Abstract
A conserved family of herpesvirus protein kinases plays a crucial role in herpesvirus DNA replication and virion production. However, despite the fact that these kinases are potential therapeutic targets, no systematic studies have been performed to identify their substrates. We generated an Epstein-Barr virus (EBV) protein array to evaluate the targets of the EBV protein kinase BGLF4. Multiple proteins involved in EBV lytic DNA replication and virion assembly were identified as previously unrecognized substrates for BGLF4, illustrating the broad role played by this protein kinase. Approximately half of the BGLF4 targets were also in vitro substrates for the cellular kinase CDK1/cyclin B. Unexpectedly, EBNA1 was identified as a substrate and binding partner of BGLF4. EBNA1 is essential for replication and maintenance of the episomal EBV genome during latency. BGLF4 did not prevent EBNA1 binding to sites in the EBV latency origin of replication, oriP. Rather, we found that BGLF4 was recruited by EBNA1 to oriP in cells transfected with an oriP vector and BGLF4 and in lytically induced EBV-positive Akata cells. In cells transfected with an oriP vector, the presence of BGLF4 led to more rapid loss of the episomal DNA, and this was dependent on BGLF4 kinase activity. Similarly, expression of doxycycline-inducible BGLF4 in Akata cells led to a reduction in episomal EBV genomes. We propose that BGLF4 contributes to effective EBV lytic cycle progression, not only through phosphorylation of EBV lytic DNA replication and virion proteins, but also by interfering with the EBNA1 replication function.
Herpesviruses encode two families of serine/threonine protein kinases, one of which, the BGLF4 (Epstein-Barr virus [EBV])/UL97 (human cytomegalovirus)/UL13 (herpes simplex virus)/ORF36 (Kaposi's sarcoma-associated herpesvirus)/ORF47 (varicella-zoster virus) family, is the sole protein kinase encoded by beta and gamma herpesviruses. The protein kinases phosphorylate both viral and host proteins (16, 21, 42) and are necessary for efficient virus lytic replication. Consequently, these kinases have been of interest as potential targets for antiviral drug development (37), and the compound 1263W94 (maribavir), which inhibits the cytomegalovirus UL97 protein (3), has been used in phase I clinical trials (27, 31, 47).
EBV infection is prevalent worldwide, and primary infection in adolescence or early adulthood is associated in 30 to 40% of cases with infectious mononucleosis. EBV efficiently infects B cells in the lymphoid tissues of the Waldeyer ring (43). EBV infection of B cells is biased toward establishment of latency with limited viral-gene expression (49). During latent infection, EBV genomes are maintained as extrachromosomal episomes. Replication of episomal genomes utilizes the latency origin of replication, oriP. The only EBV-encoded protein required is the origin binding protein EBNA1. All other essential replication factors are provided by the cell. Expression of the EBV replicative cycle and production of progeny virus take place in terminally differentiated plasma B cells (11, 29), and epithelial cells may also contribute to the cycle of virus replication and spread that is an important component of both persistent infection of the individual and transmission of virus from one individual to the next (4, 22). Lytic DNA replication initiates at separate origins, oriLyt. EBV encodes a set of six core lytic replication proteins, along with ancillary proteins, such as thymidine kinase (TK), that are involved in nucleotide metabolism (13, 44).
Several substrates have been described for the EBV BGLF4 protein kinase, including the core lytic EBV replication protein BMRF1, the polymerase processivity factor (8, 17). BGLF4 has also been found to locate to sites of lytic viral replication (46), to be required for efficient lytic DNA replication and release of nucleocapsids from the nucleus (18), and to contribute to the compaction of cell chromatin seen in cells undergoing lytic replication (32). Protein chip technology provides a new tool for global analysis of activities for biologically important enzymes, such as ubiquitin ligases, DNA repair enzymes, and kinases (7, 19, 36, 38, 52). Using an EBV protein array for unbiased screening, we identified multiple new BGLF4 substrates involved in lytic DNA replication, capsid assembly, and DNA packaging. Unexpectedly, we also identified EBNA1 as a substrate and binding partner for BGLF4. The data suggest that the contribution of BGLF4 to the EBV lytic cycle extends beyond the previously recognized contributions to lytic DNA replication and virion production and includes facilitating the switch from latent to lytic DNA replication by downregulating the EBNA1 replication function.
MATERIALS AND METHODS
Cloning of EBV ORFs.
EBV open reading frames (ORFs) were amplified by PCR using Platinum Taq DNA Polymerase High Fidelity (Invitrogen) and the Akata BXI bacterial artificial chromosome (BAC) (a gift from L. Hutt-Fletcher) as a template. Primers were designed based on the EBV sequence from GenBank accession numbers V01555 and AJ507799. The 5′ primers contained the attB1 recombination site, and the 3′ primers contained the attB2 recombination site (attB1, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCC-3′; attB2, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTC-3′). ORFs were amplified without the translational stop codon. To circumvent problems arising from PCR amplification across repeat sequences, some ORFs were amplified in segments. PCR products of the correct size were purified by gel electrophoresis and recombined into the Gateway vector pDONR201 using BP Clonase (Invitrogen). Escherichia coli bacteria were transformed with the reaction products, and three individual bacterial colonies were picked for each ORF. The DNA inserts were analyzed by digestion with BsrGI, and the inserts were sequenced in both directions. ORF-containing plasmid DNAs were recombined into a destination vector, pEGH-A, using LR Clonase (Invitrogen). pEGH-A is a derivative of the yeast glutathione S-transferase (GST) vector, pEGH, modified for compatibility with the Gateway system and by the introduction of a stop codon. The recombined products were transformed into E. coli, and four individual colonies were picked for each ORF. DNA inserts were analyzed by digestion with BamHI and HindIII. ORF-containing pEGH-A plasmid DNAs were transformed into yeast for protein expression and purification.
For expression of V5 epitope-tagged protein in bacteria and mammalian cells, EBNA1(386-641) (EBNA1c) and TK (BXLF1) were recloned into pDEST42 and pDEST40, respectively (Invitrogen). GST-EBNA1(408-641)(pYW13), GST-EBNA1(386-641)(pMRC92), EBNA1(386-641)-V5-six-His(pJZ42), EBNA1(392-641)-V5-six-His(pGL451D), and EBNA1(392-641S393R)-V5-six-His(pGL452D) were expressed in bacteria. For expression in mammalian cells, full-length EBNA1(pDY16) was expressed as a green fluorescent protein (GFP) fusion from the vector pEGFP (Clontech). BGLF4 was moved into the pSG5-Flag vector pJH253 to generate pJZ01, and wild-type and mutant BGLF4 ORFs were also ligated into the BglII site of the pSG5-HA vector (pHYC66) to generate plasmids pLS101 and pLS102, respectively. Kinase-null BGLF4(K102Q) mutated in the catalytic domain (17, 25) was generated using the QuikChange II site-directed mutagenesis kit (Stratagene). For episomal-maintenance experiments, BGLF4 and BGLF4(K102Q) were amplified by PCR from the hemagglutinin (HA)-BGLF4 plasmids and ligated into BamHI- and HindIII-cleaved pCEP4 (Invitrogen) to generate pLS103 and pLS104, respectively.
Protein purification and protein arrays.
EBV ORFs were expressed as GST fusion proteins in yeast. Cultures (50 ml) were grown at 30°C to an optical density at 600 nm of 0.7 to 0.9 and induced with 2% galactose for 4 to 6 h. Harvested cells were lysed with glass beads in lysis buffer (100 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EGTA, 0.1% 2-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% Triton X-100 plus protease inhibitor cocktail [Roche]). GST fusion proteins were bound to glutathione beads (GE Healthcare) for 1 h at 4°C and washed four times for 15 min each time with wash I buffer (50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EGTA, 0.1% Triton X-100, 0.1% β-mercaptoethanol, and 0.5 mM PMSF) and two times with wash II buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, 1 mM EGTA, 10% glycerol, 0.1% β-mercaptoethanol, and 0.5 mM PMSF) and eluted in elution buffer (100 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10 mM MgCl2, 20 mM glutathione, and 20% glycerol). The eluate was collected through a filter unit and stored in 384-well plates. Sixty EBV protein products (see Table S1 in the supplemental material) were successfully purified based on immunoblot analysis and printed in duplicate on modified glass (Full Moon Biosystems) microscope slides using a 48-pin contact printer (Bio-Rad).
Protein array phosphorylation assays.
