Abstract
Here, we report the sequencing and classification of Nyamanini virus (NYMV) and Midway virus (MIDWV), two antigenically related viruses that were first isolated in 1957 and 1966, respectively. Although these viruses have been cultured multiple times from cattle egrets, seabirds, and their ticks, efforts to classify them taxonomically using conventional serological and electron microscopic approaches have failed completely. We used a random shotgun sequencing strategy to define the genomes of NYMV and MIDWV. Contigs of 11,631 and 11,752 nucleotides, representing the complete genome of NYMV and the near-complete genome of MIDWV, respectively, were assembled. Each virus genome was predicted to carry six open reading frames (ORFs). BLAST analysis indicated that only two of the ORF proteins of each virus, the putative nucleocapsid and polymerase, had detectable sequence similarity to known viral proteins. Phylogenetic analysis of these ORF proteins demonstrated that the closest relatives of NYNV and MIDWV are negative-stranded-RNA viruses in the order Mononegavirales. On the basis of their very limited sequence similarity to known viruses, we propose that NYMV and MIDWV define a novel genus, Nyavirus, in this order.
Nyamanini virus (NYMV) was first isolated in 1957 from a cattle egret (Bubulcus ibis) in South Africa (24). It has subsequently been isolated in Nigeria, Egypt, India, and Thailand from cattle egrets and Argas walkerae ticks (14, 16, 24). Although there has been no recognized human infection or disease associated with NYMV, suckling mice succumbed to NYMV infection 7 or 8 days after intracerebral inoculation (14). NYMV has not been definitively characterized or classified to date (10).
Midway virus (MIDWV) was first isolated in 1966 from seabird ticks of two species [Ornithodoros (Alectorobius) spp.] collected on the Midway, Kure, and Manana islands in the Central Pacific and from northern Honshu, Japan. In addition, on Aomatsushima Island, nestling seabirds of two seabird species, Larus crassirostris and Nycticorax nycticorax, were found to have antibody to MIDWV. This virus is pathogenic for newborn Swiss mice but not 4-week-old Swiss mice injected intracranially. Also, the virus is cytopathic in BHK-21 cells and produces plaques in Vero cells. Efforts to classify MIDWV have revealed only that MIDWV is antigenically related to NYMV in cross-box complement fixation (CF) assays (22).
To date, conventional approaches, such as serological analysis and electron microscopy (EM), have not yielded definitive characterization of MIDWV or NYMV. Heretofore, it was not known whether MIDWV and NYMV are DNA viruses or RNA viruses or to which virus family they belong. In this paper, we describe the application of unbiased high-throughput sequencing to define the genome sequences of NYMV and MIDWV. By analysis of their genomes, NYMV and MIDWV were determined to be negative-stranded RNA viruses highly divergent from all known viruses but most closely related to viruses in the order Mononegavirales. On the basis of the analysis presented herein, we propose that NYMV and MIDWV define a novel taxon within the order Mononegavirales.
MATERIALS AND METHODS
Isolation and propagation of viruses.
NYMV (isolate tick 39) that originated from pooled ticks collected in Thailand was propagated in Vero cells. MIDWV (isolate RML47153) was originally isolated from pooled ticks of the species Ornithodoros capensis collected on Midway Island and inoculated into suckling mouse brain and Vero cells. Both viruses were obtained from the World Reference Center for Arboviruses and Emerging Viruses (WRCAEV) at the University of Texas Medical Branch, Galveston.
Transmission EM.
Infected monolayers of BHK-21 cells were fixed 6 days after infection in a mixture of 2.5% formaldehyde, 0.1% glutaraldehyde in 0.05 M cacodylate buffer (pH 7.2) containing 0.03% trinitrophenol and 0.03% CaCl2. The monolayers were washed in 0.1 M cacodylate buffer (pH 7.2), scraped off the plastic, pelleted, and further processed as a pellet. The cells were postfixed in 1% OsO4 in 0.1 M cacodylate buffer. Infected cells were stained en bloc with 1% uranyl acetate in 0.1 M maleate buffer, dehydrated in ethanol, and embedded in Poly/Bed 812 epoxy resin (Polysciences, Warrington, PA). Ultrathin sections were cut on a Reichert-Leica Ultracut S ultramicrotome, stained with 2% aqueous uranyl acetate and lead citrate, and examined with a Philips 201 or CM 100 EM at 60 kV.
CF test.
Suckling mouse brain, sucrose-acetone extracted antigens, and hyperimmune mouse ascitic fluids were prepared for NYMV and MIDWV as described previously (25). CF tests were performed by the microtiter technique (8), using 2 units of guinea pig complement and overnight incubation of the antigen and antibody at 4°C. The CF titers were recorded as the highest dilutions giving CF levels of 3+ or 4+ on a scale of 0 to 4+.
