Abstract
The timely development of safe and effective vaccines against avian influenza virus of the H5N1 subtype will be of the utmost importance in the event of a pandemic. Our aim was first to develop a safe live vaccine which induces both humoral and cell-mediated immune responses against human H5N1 influenza viruses and second, since the supply of embryonated eggs for traditional influenza vaccine production may be endangered in a pandemic, an egg-independent production procedure based on a permanent cell line. In the present article, the generation of a complementing Vero cell line suitable for the production of safe poxviral vaccines is described. This cell line was used to produce a replication-deficient vaccinia virus vector H5N1 live vaccine, dVV-HA5, expressing the hemagglutinin of a virulent clade 1 H5N1 strain. This experimental vaccine was compared with a formalin-inactivated whole-virus vaccine based on the same clade and with different replicating poxvirus-vectored vaccines. Mice were immunized to assess protective immunity after high-dose challenge with the highly virulent A/Vietnam/1203/2004(H5N1) strain. A single dose of the defective live vaccine induced complete protection from lethal homologous virus challenge and also full cross-protection against clade 0 and 2 challenge viruses. Neutralizing antibody levels were comparable to those induced by the inactivated vaccine. Unlike the whole-virus vaccine, the dVV-HA5 vaccine induced substantial amounts of gamma interferon-secreting CD8 T cells. Thus, the nonreplicating recombinant vaccinia virus vectors are promising vaccine candidates that induce a broad immune response and can be produced in an egg-independent and adjuvant-independent manner in a proven vector system.
Avian H5N1 influenza viruses, currently circulating mainly in southeast Asia, are likely to cause the next influenza pandemic (18, 26, 37). The supply of embryonated eggs for traditional influenza vaccine production may be endangered in this case. Efforts to produce inactivated H5N1 vaccines in permanent cells have resulted in large-scale manufacturing, for instance, in Vero cells (21). This approach, based either on fermentation of H5N1 wild-type (wt) viruses (21) or on viruses attenuated by reverse genetics (9, 31), is the most straightforward strategy for egg-independent, rapid vaccine production.
A further approach that may result in more widely available, egg-independent H5 vaccines is the use of recombinant viral vectors expressing protective antigens. Promising protection results were obtained so far with adenovirus-based vectors in mouse models (13, 14). Adenovirus vectors are usually produced in permanent complementing cell lines (11) and have been widely used in clinical trials. Cancellation of a recent trial involving human immunodeficiency virus adenovirus vectors due to suspected enhancement of disease, however, may complicate the future use of these vectors (41). Poxvirus vectors, including recombinant modified vaccinia virus Ankara (MVA) (1, 43), constitute a further class of vectors that have been used to express H5N1 influenza virus antigens (5, 22, 44, 46). Usually, however, the large-scale production of MVA is carried out in primary chicken cells, since these are the most efficient production substrates and are also accepted by regulators. In a pandemic, this production platform may not be available because permanent nontumorigenic avian cell lines are currently not available for production.
In this study, we used a permanent cell line, modified Vero cells, to produce nonreplicating vaccinia virus vectors expressing the H5 hemagglutinin (HA), the major influenza virus protective antigen. The defective vaccinia virus (dVV) vectors are safe due to their lack of replication capacity in normal hosts, while they share the superior immunizing properties of poxviral live vaccines (15, 33). Previously, a permanent cell line based on rabbit kidney cells was engineered to express the essential vaccinia virus D4R gene encoding the enzyme uracil-DNA-glycosylase. This cell line allowed the construction of replication-deficient vaccinia virus vectors (15). In this work, a complementing system based on Vero cells was established and used to produce the defective vaccinia virus vector dVV-HA5. The vector was used to immunize mice and was compared to an inactivated whole-virus (whv) vaccine and to replicating control viruses. The dVV-HA5 candidate vaccine induced neutralizing antibodies and full protection, similar to results with an inactivated whv vaccine. Further, it is important to ensure that the immune responses generated by a pandemic influenza vaccine give long-lived, broad, cross-clade protection. While antibody responses to influenza virus provide protective immunity, T-cell responses are also thought to play an important role in clearance of and recovery from infections. Thus, a vaccine which can produce both effective humoral and T-cell responses would be advantageous. A vaccinia virus vector-based pandemic influenza vaccine has the potential to provide this advantage.
MATERIALS AND METHODS
Viruses and cell lines.
Influenza virus strains. The following strains were used (abbreviations and source in parentheses): A/Vietnam/1203/2004(H5N1) (VN1203; CDC no. 2004706280) and A/Indonesia/05/2005(H5N1) (IN5/05; CDC no. 2005740199). Viruses were provided by the Centers for Disease Control and Prevention (Atlanta, GA) and grown under biosafety level 3 (BSL3) conditions.
(i) Vaccinia viruses.
The African green monkey kidney cell line CV-1 (CCL-70) and the vaccinia virus strains Western Reserve (VR119) and Lister/Elstree (VR-862) were obtained from the American Type Culture Collection. The basis of the Lister constructs was the subcloned virus vpDW-862/Elstree (33).
(ii) Cell lines.
The African green monkey kidney cell line CV-1 (CCL-70) was cultivated in Dulbecco's modified Eagle medium containing 5% fetal calf serum (FCS). Vero (CCL-81) and D4R complementing cVero22 cells were cultivated in serum protein-free medium as described previously (9). HT1080 (CCL-121), a human fibrosarcoma cell line containing an activated N-ras oncogene, was grown in Dulbecco's modified Eagle medium containing 5% FCS. The retrovirus packaging cell line PT67 was obtained from Clontech Laboratories, Inc.
Establishment of the D4R complementing cell line cVero22.
Generation of defective retrovirus vector particles, transduction, and screening for complementing cells. To obtain retrovirus supernatant, the packaging cell line PT67 (29) was transfected with the plasmid pLXSN-D4R#14 using a liposomal reagent (DOTAP; Roche catalog no. 1202375). The supernatant was harvested 48 h later and filtrated through a 0.45-μm filter unit to remove residual cells. Polybrene was added to detach retroviral complexes. Vero cells (cell bank 0004A, passage 141) were transduced with the retrovirus vectors. Serial dilutions from 10−1 to 10−5 were carried out in six-well plates on cells that were 50% confluent. Four days after the transductions, cells were confluent, and all dilutions were subdivided into petri dishes using TC-Vero medium (Baxter) containing 600 μg G418/ml. Three weeks later, 48 G418-resistant clones were picked and further grown in 24-well plates. After a further split into six-well plates, screening was performed with the defective vaccinia virus dVV-L at a multiplicity of infection (MOI) of 0.01. After 72 h, monolayers were stained with crystal violet. Complementation of growth was indicated by cytopathic effect and plaque formation. In 18 of the G418-positive clones, all cells were lysed, indicating excellent complementation; 14 clones showed many individual plaques, 13 allowed growth of only few plaques, and 3 cell clones did not allow plaque formation. Vero cell clone no. 22 was chosen and named cVero22. The defective virus dVV-L, originally derived in RK-D4R-44.20 cells, was passaged in cVero22 cells four times; a titer of 1.5 × 109 PFU/ml was achieved in the fourth passage. In a typical sucrose cushion preparation with 20 T-flasks (175 cm2), yields of 1 × 1010 PFU/ml were obtained.
Growth curves.
Vero and cVero22 cells were infected with different vaccinia viruses at a MOI of 0.01. At the time points 4, 24, 48, 72, and 96 h postinfection, cells were scraped into the medium, disrupted by ultrasonication, and titrated. Fifty percent tissue culture infective dose (TCID50) values were determined through double titrations on corresponding cell lines.
Tumorigenicity testing.
The cVero22 cell line and the positive control cell line HT-1080 (CCL-121) were grown in T-flasks. Cells were counted, and viability was determined twice using a Casy Technology cell counter system. The cell line cVero22 was tested in the 149th passage and showed a viability of 93%. The HT1080 positive control was in the 25th passage with a viability rate of 92%. After viability testing, the absolute living cell number of 1 × 107 was used for the injections into the mice. Female nude mice (HsdOla: ICRF-nu), 4 weeks old, were purchased from Harlan-Winkelmann GmbH (Germany). All cages, bedding, enrichment, food, and water were autoclaved before use. Food and water were ad libitum. Cages, bedding, and enrichment were changed once per week. Groups of five mice were used for the test cells and three to five animals for the control cells. Mice were inoculated intramuscularly in the right thigh with a 0.2-ml suspension containing 1 × 107 live cVero22 or HT1080 cells. Control animals were additionally injected in the left thigh with 0.2 ml cell culture medium. The animals were observed daily, and the injection sites were palpated during 30 days for evidence of tumor or nodule formation. Once identified, any nodule was measured every second day to determine whether there was any progressive growth. Animals that showed rapidly growing or large tumors with a size of at least 10 by 10 mm were sacrificed to avoid unnecessary pain and suffering. These animals were necropsied. Tumors and organs (muscle, liver, spleen, kidneys, and lungs) were isolated and fixed in 4.5% formaldehyde for histological analysis. Animals with nonprogressive nodules or without any nodule were observed for 60 to 90 days with once-a-week palpation. All animals were euthanized at the end of experiment. Necropsy and histological analysis of organs were performed as described below.
Histology. (i) Hematoxylin-eosin stain.
