL-VR::ADE2 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ura3-52 sir3Δ::kanMX6 sir1Δ::natMX hht1Δ-hhf1Δ::S.p.his5+hht2Δ-hhf2Δ::hphMX4; pHHT2 D77N-HHF2 LEU2 CEN |
| VSY31 | MATα HML::URA3 TEL-VR::ADE2 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ura3-52 sir3Δ::kanMX6 sir1Δ::natMX hht1Δ-hhf1Δ::S.p.his5+hht2Δ-hhf2Δ::hphMX4; pHHT2-HHF2 H75Y LEU2 CEN |
| UCC7014 | W303-1a dot1Δ::kanMX6 |
| PYY12 | MATα HML::URA TEL-VR::ADE2 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ura3-52 sir3Δ::kanMX6 sir1Δ::natMX hht1Δ-hhf1Δ::S.p.his5+hht2Δ-hhf2Δ:: hphMX4; pHHT1-HHF1 K16R LEU2 CEN |
| VSY38 | W303-1a hht1Δ-hhf1Δ::S.p.his5+hht2Δ-hhf2Δ::hphMX4; pHHT2-HHF2 LEU2 CEN |
| VSY39 | W303-1a hht1Δ-hhf1Δ::S.p.his5+hht2Δ-hhf2Δ::hphMX4; pHHT2 D77N-HHF2 LEU2 CEN |
| VSY41 | W303-1a hht1Δ-hhf1Δ::S.p.his5+TEL-VIIL-ADH4::URA3 sir3Δ::kanMX6 |
| VSY43 | W303-1a hht1Δ-hhf1Δ::S.p.his5+TEL-VIIL-ADH4::URA3 sir3Δ::kanMX6 dot1Δ::hphMX4 |
S.p.his5+, Schizosaccharomyces pombe his5+ gene.
The plasmid pPY41 was generated by sequential cloning of the SIR3 promoter region (−300 to +1), the SIR3 ORF in frame with LexA, and the TADH1 in the plasmid pRS314 (TRP1 CEN). pPY17 expresses the coding region corresponding to amino acids (aa) 94 to 566 of Rad7 as a fusion to the Gal4 activation domain (Gal4AD) in pGAD424 (Clontech, United States). The mutants generated from the PCR-based mutagenesis of the Sir3 ORF were subcloned as BamHI-PstI fragments into pXR58 (PSIR3-SIR3-TSIR3 TRP1 CEN) to replace the wild-type BAH domain with the mutant BAH domain. The mutant plasmids are pPY122 (sir3A2V), pPY125 (sir3N80D), pPY120 (sir3F94L), pPY123 (sir3F123P), pPY118 (sir3A136T), pPY117 (sir3C177R), pPY119 (sir3A181V), pPY126 (sir3S204P), pPY127 (sir3Y207C), and pPY124 (sir3K209R). Mutant plasmids pXR61 (sir3A2Q), pXR62 (sir3A2T), pXR63 (sir3A2S), and pXR64 (sir3A2G) were generated by site-directed mutagenesis of pXR58, as described before (48). Plasmids pPY128 (sir3T4F) and pPY129 (sir3L5A) were generated by site-directed mutagenesis of pPY41 and subcloned into the pXR58 background. The 1.6-kb histone HHT2-HHF2 fragment was cloned by PCR amplification from the wild-type strain or the D77N or H75Y suppressor into pCR2.1 Topo using primers 5′ ATGTCCCCCCAGTCTAAAT 3′ and 5′ GGTTCTATTATATTCCCAA 3′. The SpeI-XhoI HHT2-HHF2 fragment was subcloned into pRS315 (LEU2 CEN) to generate pVS11 (wild type), pVS12 (D77N), and pVS16 (H75Y). The HHT2-HHF2 K16R plasmid (LEU2 CEN) has been described before (24).
pJC82 has the glutathione S-transferase (GST) ORF subcloned from plasmid pDS472a (ATCC) into pET28a NotI-XhoI sites to allow for C-terminal GST tagging of proteins. The coding region of Sir3 aa 1 to 219 was cloned as an NcoI-BamHI fragment in frame with the GST ORF in pJC82 to generate pEP14. Mutants in this background were pVS32 (sir3A2Q), pPY136 (sir3N80D), pPY135 (sir3F94L), pPY116 (sir3A136T), pPY115 (sir3C177R), pPY134 (sir3A181V), pPY114 (sir3S204P), and pPY111 (sir3D205N).
For protein expression and purification from yeast, the Sir3 ORF fragments tagged with GST at their C termini were cloned as SpeI-PstI fragments in p425TEF (PTEF LEU2 CEN). pVS33 contained the coding region of Sir3 aa 1 to 219. pVS34 and pVS35 expressed sir3A2G and sir3A2Q mutants from the pEP14 background described above.
EMS mutagenesis screen for second-site suppressors of the Sir3 A2G mutant.
Strain XRY36 was grown overnight to a density of 2 × 108 cells/ml. One milliliter of cells was pelleted, washed once with sterile water and once with 0.2 M Na3PO4, pH 7, and then resuspended in 1 ml of 0.2 M Na3PO4, pH 7. Thirty microliters of EMS (ethyl methanesulfonate) was added, and the mixture vigorously vortexed for 30 min at 30°C. Cells were collected by centrifugation and washed twice with 1 ml of 5% NaS2O3 and once with sterile water before being resuspended in sterile YPD medium. An untreated control was processed as described above but with the EMS treatment omitted. Fractions of the treated and untreated samples were diluted and plated on the appropriate medium to calculate killing rate. Two rounds of EMS mutagenesis, one at the 90% killing rate and the other at the 50% killing rate, were performed (4).
The EMS-treated samples were grown overnight in YPD medium and transformed with plasmid pXR64 (Sir3 A2G TRP1 CEN). Strain XRY36 has two reporter genes, TEL-VR::ADE2 and HML::URA3. Suppressors of the Sir3 A2G silencing defect were screened for 5-fluorootic acid (5-FOA) resistance and pink color, indicating good suppression of both reporter genes. Twelve candidates from the first round and 18 candidates from the second round of mutagenesis were screened further.
Screen for random mutations in the BAH domain.
Mutants with random mutations in the first 757 bp of the SIR3 ORF were generated during routine PCR with Taq DNA polymerase using the 50-base primers 5′ TTACAGGGGTTTAAGAAAGTTGTTTTGTTCTAACAATTCGATTAGCTAAAGGATCC 3′ and 5′ TGTTGACGTTCCTCGCTGAGACGGTATATTTCCATTATTTACGTCATCAT 3′. pPY41 was used to generate a gapped plasmid lacking bp 1 to 757 of Sir3. The PCR-mutagenized pool and the gapped plasmid were transformed into the background strain PPY4 containing the Rad7-Gal4AD plasmid, pPY17. In vivo recombination between the gapped plasmid and the PCR fragment generated Trp+ transformants. PPY4 has a TEL-URA3 reporter and a LexAop-LacZ reporter. In the first round of screening, β-galactosidase (β-gal) assays were performed to monitor the ability of the mutant Sir3-LexA to interact with Rad7-Gal4AD, indicating an absence of stop codon in the mutant Sir3 ORF. β-Gal-positive colonies were further screened for their ability to silence the TEL-URA3 reporter by monitoring their growth on 5-FOA plates for 3 days. The mutants that did not show any growth on 5-FOA plates were screened further by other silencing assays.
Silencing assays.