RNA extraction.
Total RNA was extracted from Vero cells mock infected or infected with MIDWV or NYMV by using TRIzol-LS (Invitrogen, Carlsbad, California). The sample was mixed with chloroform (4 volumes chloroform:1 volume original sample) and centrifuged at 12,000 × g for 15 min at room temperature. The aqueous phase was transferred and mixed with an equal volume of isopropyl alcohol and centrifuged at 12,000 × g for 10 min at room temperature. The RNA pellet was washed with 70% ethanol and then dissolved in RNase-free water. Extracted total RNA was quantified using UV spectrophotometry.
Reverse transcription, amplification, and shotgun sequencing.
Random amplification of the extracted RNA was performed as described previously (29). In brief, RNA was reverse transcribed with primer A (5′-GTTTCCCAGTCACGATANNNNNNNNN), and second-strand DNA synthesis was carried out with Sequenase (United States Biochemical, Cleveland, OH). Subsequently, this material was used as the template for 40 cycles of PCR with primer B (5′-GTTTCCCAGTCACGATA) by using the following profile: 30 s at 94°C, 30 s at 40°C, 30 s at 50°C, and 60 s at 72°C. Primer B-amplified nucleic acid was cloned into pCR4-TOPO and sequenced using standard Sanger chemistry with an ABI 3730xl (Applied Biosystems, Foster City, CA) sequencer. The resulting sequence reads were trimmed to remove vector and primer B sequences and then assembled using Phred/Phrap. Contigs with sequence similarity to known viral sequences were identified using tBLASTx (1).
Genome sequencing.
Assembled contigs from the shotgun reads were confirmed by reverse transcription-PCR (RT-PCR), followed by sequencing. Gaps between contigs were closed by designing PCR primers from the existing contigs to span each gap. PCR amplicons were cloned into pCR4 and sequenced. To extend the sequences, 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE were performed. The virus-specific primers used can be found in Table S1 in the supplemental material.
Northern blot analysis of MIDWV.
Five micrograms of total RNA from MIDWV-infected Vero culture was electrophoresed through a 1% agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized under standard conditions to a radiolabeled double-stranded DNA probe generated by PCR amplification of a fragment of the MIDWV L (polymerase) gene, using the primers MIDWV_5F (5′-GTCACGAAAGGAGGGTTGTTGAGCT-3′) and MIDWV_5R (5′-CCCTGAGGAAGATCCTTACTCTCAA-3′).
Gene prediction and analysis.
The open reading frames (ORFs) of NYMV and MIDWV were predicted using the NCBI ORF finder (30). The initial predictions were then refined based on the identification of putative transcription initiation and termination signals (see “Noncoding region analysis” below). For each ORF, signal peptides were predicted using SignalP 3.0 (5). Transmembrane regions were predicted using PolyPhobius (15). Predicted ORFs were analyzed via ProtParam (11) to determine the isoelectric points of their products, NetPhos 2.0 (6) to determine potential phosphorylation sites, and NetOGlyc 3.1 (13) and NetNGlyc 1.0 (R. Gupta, E. Jung, and S. Brunak, unpublished data) to determine potential glycosylation sites.
Noncoding region analysis.
The presence of conserved transcription termination and initiation sequences was determined using MEME (2). To define any potential transcriptional termination motifs, the intervening sequences from each putative stop codon to the first downstream poly-U tract were used as input for MEME. For the terminal ORF VI (L or polymerase) gene, 50 bp of sequence upstream of the stop codon was included in the analysis, since it is known for other viruses in the Mononegavirales that the transcriptional termination signals for the polymerase genes are within the coding region. In order to search for any conserved transcriptional initiation motifs, MEME was used to analyze the following set of sequences from each ORF: (i) 1 nucleotide (nt) downstream of the previous poly-U tract to the 1st in-frame ATG; (ii) the first in-frame ATG to the second in-frame ATG; and (iii) the second in-frame ATG to the third in-frame ATG. Thus, the first three in-frame ATGs were considered candidates for the true ORF start.
Phylogenetic analysis.
Amino acid sequence alignments were performed using ClustalX V1.83 (26) with default parameters. Maximum parsimony phylogenetic trees were constructed from the ClustalX alignments by using PAUP 4.0b with 1,000 bootstrap repetitions, using default settings (21). Maximum likelihood analyses were performed using RAxML with 100 repetitions, using BLOSUM matrices.
Nucleotide sequence accession numbers.
The sequences for MIDWV and NYMV have been deposited in GenBank under accession numbers FJ554525 and FJ554526, respectively.
RESULTS
Observation of NYMV and MIDWV viral particles by EM.