Trimming, embedding in paraffin, and cutting were carried out according to standard procedures. Deparaffinized sections were stained using an automated slide stainer according to standard hematoxylin-eosin staining procedures.
(ii) Cytokeratin stain.
Paraffin-embedded sections were subjected to an antigen retrieval procedure to uncover antigenic sites from tissue sections. Sections were digested with proteinase K (10 min, room temperature) and then rinsed with buffer. The primary antibody, mouse monoclonal antibody anti-human cytokeratin (clone MNF 116, Dako), was applied in a dilution of 1:50. As isotype control mouse immunoglobulin G (IgG) was used. A Mouse-on-Mouse kit for staining murine tissues with murine antibodies was used according to the instruction sheet (Dako, Arkansas).
(iii) Immunofluorescence.
For fluorescence staining, the sections were incubated with the antibody or buffer (negative control) after a protein block. A fluorescein-labeled secondary antibody was applied subsequently. The bound antibody was detected by using a confocal microscope (LSM 510; Zeiss).
Cloning and sequencing of HA genes. (i) VN1203 and IN5/05 influenza virus strains.
Viral RNA was extracted using the Trizol procedure and transcribed into cDNA using the primer oFLURT-2 (5′-AGC AAA AGC AGG GGT ATA ATC TGTC-3′). The primers oHAv-10 (5′-AAC CAT GGA GAA AAT AGT GCT TC-3′) and oHAu-2 (5′-GTC GAC TTA AAT GCA AAT TCT GCA TTG TAAC-3′) were used for PCR amplification. The resulting HA genes were cloned, resulting in the plasmids pDD4mH55TNT-VN-HA (VN1203 insert) and pPCR-Script-HA-INa#13 (IN5/05 insert), and sequenced using standard procedures. The VN1203 and IN5/05 inserts were compared with the master sequences (GenBank accession no. AY818135 for VN1203 and Los Alamos database number ISDN125873 for IN5/05).
(ii) Hatay strain.
A plasmid containing the HA5 gene of A/chicken/Hatay/2004(H5N1) (VNHY/04; GenBank accession no. AJ867074) was kindly provided by K. Dinh Duy (Institute of Biotechnology, Hanoi, Vietnam) and S. K. Lal (International Centre for Genetic Engineering & Biotechnology, New Delhi, India).
Construction of plasmids. (i) pLXSN-D4R.
In order to obtain pLXSN-D4R#14, a SmaI-EcoRI cassette carrying the vaccinia virus D4R gene was excised from plasmid pCR-D4R(1) (15) and inserted into the HpaI-EcoRI sites of pLXSN (29). In this plasmid, the D4R gene is under the control of the murine leukemia virus long terminal repeat promoter, while the neomycin resistance gene, allowing screening with the antibiotic G418, is controlled by the simian virus 40 promoter.
(ii) pDD4-mH5-mNP-VN.
The influenza virus nucleoprotein cDNA was made from A/VietNam/1203/2004(H5N1) (accession no. AY818138) using the primer o.Flurt-4 (5′-ATGGCGTCTCAAGGCAC-3′). PCR was carried out using KOD Hot Start DNA polymerase (Novagen) from the cDNA using the primers o.NP1 (5′-CCA TGG CGT CTC AAG GCA CCA-3′) and o.NP2 (5′-GTC GAC TTA ATT GTC ATA CTC CTC TGC ATTG-3′). This PCR product was cloned in pPCR-Script (PCR-Script Amp cloning kit; Stratagene) and then subcloned in pDD4-mH5 using the NcoI and SalI sites, resulting in the plasmid pDD4-mH5-NP-VN; the resistant plasmid contains the unmodified nucleoprotein gene cassette. To substitute the vaccinia virus early transcription stop signal, TTTTTNT (51), PCR mutagenesis using the primers oNP-5 (5′-CTC CAT GGC GTC TCA AGG CA-3′), oNP-6 (5′-GAG TGG CAT GCC ATC CACAC-3′), oNP-7 (5′-GAA GAT CTC ATA TTT CTG GCA CGG TCT GC-3′), and oNP-8 (5′-GCA GAC CGT GCC AGA AAT ATG AGA TCT TC-3′) primers was performed. The PCR fragment was inserted into the plasmid pDD4-mH5-NP-VN using the NcoI and SphI sites, resulting in the plasmid pDD4-mH5-mNP-VN; this plasmid contains the modified nucleoprotein gene cassette.
(iii) pDD4-mH5-HA5.
The HA5 gene was amplified by PCR using the primers H5N1-HA-SphI-F (5′-ACA TGC ATG CAT GTA CCA TGG AGA AAA TAG TGC TTC TTC TT −3′) and H5N1-HA-XhoI-R (5′-CCG CTC GAG CGG TTA AAT GCA AAT TCT GCA TTGT-3′). The PCR fragment was inserted into the SphI and XhoI sites of the pDW-2-mH5 vector, resulting in pDW2-mH5-HA5. The pDW-2-mH5 vector was modified from the original pDW-2 vector by the insertion of an mH5 promoter sequence at the PacI site. The same gene was also amplified by PCR using the primers H5-F-NcoI (5′-TCC ACC ATG GAG AAA ATA GTG CTT CT-3′) and H5-R-SalI (5′-ATA CGT CGA CTT AAA TGC AAA TTC TGC ATT GTA-3′). The PCR product was cloned into pDD4-mH5 at the NcoI and SalI sites, resulting in the plasmid pDD4-mH5-HA5.
Construction of recombinant vaccinia viruses.
For vDD4-mNP-VN, 20 μg of pDD4-mH5-mNP-VN plasmid DNA (or pDD4-mH5-5TNT-VN-HA#1, as needed) was transfected into vaccinia virus Lister-infected CV-1 cells by calcium phosphate precipitation and further processed as described previously (16). Plaque isolates were purified three times and expanded for large-scale preparations. For rVV-L-HA-VN, 20 μg of pDD4-mH5-5TNT-VN-HA (clone no. 1) plasmid DNA was transfected into vaccinia virus Lister-infected Vero cells (0008A bank) and processed as described above. To generate the rVV-HA5 virus, CV-1 cells were infected with dVV-L virus (Lister strain) for 1 h, followed by transfection of the plasmid pDD4-mH5-HA5 as described above. The cells were harvested, and a recombinant named rVV-HA5 was selected by plaque screening in CV-1 cells. The rVV-HA5 was plaque purified three times before further characterization. To generate the dVV-HA5 virus, CV-1 cells were infected with DryVax virus (Wyeth strain) for 1 h, followed by transfection of the plasmid pDW2-mH5-HA5 using Effectene reagents (Qiagen) according to the manufacturer's protocol. The target genes were under the control of the mH5 promoter and flanked by the D3R and D5R genes of vaccinia virus at the position of the deleted D4R gene, encoding the essential DNA repair enzyme uracil DNA glycosylase. The Guanine phosphoribosyl transferase gene (gpt) and lacZ were also packaged in with the target genes as selection markers. The recombinant viruses were harvested by three cycles of freezing and thawing of infected cells and used for plaque purification. After four rounds of plaque purification in RK44 cells (D4R complementing rabbit kidney cell line), a blue plaque was picked and further plaque to plaque purified three times in RK44 cells without selection pressures. A white plaque, which grew in RK44 cells but not in CV-1 cells, without a selection marker was isolated and named dVV-HA5.
Cloning of Dryvax vaccinia virus clone B/8A.
The Dryvax smallpox vaccine (used in the United States and elsewhere in the smallpox eradication program) is a mixture of vaccinia viruses produced on calf skin. In order to start the derivation of defective viruses from a homogenous cloned virus source, this virus mixture was subcloned and the isolate growing best in two different Vero working cell banks was used for further experiments. For subcloning, an original vial of Dryvax smallpox vaccine (control no. 4998391; Wyeth, Inc) was thawed and serial dilutions (ranging from 1:10 to 1:106 on a six-well plate) of infected Vero cells were overlaid with an agarose-medium mixture. Five initial plaques were picked. In the second plaque purification step, again five plaques were picked from each initial plaque (a total number of 25). A third round of plaque purification was done on a Vero working cell bank (WCB 0006B/576) starting in passage 137. Single plaque amplification was done twice on WCB 0006B, followed by a third and fourth amplification step in both the WCB 0006B and WCB 0008A Vero cell banks. DryVax virus clone B/8A (lot 0008A/406/142/050404) was chosen for further experiments; this clone has excellent growth properties in Vero cells and is therefore a good candidate for recombinant defective viruses based on the U.S. smallpox vaccine strain.
Derivation of defective virus dVV-Y.
The plasmid pDW-2 (17) can be used to derive D4R-defective vaccinia viruses. It contains the vaccinia virus genes D3R and D5R and a lacZ/gpt marker gene cassette located between DNA repeats, allowing transient selection and blue plaque screening. Release of the selective pressure results in a loss OF the marker gene and the generation of a D4R-defective virus. To generate dVV-Y by in vivo recombination, cVero22 cells were infected with the DryVax virus clone B/8A, followed by transfection of the plasmid pDW-2. Plaque purifications were done as described earlier (15). After six rounds of plaque purification in cVero22 cells, a defective virus clone was isolated and named dVV-Y (clone no. F4ba). A screening of the isolates on noncomplementing CV1 cells performed in parallel showed no plaque formation, indicating freedom from wt virus. Clone no. F4ba was further propagated and passaged three times on cVero22 for amplification reasons. A typical sucrose cushion preparation with 24 T-flasks (175 cm2) yielded 9 × 109 PFU/ml. A one-stage passage reversion test was done by infection of CV1 with 2 × 107 PFU. Six days after infection, cells were scraped and harvested by low-spin centrifugation. The pellet was frozen and thawed three times, and dilutions −1 to −6 were titrated on CV1 cells. In crystal violet stains no plaques were detectable, indicating a pure stock of defective virus.