For telomeric and HML reporter assays, strains with plasmids were grown overnight in appropriate SC medium. Tenfold serial dilutions were generated from 2 optical density units of cells and spotted onto 5-FOA-containing plates (for TEL-URA3 and HML-URA3 reporter assays) or SC medium plates containing low levels of adenine (for the TEL-ADE2 reporter assay). The ability of the strains to grow was recorded after 3 days at 30°C for 5-FOA plates. To enhance the visualization of the pink color on low-adenine plates, the plates were incubated for 3 days at 30°C, followed by 2 days at 4°C, before being photographed.
For mating assays, 10-fold serial dilutions generated as described above were spotted onto newly replicated lawns of mating type tester strains (DC16/DC17) in YPD and incubated overnight at 30°C, followed by replica plating onto synthetic defined medium lacking any amino acids to select for diploids. The plates were incubated at 30°C for 2 days and photographed.
ChIPs.
Chromatin immunoprecipitation (ChIP) assays were performed from strains VSY29, VSY30, and VSY31 carrying pRS314, pXR58 (wild-type Sir3), or pXR64 (Sir3 A2G) as described before (21). Briefly, cell extracts from cross-linked cultures were sonicated for 12 cycles of 20 s each in an Ultrasonics, Inc., sonicator (model W220-F). One microgram of anti-Sir3 antibody (generated in our laboratory) was used to immunoprecipitate Sir3 from 2.5 mg of total protein extract. Input and immunoprecipitated DNA samples were analyzed by real-time PCR using a Mastercycler ep realplex thermal cycler (Eppendorf AG, GmBH) and LightCycler SYBR green master mix (Roche Applied Science, United States). The primers used were for HMR-a1 (5′ CAGTTTCCCCGAAAGAACAA 3′ and 5′ CCATCCGCCGATTTATTTT 3′), HMR-E (5′ ACCAGGAGTACCTGCGCTTA 3′ and 5′ TGCAAAAACCCATCAACCTGG 3′), 0.67-kb TEL-VIR (5′ CAGGCAGTCCTTTCTATTTC 3′ and 5′ GCTTGTTAACTCTCCGACAG 3′), and 5-kb TEL-VIR (5′ CGGACATGAATACTGGGTTCGTGA 3′ and 5′ CGAGACCCACTTGTATTCTTAGTGC 3′). The results presented are the averages and standard deviations of the results of duplicates from one representative experiment.
Protein purification from Escherichia coli.
Sir31-219-GST proteins were expressed from plasmid pEP14 (wild type) and its mutants (pVS32, pPY111, pPY114-116, and pPY134-136). Expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside and 3% ethanol for 5 or 6 h at room temperature. The GST-tagged proteins were purified by using glutathione Sepharose 4 fast flow resin (GE Healthcare, United States), following the manufacturer's instructions. Purified proteins were dialyzed against a buffer containing 50 mM HEPES, pH 7.5, 25 mM KCl, 1 mM EDTA, 1 mM EGTA, 5 mM magnesium acetate. Protein concentrations were estimated by comparison of Coomassie blue staining of samples to that of bovine serum albumin standards, as well as quantitation using a Bio-Rad protein assay (Bio-Rad, United States).
Protein purification from yeast.
W303-1a transformed with the plasmids expressing wild-type Sir31-219-GST and its mutants (pVS33 to pVS35) were grown in appropriate synthetic medium to an optical density of 1.2. Cells were lysed by using Yeast Buster reagent (EMD Biosciences, United States) according to the manufacturer's instructions. The supernatant was mixed with glutathione-Sepharose beads at 4°C overnight. The beads were washed with 1× PBS, and the GST proteins bound to beads eluted in 10 mM Tris, pH 8, 40 mM glutathione. The eluted proteins were dialyzed into buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 5 mM KCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM benzamidine, and other protease inhibitors) used for nucleosome binding experiments. Proteins were quantitated by using bovine serum albumin standards on a Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gel.
Nucleosome purification.
Partially purified nucleosomes were purified as described previously with some modifications (10, 33). Briefly, spheroplasts were isolated from 1-liter cultures of W3031-a and lysed by Dounce homogenization in lysis buffer (10 mM Tris, pH 7.5, 18% Ficoll, 20 mM KCl, 5 mM MgCl2, 1 mM EDTA, 0.25 mM EGTA, 0.5 mM spermidine, 3 mM dithiothreitol, 2 mM benzamidine, 2 mM sodium metabisulfite, 1 mM PMSF, protease inhibitors). The nuclei generated were washed in buffer A with and without 0.5% NP-40 (10 mM Tris-HCl, pH 7.5, 75 mM NaCl, 1 M sorbitol, 2 mM benzamidine, 2 mM sodium metabisulfite, 1 mM PMSF, and other protease inhibitors). Micrococcal nuclease (Worthington Biochemicals, United States) at 200 U/ml was added to nuclei resuspended in buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM KCl, 1 mM EDTA, 1 mM PMSF, 2 mM benzamidine, and other protease inhibitors) in the presence of 5 mM CaCl2 and incubated at 37°C for 15 min. Reactions were stopped by the addition of EGTA to 20 mM, and supernatant used as the source of partially purified nucleosomes. The presence of nucleosomes of multiple sizes was confirmed by digesting the purified material with 0.5 mg/ml Proteinase K at 50°C for 1 h, followed by electrophoresis on a 2% agarose gel and staining with ethidium bromide.
Nucleosome binding assay and Western blotting.
The purified GST-tagged proteins from yeast (different concentrations) or E. coli (20 μg) were incubated with glutathione Sepharose beads on ice and mixed with 200 to 400 μl of partially purified nucleosomes for 2 h at 4°C. The beads were spun at 3,000 rpm for 1 min, and the supernatant removed by using a syringe with a 30-guage needle. The beads were washed twice with 300 μl of wash buffer (20 mM Tris, pH 8, 25 mM NaCl, 1 mM dithiothreitol). The bound proteins were released by boiling in 2× SDS-gel loading buffer. A fraction of the bound proteins were analyzed on a 15% SDS-polyacrylamide gel electrophoresis gel. The gel was cut at a position corresponding to 25 kDa, and the top portion stained by Coomassie blue staining to visualize the ∼50-kDa Sir3-GST fragments. The lower portion of the gel was immunoblotted, using anti-H3 antibody (Abcam ab1791) as the primary antibody and anti-rabbit immunoglobulin G-horseradish peroxidase as the secondary antibody, and visualized by using an ECL Plus system (GE Healthcare, United States). The anti-dimethyl K79 antibody used for Western blotting was generated in the van Leeuwen laboratory.
RESULTS
We used two approaches to understand the function of the N-terminal BAH domain of Sir3 in silencing. First, we performed a genetic screen to isolate second-site suppressors of the previously characterized Sir3 A2G mutant (48). Second, we used PCR-based random mutagenesis to identify residues in the BAH domain that are important for telomeric silencing.
Genetic screen for suppressors of the Sir3 A2G silencing defect.
As mentioned above, previous work from our laboratory showed that the N-terminal alanine residue of Sir3 is important for silencing at both the HM and TEL loci (48). This analysis led us to hypothesize that Sir3 interacts with other proteins through the extreme N terminus. In this work, we used a suppressor screen to identify such interacting proteins.