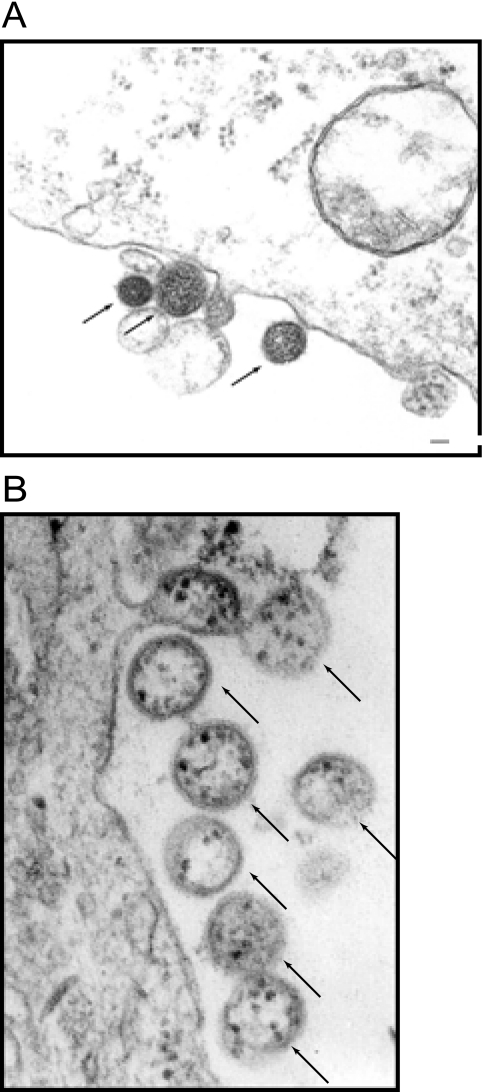
By use of thin-section EM of monolayers of Vero cells infected with NYMV or BHK-21 cells infected with MIDWV, putative virions for NYMV and MIDWV were observed. Three pleomorphic NYMV (Fig. 1A) and multiple pleomorphic MIDWV (Fig. 1B) virions could be seen upon their release, budding from the plasma membrane of infected cells. The virions were spherical and measured 100 to 130 nm in diameter.
FIG. 1.
EM of NYMV and MIDWV. (A) EM of extracellular NYMV. Shown is an ultrathin section of BHK cells infected with NYMV embedded in epoxy resin. Arrows indicate the viral particles. (B) EM of extracellular MIDWV. Shown is an ultrathin section of BHK cells infected with MIDWV embedded in epoxy resin. Arrows indicate the viral particles.
Antigenic relationship between NYMV and MIDWV.
Prior reports indicated that NYMV and MIDWV are serologically related viruses (22). In order to confirm that the isolates of NYMV and MIDWV in this study were antigenically related, cross-box CF tests were performed using NYMV and MIDWV antigens and hyperimmune mouse ascitic fluids (Table 1). The MIDWV antigen reacted both with its homologous mouse ascitic fluid and with NYMV immune ascitic fluid, confirming the antigenic relationship between the two viruses.
TABLE 1.
Results of CF tests showing serologic relationship between NYMV and MIDWV
| Antigen | Resulta for:
|
||
|---|---|---|---|
| NYMV | MIDWV | Controlb | |
| Nyamanini | 512/128 | 32/32 | 0 |
| Midway | 128/512 | 1,024/≥512 | 0 |
| Controlb | 0 | 0 | |
Reciprocal of highest antibody titer/reciprocal of highest antigen titer.
Normal (uninfected) mouse brain and ascitic fluid served as controls.
Identification of NYMV and MIDWV as members of the order Mononegavirales.
Because all previous efforts to classify NYMV and MIDWV by using classical virologic methods had failed, we used a molecular approach to characterize these viruses. Partial sequences of the NYMV and MIDWV genomes were obtained by shotgun sequencing of NYMV and MIDWV viral RNA which had been reverse transcribed and randomly amplified. The sequence reads were assembled into contigs and analyzed using BLAST (1). No similarity to any known virus was detected at the nucleotide level. However, tBLASTx analysis revealed low levels of amino acid identity between the NYMV and MIDWV contigs and the RNA polymerases from a number of viruses in the families Bornaviridae, Filoviridae, and Rhabdoviridae, which are all members of the order Mononegavirales, suggesting that NYMV and MIDWV were likely to be negative-stranded RNA viruses.