The protocol for phosphorylation assays on the EBV protein chips was modified from that of Ptacek et al. (38). Briefly, the protein chips were preincubated in Superblock (Pierce) containing 0.2% bovine serum albumin and 0.1% Triton X-100 for 1 h at room temperature. GST-BGLF4 or CDK1/cyclin B (Upstate) was diluted in kinase buffer (50 mM HEPES [pH 7.4], 10 mM MgCl2, 10 mM MnCl2, 300 mM KCl, and 0.5% Nonidet P-40) containing 0.5 mg/ml bovine serum albumen, 0.1% Triton X-100, and 2 μl of [γ-32P]ATP (33.3 nM final concentration). Duplicate protein arrays were incubated with the kinase mixture in a humidified chamber, with shaking, for 30 min at 30°C. Slides were subjected to three 10-min washes with TBST buffer (25 mM Tris-HCl [pH 7.4], 3 mM KCl, 150 mM NaCl, 0.05% Tween 20) and three 10-min washes with 10 mM Tris-HCl (pH 7.4) and 0.5% sodium dodecyl sulfate (SDS), rinsed briefly with doubly distilled water, and spun dry. Control slides were incubated with kinase reaction mixture without kinase and processed in parallel. The slides were exposed to X-ray film (Kodak), which was scanned at 1,800 pixels/inch and analyzed using Genepix 3.0 (Molecular Devices).
Substrate validation.
GST-BGLF4 and potential EBV substrates were purified from yeast using glutathione beads. The bound proteins were washed with lysis buffer containing 250 mM NaCl and eluted into kinase buffer. Purified GST-BGLF4 (0.1 μg) or CDK1/cyclin B (Upstate) was mixed with 40 nM [γ-32P]ATP and substrate protein and incubated in 25 μl kinase buffer for 30 min at 30°C. The reaction was terminated by heating the mixture at 90°C for 5 min, and the proteins were separated on NuPAGE gels (4 to 12% bis-Tris or 4 to 20% Tris-HCl; Invitrogen). The gels were dried and exposed to MP Hyperfilm (GE Healthcare). Validated substrates were tested at least two times. Control reactions lacked GST-BGLF4.
EMSA.
Biotinylated double-stranded DNA probes were made by incubating the oligonucleotide 5′-GTACCCGGGGATCCTATCTGGGTAGCATATGCTATCCTAATGGATCCTCTAGAGTCGACCCCCTATAGTGAGTCGTATTAGGATCC-3′ and the 5′-biotin-labeled primer 5′-GGATCCTAATACGACTCACTATAGGG-3′ with Klenow polymerase at 55°C for 1 min and 72°C for 10 min. The double-stranded DNA was separated from single-stranded DNA using the QiaEXII kit (Qiagen). GST-BGLF4 and EBNA1 extract (1 μl; Light Shift Chemiluminescent EMSA [electrophoretic mobility shift assay] Kit; Pierce) were added individually or together to 30 μl of kinase buffer with or without 1 mM ATP or adenosine 5′(gamma-thio) triphosphate (γ-S-ATP) and incubated at 30°C for 30 min. Biotin-labeled double-stranded EBNA1 probe DNA (20 fmol) in binding buffer [10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 50 ng/μl poly(dI-dC), 1 mM dithiothreitol, 2.5% glycerol, 0.05% Nonidet P-40] was added, and the mixture was incubated for 30 min at room temperature. Samples were analyzed by electrophoresis through a 5% DNA-Retardation Gel (Invitrogen) at 4°C for 45 min. DNA was transferred to Hybond-N nylon membranes (GE Healthcare) presoaked in TBE (89 mM boric acid, 2 mM EDTA, pH 8.3) for 10 min and cross-linked at 120 mJ/cm2 using a UV Stratalinker (Stratagene). Membrane-linked, biotinylated DNA was detected using the LightShift Chemiluminescent EMSA Kit (Pierce). The membrane was processed following the manufacturer's protocol and exposed to MP Hyperfilm (GE Healthcare).
Phosphorylation of EBNA1 and TK in transfected cells.
GFP-EBNA1 or V5-TK and Flag-BGLF4 were transfected individually or together using Lipofectamine 2000 (Invitrogen) into HeLa cells grown in six-well plates. Two days later, the cells were washed with phosphate-buffered saline (PBS) and incubated in phosphate-free Dulbecco's modified Eagle's medium at 37°C for 1 h. The cells were then labeled by incubation at 37°C for 2 h in 1 ml of phosphate-free Dulbecco's modified Eagle's medium containing 10% dialyzed fetal bovine serum and 0.1 mCi/ml [32P]orthophosphate (GE Healthcare). The cells were rinsed with 1 ml of ice-cold phosphate buffered saline and lysed in 1 ml RIPA buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.25% Na deoxycholate, 150 mM NaCl, and 1 mM EDTA) containing protease inhibitor cocktail (1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin) and phosphatase inhibitor cocktail (Sigma). After sonication (Branson Sonifier 450) on ice for 1 min, the cell lysates were centrifuged at 10,000 × g for 10 min. The supernatants were precleared by incubation, with rotation, with 20 μl of a 50% protein A-Sepharose bead slurry (Amersham) at 4°C for 30 min. The mixture was centrifuged, and the supernatant (400 μl) was incubated with 1 μl of EBNA.OT1x (a gift of J. Middeldorp) or anti-V5 (Invitrogen) antibody for 4 h at 4°C. Protein A-Sepharose beads (20 μl) were added, and the mixture was rotated at 4°C for 2 h. The beads were washed four times with 500 μl of ice-cold lysis buffer. Samples were resuspended in 60 μl of 2× Laemmli sampling buffer (Bio-Rad) and heated at 95°C for 5 min. The samples were resolved on two parallel 4 to 20% SDS-polyacrylamide gel electrophoresis gels (Invitrogen). One gel was dried for autoradiographic analysis, and the proteins on the second gel were transferred to a nitrocellulose membrane and subjected to immunoblot analysis with EBNA.OT1x (9) or anti-V5 antibodies.
GST-affinity assay.
GST-BGLF4 produced in yeast was captured on glutathione beads and washed with wash I and wash II buffers. Six-His-V5-tagged EBNA1c was purified from bacteria using a nickel affinity column (USB Corporation). Eluted six-His-V5-tagged EBNA1c was incubated with glutathione beads with or without GST-BGLF4 for 1 h at 4°C. The beads were pelleted and washed six times with TBST buffer. The protein/bead complex was boiled in Laemmli sample buffer for 5 min at 95°C and then subjected to gel electrophoresis and immunoblot analysis using anti-GST and anti-V5 antibodies.
Indirect immunofluorescence.
HeLa cells grown on two-well slides were transfected with V5-EBNA1c alone or together with HA-BGLF4 or HA-BGLF4 mutated in the kinase active site (HA-mtBGLF4). Two days after transfection, the cells were fixed for 10 min in 1% paraformaldehyde and the slides were incubated in blocking buffer (2.5% normal goat serum, 0.3% Triton X-100, and 2% bovine serum albumin). Primary antibody (mouse anti-V5 [Invitrogen] or rabbit anti-HA [Sigma]) was diluted 1:100 in blocking buffer and incubated with the cells for 2 h at room temperature. The cells were subjected to three 5-min washes with PBS. Secondary antibody (rhodamine anti-mouse immunoglobulin or fluorescein isothiocyanate-anti-rabbit immunoglobulin [Jackson]) was used at a 1:500 dilution in blocking buffer and incubated with the cells for 1 h at room temperature. The cells were finally subjected to three 5-min washes with PBS, and the slides were mounted in Vectashield containing DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories).
Lytic induction of EBV-positive cells.
EBV-positive Akata B cells were cultured in RPMI 1640 containing 10% fetal bovine serum and grown in a humidified 5% CO2 incubator at 37°C. To induce the EBV lytic cycle, the Akata cells were treated with 50 μg/ml of goat anti-human immunoglobulin G (IgG) (MP Biomedicals) for 24 h before being harvested.
Doxycycline-inducible Akata-BGLF4 cell line.
To generate Akata-BGLF4 cells, Akata cells were first transduced with pRetroX-Tet-ON-Advanced (Clontech) and selected by growth in G418. A doxycycline-responsive population was subcloned by transfecting pRetroX-Tight-Pur-Luc (Clontech) and selecting for highly inducible luciferase expression. BGLF4 was cloned into pRetroX-Tight-Pur (Clontech), and transfection of Akata (pRetroX-Tet-ON-Advanced) cells with the empty vector or the BGLF4-expressing vector followed by selection in puromycin yielded the doxycycline-inducible Akata-BGLF4 and Akata-vector cell lines. BGLF4 was induced using 1 μg/ml doxycycline. The episomal EBV genome content of the Akata cell lines was analyzed using gel electrophoresis and Southern blotting (14). Briefly 2.5 × 106 cells from each cell line were resuspended in TBE buffer containing 15% Ficoll, 10 μg/ml RNase A, and 0.01% bromophenol blue. Cells were loaded into wells in a 2.5-cm, 0.8% agarose overlay of a 0.75% agarose gel. The overlay contained 2% SDS and 1 mg/ml nuclease-free pronase (CalBiochem). Electrophoresis was carried out at 4°C and 30 V for 3 h, followed by 100 V for 16 h. The separated DNA was transferred to a nitrocellulose membrane and hybridized with an EBV BamHI-W restriction enzyme fragment DNA probe that was labeled with [32P]dCTP using the Promega Prime-a-Gene Labeling System. The DNA was visualized by autoradiography.