After EMS mutagenesis, we looked for mutants that restored silencing to a Sir3 Ala-to-Gly (A2G) mutant. We used this mutant because its phenotypes were moderate, it lacked acetylation at its N terminus, and we presumed that the small size of the glycine residue would be readily suppressed by second-site mutants. The strain used for the screen (XRY36) was MATα sir3Δ sir1Δ, carrying a CEN plasmid expressing the Sir3 A2G mutant protein. The reason for the sir1Δ mutation was that Sir3 A2G only shows defects in silencing the HM loci when Sir1 is absent (48). The strain also had a deletion of one of the copies of the histone H3/H4 genes (hht1Δ-hhf1Δ), as we thought having both copies might mask any suppressor mutations in these genes. Silencing in this strain was assessed by using HML::URA3 and TEL-VR::ADE2 reporter genes. In the presence of wild-type Sir3, these genes were silenced, as monitored by growth on 5-FOA and the formation of pink colonies on low-adenine medium (Fig. 1A). On the other hand, the Sir3 A2G mutant was 5-FOA sensitive and formed white colonies. When the mutagenesis was done on the strain carrying the Sir3 A2G plasmid, the only mutations obtained were intragenic suppressors that changed Sir3 residue 205 from Asp to Asn. This is a well-known suppressor of many silencing defects (17, 22, 32, 42). To avoid intragenic suppressors, we mutagenized the strain without the Sir3 A2G plasmid and then transformed the plasmid into the mutagenized cells. With this method, we identified 20 putative suppressors that formed 5-FOA-resistant and pink colonies.
FIG. 1.
(A) A screen for second-site suppressors of the Sir3 A2G silencing defect. Expected results for wild-type Sir3, the Sir3 A2G mutant, and a putative suppressor (sup X) of Sir3 A2G are depicted. Strain XRY36 was used for the screen. 5-FOAr, 5-FOA resistant. (B) H3 D77N and H4 H75Y mutants can suppress the silencing defects of some Sir3 Ala2 mutants. MATα sir3Δ sir1Δ strains carrying the indicated alleles of Sir3 Ala2 in combination with either a plasmid encoding wild-type (WT) H3/H4 (top panels) or H3 D77N/H4 (middle panels) or H3/H4 H75Y (bottom panels) were assayed for silencing at HML and HMR, using strains VSY29, VSY30, and VSY31, respectively. Silencing at HML::URA3 was assessed on 5-FOA medium, and silencing at HMR by a mating assay. Silencing at a telomere was assessed by pink colony color on low-adenine medium.
Crossing each of these mutants with an appropriate strain of opposite mating type showed that almost all the mutations were dominant. This would have made it difficult to clone the corresponding wild-type gene by complementation. For this reason and because we suspected that some of these mutations might be in the genes for H3 or H4, we sequenced the HHT2-HHF2 locus (the only H3/H4 locus present) in each of the mutants. Interestingly, of 18 suppressors characterized, 15 had mutations in the gene for histone H3 or H4. Thirteen mutants had an H3 D77N mutation, and two had an H4 H75Y mutation. At least some of these 15 histone mutants were independent isolates, as they were recovered from separate rounds of EMS mutagenesis or had different growth properties due to secondary mutations. The three suppressors that did not have mutations in the genes for H3 or H4 did not segregate as single mutations in crosses and were not characterized further.
H3 D77N and H4 H75Y mutants were bona fide suppressors of the silencing defects of Sir3 A2G.
To verify that the observed suppression of the silencing defect was due to the histone H3/H4 mutations, we cloned the HHT2-HHF2 locus from these suppressors onto plasmids and analyzed whether the histone mutations could suppress Sir3 A2G and other Sir3 Ala2 mutants in strains that had not been subjected to mutagenesis. To do this, we generated strains carrying the wild-type or mutant HHT2-HHF2 genes on a plasmid as the only source of histone genes. As with the strain used for the screen, these strains were MATα sir3Δ sir1Δ and carried the same two reporter genes, HML::URA3 and TEL-VR::ADE2. In addition, silencing at HMR could be tested by mating. The strain with the wild-type histone H3/H4 genes gave the expected results with the different Sir3 Ala2 mutants; only wild-type Sir3 and the A2S mutant were capable of silencing both HM loci and the telomere (Fig. 1B, top panel). As reported previously, the Sir3 A2Q, A2T, and A2G mutants were silencing defective with wild-type histones (48). As seen in Fig. 1B, middle and bottom panels, the plasmid-borne H3 D77N and H4 H75Y histone mutations each suppressed the silencing defect of the Sir3 A2G mutant at both HM loci and the telomere, thus demonstrating that they were the bona fide suppressors found in the original screen. The histone mutations also suppressed the silencing defects of a Sir3 A2T mutant but not a Sir3 A2Q mutant. This indicated that the histone mutations could suppress silencing defects of weak mutants, like Sir3 A2G or A2T, but not a strong mutant, like A2Q.
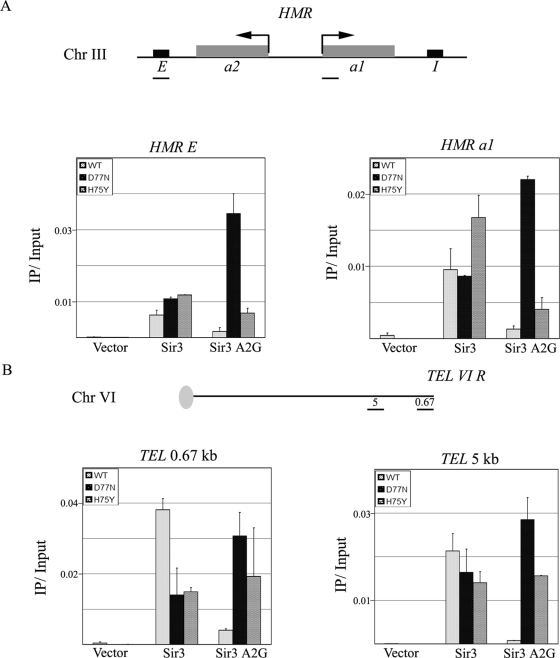
We performed ChIP of Sir3 at HMR and at two locations near TEL-VIR. There was decreased occupancy of Sir3 A2G compared to the occupancy of wild-type Sir3 at both HMR and TEL-VIR in strains with wild-type histones (Fig. 2). In the presence of the H3 D77N mutation, there was a striking increase in occupancy of Sir3 A2G at both loci. This was also true for strains with H75Y nucleosomes, but the effect was less pronounced. The correlation between the results of the in vivo silencing assays and the ChIP data suggested that the suppression by the histone H3 D77N and H4 H75Y mutations was due to increased recruitment of the Sir3-containing silencing complex to silent loci.
FIG. 2.
Sir3 occupancy at silent regions increases in the presence of the H3 D77N mutant. (A) Anti-Sir3 antibodies were used in ChIP experiments to determine localization of Sir3 at HMR E and HMR-a1 (histograms). A schematic representation of the HMR locus and the regions analyzed is shown. The bar graphs show average Sir3 occupancy with vector, wild-type Sir3, or Sir3 A2G plasmids in strains with wild-type H3 (VSY29), H3 D77N (VSY30), or H4 H75Y (VSY31). (B) A schematic representation of TEL-VIR and the regions analyzed is shown. Sir3 occupancy at 0.67 kb and 5 kb from the telomere is presented in the bar graphs. Strains used were the same as for the experiment whose results are shown in panel A. The data shown are the averages and standard deviations of the results of two PCR amplifications of one representative IP. Sir3 localization in the presence of wild-type histone H3, the histone H3 D77N mutant, or the histone H4 H75Y mutant is depicted. WT, wild type.
Loss of H3 K79 methylation did not suppress the silencing defects of Sir3 Ala2 mutants.
