Abstract
The Epstein-Barr virus (EBV) alkaline exonuclease BGLF5 has previously been recognized to contribute to immune evasion by downregulating production of HLA molecules during virus replication. We have constructed a BGLF5-null virus mutant to determine BGLF5's functions during EBV viral replication. Quantification of virus production in permissive 293 cells carrying a ΔBGLF5 genome identified a 17- to 21-fold reduction relative to complemented or wild-type controls. Detailed monitoring of ΔBGLF5 replication evidenced an impaired virus nucleocapsid maturation, a reduced primary egress and a 1.4-fold reduction in total viral DNA synthesis. ΔBGLF5 single-unit-length viral genomes were not only less abundant but also migrated faster than expected in gel electrophoresis. We concluded that BGLF5 pertained both to the generation and to the processing of viral linear genomes. ΔBGLF5 phenotypic traits were reminiscent of those previously identified in a mutant devoid of UL12, BGLF5's homolog in herpes simplex virus type 1, and indeed UL12 was found to partially complement the ΔBGLF5 phenotype. However, BGLF5-specific functions could also be identified; the nuclear membrane of replicating cells displayed images of reduplication and complex folding that could be completely corrected by BGLF5 but not UL12. Similar nuclear abnormalities were previously observed in cells transfected with BFLF2 and BFRF1, two viral proteins crucial for EBV nuclear egress. Interestingly, ΔBGLF5 cells produced more BFLF2 than wild-type or complemented counterparts. The present study provides an overview of BGLF5's functions that will guide future molecular studies. We anticipate that the 293/ΔBGLF5 cell line will be instrumental in such developments.
The Epstein-Barr virus (EBV) is a predominantly B lymphotropic member of the gammaherpesvirus subfamily whose host spectrum is physiologically restricted to humans (35). EBV possesses a large genome that encodes for more than 100 genes, the majority of which are required for efficient virus replication and propagation. Although strictly dependent on its host cells for replication, EBV encodes several proteins endowed with enzymatic activities. Some enzymes, such as the viral DNA polymerase (Pol) BALF5, are directly involved in virus construction, but others interact with the cellular host (21). One example is provided by viral proteins, first identified in the alphaherpesviruses, that serve a host shutoff function (HSO), i.e., act as negative regulators of cellular protein production to the benefit of the virus. In herpes simplex virus (HSV), HSO leads to preferential synthesis of viral proteins and to downregulation of cellular proteins crucial for immune response against the virus (for a review, see reference 14). Previous genetic and biochemical studies have identified the UL41 gene product vhs (for viral host shutoff) as one essential mediator of HSV-1-induced HSO (24, 30); vhs is thought to curb cellular protein production through its RNase activity (6, 23, 41, 45). More recent work has shown that this function extends to the two human gammaherpesviruses EBV and Kaposi's sarcoma-associated virus (12, 13, 38). However, the molecular mechanisms that underlie HSO in these viruses appear to be distinct from those at work in HSV, since EBV and Kaposi's sarcoma-associated virus do not have vhs homologs. HSO seems to have been taken over, at least in part, by the alkaline exonucleases BGLF5 and SOX (shutoff and exonuclease), respectively. SOX enhances mRNA turnover without affecting de novo gene transcription: it reduces both a green fluorescent protein (GFP) reporter mRNA and actin half-lives and small interfering RNAs against SOX are able to prevent HSO (13). Although both SOX and BGLF5 display DNase activities in vitro as exonucleases (2, 13, 42, 46), direct evidence that SOX and/or BGLF5 have intrinsic RNase activities is thus far lacking. Therefore, the precise molecular mechanisms that lead to the increased mRNA turnover are still unknown. However, introduction of mutations in either SOX or BGLF5 by random PCR has allowed identification of mutants that either retain the DNase or the HSO functions, demonstrating that both functions are distinct (11, 47). Further evidence for such a dichotomy emerged from the observation that SOX and BGLF5 are located both in the nucleus and the cytoplasm but that SOX enhances mRNA degradation exclusively in the cytoplasm (11).
BGLF5's physiological contribution to viral replication has thus far been attributed to its ability to facilitate immune evasion (38, 47). BGLF5-mediated HSO leads to a block in HLA class I and II protein production during lytic replication in EBV-positive Akata cells; neither mature nor immature class I molecules can be recovered from them. The shutoff effect also negatively affects the β2 microglobulin, HLA-DRα, and TAP1 genes whose RNA levels were found to be substantially reduced by the viral exonuclease (38). 293 cells transfected with a BGLF5 expression plasmid similarly displayed a reduction in HLA class I levels and the viral exonuclease was found to inhibit T-cell recognition of HLA-A2-positive 293 cells cotransfected with the EBV protein SM and GFP (38, 47).
Whether BGLF5 directly contributes to viral DNA replication and virion assembly is unknown. To address this question, we constructed a viral EBV mutant that lacks the BGLF5 gene and report here its general phenotypic traits.
MATERIALS AND METHODS
Primary cells and cell lines.
HEK293 is a neuroendocrine cell line obtained by transformation of embryonic epithelial kidney cells with adenovirus (17, 40). Raji is a human EBV-positive Burkitt's lymphoma cell line (29). WI38 are primary human lung embryonic fibroblasts. Mononuclear cells were purified from fresh blood buffy coats by density gradient centrifugation. CD19 positive B cells were isolated from total lymphocytes using M-450 CD19 (PanB) magnetic beads (Dynal). All cell lines were routinely grown in RPMI 1640 medium supplemented with 10% fetal calf serum (Biochrom).
Recombinant plasmids.
A BZLF1 expression plasmid (p509) was used to initiate the viral lytic cycle (37). The entire BGLF5 open reading frame (ORF; B95.8 coordinates 120930 to 122341) was PCR amplified and cloned into the EcoRI-HindII site of pCDNA3.1(+). Sequencing confirmed the integrity of the BGLF5 sequence. The UL12 gene was PCR amplified from a HSV-1 genome cloned onto a bacterial artificial chromosome (22). The UL12 ORF fused to a 3′ hemagglutinin (HA) tag was cloned into the EcoRI/HindIII sites of pCDNA3.1(+). Sequencing confirmed the identity of the UL12 ORF with published sequences.
Recombinant EBV genomes.
