Abstract
Hypomorphic mutation in one allele of ribosomal protein l24 gene (Rpl24) is responsible for the Belly Spot and Tail (Bst) mouse, which suffers from defects of the eye, skeleton, and coat pigmentation. It has been hypothesized that these pathological manifestations result exclusively from faulty protein synthesis. We demonstrate here that upregulation of the p53 tumor suppressor during the restricted period of embryonic development significantly contributes to the Bst phenotype. However, in the absence of p53 a large majority of Rpl24Bst/+ embryos die. We showed that p53 promotes survival of these mice via p21-dependent mechanism. Our results imply that activation of a p53-dependent checkpoint mechanism in response to various ribosomal protein deficiencies might also play a role in the pathogenesis of congenital malformations in humans.
Nascent ribosome biogenesis is required during cell growth, proliferation and differentiation (42, 47). It is temporally and spatially organized within the nucleolus, where rRNAs are transcribed, processed, modified, and assembled with ribosomal proteins (RPS) to generate the mature 40S and 60S ribosomal subunits (13). RPS participate in additional steps in ribosome biogenesis in the nucleoplasm and the cytoplasm, such as the transport of ribosomal precursors, stabilization of ribosome structure, and regulation of different steps in protein synthesis (15).
The critical role of at least some RPS in mammals is underscored by the pathological or lethal consequences of the deficiency of just one allele. Only a few heterozygous mutations of RP genes have been shown to be viable in mammals, and each of them has shown a relatively specific phenotype. Germ line heterozygous mutation for RPS19, RPS24, RPS17, RPL35a, RPL5, and RPL11 genes have been found in patients with Diamond-Blackfan anemia, which is characterized by absent or decreased erythropoiesis, and less frequently by small stature and various somatic malformations that mostly occur in the cephalic region, as well as an increased incidence of leukemia, osteogenic sarcoma, and myelodisplastic syndrome (7, 9, 12, 17, 18). Recently, a link between heterozygous mutations in Rps19 and Rps20 and dark skin phenotype in mice has been demonstrated (29).
Another heterozygous RP mutant is the Belly Spot and Tail (Bst) mouse (34). This is a semidominant, hypomorphic mutation caused by an intronic deletion in the Rpl24 gene, affecting Rpl24 mRNA splicing. Rpl24Bst/+ mice are characterized by reduced body size, a white ventral middle spot, white hind feet, retinal abnormalities, a kinked tail, and other skeletal abnormalities. Since Rpl24Bst/+ mouse embryonic fibroblasts from these mice showed a significant reduction in the rate of overall protein synthesis, it has been suggested that their phenotype result exclusively from faulty translation of mRNAs in tissues that depend on rapid and flawless protein synthesis (34).
It has been argued that the differential phenotypes of heterozygous mutants of RP genes in mammals might be attributable to the expression levels of the respective RP and the consequent decrease in the amount of ribosomes, impairment of specific steps in protein synthesis, and potential extraribosomal function. However, it should be pointed out that the relative contribution of the impaired protein synthesis or extraribosomal function to these phenotypes remains to be determined (9, 17, 34, 36, 37, 43, 57).
Recent evidence indicates that deficiencies in individual RPS could lead to pathological consequences via activation of a p53-dependent checkpoint regulatory mechanism. We demonstrated that inducible deletion of the Rps6 gene in the liver of adult mice inhibits the synthesis of the 40S ribosomal subunit, as well as proliferation of liver cells, after partial hepatectomy, despite seemingly unaffected protein synthesis (54). These observations suggested the existence of a novel checkpoint, downstream of the deficiency in ribosome biogenesis. Likewise, the perigastrulation lethality of Rps6 heterozygote embryos appears to reflect the triggering of a p53-dependent checkpoint response rather than a deficit in protein synthesis (37). We have assumed that activation of a p53-dependent checkpoint is triggered by impaired rRNA processing in the nucleolus in Rps6-deficient cells, since the nucleolar structure and function are compromised by almost all known p53-inducing stresses (37, 41, 50). Based on all of these observations, it could be speculated that the rare occurrence of RP heterozygosity in mammals reflects the fatal consequences of p53-dependent checkpoint activation (36, 37). In addition to function in development, this checkpoint may also play a role in other processes. Since various RP deficiencies in Drosophila melanogaster, zebrafish, and humans pose a great risk for development of malignant tumors, it is possible that induction of a p53-dependent checkpoint response prevents expansion of such potentially hazardous cells (1, 9, 11, 56).
Recently, we initiated an RNA interference screen for RP deficiencies that upregulate the p53 tumor suppressor in A549 cells. It has been previously suggested that a defect in ribosome biogenesis in the nucleolus caused by a RP deficiency triggers the p53 response (36, 37, 50). A number of studies in yeast showed that Rpl24 does not participate in ribosome biogenesis in the nucleolus, but it assembles late with the nascent 60S ribosomes in the cytoplasm and regulates the 60S subunit joining step during translation initiation and other steps in protein synthesis (10, 25, 45). Thus, it was surprising to observe that RPL24 deficiency triggered the p53 response in our screen. This observation led us to consider the possibility that p53 is upregulated in Rpl24Bst/+ mice. In contrast to previous opinion that the phenotype of these mice results exclusively from impaired protein synthesis (34), we demonstrate here that it is largely caused by the aberrant upregulation of p53 protein expression during embryonic development.
MATERIALS AND METHODS
Mice and embryo collection.
Rpl24Bst/+, p53−/−, and p21−/− mice were purchased from the Jackson Laboratory (4, 21, 34) (Bar Harbor, ME). All mice were kept on the C57BL/6 genetic background for more than eight generations. Embryo collection and preparation were performed as described previously (31). Briefly, noon on the day of the vaginal plug was taken as day 0.5 of gestation (E0.5). Pregnant females were sacrificed at different time points of gestation, and postimplantation embryos were separated from maternal tissue under a dissecting microscope (Stereomicroscope SZX 12; Olympus) in Dulbecco modified Eagle medium containing 25 mM HEPES (pH 7.4) and 10% fetal bovine serum by using fine forceps. All mice and embryos were genotyped by PCR analysis using specific primers (4, 21, 34). All procedures with mice were conducted with the approval of the ethical committee of the School of Medicine, University of Rijeka, in accordance with relevant guidelines and regulations.
Pupillary reflex.
The direct pupillary light response was examined by shining a pen light into the eye of dark-adapted nonanesthetized mice. An abnormal pupilary response was defined as one that did not constrict to a diameter comparable to that in wild-type (wt) mice (∼1 mm).
Histological analysis and immunohistochemistry.
Embryos and mouse adult tissues were fixed overnight in 10% formalin and embedded in paraffin. Sections 4 μm thick were cut and stained with hematoxylin and eosin. p53 protein in embryo sections was detected with anti-p53 antibody (Novocastra Laboratories). Apoptotic cells were identified by immunohistochemistry using antibody against activated caspase-3 (Cell Signaling Technology). Horseradish peroxidase-labeled anti-rabbit antibody was used as the secondary antibody, and detection was done by using diaminobenzidine substrate (Dako Cytomation). For identification of bromodeoxyuridine (BrdU)-labeled cells, 5-bromo-2′-deoxyuridine (5-BrdU) (100 μg/g of body weight) was injected intraperitoneally into pregnant females (Sigma-Aldrich Chemie GmbH). The females were sacrificed 1 h after injection, and deciduas were processed for immunohistochemistry. 5-BrdU-positive cells were detected by using a rat monoclonal to BrdU (Abcam), and detection was done by using the ABC system with biotinylated anti-rat secondary antibody (Vector Laboratories). BrdU-positive cells and apoptotic cells were counted on serial sections, and the number was compared to the total number of cells. Mitotic cells were counted on hematoxylin and eosin-stained histological sections of embryos, and the number was compared to the total number of cells.
Total RNA isolation and Northern blot analysis.
Total RNA was extracted from human lung carcinoma A549 cells (American Type Culture Collection), embryos, or adult mouse tissues using TRIzol (Invitrogen Life Technologies). RNA was separated on 1% agarose-formaldehyde gel and transferred to Hybond N+ membrane (Amersham Biosciences), hybridized at 54°C with cDNA probes labeled, and detected with an AlkPhosDirect kit (Amersham Biosciences).
Real-time RT-PCR analyses.
For p53 Signaling Pathway PCR Array analysis RNAs from two embryos of each genotype were pooled and treated with DNase (Turbo DNA-Free; Ambion). A total of 1.2 μg of pooled RNA was retrotranscribed by reverse transcription-PCR (RT-PCR) kit (SABiosciences Corp.). The quality of cDNAs was tested by using RT2 RNA QC-Array (SABiosciences). Mouse p53 RT2 PCR Array and RT2 Real-Time SyBR Green/ROX PCR mix were purchased from SABioscience. PCR was performed according to the manufacturer's instructions on an ABI Prism 7300 sequence detector (Applied Biosystems). PCR amplification was conducted with an initial 10-min step at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The fluorescent signal from SYBR green was detected immediately after the extension step of each cycle, and the cycle at which the product was first detectable was recorded as the cycle threshold. The data were imported into an Excel database and analyzed using the comparative cycle threshold method with normalization of the raw data to β-actin. The results are presented as n-fold changes versus the values in wt control embryos.
For quantification of 47S pre-rRNA precursor cDNA was amplified by using a LightCycler (Roche) and a LightCycler FastStart DNA Master SYBR green I kit (Roche) with the primers 5′-GCCTGTCACTTTCCTCCCTG-3′ (forward) and 5′-GCCGAAATAAGGTGGCCCTC-3′ (reverse). For an internal control, we used the primers 5′-GTGGGCCGCTCTAGGCACCAA-3′ (forward) and 5′-CTCCTTAATGTCACGCACGATTTC-3′ (reverse) for β-actin. The relative quantification of gene expression was calculated as described by the manufacturer and relative to β-actin. The mean value was calculated from three independent measurements.
Analysis of pre-rRNA processing.
A549 cells were starved for 30 min in methionine-free medium, incubated for 30 min with 25 μCi of l-[methyl-3H]methionine (Amersham Biosciences)/ml, and then chased in medium containing nonradioactive methionine for 0, 15, 30, and 90 min. Total RNA was separated on a 1% agarose-formaldehyde gel and blotted onto the Hybond N+ membrane, which was dried and treated with En3Hance (New England Nuclear) and exposed to Kodak BioMax MS film at −80°C (Sigma).
Preparation of cytosolic and nuclear fractions.
A549 cell pellets were resuspended in buffer A (25 mM HEPES [pH 7.4], 50 mM KCl, 1 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol, 25 mM NaF, 1 mMNaO3V, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of aprotinin/ml,10 μg of leupeptin/ml, and 1% [vol/vol] phosphatase inhibitor cocktail I) and incubated on ice for 30 min. Samples were then centrifuged at 500 × g for 1 min, and the supernatants were collected as cytosolic fractions. The pellets were washed in buffer B (buffer A without NP-40) and centrifuged at 500 × g for 1 min. The recovered nuclear pellets were resuspended in nuclei extraction buffer (buffer B, 450 mM KCl, 50% glycerol) and frozen at −70°C for 30 min. The nuclear suspensions were then thawed and clarified by centrifugation (16,000 × g for 15 min), and the supernatants were retained as the nuclear fraction.
Ribosome fractionation.