H3 D77 and H4 H75 localize to a region around H3 K79 in the nucleosome core (see Fig. 7B). H3 K79 methylation is one of the factors distinguishing euchromatin from heterochromatin (45). The high levels of H3 K79 methylation in euchromatin are thought to inhibit binding of the Sir complex, whereas heterochromatin, with lower levels of K79 methylation, permits binding of the Sir complex and allows silencing to occur at these loci (18, 27, 44).
FIG. 7.
Location of the mutated residues in Sir3 BAH domain and in the nucleosome. (A) Sir3 BAH domain crystal structure (Protein Data Bank accession number 2FVU) was used to pinpoint the location of the mutated residues by using PyMOL. The region 1 mutant is colored orange, region 2 mutants are colored blue, and region 3 mutants are colored red. (B) The crystal structure of the nucleosome from Xenopus laevis (Protein Data Bank accession number 1KX5) was used to visualize the location of the suppressor mutations in histones H3 and H4 by using PyMOL. H3 is colored blue, H4 yellow, and H2A and H2B green. Residues depicted as spheres are H3 D77 in light pink, H3 K79 in brown, H4 H75 in red, and H4 K16 in purple.
Thus, we considered the possibility that the D77N mutation resulted in a loss of K79 methylation, leading to better binding of the Sir complex and, hence, suppression. However, as evaluated by a Western blot assay, H3 D77N mutants retained K79 dimethylation even though the levels of methylation were somewhat lower than in the wild-type histone control (Fig. 3A). This reduction could be reflective of the reduced ability of the antibody raised to an epitope containing D77 to recognize N77. Some commercial antibodies to different H3 K79-methylated states did not recognize H3 with the N77 mutation and, hence, the levels of mono- or trimethylated K79 could not be evaluated.
FIG. 3.
Suppression by histone H3 D77N mutant is not mimicked by a loss of K79 methylation. (A) Strains W303-1a, UCC7014 (dot1Δ), VSY38 (wild-type H3), and VSY39 (H3 D77N) were assayed for levels of dimethyl K79 and H3 by Western blotting. The lower band marked by an asterisk in the Western blot for dimethylated K79 (me2 K79) is a nonspecific band recognized by the antibody and used as an internal loading control. α, anti. (B) Genetic interaction between dot1Δ and H3 D77N mutations. A DOT1 sir3Δ strain (VSY41) and a dot1Δ sir3Δ strain (VSY43) carrying various plasmid-borne alleles of Sir3 in combination with either a wild-type histone H3/H4 plasmid or a histone H3 D77N/H4 plasmid were assayed for telomeric silencing by growth on 5-FOA plates. WT, wild type.
To confirm that D77N suppression was not due to a lack of K79 methylation, we compared the suppression in DOT1 to that in dot1Δ strains. For these experiments, we tested suppression of the silencing defect of various Sir3 Ala2 mutants by using a URA3 reporter gene at a telomere. As seen in Fig. 3B, top panels, in a DOT1 strain, there is no suppression of Sir3 A2G or A2T mutants in the presence of wild-type histones but there is in the presence of H3 D77N histones. This confirms the initial results shown in Fig. 1 but with a different reporter gene and a different strain background. It should be pointed out that these experiments were done in a strain in which a wild-type H3/H4 locus (HHT2-HHF2) was present on the genome, thus confirming the dominance of the D77N mutation. The results for the dot1Δ strain in the presence of wild-type or D77N histones are shown in the bottom two panels of Fig. 3B. They show that lack of K79 methylation does not lead to suppression of Sir3 A2G and A2T mutants in the presence of wild-type histones. Thus, the suppression ability of D77N cannot be due to an inhibition of K79 methylation. Fig. 3B also shows that in the dot1Δ mutants, silencing with wild-type Sir3 and the Sir3 A2S mutant is poor with wild-type H3 and not detected with H3 D77N. The results with wild-type H3 confirm what was known previously, that dot1Δ mutants have a telomere silencing defect (although we observed more silencing than has been seen previously for dot1Δ strains, perhaps because more Sir3 is produced from a CEN plasmid than from the chromosomal locus). The presence of H3 D77N exacerbates this defect. In both cases, the loss of silencing is presumably due to the spreading and, hence, dilution of Sir proteins from the telomeres into euchromatin due to lack of methylation of K79 in dot1Δ strains. The dot1Δ mutant also did not suppress Sir3 Ala2 mutants at HMR (data not shown). Suppression could not be tested at HML because a dot1 sir1 double mutant is silencing defective even with wild-type Sir3 (46).
Random mutagenesis of the Sir3 BAH domain identified residues important for silencing.
Another goal of this work was to screen for mutations in the BAH domain that affected silencing. We used mutagenic PCR and screened for mutants as outlined in Fig. 4A and described in detail in Materials and Methods. Briefly, the DNA sequence corresponding to aa 1 to 253 of Sir3 was mutagenized by PCR. Gap repair was used to recombine the mutagenized fragments into a plasmid encoding full-length Sir3 with LexA fused to its C terminus. Plasmids were transformed into a strain with a two-hybrid lacZ reporter gene and assayed for the ability to interact with a Rad7-Gal4AD hybrid. It is known that Rad7 binds to the C-terminal region of Sir3 (30). Therefore, a positive two-hybrid signal indicated that no stop codon was introduced during the mutagenesis. The β-gal-positive colonies were assayed for their ability to silence a TEL-URA3 reporter gene that is present in the strain. Of 24,000 transformants screened, 9 silencing-defective sir3 mutants with single amino acid changes in the BAH domain were identified. The nine mutants, plus one that was made by site-directed mutagenesis (F123P), were subcloned into plasmids expressing full-length Sir3 from its own promoter and terminator without the LexA fusion. These mutants were then assessed for their silencing ability at a telomere and the HM loci. As seen by the results shown in Table 2, all the mutants except one were completely defective at telomeric silencing; the Y207C mutant was only partially defective. Data for four representative mutants are shown in Fig. 4B and C. The mutants showed various abilities to silence the HML locus, but none of them showed any observable HMR silencing defect (Fig. 4C and Table 2). We categorized the 10 mutants as strong (defective at TEL and HML), moderate (defective at TEL, moderately affected at HML), and weak (defective at TEL but not at HML or HMR). A Western blot assay with an antibody to Sir3 showed that the mutant proteins were expressed at levels similar to that of wild-type protein (data not shown).
FIG. 4.
(A) Schematic representation of the PCR mutagenesis screen used to isolate mutants with mutations in the Sir3 BAH domain. In vivo recombination between PCR-mutagenized DNA fragments and a gapped plasmid generated mutants with mutations in the Sir3 BAH domain. The mutants were screened for a telomeric silencing defect by using a TEL-URA3 reporter and identifying 5-FOA-sensitive colonies. For details, see Materials and Methods. 5-FOAs, 5-FOA sensitive; 5-FOAr, 5-FOA resistant; GAD, Gal4AD. (B) The telomeric silencing defect in strain PYY4 is shown for four representative mutants by lack of growth on 5-FOA medium. Sir3 Y207C is the only mutant that showed some growth on 5-FOA. (C) The results of mating tests in strains JCY3 and JCY4 show that some of the BAH domain mutants have a silencing defect at HML, while none are defective in silencing at HMR. WT, wild type.
TABLE 2.