The wild-type EBV recombinant plasmid (p2089) used in the present study consists of a B95.8 genome cloned onto the prokaryotic F factor origin of replication. This recombinant virus carries the green fluorescent protein (GFP), chloramphenicol resistance, and hygromycin resistance genes (5). The EBV BGLF5-negative mutant was constructed by replacing parts of the BGLF5 gene (B95.8 coordinates 121348 to 122304) with the kanamycin resistance gene by homologous recombination (27). To this aim, composite primers were used whose internal parts (24 bp; underlined below) are specific for the kanamycin resistance gene, and whose external parts (40 bp) are specific for the BGLF5 gene. To ensure termination of the kanamycin resistance gene, additional stop codons were introduced between the kanamycin resistance and the BGLF5-specific sequence. These primers (BGLF5-up [5′-TTGACTGGGACCCGGTCTTTAATACCAATGCGCCCCGCATTTACTCAGCTAAACAGCTATGACCATGATTACGCC-3′] and BGLF5-down [5′-CAGATGGCCGACGTGGATGAGCTCGAGGATCCCATGGAGGCCAGTCACGACGTTGTAAAACGAC-3′]) allowed PCR-mediated amplification of the kanamycin resistance gene through their internal sequences and then homologous recombination of the amplified PCR product with the EBV wild-type genome via their external sequences. PCR amplification products were incubated with the restriction enzyme DpnI to remove traces of the parental plasmid and introduced by electroporation (1,000 V, 25 μF, 100 Ω) into Escherichia coli DH10B cells carrying the recombinant virus p2089 and the temperature-sensitive pKD46 helper plasmid that encodes the phage lambda red recombinase to foster homologous recombination. Cells were grown in Luria broth (LB) with cam (15 μg/ml) at 37°C for an hour and then plated onto LB agar plates containing chloramphenicol (15 μg/ml) and kanamycin (10 μg/ml). Incubation at 42°C led to a progressive loss of the helper plasmid. After double selection, DNA of positive clones was purified and analyzed with the EcoRI restriction enzyme to confirm correct recombination. Homologous recombination via the flanking regions resulted in the replacement of parts of the BGLF5 gene with the kanamycin resistance cassette and introduction of an additional EcoRI restriction site.
Stable clone selection.
Recombinant EBV plasmid DNA was transfected into 293 cells using the Lipofectamine reagent (Invitrogen) as described previously (20). At 1 day posttransfection, the cells were transferred to a 150-mm cell culture dish, and hygromycin (100 μg/ml) was added to the culture medium for selection of stable 293 clones carrying the EBV recombinant plasmid. Fifteen outgrowing GFP-positive colonies were expanded for further investigation. The BGLF5 knockout cell clone used in the present study is referred to as 293/ΔBGLF5.
Plasmid rescue in E. coli.
Circular plasmid DNA from 293/ΔBGLF5 cells was extracted by using a denaturation-renaturation method as described previously (19). E. coli strain DH10B was transformed with the viral recombinant DNA by electroporation as described before (27), and clones were selected on LB plates containing chloramphenicol (15 μg/ml). Single bacterial colonies were expanded, and DNA plasmid preparations were digested with EcoRI and BamHI restriction enzymes.
Virus induction and infection of target cells.
Producer cell clones 293/EBV-wt (carrying p2089) and 293/ΔBGLF5 were transfected with a BZLF1 expression plasmid (0.5 μg/well) to induce the lytic cycle using lipid micelles (Metafectene; Biontex). In transcomplementation assays, 293/ΔBGLF5 were cotransfected with a BGLF5 or UL12 expression plasmid (1 μg/well). At 2 days posttransfection, cells were harvested for analysis of gene expression. Virus supernatants were harvested 4 days posttransfection, filtered through a 0.8-μm-pore-size filter, and stored at 4°C. Virus titers were determined by infecting 104 Raji cells with increasing dilutions of supernatants. At 3 days after infection, GFP-positive cells were counted by using a fluorescence microscope. For immortalization experiments, primary B cells were infected at a multiplicity of infection (MOI) of 10 genome equivalents per cell (as calculated by quantitative real-time PCR [qPCR]) with infectious supernatants and seeded into U-bottom 96-well microtiter plates coated with 103 gamma-irradiated WI38 feeder cells at a concentration of 102 cells per well. Wells containing outgrowing LCL clones were visually counted. 293 cells (5 × 103) were infected with 150 μl of undiluted supernatants from various virus producer cell lines. The number of GFP-positive cells was assessed 3 days after infection.
Immunostaining.
Mouse monoclonal antibodies against the following viral proteins were used for staining: gp350/220 (clone 72A1), BFRF1 (clone E10), BFLF2 (clone C1). We further used a rabbit polyclonal antibody against BGLF5 (3). Cells were washed three times in phosphate-buffered saline (PBS) and air-dried onto glass slides. Fixation was carried out in pure acetone (for gp350) for 20 min at room temperature or in 4% paraformaldehyde also for 20 min at room temperature followed by a 2-min treatment with 0.1% Triton X-100 to permeabilize cells (for BFRF1, BFLF2, and BGLF5). The slides were incubated with the respective primary antibodies for 30 min, washed three times in PBS, and incubated with a secondary antibody conjugated with Cy3 fluorochrome (Dianova). Cells were then washed three times in PBS and embedded in 90% glycerol. Counterstaining of the cell nuclei was obtained by incubation with Hoechst 33258 (1:10,000) before embedding. Immunofluorescence was evaluated and recorded by using a confocal fluorescence microscope (Nikon A1R).
Electron microscopy.
Lytically induced producer cells or virus pellet obtained after centrifugation of 5 ml of virus supernatant for 2 h at 30,000 × g were fixed with 2.5% glutaraldehyde, and further preparation was carried out as described previously (8). Ultrathin sections were examined by electron microscopy (Zeiss).
Quantitative real-time PCR.
Genomic DNA was extracted from induced cells by using a DNeasy tissue kit (Qiagen). Detection of viral DNA and calculation of virus titers was carried out by qPCR as previously described (8). Supernatants were first digested with DNase I (5 U/50 μl of supernatant) at 37°C for 1 h. After DNase I heat inactivation (10 min at 70°C), supernatants were mixed (1:1 [vol/vol]) with lysis buffer (0.1 mg/ml of proteinase K in water) and incubated for 60 min at 50°C, after which the enzyme was heat-inactivated (75°C for 20 min). Water diluted supernatants (1:10) or genomic DNA (350 pg of input DNA) were subjected to qPCR using primers and probe specific to the BALF5 DNA Pol gene. Amplification reactions were performed in 25-μl volumes containing 12.5 μl of TaqMan Universal master mix (Applied Biosystems), 2.5 μl of forward and reverse Pol primers (2 μM), 1 μl of 5 μM FAM-labeled Pol probe, 1.5 μl of water, and 5 μl of virus supernatant or DNA. After activation of the AmpliTaq Gold DNA Pol (10 min at 95°C), the reaction mixtures were amplified for 40 cycles (15 s at 95°C and 60 s at 60°C), and the fluorescent signals detected using an ABI 7300 sequence detection system (Applied Biosystems). The primer and probe sequences were as follows: reverse Pol primer, 5′-AGTCCTTCTTGGCTAGTCTGTTGAC-3′; forward Pol primer, 5′-CTTTGGCGCGGATCCTC-3′; and EBV Pol probe, 5′-FAM-CATCAAGAAGCTGCTGGCGGCC-TAMRA-3′. The DNA content was calculated by using a serial dilution of Namalwa DNA, a human Burkitt's lymphoma cell line that contains two EBV genome copies per cell, as a reference for the standard curve.
Gardella gel electrophoresis and Southern blot analysis.
Genomic DNA extraction and Gardella gel electrophoresis from induced producer cells, DNA digestion and hybridization were carried out as described previously (5, 20). DNA from cells carrying different viral genomes were digested with the BamHI restriction enzyme, blotted onto a Hybond XL membrane (Amersham), and hybridized with a 32P-labeled DNA fragment specific to EBV gp350 or to EBV terminal repeats (TR).
Western blot analysis.