The assembled viral contigs were experimentally confirmed by sequencing of RT-PCR products generated by contig-specific primers. The contigs were then joined by RT-PCR with primers designed to span the gaps between the contigs. The contigs were extended first through 3′ RACE reactions targeting the viral mRNAs. Finally, the sequences at the extreme 5′ and 3′ ends of the genome were obtained through a series of 5′ RACE reactions to obtain the noncoding sequence from the viral genome and antigenome. Multiple 5′ RACE reactions for NYMV yielded consensus sequences suggesting that the entire genome of NYMV was 11,631 nt. For MIDWV, 5′RACE targeting the genome strand yielded a consensus sequence for the 5′ untranslated region of MIDWV. Multiple 5′RACE reactions targeting the antigenomic polarity strand of MIDWV gave multiple products containing various deletions. Despite many efforts, we were unable to resolve these discrepancies in the 3′ untranslated region of MIDWV. The 3′ end of the existing contig was defined using the regions of the 5′ RACE products that were in agreement in multiple experiments. The near-complete genome of MIDWV is 11,752 nt.
Experimental confirmation of MIDWV genome size.
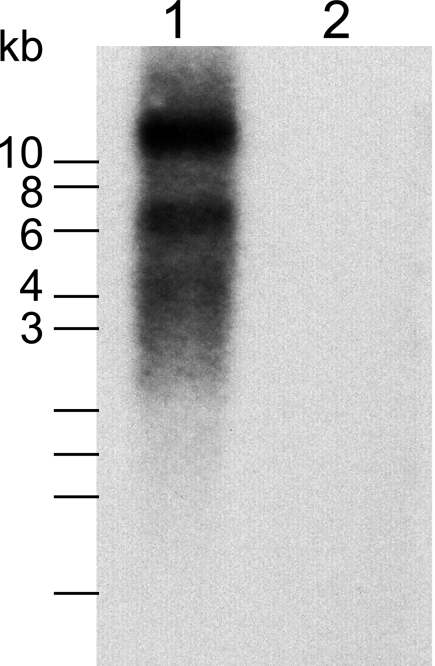
To experimentally confirm the size of the MIDWV genome, we performed Northern blot analysis on total RNA from MIDWV-infected Vero cell culture. The MIDWV probe, which targets a portion of the MIDWV RNA polymerase, yielded two bands of approximately 11 kb and 6 kb (Fig. 2, lane 1). The observation of a band of approximately 11 kb was consistent with the 11,752-nt contig assembled to date, suggesting that we had sequenced the majority of the MIDWV genome. The smaller, 6-kb band was presumed to represent the mRNA for ORF VI.
FIG. 2.
Northern blot hybridization using radiolabeled MIDWV cDNA as a probe. Lane 1, total RNA from Vero cells infected with MIDWV; lane 2, total RNA from uninfected Vero cells. The position of the single-stranded RNA marker segments are indicated.
Analysis of viral ORFs.
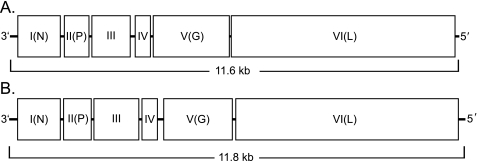
Both NYMV and MIDWV were predicted to contain six ORFs (Fig. 3). All known members of the order Mononegavirales carry at least five ORFs, with a conserved genomic orientation as follows: 3′ leader, nucleoprotein (N) gene, phosphoprotein (P) gene, glycosylated matrix protein (M) gene, transmembrane glycoprotein (G) gene, RNA-dependent RNA polymerase (L) gene, and 5′ trailer. In addition, some members of this order encode additional accessory proteins, such as the nonstructural proteins (NS) found in paramyxoviruses (9, 10). The product of ORF I had detectable amino acid similarity to Borna disease virus nucleocapsid proteins, and that of ORF VI had amino acid similarity to RNA polymerases of various member viruses of the order Mononegavirales (see Fig. 5). While products of the other four ORFs predicted to occur in MIDWV and NYMV did not have any detectable similarity to other viral proteins, there was strong conservation between NYMV and MIDWV in each of these ORF proteins. The NYMV ORF proteins shared at least 46% amino acid identity with the corresponding MIDWV ORF proteins (Table 2). In order to identify potential functional motifs in the internal ORF proteins of MIDWV and NYMV, computational sequence analysis was applied. ORFs II, III, IV, and V were analyzed to determine the isoelectric points, potential phosphorylation sites, presence of a signal peptide, presence of transmembrane domains, and potential glycosylation sites in their products (Table 3).
FIG. 3.
Genome organization of NYMV and MIDWV. Diagrammatic representation of the 3′-to-5′ arrangement of ORFs of the genomes of NYMV (A) and MIDWV (B). The putative ORFs are shown as open rectangles.