Silencing phenotypes of Sir3 BAH domain mutantsa
| Sir3 | Silencing ability of indicated mutant at indicated locus
|
Expression level | Phenotype | ||||
|---|---|---|---|---|---|---|---|
| sir3Δ, telomere | sir3Δ, HML | sir3Δ, HMR | sir3Δ sir1Δ, HML | sir3Δ sir1Δ, HMR | |||
| WT | +++ | +++ | +++ | +++ | +++ | +++ | |
| A2V | − | +++ | +++ | − | − | +++ | Weak |
| N80D | − | ++ | +++ | − | − | +++ | Moderate |
| F94L | − | +++ | +++ | − | − | +++ | Weak |
| F123Pb | − | +++ | +++ | − | − | ++ | Weak |
| A136T | − | +/− | ++ | − | − | +++ | Strong |
| C177R | − | +/− | ++ | − | − | +++ | Strong |
| A181V | − | +++ | +++ | − | − | ++ | Weak |
| S204P | − | +/− | ++ | − | − | +++ | Strong |
| Y207C | +/− | +++ | +++ | − | − | ++ | Weak |
| K209R | − | +++ | +++ | − | − | +++ | Weak |
| Vector | − | − | − | − | − | − | |
Different levels of silencing are indicated as +++ to − based on comparison to wild type (+++) or vector (−) phenotypes. Strain XRY16 was used to measure telomeric silencing, and strains JCY3, JCY4, JCY8, and JCY9 were used to assess HM silencing.
The F123P mutant was generated by site-directed mutagenesis.
Several sir3 mutations were identified previously that caused no silencing defect by themselves but completely lost silencing, as judged by mating, in the absence of SIR1 (38). Many of these mutations caused single amino acid changes in the Sir3 BAH domain (38). To test whether any of our BAH mutants were also more defective in the absence of SIR1, their mating ability was analyzed in sir3Δ sir1Δ strains of both mating types. All 10 Sir3 BAH mutants lost the ability to mate in these strains and thus were completely defective at silencing both HML and HMR in the absence of SIR1 (Table 2).
We previously noted that mutations that changed the N-terminal Ala2 residue of Sir3 were detrimental to silencing (48) (Table 2). A comparison of the N-terminal sequences of S. cerevisiae and related Saccharomyces species revealed that Thr4, Leu5, and Asp7 were identical in all species. To test whether these N-terminal residues were also important for silencing, they were mutated in SIR3 and assayed for silencing. Tests showed that only Thr4 and, to a lesser extent, Leu5 mutants weakened silencing. Mutations of Asp7 to Ala, Lys, or Asn did not affect silencing (Table 3). Deletion of residues 3 to 6 or 3 to 10 affected the stability of Sir3 (data not shown), suggesting that these residues in the N terminus play a role in maintaining the structure of the protein.
TABLE 3.
Silencing phenotypes of N-terminal Sir3 BAH domain mutantsa
| Sir3 | Silencing ability of indicated mutant at indicated locus
|
Expression level | Phenotype | ||||
|---|---|---|---|---|---|---|---|
| sir3Δ, telomere | sir3Δ, HML | sir3Δ, HMR | Sir3Δ sir1Δ, HML | sir3Δ sir1Δ, HMR | |||
| WT | +++ | +++ | +++ | +++ | +++ | +++ | |
| T4F | − | +++ | +++ | − | − | +++ | Weak |
| L5A | − | +++ | +++ | +/− | − | +++ | Weak |
| D7A | +++ | +++ | +++ | +++ | +++ | ND | None |
| D7K | +++ | +++ | +++ | +++ | +++ | ND | None |
| D7N | +++ | +++ | +++ | +++ | +++ | ND | None |
| Vector | − | − | − | − | − | − | |
Different levels of silencing are indicated as +++ to − based on comparison to wild type (+++) or vector (−) phenotypes. The strains used are those listed for Table 2.
Histone mutations could suppress silencing defects of many Sir3 BAH domain mutants.
Previous reports identified the region around histone H3 K79 to be important for silencing, as various mutations in this region either weakened or improved silencing (31, 42, 49). For example, H3 D77N and H4 H75Y were previously identified as suppressors of the weak silencing defect of a cac1 mutant (37). H3 residues Q76, D77, and D81 have also been shown to be important for preventing the ectopic spread of Sir3 away from the HMR locus (43). Since H3 D77N and H4 H75Y mutations could suppress the Sir3 A2G mutant, we checked whether they could suppress the silencing defects of the Sir3 BAH domain mutants isolated in the screen described above. As mentioned before, all the Sir3 BAH domain mutants lost silencing ability at both HM loci in the absence of Sir1. As shown in Fig. 5A and Table 4, in a sir3Δ sir1Δ strain, H3 D77N and H4 H75Y could suppress the silencing defects of all the weak BAH mutants at both HML and HMR loci. However, they could not suppress the silencing defects of the moderate and strong mutants. The degree of suppression varied among the weak BAH mutants, with F94L, F123P, and A181V being suppressed more efficiently than A2V. Both H3 D77N and H4 H75Y had similar patterns of suppression of the Sir3 mutants, supporting the idea that the region of the nucleosome around H3 K79 is important for silencing.
FIG. 5.
(A) Suppression of the silencing defect of the Sir3 BAH domain mutants is due to the H3 D77N and H4 H75Y mutations. Results for six representative Sir3 mutants are depicted. Silencing was measured at HML::URA3 by growth on 5-FOA and at HMR by mating in the wild-type H3/H4 (strain VSY29), the H3 D77N mutant (VSY30), and the H4 H75Y mutant (VSY31). (B) Suppression of the silencing defect at HMR of some of the Sir3 BAH domain mutants by a histone H4 K16R mutant (strain PYY12). WT, wild type.
TABLE 4.
Suppression of Sir3 BAH domain mutations by histone mutationsa
| Sir3 | WT H3/H4 | H3 D77N | H4 H75Y | H4 K16R |
|---|---|---|---|---|
| WT | +++ | +++ | +++ | +++ |
| A2V | − | +/− | +/− | + |
| N80D | − | − | − | + |
| F94L | − | +++ | +++ | ++ |
| F123P | − | +++ | +++ | ++ |
| A136T | − | − | − | − |
| C177R | − | − | − | − |
| A181V | − | ++ | ++ | +++ |
| S204P | − | − | − | − |
| Y207C | +/− | +++ | +++ | +++ |
| K209R | − | +++ | +++ | +++ |
| Vector | − | − | − | − |
Different levels of silencing are indicated as +++ to − based on comparison to wild type (+++) or vector (−) phenotypes. Strains used are VSY29, VSY30, VSY31, and PYY12.
The N-terminal tails of histones H3 and H4 are known binding sites for the C-terminal region of Sir3 (15). Mutations around H4 K16 that alter the basic nature of that region diminish silencing (17). This silencing defect can be suppressed by mutations in the Sir3 BAH domain (W86R and D205N), suggesting an interaction between the BAH domain and the N-terminal tail of H4 (17). More recently, direct evidence for binding of the BAH domain to unacetylated H4 K16 has been obtained (29). Consistent with previous results, we found that an H4 K16R mutation that mimicked the unacetylated state of K16 could suppress the silencing defects of the weak and moderate but not the strong BAH domain mutants at HMR in a sir1Δ strain (Fig. 5B and Table 4). In fact, the pattern of suppression by H4 K16R was very similar to that of the H3 D77N and H4 H75Y mutations (Fig. 5A), suggesting a link between these two regions of the nucleosome.
Sir3 BAH domain binds to nucleosomes.