Cells were resuspended in PBS and lysed by sonication. Then, 20 μg of proteins were denatured in Laemmli buffer for 5 min at 95°C, separated on a 12.5% sodium dodecyl sulfate-polyacrylamide gel, and electroblotted onto a Hybond ECL membrane (Amersham). After preincubation for 30 min in 5% milk powder in PBS, blots were incubated with a rabbit polyclonal serum against BGLF5 (3) or mouse monoclonal antibodies against BFLF2 (clone C1), BFRF1 (clone E10), or actin (clone ACTN05; Dianova) for 1 h at room temperature. After several washings in 0.1% Tween in PBS, blots were incubated for 1 h with goat anti-mouse antibody coupled with horseradish peroxidase (Promega) or protein A coupled with horseradish peroxidase (for BGLF5 detection). HA-tagged UL12 protein was detected using a peroxidase-conjugated mouse monoclonal anti-HA antibody (clone 12CA5; Roche). Antibody binding was revealed by using an ECL detection reagent (Amersham).
RESULTS
Construction of a BGLF5 mutant.
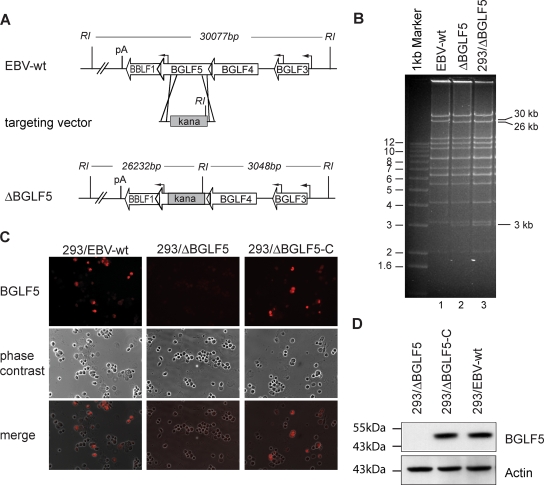
The BGLF5 gene is located in a complex gene locus of the EBV genome. It partly overlaps with the BGLF4 and BBLF1 genes, and all three genes share the same polyadenylation site (1) (Fig. 1A). The mRNA of the BGLF5 gene has been sized to 1.7 kb (4), but its promoter region has not been precisely mapped. To generate a BGLF5 mutant, 956 bp (B95.8 coordinates 121348 to 122304) of the BGLF5 ORF were exchanged against the kanamycin resistance gene sequence by homologous recombination in E. coli, leaving the BGLF4 and BBLF1 ORFs and their respective promoters intact (Fig. 1A). The DNA of the resulting recombinant EBV genomes was analyzed by EcoRI restriction enzyme analysis. As expected from the cloning strategy, this experiment confirmed that the 30-kbp EcoRI fragment that encompasses the BGLF5 gene sequence in the EBV-wt configuration had been exchanged against a 26-kbp fragment and a 3-kbp fragment in the mutant. This pattern alteration results from the introduction of an additional EcoRI site located within the kanamycin resistance gene (Fig. 1B, lanes 1 and 2). With the aim of generating a mutant producer cell line, DNA was prepared from the recombinant clone and transfected into 293 cells. Fifteen of the hygromycin-resistant and GFP-positive cell clones that survived selection were expanded for further analysis. First, we screened these clones for permissivity to lytic replication. At 3 days after transient transfection of the viral transactivator BZLF1, cells were stained for the late viral structural protein gp350. Seven of the tested clones stained positive for this late marker, and we selected the cell clone with the highest percentage of gp350-positive cells (ca. 10%) to perform further experiments (data not shown). This clone is referred to as 293/ΔBGLF5 here. Next, we assessed the integrity of the viral genome present in 293/ΔBGLF5 cells. To this aim, viral episomes were purified from 293/ΔBGLF5 cells and retransformed into E. coli cells. Restriction enzyme analysis of plasmid preparations obtained from these bacterial clones demonstrated that the viral genome in 293/ΔBGLF5 cells was complete (Fig. 1B, lane 3). To confirm the disruption of the BGLF5 function, lytically induced 293/ΔBGLF5 cells were stained with a BGLF5-specific polyclonal antibody. The BGLF5-specific immunostaining was negative in induced 293/ΔBGLF5 cells. In contrast, BGLF5 could be detected 2 days after induction in 293/EBV-wt or 293/ΔBGLF5 cells transcomplemented with a BGLF5 expression plasmid (referred to as 293/ΔBGLF5-C) (Fig. 1C). Furthermore, a BGLF5-specific Western blot analysis performed with extracts from lytically induced 293/EBV-wt, 293/ΔBGLF5, and 293/ΔBGLF5-C cells corroborated the data obtained by immunostaining (Fig. 1D). From these results we inferred that the 293/ΔBGLF5 cells are deficient for BGLF5 expression.
FIG. 1.
Construction of a ΔBGLF5 recombinant virus. (A) Schematic map of the EBV genome segment that encompasses the BGLF5 gene in EBV-wt and after homologous recombination with the targeting vector carrying the kanamycin resistance gene. The overlapping BGLF4 and BBLF1 coding regions are not affected by the recombination. The cleavage sites for EcoRI (RI) and the expected fragment sizes for cleavage of EBV-wt and ΔBGLF5 genomes are given. pA, polyadenylation site, kana, kanamycin. (B) EcoRI restriction fragment analysis of EBV-wt (lane 1) and ΔBGLF5 mutant genomes (lane 2) after construction in E. coli or after rescue from stably transfected 293 cells (293/ΔBGLF5) (lane 3). The result is consistent with the predicted restriction pattern (see panel A). (C) Immunofluorescence analysis of lytically induced 293/ΔBGLF5, 293/EBV-wt and transcomplemented 293/ΔBGLF5 cells (293/ΔBGLF5-C) with a BGLF5-specific antibody. No BGLF5 protein was detected in 293/ΔBGLF5 mutant cells 2 days after induction. (D) Western blot analysis on extracts from induced 293/EBV-wt, 293/ΔBGLF5, and 293/ΔBGLF5-C cells, 48 h after induction. The transcomplemented 293/ΔBGLF5-C and 293/EBV-wt producer lines express BGLF5 at comparable levels, 293/ΔBGLF5 cells were BGLF5 negative. Staining of the same blot with an actin-specific antibody served as a loading control.
Induced 293/ΔBGLF5 cells exhibit reduced virus production.