Western blots.
Expression of the HA proteins by recombinant vaccinia virus was detected by Western blotting. Vero cells (5 × 106) or in the case of defective recombinant vaccinia virus, cVero cells were infected at a MOI of 0.01 or at 5.0, respectively, for 72 h. Sonicated cell lysates were loaded on a 12% polyacrylamide gel (Bio-Rad, Inc). To detect HA protein, a sheep antiserum against A/Vietnam/1194/04 was used. Donkey anti-sheep alkaline phosphatase-conjugated IgG (Sigma Inc.) was used as a secondary antibody in a dilution of 1:2,000. To detect vaccinia virus protein, a polyclonal rabbit anti-vaccinia virus serum was used (lot no. aVVSKP26012006; Baxter). The second antibody was a 1:2,000-diluted goat anti-rabbit alkaline phosphatase-conjugated IgG (Sigma Inc.). Inactivated VN1203 bulk material served as a positive control.
Preparation of whv vaccine.
Inactivated influenza virus was purified by sucrose gradient ultracentrifugation followed by ultra/diafiltration and sterile filtration to produce the monovalent bulk material as described recently (21). Influenza virus antigen content was determined by a single radial immunodiffusion assay (20).
Immunization and challenge of animals.
With vaccinia virus constructs, mice were immunized by intramuscular injections of the indicated doses in a volume of 50 μl in Tris-buffered saline-0.01% human serum albumin buffer. Using the inactivated whv influenza virus vaccine, mice were immunized by intramuscular injection of 50 μl as described herein. Mice were challenged intranasally with 20 μl containing 1 × 105 TCID50 of virus and monitored for clinical parameters and survival over a period of 14 days. H5N1 strains used for challenge were grown and titrated via TCID50 assays in Vero cells. Prior to the challenge studies, the virus dose that kills 50% of the BALB/c mice was determined for the H5N1 strains HK156, VN1203, and IN5/05 to be 11, 24, and 18 TCID50, respectively.
Microneutralization assays.
For the standard microneutralization assay (done with wt viruses which have the polybasic cleavage sites in the HA, allowing trypsin-independent growth in Vero cells), heat-inactivated serum samples were serially diluted with cell culture medium in twofold steps. The dilutions were mixed at a ratio of 1:1 with VN1203 virus (100 TCID50 per well), incubated for 1 h at room temperature, and transferred to a microtiter plate with a Vero cell monolayer. After 5 to 7 days of incubation at 37°C, the cultures are inspected for cytopathic effect. The neutralizing titer (NT), expressed as the reciprocal of the antiserum dilution at which virus growth is 50% inhibited, was calculated by determining the number of virus-negative wells and the serum dilution according to the method in reference 40.
Viral infectivity (TCID50) assays.
The H5N1 virus titers of samples (TCID50) were determined by titration on standard Vero cells by serial 10-fold dilutions of samples inoculated into 96-well microtiter plates as described earlier (20).
Detection of gamma interferon (IFN-γ) in CD8 and CD4 T cells by flow cytometry after antigen-specific stimulation of splenocytes. (i) Preparation of mouse splenocytes.
Spleens were ground on a metal mesh to prepare single-cell suspension in culture medium (45% RPMI 1640 [Gibco], 45% CLICKs medium [Sigma], 10% FCS [Gibco], penicillin-streptomycin [Gibco], 2 mM l-glutamine [Gibco]). Viability of the splenocytes was assessed by propidium iodide staining (Sigma). Splenocytes either were used immediately or were frozen in Cryostor CS-10 (VWR) at 2 × 107 viable cells per ml.
(ii) Cytokine flow cytometry.
Viable splenocytes (1 × 106 to 3 × 106, fresh or thawed) from at least two mice per group were dispensed into 96-well round-bottom-plates (Costar). Protein antigens (3 μg/ml HA), peptides (2 μg/ml), or medium was added to a final volume of 220 μl. After 2 h at 37°C, 10 μg/ml brefeldin A (Sigma) was added to inhibit secretion of cytokines, and incubation was continued for approximately 14 h. Then, cells were resuspended in 50 mM phosphate-buffered saline (PBS)-EDTA, washed, and incubated on ice with ethidium monoacide (1 μg/ml; Molecular Probes) and with rat anti-mouse CD4 allophycocyanin and CD8 phycoerythrin antibodies (0.5 μg/ml; BD Biosciences) under UV light. After fixation with 1% paraformaldehyde (Merck), cells were permeabilized in PBS, supplemented with 0.08% saponin (Sigma), and incubated with rat anti-mouse IFN-γ fluorescein isothiocyanate antibody, (0.5 μg/ml; BD Biosciences) for 30 min at room temperature. Finally, cells were washed and fixed with 1% paraformaldehyde. At least 100,000 viable cells were applied on a FACSCalibur flow cytometer (BD BioSciences), and data analysis was performed using the FlowJo software program (Tree Star, Inc.). Percentages of IFN-γ-producing T cells were calculated after gating on ethidium monoacide-negative, CD4- or CD8-positive lymphocytes.
Flow cytometric analysis of killing of peptide-pulsed target cells by specific CD8 T cells.
The cytotoxic capacity of influenza virus-specific CD8 T cells was assessed by the Vital assay (12). Briefly, target cells were fluorescence labeled by incubation of 2 million splenocytes from nonimmunized control animals with 0.1 μM 5-(and-6)-carboxyfluorescein diacetate succimidyl ester (CFSE) or 9 μM 5-(and-6)-(((4-chloromethyl)benzoyl)amino) tetramethylrhodamine (CMTMR). Next, labeled cells were pulsed with either 2 μM HA534-543 peptide, 2 μM NP147-155 peptide, or 2 μM M1 58-66 peptide for 60 min at 37°C. Then, 10,000 of both CFSE- and CMTMR-labeled splenocytes, representing specific peptide- and control peptide-loaded target cells, were incubated in 5 ml polystyrene round-bottom tubes (BD Falcon) for 64 h together with 2 million pooled effector splenocytes obtained from immunized mice. Finally, propidium iodide was added to the cell cultures at 3 μg/ml and analyzed by flow cytometry. At least 100,000 viable cells were evaluated using a FACSCalibur (BD BioSciences) instrument, and data analysis was performed using the FlowJo software program (Tree Star Inc.). Percentages of CFSE- and CMTMR-positive cells were calculated after gating on propidium iodide-negative viable lymphocytes. The percent specific killing was calculated as 100 × (1 − percent target cells/percent control cells).
ELISPOT assay.
The frequency of IFN-γ- or interleukin-4 (IL-4)-secreting cells was analyzed using mouse IFN-γ and IL-4 enzyme-linked immunospot (ELISPOT) kits (Mabtech AB, Nacka, Sweden), following the instructions of the manufacturer and as described recently (21).
Statistical analysis.
Data were analyzed using the Statistica software program (version 7.0; Statsoft, Tulsa, OK). The numbers of protected mice in vaccinated and control groups were compared by using Fisher's exact test (two-tailed). Results were considered statistically significant with P values of <0.05.
RESULTS
Generation of complementing Vero cell system.
Due to their restricted interferon system (8), Vero cells are among the most productive cells for the propagation of viruses and thus are one of the most promising cell culture systems for production of viral vaccines. We therefore chose Vero cells to derive cell lines that complement the growth of defective vaccinia virus. Since generation of recombinant cell clones by plasmid transfection and selection turned out to be difficult with Vero cells, retrovirus vectors were used to generate complementing cell clones. For the construction of the retrovirus vectors, the plasmid pLXSN (29) was used to insert the vaccinia virus D4R gene (resulting in the plasmid pLXSN-D4R). The provirus-containing plasmid was transfected into packaging cells, and retroviral vectors carrying the complementing vaccinia virus gene and the neomycin selection marker were obtained. The retroviral vectors were used to transduce Vero cells. Neomycin selection resulted in cell clones which were screened for complementation of defective vaccinia virus. A cell line with acceptable growth and complementation properties was finally obtained and was named cVero22 (see Materials and Methods). This cell line provides the viral enzyme uracil-DNA-glycosylase, required for replication of vaccinia virus in trans. The defective virus dVV-HA5 grew in this cell line to titers equivalent to those obtained for the replicating counterpart rVV-HA5 in normal Vero cells (see below), indicating full complementation.
cVero22 cell line is nontumorigenic in nude mouse model.