A549 cells (5 × 108 cells) were homogenized in 5 ml of buffer containing 20 mM Tris [pH 7.4], 10 mM MgCl2, 300 mM KCl, 10 mM dithiothreitol, 100 U of RNasin/ml, and 100 μg of cycloheximide/ml. After centrifugation at 10,000 × g for 15 min to remove mitochondria and debris, the supernatant was layered over a sucrose (20% [wt/vol]) cushion containing cycloheximide and centrifuged at 149,000 × g for 2 h. The ribosome-containing pellet and nonribosomal supernatant were collected. To remove any ribosome contaminants, the supernatant was subjected to a second centrifugation at 149,000 × g. For immunoblot analysis, ribosome pellets were resuspended in Laemmli buffer, and cytosolic fractions were concentrated by precipitation with 10% trichloroacetic acid and mixed with Laemmli buffer.
Immunoblotting.
Lysates of A549 cells, mouse embryos, or tissues from adult mice were prepared with radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate) containing protease and phosphatase inhibitors (1 μg of leupeptin/ml, 1 μg of aprotinin/ml, 50 μg of PMSF/ml, 1 mM Na3VO4, 1 mM NaF), lysed on ice for 30 min, and centrifuged at 13,000 rpm for 20 min. The protein concentration was determined by the Bio-Rad protein assay according to the manufacturer's instructions (Bio-Rad). Portions (50 μg) of total proteins were electrophoresed on sodium dodecyl sulfate-polyacrylamide gels. Western blots were probed with antibodies against the following proteins: actin (Chemicon International), p53 (Novocastra Laboratories), p21 (Santa Cruz Biotechnology), RPS6, RPL7a (kindly provided by Stefano Fumugalli and George Thomas), HDM2 (SMP-14; Santa Cruz Biotechnology), and RPL11 (50). Polyclonal antibodies against Rpl24 and Rps3a were produced by immunization of rabbits with His-tagged mouse Rpl24 and Rps3a proteins in our laboratory. Primary antibodies were detected by using horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) and an enhanced chemiluminescence kit (Amersham Biosciences).
Immunoprecipitation.
For coimmunoprecipitation of HDM2-RPL11 complexes, cells were lysed in buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.8% NP-40, 1 mM dithiothreitol, 1 mM EDTA, 1 μg of leupeptin/ml, 1 μg of aprotinin/ml, and 50 μg of PMSF/ml. For each sample, 1.5 mg of protein extract was precleared with protein A-Sepharose (Pharmacia Biotech) for 30 min and then incubated overnight at 4°C with rabbit polyclonal antibody (H-221 anti-HDM2; Santa Cruz Biotechnology). The immunoprecipitates were washed four times in lysis buffer and resuspended in protein sample buffer containing 50% glycerol, 300 mM Tris-HCl (pH 6.8), 10% sodium dodecyl sulfate, 715 mM β-mercaptoethanol, 10 mM EDTA, and 0.02% bromophenol blue.
In vitro BrdU labeling studies.
A549 cells were incubated with 10 μM BrdU for 15 min and stained with BrdU-fluorescein isothiocyanate (BD Biosciences) according to the manufacturer's instructions. Briefly, cells were washed with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS), fixed in 70% ethanol at 4°C, resuspended in 2 N HCl-0.5% Triton X-100, incubated for 30 min at room temperature, neutralized with 0.1 M sodium tetraborate (pH 8.5), rinsed PBS containing 1% BSA and 0.5% Tween 20, stained with the above-mentioned anti-BrdU antibody, and then analyzed by fluorescence-activated cell sorting (50). Flow cytometry was performed with a FACScan (Becton Dickinson) using CellQuest software. Ten thousand events were collected.
Stimulation of T lymphocytes in vitro.
Experiments were performed essentially as described previously (50). Briefly, lymph node T cells were plated in a flat-bottom six-well plate in RPMI 1640 medium containing 12% fetal calf serum, 2 mM l-glutamine, 5 μM 2-mercaptoethanol, 105 U of penicillin/liter, 0.1 g of streptomycin/liter, and 0.035 g of gentamicin/liter and stimulated with various concentrations of soluble anti-CD3 (1 μg/ml) and anti-CD28 (0.1 μg/ml) monoclonal antibodies (both purchased from BD Biosciences).
RNA interference.
A549 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and 2 mM l-glutamine. We used the Whitehead siRNA Selection Web Server (http://jura.wi.mit.edu/bioc/siRNA) to design short oligonucleotides that specifically “knock down” expression of target genes (58). These analyses did not identify any unwanted off-target sites for small interfering RNAs (siRNAs) used in the present study. Still, in our experiments we used two different siRNAs for each of the candidate genes. High-pressure liquid chromatography-purified and annealed oligonucleotides targeting the following sequences relative to translation initiation codon were used (Ambion): human RPL24, target sequence 1 (223-245, 5′-AAAGGGACAGTCGGAAGAAAT-3′) and target sequence 2 (392-414, (5′-AAGGCTAAGCAAGCATCTAAA-3′); human RPL11, target sequence 1 (282-304, (5′-AAGGTGCGGGAGTATGAGTTA-3′) and target sequence 2 (396-418, 5′-TACGGCCTGGACTTCTATGTG-3′); human p53, target sequence 825-845 (5′-CAGCATCTTATCCGAGTGGAA-3′); and human S6, target sequence 516-538 (5′-AAGAAAGCCCTTAAATAAAGA-3′). As a nonsilencing control we used scrambled siRNA also from Ambion. Transfection of siRNAs was performed with Lipofectamine RNAiMAX transfection reagent (Invitrogen) at a final siRNA concentration of 80 nM according to the manufacturer's instructions.
Immunofluorescence.
A549 cells were seeded on coverslips and transfected with different siRNAs as described above. After 48 h, coverslips were washed in PBS, fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.5% Triton X-100-PBS for 7 min, and then blocked for 30 min with 3% BSA-0.2% Triton X-100-PBS. Primary rabbit anti-p53 (Santa Cruz Biotechnology) was diluted 1:100 in above-mentioned blocking solution and incubated with cells for 1 h at room temperature. Embryos were fixed overnight in 10% formalin and embedded in paraffin. Paraffin sections were blocked for 30 min with 2% goat serum in PBS-1% BSA. Paraffin sections of embryos were stained with primary rabbit polyclonal c-kit (Dako), goat polyclonal Bax (Santa Cruz Biotechnology), and rabbit polyclonal anti-p53 (Santa Cruz Biotechnology) antibodies at appropriate dilution in PBS-1% BSA overnight.
Secondary anti-rabbit antibody was conjugated with Alexa 594, while secondary anti-goat antibody was conjugated with Alexa 488, both from Molecular Probes. These antibodies were used in a 1:1,000 dilution for 45 min at 37°C. DAPI (4′,6′-diamidino-2-phenylindole) was added for 5 min in the dark before inclusion in Mowiol (Calbiochem). Slides were analyzed by using either fluorescence microscopy (Olympus BX51) or a Olympus FV300 confocal laser scanning microscope.
Statistical analyses.
All data are presented as the means ± the standard deviation (SD). unless otherwise indicated. We analyzed numeric data for statistical significance by using a Mann-Whitney or a Pearson chi-square (χ2) test. We considered P values of <0.05 statistically significant.
RESULTS
RPL24 silencing activates a p53-dependent checkpoint in the absence of pre-rRNA processing defect.
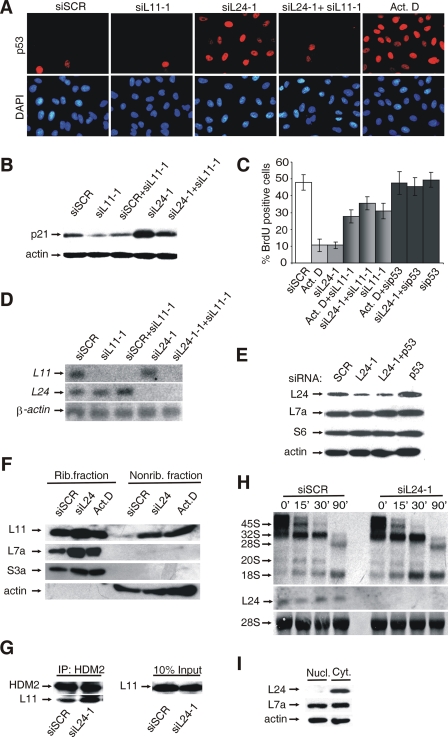
siRNA against RPL24 (siRPL24-1) but not the scrambled (SCR) siRNA induced p53 (Fig. 1A) and p21 (Fig. 1B) protein expression and inhibited the entry of cells into the S phase of the cell cycle (Fig. 1C). As a positive control, these cells were treated with 5 nM actinomycin D, which selectively inhibits RNA polymerase I and so disrupts ribosome biogenesis, leading to p53 activation and G1/S block (26) (Fig. 1A and C). The effects of actinomycin D and RPL24-1 siRNA on the ability of cells to enter S phase were p53 dependent, since cotransfection of p53 siRNA almost abolished these responses (Fig. 1C). It has been previously shown that the treatment of cells with 5 nM actinomycin D stabilize p53, at least in part, by enhancing the interaction between RPL11 and p53 negative regulator, HDM2/MDM2 (3, 26, 61). To test whether p53 activation in response to RPL24 deficiency is also dependent on RPL11, A549 cells were treated with RPL24-1 siRNAs in combination with either the SCR or RPL11-1 siRNAs. Silencing of RPL11 mRNA did not upregulate p53 and p21 proteins, but independently of that partially inhibited cell cycle progression (Fig. 1A to C). RPL11-1 siRNA almost completely inhibited p53 and p21 induction in response to actinomycin D treatment or knockdown of RPL24 mRNA (Fig. 1A and B and data not shown). Furthermore, it rescued the entry of actinomycin D- or RPL24-1 siRNA-treated cells into S phase to the level seen with RPL11-1 siRNA alone (Fig. 1C). The effective silencing of RPL24 and RPL11 mRNAs was confirmed by Northern blot analysis (Fig. 1D). RPL24 mRNA silencing also led to a dramatic reduction of the RPL24 protein levels without affecting the expression of 40S and 60S ribosomal subunit proteins RPS6 and RPL7a, respectively (Fig. 1E). The ribosomal fraction from these cells contain significantly less RPL24, suggesting that ribosomes made during RPL24-1 siRNA treatment had no RPL24 (data not shown). A549 cells in which RPL24 and p53 were cosilenced incorporate BrdU normally despite markedly reduced levels of RPL24, suggesting that RPL24 is not essential for this response in vitro (Fig. 1C and E). To control for possible off-target effects of individual siRNAs, we used two different siRNAs for each of the candidate genes (see Fig. S1A in the supplemental material). Since actinomycin D induces nucleolar disruption, it was suggested that RPL11 is released from the nucleolus to inhibit MDM2/HDM2, leading to p53 stabilization. To determine whether RPL24 silencing also increases the amount of nonribosomal pool of RPL11, we separated the soluble, nonribosome fraction from a particulate pellet containing ribosomes by ultracentrifugation of lysates from actinomycin D-, SCR siRNA-, or RPL24 siRNA-treated A549 cells. Both fractions were subjected to Western blot analysis with antibodies against RPL11, RPL7a, RPS3a, and actin (Fig. 1F). RPL7a and RPS3a were exclusively associated with ribosomes, while actin was present only in nonribosome fraction after all treatments. We detected a significant increase in the amount of ribosome-free RPL11, without a corresponding decrease in its amount in the ribosome fraction after both actinomycin D and RPL24 siRNA treatments. To determine whether RPL11 could be found associated with HDM2 after silencing of RPL24 mRNA, HDM2 immunoprecipitates were analyzed by Western blotting with anti-RPL11 antibody (3, 26, 61). We observed increased levels of RPL11 being coimmunoprecipitated with HDM2 in cells treated with RPL24-1 siRNA, compared to cells treated with the SCR siRNA (Fig. 1G). Thus, RPL24 deficiency elicits RPL11-mediated p53 activation, similar to actinomycin D treatment (3, 20, 50). Since it has been assumed that activation of the p53-dependent checkpoint in response to a RP deficiency is triggered by impaired ribosome biogenesis in the nucleolus (36-38, 50, 59), we decided to analyze this process in A549 cells after transfection of RPL24-1 siRNA by pulse-chase labeling of rRNA with l-[methyl-3H]methionine (Fig. 1H). There was no difference in the rate of appearance and disappearance of rRNA processing intermediates with different chase time between cells that were treated with the SCR or RPL24-1 siRNAs. The effective depletion of RPL24 mRNA in this experiment was monitored by Northern analysis (Fig. 1H). These results are consistent with the exclusive cytoplasmic expression of RPL24 (Fig. 1I).