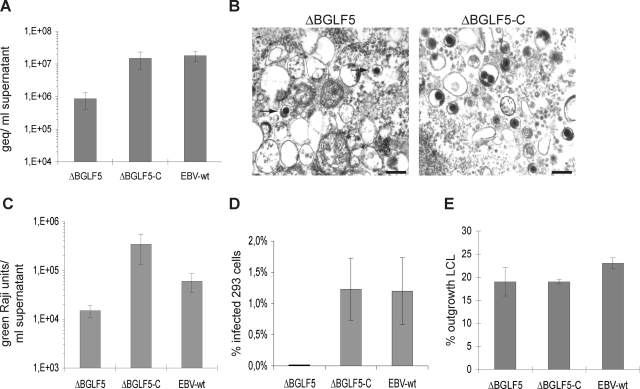
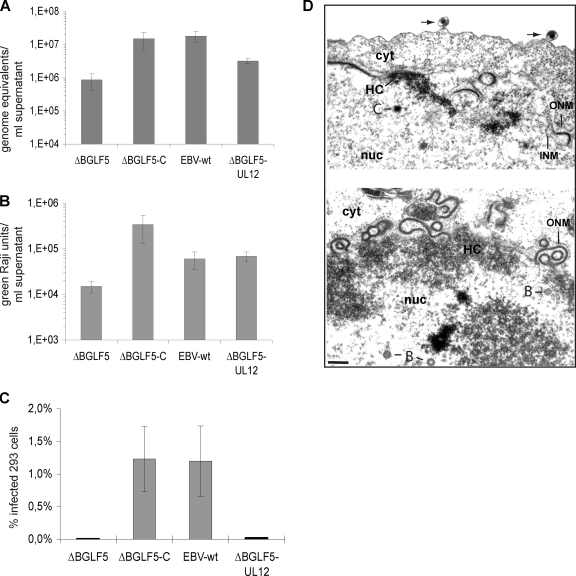
To understand the role of BGLF5 during virus replication, we first quantified viral DNA production in supernatants from induced 293/ΔBGLF5, 293/ΔBGLF5-C and 293/EBV-wt cells by qPCR. Supernatants harvested from 293/ΔBGLF5 cells exhibited a 17- and 21-fold reduction in viral genome equivalents (geq) (mean value from eight experiments, 8.6 × 105 geq/ml) compared to ΔBGLF5-C (mean value from eight experiments, 1.5 × 107 geq/ml) or EBV-wt supernatants (mean value from eight experiments, 1.8 × 107 geq/ml), respectively (Fig. 2A). The hypothesis that 293/ΔBGLF5 cells produced less virus was further validated by direct electron microscopic analysis of pelleted virus supernatants. The number of mature virus particles observed in ΔBGLF5 supernatants was much lower than in EBV-wt or ΔBGLF5-C5 supernatants. The rare ΔBGLF5 viruses that could be found, however, displayed normal morphological features (Fig. 2B). We then assess the ability of the BGLF5 mutant to infect the Raji and HEK293 cell lines. Raji cells were incubated with serial dilutions of supernatants from induced 293/ΔBGLF5 cells, and the number of gfp-positive cells was evaluated 3 days after infection. The infection efficiency of ΔBGLF5 supernatants was reduced by 23-fold relative to ΔBGLF5-C and by 4-fold relative to EBV-wt supernatants (Fig. 2C). HEK293 cell infection with ΔBGLF5 supernatants was 470 times less efficient than with wild-type or complemented supernatants (0.0026% versus 1.2%) (Fig. 2D). Taken together, these results provide solid evidence for a reduced virus production in induced 293/ΔBGLF5 cells. This phenotypic trait could be readily complemented with a BGLF5 expression plasmid. The 293/ΔBGLF5 cells therefore represent a suitable experimental system to assess BGLF5's contribution to EBV replication.
FIG. 2.
BGLF5 is required for virus production but not B-cell immortalization. (A) qPCR analysis with supernatants from lytically induced 293/ΔBGLF5, 293/ΔBGLF5-C and 293/EBV-wt cells. The concentration of viral geq is reduced in ΔBGLF5 supernatants compared to EBV-wt and ΔBGLF5-C supernatants. Presented are the mean values from eight independent experiments. (B) Electron micrographs of pelleted ΔBGLF5 and ΔBGLF5-C virus supernatants. ΔBGLF5 supernatants contained a lower number of mature virions in comparison with ΔBGLF5-C supernatants. Arrows indicate mature virions. Bar, 0.2 μm. (C) Raji B-cell infection assay. The concentration of infectious particles in ΔBGLF5 supernatants (as measured by green Raji cells) is reduced compared to EBV-wt and ΔBGLF5-C supernatants (mean value from eight independent experiments). (D) Multiple infections were carried out with 293 cells as target cells. Mean values of the percentages of infected cells are given. (E) Immortalization experiments with primary B cells from different donors. The results are given as mean values of the percentage of wells with cell outgrowth.
BGLF5 is not required for primary B-cell transformation.
We completed the infection study by exposing primary B cells to ΔBGLF5 and control supernatants at an MOI of 10 viruses per cell as determined by qPCR. The number of outgrowing B-cell clones was taken as a read-out of the ability of the various viruses to infect and transform primary B lymphocytes into lymphoblastoid cells (LCL). Infections with ΔBGLF5 and ΔBGLF5-C viruses were found to yield very similar results (19% outgrowing cell clones); the EBV wild-type positive control was marginally more efficient (23% outgrowing cell clones) (Fig. 2E).
BGLF5 is required for efficient virus maturation and egress.
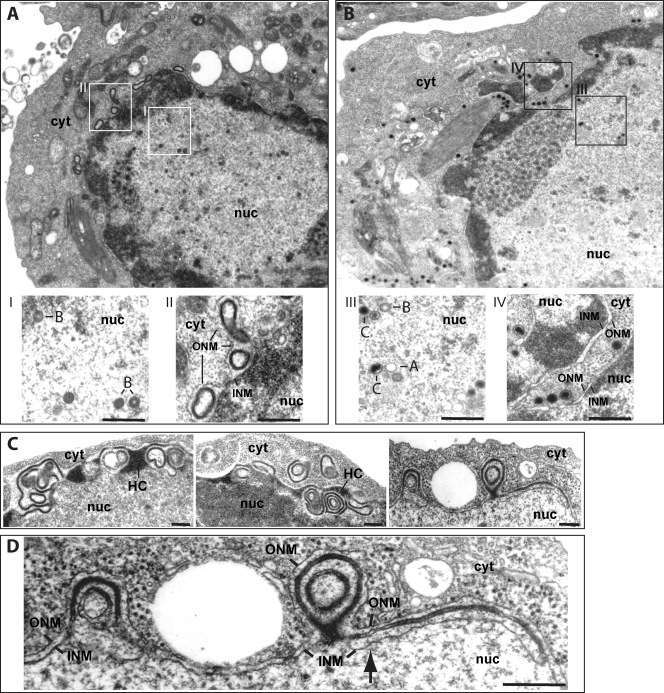
The observation that BGLF5 mutant cells fail to produce infectious virions efficiently indicated that one or more steps of virus maturation were defective in the absence of BGLF5. By examining electron micrographs of induced 293/ΔBGLF5 cells, we obtained a good overview of the replication process, as presented in Fig. 3. While numerous A and B capsids were easily identified (86% of all nucleocapsids), only a reduced number of C capsids with packaged viral DNA (14%) were visible in the nuclei of induced 293/ΔBGLF5 cells (401 A and B capsids and 64 C capsids in 36 replicating cells out of 344 examined cells) (Fig. 3A and inset I). In contrast, all three capsid types were present in normal proportions in the nuclei of induced 293/ΔBGLF5-C cells (67% A and B capsids, 33% C capsids) (Fig. 3B and inset III). Further, virtually no mature capsids in the cytoplasm and no virus particles in the extracellular space could be detected in induced 293/ΔBGLF5 cells; this contrasted with 293/ΔBGLF5-C cells in which mature capsids and released virus particles were easily detectable in the cytoplasm and extracellular milieu (Fig. 3, inset IV, and data not shown). We concluded from these findings that in the absence of BGLF5 the number and proportion of mature capsids was reduced and that nuclear egress was impaired. In addition to the described defect in virus maturation, morphological changes of the nuclear membrane were observed in induced 293/ΔBGLF5 cells (Fig. 3A, inset II, and Fig. 3C and D). There were images of duplication of both the inner and the outer nuclear membrane, which resulted in a stacking of four parallel membranes (Fig. 3C and D). In addition, the perinuclear space between inner and outer nuclear membrane frequently contained concentric invaginations of electron-dense structures that, by places, were seen to be contiguous with heterochromatin. These alterations were not observed in 293/ΔBGLF5-C or EBV-wt cells, where the nuclear envelope exhibited a perfectly regular structure (Fig. 3B, inset IV, and data not shown).