Immunoprecipitation.
Endogenous immunoprecipitations were performed on Akata EBV-positive cells with or without lytic induction and on doxycycline-treated Akata-BGLF4 cells. Protein A- and protein G-Sepharose beads were added to the cell lysates, and the mixture was incubated for 2 h at 4°C with goat anti-EBNA1 antibody (ViroStat) or goat IgG (GE Healthcare). The beads were washed six times with RIPA buffer, and the pellet was resuspended in 50 μl of sample buffer. The samples were subjected to Western blotting using mouse anti-BGLF4 antibody (46). HeLa cells were cotransfected with GFP-EBNA1(pDY16) and either pCEP4-HA-BGLF4 or pCEP4-HA-mtBGLF4. Cells were harvested 3 days posttransfection. The cell lysate was incubated at 4°C with 2 μl of mouse EBNA.OT1x antibody (1:4,000) or 2 μl of mouse anti-HA antibody (Sigma). The immunoprecipitated proteins were subjected to Western blotting using rabbit anti-HA antibody (1:5,000; Sigma) to detect HA-BGLF4.
Chromatin immunoprecipitation (ChIP).
HeLa cells grown in six-well plates were transfected with pCEP4 plus V5-EBNA1c or pCEP4 plusV5-EBNA1c and Flag-BGLF4. Two days after transfection, formaldehyde was added to the culture medium to a final concentration of 1%, and the cells were incubated for 10 min at 37°C. The cells were washed twice with ice-cold PBS and lysed in 1 ml of RIPA buffer containing protease inhibitors. To shear the DNA, the cells were sonicated on ice for four 30-s intervals with a Sonifier 450 sonicator (Branson; output, 3.0; duty cycle, 30%). Sonicated samples were centrifuged at 4°C for 10 min at 13,000 rpm. Supernatant (100 μl) was transferred to a new tube and diluted with 900 μl RIPA buffer. The supernatant was precleared with 50 μl of protein A-Sepharose that had been preincubated at 4°C for 1 h with 0.5 mg/ml of sonicated herring sperm DNA (Invitrogen). The beads were pelleted by centrifugation, and the supernatant was transferred to a new tube. The precleared supernatant was incubated with anti-V5 or anti-Flag antibody overnight at 4°C. Mouse IgG (mIgG) was added to negative control samples. Supernatants were then incubated with 60 μl of the herring sperm DNA/protein A-Sepharose slurry for 1 h at 4°C with rotation. The beads were pelleted by centrifugation and washed six times with RIPA buffer at 4°C. The protein complex was eluted using two 150-μl volumes of elution buffer (1% SDS and 0.1 M NaHCO3) and incubation at room temperature for 15 min with shaking. After the addition of 12 μl of 5 M NaCl to the combined eluates (300 μl), the protein-DNA cross-linking was reversed by heating the mixture at 65°C for 4 h. An aliquot (5 μl) was then used in the PCR. The primer pair 5′-GACTGGATGTCCATCTCACACG-3′ and 5′-CAATCAGCCAAGTTATTAC-3′ was used to amplify a DNA fragment adjacent to the EBNA1 binding sites on pCEP4.
For the endogenous ChIP assay, Akata cells with or without lytic induction were cross-linked with 1% formaldehyde for 10 min at 37°C before being harvested. Cell extracts were processed as described above. The endogenous EBNA1 and BGLF4 were immunoprecipitated with mouse EBNA.OT1x antibody and mouse BGLF4 antibody (46). The primer pairs used for DNA amplification were as follows: FR, 5′-GACTCTGCTTTCTGCCGTCTTCG-3′ and 5′-GGCAAAAGGATGGTTAGGGGAG-3′; DS, 5′-CCGTGACAGCTCATGGGGTGGGAG-3′ and 5′-CAGAGGGGCCTGTGTAGCTACCG-3′; BBLF1, 5′-TCGGCCGCTATTAGCTTAGT-3′ and 5′-GGTGCCCTCTGGTCTCTTT-3′; BOLF1, 5′-GGTGACTGTGATCTGTTCCG-3′ and 5′-AGCATGGACGACATCCTGTA-3′.
Episome retention assays.
HeLa cells seeded into 10-cm plates were transfected with 24 μg/well of pCEP4 (Invitrogen), pCEP4-BGLF4, or pCEP4-mtBGLF4. At 3, 6, 9, 12, and 15 days posttransfection, cells were harvested and DNA was extracted using the Wizard genomic-DNA purification kit (Promega). PCR primers for the detection of EBNA1 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were designed using Primer3 software as follows: EBNA1, 5′-ACGTAGAAAGGACTACCGACGA-3′ and 5′-GGTGTAAGACGACATTGTGGAA-3′; GAPDH, 5′-GCCTCTTGTCTCTTAGATTTGGTC-3′ and 5′-TAGCACTCACCATGTAGTTGAGGT-3′. PCR mixtures (25 μl) contained primers (100 nM), cell DNA (50 ng), and SYBR Green PCR Master Mix (Applied Biosystems). Cycle threshold values were measured using a 7500 Real-Time PCR System (Applied Biosystems). Relative levels of EBNA1(pCEP4) DNA were normalized to cellular GAPDH for each sample. EBV Akata genomes were quantitated using the BALF5 primers 5′-AGTCCTTCTTGGCTACTCTGTTGAC-3′ and 5′-CTTTGGCGCGGATCCTC-3′.
RESULTS
Identification of substrates for the EBV protein kinase BGLF4.
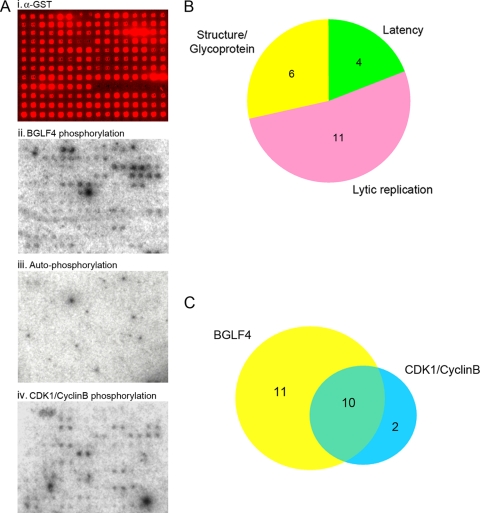
Five EBV proteins have been identified as substrates for the EBV protein kinase BGLF4 (2, 8, 17, 24, 25, 50). To systematically identify EBV protein substrates and to better understand the contributions of BGLF4 to EBV biology, we turned to a proteomic approach. EBV ORFs were amplified by PCR using Akata BX1 BAC DNA as a template and recombined into the Gateway vector pDONR201. A master set of entry clones with the correct insert sizes was assembled, and the EBV inserts were sequenced in both directions. Inserts from the sequence-confirmed entry clones were shuttled into the yeast expression plasmid pEGH-A using LR Clonase recombination. Successful subcloning was confirmed by restriction digestion, and the plasmids were then transformed into yeast cells for protein expression and purification. Sixty proteins were obtained that had the appropriate size and could be obtained in satisfactory yields, and these 60 proteins were purified and printed onto Full Moon slides. The printing quality and the quantity of the immobilized proteins on the chip were monitored using anti-GST antibody followed by Cy5-labeled secondary antibody (Fig. 1A, i), and the signal intensity of each spot was quantified using GenePix software.
FIG. 1.
Identification of BGLF4 and CDK1/cyclin B substrates. (A) Phosphorylation assays on EBV chips. (i) Validation of protein printing using anti-GST (α-GST) primary antibody and Cy5-conjugated secondary antibody. (ii) BGLF4 phosphorylation. (iii) Phosphorylation of the array proteins by [γ-32P]ATP in the absence of added kinases (autophosphorylation). (iv) CDK1/cyclin B phosphorylation. (B) Distribution of BGLF4 targets between EBV protein categories. (C) Overlap between the BGLF4 and CDK1/cyclin B protein array substrates.
GST-BGLF4 protein purified from yeast was incubated with the EBV protein chip in kinase reaction buffer containing [γ-32P]ATP. After the kinase reaction, the chips were washed under denaturing conditions in buffer containing 0.5% SDS to remove added BGLF4 and unincorporated [γ-32P]ATP and exposed to X-ray film (Fig. 1A, ii). Reactions were performed in duplicate and repeated twice to ensure reproducibility. Potential BGLF4 substrates were identified using GenePix software. GST-BGLF4 was omitted from the control parallel reaction (Fig. 1A, iii). In this negative control, the lack of reproducible signals indicated that there was minimal contamination of the printed proteins with yeast kinases. A total of 21 EBV proteins consistently scored as BGLF4 substrates in the chip assays (Table 1). Both latent and lytic cycle EBV proteins were identified as BGLF4 substrates. The lytic cycle proteins included those involved in lytic DNA replication and virion structure (Fig. 1B and Table 1). BGLF4 is known to phosphorylate consensus sites for the cell CDK1 kinase. To compare these two activities, a kinase reaction was also performed on the EBV protein array using commercial CDK1 complexed with cyclin B (Fig. 2B). Ten of the 21 BGLF4 substrates were also substrates for CDK1/cyclin B, and 10 of 12 CDK1/cyclin B substrates were BGLF4 substrates (Fig. 1C). This suggests that BGLF4 has a broader substrate specificity than CDK1/cyclin B.