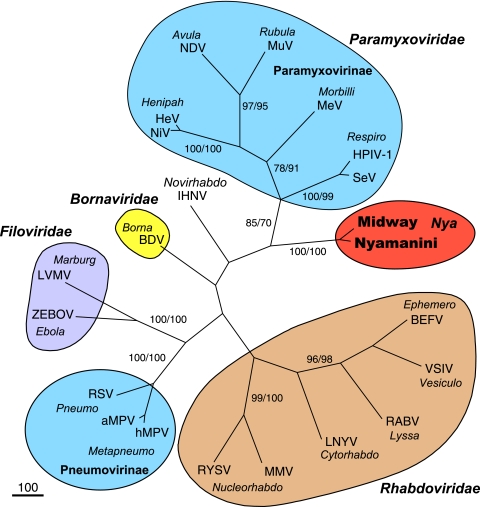
FIG. 5.
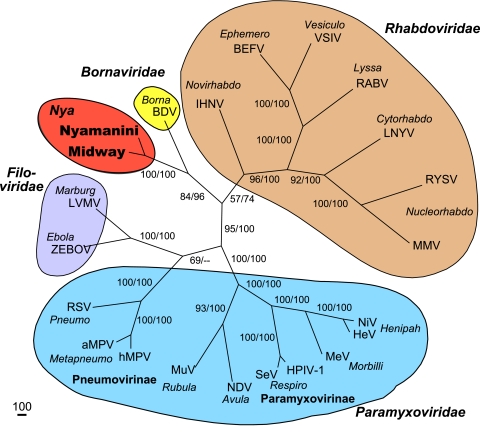
Phylogenetic analysis of the viral polymerase (L) proteins. Phylogenetic trees were constructed with the protein sequences by using the maximum parsimony method. The first and second numbers at the branches show the bootstrap percentage values obtained from maximum parsimony (1,000 repetitions) and maximum likelihood (100 repetitions) analyses, respectively. The type species for each genus in the order Mononegavirales was included in the analysis. Genus names are shown in italics. Abbreviations: BDV, Borna disease virus; ZEBOV, Zaire Ebola virus; LVMV, Lake Victoria marburgvirus; NDV, Newcastle disease virus; HeV, Hendra virus; NiV, Nipah virus; MeV, measles virus; HPIV-1, human parainfluenza 1 virus; SeV, Sendai virus; MuV, mumps virus; aMPV, avian metapneumovirus; hMPV, human metapneumovirus; RSV, respiratory syncytial virus; LNYV, lettuce necrotic yellows virus; BEFV, bovine ephemeral fever virus; VSIV, vesicular stomatitis Indiana virus; RABV, rabies virus; IHNV, infectious hematopoetic necrosis virus; MMV, maize mosaic virus; RYSV, rice yellow stunt virus.
TABLE 2.
Predicted ORFs of NYMV and MIDWVa
| ORF protein | Predicted size (aa)
|
% Identity | |
|---|---|---|---|
| NYMV | MIDWV | ||
| I (N) | 370 | 370 | 80 |
| II (P) | 214 | 230 | 48 |
| III | 383 | 426 | 46 |
| IV | 131 | 132 | 66 |
| V (G) | 658 | 597 | 59 |
| VI (L) | 1,936 | 1,935 | 78 |
The amino acid (aa) sequences of each pair of ORFs were aligned globally using BioEdit (12).
TABLE 3.
Calculated and predicted properties of NYMV and MIDWV ORF II, III, IV, and V proteins
| ORF protein | Molecular weight | pI | No. of phosphorylation sites
|
No. of glycosylation sites
|
|||
|---|---|---|---|---|---|---|---|
| Ser | Thr | Tyr | O-linked | N-linked | |||
| NYMV II (P) | 24.4982 | 4.27 | 7 | 6 | 1 | 8a | 0 |
| NYMV III | 44.5756 | 9.08 | 18 | 4 | 1 | 0 | 0 |
| NYMV IV | 15.1427 | 9.84 | 5 | 3 | 0 | 0 | 0 |
| NYMV V (G) | 74.0621 | 6.57 | 15 | 8 | 9 | 1b | 8b |
| MIDWV II (P) | 25.7313 | 4.20 | 18 | 5 | 1 | 9a | 0 |
| MIDWV III | 49.0652 | 8.30 | 21 | 7 | 2 | 3a | 0 |
| MIDWV IV | 15.3308 | 9.23 | 3 | 1 | 0 | 0 | 0 |
| MIDWV V (G) | 67.3509 | 7.46 | 20 | 8 | 5 | 0 | 6b |
Does not contain a predicted signal peptide, so these sites may not be glycosylated in vivo.
Contains a predicted signal peptide.
The phosphoprotein (P) of mononegaviruses is an acidic protein which is phosphorylated (7). All of the ORF proteins of NYMV and MIDWV were predicted to have multiple potential phosphorylation sites, but only the ORF II proteins were acidic, whereas all other ORF proteins were basic or neutral (Table 2). This, coupled with its position in the genome, suggested that ORF II encodes the viral P protein.