FIG. 3.
Electron micrographs of induced 293/ΔBGLF5 (A, C, and D) and 293/ΔBGLF5-C cells (B). (A) Overview of a replicating 293/ΔBGLF5 cell at low (×8,000) magnification. The nucleus contains numerous immature B capsids with scaffolding structures but only rare mature C-capsids carrying electron-dense DNA (inset I). No viral structures could be detected in the cytoplasm. The nuclear membrane was highly irregular in shape and displayed electron-dense membrane foldings and/or duplications within the perinuclear space resulting in the formation of nuclear pockets, some of which were deeply located in the cytoplasm (inset II). (B) Complementation of 293/ΔBGLF5 cells corrected these abnormalities; all three types of nucleocapsids (inset III) and mature capsids were visible in the cytoplasm, and the nuclear membrane displayed normal morphological features (inset IV). (C and D) Several micrographs provide a more detailed view of the nuclear membrane from induced 293/ΔBGLF5 cells at lower and high magnifications. The nuclear membrane appears thickened and by places connected to heterochromatin (panel C, left and middle images). The cellular material accumulated in the nuclear membrane has a similar density as heterochromatin. The remaining pictures (panel C, right image, and panel D) exemplify what are suspected to be earlier events; the nuclear membrane appears duplicated with four stacked membrane layers visible (arrow). The electron-dense material appears to progressively fill in the space between nuclear membranes to form multiple projections of nuclear envelope that produce an appearance of concentric nuclear pockets on these two-dimensional sections. Note that some nuclear pockets enclose a profile of cytoplasm (panel B, middle image). INM, inner nuclear membrane; ONM, outer nuclear membrane; nuc, nucleus; cyt, cytoplasm; HC, heterochromatin; A, B, and C, A-, B-, and C-type capsids. Bar, 0.2 μm.
Induced 293/ΔBGLF5 cells exhibit mildly reduced viral DNA replication coupled with the generation of abnormal linear genomes.
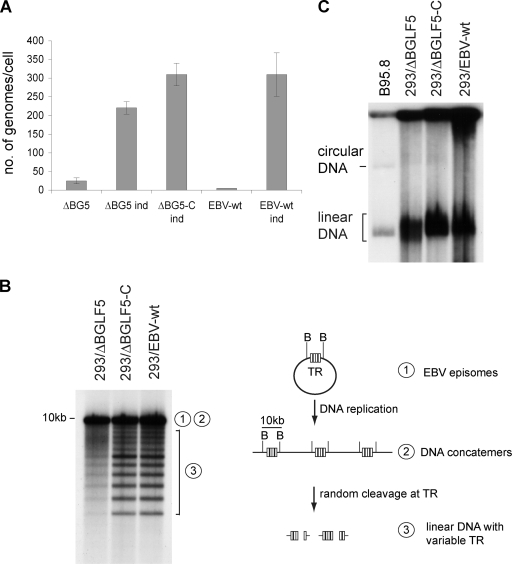
The herpesviral genome is replicated upon induction of the lytic cycle to form highly branched concatemeric DNA molecules that are then resolved into monomeric linear genomes obtained by introduction of a double-strand break between the packaging sequences (also called TR in EBV) that are used for incorporation of the linear genomes into preassembled B capsids (36). To examine whether a defect in viral DNA replication and/or in the packaging of viral DNA into capsids was responsible for the reduced number of packaged capsids seen in the nucleus of induced 293/ΔBGLF5 cells, we first quantified the viral genome numbers in induced cells by qPCR. Lytically induced 293/ΔBGLF5 cells exhibited a 29% reduction in the total number of intracellular viral DNA molecules compared to 293/EBV-wt or 293/ΔBGLF5-C cells (Fig. 4A). The production of newly replicated linear viral DNA was further gauged by Southern blot analysis of BamHI-cleaved DNA with a probe specific to the EBV TR. This technique distinguishes genomes in a concatemeric or circular form, in which all of the TR are located on the same 10-kb BamHI Nhet fragment, from linear genomes in which the TR are parted into two different noncovalent BamHI fragments (Fig. 4B, schematic). Full-length BamHI fragments are therefore indicative of concatemeric or circular genomes, shorter fragments of linear ones. The Southern blot analysis did not find evidence that the synthesis of DNA concatemers was substantially affected by the absence of BGLF5. However, this assay also evidenced a moderate reduction in the production of linear BamHI Nhet fragments from linear genomes in induced 293/ΔBGLF5 cells compared to 293/EBV-wt or 293/ΔBGLF5-C cells (Fig. 4B, left panel). These data prompted us to assess newly replicated linear DNA by Gardella gel analysis, a technique that allows visualization of unit-length circular and linear viral genomes. This test revealed an aberrant migration pattern of the linear ΔBGLF5 genomes extracted from 293/ΔBGLF5 cells; these genomes appeared to migrate faster than recombinant EBV-wt genomes or viral genomes extracted from induced complemented 293/ΔBGLF5 cells, although all three genomes are of the same size. Indeed, nonrecombinant B95.8 genomes that were loaded on the same gel as a control were found to migrate as fast as the recombinant ΔBGLF5 genomes (that are derived from the same B95.8 genome), even though the latter are 11 kb larger than the former.
FIG. 4.
BGLF5's functions in viral lytic DNA replication. (A) Viral DNA replication in uninduced and induced (ind) cells was quantitated by qPCR. Mean values and standard deviations from three independent experiments are presented. (B) Southern blot analysis of BamHI-cleaved DNA fragments from induced cells hybridized with a TR-specific probe. The 10-kb fragment results from restriction of complete BamHI Nhet fragments that are present only in circular genomes or genome concatemers. In contrast, the smaller fragments are generated by restriction of single unit-length linear genomes (see the adjacent schematic). The “1” and “2” represent circular and concatemeric DNA; the “3” corresponds to linear DNA segments. (C) Gardella gel electrophoresis coupled to Southern blot analysis using a nonrepetitive EBV-specific probe. The linear B95.8 genomes are 172 kb, whereas the ΔBGLF5 and EBV-wt genomes are 183 kb.
Taken together, these data demonstrate that BGLF5 significantly contributes to the generation of monomeric linear genomes and to a certain extent to total viral DNA synthesis. The aberrant migration behavior of linear ΔBGLF5 genomes further supports a role for BGLF5 in the processing of linear viral DNA.