FIG. 1.
Silencing of RPL24 triggers a p53-dependent checkpoint response without affecting pre-rRNA processing. A549 cells were transfected with either scrambled (SCR) siRNA or siRNAs specific for indicated RPS for 48 h unless otherwise stated. (A) Transfected cells were fixed and subjected to immunofluorescence staining with anti-p53 antibody (red) and DAPI (blue). Cells that were treated with actinomycin D (5 nM) for 12 h were used as a positive control for p53 staining. (B) Induction of p21 was verified by immunoblot analysis of lysates of transfected cells. The membrane was reprobed with actin antibody as a loading control. (C) BrdU incorporation into DNA of transfected or actinomycin D-treated cells was measured by flow cytometry. Each value represents the mean ± the SD. The SD was calculated on the basis of three independent experiments. (D) The efficacy of silencing in the above experiments was monitored by Northern blot analysis of total RNA using specific cDNA probes. (E) The levels of the indicated RPS and actin in lysates of transfected cells were determined by immunoblotting. (F) Subcellular fractions were prepared from cells that were treated either with SCR siRNA, RPL24 siRNA, or actinomycin D. Equal amounts of ribosome and nonribosome fractions were immunoblotted with antibodies against RPL11, RPL7a, RPS3a, and actin. (G) Lysates of transfected cells were immunoprecipitated with anti-HDM2 antibodies, followed by immunoblotting with anti-RPL11 antibody. Ten percent of the lysates loaded as input was also blotted with anti-RPL11 antibody. (H) Cells were transfected with either SCR or RPL24-1 siRNAs for 24 h and then pulse-chased with l-[methyl-3H]methionine. The positions of the major newly synthesized pre-rRNA intermediates and mature 28S and 18S rRNA species are indicated on the left (upper panel). The efficacy of RPL24 silencing was monitored by Northern blot analysis of total RNA using RPL24 cDNA probe (middle panel). RNA loading was normalized to the expression of 28S rRNA (lower panel). This experiment is a representative of four. (I) Cytosolic (Cyt.) and nuclear fractions (Nucl.) were prepared from untransfected A549 cells and immunoblotted with indicated antibodies.
In agreement with our previous in vivo studies, silencing of RPS6 led to activation of a p53-dependent checkpoint in A549 cells (36, 37, 50) (see Fig. S1B to D in the supplemental material). We previously suggested that activation of a p53-dependent checkpoint in response to RPS6 deficiency is triggered by impaired rRNA processing in the nucleolus (37, 50). The observation that RPL24 deficiency upregulates p53 protein expression in the absence of detectable defects in rRNA processing and without apparent nucleolar disruption (see Fig. S1E in the supplemental material) suggests that these events are not a prerequisite for p53 activation in response to a RP deficiency.
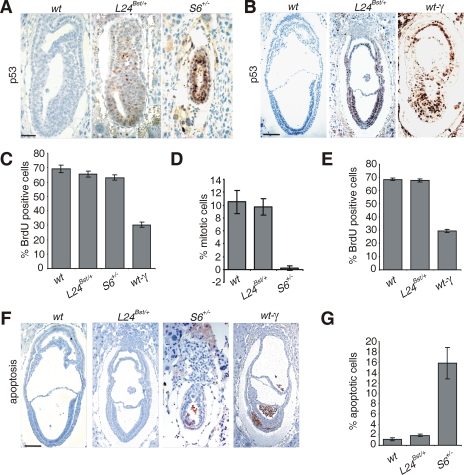
Rpl24Bst/+ mutation leads to p53 accumulation in gastrulating embryos.
All Rps6 heterozygote embryos die during gastrulation as a result of the triggering of a p53-dependent checkpoint response (36, 37). Genetic inactivation of p53 bypassed this checkpoint and prolonged development until embryonic day 12.5 (E12.5) (36, 37). In contrast, the majority of Rpl24Bst/+ mutants complete embryonic development (51). Since RPL24 deficiency triggered a p53-dependent checkpoint in A549 cells (Fig. 1A to C), we set out to determine whether Rpl24Bst/+ mutation also leads to p53 upregulation in the mouse and determine its contribution to the Bst phenotype. We first compared the expression of p53 protein by immunohistochemistry in Rpl24Bst/+ and Rps6+/− embryos during gastrulation. No p53 protein expression was detected in E6.5 wt embryos, whereas the majority of Rps6+/− cells strongly stained with p53 antibody as expected (37) (Fig. 2A). A great majority of cells in E6.5 Rpl24Bst/+ embryos were stained with anti-p53 antibody. Even though immunohistochemistry is not an absolutely quantitative method, we observed that the intensity of p53 staining in all analyzed E6.5 Rpl24Bst/+ embryos (n = 15) was markedly weaker than in Rps6+/− embryos (n = 13) (Fig. 2A and data not shown). Although no p53 protein expression was detected in E7.5 wt embryos, the majority of cells in Rpl24Bst/+ embryos expressed p53 at this stage (Fig. 2B).
FIG. 2.
Rpl24Bst/+ mutation leads to p53 protein accumulation in gastrulating embryos. (A and B) Immunohistochemistry of sections of representative E6.5 (A) and E7.5 (B) embryos of the indicated genotypes with anti-p53 antibody. (C and E) Percentage of cells that were BrdU labeled in E6.5 (C) and E7.5 (E) embryos of the indicated genotypes. (D) Percentage of cells that were in mitosis in the embryonic region of E6.5 wt, Rpl24Bst/+, and Rps6+/− embryos. (F) Apoptosis in section of E7.5 wt, Rpl24Bst/+, Rps6+/−, and gamma-irradiated wt embryos. (G) Percentages of apoptotic cells in the embryonic part of E7.5 embryos of the indicated genotypes. wt embryos were gamma irradiated (5 Gy) and analyzed 5 h thereafter in panels B, C, E, and F. The error bars in panels C to E and G denote the SD. In panels C to E and panel G, n = 8 per each genotype. Scale bars: 50 μm (A) and 100 μm (B and F).
Accumulation of p53 after different stresses does not necessarily translate into specific p53-dependent biological responses (55). Since p53 can inhibit various steps of the cell cycle progression (53), we first analyzed G1/S transition by comparing the incorporation of 5-BrdU into DNA of E6.5 Rpl24Bst/+ embryos with Rps6+/−, wt, and gamma-irradiated wt embryos. Gamma irradiation dramatically inhibited incorporation of 5-BrdU into DNA of wt embryos. The percentages of 5-BrdU-labeled cells were only slightly decreased in Rps6+/− and Rpl24Bst/+ embryos at this stage, despite p53 upregulation (37) (Fig. 2C; also see Fig. S2A in the supplemental material). We previously demonstrated that the percentage of mitotic figures was dramatically decreased in the embryonic region of E6.5 Rps6+/− embryos in a p53-dependent manner (37). In contrast to Rps6+/− embryos, the percentage of mitotic figures was only marginally decreased in E6.5 Rpl24Bst/+ embryos (Fig. 2D; also see Fig. S2B in the supplemental material). At E7.5 gamma-irradiated wt embryos show a significant reduction in BrdU incorporation. Despite the increased p53 protein expression in E7.5 Rpl24Bst/+ embryos, the percentage of BrdU-labeled cells was similar to wt embryos (Fig. 2E; also see Fig. S2C in the supplemental material). To find out whether apoptosis was triggered by p53 in E7.5 Rpl24Bst/+ embryos, we tested the expression of the activated form of caspase-3 by immunohistochemistry. Consistent with published results, a significant increase in the number of apoptotic cells was observed in the embryonic region of E7.5 Rps6+/− and gamma-irradiated wt embryos (37), while a negligible number of apoptotic cells was detected in E7.5 Rpl24Bst/+ embryos (Fig. 2F and G).
Thus, both, Rpl24Bst/+ and Rps6+/− mutations increase the protein expression of p53 during gastrulation. While Rps6-heterozygosity led to p53-dependent perigastrulation lethality (37), Rpl24Bst/+ embryos survived (34), most likely because levels of p53 protein were not sufficiently high (Fig. 2A) to trigger a significant apoptosis and cell cycle block at this developmental period, although of course other explanations are possible (55).
Upregulation p53 protein in response to Rpl24Bst/+ mutation is temporally and spatially controlled during postgastrulation embryogenesis.
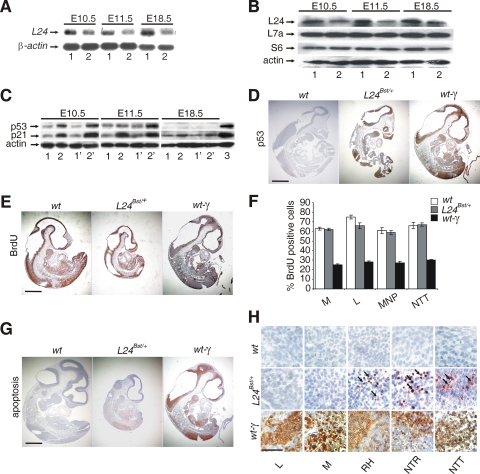
The Rpl24Bst/+ embryos are noticeably smaller then wt littermates from gastrulation onward (51). The pathological features in the developing retina of these embryos are already apparent at E10.5, while shortened tail is obvious at E11.5, and it begins to show kinks by E12.5 (51). Consistent with published results, Rpl24Bst/+ embryos showed significant decrease in the expression of Rpl24 mRNA during different stages of embryonic development (34) (Fig. 3A). In concordance with Rpl24 mRNA expression data, the Rpl24 protein levels were decreased in E10.5, E11.5, and E18.5 Rpl24Bst/+ embryos (Fig. 3B), whereas there was no significant effect on the expression of Rps6 and Rpl7a (Fig. 3B).
FIG. 3.