The matrix protein (M) of mononegaviruses is a basic, nonglycosylated membrane protein that ranges in size from 16.2 kDa (for Borna disease virus) to 43.1 kDa (for human parainfluenza 4a virus). Either ORF III or ORF IV of NYMV and MIDWV could potentially encode the matrix protein. They are all basic proteins, but the ORF III proteins are larger than the known M proteins (NYMV, 44.6 kDa; MIDWV, 49.1 kDa), while ORF IV proteins are smaller than the known M proteins (NYMV, 15.1 kDa; MIDWV, 15.3 kDa). None of these ORF proteins were predicted to be glycosylated, as only MIDWV III had any potential glycosylation sites and none of the ORF proteins had the signal peptide necessary for targeting to the N or O glycosylation machinery. No transmembrane region was predicted for either ORF protein, which is consistent with the role of Borna disease virus M as a peripheral membrane-associated protein (17). We cannot conclude at this time whether ORF III or ORF IV encodes the M protein.
The glycoproteins of rhabdoviruses, filoviruses, and bornaviruses are classical type I transmembrane proteins containing a cleavable signal sequence and an exterior N terminus and cytoplasmic C terminus, while the glycoproteins of paramyxoviruses are type II transmembrane proteins with a cytoplasmic N terminus, an exterior C terminus, and a noncleavable signal peptide (7). Using SignalP 3.0 (5) and PolyPhobius (15), we looked for the presence and cleavability of a signal peptide and the presence and orientation of transmembrane regions in the NYMV and MIDWV ORF proteins. Only the NYMV and MIDWV ORF V proteins were predicted to possess a cleavable signal peptide and to be type I transmembrane proteins. In addition, ORF V proteins contained multiple predicted glycosylation sites (Table 2). These data, coupled with the observation that these proteins were located immediately preceding the viral polymerase, suggested that ORFs V of NYMV and MIDWV encode the viral glycoproteins (G).
Analysis of noncoding viral RNA.
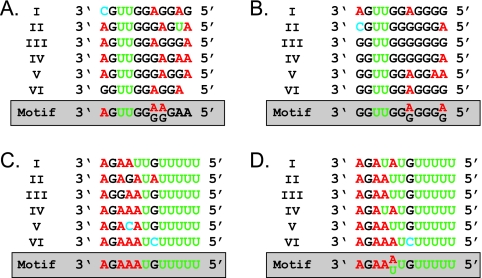
To better assess the relationship between NYMV, MIDWV, and the viral families in the order Mononegavirales, we compared the 5′ and 3′ untranslated regions and intergenic regions of NYMV and MIDWV to those of members of the Mononegavirales. Specifically, we searched for the putative transcription initiation and termination signals for each ORF in each virus. By use of MEME (2), a computational algorithm for identifying conserved sequence motifs, a transcription initiation motif of 3′-AGUUGG(G/A)(G/A)GAA-5′ was identified upstream of every putative ORF for NYMV (Fig. 4A). The motif was present 3 to 68 nt before the start codon. MEME also detected a transcription initiation motif of 3′-GGUUGG(A/G)GGG(G/A) −5′ in MIDWV (Fig. 4B). This motif was positioned 1 to 68 nt upstream of every putative ORF. Similarly, conserved transcription termination motifs of 3′-AGAAAUGUUUUU-5′ and 3′-AGAA(U/A)UGUUUUU-5′ were detected for NYMV (Fig. 4C) and MIDWV (Fig. 4D), respectively.
FIG. 4.
Analysis of the noncoding regions of NYMV and MIDWV. Putative transcription initiation sequences for NYMV (A) and MIDWV (B) and transcription termination sequences for NYMV (C) and MIDWV (D) are shown. The consensus motif is shown in the 3′-to-5′ orientation below the sequences for each viral ORF.
Phylogenetic analysis of NYMV and MIDWV genomes.
To determine the relationship of NYMV and MIDWV to the other viruses in the Mononegavirales order, we conducted a phylogenetic analysis of the polymerase (L; ORF VI) and nucleocapsid (N; ORF I) genes, the only genes that had detectable identity to those encoding other viral proteins. The type species for each genus in Mononegavirales was included in the phylogenetic tree. Analysis of both the polymerase (Fig. 5) and the nucleocapsid (Fig. 6) proteins by both maximum parsimony and maximum likelihood demonstrated that NYMV and MIDWV are clearly most related to each other but only distantly related to all other members of the order Mononegavirales. Analysis of the polymerase (L) gene supported the interpretation that NYMV and MIDWV are most closely, albeit distantly, related to Borna disease virus, but the nucleocapsid (N) gene analysis was more ambiguous and did not preferentially associate NYMV and MIDWV with any specific viral family.
FIG. 6.