Cell lines used for the production of live vaccines should be nontumorigenic. Vero cells below a limited passage number fulfill this requirement and are suitable for vaccine production (25, 42). The next step was therefore to examine the tumorigenic potential of the cVero22 cell line. The cells were injected intramuscularly into nude mice, and the animals were observed for 90 days for tumor growth. The intramuscular route is among the most efficient routes to support growth of tumorigenic cell lines and was found to be superior to subcutaneous injection (24). As a positive control, the human fibrosarcoma cell line HT1080 was chosen. This cell line grew progressively and induced large visible tumors within 3 weeks, and animals were euthanized for humane reasons. After injection of the cVero22 cells, no macroscopic tumor growth was seen. Histochemical analysis at day 90, however, revealed some residual surviving Vero cells in intermuscular spaces in two of five mice (Fig. 1). Since Vero cells are of epithelial origin, cytokeratin (intermediate filament) staining (Fig. 1A) and immunofluorescence (Fig. 1C) were used to show the atopic presence of the Vero cells in muscle. Thus, tumor formation, characterized by progressive growth, capsule formation, and densely growing cells, was not observed with the cVero22 cell line; however, residual Vero cells were identified at late times in some of the animals, consistent with the notion that mice without functional immune systems have difficulty clearing a permanent cell line injected in huge amounts. The parental Vero cell line was found to be nontumorigenic when the subcutaneous route was used in previous studies.
FIG. 1.
Histology of muscle tissue of nude mice. (A) Cytokeratin staining around the injection sites for epithelial tissue indicates residual Vero cells. (B) Murine muscle tissue stained with hematoxylin/eosin. (C) Immunofluorescence for Vero proteins in muscle cryosections. (D) cryosections of muscle tissue, negative control.
Construction and characterization of vaccinia viruses.
Next, the plasmids and vaccinia viruses encoding the H5 HA were constructed. The HA genes (cDNAs) of the influenza virus strains A/Vietnam/1203/2004(H5N1) (VN1203) and A/chicken/Hatay/2004(H5N1) (VNHY/04) were placed downstream of a strong vaccinia virus early/late promoter (48). The resulting plasmids (Table 1) (see Methods) were used to construct the nonreplicating virus dVV-HA5 and the replicating controls rVV-HA5 (expressing Hatay HA) and rVV-HA-VN (expressing VN1203 HA). The HAs of the two strains are practically identical; they share >98% amino acid identity, belong to the same clade, and have the same polybasic cleavage site, characteristic for highly virulent H5N1 strains. One of the mutations in the Hatay sequence, the N154S(HA2) mutation (49), results in the loss of a potential glycosylation site in the HA2 molecule (see Discussion).
TABLE 1.
Vaccinia virus constructs and vaccines
| Vaccinia virus/vaccine | Inserted flu genea/strain | Plasmid construct | Titerb (PFU/ml) | HA expc (%) |
|---|---|---|---|---|
| dVV-HA5 | HA/VN-HY04 | pDW2-mH5-HA5 | 4.9 × 109 | 100 |
| rVV-HA5 | HA/VN-HY04 | pDD4-mH5-HA5 | 2.5 × 109 | 100 |
| rVV-L-HA-VNd | HA/VN1203 | pDD4-mH5-HA-VN | 2.3 × 1011 | 100 |
| vDD4-mNPe | NP/VN1203 | pDD4-mH5-NP-VN | 1.9 × 109 | NA |
| VV-WT (Lister) | Empty vector | NAg | 1.7 × 1011 | NA |
| dVV-Y (Wyeth) | Empty vector | NA | 9.0 × 109 | NA |
| Inact. whvvf | None/VN1203 | NA | NA | NA |
All foreign genes are controlled by the vaccinia virus mH5 promoter. Flu gene, influenza virus gene.
Titers of sucrose-purified virus preparations from Vero cells (see Methods).
HA expression as determined by immunostaining.
Virus grown in BHK cells.
Replicating control virus expressing the influenza virus nucleoprotein.
Inactivated whv vaccine.
NA, not applicable.
An outline of the genomic structures of the generated viruses is shown in (Fig. 2). In the defective vector dVV-HA5 (Fig. 2C), the influenza virus HA gene is inserted between the D3 and D5 genes, replacing the essential D4R gene. In the replicating vaccinia virus controls, the HA gene is in a similar location, in the D4/D5 intergenic region (Fig. 2D and E). The parental strain of the virus dVV-HA5 is the Wyeth smallpox vaccine strain, used to eradicate smallpox in the United States and elsewhere. The structures were confirmed by PCR analysis. A primer set flanking the insertion site of the foreign gene was used to show that the recombinant viruses had the expected enlarged fragment and were free of wt virus (not shown). Further, a control virus for the T-cell experiments, vDD4-mNP-VN, was constructed by inserting the nucleoprotein of the VN1203 strain into the vaccinia virus D region (Fig. 2F).
FIG. 2.
Outline of the genomic structures of the wt and recombinant vaccinia viruses. VV-WT, wt vaccinia virus; D3R, D4R, and D5R, vaccinia virus genes; P, promoter; HA-HY, HA gene, Hatay strain; HA-VN, HA gene, VN1203 strain; mNP, modified gene of the influenza virus nucleoprotein; dVV, vaccinia virus defective in D4R open reading frame; mH5, modified vaccinia virus H5 promoter (see the text). For further description of the viruses, see Table 1.
Growth behavior of viruses in Vero cells.
To further characterize the viruses and the modified Vero cell line, one-step growth curves were performed. The defective dVV-HA5 viruses and the corresponding empty vector control, dVV-Y, were grown in complementing cVero22 cells. The replicating viruses rVV-HA5 and rVV-L-HA-VN were grown in normal Vero cells. In addition, the Lister vaccinia virus strain (rVV-L) was used as a control. Cell monolayers were infected with the viruses at a MOI of 0.01 and grown for 92 h. Titers of virus were determined at regular intervals (Fig. 3). The viruses with the influenza virus HA gene insert reached similar endpoint titers (range, 2 × 107 to 6 × 107 PFU/ml); the empty replicating virus rVV-L reached a higher titer of 2 × 108 PFU/ml. Maximal titers were achieved after 72 h. The growth experiment demonstrates that defective viruses grown in cVero22 reach titers similar to those of corresponding replicating viruses in Vero cells. Thus, the modified cells allow full complementation of the defective viruses providing the D4R gene in trans.
FIG. 3.
Growth of vaccinia viruses in the cVero22 cell line compared to that of replicating viruses in normal Vero cells. Viruses with HA gene inserts (dVV-HA5, rVV-HA5, and rVV-L-HA-VN) and empty defective viruses (dVV-L and dVV-Y) reach similar endpoint titers. The Lister wt strain (VV-WT) grew to a higher titer. The titers are means of two determinations; error bars indicate the standard errors of the means.
Next, the viruses were grown on a larger scale. The dVV-HA5 virus was grown in the cVero22 cell line, while rVV-HA5 was grown in normal Vero cells. Both viruses were purified by sucrose cushion centrifugation. Concentrated virus stocks had similar titers, 4.9 × 109 and 2.5 × 109 PFU/ml, respectively, confirming again that the complementing cell line supported full growth.
Expression of influenza virus H5 HA genes in permissive and nonpermissive Vero cells.
In live vaccines, parameters such as expression levels and processing of the protective antigen influence the immune response. During protein maturation, the influenza virus HA precursor (H0) is cleaved by intracellular proteases with furin or furin-like activity into a heterodimeric receptor molecule (H1 and H2), resulting in proper folding and full infectivity of influenza virus. In order to assess HA antigen levels and processing, expression experiments in complementing and normal Vero cells were carried out. For permissive infections, cells were infected at a MOI of 0.01; for nonpermissive conditions (dVV in Vero), a MOI of 5 was used. Infected cells were incubated for 72 h, and total cell lysates were analyzed by polyacrylamide gel electrophoresis and Western blotting using anti-HA and anti-vaccinia virus antisera.
As shown in Fig. 4A, the viruses with HA gene inserts induced high-level expression of HA. The largest band, at around 80 kDa, represents the uncleaved H0 HA precursor; the two bands at approximately 64 and 26 kDa represent the subunits HA1 and HA2 (lanes 1 to 5). Thus, Vero cells process the HA antigen; however, processing is incomplete despite the presence of the polybasic cleavage site. Processing seems to be more efficient with defective vectors when nonpermissive cells are infected at a high MOI. In this case, HA0 is a minor band, while the HA1 and HA2 bands get more prominent (lane 5). Efficient processing results in a correct display of the HA receptor molecule on the cell surface and should also result in good neutralizing antibodies. Interestingly, HA2 of the Hathay strain migrates faster, suggesting that in fact the N154S(HA2) mutation results in the loss of a glycosylation site in the HA2 molecule. Lanes 6 to 9 are appropriate negative controls.
FIG. 4.
Western blots of cell lysates probed for influenza virus HA expression (A) or vaccinia virus antigen expression (B). Lane 1, formalin-inactivated purified influenza virus. Lane 2, replicating virus rVV-HA-VN (VN1203) grown in Vero, MOI of 0.01. Lane 3, replicating virus rVV-HA5 (VNHY/04), Vero, MOI of 0.01. Lane 4, defective virus dVV-HA5 (VNHY/04), cVero22, MOI of 0.01. Lane 5, dVV-HA5 (VNHY/04), Vero, MOI of 5; lane 6, negative control dVV-L, Vero, MOI of 5.0; lane 7, dVV-L, cVero 22, MOI of 0.01; lane 8, Lister wt, MOI of 0.01; lane 9, negative control, uninfected Vero cell lysate. (C) HA expression in wt Vero cells infected with the different vectors at a constant MOI of 5. Lane 1, size markers; lane 2, positive control, 0.5 μg of the formalin-inactivated influenza vaccine; lane 3, replicating virus rVV-HA-VN; lane 4, replicating virus rVV-HA5; lane 5, defective virus dVV-HA5; lane 6, empty vector dVV-L; lane 7, empty vector VV-L; lane 8, uninfected Vero cell lysate.