TABLE 1.
BGLF4 and CDK1 substratesa
| EBV ORF | BGLF4 | CDK1/cyclin B |
|---|---|---|
| Latency genes | ||
| BNLF1 (LMP1 exon 3) | + | − |
| BYRF1 (EBNA2) | + | + |
| A73 (BART ORF) | + | + |
| BKRF1 (EBNA1 aa 386-641) | + | + |
| LMP2A | − | + |
| Lytic genes | ||
| BGLF5 (alkaline exonuclease) | + | + |
| BXLF1 (TK) | + | + |
| BALF2 (single-stranded DNA binding protein) | + | − |
| BBLF4 (helicase) | + | − |
| BBRF3 (glycoprotein M) | + | + |
| BLLF1 (gp350) | + | − |
| BALF1 (antiapoptotic) | + | − |
| BILF1 (G-protein-coupled receptor) | + | + |
| BORF1 (capsid) | + | − |
| BORF2 (ribonucleotide reductase, large subunit) | + | − |
| BGLF2 (capsid maturation) | + | − |
| BKRF3 (uracil DNA glycosylase) | + | + |
| BDLF1 (capsid) | + | + |
| BMLF1 (MTA, post-transcriptional regulator) | + | − |
| BMRF1 (polymerase processivity factor) | + | − |
| BLRF1 (glycoprotein N) | + | − |
| BaRF1 (ribonucleotide reductase, small subunit) | − | + |
| BDRF1 (terminase exon 2) | + | + |
+, phosphorylated; −, not phosphorylated; BART, BamHI-A rightward transcript.
FIG. 2.
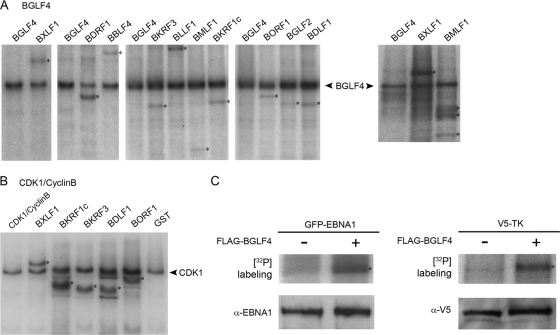
Validation of BGLF4 and CDK1/cyclin B substrates. (A and B) In vitro kinase assays using GST-BGLF4 (A) or CDK1/cyclin B (B) and the indicated GST target proteins purified from yeast and incubated in kinase buffer containing [γ-32P]ATP. Samples were subjected to gel electrophoretic separation, and the dried gels were exposed to X-ray film. Phosphorylated target proteins are indicated by asterisks. (C) Phosphorylation of EBNA1 (BKRF1) and TK (BXLF1) by BGLF4 in HeLa cells transfected with GFP-EBNA1 or V5-TK alone or together with Flag-BGLF4 and incubated for 2 h in medium containing [32P]orthophosphate prior to being harvested. (Top) Phosphorylation of the EBNA.OT1x and anti-V5 immunoprecipitates visualized by autoradiography. (Bottom) Verification of GFP-EBNA1 and V5-TK expression by Western blotting of EBNA.OT1x and anti-V5 immunoprecipitates.
Validation of selected BGLF4 and CDK1/cyclin B substrates.
Phosphorylation of selected EBV targets by BGLF4 was confirmed using a standard in vitro kinase assay with [γ-32P]ATP, GST-BGLF4, and GST-EBV fusion proteins purified from yeast. After the kinase reaction, the proteins were separated by gel electrophoresis, and the dried gel was exposed to X-ray film (Fig. 2A). Nine of the 10 phosphorylated products in this assay were of the expected size. The exception was GST-BMLF1, which is difficult to obtain in intact form (40) and was degraded. GST-BMLF1 purified from bacteria was retested in a separate kinase reaction along with BXLF1 (Fig. 2A, far right). Phosphorylation of BMLF1 was confirmed, although again, the fusion protein generated breakdown products. BXLF1 (TK) was further confirmed in this assay as a BGLF4 substrate. Five CDK1/cyclin B substrates that were also BGLF4 substrates were selected and tested in the in vitro kinase assay. Each was confirmed as a CDK1/cyclin B substrate (Fig. 2B).
One of the new BGLF4 targets identified in these assays was EBNA1. EBNA1c (amino acids [aa] 386 to 641) was phophorylated by BGLF4 in the protein array phosphorylation assay, while the N terminus of EBNA1 (aa 1 to 87) was not a BGLF4 substrate. Another substrate was BXLF1 (TK), which had previously been found to interact with BGLF4 in a yeast two-hybrid screen (5) but had not been shown to be phosphorylated by BGLF4. To further validate EBNA1 and BXLF1 as BGLF4 substrates, HeLa cells were transfected with full-length GFP-tagged EBNA1 or V5-tagged TK in the presence or absence of Flag-BGLF4. Two days posttransfection, cells were metabolically labeled for 2 hours in medium containing [32P]orthophosphate and then harvested. GFP-EBNA1 and V5-TK were immunoprecipitated from the transfected cell lysates with EBNA.OT1x and anti-V5 antibodies, respectively, and the immunoprecipitates were subjected to gel electrophoresis. The gel containing the EBNA1 and V5 immunoprecipitates was dried and exposed to X-ray film to examine the phosphorylation status of EBNA1 and TK. A marked increase in isotopic labeling of EBNA1 and TK occurred in the presence of cotransfected Flag-BGLF4, indicating that EBNA1 and TK were BGLF4 substrates in cultured cells (Fig. 2C, top). The precipitates were also immunoblotted with EBNA.OT1x and anti-V5 antibodies to demonstrate equal expression of EBNA1 and TK in samples with and without BGLF4 (Fig. 2C, bottom).
Mapping the phosphorylation site in EBNA1c.
We focused on the unexpected substrate, EBNA1, and sought to map the region of EBNA1c required for BGLF4 phosphorylation. A kinase assay was performed using equal amounts of GST-EBNA1c fusions commencing at aa 386, 408, and 459 (Fig. 3A). Only the construction commencing at aa 386 was a substrate for BGLF4. Coomassie blue staining was used to show equal loading of the EBNA1c proteins (Fig. 3A, bottom). To eliminate any possible contribution of the GST tag to the phosphorylation signal, the in vitro kinase assay was repeated using six-His-V5-tagged EBNA1c. BGLF4 also phosphorylated this EBNA1c protein (Fig. 3B, left). Coomassie blue staining of the same gels was used to show equal loading of the EBNA1c proteins (Fig. 3B, Input). Examination of the EBNA1 sequence between aa 386 and 408 revealed a consensus CDK1 phosphorylation site between aa 393 and 396 (SPPR) (Fig. 3A). BGLF4 is known to phosphorylate CDK1 sites (24, 26). To further define the phosphorylation site, in vitro kinase assays were performed on six-His-V5-tagged EBNA1c proteins commencing at aa 392, with the 393 serine being either wild type or mutated to an arginine residue (S393R). Both BGLF4 and CDK1/cyclin B phosphorylated the proteins containing serine at amino acid 393, and in each case, phosphorylation was eliminated by mutation of serine 393 (Fig. 3B, right). Thus, EBNA1 serine 393 is a substrate for BGLF4 and for cellular CDK1/cyclin B.
FIG. 3.
Mapping the BGLF4 phosphorylation site in EBNA1c. (A) In vitro kinase assay comparing BGLF4 phosphorylation of three GST-EBNA1 fusion proteins; EBNA1c(386-641), EBNA1c-1(408-641), and EBNA1c-2(459-641). Equal amounts of EBNA1c proteins were present in the assays, as illustrated by Coomassie blue staining (bottom). The cartoon shows the location of the consensus CDK1 site relative to key EBNA1c functional domains. GA, glycine and alanine. (B) In vitro kinase assays showing that six-His-V5-tagged EBNA1 is also a BGLF4 substrate (left) and that mutation of EBNA1 aa 393 from S to R in EBNA1c(392-641) causes loss of both CDK1 and BGLF4 phosphorylation (right). Coomassie blue staining of the bacterially expressed EBNA1 proteins (Input) shows equality of substrate loading.
BGLF4 interacts with EBNA1.