Accumulation of p53 and p21 proteins in Rpl24Bst/+ embryos during postgastrulation development. (A) Northern blot analysis of total RNA isolated from E10.5, E11.5, and E18.5 wt (lanes 1) and Rpl24Bst/+ (lanes 2) embryos using Rpl24 cDNA as a probe. RNA loading was normalized to the expression of β-actin mRNA. (B) The levels of Rpl24 protein in lysates of E10.5 and E11.5 wt (lanes 1) and Rpl24Bst/+ (lanes 2) embryos were determined by immunoblotting with anti-Rpl24 rabbit polyclonal antibody. Membranes were reprobed with antibodies to Rps6, Rpl7a, and actin. (C) Western blots showing the levels of p53 and p21 proteins in lysates from E10.5, E11.5, and E18.5 wt (lanes 1 and 1′) and Rpl24Bst/+ (lanes 2 and 2′) embryos. For each genotype, lysates of two littermates were analyzed. Reprobing with antibodies to actin served as a loading control. Lysates of E10.5, E11.5, and E18.5 samples were run on different gels but the membranes were immunoblotted at the same time. Actinomycin D-treated wt mouse embryonic fibroblasts served as a positive control for p53 and p21 antibodies (3). (D) Immunohistochemistry of sagittal sections of the entire E10.5 wt, Rpl24Bst/+, and gamma-irradiated wt embryos with anti-p53 antibody. (E) Sagittal sections of E10.5 embryos of the indicated genotypes were stained with anti-BrdU antibody. (F) Percentage of cells that were BrdU labeled in the liver (L), mandibula (M), medial nasal process (MNP) and neural tube in the tail (NTT) from E10.5 embryos of the indicated genotypes (n = 8 per each genotype). Error bars denote the SD. (G) Apoptosis in sagittal sections of the entire E10.5 wt, Rpl24Bst/+, and gamma-irradiated wt embryos. (H) Apoptosis in the liver (L), mandibula (M), roofs of hindbrain (RH), neural tube in the trunk (NTR), and neural tube in the tail (NTT) from embryos of the indicated genotypes (higher magnification of sections from panel G). Apoptotic cells were shown by arrows. wt embryos were gamma-irradiated (5 Gy) and analyzed 5 h thereafter in panels D to H. Scale bars: 500 μm (D, E, and G) and 100 μm (H).
To determine whether the protein expression of p53 is upregulated in RPL24Bst/+ embryos during postgastrulation development, lysates of wt and Rpl24Bst/+ embryos at various stages of embryonic development were subjected to immunoblotting with anti-p53 antibodies. E10.5 and E11.5 RPL24Bst/+ embryos showed a significant increase in the p53 protein levels (Fig. 3C). We observed small differences in the level of p53 protein among Rpl24Bst/+ littermates at E10.5 or E11.5, most likely due to small variations in their developmental stage. The p53 protein was not only upregulated, but it was also functionally active, as indicated by the concomitant upregulation of a p53 transcriptional target, the cell cycle inhibitor p21 (48) (Fig. 3C). The p53 protein expression in lysates of Rpl24Bst/+ embryos gradually decreased from E11.5 to E16.5 (data not shown), and it was undetectable at E18.5 (Fig. 3C). However, we observed a slight increase in the expression of p21 in E18.5 embryos, most likely because antibody against p21 was more sensitive in this assay than anti-p53 antibody (Fig. 3C). Alternatively, this may reflect a higher stability of p21 than p53. At this developmental stage, restrictions may be imposed on p53 upregulation in response to Rpl24Bst/+ mutation by limited p53 mRNA expression (46).
To determine tissue distribution of p53 protein during postgastrulation development of Rpl24Bst/+ embryos, their sections were stained with anti-p53 antibody. At E8.5, all embryonic tissues except the primitive heart showed a significant upregulation of p53 protein (data not shown). At E10.5 stage Rpl24Bst/+ embryos the pattern of p53 induction becomes more selective than at E8.5 (Fig. 3D). The brain tissue, trunk neural tube, and tail neural tube were strongly stained with anti-p53 antibody (Fig. 3D). Furthermore, some p53 protein expression was detected in brachial arches, maxillary areas, mandibula, lung bud, liver, hindgut, and limb buds (Fig. 3D and data not shown). In contrast, the intensity of p53 staining was weak in the anterior portion of the trunk, cephalic mesenchyme tissue and in the heart (Fig. 3D). In conclusion, Rpl24Bst/+ mutation increases p53 protein expression in rapidly proliferating and relatively undifferentiated cells. Gamma irradiation of E10.5 wt embryos at midgestation led to upregulation of p53 in the same tissues as Rpl24Bst/+ mutation (20, 24). However, p53 protein induction was much stronger in gamma-irradiated wt embryos than in Rpl24Bst/+ embryos. To find out whether p53 inhibited cell cycle progression in E10.5 Rpl24Bst/+ embryos, we compared the incorporation of 5-BrdU into DNA of Rpl24Bst/+, wt and gamma-irradiated wt embryos (Fig. 3E and F). As expected, gamma irradiation dramatically inhibited BrdU incorporation in many tissues of wt embryos. Despite upregulation of p53 protein expression in Rpl24Bst/+ embryos, the percentage of 5-BrdU-labeled cells was similar to wt embryos, although we cannot formally rule out the possibility that some proliferation defects exist in specific cells in these embryos.
Next, we analyzed apoptosis in E10.5 Rpl24Bst/+ embryos. Apoptotic cells were not observed in any tissue from control wt embryos, except for very rare apoptotic cells in the brain (Fig. 3G and data not shown). Not all regions of E10.5 Rpl24Bst/+ embryo that expressed p53 protein contain apoptotic cells (Fig. 3D, G, and H). Apoptosis is evident in E10.5 Rpl24Bst/+ embryo neuronal tissues: roofs of hindbrain, neural tube in the trunk and neural tube in the tail. Gamma irradiation induced a dramatic apoptosis in wt embryos at sites that display apoptotic cells in Rpl24Bst/+ embryos, as well as other sites (Fig. 3G and H).
Approximately a half of total adult Rpl24Bst/+ mice have deficient pupillary light reflex (40). A higher percentage of females than male Rpl24Bst/+ mice show this phenotype (40). This functional defect correlates with a reduction in the number of retinal ganglion cells (40). To test the possibility that p53 is responsible for this phenotype, we first analyzed the expression of p53 protein in E10.5 Rpl24Bst/+, wt and gamma-irradiated wt retinas. Gamma irradiation induced p53 protein expression in retinal ganglionar cells and massive apoptosis in the retina, whereas there was no p53 protein and apoptosis in Rpl24Bst/+ and wt retinas (see Fig. S3A in the supplemental material). These results suggest that p53-dependent responses may not be responsible for the retinal phenotype of Rpl24Bst/+ mice.
At E11.5 no detectable expression of p53 protein was observed in tissue sections of wt embryos, while Rpl24Bst/+ embryos expressed p53 protein in the liver, heart, and posterior part of neural tube, although the intensity of staining was typically weaker than in E10.5 Rpl24Bst/+ embryos (see Fig. S4A in the supplemental material). Only a small number of apoptotic cells was detected in the posterior part of the neural tube (see Fig. S4B in the supplemental material). The incorporation of BrdU in E11.5 Rpl24Bst/+ embryos was slightly inhibited in the indicated tissues (see Fig. S4C and D in the supplemental material). Similar to E10.5 Rpl24Bst+/ embryos, there were no p53-positive or apoptotic cells in E11.5 retinas of these embryos (see Fig. S3B in the supplemental material). These results hint at the possibility that the observed aberrant p53 protein expression in E10.5 and E11.5 Rpl24Bst/+ embryos may contribute to some of the Bst phenotypes.
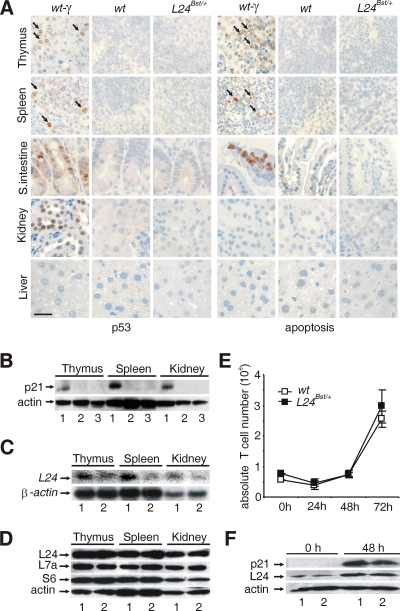
Rpl24Bst/+ mutation does not elicit p53-dependent checkpoint response in adult mice.
Since Rpl24Bst/+ mutation increased p53 protein expression at specific stages of embryonic development (Fig. 2A and B and Fig. 3C and D; see also Fig. S4A in the supplemental material), we were interested to test whether the same response is activated in tissues from adult Rpl24Bst/+ mice. The p53 protein expression was analyzed by immunohistochemistry in the various tissues from Rpl24Bst/+, wt, and whole-body gamma-irradiated wt mice (Fig. 4A). In agreement with previous reports gamma irradiation upregulated p53 protein expression in the thymus, spleen, small intestine, and kidney, but not in the liver (16, 27). Interestingly, induction of p53 protein by gamma irradiation was accompanied by increased apoptosis in the thymus, spleen, and small intestine, while this response was absent in the kidney (Fig. 4A). In contrast to gamma irradiation, no induction of p53 or apoptosis was observed in the same tissues from Rpl24Bst/+ or wt mice. Next, we analyzed the protein expression of p21 by Western blot analysis in the indicated tissues from wt, gamma-irradiated wt, and Rpl24Bst/+ mice as a surrogate indicator of p53 functionality (Fig. 4B). This analysis is extremely sensitive to the variations in p53 gene activity. The protein levels of p21 were dramatically increased following gamma irradiation of wt mice in the spleen, thymus, and kidney. Consistent with p53 protein expression results, there was no p21 upregulation in any of these tissues from Rpl24Bst/+ mice (Fig. 4B).
FIG. 4.
Rpl24Bst/+ mutation does not trigger a p53-dependent checkpoint response in adult mice. (A) Sections of the thymus, spleen, small intestine, kidney, and liver from 6-week-old whole-body gamma-irradiated (5 Gy) wt, nonirradiated wt, and Rpl24Bst/+ mice were stained with either anti-p53 (left) or anti-activated caspase-3 antibody (right). p53-positive or activated caspase-3-positive cells are indicated by arrows. Scale bar, 100 μm. (B) The protein expression of p21 was determined by immunoblot analysis of lysates of the indicated tissues from whole-body gamma-irradiated wt (lanes 1), nonirradiated wt (lanes 2), and Rpl24Bst/+ (lanes 3) mice. (C) Northern blot analysis of total RNA was isolated from the indicated tissues from wt (lanes 1) and Rpl24Bst/+ (lanes 2) mice by using Rpl24 cDNA probe. RNA loading was normalized to the level of β-actin. (D) The levels of the indicated ribosomal proteins in lysates of the indicated tissues from wt (lanes 1) and Rpl24Bst/+ (lanes 2) mice were determined by immunoblotting. (E) Normal proliferation of Rpl24Bst/+ T cells. Absolute number of stimulated wt and Rpl24Bst/+ T cells during 72 h in vitro. Error bars indicate the SD. This experiment is a representative of four. (F) The levels of Rpl24 and p21 proteins in lysates of stimulated and unstimulated wt and Rpl24Bst/+ T cells. Reprobing with antibody to actin served as a loading control in panels B, D, and F.