Sir3 has been shown to bind to the H3 and H4 N-terminal tails through its C-terminal region (aa 623 to 910). As mentioned above, more-recent work has demonstrated that the N-terminal BAH domain interacts with both the nucleosome core near H3 K79 and the H4 tail near K16 (29). Our analysis of genetic interactions between the Sir3 BAH domain mutants and the histone mutants led us to investigate whether the N-terminal Sir3 BAH domain could interact directly with nucleosomes. We had previously shown that certain Sir3 N-terminal fragments could bind to nucleosomes and DNA (7). Specifically, a Sir3 aa 1-to-380 fragment had nucleosome and DNA binding activities, while a shorter fragment with aa 1 to 214 had no nucleosome or DNA binding ability unless the hypermorphic mutation D205N was introduced.
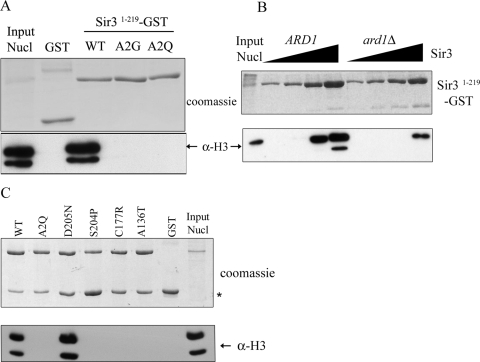
In this study, we purified Sir3 aa 1 to 219 C-terminally fused to GST from E. coli or from yeast. First, Sir3 aa 1-to-219 GST fusions were purified from yeast, immobilized on glutathione beads, and mixed with partially purified nucleosomes. We used three different versions of Sir3 aa 1 to 219: the wild type and the A2G and A2Q mutants. The amount of nucleosomes retained on the beads was measured by a Western blot assay using anti-H3 antibody. As seen by the results shown in Fig. 6A, only the wild-type Sir3 fragment bound to nucleosomes in this assay, while Sir3 A2G and A2Q did not. Next we tested whether the acetylation state of the N terminus influenced binding by purifying the wild-type Sir3 fragment from ARD1 and ard1Δ strains. Onishi et al. had previously shown that Sir3 BAH isolated from an ard1Δ mutant did not bind to nucleosomes (29). As seen by the results in Fig. 6B, we found that both the N-terminally acetylated and unacetylated proteins from yeast bound to nucleosomes, but a much greater concentration of the unacetylated Sir3 from the ard1Δ strain was required to detect binding. This shows that acetylation of Ala2 by Ard1, the catalytic subunit of the NatA acetyltransferase, is important but not absolutely required for binding. We had shown previously that Sir3 N-terminal fragments expressed in E. coli are not acetylated on the N-terminal alanine, while those from yeast are (48). Consistent with the results shown in Fig. 6B, high concentrations of Sir3 aa 1 to 219 purified from E. coli can also bind to nucleosomes (Fig. 6C, lane 1).
FIG. 6.
Sir3 BAH domain binds to nucleosomes. (A) The N terminus of Sir3 is important for binding to nucleosomes. Wild-type (W303-1a) nucleosomes were mixed with GST, Sir31-219-GST, Sir3A2G 1-219-GST, or Sir3A2Q 1-219-GST bound to glutathione-Sepharose beads. One-third of the nucleosomes retained by the GST-tagged protein on the beads was visualized by Western blotting with anti-H3 antibody. “Input Nucl” refers to 1% of nucleosomes used for the binding reaction. (B) N-terminal acetylation of Sir3 stimulates binding to nucleosomes. Wild-type (W303-1a) nucleosomes were incubated with increasing concentrations of wild-type Sir31-219-GST purified from ARD1 (W303-1a) and ard1Δ (JCY5) strains. One-third of the nucleosomes retained on the beads was visualized as described for panel A. (C) Loss-of-function Sir3 BAH domain mutants are unable to bind to nucleosomes, whereas the D205N mutant binds well. Nucleosomes from W303-1a were incubated with wild-type Sir31-219-GST purified from E. coli and with the indicated Sir3 mutants. One-fourth of the nucleosomes retained on the beads was visualized as described for panel A. “Input Nucl” refers to 0.5% of nucleosomes used for the binding reaction. The asterisk indicates the position of a Sir3-GST degradation product. WT, wild type; α, anti.
Some of the BAH domain mutants isolated from our screen were also tested for their ability to bind to nucleosomes, using proteins purified from E. coli. No binding was detected for the three strongest mutants (S204P, C177R, and A136T), suggesting that regions apart from the extreme N terminus are also important for nucleosome binding (Fig. 6C). Furthermore, two other mutants tested, of which one was classified as moderate (N80D) and one as weak (F94L) (Fig. 4 and Table 2), also showed no binding (data not shown). As noted before (7), Sir3 D205N bound to nucleosomes more strongly than the wild type (Fig. 6C).
DISCUSSION
Three distinct regions of the Sir3 BAH domain are important for silencing.
A goal of this study was to understand the role of the N-terminal Sir3 BAH domain in silencing. Our previous work had shown that overexpression of this domain could bring about significant silencing in the complete absence of the rest of the protein (7). This silencing required Sir1, Sir2, and Sir4 and was greatly enhanced by overexpression of Sir1, thus indicating that the Sir3 BAH domain plays a crucial role in the function of full-length Sir3.
The mutational analysis of the BAH domain described here revealed three different regions that were important for silencing (Fig. 7A). Many of the mutants clustered in or near an α-helix in the structure called helix F (region 1 in Fig. 7A). Helix F is located at the surface of the structure and thus is a potential site for interaction with other proteins. Two mutations, F94L and F123P, were near the so-called H-domain of Sir3 BAH, called region 2 in Fig. 7A. The H-domain of Orc1, another BAH domain-containing protein, interacts with Sir1 (50). Although there is no evidence that the Sir3 BAH domain interacts with Sir1, its H-domain is likely be a site for interaction with other proteins involved in silencing. The results for the A2V mutant (Fig. 7A, region 3), as well as the evidence that acetylation of the N-terminal Ala residue is important for nucleosome binding (Fig. 6C), confirm the importance of the extreme N terminus of Sir3 for silencing, as shown before (48). Site-directed mutagenesis showed that Thr4 and Leu5 are also important for function.
Interestingly, a previous screen for Sir3 mutants whose silencing defect was enhanced by a sir1Δ mutation identified eight mutants with mutations in the Sir3 BAH domain, all of which had phenotypes similar to the ones we found (38). Six of those eight mutations localized to the three regions highlighted in Fig. 7, reiterating the importance of those domains for Sir3 function. The three strongest mutations from that screen localized to region 3, as did all of our strong mutations. More recently, Moazed's group looked for dominant Sir3 mutations that disrupted telomeric silencing. Most of the mutations they found were in the BAH domain, and they were either at or near the residues we identified in our screen (3).
The Sir3 BAH domain interacts with nucleosomes.
Sir3 interacts with many silencing proteins, like Sir4, Rap1, Abf1, and histone H3/H4, through its C-terminal region (aa 440 to 978) (9). More recently, it has been demonstrated that the N-terminal BAH domain interacts directly with the nucleosome (29). Moreover, it has been shown that silencing-defective BAH mutants disrupt the nucleosome interaction (3). Our quest to identify interacting partners of the Sir3 BAH domain also uncovered genetic and biochemical evidence that the nucleosome core provides an interaction surface for the Sir3 BAH domain. We found that it interacted with partially or fully purified nucleosomes from yeast, as well as with reconstituted mononucleosomes (Fig. 6 and data not shown). It is noteworthy that the Sir3 aa 1-to-219 fragment binds to nucleosomes, whereas we reported previously that the Sir3 aa 1-to-214 fragment does not (7). It is possible that the crucial α-helix starting at residue 202 is less stable in the aa 1-to-214 protein than in the aa 1-to-219 protein, accounting for their different binding properties. Although the N-terminal Ala residue is important for binding, Sir3 BAH purified from an ard1Δ strain or from E. coli (and therefore unacetylated) can interact with nucleosomes at high concentrations in vitro, suggesting that regions of the BAH domain other than the N terminus can also interact with nucleosomes (Fig. 6). Consistent with this, none of the BAH mutant proteins with mutations in region 1 or 2 purified from E. coli were able to interact with nucleosomes in vitro (Fig. 6C and data not shown). It should be emphasized that the mutant recombinant proteins lacked acetylation at their N termini and thus, in effect, carried two lesions that hindered binding to nucleosomes.