Since EBNA1c was a substrate for BGLF4, we also tested for interaction between EBNA1c and BGLF4. Three approaches were taken. First, a GST affinity assay was performed using GST-BGLF4 purified from yeast and six-His-V5-tagged EBNA1c purified from bacteria. Binding of six-His-V5-EBNA1c to GST-BGLF4 immobilized on glutathione beads was detected by immunoblotting using anti-V5 antibody. Interaction did not require ATP or γ-S-ATP, suggesting that the interaction was not dependent on BGLF4 kinase activity or fixation of BGLF4 in an enzymatically active conformation (Fig. 4A, top). Negative control assays lacked BGLF4. Equal loading of GST-BGLF4 was demonstrated by immunoblotting with anti-GST antibody (Fig. 4A, bottom).
FIG. 4.
BGLF4 interacts with EBNA1. (A) GST affinity assay. GST-BGLF4-bound glutathione beads were incubated with purified, bacterially expressed V5-EBNA1c in kinase buffer with or without ATP or γ-S-ATP. The control reaction lacked GST-BGLF4. Bound V5-EBNA1c was detected by Western blotting using anti-V5 antibody. The blots were probed with anti-GST (α-GST) antibody to show loading of GST-BGLF4. (B) Relocation of V5-EBNA1c into the nucleus by BGLF4. Immunofluorescence assays in transfected HeLa cells are shown. EBNA1c lacks a nuclear localization signal and has a cytoplasmic localization (RHOD) (top). In the presence of HA-BGLF4 or HA-mtBGLF4 (FITC) (middle and bottom), EBNA1c was relocated into the nucleus (RHOD and Merge) (middle and bottom). Nuclei were visualized with DAPI. (C) Coprecipitation of GFP-EBNA1 with HA-BGLF4 and HA-mtBGLF4. HA-BGLF4 proteins present in the direct α-HA and indirect α-EBNA1 immunoprecipitates generated from extracts of cotransfected HeLa cells were detected by probing a Western blot (WB) with α-HA antibody. IP, immunoprecipitation. (D) Coprecipitation of endogenous EBNA1 with BGLF4 using extracts from a doxycycline (Dox)-inducible BGLF4-expressing Akata cell line. (Left) Western blots showing expression of EBNA1 and BGLF4 before and after treatment with doxycycline. (Right) Western blot showing BGLF4 protein present in the direct anti-BGLF4 immunoprecipate, the cell extract, and the indirect anti-EBNA1 immunoprecipitate. BGLF4 was detected with anti-BGLF4 antibody. (E) Western blots showing coprecipitation of BGLF4 with EBNA1 in lytically induced Akata cells.
The second assay used indirect immunofluorescence to demonstrate interaction through relocalization of EBNA1c by cotransfected BGLF4. EBNA1c lacks the EBNA1 nuclear localization signal, which maps between aa 379 and 386 (1). Consequently, V5-EBNA1c has a cytoplasmic localization in transfected HeLa cells (Fig. 4B, top). BGLF4 localizes to the nucleus (Fig. 4B) (2, 15). When HeLa cells were cotransfected with HA-BGLF4, V5-EBNA1c became relocalized into the nucleus (Fig. 4B, middle). We generated a kinase-null HA-mtBGLF4 carrying a previously described K102Q mutation (17, 25). Mutant BGLF4 also mediated nuclear localization of V5-EBNA1c (Fig. 4B, bottom), reinforcing the evidence that BGLF4 kinase activity is not needed for physical interaction between BGLF4 and EBNA1. In the immunofluorescence assays, 100 fluorescent cells were counted for each sample. The intracellular localization shown in Fig. 4C was observed in >75% of the positive cells.
Third, interaction between BGLF4 and EBNA1 was demonstrated in transfected cells and in EBV-positive Akata cells. Coimmunoprecipitation assays were performed on HeLa cells cotransfected with GFP-EBNA1 and either wild-type HA-BGLF4 or HA-mtBGLF4 carrying the K102Q mutation. HA-BGLF4 and HA-mtBGLF4 were directly precipitated by anti-HA antibody and coprecipitated with EBNA1 in the EBNA.OT1x immunoprecipitates and were not precipitated with control IgG (Fig. 4C). The coprecipitation of both BGLF4 and mtBGLF4 with EBNA1 further confirmed that the kinase activity of BGLF4 is not essential for their physical interaction. Coprecipitation of endogenous EBNA1 with BGLF4 was also demonstrated using Akata cells. A pair of EBV-positive Akata cell lines were generated carrying the doxycycline regulatory element and an empty expression vector or carrying the doxycycline regulatory element plus a BGLF4-expressing vector. Inducible expression of BGLF4 in the Akata-BGLF4 cells and expression of endogenous EBNA1 are illustrated in the Western blots in Fig. 4D (left), where BGLF4 was detected using anti-BGLF4 antibody (46) and EBNA1 was detected with anti-EBNA1 antibody. After doxycycline induction of BGLF4, the cell lysate was immunoprecipitated with anti-EBNA1 antibody (ViroStat), control anti-goat IgG, or anti-BGLF4 antibody. The immunoprecipitates and cell extract were subjected to SDS-polyacrylamide gel electrophoresis, and Western blotting was performed using anti-BGLF4 antibody (Fig. 4D, right). BGLF4 was detected in the extract, in the direct anti-BGLF4 immunoprecipitate, and as a coprecipitating protein in the anti-EBNA1 precipitate. BGLF4 was not present in the control IgG immunoprecipitate. The coprecipitation of BGLF4 with endogenous EBNA1 was also detected after, but not before, treatment of the parental Akata cells with anti-human IgG to induce the EBV lytic cycle (Fig. 4E, bottom). These experiments demonstrate that BGLF4 can interact with endogenous EBNA1 in Akata cells.
BGLF4 does not prevent EBNA1 DNA binding.
To evaluate how phosphorylation of EBNA1 by BGLF4 might affect EBNA1 function, we first examined EBNA1 DNA binding activity. An EMSA was performed using a biotin-labeled, double-stranded DNA probe containing a single EBNA1 binding site. Preincubation with GST-BGLF4 purified from yeast did not prevent EBNA1 binding to the DNA probe even when ATP was included in the incubation to allow phosphorylation of EBNA1 (Fig. 5A). The specificity of the binding was confirmed by using excess nonlabeled DNA probe to compete away the shifted DNA bands (data not shown).
FIG. 5.
EBNA1 recruits BGLF4 to oriP. (A) EMSA showing that EBNA1 DNA binding activity is not inhibited by BGLF4. EBNA1 nuclear extract (ext) was incubated with biotin-labeled DNA probe with (+) or without (−) preincubation with GST-BGLF4, as indicated. (B) ChIP assay examining the recruitment of BGLF4 to oriP. HeLa cells were transfected with an oriP-containing plasmid, pCEP4, and either V5-EBNA1c or V5-EBNA1c plus Flag-BGLF4. In cotransfected cells, pCEP4 oriP sequences were ChIPed by both anti-Flag (α-Flag) and α-V5 antibodies, indicating recruitment of Flag-BGLF4 to oriP in the presence of V5-EBNA1c. mIgG, control nonspecific mouse antibody. (C) ChIP assays performed on uninduced (left) and lytically induced (right) Akata cells. BGLF4 was recruited to oriP after lytic induction. mIgG served as the control antibody. FR and DS primer pairs, but not the BBLF1 and BOLF1 primer pairs, amplify the oriP region of the EBV genome.
BGLF4 associates with oriP in the presence of EBNA1.
The EMSAs indicated that BGLF4 did not impair binding of EBNA1 to a consensus binding site in vitro. A ChIP assay was performed to examine EBNA1 binding to oriP in cultured cells. HeLa cells were cotransfected with the oriP-containing vector pCEP4 plus either V5-EBNA1c or V5-EBNA1c and Flag-BGLF4. Two days posttransfection, the cells were fixed and lysed, and V5-EBNA1c and Flag-BGLF4 complexes were immunoprecipitated from the extract using anti-V5 and anti-Flag antibodies. The protein-DNA cross-linking was reversed, and DNA associated with the immunoprecipitates was amplified by PCR using primers complementary to sequences adjacent to the oriP EBNA1 binding sites. No DNA signal was obtained from the immunoprecipitates generated from the pCEP4- and V5-EBNA1c-transfected cells (Fig. 5B). This was to be expected. EBNA1c localizes to the cytoplasm (Fig. 4B) and therefore would not be available in the cell nucleus to bind to oriP. However, when V5-EBNA1c was cotransfected with Flag-BGLF4, not only were pCEP4 sequences amplified from the anti-V5 immunoprecipitates, they were also amplified from the anti-Flag immunoprecipitates (Fig. 5B). Therefore, BGLF4 associates with oriP in the presence of EBNA1 in transfected cells.