In a vaccinated host, the dVV vectors express exclusively early antigens for a prolonged period of time (15). In order to also analyze the expression of vaccinia virus structural antigens, the Western blot was now probed with a rabbit anti-vaccinia virus antibody (Fig. 4B). The analysis showed that only under replicative conditions could strong vaccinia virus-specific protein expression be observed (lanes 2 to 4, 7, and 8), reflecting the pattern of vaccinia virus structural proteins. Infection of nonpermissive Vero cells with the defective virus at a high MOI resulted in efficient HA expression (Fig. 4A, lane 5) but low vaccinia virus antigen expression (Fig. 4B, lane 5). With the empty vector, no HA is seen (Fig. 4A, lane 6), and vaccinia virus bands were also strongly reduced (Fig. 4B, lane 6). In permissive infections, the dVV-L virus induces strong vaccinia virus-specific bands (Fig. 4B, lane 7). The total amounts loaded on the Western blot were the same, as controlled by Ponceau staining determination of the transfer efficiency (not shown). In sum, the HA was properly cleaved in Vero cells, consistent with excellent growth of the wt H5N1 influenza virus strains in this cell line (21). Expression levels under permissive conditions were high, and under nonpermissive conditions, better processing and strongly reduced amounts of vaccinia virus structural proteins were observed. Thus, in the host vaccinated with this nonreplicating vector, similar expression patterns are expected: high levels of the foreign gene and reduced levels of vector structural proteins, a property that may prevent the diversion of the immune system to unwanted vector targets.
To study HA expression induced by the different vectors under comparable conditions in the same cell line, wt Vero cells were infected at a MOI of 5 and lysates were analyzed by Western blotting. As shown in Fig. 4C, comparable HA expression levels were obtained under these conditions with the different vectors (lanes 3 to 5). The amount of 0.5 μg of HA of the inactivated whv vaccine was used as the positive control (lane 2).
Protection studies with mice.
Subsequently, protection studies with BALB/c mice were carried out, and the dVV-HA5 construct was compared to a formalin-inactivated whv vaccine and a corresponding replicating vaccinia virus control, rVV-HA5. A number of other replicating virus controls were included, as described in Table 1. The live virus vaccines were given by intramuscular injection in a dose of 1 × 106 PFU per animal. The live vaccines were diluted from sucrose-purified high-titer stocks (for titers, see Table 1) and thus contained only minor amounts of residual host cell protein, minimizing a possible immunizing effect of residual HA antigen present in the preparation. The antigen content of the whv vaccine was determined using single radial diffusion assays with anti-HA standard antiserum and purified HA standard antigen. Therefore, it is not possible to directly compare dosing of the inactivated whv vaccine with that of the live vaccines. Negative controls included groups immunized with the empty vaccinia virus vector (VV-WT) and with PBS. Single- and double-dose immunizations were carried out. The single-dose groups were immunized at day 0 and challenged at day 42. The double-dose groups received a booster immunization at day 21. The challenge virus was the highly virulent H5N1 VN1203 strain (27), given at the dose of 1 × 105 TCID50 per animal, a high dose, corresponding to an approximately 4,000-fold LD50 for the BALB/c mouse.
In the single-dose groups (Table 2), complete protection was seen with all viral constructs expressing the HA gene and with the inactivated whv vaccine after homologous high-dose challenge. All negative control animals, injected with the empty vaccinia virus vector, the NP-expressing vector, or PBS, died (Table 2, bottom). Prechallenge sera of all groups were analyzed for anti-HA reactivity in an enzyme-linked immunosorbent assay (ELISA) and for neutralizing antibodies in a microneutralization assay, in which wt VN1203 virus growth in Vero cells is assessed after incubation with mouse sera. With the whv vaccine, ELISA geometric mean titers of >16,000 were achieved. With the dVV-HA5 and replicating HA vectors, the ELISA titers were lower. Neutralizing antibodies were detected in the groups receiving the whv vaccine and in the groups with the poxviral HA constructs. With the NP control virus (vDD4-mNP), ELISA titers and NTs were not detectable.
TABLE 2.
Protection, ELISA, and microneutralization results
| Vaccine/construct | Imm dosea | No. of survivors/total (%)b | rH5 ELISA resultc (GMTd) | μ-NTc (GMT) |
|---|---|---|---|---|
| Single dose | ||||
| Inact. whvv | 3 μg | 12/12 (100) | 16,127 | 49.7 |
| dVV-HA5 | 1 × 106 | 12/12 (100) | 3,447 | 45.5 |
| rVV-HA5 | 1 × 106 | 12/12 (100) | 1,600e | 83.0 |
| rVV-HA-VN | 1 × 106 | 12/12 (100) | 12,800e | 41.7 |
| vDD4-mNP | 1 × 106 | 0/12 (0) | <100 | <28 |
| Double dose | ||||
| Inact. whvv | 3 μg | 12/12 (100) | 144,815 | 246.8 |
| dVV-HA5 | 1 × 106 | 12/12 (100) | 16,127 | 257.7 |
| rVV-HA5 | 1 × 106 | 6/6 (100) | 25,600e | 226.0 |
| rVV-HA-VN | 1 × 106 | 12/12 (100) | 40,637e | 123.4 |
| vDD4-mNP | 1 × 106 | 0/12 (0) | <100 | <28 |
| VV-wtf | 1 × 106 | 0/12 (0) | <100 | <28 |
| PBSf | 0/12 (0) | <100 | <28 |
Immunization dose. Values are PFU except where otherwise indicated. See Table 1 for vaccine abbreviations.
Challenge dose, 1 × 105 TCID50, VN1203 strain; total animals of two subsequent experiments.
ELISA and microneutralization titer (μ-NT) results of three separate experiments are shown unless otherwise indicated. rH5, recombinant H5 hemagglutinin.
GMT, geometric mean titer of pooled prechallenge sera (week 6).
Results obtained from two experiments; protection of mice was highly significant in the groups immunized with HA-containing vaccines compared to that for the controls (P < 0.001).
For animal care reasons, the empty-vector and PBS challenge controls were performed only in the double-dose experiment.
After the two-dose regimen, as expected, all HA-containing vaccines again induced full protection (Table 2). The ELISA titers were on average ninefold higher with the whv vaccine and increased approximately fivefold with dVV-HA5. A large increase was also seen with the replicating controls. The neutralizing antibodies were elevated severalfold with the whv vaccine and also with the experimental dVV-HA5 and rVV-HA5 vaccines. NT levels comparable to those with the whv vaccine were achieved. An interesting observation was seen during clinical monitoring of the mice of the single-dose groups. Although complete protection was seen, mice usually displayed some clinical symptoms (including ruffled fur and arched posture, with scoring as described in Fig. 7), which were usually less severe in the dVV-vaccinated groups than in mice receiving replicating constructs. The number of protected mice in the groups immunized either with inactivated vaccine, dVV-HA5, or rVV-HA5 was significantly higher than that for the controls (P < 0.001). In summary, all HA-containing vaccines induced full protection; however, there were qualitative differences in antibody titers and the clinical outcome. The dVV vectors were equivalent to the whv vaccine in inducing neutralizing antibodies; however, the ELISA titers were somewhat lower.
FIG. 7.
Clinical disease scoring after immunization with clade 1 vaccines and challenge with wt H5N1 strains of different clades. Groups of BALB/c mice (n = 6) were intramuscularly immunized with a single dose (1 × 106 TCID50 or 3 μg) of clade 1 vaccines. Animals were challenged 6 weeks later with 1 × 105 TCID50 of VN1203, clade 1 (A); IN5/05, clade 2.1 (B); or HK156, clade 0 (C). Mice were clinically monitored for 14 days. Clinical scoring for disease development was performed according to the following scale: no disease symptoms, 0; ruffled fur, 2; ruffled fur and arched posture, 4; ruffled fur, arched posture, and apathy, 6; death, 8. Each data point represents the arithmetic mean for six animals. For abbreviations of experimental vaccines, see Table 1.
Live vaccines induce H5-specific CD8 T-cell responses.