BGLF5 influences the expression levels of viral proteins involved in nuclear egress.
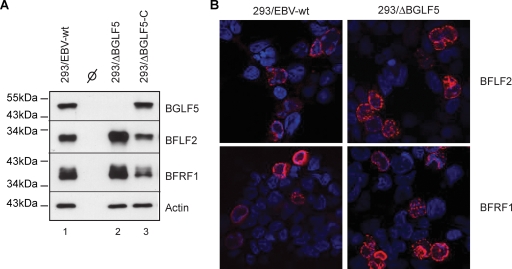
The observation of both an abnormal nuclear envelope and of a marked reduction of virion numbers both in the cytoplasm and the extracellular milieu from induced ΔBGLF5 cells prompted us to assess the status of BFRF1 and BFLF2, two nuclear proteins expressed early during lytic replication, that were previously deemed essential for nuclear egress during EBV maturation (7, 18). Overexpression of both proteins in the absence of the viral genome in HEK293 cells was previously found to result in highly irregular nuclear membrane duplications and convolutions that were not dissimilar from our own observations (16). We therefore performed a Western blot analysis of induced 293/ΔBGLF5 cells using BFRF1- or BFLF2-specific antibodies that revealed an enhanced production of BFLF2 and possibly of BFRF1, compared to 293/EBV-wt cells (Fig. 5A, lanes 1 and 2). Reintroduction of BGLF5 reverted this effect (Fig. 5A, lane 3). We then determined the topological distribution of BFRF1 and BFLF2 in induced 293/ΔBGLF5 cells by analyzing confocal images of immunostained cells, an example of which is illustrated in Fig. 5B. These experiments confirmed the relatively enhanced production of BFLF2; localization of BFRF1 and BFLF2 at the nuclear membrane was, however, not altered in the absence of BGLF5.
FIG. 5.
Expression of BFLF2/BFRF1 nuclear proteins in ΔBGLF5 cells. (A) Western blot analysis of protein extracts from lytically induced 293/EBV-wt (lane 1), 293/ΔBGLF5 (lane 2), and 293/ΔBGLF5-C (lane 3) cells. The same blot was sequentially immunostained with antibodies specific to BFLF2, BFRF1, BGLF5, or actin as a loading control. (B) Induced cells were immunostained with antibodies specific to BFLF2 or BFRF1, and nuclei were counterstained with Hoechst dye (blue). Immunofluorescence was recorded by using a confocal microscope using identical exposure times to analyze both the distribution and the intensity of the fluorescent signals. Localization of both proteins at the nuclear rim is not altered in the absence of BGLF5.
The UL12 HSV-1 protein can partially complement the ΔBGLF5 defective phenotype.
Several phenotypic traits identified in the 293/ΔBGLF5 mutant were congruent with those previously identified in the HSV-1 UL12 mutant, i.e., the moderately reduced viral DNA synthesis, abnormal processing of the linear genomes, and defective packaging and nuclear egress (15, 25, 28, 39). This led us to test the ability of this exonuclease to fulfill the functions of its EBV homologue. To this aim, 293/ΔBGLF5 were induced by cotransfection of BZLF1 and of a HA-tagged UL12 expression plasmid. The adjunction of a HA tag has previously been shown to be neutral for the UL12 functions (31). UL12 protein expression was assessed by Western blot analysis with an HA-specific antibody (data not shown). Virus titers in the UL12-complemented supernatants were fourfold higher than in those from induced 293/ΔBGLF5 cells; they were, however, themselves fourfold lower than those obtained by complementation with BGLF5 (Fig. 6A). In accordance with these data, Raji cell infection experiments revealed a fourfold-higher infection rate after exposure to supernatants from UL12-complemented 293/ΔBGLF5 cells than to supernatants from 293/ΔBGLF5 cells. Complementation with UL12 nevertheless remained fivefold less efficient than complementation with BGLF5 (Fig. 6B). The infection rate of 293 cells was sevenfold enhanced with UL12 complemented supernatants (Fig. 6C). Electron micrographs of induced 293/ΔBGLF5 cells complemented with an UL12 expression plasmid exhibited improved capsid maturation. However, the nuclear membrane remained abnormal in the presence of UL12 (Fig. 6D).
FIG. 6.
Complementation assays with UL12. UL12 was transfected into induced 293/ΔBGLF5 cells. Genome DNA equivalents (A) and green Raji titers (B) were assessed and compared to those obtained with noncomplemented 293/ΔBGLF5 cells, BGLF5-complemented 293/ΔBGLF5 cells, and induced 293/EBV-wt cells (i.e., the same data as presented in Fig. 2). (C) The same experiments were conducted with 293 cells as target cells. (D) Two electron micrographs of induced 293/ΔBGLF5 cells complemented with UL12. Note the irregular nuclear envelope and the presence of C-type nucleocapsids and of extracellular mature virions (arrows). INM, inner nuclear membrane; ONM, outer nuclear membrane; nuc, nucleus; cyt, cytoplasm; HC, heterochromatin; B and C, B- and C-type capsids. Bar, 0.2 μm.
DISCUSSION
Recent interest in the EBV alkaline exonuclease BGLF5 stems from its identification as the HHV8 SOX positional homolog. Reports on BGLF5's functions confirmed the functional homology between SOX and BGLF5 as the latter protein was identified as a host shutoff inducer during EBV lytic replication (38, 47). Here, we more specifically addressed BGLF5's functions during virus replication. Detailed phenotypic analysis of a recombinant BGLF5-null mutant identified a wealth of viral functions targeted by BGLF5. However, despite these multiple defects, a few morphologically intact ΔBGLF5 viruses could be detected in supernatants from induced 293/ΔBGLF5 cells, suggesting that BGLF5 modulates rather than directly fulfills essential viral functions. This establishes a contrast with other replication proteins that were identified as members of the minimal set of factors required for efficient virus replication (10). Viruses lacking proteins from this group are expected to be completely replication deficient as was, for example, demonstrated for BMRF1 (26).
We constructed a BGLF5 mutant by deleting most of the BGLF5 ORF. Western blots or immunostains of 293 cells carrying this mutant genome with antibodies directed against BGLF5 were negative. We can therefore be confident that the 293/ΔBGLF5 has completely lost any full-length or truncated BGLF5 activity. Such an approach runs the risk of inadvertently deleting additional genes or regulatory elements required by adjacent genes. However, we deem this hypothesis unlikely since this type of defect would not have been efficiently transcomplemented with a BGLF5 ORF encoded from an expression vector (Fig. 1). We cannot, however, formally exclude that an as-yet-uncharacterized viral element, such as a small RNA, was inadvertently deleted concomitantly with BGLF5.