A possibility exists that the differential upregulation of p53 in response to Rpl24Bst/+ mutation during embryonic development and adulthood is caused by differences in Rpl24 production. Despite a significant decrease in the expression level of Rpl24 mRNA in all Rpl24Bst/+ tissues tested (Fig. 4C), we observed a normal Rpl24 expression (Fig. 4D and data not shown), suggesting that the Rpl24Bst/+ gene is sufficient to allow normal Rpl24 production in adult mice, where rates of ribosome biogenesis, cell growth, and cell division are much slower than in the embryo. Therefore, we tested whether the p53 response could be triggered by Rpl24Bst/+ mutation in highly proliferating T cells. To that end, purified lymph node T cells from Rpl24Bst/+ and wt mice were stimulated with anti-CD3 and anti-CD28 antibodies in vitro, and the relative numbers of live T cells were determined at 24, 48, and 72 h thereafter (Fig. 4E). The number of both control and mutant T cells decreased during the first 24 h of stimulation. There was a similar increase in the number of control and mutant T cells at 48 h and after 72 h. Western blot analysis revealed no p21 protein induction in proliferating Rpl24Bst/+ T cells relative to the wt control (Fig. 4F). In the same experiment, p21 protein was markedly upregulated in control T cells after treatment with actinomycin D or etoposide (data not shown). Thus, the proliferative state does not determine whether or not Rpl24Bst/+ mutation will trigger a p53-dependent checkpoint in T cells per se. A likely explanation for the absence of a p53-dependent checkpoint response in proliferating Rpl24Bst/+ T cells is a normal level of Rpl24 expression (Fig. 4F).
p53 promotes the survival of Rpl24Bst/+ animals.
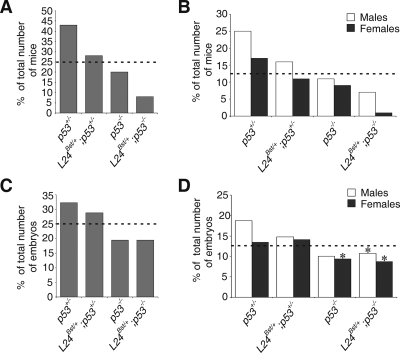
The capacity to detect and appropriately respond to stresses that interfere with functional homeostasis is essential for organismal survival. Upregulated p53 might regulate adaptation to Rpl24 deficiency in Rpl24Bst/+ embryos and thus promote their survival. To rigorously address this possibility, we introduced Rpl24Bst/+ mutation into the p53-negative genetic background by appropriate breeding (21). First, Rpl24Bst/+ females were bred with p53−/− males. F1 Rpl24Bst/+; p53+/− females were then crossed to p53−/− males. The expected frequency of each of four categories of progeny—(i) p53+/−, (ii) Rpl24Bst/+; p53+/−, (iii) p53−/−, and (iv) Rpl24Bst/+; p53−/−—is 25%. The frequency of p53−/− mutants recovered at weaning was ca. 21%, slightly less than the anticipated value (2, 44) (Fig. 5A). Furthermore, the frequency of p53+/− mice recovered was 43%, higher than anticipated value, whereas there was only a slightly higher percentage of Rpl24Bst/+; p53+/− mice (28%). The percentage of Rpl24Bst/+; p53−/− mice recovered at the same stage was ca. 8%, which is threefold less than the anticipated value (Fig. 5A), suggesting that Rpl24Bst/+ animals have a significantly lower chance of survival in the absence of p53.
FIG. 5.
Frequency of adult or embryonic progeny from p53−/− males and Rpl24Bst/+; p53+/− females. For each genotype, percentages of the total number of recovered animals at weaning (A) or E13.5 (C) are indicated. The ratio of males and females of the indicated genotypes at weaning (B) or E13.5 (D) is shown. The numbers of animals recovered (n) were 192 (A and B) and 149 (C and D). Average litter size, five (A and B) and eight (C and D). At E13.5, 5 of 13 Rpl24Bst/+; p53−/− females, 1 of 16 Rpl24Bst/+; p53−/− males, and 4 of 14 p53−/− females were exencephalic (*). Dotted lines indicate the expected percentages of mice of each genotype.
We also noted a slightly higher ratio of males to females among (i) p53+/−, (ii) Rpl24Bst/+; p53+/−, and (iii) p53−/− animals at weaning. However, a dramatic skewing of ratio of males to females among Rpl24Bst/+; p53−/− animals at this stage was observed, as evident from 1.7- and 12-fold reductions in the percentages of Rpl24Bst/+; p53−/− males and females, respectively (Fig. 5B). These results suggest that a large fraction of Rpl24Bst/+; p53−/− mice, mostly females, either do not survive gestation or die sometime after birth prior to weaning.
To address whether some of these animals fail during embryonic development, E13.5 and E16.5 embryos from the same cross were examined (Fig. 5C and data not shown). A small increase in the percentage of E13.5 p53+/− embryos was observed. The percentage of E13.5 Rpl24Bst/+; p53+/− embryos was close to the expected value. The frequency of E13.5 p53−/− and Rpl24Bst/+; p53−/− mutants was slightly less than the anticipated value, suggesting that their survival rate is similar until this embryonic stage (2, 44). PCR amplification of Y-chromosome-specific fragment from the Zfy gene was used to determine sex (35). There was no apparent change in the sex ratio of E13.5 Rpl24Bst/+; p53−/− embryos toward males (Fig. 5D). We observed 38% (5 of 13) and 7% (1 of 15) exencephalic E13.5 Rpl24Bst/+; p53−/− female and male embryos, respectively. On the other hand, there was 28% (4 of 14) of exencephalic E13.5 p53−/− female embryos. No significant sex and genotype differences between E13.5 and E16.5 were found (data not shown), suggesting that the majority of Rpl24Bst/+; p53−/− females die after E16.5. Significantly, we observed massive death of pups within the first 2 days after birth. PCR genotyping of recovered genomic DNAs from death pups (n = 30) revealed that 60% of them were Rpl24Bst/+; p53−/−. Altogether, these results strongly suggest that p53 improves the survival rate of Rpl24Bst/+ animals. If the change in average litter size from E13.5 (eight) to weaning (five) is taken into account, the survival rate of Rpl24Bst/+; p53−/− animals is even more dramatically reduced.
p53 is responsible for the Bst phenotype.
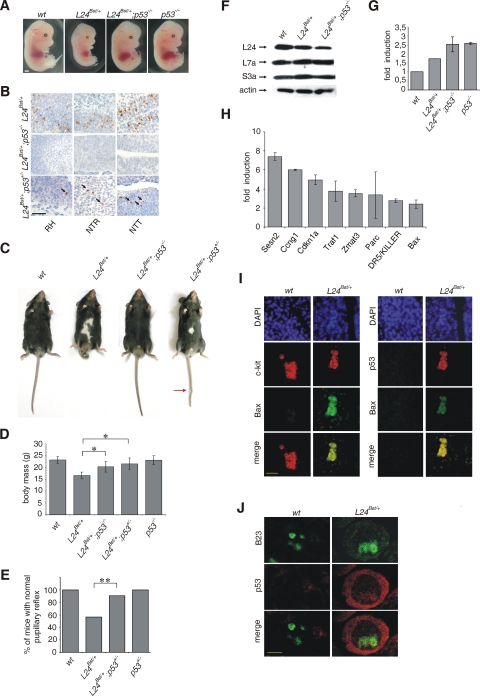
To address whether the pathological manifestations in Rpl24Bst/+ mice are caused by p53-dependent biological responses during embryonic development, the Rpl24Bst/+mutation was introduced into the p53-null genetic background (21). We first compared morphology of E13.5 (i) wt, (ii) Rpl24Bst/+, (iii) Rpl24Bst/+; p53−/−, and (iv) p53−/− embryos. The morphology of wt and p53−/− embryos was indistinguishable, except that a small fraction of p53−/− embryos are presented with exencephaly at this stage (2, 44) (Fig. 6A and data not shown). In contrast, E13.5 Rpl24Bst/+ embryos are noticeably smaller than their wt counterparts, their livers are externally marginally evident, and they have the diminutive tail (Fig. 6A). E13.5 Rpl24Bst/+; p53−/− embryos showed a normal size, their livers are prominent and externally easily visible organs, and they have normal tails, strongly arguing that the p53 protein upregulation in Rpl24Bst/+ embryos is responsible for the described phenotype (Fig. 6A). The upregulation of p53 protein in E10.5 Rpl24Bst/+ embryos was accompanied by the increased apoptosis in specific tissues (Fig. 3D, G, and H). To determine the dependence of this apoptotic response on p53, we compared the expression of activated caspase-3 in sections of (i) Rpl24Bst/+, (ii) Rpl24Bst/+; p53−/−, and (iii) Rpl24Bst/+; p53+/− embryos by immunohistochemistry (Fig. 6B). Genetic inactivation of the p53 gene abolished apoptosis in neural tissues of E10.5 Rpl24Bst/+ embryos, demonstrating its dependence on the p53 protein expression. On the other hand, the presence of one p53 allele was insufficient to trigger a significant apoptotic response in neural tissues of E10.5 Rpl24Bst/+ embryos, suggesting that p53 is haploinsufficient for this response (Fig. 6B).
FIG. 6.
p53 inactivation suppresses the phenotype of Rpl24Bst/+ mice. (A) Morphology of representative (i) wt, (ii) Rpl24Bst/+, and (iii) Rpl24Bst/+; p53−/− embryos at E13.5. (B) Apoptosis in roofs of hindbrains (RH), neural tubes in the trunks (NTR), and neural tubes in the tails (NTT) of E10.5 (i) Rpl24Bst/+, (ii) Rpl24Bst/+; p53−/−, and (iii) Rpl24Bst/+; p53+/− embryos. Apoptotic cells are indicated by arrows. (C) The phenotype of adult Rpl24Bst/+ mice is suppressed by p53 inactivation. Shown are photographs of representative (i) wt, (ii) Rpl24Bst/+, and (iii) Rpl24Bst/+; p53−/− mice (n = 16) and Rpl24Bst/+; p53+/− mice (n = 54) at 6 weeks of age. A kink in the tail is indicated by the red arrow. (D) Mean body masses of wt (n = 18), Rpl24Bst/+ (n = 10), Rpl24Bst/+; p53−/− (n = 14), Rpl24Bst/+; p53+/− (n = 23), and p53−/− (n = 17) male mice at 6 weeks of age. We analyzed males because only two female Rpl24Bst/+; p53−/− mice were recovered. *, P < 0.05 (Mann-Whitney test). (E) Percentage of wt (n = 32), Rpl24Bst/+ (n = 30), Rpl24Bst/+; p53+/− (n = 32), and p53+/− (n = 32) mice with the normal pupillary reflex. **, P = 0.006 (chi-square test). Equal numbers of males and females were analyzed. (F) The levels of Rpl24 protein in lysates of E10.5 wt, Rpl24Bst/+, and Rpl24Bst/+; p53−/− embryos were determined by immunoblotting with Rpl24 rabbit polyclonal antibody. Membranes were reprobed with antibodies to Rpl7a, Rps3a, and actin. (G) RNA was isolated from E10.5 embryos of the indicated genotypes. A real-time PCR using 47S rRNA probes was performed in triplicate. The SD was calculated on the basis of three independent experiments. (H) Real time RT-PCR analyses of 84 selected p53 target genes. Eight genes were upregulated more than twofold in E10.5 Rpl24Bst/+ embryos compared to wt embryos. The SD was calculated on the basis of three independent experiments. (I) Transversal sections of the trunk neural tube of E10.5 wt and Rpl24Bst/+ embryos were costained with a c-kit and anti-Bax (left panel) or anti-p53 and anti-Bax (right panel) antibodies and analyzed by fluorescence microscopy. Nuclei are stained with DAPI (blue). (J) Transversal sections of the trunk neural tube of E10.5 wt and RPL24Bst/+ embryos were stained with p53 antibody (red) and B23 (green) and analyzed by confocal laser scanning microscope. Error bars in panels D, G, and H indicate the SD. Scale bars: 1 mm (A), 100 μm (B), 50 μm (I), and 5 μm (J).