To examine this association in EBV-infected cells, a ChIP analysis was next performed using extracts from untreated Akata cells and Akata cells treated with anti-IgG to induce the lytic cycle. The ChIP assay results showed that EBNA1 was associated with the family of repeats (FR) and the dyad symmetry (DS) regions of oriP in both untreated (Fig. 5C, left) and lytically induced (Fig. 5C, right) cells. The continued detection of oriP-bound EBNA1 after lytic induction is consistent with previously published data (12). BGLF4 was not associated with the FR or DS region of oriP prior to lytic induction (Fig. 5C, left) or with the non-replication-associated BBLF1 or BOLF1 region of the EBV genome after lytic induction (Fig. 5C, right). BGLF4 association with the FR and DS regions was detected after lytic induction (Fig. 5C, right).
BGLF4 inhibits oriP replication/maintenance.
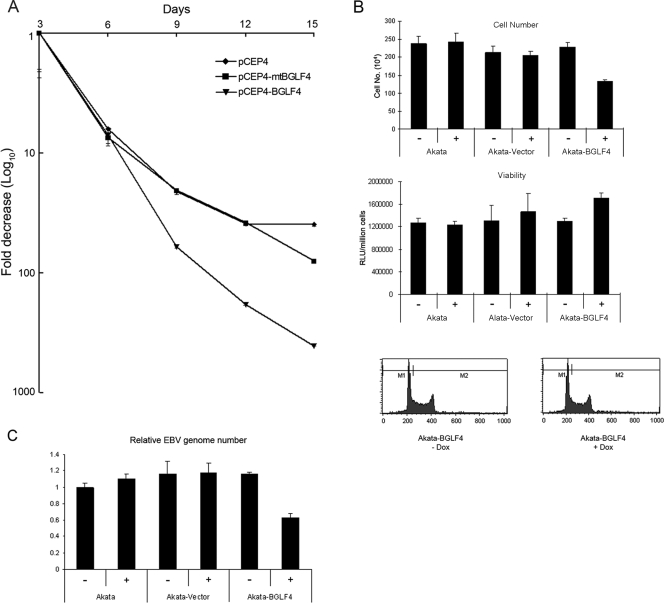
Since EBNA1 still bound DNA in the presence of BGLF4, we sought another EBNA1 activity that could be impacted by BGLF4. The effect of BGLF4 on the episomal DNA copy number was examined using pCEP4, which is replicated episomally in the presence of EBNA1. The amount of pCEP4 DNA in transfected HeLa cells was measured by real-time PCR using cellular GAPDH as the internal reference (Fig. 6A). Transfected oriP-containing plasmids are rapidly lost upon serial passage in the absence of selection (34). The numbers of copies of pCEP4, pCEP4-BGLF4, and pCEP4-mtBGLF4 in the transfected cells decreased at equal rates up to day 6 posttransfection. From day 6 onward, there was a greater loss of pCEP4-BGLF4 plasmids than of pCEP4 or pCEP4-mtBGLF4 plasmids. At day 15, the number of copies of pCEP4 was 13-fold greater than that of pCEP4-BGLF4. This transfection experiment suggested that recruitment of BGLF4 to oriP impaired the functioning of EBNA1 in episomal DNA replication or maintenance.
FIG. 6.
BGLF4 interferes with EBNA1 function. (A) Retention of pCEP4 DNA in HeLa cells transfected with pCEP4, pCEP4-BGLF4, or pCEP4-mBGLF4 and harvested at the indicated times posttransfection. pCEP4 DNA was quantified by real-time PCR using GAPDH as an internal reference. (B) Growth properties of Akata-BGLF4 cells. The cells were grown for 3 days in the presence (+) or absence (−) of doxycycline (Dox). (Top and middle) Live-cell number determined after trypan blue staining (top) or measured using the Cell Titer-Glo luciferase assay (middle). RLU, relative light units. (Bottom) Cell cycle distribution of Akata-BGLF4 cells with (+) and without (−) doxycycline treatment using fluorescence-activated cell sorter analysis of formaldehyde-fixed, Hoechst 33258-stained cells. (C) Real-time PCR analysis of relative EBV genome copy numbers using GAPDH as an internal reference. Akata cells lines were grown for 3 days in the presence (+) or absence (−) of doxycycline.
To further explore the effect of BGLF4 on the episomal EBV genome copy number, we turned to the doxycycline-regulated Akata cell lines. The Akata, Akata-BGLF4, and Akata-vector cells were grown for 3 days with or without doxycycline. Doxycycline treatment of the parental Akata cells or the Akata-vector cells did not affect cell growth or viability (Fig. 6B). Untreated Akata-BGLF4 cells had the same growth rate and viability as Akata-vector cells (Fig. 6B). After doxycycline treatment, the Akata-BGLF4 cells grew 39% more slowly than the treated Akata-vector cells (Fig. 6B, top), but there was no decrease in cell viability and no gross change in cell cycle distribution as determined by fluorescence-activated cell sorter analysis (Fig. 6B, middle and bottom). The EBV genome copy numbers in these cells were examined by real-time PCR. The EBV genome number was normalized relative to cell GAPDH, and the value for the parental Akata cells without doxycycline treatment was set at 1. Akata-vector and Akata-BGLF4 cells showed no difference in EBV genome copy numbers in the absence of doxycycline treatment, and 3 days of doxycycline treatment did not change the genome copy number in the Akata-vector cells (Fig. 6C). However, a 53% reduction in EBV copy numbers was detected in the doxycycline-treated Akata-BGLF4 cells compared to the untreated Akata-BGLF4 cells (Fig. 6C).
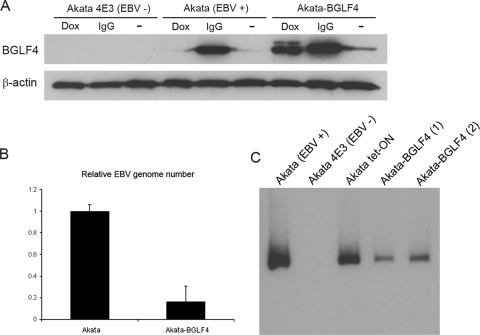
Doxycycline treatment increased BGLF4 expression to approximately 50% of the level seen after immunoglobulin cross-linking of Akata cells (Fig. 7A). However, we also detected low-level breakthrough BGLF4 expression in Akata-BGLF4 cells grown in medium without doxycycline (Fig. 4D and 7A). Akata-BGLF4 cells were passaged continuously in standard medium in the absence of doxycycline for 3 months to determine the effect on the EBV genome content. Real-time PCR analysis using primers to the BALF5 ORF and GAPDH as a reference revealed that the passaged Akata-BGLF4 cells also showed a significant reduction in EBV genome copy number compared to the original Akata cells (Fig. 7B). To examine whether the Akata genomes remaining in the passaged Akata-BGLF4 cells were episomal, DNA from the Akata cell lines was analyzed by Gardella gel electrophoresis and Southern blotting. Equal numbers of cells were lysed in each well of the gel. The released episomal DNA was visualized using a probe to the BamHI-W region of the EBV genome. This analysis revealed that two passaged cultures of Akata-BGLF4 cells contained fewer episomal DNA genomes than either the original Akata cells or the parental Akata Tet-ON cell line (Fig. 7C).
FIG. 7.
Long-term BGLF4 expression causes episomal DNA loss. (A) Western blot comparing BGLF4 expression in Akata 4E3 (EBV −), Akata (EBV +), and Akata-BGLF4 cells with and without treatment with doxycycline (Dox) or anti-human IgG (IgG). (B) Real-time PCR analysis of EBV genome copy numbers in Akata and Akata-BGLF4 cells passed continuously for 3 months in the absence of doxycycline. The error bars indicate standard deviations. (C) Southern blot of episomal EBV DNA separated by Gardella gel electrophoresis (14) and probed with the EBV BamHI-W fragment to detect episomal EBV DNA.
DISCUSSION
In this study, we used a protein microarray approach to identify potential substrates for the EBV-encoded serine/threonine protein kinase BGLF4. An EBV protein chip that covered 73% of the EBV proteome was generated for these experiments. Previous studies had identified EBNA2 (BYRF1) (50) and the polymerase processivity factor (BMRF1) (8, 17) as BGLF4 substrates and TK (BXLF1) as a BGLF4-interacting protein (5). These proteins were also identified in our protein array screen. One of the other described BGLF4 substrates, EBNA-LP (25), was not present on the EBV chip, and the other described substrate, BZLF1 (2), was present but was not detected in our assay. Twenty-one of the 60 (35%) EBV proteins on the array were phosphorylated by BGLF4, and approximately half of these were also substrates for CDK1/cyclin B. The number of EBV proteins phosphorylated by BGLF4 seems surprisingly high. However, a phosphorylation assay performed with BGLF4 on a 5,000-human-protein array identified only 2.1% of the human proteins as in vitro BGLF4 substrates (unpublished data). This differential in the percentages of array proteins phosphorylated suggests that the EBV substrates identified are likely to be biologically relevant. However, these substrates will need further experimental validation in EBV-infected cells.