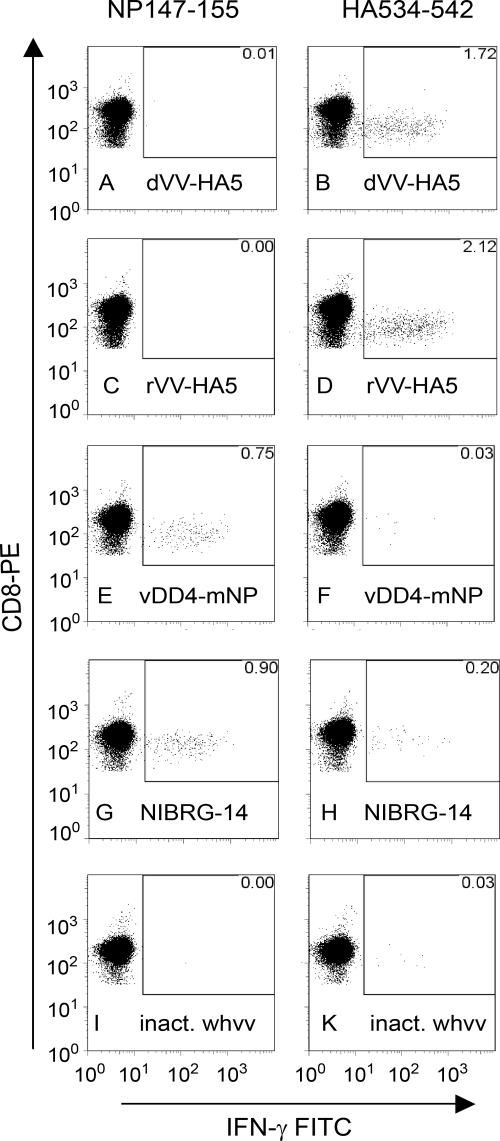
Earlier work (50) indicates that CD8 T cells contribute to viral clearance of H5N1 influenza virus in the lungs of mice and thus contribute to protective immunity. Furthermore, work using adenoviral vectors has shown that efficient CD8 T-cell responses are induced against H5N1 viruses (13). In order to investigate T-cell responses, mice were immunized twice (days 0 and 21) with the inactivated vaccine, the nonreplicating live vaccine dVV-HA5, the replicating vector rVV-HA5 or rVV-HA-VN, or the controls, including NIBRG-14 (a reverse-genetic influenza virus expressing the HA of the H5N1 Vietnam 1194 strain [32]) and vDD4-mNP (a vaccinia virus expressing the VN1203 nucleoprotein). Splenocytes were prepared on days 8 and 28 and stimulated in vitro with CD8-specific peptides. The numbers of IFN-γ-producing cells were then determined by fluorescence-activated cell sorting (FACS)-based intracellular cytokine assays.
CD8 T-cell responses to influenza virus infections in BALB/c mice are directed mainly against common epitopes on non-surface-exposed core proteins, such as the nucleoprotein, and less frequently against surface-exposed proteins, such as the highly variable HA. In this study, the CD8 responses induced against these two proteins by the vaccines indicated above were compared and contrasted. The NP immunodominant epitope, NP147-155, was used to monitor NP-specific CD8 responses, while an epitope conserved in the HAs of influenza virus subtypes 1, 5, and 9 (the 9-mer HA534-542) was used to study the HA-specific T-cell responses.
The results for the intracellular IFN-γ cytokine responses of CD8 cells induced by the different vaccines upon in vitro stimulation with the NP and HA peptides are shown in Fig. 5. As expected, only the NP-expressing vaccinia virus control (panel E) and NIBRG-14 (panel G) induced NP-specific CD8 responses. HA-specific CD8 responses were observed for the dVV-H5, rVV-H5, and NIBRG-14 live vaccines, although the responses seen for the two vaccinia virus constructs (1.72% and 2.12% of total CD8 cells for dVV-HA and rVV-HA, respectively) were considerably higher than that observed for the NIBRG-14 vaccine (0.20% of total CD8 cells). In general, the level of H5-specific T cells induced by dVV-H5 and rVV-H5 was higher than the level of NP-specific T cells induced by the vDD4-mNP vector, although the HA- and NP-specific CD8 cell frequencies were reversed in the case of the NIBRG-14 control, in agreement with the known immunodominance of the NP epitope over the HA epitope when both antigens are expressed. As expected, only minor HA-specific CD8 responses were seen with the inactivated whv vaccine, which also contains both proteins.
FIG. 5.
Intracellular cytokine analysis by FACS of influenza virus HA or NP-specific CD8 T cells from the spleens of immunized mice. The fraction of IFN-γ-producing cells within the CD8 T-cell population is shown. The first column (panels A to I) shows the results after stimulation of splenocytes with the CD8-specific peptide NP147-155 (derived from the influenza virus nucleoprotein). The second column (panels B to K) shows the results obtained with the CD8-specific peptide HA534-542 (derived from the influenza virus HA). In the NP column, only the vaccinia NP virus and the influenza virus NIBRG-14 control (panels E and G) induce IFN-γ-producing CD8 T cells; 0.75% and 0.90% of all CD8 T cells are NP specific. In the HA column, the live viruses expressing HA (dVV-HA5 [B] and rVV-HA5 [D]) induce substantial amounts of specific CD8 T cells, 1.72 and 2.12%, respectively. The vaccinia NP virus is negative (F); however, in the NIBRG-14 control, 0.20% positive T cells are found. The inactivated whv vaccine (inact. whvv) does not induce detectable levels of CD8 T cells in this system (I and K). Percentages of specific T cells are indicated in the upper-right corners.
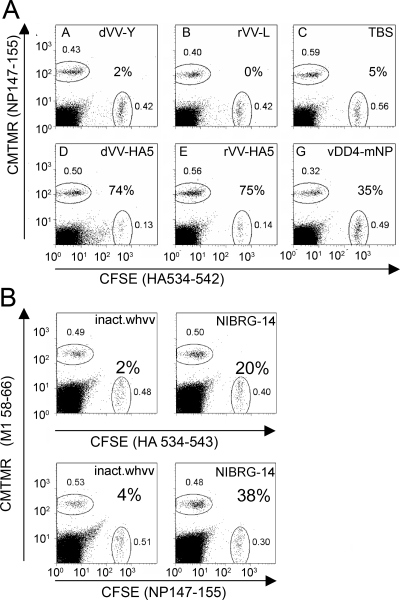
To verify that the CD8 cells were functionally competent and can kill target cells presenting specific influenza virus peptides, a cytotoxic-T-lymphocyte (CTL) killing assay based on fluorometric techniques was used (12). In this assay, splenocytes are coincubated with peptide-pulsed and dye-labeled target and control cells. A reduction in the number of target versus control cells after incubation indicates the presence of functional T cells. More specifically, splenocytes from mice immunized with influenza virus antigen-expressing and -nonexpressing vaccinia virus vaccines, NIBRG-14 or inactivated whv vaccine, were incubated with a 1:1 mixture of fluorescence-labeled target cells pulsed with specific or control peptides, and the numbers of specific target and control cells remaining after 64 h were counted and compared. For the groups immunized with vaccinia virus vaccines, CMTMR-positive (CMTMR+) target cells were loaded with the NP147-155 peptide and CSFE+ target cell were loaded with HA534-542 (Fig. 6A). The target cell populations remained equivalent after 48 h when no epitope-specific CD8 cells were induced, which was the case with the empty vaccinia virus vectors (dVV-Y and rVV-L) and buffer controls. However, the HA peptide-pulsed CSFE+ target cell populations were clearly reduced when splenocytes from mice immunized with dVV-HA5 and rVV-HA5 remaining after 64 h were counted (74 and 75% specific killing, respectively), indicating that the HA peptide-specific CD8 cells were functionally highly competent. With the splenocytes from NP vaccinia virus-vaccinated mice, 35% of the NP peptide-pulsed CMTMR+ target cells were specifically killed, indicating that the NP peptide-specific CD8 cells were also functionally competent but to a lesser extent. Because HA- and NP-specific immune responses can be induced at the same time in the groups immunized with inactivated whv vaccine or NIBRG-14, individual experiments were performed with the NP and HA peptides to assess the functionality of CD8 T cells. In this case, M1 58-66, a peptide binding to human HLA-A2, served as a control (Fig. 6B). While no significant reduction of target cells was observed for the animals immunized with inactivated whv vaccine, both HA and NP peptide-pulsed target cells were efficiently killed by T cells induced after immunization with NIBRG-14. The higher percentage of reduction with the NP peptide (38% compared to 20%) is in good agreement with the relatively higher frequency of NP peptide-specific CD8 T cells induced by NIBRG-14 (see Fig. 5).
FIG. 6.
Flow cytometric analysis of killing of peptide-pulsed target cells by specific CD8 T cells. Peptide-loaded CMTMR+ and CSFE+ splenocytes are shown as a fraction of total lymphocytes in the culture (small numbers near region gates). Percent specific killing was calculated as 100 × (1 − [cells with specific peptide/cells with control peptide]). (A) Vaccinia virus vaccine groups. (B) Control vaccine groups. For abbreviations of experimental vaccines, see Table 1.
Finally, CD4 T-helper-cell responses were also assessed by ELISPOT analysis. IFN-γ- and IL-4-producing T cells—when stimulated in vitro with either the inactivated homologous H5N1 VN1203 (clade 2.1) virus or the clade 2.2 H5N1/IN5/05 strain—were measured at day 28 (data not shown). These responses are derived primarily from influenza virus antigen-specific Th1 (IFN-γ) and Th2 (IL-4) CD4 T-helper cells. All the live vaccines induced strong and predominantly Th1 response to both strains. The inactivated whv vaccine induced a mixed-type response: both Th1 and Th2 responses were seen, although the Th2 response was dominant. The almost comparable responses following stimulation with the IN5/05 and VN1203 strains, of all the vaccines, indicated that the live and inactivated whv vaccines were both highly efficient in inducing cross-clade T-cell responses.
Cross-protection from virulent human H5N1 strains.