Deletion of BGLF5 resulted in a 17- to 21-fold reduction in DNA-equivalent virus titers in supernatants from induced 293/ΔBGLF5 cells relative to 293/ΔBGLF5-C and 293/EBV-wt supernatants, respectively (Fig. 2A). These two control cell lines produced very similar virus titers. It should nevertheless be stressed that different 293/EBV-wt clones will deliver different titers and that there is no absolute reference for a wild-type control. Infection experiments with HEK293 or Raji cells as target cells confirmed the decrease in the concentration of infectious particles present in supernatants from induced 293/ΔBGLF5 cells (Fig. 2D and E). Viruses complemented with BGLF5, however, overshot wild-type virions in their ability to infect Raji cells. The mechanisms that underlie these findings are unclear but the fact that HEK293 cell infection did not reproduce this effect suggests that it does not reflect a general property of the ΔBGLF5 viruses. The Raji cell line carries a replication-defective EBV strain but can support virus production upon superinfection with a wild-type virus (44). It is conceivable, although unproven, that in contrast to what is observed with wild-type viruses, superinfection with ΔBGLF5 viruses did not lead to lytic replication in the Raji cells and therefore inhibited lysis of superinfected cells. In any case, exposure of primary B cells to mutant or control supernatants at the same MOI led to the generation of immortalized B-cell lines with the same efficiency (Fig. 2E). We interpreted this finding as evidence that BGLF5 does not influence B-cell infection. Here again, we noted a mild discrepancy between immortalization efficiencies of the two positive controls: wild-type viruses were slightly more efficient than complemented supernatants. At this stage of the study we cannot rule out a minor effect of BGLF5 at the early stages of primary B-cell infection.
Direct examination of induced 293/ΔBGLF5 cells by electron microscopy (Fig. 3) revealed three major alterations in the viral replication cycle: (i) nucleocapsid maturation was left-shifted, i.e., DNA-filled C-capsid numbers were under-represented; (ii) the ratio of intranuclear capsids to intracytoplasmic virions was abnormally high, suggesting that primary egress was impaired; (iii) the nuclear envelope displayed strikingly abnormal morphological features. These observations prompted us to study lytic DNA replication which was indeed negatively influenced by the absence of BGLF5; total viral DNA synthesis and generation of single unit-length linear genomes were found to be slightly lower in the ΔBGLF5 mutant (Fig. 4). It is currently unclear whether BGLF5 acts on both these processes or whether one defect is a consequence of the other; it is conceivable that a reduction in the generation of individual linear viral genomes negatively influences the DNA replication machinery or vice versa. Interestingly, the reduced pool of linear genomes displayed an aberrant electrophoretic migration pattern, whereas the same genome generated in complemented 293/ΔBGLF5 cells did not exhibit this defect (Fig. 4C). All of these features were previously reported in cells infected by a HSV-1 mutant devoid of the alkaline nuclease UL12 (15, 25, 43). UL12 possesses structural homologies with the lambda phage red alpha protein UL12 and can degrade both single- and double-stranded DNA. UL12 has been shown to act together with ICP8, a single-stranded DNA-binding protein that possesses single-stranded DNA annealing activities, to effect strand exchange between double-stranded DNA molecules and unrelated single-stranded DNA in vitro (32, 34). ICP8 itself seems to increase UL12 processivity and to modulate its exonuclease functions; in the presence of ICP8 UL12 predominantly degrades double-stranded DNA (33). The current view is that the absence of UL12 leads to a lack of resolution of branched structures that are generated during lytic replication which can be evidenced by a diffuse migration pattern of linear genomes in pulsed-field gel electrophoresis (25, 28). Although UL12 lacks the specific structure of a resolvase, it is thought to be required to resolve recombination intermediates which arise during lytic DNA replication. Theses branched structures have been suggested to hamper stable packaging of linear genomes into preassembled capsids. Were these molecular mechanisms to extend to EBV and BGLF5, the combination of reduced linear genome generation with impaired packaging ability would be expected to lead to a substantial reduction in the proportion of mature capsids. However, additional direct effects of BGLF5 on virus packaging cannot currently be ruled out.
The observed impairment in primary egress could itself result from the reduced packaging efficiency. However, there are two reasons to suspect that BGLF5 directly influences primary egress. First, previous analysis of a mutant devoid of the EBV TR (293/ΔTR), that as a consequence cannot be packaged, revealed that DNA-free immature capsids can undergo primary egress and can be found in large amounts in the viral supernatants of induced 293/ΔTR cells (9). Therefore, a defective packaging does not impair primary egress. Second, the nuclear membrane abnormalities are unlikely to result from a deficient virus packaging activity. Such defects were observed neither in 293/ΔTR nor in a ΔUL12 HSV-1 mutant, two mutants that have a reduced ability to produce mature virions (9, 39). The nature of the electron-dense material surrounded by the duplicated bilayered nuclear membrane in induced 293/ΔBGLF5 cells is unknown. Some arguments suggest that it could be heterochromatin; both structures are located beneath the nuclear membrane, both possess the same electron density, and there are areas in which both are in continuity with each other (Fig. 3).
The relationship between the UL12 and the BGLF5-null mutants were further analyzed by conducting complementation experiments of the BGLF5 phenotype with UL12; introduction of the HSV-1 nuclease led to a substantial (fourfold) but not complete virus titer correction (Fig. 6). UL12 was further found to partially reestablish proper capsid maturation; the abnormal nuclear membrane folding were, however, unchanged by the herpes simplex nuclease. This experiment validated the previously suggested functional homology between UL12 and BGLF5 but further suggested that the latter protein possesses a broader spectrum of functions than those thus far identified for UL12.
A first hint of the molecular mechanisms that could underlie the nuclear membrane duplication observed in induced 293/ΔBGLF5 cells came from a previous report in which BFLF2 and BFRF1 were found to induce similar membrane alterations when transfected in EBV-negative 293 cells (16). Interestingly, both immunostains and Western blots with antibodies against BFLF2 indeed revealed an increased production of BFLF2 in induced 293/ΔBGLF5 cells, compared to their wild-type homologs (Fig. 5). Reintroduction of BGLF5 reversed this effect and even led to the production of BFLF2 at levels slightly lower than in wild-type cells, probably reflecting the inadequacy of transfected plasmids to reproduce physiological expression levels. Nevertheless, as shown in Fig. 2A, virus production was perfectly normal in 293/ΔBGLF5-C cells, a finding from which we infer that excess rather than reduction in BFLF2 synthesis levels has deleterious effects on virus production. Analysis of the BFRF1 production pattern led to inconclusive results; BGLF5 might also repress its production but, if it does, the effect is rather modest. What could be the link between BFLF2 and BGLF5? An obvious possibility would be that BGLF5 downregulates BFLF2 through its shutoff function. A similar effect has previously been ascribed to HSV-1 where the Vhs protein helps clearing viral proteins to facilitate transition between the successive waves of viral gene expression (14, 30). However, at the present stage we cannot exclude that the observed effects on BFLF2 result from more complex molecular circuitry that might be mediated by BGLF5 shutoff property or be an additional, as-yet-unidentified function.
In conclusion, the present study has used a genetic approach to identify broad BGLF5 functions during virus replication. A deeper insight in the molecular mechanisms that underlie these functions will require different experimental approaches, but we anticipate that the 293/ΔBGLF5 cells will be instrumental in this regard. A fruitful direction might be to identify BGLF5's functions that depend on its nuclease or its shutoff properties using previously reported BGLF5 mutants that possess only one of these properties (47).