BGLF4 is expressed during the EBV lytic cycle, and most previous work has focused on the effects of BGLF4 on lytic virus replication. BGLF4 is detected in viral-replication compartments (46), and reduction of BGLF4 expression with short interfering RNA partially inhibited viral-DNA replication (18). Virion production is also enhanced by BGLF4-induced disassembly of the nuclear lamina (33). The protein chip identified two other essential viral replication proteins (13), BBLF4 (DNA helicase) and BALF2 (single-stranded DNA binding protein), as BGLF4 substrates and also identified several ancillary replication proteins, BXLF1 (TK), BORF2 and BARF1 (ribonucleotide reductase subunits), BGLF5 (alkaline exonuclease), and BKRF3 (uracil DNA glycosylase). Knockdown of BGLF4 also results in a reduction in the production of infectious virions (18). Structural proteins identified by the protein chip analysis were the capsid proteins BDLF1 and BORF1 (10), the tegument protein BGLF2, and the envelope glycoproteins gp350, gM, and gN (23, 30, 41). Other BGLF4 substrates included the herpes simplex virus terminase homolog BGRF1/BDRF1 (48) that is required for DNA cleavage and packaging. Further, expression of DNA replication and virion proteins is dependent on the three EBV lytic transactivators, and we identified one of these, Mta (BMLF1 or SM) (20, 40), as a BGLF4 substrate. These data suggest that the BGLF4 kinase has multiple roles in EBV lytic DNA replication and virion production.
During latent infection, the episomal EBV genome is replicated using the plasmid origin of replication and a single EBV-encoded protein, EBNA1, which also associates with cell chromosomes to ensure effective segregation of EBV genomes to daughter cells at the time of cell division (45). The identification of EBNA1 as a BGLF4 substrate and interactor was unexpected, since EBNA1 is a latency protein and BGLF4 is expressed in the lytic cycle. BGLF4 is structurally related to the cellular CDK1 kinase and phosphorylates at CDK1 consensus sites (24, 26, 39). A single consensus CDK1 kinase site (S/T P × K/R, where x is any amino acid) occurs in EBNA1 at aa 393 to 396 (SPPR). EBNA1 proteins commencing at aa 408 or 459, and thus lacking this CDK1 site, were not substrates for BGLF4, and mutation of Ser393 in EBNA1c abolished phosphorylation by both CDK1 and BGLF4.
EBNA1 binds to the family of repeats and to the dyad symmetry sites located in oriP, and DNA binding is essential for EBNA1 to function in oriP replication. ChIP assays provided evidence that BGLF4 was also associated with oriP in transfected cells in the presence of EBNA1 and in EBV-positive Akata cells induced to express BGLF4. Experiments to determine the biological consequences of BGLF4 recruitment showed that in transfected cells the oriP-containing plasmid pCEP4 was lost more rapidly in cells that also expressed BGLF4 and that this effect was dependent upon BGLF4 kinase activity. Second, Akata B cells modified for inducible expression of BGLF4 showed a reduction in the EBV genome copy number per cell after short-term induction of BGLF4 and after continuous passage with low-level BGLF4 expression. This loss is consistent with BGLF4 interference with EBNA1 function in oriP replication or episomal maintenance. EBNA1 is a long-lived protein whose central Gly-Gly-Ala repeat region limits proteasomal degradation (35). Daikoku et al. (12) found by ChIP analyses that EBNA1 remained associated with oriP after lytic induction. The continued presence of oriP-bound EBNA1 in the cell after the switch into the lytic cycle may necessitate a means of repressing EBNA1 replication activity. Latent and lytic DNA replication not only initiate at different origins, but also proceed by different mechanisms, and conversion into the lytic rolling-circle form of DNA replication may require effective termination of competing oriP-initiated replication.
The mechanism of BGLF4 action is not known. Constitutive phosphorylation of EBNA1 at Ser393 may affect EBNA1 recruitment of cellular replication factors, or BGLF4 recruited to oriP may directly phosphorylate cellular replication proteins to modify their functioning, as suggested in the summary model in Fig. 8. BGLF4 has previously been shown to phosphorylate MCM4, a component of the minichromosome maintenance (MCM) complex, and to inactivate the helicase activity of MCM4-6-7 hexamers (28). The MCM complex is part of the licensing machinery that limits reinitiation of DNA replication. EBV lytic replication takes place in cell cycle-arrested cells, and phosphorylation of MCM4 by BGLF4 was proposed to serve as one of the viral mechanisms for inhibition of cellular DNA synthesis. This property may contribute to the slower cell growth seen in our BGLF4-expressing cells. However, components of the licensing machinery also bind to oriP (6, 51), and thus, interference with MCM protein function could have an even greater impact on reinitiation of oriP replication. The discovery that EBNA1 is a BGLF4 substrate reinforces a recurring theme in herpesvirus biology, namely, that the viral latent and lytic cycles are antagonistic in their regulation, with factors that promote the lytic cycle frequently negatively impacting on virus latency and vice versa.
FIG. 8.
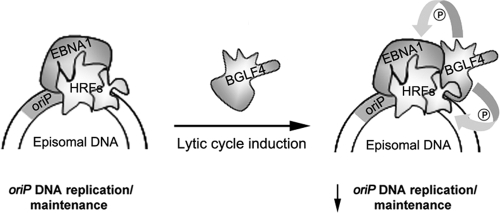
BGLF4 regulation of EBNA1 function. Shown is a model in which BGLF4 is recruited to oriP by EBNA1 after induction of the lytic cycle. BGLF4 constitutively phosphorylates EBNA1 or cell replication proteins, such as MCM4 (28), that are normally transiently phosphorylated by CDK1/cyclin complexes. Constitutive phosphorylation is proposed to interfere with the activities of host replication factors (HRFs) and to downregulate oriP replication.
Supplementary Material
Acknowledgments
We thank Lindsey Hutt-Fletcher for the Akata BX1 BAC used for amplification of EBV ORFs, G. Sanford and J. Nicholas for the tet-converted Akata cell line, and J. Liu and J. H. Ahn for technical advice.
The work was funded by PHS grants R21 AI070740 to H.Z. and P.D., R01 AI033077 to P.D., and R01 CA30356 to S.D.H. and by NIH SPORE grant 1P50 CA96888. J.Z. was supported by American Heart Association Predoctoral Fellowship 0715295U.
J.Z., L.S., and G.L. performed the experiments. J.Z., J.Z., L.S., G.L., G.S.H., and M.-R.C. generated reagents. J.Z., S.D.H., P.D., and H.Z. designed experiments. J.Z., S.D.H., and H.Z. wrote the manuscript.