We further compared cross-protection induced by the experimental vaccines against H5N1 strains of different clades. The aim of this experiment was to evaluate differences in the protective potential of the clade 1-based vaccines after challenge with more divergent strains. Therefore, mice were immunized only once with the experimental vaccines and were then challenged with the different strains, including the A/HongKong/156/97[H5N1] (HK156), A/Vietnam/1203/2004[H5N1] (VN1203), and A/Indonesia/05/2005(H5N1) (IN5/05) strains. The HK156 strain (clade 0), the first virus of the H5N1 subtype isolated from a human host, is highly virulent in the mouse model and can infect mouse lungs without requiring adaptation. VN1203 is a clade 1 virus and IN5/05 a clade 2.1 virus, both highly pathogenic for humans and mice. As shown in Table 3, all mice survived, whereas the controls died. Weight monitoring, which usually permits observation of subtle differences in vaccine efficacies, was not done due to safety reasons because it requires more handling of animals under BSL3 conditions. However, the clinical outcome was monitored (Fig. 7). Challenge with the homologous VN1203 strain resulted in full protection of the animals by all vaccines; some minor clinical signs (ruffled fur) were seen with the whv vaccine (Fig. 7A). After challenge with the IN5/05 strain, animals vaccinated with dVV-HA5, rVV-HA5, and inactivated vaccine showed signs of minor to intermediate sickness (Fig. 7B). Similar results were seen after challenge with the HK156 strain (Fig. 7C). Furthermore, the CD4 T-cell responses were shown to be cross clade reactive when the IN5/05 clade 2.2 strain was used as a stimulant, consistent with the good protection against IN5/05 virus challenge (data not shown). In summary, the cross-protection experiment carried out under suboptimal (single-dose vaccination) conditions revealed good protection; however, subtle differences in protective potential against heterologous challenge virus were found by clinical monitoring.
TABLE 3.
Cross-protection results after single-dose immunizations and challenge with different H5N1 strains
| Immunizationa | Protection [no. of survivors/total (%)] against challenge withb:
|
||
|---|---|---|---|
| VN1203 | HK156 | IN5/05 | |
| Inact. whvv | 6/6 (100) | 6/6 (100) | 6/6 (100) |
| dVV-HA5 | 6/6 (100) | 6/6 (100) | 6/6 (100) |
| rVV-HA5 | 6/6 (100) | 6/6 (100) | 6/6 (100) |
| rVV-HA-VN | 6/6 (100) | 6/6 (100) | 6/6 (100) |
| VV-Lister | 0/6 (0) | 0/6 (0) | 0/6 (0) |
| PBS | 0/6 (0) | 0/6 (0) | 0/6 (0) |
See Table 1 for vaccine abbreviations.
Challenge dose, 1 × 105 TCID50; the LD50s for BALB/c mice for the HK156, VN1203, and IN5/05 strains were 11, 24, and 18 TCID50, respectively. Protection of mice was highly significant in the groups immunized with HA-containing vaccines compared to that for the controls (P < 0.001).
DISCUSSION
The majority of influenza vaccines, including the pandemic vaccine candidates, are egg-derived inactivated virus preparations. Since the egg supply may be endangered in a pandemic caused by an avian virus, production in permanent cell lines, such as Vero cells, offers an excellent alternative. Vero cells have been widely used for human vaccine production (38) and are fully accepted by regulatory authorities. The system is extremely robust, and scale-up of production of H5N1 influenza virus to 6,000 liters has been achieved (21). Furthermore, hundred of millions of doses of a second-generation smallpox vaccine were produced in Vero cells (30), supporting the use of the technology for poxviral vector production as well. Based on this experience, we chose to also develop Vero production technologies for recombinant nonreplicating poxviral vectors. To achieve growth of nonreplicating poxviruses in this cell line, the previously described defective vaccinia virus system, which was originally based on complementing rabbit kidney cells (15), was adapted to Vero cells. Complementing rabbit kidney cells, although originally derived from the kidney of a healthy rabbit (28), turned out to be tumorigenic (S. Coulibaly and F. G. Falkner, unpublished) and therefore not usable for live vaccine production. The cVero22 cell line was nontumorigenic using the most sensitive route of injection, the intramuscular route. The cell line was also negative in a PCR-enhanced reverse transcriptase assay, qualifying it for live vaccine production. Thus, a cell line suitable for production of nonreplicating vaccinia virus vectors was generated. The ability to produce the dVV-HA5 live vaccine at BSL2 is a further advantage compared to the need to grow and inactivate H5N1 virus at higher biosafety levels (21).
One of the reasons to focus on live vaccines is the good induction of cytotoxic T-cell responses by this type of vaccines. Among the H5N1 live vaccines that are currently explored are those based on poxviruses, such as modified vaccinia virus Ankara (22), and those based on adenovirus (13, 14). A replication-incompetent, adenoviral vector-based, HA subtype 5 influenza vaccine induced both humoral and cell-mediated immune responses and seems to be a promising candidate vaccine. It induced levels of HA-specific CTLs in BALB/c mice similar to those induced by dVV-HA5 (range, 1.3 to 1.7%); however, a much higher immunization dose was used (13).
Among the poxviral vectors, MVA is used mainly in live vaccine development and is as an exceptionally safe live vector. MVA has recently been used to successfully express H5N1 antigens (22, 46). This strain, however, is strongly host range restricted and is usually grown in primary chicken cells, which may be in short supply in the case of a pandemic H5N1 influenza outbreak. MVA also grows reasonably well in baby hamster kidney cells (3, 6), which are, unfortunately, tumorigenic and not usable for live vaccine production. Since MVA does not grow well in Vero cells, there are currently no methods for egg-independent production using this vector.
In this report, we have focused on an H5N1 live vaccine based on a nonreplicating poxviral vector that has a safety profile similar to that of MVA (33) and can be produced in an egg-independent way in a modified permanent Vero cell line. In the present study, BALB/c mice were immunized with low doses of the vaccinia virus constructs and challenged with a very high dose of virulent H5N1 virus (105 TCID50 per animal). Even after a single dose of dVV-HA5, mice were protected from death and severe clinical signs against homologous challenge, comparable to a results with successful inactivated whv vaccine (7). Good protection correlated with neutralizing antibodies. Prime-boost immunizations led to robust protection with all experimental vaccines. As expected, the live vaccines induced substantial levels of HA-specific CD8 CTL responses, while the inactivated whv vaccine gave rise to poor HA-specific CD8 responses but induced considerable Th1 and even higher Th2 CD4 responses. The CD8 responses, especially the HA-peptide-specific response induced by the dVV-HA5 and rVV-HA5 vaccines, were shown to be functionally highly competent, confirming the supportive role of cytotoxic T cells in fighting disease by killing infected cells.
Cross-protection was also studied. After single-dose immunization with a clade 1 vaccine, full cross-protection against death with clade 0 and 2.1 challenge viruses was obtained. Some clinical signs of sickness were observed with the nonreplicating vaccines after cross-clade challenge, but this was not seen with the replicating controls. Complete cross-protection from death and disease is usually obtained in this system by vaccinations with two doses.
Temporal and quantitative gene expression of relevant antigens, induction of apoptosis in host cells, including dendritic cells, and de novo presentation and cross-presentation are some of the parameters determining the outcome of immunizations with viral vectors. Strong and sustained viral early gene expression is important for a good CD8 T-cell response (34), and high-level expression in general is important for an overall good immune response (47). Anti-vaccinia virus CTL responses (in humans and mice) are directed mainly against early gene products (36, 45). In nonpermissive (normal) host cells, defective vaccinia virus induces early genes that are expressed for a prolonged period of time. Late gene expression does not occur. Therefore, despite the nonreplicating nature, induction of T cells is very efficient and is directed against the relevant targets. Ideally, in a vectored vaccine, the response to the foreign gene should be dominant, while antivector responses should remain in the background. This goal is difficult to achieve with complex viral vectors. One step in this direction is the use of nonreplicating vectors deficient in expressing a whole class of antigens. However, the most efficient viral CD8 targets in mice and humans are early genes, mainly involved in virulence and gene regulation (36, 45), implying that the antivector response to early antigens also is strong. For a reasonable balance between the vector and the foreign antigen, presumably the presence of strong CD8 epitopes in the foreign gene and strong early/late expression is required to confer solid protection. The HA gene controlled by the strong early/late promoter (48) seems to fulfill these criteria. Moreover, since vaccinia virus inhibits maturation of dendritic cells and induces rapid apoptosis and thus direct presentation of antigen, cross-presentation of antigen by infected cells plays a major role in induction of immunity (10, 23). Due to its unique features, including temporal gene expression, dVV vectors seem to focus the immune response to the relevant antigens and epitopes either by directly presenting or by cross-presenting early antigen-enriched material.
Preexisting immunity to vaccinia virus may further influence the outcome of vaccinations with recombinant vaccinia virus vectors (see, for instance, references 2, 4, 19, 35, and 39). This is relevant mainly with replicating vaccines administered by scarification, which require local replication for induction of immunity. In this case, preexisting neutralizing antibodies prevent viremic spread and vaccine efficacy, resulting in no “take” after smallpox revaccination (35) or no detectable antibodies to the foreign gene with recombinant vectors (4). Further, in a previous clinical study, several vaccinia virus immune volunteers did not seroconvert for neutralizing antibodies after booster injection with a chicken whole-cell lysate vaccine (containing low titers of vaccinia virus and relatively high titers of residual antigen) (19). Currently manufactured, nonreplicating poxviral vaccines typically contain 100- to 1,000-fold-higher titers and contain far less residual protein; therefore, the outcome of this study may not represent the performance of current vaccines. Moreover, nonreplicating vectors, such as MVA, seem to tolerate preexisting immunity. Although vaccine efficacy was somewhat limited, presumably by a reduced virus input that could reach the target tissues during secondary vaccination, reuse of MVA was possible (39). In summary, preexisting immunity against vaccinia virus may be overcome by using the optimal vaccination route, higher dosing, and state-of-the-art nonreplicating vaccines.