Next, we tested to what degree the phenotype of adult Rpl24Bst/+ mice is dependent on the aberrant p53 protein expression during embryonic development. Strikingly, genetic inactivation of both p53 alleles in Rpl24Bst/+ animals completely rescued a white ventral middle spot, white hind feet, curly tail phenotype (Fig. 6C) and partially rescued postnatal body mass deficits in all recovered animals (Fig. 6D). Genetic inactivation of one p53 allele in Rpl24Bst/+ mice was sufficient to largely, but not completely, suppress a white ventral middle spot, white hind feet, and a curly tail phenotypes (Fig. 6C), as well as postnatal body mass deficits in all recovered animals (Fig. 6D). In addition, inactivation of one p53 allele rescued pupillary light reflex in a large majority of Rpl24Bst/+ mice (Fig. 6E), suggesting that p53 is responsible for a defective pupillary reflex in Rpl24Bst/+ mice. In support of that suggestion is the observation that all 16 recovered Rpl24Bst/+; p53−/− adult mice showed normal pupillary reflex (data not shown). As mentioned above, we failed to detect increased p53 protein expression in retinal ganglionar cells at E10.5 and E11.5 in Rpl24Bst/+ embryos (see Fig. S3 in the supplemental material). However, we cannot formally rule out the possibility that the p53 expression is upregulated in the retinas of Rpl24Bst/+ embryos during other stages of the development. Alternatively, the developmental delay brought about by the aberrant p53 expression in other tissues during development of Rpl24Bst/+ embryos could potentially alter retinal differentiation.
To test the possibility that inactivation of p53 rescues the phenotype of Rpl24Bst/+ mice by upregulating the levels of Rpl24, lysates of E10.5 (i) wt, (ii) Rpl24Bst/+, and (iii) Rpl24Bst/+; p53−/− embryos were subjected to Western blot analysis by using anti-Rpl24 antibodies (Fig. 6F). Consistent with the results shown in Fig. 3B, the Rpl24 protein levels were decreased in Rpl24Bst/+ embryos relative to wt embryos, and genetic inactivation of p53 did not upregulate the expression of Rpl24 in Rpl24Bst/+ embryos, arguing against possibility that p53 rescues their phenotype by upregulating Rpl24 (Fig. 6F). To compare the transcription rate of rRNA between E10.5 (i) wt, (ii) Rpl24Bst/+, (iii) Rpl24Bst/+; p53−/−, and (iv) p53−/− embryos, we performed a real-time PCR using 47S rRNA-specific primers (Fig. 6G). Since 5′ primer recognizes the leader sequence, which is processed rapidly during transcription, the abundance of the 47S precursor reflects the rate of rRNA transcription. Despite the increased levels of p53 protein in Rpl24Bst/+ embryos, which is known to suppress Pol I transcription (6, 60), a 1.74-fold increase in the amount of the 47S rRNA was observed. Genetic inactivation of p53 in Rpl24Bst/+ embryos increased the amount of 47S rRNA to the level seen in p53−/− embryos (Fig. 6G). A similar result was obtained when primers that recognize the 5′-external transcribed spacer were used (data not shown). Thus, Rpl24 deficiency upregulates rRNA transcription in E10.5 embryos.
To understand how p53 contributes to congenital malformations of Rpl24Bst/+ mice as well as their increased survival, we profiled the expression of 84 genes p53 target genes that are involved in the processes of apoptosis, the cell cycle, cell growth, proliferation, and differentiation, and DNA repair in E10.5 wt and Rpl24Bst/+ embryos by using a real-time RT-PCR array from SABiosciences (Fig. 6H). Eight were significantly upregulated by a factor of ≥2.0: Sesn2 (7.41), Ccng1 (6.00), Cdkn1a (4.95), Traf1 (3.77), Zmat3 (3.53), Parc (3.36), DR5/KILLER (2.76), and Bax (2.41) (Fig. 6H). Sesn2 (encoding sestrin 2) has been previously suggested to inhibit mammalian target of rapamycin (mTOR) signaling network (5, 14), Ccng1 (encoding cyclin G1), and Cdkn1a (encoding the cell cycle inhibitor p21) are inhibitors of cell cycle progression (55, 62), Zmat3 (zinc finger martin type 3) binds double-stranded RNA as a part of a p53-dependent stress response (30), Parc (for p53-associated parkin-like cytoplasmic protein) regulates cytoplasmic localization of p53 (32), while Bax (Bcl2-associated X protein), Traf1 (encoding TNF receptor-associated factor 1), and DR5/KILLER (encoding tumor necrosis factor superfamily, member 10b) regulate apoptosis (55). It is likely that these genes play a role in the phenotype of Rpl24Bst/+ mice. However, understanding how each of these genes regulate a p53-dependent phenotype of Rpl24Bst/+ mice will require further investigation.
C-kit-positive neural crest cells at the dorsalmost region of the trunk neural tube that are committed to become melanocytes migrate into the ectoderm and move through the skin toward the ventral midline of the belly during embryonic development (19). A potential decrease in the number of these cells in Rpl24Bst/+ embryos could be responsible for a white ventral middle spot in adult mice (19, 34, 49). To address this possibility, transversal sections of the trunk neural tubes of E10.5 wt and Rpl24Bst/+ embryos were subjected to immunofluorescence analysis by using a c-kit, p53 and Bax antibodies (Fig. 6I). Sections of wt embryos stained with the c-kit, but not with p53 and Bax antibodies, while sections of Rpl24Bst/+ embryos stained with c-kit, Bax, and p53 antibodies. Genetic inactivation of both p53 alleles in Rpl24Bst/+ embryos prevented Bax expression in c-kit-positive cells (data not shown). These results strongly suggest that a p53- and Bax-dependent apoptosis of c-kit-positive melanocytes in the trunk region of neural tube (Fig. 6B and I) is responsible for a ventral middle spot hypopigmentation in adult Rpl24Bst/+ mice (34).
Interestingly, p53 was largely expressed in cytoplasm of cells in the trunk neural tube of E10.5 Rpl24Bst/+ embryos (Fig. 6J). A very small portion of p53 was present in the nucleus and some of it colocalizes with the nucleolar marker B23.
p53 promotes survival of Rpl24Bst/+ mice via p21-dependent mechanism(s).
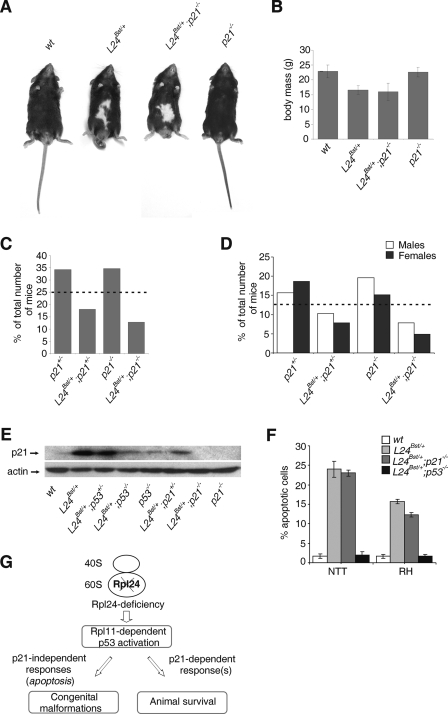
To gain insight into the importance of an aberrant p21 upregulation in the developing Rpl24Bst/+ embryos to the phenotype of adult mice (Fig. 3C and Fig. 6H), we introduced Rpl24Bst/+ mutation into the p21-negative genetic background (4). All recovered adult Rpl24Bst/+; p21−/− mice were macroscopically indistinguishable from Rpl24Bst/+mice (Fig. 7A and B). These results demonstrate that p21 does not contribute to p53-mediated congenital malformations and body mass deficits in Rpl24Bst/+mice. To test whether p21 promotes survival of Rpl24Bst/+mice, Rpl24Bst/+; p21+/− females were then crossed to p21−/− males. The expected frequency of each of the four categories of progeny—(i) p21+/−, (ii) Rpl24Bst/+; p21+/−, (iii) p21−/−, and (iv) Rpl24Bst/+; p21−/−—is 25%. The expected frequency of each of four categories of progeny was observed E13.5 (data not shown). The percentage of Rpl24Bst/+; p21−/− mice recovered at weaning was ∼2-fold less than anticipated value (Fig. 7C), suggesting that Rpl24Bst/+animals have a significantly lower chance of survival in the absence of p21. We also noted a higher ratio of males to females among Rpl24Bst/+; p21−/− animals at weaning. A 28% decrease in the number of Rpl24Bst/+; p21+/− mice recovered at weaning was observed (Fig. 7D). If p21 is an important regulator of p53-mediated survival of Rpl24Bst/+ mice, it would be expected that the 21 protein expression levels in (i) Rpl24Bst/+, (ii) Rpl24Bst/+; p53+/−, (iii) RPL24Bst/+; p53−/−, (iv) Rpl24Bst/+; p21+/−, and (v) Rpl24Bst/+; p21−/− embryos correlate with the survival rate of these mice (Fig. 5A and Fig. 7C). Western blot analysis of p21 (shown in Fig. 7E) confirmed that prediction, further supporting the role of p21 in promoting the survival of Rpl24Bst/+ mice. It has been shown that cancer cells lacking p21 display enhanced sensitivity toward apoptosis induced by DNA-damaging agents (52). Thus, reduced survival of Rpl24Bst/+; p21−/− mice could be the result of enhanced apoptosis during embryonic development. This does not seem to be the case, as p21 inactivation did not enhance apoptosis in neuronal tissues of E10.5 Rpl24Bst/+ embryos.
FIG. 7.
p21 promotes survival of Rpl24Bst/+ mice. (A) The phenotype of adult Rpl24Bst/+ mice is not suppressed by p21 inactivation. Shown are photographs of representative (i) wt, (ii) Rpl24Bst/+, and (iii) Rpl24Bst/+; p21−/− mice (n = 13) and p21−/− mice (n = 29) at 6 weeks of age. (B) Mean body masses of wt (n = 18), Rpl24Bst/+ (n = 10), Rpl24Bst/+; p21−/− (n = 13), and p21−/− (n = 29) male mice at 6 weeks of age. (C and D) Frequency of adult progeny from p21−/− males and Rpl24Bst/+; p21+/− females. The number of animals recovered (n) was 204. Average litter size, five. (C) For each genotype the percentages of the total number of recovered animals at weaning are indicated. (D) Ratio of males and females of the indicated genotypes at weaning. Dotted lines indicate the expected percentages of mice of each genotype. (E) Western blot showing the levels of p21 protein in lysates from embryos of the indicated genotypes at E10.5. The membrane was reprobed with antibodies to actin. (F) Percentages of apoptotic cells in the neural tube in the tail (NTT) and roofs of hindbrain (RH) from embryos of the indicated genotypes (n = 8, for each genotype). Error bars in panels B and F indicate the SD. (G) Model explaining our results. Rpl24-deficient ribosomes in the mouse embryos trigger Rpl11-dependent p53 induction. Activated p53 promotes survival of these mice via p21-dependent mechanism and causes congenital malformations independently of p21, most likely by inducing apoptosis.