Acknowledgments
We thank Birgit Hub for expert help with the electron micrographs and Charles Knopf for providing the HSV-1 BAC. Confocal microscope images were recorded, processed, and analyzed at the Nikon Imaging Center at the University of Heidelberg. We thank Christian Ackermann and Ulrike Engel for training with this equipment.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tufnell, and B. G. Barell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310207-211. [DOI] [PubMed] [Google Scholar]
- 2.Baylis, S. A., D. J. Purifoy, and E. Littler. 1989. The characterization of the EBV alkaline deoxyribonuclease cloned and expressed in Escherichia coli. Nucleic Acids Res. 177609-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H. F., M. Sauter, P. Haiss, and N. Muller-Lantzsch. 1991. Immunological characterization of the Epstein-Barr virus phosphoprotein PP58 and deoxyribonuclease expressed in the baculovirus expression system. Int. J. Cancer 48879-888. [DOI] [PubMed] [Google Scholar]
- 4.Chen, M. R., T. Y. Hsu, J. Y. Chen, and C. S. Yang. 1990. Molecular characterization of a cDNA clone encoding the Epstein-Barr virus (EBV) DNase. J. Virol. Methods 29127-141. [DOI] [PubMed] [Google Scholar]
- 5.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 958245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everly, D. N., Jr., P. Feng, I. S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 768560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H. J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 793703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feederle, R., B. Neuhierl, G. Baldwin, H. Bannert, B. Hub, J. Mautner, U. Behrends, and H. J. Delecluse. 2006. Epstein-Barr virus BNRF1 protein allows efficient transfer from the endosomal compartment to the nucleus of primary B lymphocytes. J. Virol. 809435-9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feederle, R., C. Shannon-Lowe, G. Baldwin, and H. J. Delecluse. 2005. Defective infectious particles and rare packaged genomes produced by cells carrying terminal-repeat-negative Epstein-Barr virus. J. Virol. 797641-7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-Acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 665030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaunsinger, B., L. Chavez, and D. Ganem. 2005. The exonuclease and host shutoff functions of the SOX protein of Kaposi's sarcoma-associated herpesvirus are genetically separable. J. Virol. 797396-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaunsinger, B., and D. Ganem. 2004. Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral pathogenesis. J. Exp. Med. 200391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaunsinger, B., and D. Ganem. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13713-723. [DOI] [PubMed] [Google Scholar]
- 14.Glaunsinger, B. A., and D. E. Ganem. 2006. Messenger RNA turnover and its regulation in herpesviral infection. Adv. Virus Res. 66337-394. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein, J. N., and S. K. Weller. 1998. In vitro processing of herpes simplex virus type 1 DNA replication intermediates by the viral alkaline nuclease, UL12. J. Virol. 728772-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonnella, R., A. Farina, R. Santarelli, S. Raffa, R. Feederle, R. Bei, M. Granato, A. Modesti, L. Frati, H. J. Delecluse, M. R. Torrisi, A. Angeloni, and A. Faggioni. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 793713-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 3659-74. [DOI] [PubMed] [Google Scholar]
- 18.Granato, M., R. Feederle, A. Farina, R. Gonnella, R. Santarelli, B. Hub, A. Faggioni, and H. J. Delecluse. 2008. Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 824042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, B. E., E. Bjorck, G. Bjursell, and T. Lindahl. 1981. Sequence complexity of circular Epstein-Bar virus DNA in transformed cells. J. Virol. 4011-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 7410142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieff, E., and A. B. Rickinson. 2007. Epstein-Barr virus and its replication, p. 2603-2654. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 22.Knopf, C. W., O. Zavidij, I. Rezuchova, and J. Rajcani. 2008. Evaluation of the T-REx transcription switch for conditional expression and regulation of HSV-1 vectors. Virus Genes 3655-66. [DOI] [PubMed] [Google Scholar]
- 23.Krikorian, C. R., and G. S. Read. 1991. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J. Virol. 65112-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 841926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 702075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuhierl, B., and H. J. Delecluse. 2006. The Epstein-Barr virus BMRF1 gene is essential for lytic virus replication. J. Virol. 805078-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuhierl, B., and H. J. Delecluse. 2005. Molecular genetics of DNA viruses: recombinant virus technology. Methods Mol. Biol. 292353-730. [DOI] [PubMed] [Google Scholar]
- 28.Porter, I. M., and N. D. Stow. 2004. Virus particles produced by the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12 contain abnormal genomes. J. Gen. Virol. 85583-591. [DOI] [PubMed] [Google Scholar]
- 29.Pulvertaft, R. J. V. 1964. Cytology of Burkitt's lymphoma (African lymphoma). Lancet i238-240. [DOI] [PubMed] [Google Scholar]
- 30.Read, G. S., and N. Frenkel. 1983. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate early) viral polypeptides. J. Virol. 46498-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuven, N. B., S. Antoku, and S. K. Weller. 2004. The UL12.5 gene product of herpes simplex virus type 1 exhibits nuclease and strand exchange activities but does not localize to the nucleus. J. Virol. 784599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuven, N. B., A. E. Staire, R. S. Myers, and S. K. Weller. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 777425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reuven, N. B., and S. K. Weller. 2005. Herpes simplex virus type 1 single-strand DNA binding protein ICP8 enhances the nuclease activity of the UL12 alkaline nuclease by increasing its processivity. J. Virol. 799356-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuven, N. B., S. Willcox, J. D. Griffith, and S. K. Weller. 2004. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J. Mol. Biol. 34257-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 36.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses, p. 2501-2601. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 37.Rooney, C. M., D. T. Rowe, T. Ragot, and P. J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 633109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowe, M., B. Glaunsinger, D. van Leeuwen, J. Zuo, D. Sweetman, D. Ganem, J. Middeldorp, E. J. Wiertz, and M. E. Ressing. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. USA 1043366-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao, L., L. M. Rapp, and S. K. Weller. 1993. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196146-162. [DOI] [PubMed] [Google Scholar]
- 40.Shaw, G., S. Morse, M. Ararat, and F. L. Graham. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16869-871. [DOI] [PubMed] [Google Scholar]
- 41.Smiley, J. R., M. M. Elgadi, and H. A. Saffran. 2001. Herpes simplex virus Vhs protein. Methods Enzymol. 342440-451. [DOI] [PubMed] [Google Scholar]
- 42.Stolzenberg, M. C., and T. Ooka. 1990. Purification and properties of Epstein-Barr virus DNase expressed in Escherichia coli. J. Virol. 6496-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weller, S. K., M. R. Seghatoleslami, L. Shao, D. Rowse, and E. P. Carmichael. 1990. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J. Gen. Virol. 712941-2952. [DOI] [PubMed] [Google Scholar]
- 44.Yajima, Y., and M. Nonoyama. 1976. Mechanisms of infection with Epstein-Barr virus. I. Viral DNA replication and formation of noninfectious virus particles in superinfected Raji cells. J. Virol. 19187-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zelus, B. D., R. S. Stewart, and J. Ross. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 702411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, C. X., G. Decaussin, M. de Turenne Tessier, J. Daillie, and T. Ooka. 1987. Identification of an Epstein-Barr virus-specific desoxyribonuclease gene using complementary DNA. Nucleic Acids Res. 152707-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuo, J., W. Thomas, D. van Leeuwen, J. M. Middeldorp, E. J. Wiertz, M. E. Ressing, and M. Rowe. 2008. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J. Virol. 822385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]