Phylogenetic analysis of the viral nucleocapsid (N) proteins. Phylogenetic trees were constructed with the protein sequences by using the maximum parsimony method. The first and second numbers at the branches show the bootstrap percentage values obtained from maximum parsimony (1,000 repetitions) and maximum likelihood (100 repetitions) analyses, respectively.
DISCUSSION
In this paper, we taxonomically classified NYMV and MIDWV on the basis of analysis of their genome sequences. While the existence of these viruses was initially described in the literature over 25 and 40 years ago, respectively (14, 22-24), they have defied all classification efforts to date. We sequenced what we believe to be the complete genome of NYMV and the near-complete genome of MIDWV. By BLAST analysis, the putative polymerase proteins of NYMV and MIDWV shared only 28 to 29% amino acid identity to their top hit in GenBank, the L protein of Borna disease virus, a negative-sense monosegmented RNA virus. Annotation of the predicted ORFs in the genomes of NYMV and MIDWV demonstrated that both viruses contained six ORFs, organized in the canonical fashion of viruses in the order Mononegavirales. Currently, this order contains the four virus families Paramyxoviridae, Filoviridae, Bornaviridae, and Rhabdoviridae. Phylogenetic analysis demonstrated that NYMV and MIDWV were closely related to each other but highly divergent from all other known viruses. Analysis of the L (ORF VI) protein (Fig. 5), but not the N (ORF I) protein (Fig. 6), showed significant bootstrap support for a closer relationship to Borna disease virus than to viruses in other families within the Mononegavirales.
Additional analyses of the noncoding regions of MIDWV and NYMV identified conserved transcription initiation and termination motifs among the ORFs of each virus. Typical viruses in the order Mononegavirales have transcriptional termination signals that contain several features, including a U tract of 4 to 8 nt, which serves as a template for the addition of the poly(A) tail to the nascent mRNAs through reiterative transcription, a conserved C residue immediately preceding this poly(U) tract and an A/U-rich tract (31). In some rhabdoviruses and paramyxoviruses, the A/U-rich tracts are conserved among the genes within a particular virus (3, 4, 18, 20, 27, 28). In contrast, within the Borna disease virus genome, the transcriptional termination motif contains an A followed by six or seven U's (19). Although this motif is preceded by an A/U-rich tract, the residues within the tract are not conserved among the Borna disease virus genes. Thus, the presence of these conserved motifs in each ORF of MIDWV and NYMV was more similar to the pattern found in the rhabdoviruses and paramyxoviruses than to that found in the bornaviruses.
Other physical properties of MIDWV and NYMV were not useful classifiers of these viruses. The genome sizes of NYMV (11.6 kb) and MIDWV (∼11.8 kb) were most consistent with that previously described for rhabdoviruses (11 to 15 kb) but are not significantly greater than that for Borna disease virus (9 kb), while those for the paramyxoviruses (15.1 to 19.2 kb) and the filoviruses (∼19 kb) are generally much greater (10). The shape of the virions seen by EM was more similar to those typically seen with some paramyxoviruses or Borna disease virus (9, 32) than to those seen with the classic bullet-shaped virions usually associated with rhabdoviruses or with the thread-like particles of filoviruses. Clearly, experimental data are needed to definitively classify NYMV and MIDWV. For example, key properties of Borna disease virus that differentiate it from the rhabdoviruses include the facts that it replicates in the nucleus and that its genes undergo extensive splicing. It is unknown at this point where NYMV and MIDWV replicate and whether splicing of their transcripts occurs.
In conclusion, we have sequenced the genomes of NYMV and MIDWV, two previously unclassified viruses. Based on our analysis, MIDWV and NYMV clearly define a novel genus, tentatively named Nyavirus, in the order Mononegavirales. Because they are highly divergent from all members of known virus families, it is also possible that they define a novel family, although additional data will be necessary to clarify the relationship of the nyaviruses to other mononegaviruses. This study underscores the power of high-throughput sequencing strategies to identify highly divergent viruses and to resolve long-standing mysteries such as the identities of NYMV and MIDWV as well as the tremendous virus diversity in the world that remains unexplored to date.
Supplementary Material
Acknowledgments
This study was supported in part by National Institutes of Health grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research and by NIH contract N01-AI30027 to the World Reference Center for Emerging Viruses and Arboviruses.
We thank Fred Murphy for assistance with interpretation of the electron micrographs and for reading the manuscript.
Footnotes
Published ahead of print on 11 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 228-36. [PubMed] [Google Scholar]
- 3.Barr, J. N., S. P. Whelan, and G. W. Wertz. 1997. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J. Virol. 718718-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, J. N., S. P. Whelan, and G. W. Wertz. 1997. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J. Virol. 711794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 6.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 2941351-1362. [DOI] [PubMed] [Google Scholar]
- 7.Bock, J. O., T. Lundsgaard, P. A. Pedersen, and L. S. Christensen. 2004. Identification and partial characterization of Taastrup virus: a newly identified member species of the Mononegavirales. Virology 31949-59. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, D. H., and J. Casals. 1958. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg. 7561-573. [DOI] [PubMed] [Google Scholar]
- 9.Conzelmann, K. K. 1998. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu. Rev. Genet. 32123-162. [DOI] [PubMed] [Google Scholar]
- 10.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, Amsterdam, The Netherlands.