The successful rescue of the pathological phenotypes of Rpl24Bst/+ mice by genetic inactivation of p53 demonstrates that the aberrant upregulation of p53 protein expression, likely during restricted period of embryonic development, is the major cause of congenital malformation in these mice. Our results strongly suggest that the effect of p53 on increased survival of Rpl24Bst/+mice is mediated via p21 (Fig. 7G).
DISCUSSION
In contrast to the previous opinion that faulty protein synthesis is the exclusive cause of the Bst phenotype (34), in the present study we convincingly demonstrate that the aberrant upregulation of p53 protein expression in response to Rpl24Bst/+ mutation during embryonic development plays an important role in the phenotype of these mice.
We demonstrated here that the RPL24 knockdown induced an increase in the p53 protein level in A549 cells independently of defects in pre-rRNA processing in the nucleolus. Why and how would RPL24 deficiency trigger the p53 response? It could be speculated that a change in the 60S ribosome subunit structure in the cytoplasm is sensed by a p53-dependent checkpoint in RPL24-deficient cells to prevent the imprecise execution of the genetic program at the level of protein synthesis. Since RPL24 resides at the surface of the 60S subunits (28), it could be hypothesized that RPL24 deficiency might indirectly elicit the p53 checkpoint response by promoting the release RPL11 that also reside at the surface of the 60S subunit and lack the long rRNA-penetrating extensions that are characteristics of many other RPS (36, 39). However, our preliminary experiments in RPL24 siRNA-treated A549 cells suggest that this does not seem to be the case, since we detected a significant increase in the amount of ribosome-free RPL11, without a corresponding decrease in the amount of RPL11 in the ribosomal fraction. Thus, in the absence of nucleolar disruption and release of RPL11 from ribosomes in RPL24-deficient cells, the accumulation of ribosome-free RPL11 could be due to upregulation of its synthesis or protein stabilization. Further experiments will be required to determine the source of ribosome-free RPL11 in this model.
All Rps6 heterozygote embryos die during gastrulation as a result of the triggering of a p53-dependent checkpoint (36, 37). In contrast, the majority of Rpl24Bst/+ mutants survive most likely because levels of p53 protein in these embryos at E6.5 were significantly lower than in Rps6 heterozygote embryos and thus insufficient to trigger apoptosis and inhibit cell cycle progression (37, 53, 55). In agreement with this model, a complete abrogation of Rpl24 or Rps6 mRNA expression by specific siRNAs leads to a comparable induction of p53 and p21 and to G1/S cell cycle arrest in A549 cells. The Bst mutation is a hypomorphic heterozygous mutation. The possibility exists that the expression level of Rpl24 is above the required threshold for triggering a significant p53-dependent apoptosis and cell cycle block at gastrulation. Thus, it would be of great interest to test whether a true Rpl24 heterozygosity will trigger a p53-dependent checkpoint and consequently perigastrulation lethality, similar to Rps6 heterozygosity. The possibility also exists that differences in p53 modifications and regulation of its cooperating factors could also contribute to a differential regulation of p53-mediated apoptosis and cell cycle progression in gastrulating Rps6+/− and Rpl24Bst/+ embryos (53, 55).
Expression profiling of 84 p53-target genes revealed that only 8 of them were significantly upregulated in E10.5 Rpl24Bst/+ embryos, suggesting they play an important role in the phenotype of Rpl24Bst/+ mice. These are Sesn2, Ccng1, Cdkn1a, Traf1, Zmat3, Parc, DR5/KILLER, and Bax. Understanding how each of these genes contributes to a p53-dependent phenotype of Rpl24Bst/+ mice will require further investigation.
In the absence of p53 a large percentage of Rpl24Bst/+animals die before weaning, most likely as a result of a defective protein translation, although we cannot exclude the possibility that a potential extra-ribosomal function of Rpl24 contributes as well. Since the percentage of recovered Rpl24Bst/+; p53−/− females at weaning was 12-fold less than anticipated value, the p53 function seems to be especially important in females. Although 38% of female Rpl24Bst/+; p53−/− embryos exhibit exencephaly, this defect can only partially explain the reduction in the number of the adult Rpl24Bst/+; p53−/− animals. Despite considerable efforts, we were not able to identify any other morphological defect in specific tissues in Rpl24Bst/+; p53−/− embryos that could easily explain their reduced survival rate (data not shown).
Our results strongly suggest that p53 promotes the survival of Rpl24Bst/+ mice via the Cdkn1a gene product, the cell cycle inhibitor p21. It might be that p21 regulates this response by slowing down cell cycle progression during the embryogenesis of Rpl24Bst/+ animals, thus allowing the developmental program to complete in the presence of impaired protein synthesis. Even though we did not observe a significant change in the incorporation of 5-BrdU in Rpl24Bst/+embryos, we cannot formally rule out the possibility that some larger defect in proliferation exists in specific tissues. Alternatively, p21 could coordinate additional, as yet uncharacterized processes that promote the survival of Rpl24Bst/+ mice. This p53- and p21-dependent prosurvival function could be cell autonomous, although we cannot exclude the existence of some systemic effects. Further studies will be required to understand this novel observation at the molecular level.
Interestingly, the loss of one allele of p53 in Rpl24Bst/+ animals was sufficient to largely suppress the Bst phenotype without significantly affecting their survival rate. How a single p53 allele protects against the lethality associated with Rpl24 deficiency is a challenging question. Our results strongly suggest that the presence of a single p53 allele in Rpl24Bst/+ embryos is sufficient for induction of high levels of p21, potentially explaining the relatively normal survival of these animals.
The neuroepithelial cells during midgestation proliferate rapidly and at the same time generate neural crest cells. These processes are highly energy consuming and require intense ribosome biogenesis (33). Under such conditions Rpl24 deficiency may become limiting for proper ribosome formation, leading to the activation of a p53-dependent checkpoint in these tissues. Furthermore, other neural-tissue-specific factors can determine whether or not this potential for rapid overproduction and accumulation of p53 in response to Rpl24 deficiency is actually materialized. One such a factor could be a high expression of p53 mRNA in these tissues (20, 24, 46).
The fact that c-kit-positive melanocyte precursors in the neural tube in the trunk of Rpl24Bst/+ embryos stained with both Bax and p53 antibodies strongly suggests that a p53- and Bax-dependent apoptosis of these cells is responsible for a ventral middle spot hypopigmentation in adult Rpl24Bst/+ mice (19, 34, 49).
Although it has been suggested that an imbalance of cell proliferation within the tail neural tube is responsible for the curly tail phenotype in curly tail mice (8), we did not observe a significant change in the incorporation of 5-BrdU in that region in E10.5 and E11.5 Rpl24Bst/+embryos. However, we cannot formally rule out the possibility that some small defect in proliferation exists in the tail neural tubes of these embryos. It is also possible that the induction of a p53-dependent apoptosis in that region during midgestation is responsible for the curly tail phenotype in Rpl24Bst/+ mice. Furthermore, potential changes in neural differentiation brought about by p53 upregulation in the spinal neural tube could contribute to this phenotype as well.
The observation that p53 inactivation significantly rescues the body mass deficit of Rpl24Bst/+ mice argues that p53 activation in these embryos is, at least partially, responsible for this phenotype. The fact that p53 inactivation does not completely rescue the body mass deficit of Rpl24Bst/+ mice implies that impaired protein synthesis could also negatively affect the size of these animals.
Since Sesn2 was dramatically upregulated in Rpl24Bst/+ embryos, it might have an important role in the phenotype. This gene has been recently implicated in the AMP-responsive kinase-dependent inhibition of mTOR signaling in response to DNA-damaging agents (5). One could speculate that p53-dependent induction of Sesn2 in response to Rpl24 deficiency also inhibits mTOR activity and consequently slows down protein synthesis. This response could be potentially beneficial to Rpl24Bst/+ embryos. However, it may also play a role in the pathological phenotype of these mice. Certainly, future studies will be required to test these exciting possibilities.
We observed that p53 was largely expressed in cytoplasm of cells in the trunk neural tube of E10.5 RPL24Bst/+ embryos. Since Parc mRNA was significantly upregulated in E10.5 RPL24Bst/+ embryos, one could speculate that Parc regulates the cytoplasmic localization of p53 in these embryos. However, this possibility has to be addressed experimentally.
One could argue that inactivation of p53 rescues the phenotype of Rpl24Bst/+ mice by upregulating the levels of Rpl24. Genetic inactivation of p53, however, did not upregulate Rpl24 expression in Rpl24Bst/+ embryos, indicating that p53 does not rescue their phenotype by upregulating the expression of Rpl24.
Unexpectedly, we observed a significant increase in the transcription rate of rRNA in Rpl24Bst/+ embryos, even in the presence of known inhibitor of this process, p53 (6, 60). It will be challenging to understand how and why rRNA transcription is upregulated in this embryos and whether it plays any role in the Bst phenotype. One can only speculate that rRNA transcription is upregulated in Rpl24Bst/+ embryos (even in the presence of p53) to increase the rate of ribosome biogenesis in Rpl24-deficient cells.
Altogether, the results presented here suggest that mutations in RP genes could be involved in the pathogenesis of neural tube closure defects, as well as a number of neurocristopathies, diseases caused by defects in the neural crest cells, via activation of a p53 checkpoint (19, 22).
Only few heterozygous mutations of RP genes are known to be viable in humans. Since various RPS have specific roles in ribosome biogenesis, protein synthesis, or outside of ribosomes, different RP deficiencies in humans may greatly differ in their phenotypes (36). In addition, the expression levels of an RP gene in different tissues, the extent of the p53-dependent checkpoint activation in response to a RP deficiency, and polymorphisms in genes involved in this checkpoint could also contribute to the specificity of the phenotype of an RP-deficient human. In a broader perspective, mutations in genes that are involved in various steps of ribosome biogenesis, as well as many stresses that interfere with this process (certain chemicals and drugs, malnutrition, hypoxia, heat shock, etc.), could potentially lead to “spontaneous abortions,” congenital malformations, and a number of other diseases in mammals via activation of a p53-dependent ribosome checkpoint during embryonic development (22, 37, 41).
Although it might be farfetched, our results suggest that appropriate pharmacologic titration of p53 function during embryonic development may provide a therapeutic opportunity for preventing malformations caused by RP deficiencies without affecting embryo survival (23). Given the fact that it would be extremely difficult to precisely titrate the p53 levels and that p53 is the key tumor suppressor, targeting downstream or upstream components of this checkpoint pathway might provide more efficient and specific means of preventing congenital malformations caused by RP deficiencies.
Supplementary Material
Acknowledgments
We thank Stefano Fumagalli for his useful discussions; Jacques Montagne, Mario Pende, and Nick Pullen for their comments; Jonathan Ashwell and Ivan Dikic for reading the manuscript; Tihomila Bušić and Miljana Uzelac for technical assistance; and Mladen Petrovečki for statistical analysis. We are grateful to George Thomas and Davor Solter for their continuous support of our work.
This study was supported by grants from the Ministry of Science, Education, and Sports of Croatia.