Footnotes
Published ahead of print on 25 February 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ambinder, R. F., M. A. Mullen, Y. N. Chang, G. S. Hayward, and S. D. Hayward. 1991. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J. Virol. 651466-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, R., A. Kato, K. Kato, M. Kanamori-Koyama, K. Sugimoto, T. Sairenji, Y. Nishiyama, and Y. Kawaguchi. 2006. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J. Virol. 805125-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biron, K. K., R. J. Harvey, S. C. Chamberlain, S. S. Good, A. A. Smith III, M. G. Davis, C. L. Talarico, W. H. Miller, R. Ferris, R. E. Dornsife, S. C. Stanat, J. C. Drach, L. B. Townsend, and G. W. Koszalka. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 462365-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornkamm, G. W., U. Behrends, and J. Mautner. 2006. The infectious kiss: newly infected B cells deliver Epstein-Barr virus to epithelial cells. Proc. Natl. Acad. Sci. USA 1037201-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderwood, M. A., K. Venkatesan, L. Xing, M. R. Chase, A. Vazquez, A. M. Holthaus, A. E. Ewence, N. Li, T. Hirozane-Kishikawa, D. E. Hill, M. Vidal, E. Kieff, and E. Johannsen. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. USA 1047606-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 9810085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. S., E. Korobkova, H. Chen, J. Zhu, X. Jian, S. C. Tao, C. He, and H. Zhu. 2008. A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat. Methods 569-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, M. R., S. J. Chang, H. Huang, and J. Y. Chen. 2000. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J. Virol. 743093-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M. R., J. M. Middeldorp, and S. D. Hayward. 1993. Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J. Virol. 674875-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu, Y. F., C. P. Tung, Y. H. Lee, W. H. Wang, C. Li, J. Y. Hung, C. Y. Wang, Y. Kawaguchi, and S. T. Liu. 2007. A comprehensive library of mutations of Epstein Barr virus. J. Gen. Virol. 882463-2472. [DOI] [PubMed] [Google Scholar]
- 11.Crawford, D. H., and I. Ando. 1986. EB virus induction is associated with B-cell maturation. Immunology 59405-409. [PMC free article] [PubMed] [Google Scholar]
- 12.Daikoku, T., A. Kudoh, M. Fujita, Y. Sugaya, H. Isomura, and T. Tsurumi. 2004. In vivo dynamics of EBNA1-oriP interaction during latent and lytic replication of Epstein-Barr virus. J. Biol. Chem. 27954817-54825. [DOI] [PubMed] [Google Scholar]
- 13.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 665030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardella, T., P. Medveczky, T. Sairenji, and C. Mulder. 1984. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 50248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gershburg, E., M. Marschall, K. Hong, and J. S. Pagano. 2004. Expression and localization of the Epstein-Barr virus-encoded protein kinase. J. Virol. 7812140-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershburg, E., and J. S. Pagano. 2007. Conserved herpesvirus protein kinases. Biochim. Biophys. Acta 17842003-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gershburg, E., and J. S. Pagano. 2002. Phosphorylation of the Epstein-Barr virus (EBV) DNA polymerase processivity factor EA-D by the EBV-encoded protein kinase and effects of the l-riboside benzimidazole 1263W94. J. Virol. 76998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gershburg, E., S. Raffa, M. R. Torrisi, and J. S. Pagano. 2007. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J. Virol. 815407-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta, R., B. Kus, C. Fladd, J. Wasmuth, R. Tonikian, S. Sidhu, N. J. Krogan, J. Parkinson, and D. Rotin. 2007. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol. Syst. Biol. 3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, Z., E. Marendy, Y. D. Wang, J. Yuan, J. T. Sample, and S. Swaminathan. 2007. Multiple roles of Epstein-Barr virus SM protein in lytic replication. J. Virol. 814058-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hume, A. J., J. S. Finkel, J. P. Kamil, D. M. Coen, M. R. Culbertson, and R. F. Kalejta. 2008. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science 320797-799. [DOI] [PubMed] [Google Scholar]
- 22.Hutt-Fletcher, L. M. 2007. Epstein-Barr virus entry. J. Virol. 817825-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 10116286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato, K., Y. Kawaguchi, M. Tanaka, M. Igarashi, A. Yokoyama, G. Matsuda, M. Kanamori, K. Nakajima, Y. Nishimura, M. Shimojima, H. T. Phung, E. Takahashi, and K. Hirai. 2001. Epstein-Barr virus-encoded protein kinase BGLF4 mediates hyperphosphorylation of cellular elongation factor 1δ (EF-1δ): EF-1δ is universally modified by conserved protein kinases of herpesviruses in mammalian cells. J. Gen. Virol. 821457-1463. [DOI] [PubMed] [Google Scholar]
- 25.Kato, K., A. Yokoyama, Y. Tohya, H. Akashi, Y. Nishiyama, and Y. Kawaguchi. 2003. Identification of protein kinases responsible for phosphorylation of Epstein-Barr virus nuclear antigen leader protein at serine-35, which regulates its coactivator function. J. Gen. Virol. 843381-3392. [DOI] [PubMed] [Google Scholar]
- 26.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1δ. J. Virol. 772359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koszalka, G. W., N. W. Johnson, S. S. Good, L. Boyd, S. C. Chamberlain, L. B. Townsend, J. C. Drach, and K. K. Biron. 2002. Preclinical and toxicology studies of 1263W94, a potent and selective inhibitor of human cytomegalovirus replication. Antimicrob. Agents Chemother. 462373-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudoh, A., T. Daikoku, Y. Ishimi, Y. Kawaguchi, N. Shirata, S. Iwahori, H. Isomura, and T. Tsurumi. 2006. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4-MCM6-MCM7 complex during Epstein-Barr virus productive replication. J. Virol. 8010064-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laichalk, L. L., and D. A. Thorley-Lawson. 2005. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 791296-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lake, C. M., S. J. Molesworth, and L. M. Hutt-Fletcher. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 725559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lalezari, J. P., J. A. Aberg, L. H. Wang, M. B. Wire, R. Miner, W. Snowden, C. L. Talarico, S. Shaw, M. A. Jacobson, and W. L. Drew. 2002. Phase I dose escalation trial evaluating the pharmacokinetics, anti-human cytomegalovirus (HCMV) activity, and safety of 1263W94 in human immunodeficiency virus-infected men with asymptomatic HCMV shedding. Antimicrob. Agents Chemother. 462969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, C. P., J. Y. Chen, J. T. Wang, K. Kimura, A. Takemoto, C. C. Lu, and M. R. Chen. 2007. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J. Virol. 815166-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, C. P., Y. H. Huang, S. F. Lin, Y. Chang, Y. H. Chang, K. Takada, and M. R. Chen. 2008. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J. Virol. 8211913-11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leight, E. R., and B. Sugden. 2001. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol. Cell. Biol. 214149-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levitskaya, J., A. Sharipo, A. Leonchiks, A. Ciechanover, and M. G. Masucci. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. USA 9412616-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, J. Y., Y. Y. Lin, J. Qian, S. C. Tao, J. Zhu, C. Pickart, and H. Zhu. 2008. Functional Dissection of a HECT ubiquitin E3 ligase. Mol. Cell Proteomics 735-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel, D., and T. Mertens. 2004. The UL97 protein kinase of human cytomegalovirus and homologues in other herpesviruses: impact on virus and host. Biochim. Biophys. Acta 1697169-180. [DOI] [PubMed] [Google Scholar]
- 38.Ptacek, J., G. Devgan, G. Michaud, H. Zhu, X. Zhu, J. Fasolo, H. Guo, G. Jona, A. Breitkreutz, R. Sopko, R. R. McCartney, M. C. Schmidt, N. Rachidi, S. J. Lee, A. S. Mah, L. Meng, M. J. Stark, D. F. Stern, C. De Virgilio, M. Tyers, B. Andrews, M. Gerstein, B. Schweitzer, P. F. Predki, and M. Snyder. 2005. Global analysis of protein phosphorylation in yeast. Nature 438679-684. [DOI] [PubMed] [Google Scholar]
- 39.Romaker, D., V. Schregel, K. Maurer, S. Auerochs, A. Marzi, H. Sticht, and M. Marschall. 2006. Analysis of the structure-activity relationship of four herpesviral UL97 subfamily protein kinases reveals partial but not full functional conservation. J. Med. Chem. 497044-7053. [DOI] [PubMed] [Google Scholar]
- 40.Semmes, O. J., L. Chen, R. T. Sarisky, Z. Gao, L. Zhong, and S. D. Hayward. 1998. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J. Virol. 729526-9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanner, J., J. Weis, D. Fearon, Y. Whang, and E. Kieff. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50203-213. [DOI] [PubMed] [Google Scholar]
- 42.Tarakanova, V. L., V. Leung-Pineda, S. Hwang, C. W. Yang, K. Matatall, M. Basson, R. Sun, H. Piwnica-Worms, B. P. Sleckman, and H. W. Virgin. 2007. Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 175-82. [DOI] [PubMed] [Google Scholar]
- 44.Tsurumi, T., M. Fujita, and A. Kudoh. 2005. Latent and lytic Epstein-Barr virus replication strategies. Rev. Med. Virol. 153-15. [DOI] [PubMed] [Google Scholar]
- 45.Wang, J., and B. Sugden. 2005. Origins of bidirectional replication of Epstein-Barr virus: models for understanding mammalian origins of DNA synthesis. J. Cell Biochem. 94247-256. [DOI] [PubMed] [Google Scholar]
- 46.Wang, J. T., P. W. Yang, C. P. Lee, C. H. Han, C. H. Tsai, and M. R. Chen. 2005. Detection of Epstein-Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J. Gen. Virol. 863215-3225. [DOI] [PubMed] [Google Scholar]
- 47.Wang, L. H., R. W. Peck, Y. Yin, J. Allanson, R. Wiggs, and M. B. Wire. 2003. Phase I safety and pharmacokinetic trials of 1263W94, a novel oral anti-human cytomegalovirus agent, in healthy and human immunodeficiency virus-infected subjects. Antimicrob. Agents Chemother. 471334-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, K., A. P. Poon, B. Roizman, and J. D. Baines. 2008. Temperature-sensitive mutations in the putative herpes simplex virus type 1 terminase subunits pUL15 and pUL33 preclude viral DNA cleavage/packaging and interaction with pUL28 at the nonpermissive temperature. J. Virol. 82487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4757-768. [DOI] [PubMed] [Google Scholar]
- 50.Yue, W., E. Gershburg, and J. S. Pagano. 2005. Hyperphosphorylation of EBNA2 by Epstein-Barr virus protein kinase suppresses transactivation of the LMP1 promoter. J. Virol. 795880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, J., C. M. Chau, Z. Deng, R. Shiekhattar, M. P. Spindler, A. Schepers, and P. M. Lieberman. 2005. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 241406-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, H., J. F. Klemic, S. Chang, P. Bertone, A. Casamayor, K. G. Klemic, D. Smith, M. Gerstein, M. A. Reed, and M. Snyder. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26283-289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.