While H3 D77N and H4 H75Y mutants were isolated as suppressors of the Sir3 A2G silencing defect, they could also silence the other weak and moderate BAH mutations found in this study (Fig. 5A). However, H3 D77N did not suppress sir1 or a sir3-8 mutation (a temperature-sensitive mutation leading to lower levels of Sir3), unlike H4 H75Y, which could suppress those two mutations (49 and data not shown). It should be pointed out that H3 D77 is on the surface of the nucleosome, while H4 H75 is somewhat buried (Fig. 7B), perhaps accounting for their different suppression properties. The H3 D77N and H4 H75Y mutants also could not suppress strong mutations in Sir3, such as A2Q, S204P, A136T, and C177R. Either these mutations alter the interaction between Sir3 and nucleosomes so drastically that they cannot be restored by second-site mutations in histones or they lead to unfolding and subsequent loss of function of the Sir3 BAH domain. On the other hand, since these mutants show almost-normal silencing at HMR, they may have a properly folded BAH domain.
A very recent report provided additional genetic evidence for an interaction between the nucleosome core and the Sir3 BAH domain (28). This work showed that the silencing defects of an H3 A75V mutant could be suppressed by the same two histone mutations we identified, H3 D77N and H75Y, as well as by mutations in the Sir3 BAH domain.
DNA binding versus nucleosome binding.
We showed previously that, unlike the full-length Sir3, the Sir3 BAH domain (aa 1 to 214) did not bind to DNA in vitro (7, 12). We have observed that a longer Sir3 fragment (aa 1 to 253) bound weakly to DNA (data not shown). The residues between aa 215 and 253 include a large number of basic amino acids (pI = 10.8), which could account for its ability to bind to DNA. H3 residue D77 and H4 H75 are not near DNA in the nucleosome structure and thus it is not clear if the observed in vitro DNA binding by N-terminal fragments is significant. It is worth noting that the previously characterized gain-of-function mutant, Sir3 D205N, also falls in helix F of the BAH structure. The D205N mutant binds with much higher affinity than wild-type Sir3 to nucleosomes and free DNA, probably accounting for its ability to suppress many different silencing defects (17, 22, 32, 42).
Role of the region around H3 K79 in spreading.
Both H3 D77 and H4 H75 are near the H3 K79 residue that is methylated by Dot1 (Fig. 7B). Silencing defects caused by mutation of DOT1 and residues around H3 K79 or by DOT1 overexpression suggested that this region of the nucleosome interacts with the Sir complex (45). Several studies have indicated that H3 K79 methylation is one of the marks distinguishing yeast euchromatin from heterochromatin by controlling the spread of the Sir complex along the chromosome (27, 45). Loss of H3 K79 methylation promotes binding of the Sir complex and its spread away from silent loci, resulting in a telomeric and HM silencing defect. Our results show that the suppression by H3 D77N and H4 H75Y is not due to lack of K79 methylation and, furthermore, that just the lack of K79 methylation, as in dot1Δ strains, cannot suppress the defects of Sir3Ala2 mutants (Fig. 3). We interpret this to mean that some region of the Sir3 BAH domain, perhaps the N terminus, binds to the region around H3 D77, and methylation at K79 blocks this interaction. The Sir3 A2G mutant and Sir3 that lacks N-terminal acetylation bind more poorly to nucleosomes (Fig. 6A and B). In a dot1Δ strain, Sir3 and, by association, the Sir complex can interact with nucleosomes throughout the genome and, thus, Sir3 is spread away from silent loci. A recent report described evidence for a genetic interaction between the N terminus of Sir3 and Dot1. Based on the results of epistasis tests and ChIP assays, the authors proposed that K79 methylation and acetylation of Sir3 Ala2 by the NatA acetyltransferase act in the same pathway to promote demarcation between euchromatin and heterochromatin (46).
We assume that nucleosomes with the H3 D77N or H4 H75Y mutation can bind the Sir3 BAH domain more tightly, thus accounting for the observed suppression of many of the Sir3 mutants. It is noteworthy that strains with the H3 D77N mutation grow more poorly when they carry wild-type Sir3 than Sir3 A2G or no Sir3 (Fig. 1). This growth defect is not present in sir2 or sir4 mutants (data not shown), suggesting that the wild-type Sir complex spreads inappropriately in cells with H3 N77 nucleosomes, silencing genes that are ordinarily in euchromatin and thus causing a growth defect. Consistent with this, strains carrying mutations at H3 D77 and nearby residues show enhanced silencing of a reporter gene located adjacent to HMR loci (43). Our attempts to document increased in vitro binding of the Sir3 BAH domain to mutant nucleosomes had inconsistent results and hence are not reported here.
Multiple points of interaction between Sir3 BAH domain and nucleosomes.
Taken together with previously published observations, our results indicate that the BAH domain and the C-terminal region of Sir3 interact with the H4 tail region and with the globular region of the nucleosome around H3 K79 (15, 29). Some of the weak mutants of the Sir3 BAH domain were completely defective for in vitro nucleosome binding ability as short fragments (aa 1 to 219) while retaining a substantial ability to silence in vivo in the context of full-length Sir3 (Table 2 and data not shown). This difference could be due to the well-documented ability of the C-terminal regions of Sir3 to bind to histone H3/H4 tails, thus allowing some interaction of full-length Sir3 with nucleosomes even when interaction between the BAH domain and nucleosome is abolished (15).
Suppression of the silencing defects of Sir3 BAH domain mutants by the H4 K16R mutation suggests that H4 K16 or residues around it are involved in binding the BAH domain. Recent reports suggest that both the BAH domain and the C-terminal region of Sir3 can bind to a region around H4 K16 and that this binding requires an unacetylated K16 residue (1, 8, 29). While the BAH domain is thought to bind to residues 20 to 34 in the H4 tail, the C-terminal region of Sir3 has been shown to bind to the basic patch around residues 17 to 20 (1, 29). It is not yet clear how two adjacent regions of H4 interact with two regions of Sir3 that are separated by more than 500 amino acids.
Our finding that all three histone mutations, H3 D77N, H4 H75Y, and K16R, resulted in the same pattern of suppression for the Sir3 BAH domain mutants can be explained in the context of multiple interaction points between Sir3 and nucleosomes. Strengthening one interaction could lead to suppression of mutations weakening another interaction.
In conclusion, our study shows that the Sir3 BAH domain interacts with nucleosomes, probably through multiple points of interaction, and that this interaction is necessary for accurate silencing at the silent loci in yeast. X-ray crystallography data will be needed for delineating the exact contact points between the Sir3 BAH domain and the nucleosome.
Acknowledgments
We thank Danesh Moazed for sharing protocols and unpublished observations. We also thank Ann Sutton and members of our laboratory for helpful advice. We thank Farihah Anwar, John Kilecki, and Huiyan Huang for technical assistance.
This work was supported by NIH grant GM28220 to R.S.