Footnotes
Published ahead of print on 9 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amsterdam, A., K. C. Sadler, K. Lai, S. Farrington, R. T. Bronson, J. A. Lees, and N. Hopkins. 2004. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. F., M. H. Kaufman, D. J. Harrison, and A. R. Clarke. 1995. High-frequency developmental abnormalities in p53-deficient mice. Curr. Biol. 5931-936. [DOI] [PubMed] [Google Scholar]
- 3.Bhat, K. P., K. Itahana, A. Jin, and Y. Zhang. 2004. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 232402-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brugarolas, J., C. Chandrasekaran, J. I. Gordon, D. Beach, T. Jacks, and G. J. Hannon. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377552-557. [DOI] [PubMed] [Google Scholar]
- 5.Budanov, A. V., and M. Karin. 2008. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budde, A., and I. Grummt. 1999. p53 represses ribosomal gene transcription. Oncogene 18:1119-1124. [DOI] [PubMed] [Google Scholar]
- 7.Cmejla, R., J. Cmejlova, H. Handrkova, J. Petrak, and D. Pospisilova. 2007. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum. Mutat. 281178-1182. [DOI] [PubMed] [Google Scholar]
- 8.Copp, A. J., F. A. Brook, and H. J. Roberts. 1988. A cell-type-specific abnormality of cell proliferation in mutant (curly tail) mouse embryos developing spinal neural tube defects. Development 104285-295. [DOI] [PubMed] [Google Scholar]
- 9.Draptchinskaia, N., P. Gustavsson, B. Andersson, M. Pettersson, T. N. Willig, I. Dianzani, S. Ball, G. Tchernia, J. Klar, H. Matsson, D. Tentler, N. Mohandas, B. Carlsson, and N. Dahl. 1999. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 21169-175. [DOI] [PubMed] [Google Scholar]
- 10.Dresios, J., I. L. Derkatch, S. W. Liebman, and D. Synetos. 2000. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 397236-7244. [DOI] [PubMed] [Google Scholar]
- 11.Ebert, B. L., J. Pretz, J. Bosco, C. Y. Chang, P. Tamayo, N. Galili, A. Raza, D. E. Root, E. Attar, S. R. Ellis, and T. R. Golub. 2008. Identification of RPS14 as a 5q− syndrome gene by RNA interference screen. Nature 451335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrar, J. E., M. Nater, E. Caywood, M. A. McDevitt, J. Kowalski, C. M. Takemoto, C. C. Talbot, Jr., P. Meltzer, D. Esposito, A. H. Beggs, H. E. Schneider, A. Grabowska, S. E. Ball, E. Niewiadomska, C. A. Sieff, A. Vlachos, E. Atsidaftos, S. R. Ellis, J. M. Lipton, H. T. Gazda, and R. J. Arceci. 2008. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood 1121582-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 14.Feng, Z., H. Zhang, A. J. Levine, and S. Jin. 2005. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. USA 1028204-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira-Cerca, S., G. Poll, P. E. Gleizes, H. Tschochner, and P. Milkereit. 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell 20263-275. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Cao, I., M. Garcia-Cao, J. Martin-Caballero, L. M. Criado, P. Klatt, J. M. Flores, J. C. Weill, M. A. Blasco, and M. Serrano. 2002. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 216225-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gazda, H. T., A. Grabowska, L. B. Merida-Long, E. Latawiec, H. E. Schneider, J. M. Lipton, A. Vlachos, E. Atsidaftos, S. E. Ball, K. A. Orfali, E. Niewiadomska, L. Da Costa, G. Tchernia, C. Niemeyer, J. J. Meerpohl, J. Stahl, G. Schratt, B. Glader, K. Backer, C. Wong, D. G. Nathan, A. H. Beggs, and C. A. Sieff. 2006. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 791110-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gazda, H. T., M. R. Sheen, A. Vlachos, V. Choesmel, M. F. O'Donohue, H. Schneider, N. Darras, C. Hasman, C. A. Sieff, P. E. Newburger, S. E. Ball, E. Niewiadomska, M. Matysiak, J. M. Zaucha, B. Glader, C. Niemeyer, J. J. Meerpohl, E. Atsidaftos, J. M. Lipton, P. E. Gleizes, and A. H. Beggs. 2008. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 83769-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, S. F. 2003. Developmental biology, 7th ed. Sinauer Associates, Inc., Sunderland, MA.
- 20.Gottlieb, E., R. Haffner, A. King, G. Asher, P. Gruss, P. Lonai, and M. Oren. 1997. Transgenic mouse model for studying the transcriptional activity of the p53 protein: age- and tissue-dependent changes in radiation-induced activation during embryogenesis. EMBO J. 161381-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 41-7. [DOI] [PubMed] [Google Scholar]
- 22.Jones, N. C., M. L. Lynn, K. Gaudenz, D. Sakai, K. Aoto, J. P. Rey, E. F. Glynn, L. Ellington, C. Du, J. Dixon, M. J. Dixon, and P. A. Trainor. 2008. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 14125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komarov, P. G., E. A. Komarova, R. V. Kondratov, K. Christov-Tselkov, J. S. Coon, M. V. Chernov, and A. V. Gudkov. 1999. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 2851733-1737. [DOI] [PubMed] [Google Scholar]
- 24.Komarova, E. A., M. V. Chernov, R. Franks, K. Wang, G. Armin, C. R. Zelnick, D. M. Chin, S. S. Bacus, G. R. Stark, and A. V. Gudkov. 1997. Transgenic mice with p53-responsive lacZ: p53 activity varies dramatically during normal development and determines radiation and drug sensitivity in vivo. EMBO J. 161391-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruiswijk, T., R. J. Planta, and J. M. Krop. 1978. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta 517378-389. [DOI] [PubMed] [Google Scholar]
- 26.Lohrum, M. A., R. L. Ludwig, M. H. Kubbutat, M. Hanlon, and K. H. Vousden. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3577-587. [DOI] [PubMed] [Google Scholar]
- 27.Lowe, S. W., E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362847-849. [DOI] [PubMed] [Google Scholar]
- 28.Marion, M. J., and C. Marion. 1987. Localization of ribosomal proteins on the surface of mammalian 60S ribosomal subunits by means of immobilized enzymes: correlation with chemical cross-linking data. Biochem. Biophys. Res. Commun. 149:1077-1083. [DOI] [PubMed] [Google Scholar]
- 29.McGowan, K. A., J. Z. Li, C. Y. Park, V. Beaudry, H. K. Tabor, A. J. Sabnis, W. Zhang, H. Fuchs, M. H. de Angelis, R. M. Myers, L. D. Attardi, and G. S. Barsh. 2008. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 40963-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendez-Vidal, C., M. T. Wilhelm, F. Hellborg, W. Qian, and K. G. Wiman. 2002. The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res. 301991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagy, A., M. Gertsenstein, K. Vintersten, and R. Behringer. 2003. Manipulating the mouse embryo: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Nikolaev, A. Y., M. Li, N. Puskas, J. Qin, and W. Gu. 2003. Parc: a cytoplasmic anchor for p53. Cell 11229-40. [DOI] [PubMed] [Google Scholar]
- 33.Nonn, L., R. R. Williams, R. P. Erickson, and G. Powis. 2003. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol. 23916-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliver, E. R., T. L. Saunders, S. A. Tarle, and T. Glaser. 2004. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development 1313907-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page, D. C., R. Mosher, E. M. Simpson, E. M. Fisher, G. Mardon, J. Pollack, B. McGillivray, A. de la Chapelle, and L. G. Brown. 1987. The sex-determining region of the human Y chromosome encodes a finger protein. Cell 511091-1104. [DOI] [PubMed] [Google Scholar]
- 36.Panic, L., J. Montagne, M. Cokaric, and S. Volarevic. 2007. S6-haploinsufficiency activates the p53 tumor suppressor. Cell Cycle 620-24. [DOI] [PubMed] [Google Scholar]
- 37.Panic, L., S. Tamarut, M. Sticker-Jantscheff, M. Barkic, D. Solter, M. Uzelac, K. Grabusic, and S. Volarevic. 2006. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol. Cell. Biol. 268880-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestov, D. G., Z. Strezoska, and L. F. Lau. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 214246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray, P. S., A. Arif, and P. L. Fox. 2007. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem. Sci. 32158-164. [DOI] [PubMed] [Google Scholar]
- 40.Rice, D. S., Q. Tang, R. W. Williams, B. S. Harris, M. T. Davisson, and D. Goldowitz. 1997. Decreased retinal ganglion cell number and misdirected axon growth associated with fissure defects in Bst/+ mutant mice. Investig. Ophthalmol. Vis. Sci. 382112-2124. [PubMed] [Google Scholar]
- 41.Rubbi, C. P., and J. Milner. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22:6068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudra, D., and J. R. Warner. 2004. What better measure than ribosome synthesis? Genes Dev. 18:2431-2436. [DOI] [PubMed] [Google Scholar]
- 43.Ruggero, D., and P. P. Pandolfi. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3:179-192. [DOI] [PubMed] [Google Scholar]
- 44.Sah, V. P., L. D. Attardi, G. J. Mulligan, B. O. Williams, R. T. Bronson, and T. Jacks. 1995. A subset of p53-deficient embryos exhibit exencephaly. Nat. Genet. 10175-180. [DOI] [PubMed] [Google Scholar]
- 45.Saveanu, C., A. Namane, P. E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 234449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmid, P., A. Lorenz, H. Hameister, and M. Montenarh. 1991. Expression of p53 during mouse embryogenesis. Development 113857-865. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt, E. V. 1999. The role of c-myc in cellular growth control. Oncogene 182988-2996. [DOI] [PubMed] [Google Scholar]
- 48.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 49.Spritz, R. A., S. A. Holmes, R. Ramesar, J. Greenberg, D. Curtis, and P. Beighton. 1992. Mutations of the KIT (mast/stem cell growth factor receptor) proto-oncogene account for a continuous range of phenotypes in human piebaldism. Am. J. Hum. Genet. 511058-1065. [PMC free article] [PubMed] [Google Scholar]
- 50.Sulic, S., L. Panic, M. Barkic, M. Mercep, M. Uzelac, and S. Volarevic. 2005. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 193070-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang, Q., D. S. Rice, and D. Goldowitz. 1999. Disrupted retinal development in the embryonic belly spot and tail mutant mouse. Dev. Biol. 207239-255. [DOI] [PubMed] [Google Scholar]
- 52.Tian, H., E. K. Wittmack, and T. J. Jorgensen. 2000. p21WAF1/CIP1 antisense therapy radiosensitizes human colon cancer by converting growth arrest to apoptosis. Cancer Res. 60679-684. [PubMed] [Google Scholar]
- 53.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408307-310. [DOI] [PubMed] [Google Scholar]
- 54.Volarevic, S., M. J. Stewart, B. Ledermann, F. Zilberman, L. Terracciano, E. Montini, M. Grompe, S. C. Kozma, and G. Thomas. 2000. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 2882045-2047. [DOI] [PubMed] [Google Scholar]
- 55.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 56.Watson, K. L., K. D. Konrad, D. F. Woods, and P. J. Bryant. 1992. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. USA 8911302-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wool, I. G. 1996. Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 21164-165. [PubMed] [Google Scholar]
- 58.Yuan, B., R. Latek, M. Hossbach, T. Tuschl, and F. Lewitter. 2004. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 32W130-W134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan, X., Y. Zhou, E. Casanova, M. Chai, E. Kiss, H. J. Grone, G. Schutz, and I. Grummt. 2005. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell 1977-87. [DOI] [PubMed] [Google Scholar]
- 60.Zhai, W., and L. Comai. 2000. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20:5930-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, Y., G. W. Wolf, K. Bhat, A. Jin, T. Allio, W. A. Burkhart, and Y. Xiong. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 238902-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao, L., T. Samuels, S. Winckler, C. Korgaonkar, V. Tompkins, M. C. Horne, and D. E. Quelle. 2003. Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways. Mol. Cancer Res. 1195-206. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.