- 11.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. University of Hertfordshire, Hatfield, Herts, United Kingdom.
- 12.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 13.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15153-164. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser, M. N. 1966. Viruses in ticks. I. Natural infections of Argas (Persicargas) arboreus by Quaranfil and Nyamanini viruses and absence of infections in A. (p.) persicus in Egypt. Am. J. Trop. Med. Hyg. 15964-975. [PubMed] [Google Scholar]
- 15.Kall, L., A. Krogh, and E. L. Sonnhammer. 2005. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21(Suppl. 1)i251-i257. [DOI] [PubMed] [Google Scholar]
- 16.Kemp, G. E., V. H. Lee, and D. L. Moore. 1975. Isolation of Nyamanini and Quaranfil viruses from Argas (Persicargas) arboreus ticks in Nigeria. J. Med. Entomol. 12535-537. [DOI] [PubMed] [Google Scholar]
- 17.Kraus, I., M. Eickmann, S. Kiermayer, H. Scheffczik, M. Fluess, J. A. Richt, and W. Garten. 2001. Open reading frame III of Borna disease virus encodes a nonglycosylated matrix protein. J. Virol. 7512098-12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rassa, J. C., G. M. Wilson, G. A. Brewer, and G. D. Parks. 2000. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology 274438-449. [DOI] [PubMed] [Google Scholar]
- 19.Schneemann, A., P. A. Schneider, R. A. Lamb, and W. I. Lipkin. 1995. The remarkable coding strategy of Borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology 2101-8. [DOI] [PubMed] [Google Scholar]
- 20.Schnell, M. J., L. Buonocore, M. A. Whitt, and J. K. Rose. 1996. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 702318-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA.
- 22.Takahashi, M., C. E. Yunker, C. M. Clifford, W. Nakano, N. Fujino, K. Tanifuji, and L. A. Thomas. 1982. Isolation and characterization of Midway virus: a new tick-borne virus related to Nyamanini. J. Med. Virol. 10181-193. [DOI] [PubMed] [Google Scholar]
- 23.Taylor, R. M., J. R. Henderson, and L. A. Thomas. 1966. Antigenic and other characteristics of Quaranfil, Chenuda, and Nyamanini arboviruses. Am. J. Trop. Med. Hyg. 1587-90. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, R. M., H. S. Hurlbut, T. H. Work, J. R. Kingston, and H. Hoogstraal. 1966. Arboviruses isolated from Argas ticks in Egypt: Quaranfil, Chenuda, and Nyamanini. Am. J. Trop. Med. Hyg. 1576-86. [DOI] [PubMed] [Google Scholar]
- 25.Tesh, R. B., A. P. Travassos Da Rosa, and J. S. Travassos Da Rosa. 1983. Antigenic relationship among rhabdoviruses infecting terrestrial vertebrates. J. Gen. Virol. 64(Pt. 1)169-176. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tordo, N., O. Poch, A. Ermine, G. Keith, and F. Rougeon. 1988. Completion of the rabies virus genome sequence determination: highly conserved domains among the L (polymerase) proteins of unsegmented negative-strand RNA viruses. Virology 165565-576. [DOI] [PubMed] [Google Scholar]
- 28.Tordo, N., O. Poch, A. Ermine, G. Keith, and F. Rougeon. 1986. Walking along the rabies genome: is the large G-L intergenic region a remnant gene? Proc. Natl. Acad. Sci. USA 833914-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, D., A. Urisman, Y. T. Liu, M. Springer, T. G. Ksiazek, D. D. Erdman, E. R. Mardis, M. Hickenbotham, V. Magrini, J. Eldred, J. P. Latreille, R. K. Wilson, D. Ganem, and J. L. DeRisi. 2003. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler, D. L., D. M. Church, S. Federhen, A. E. Lash, T. L. Madden, J. U. Pontius, G. D. Schuler, L. M. Schriml, E. Sequeira, T. A. Tatusova, and L. Wagner. 2003. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 3128-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whelan, S. P., J. N. Barr, and G. W. Wertz. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 28361-119. [DOI] [PubMed] [Google Scholar]
- 32.Zimmermann, W., H. Breter, M. Rudolph, and H. Ludwig. 1994. Borna disease virus: immunoelectron microscopic characterization of cell-free virus and further information about the genome. J. Virol. 686755-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.