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Altaf, M., R. T. Utley, N. Lacoste, S. Tan, S. D. Briggs, and J. Cote. 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 281002-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. Kingston, D. Moore, J. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 3.Buchberger, J. R., M. Onishi, G. Li, J. Seebacher, A. D. Rudner, S. P. Gygi, and D. Moazed. 2008. Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 286903-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke, D., D. Dawson, T. Stearns, and Cold Spring Harbor Laboratory. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual, 2000 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 5.Callebaut, I., J. C. Courvalin, and J. P. Mornon. 1999. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446189-193. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., and J. Widom. 2005. Mechanism of transcriptional silencing in yeast. Cell 12037-48. [DOI] [PubMed] [Google Scholar]
- 7.Connelly, J. J., P. Yuan, H. C. Hsu, Z. Li, R. M. Xu, and R. Sternglanz. 2006. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol. Cell. Biol. 263256-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fingerman, I. M., H. C. Li, and S. D. Briggs. 2007. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 212018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, C. A., and K. H. McConnell. 2005. Toward biochemical understanding of a transcriptionally silenced chromosomal domain in Saccharomyces cerevisiae. J. Biol. Chem. 2808629-8632. [DOI] [PubMed] [Google Scholar]
- 10.Fukuma, M., Y. Hiraoka, H. Sakurai, and T. Fukasawa. 1994. Purification of yeast histones competent for nucleosome assembly in vitro. Yeast 10319-331. [DOI] [PubMed] [Google Scholar]
- 11.Gasser, S. M., M. Gotta, H. Renauld, T. Laroche, and M. Cockell. 1998. Nuclear organization and silencing: trafficking of Sir proteins. Novartis Found. Symp. 214114-132. [DOI] [PubMed] [Google Scholar]
- 12.Georgel, P. T., M. A. Palacios DeBeer, G. Pietz, C. A. Fox, and J. C. Hansen. 2001. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. USA 988584-8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 151541-1553. [DOI] [PubMed] [Google Scholar]
- 14.Gotta, M., F. Palladino, and S. M. Gasser. 1998. Functional characterization of the N terminus of Sir3p. Mol. Cell. Biol. 186110-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80583-592. [DOI] [PubMed] [Google Scholar]
- 16.Hecht, A., S. Strahl-Bolsinger, and M. Grunstein. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 38392-96. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, L. M., P. S. Kayne, E. S. Kahn, and M. Grunstein. 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 876286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katan-Khaykovich, Y., and K. Struhl. 2005. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 242138-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayne, P. S., U. J. Kim, M. Han, J. R. Mullen, F. Yoshizaki, and M. Grunstein. 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 5527-39. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, A., T. Umehara, and M. Horikoshi. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32370-377. [DOI] [PubMed] [Google Scholar]
- 21.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28327-334. [DOI] [PubMed] [Google Scholar]
- 22.Liu, C., and A. J. Lustig. 1996. Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics 14381-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389251-260. [DOI] [PubMed] [Google Scholar]
- 24.Megee, P. C., B. A. Morgan, B. A. Mittman, and M. M. Smith. 1990. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247841-845. [DOI] [PubMed] [Google Scholar]
- 25.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112725-736. [DOI] [PubMed] [Google Scholar]
- 26.Moazed, D., A. D. Rudner, J. Huang, G. J. Hoppe, and J. C. Tanny. 2004. A model for step-wise assembly of heterochromatin in yeast. Novartis Found. Symp. 25948-62. [PubMed] [Google Scholar]
- 27.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 1001820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norris, A., M. A. Bianchet, and J. D. Boeke. 2008. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet. 4e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onishi, M., G. G. Liou, J. R. Buchberger, T. Walz, and D. Moazed. 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 281015-1028. [DOI] [PubMed] [Google Scholar]
- 30.Paetkau, D. W., J. A. Riese, W. S. MacMorran, R. A. Woods, and R. D. Gietz. 1994. Interaction of the yeast RAD7 and SIR3 proteins: implications for DNA repair and chromatin structure. Genes Dev. 82035-2045. [DOI] [PubMed] [Google Scholar]
- 31.Park, J. H., M. S. Cosgrove, E. Youngman, C. Wolberger, and J. D. Boeke. 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 32273-279. [DOI] [PubMed] [Google Scholar]
- 32.Park, Y., J. Hanish, and A. J. Lustig. 1998. Sir3p domains involved in the initiation of telomeric silencing in Saccharomyces cerevisiae. Genetics 150977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilon, J., A. Terrell, and P. J. Laybourn. 1997. Yeast chromatin reconstitution system using purified yeast core histones and yeast nucleosome assembly protein-1. Protein Expr. Purif. 10132-140. [DOI] [PubMed] [Google Scholar]
- 34.Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani, and D. E. Gottschling. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 71133-1145. [DOI] [PubMed] [Google Scholar]
- 35.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72481-516. [DOI] [PubMed] [Google Scholar]
- 36.Sekinger, E. A., and D. S. Gross. 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105403-414. [DOI] [PubMed] [Google Scholar]
- 37.Smith, C. M., Z. W. Haimberger, C. O. Johnson, A. J. Wolf, P. R. Gafken, Z. Zhang, M. R. Parthun, and D. E. Gottschling. 2002. Heritable chromatin structure: mapping “memory” in histones H3 and H4. Proc. Natl. Acad. Sci. USA 99(Suppl. 4)16454-16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stone, E. M., C. Reifsnyder, M. McVey, B. Gazo, and L. Pillus. 2000. Two classes of sir3 mutants enhance the sir1 mutant mating defect and abolish telomeric silencing in Saccharomyces cerevisiae. Genetics 155509-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stone, E. M., M. J. Swanson, A. M. Romeo, J. B. Hicks, and R. Sternglanz. 1991. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol. Cell. Biol. 112253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suka, N., K. Luo, and M. Grunstein. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32378-383. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. S., X. Ling, and M. Grunstein. 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369245-247. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. S., M. L. Snow, S. Giles, L. E. McPherson, and M. Grunstein. 2003. Identification of a functional domain within the essential core of histone H3 that is required for telomeric and HM silencing in Saccharomyces cerevisiae. Genetics 163447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tompa, R., and H. D. Madhani. 2007. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics 175585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109745-756. [DOI] [PubMed] [Google Scholar]
- 45.van Leeuwen, F., and D. E. Gottschling. 2002. Genome-wide histone modifications: gaining specificity by preventing promiscuity. Curr. Opin. Cell Biol. 14756-762. [DOI] [PubMed] [Google Scholar]
- 46.van Welsem, T., F. Frederiks, K. F. Verzijlbergen, A. W. Faber, Z. W. Nelson, D. A. Egan, D. E. Gottschling, and F. van Leeuwen. 2008. Synthetic lethal screens identify gene silencing processes in yeast and implicate the acetylated amino terminus of Sir3 in recognition of the nucleosome core. Mol. Cell. Biol. 283861-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkatasubrahmanyam, S., W. W. Hwang, M. D. Meneghini, A. H. Tong, and H. D. Madhani. 2007. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc. Natl. Acad. Sci. USA 10416609-16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, X., J. J. Connelly, C. L. Wang, and R. Sternglanz. 2004. Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing. Genetics 168547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, E. Y., X. Bi, M. J. Holland, D. E. Gottschling, and J. R. Broach. 2005. Mutations in the nucleosome core enhance transcriptional silencing. Mol. Cell. Biol. 251846-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, Z., M. K. Hayashi, O. Merkel, B. Stillman, and R. M. Xu. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 214600-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]