H5N1 vaccines based on recombinant nonreplicating viral vectors are promising candidate vaccines and induce a broad immune response, including cytotoxic CD8 T cells. In the case of defective vaccinia virus, a scale-up of vector production using the complementing Vero cell approach is desirable. Furthermore, it will be interesting to directly compare different categories of H5N1 vaccines, including those based on MVA and dVV vectors or adenovirus, under the same conditions.
Acknowledgments
We thank Robert Schmid and Bianca Hube for the NT assays, Nicole Hetzelt and Mandy Seugling for the ELISA assays, Karl Schmid for titrations and cell culture, D. Fritz for FACS analyses, and I. Bacanovic, E. Schweitzer, and S. Bader for expert technical assistance. We also thank K. Dinh Duy (Institute of Biotechnology, Hanoi, Vietnam) and S. K. Lal (International Centre for Genetic Engineering & Biotechnology, New Delhi, India) for providing a plasmid that contains the HA5 gene of A/chicken/Hatay/2004 (H5N1).
Footnotes
Published ahead of print on 11 March 2009.
REFERENCES
- 1.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244365-396. [DOI] [PubMed] [Google Scholar]
- 2.Belyakov, I. M., B. Moss, W. Strober, and J. A. Berzofsky. 1999. Mucosal vaccination overcomes the barrier to recombinant vaccinia immunization caused by preexisting poxvirus immunity. Proc. Natl. Acad. Sci. USA 964512-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll, M. W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 238198-211. [DOI] [PubMed] [Google Scholar]
- 4.Cooney, E. L., A. C. Collier, P. D. Greenberg, R. W. Coombs, J. Zarling, D. E. Arditti, M. C. Hoffman, S. L. Hu, and L. Corey. 1991. Safety of and immunological response to a recombinant vaccinia virus vaccine expressing HIV envelope glycoprotein. Lancet 337567-572. [DOI] [PubMed] [Google Scholar]
- 5.De, B. K., M. W. Shaw, P. A. Rota, M. W. Harmon, J. J. Esposito, R. Rott, N. J. Cox, and A. P. Kendal. 1988. Protection against virulent H5 avian influenza virus infection in chickens by an inactivated vaccine produced with recombinant vaccinia virus. Vaccine 6257-261. [DOI] [PubMed] [Google Scholar]
- 6.Drexler, I., K. Heller, B. Wahren, V. Erfle, and G. Sutter. 1998. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J. Gen. Virol. 79347-352. [DOI] [PubMed] [Google Scholar]
- 7.Ehrlich, H. J., M. Muller, H. M. Oh, P. A. Tambyah, C. Joukhadar, E. Montomoli, D. Fisher, G. Berezuk, S. Fritsch, A. Low-Baselli, N. Vartian, R. Bobrovsky, B. G. Pavlova, E. M. Pollabauer, O. Kistner, and P. N. Barrett. 2008. A clinical trial of a whole-virus H5N1 vaccine derived from cell culture. N. Engl. J. Med. 3582573-2584. [DOI] [PubMed] [Google Scholar]
- 8.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43247-252. [DOI] [PubMed] [Google Scholar]
- 9.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 739679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasteiger, G., W. Kastenmuller, R. Ljapoci, G. Sutter, and I. Drexler. 2007. Cross-priming of cytotoxic T cells dictates antigen requisites for modified vaccinia virus Ankara vector vaccines. J. Virol. 8111925-11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, F. L. 2000. Adenovirus vectors for high-efficiency gene transfer into mammalian cells. Immunol. Today 21426-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermans, I. F., J. D. Silk, J. Yang, M. J. Palmowski, U. Gileadi, C. McCarthy, M. Salio, F. Ronchese, and V. Cerundolo. 2004. The VITAL assay: a versatile fluorometric technique for assessing CTL- and NKT-mediated cytotoxicity against multiple targets in vitro and in vivo. J. Immunol. Methods 28525-40. [DOI] [PubMed] [Google Scholar]
- 13.Hoelscher, M. A., S. Garg, D. S. Bangari, J. A. Belser, X. Lu, I. Stephenson, R. A. Bright, J. M. Katz, S. K. Mittal, and S. Sambhara. 2006. Development of adenoviral-vector-based pandemic influenza vaccine against antigenically distinct human H5N1 strains in mice. Lancet 367475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoelscher, M. A., L. Jayashankar, S. Garg, V. Veguilla, X. Lu, N. Singh, J. M. Katz, S. K. Mittal, and S. Sambhara. 2007. New pre-pandemic influenza vaccines: an egg- and adjuvant-independent human adenoviral vector strategy induces long-lasting protective immune responses in mice. Clin. Pharmacol. Ther. 82665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holzer, G. W., and F. G. Falkner. 1997. Construction of a vaccinia virus deficient in the essential DNA repair enzyme uracil DNA glycosylase by a complementing cell line. J. Virol. 714997-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holzer, G. W., W. Gritschenberger, J. A. Mayrhofer, V. Wieser, F. Dorner, and F. G. Falkner. 1998. Dominant host range selection of vaccinia recombinants by rescue of an essential gene. Virology 249160-166. [DOI] [PubMed] [Google Scholar]
- 17.Holzer, G. W., J. Mayrhofer, W. Gritschenberger, and F. G. Falkner. 2005. Dominant negative selection of vaccinia virus using a thymidine kinase/thymidylate kinase fusion gene and the prodrug azidothymidine. Virology 337235-241. [DOI] [PubMed] [Google Scholar]
- 18.Horimoto, T., and Y. Kawaoka. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3591-600. [DOI] [PubMed] [Google Scholar]
- 19.Kanesa-thasan, N., J. J. Smucny, C. H. Hoke, D. H. Marks, E. Konishi, I. Kurane, D. B. Tang, D. W. Vaughn, P. W. Mason, and R. E. Shope. 2000. Safety and immunogenicity of NYVAC-JEV and ALVAC-JEV attenuated recombinant Japanese encephalitis virus-poxvirus vaccines in vaccinia-nonimmune and vaccinia-immune humans. Vaccine 19483-491. [DOI] [PubMed] [Google Scholar]
- 20.Kistner, O., P. N. Barrett, W. Mundt, M. Reiter, S. Schober-Bendixen, G. Eder, and F. Dorner. 1999. Development of a Vero cell-derived influenza whole virus vaccine. Dev. Biol. Stand. 98101-110. [PubMed] [Google Scholar]
- 21.Kistner, O., M. K. Howard, M. Spruth, W. Wodal, P. Bruhl, M. Gerencer, B. A. Crowe, H. Savidis-Dacho, I. Livey, M. Reiter, I. Mayerhofer, C. Tauer, L. Grillberger, W. Mundt, F. G. Falkner, and P. N. Barrett. 2007. Cell culture (Vero) derived whole virus (H5N1) vaccine based on wild-type virus strain induces cross-protective immune responses. Vaccine 256028-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreijtz, J. H., Y. Suezer, G. van Amerongen, G. de Mutsert, B. S. Schnierle, J. M. Wood, T. Kuiken, R. A. Fouchier, J. Lower, A. D. Osterhaus, G. Sutter, and G. F. Rimmelzwaan. 2007. Recombinant modified vaccinia virus Ankara-based vaccine induces protective immunity in mice against infection with influenza virus H5N1. J. Infect. Dis. 1951598-1606. [DOI] [PubMed] [Google Scholar]
- 23.Larsson, M., J. F. Fonteneau, S. Somersan, C. Sanders, K. Bickham, E. K. Thomas, K. Mahnke, and N. Bhardwaj. 2001. Efficiency of cross presentation of vaccinia virus-derived antigens by human dendritic cells. Eur. J. Immunol. 313432-3442. [DOI] [PubMed] [Google Scholar]
- 24.Levenbook, I., P. L. Smith, and J. C. Petricciani. 1981. A comparison of three routes of inoculation for testing tumorigenicity of cell lines in nude mice. J. Biol. Stand. 975-80. [DOI] [PubMed] [Google Scholar]
- 25.Levenbook, I. S., J. C. Petricciani, and B. L. Elisberg. 1984. Tumorigenicity of Vero cells. J. Biol. Stand. 12391-398. [DOI] [PubMed] [Google Scholar]
- 26.Lipatov, A. S., E. A. Govorkova, R. J. Webby, H. Ozaki, M. Peiris, Y. Guan, L. Poon, and R. G. Webster. 2004. Influenza: emergence and control. J. Virol. 788951-8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, X., T. M. Tumpey, T. Morken, S. R. Zaki, N. J. Cox, and J. M. Katz. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 735903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy, K., C. H. Taylor-Robinson, and S. E. Pillinger. 1963. Isolation of Rubella virus from cases in Britain. Lancet ii593-598. [DOI] [PubMed] [Google Scholar]
- 29.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7980-990. [PMC free article] [PubMed] [Google Scholar]
- 30.Monath, T. P., J. R. Caldwell, W. Mundt, J. Fusco, C. S. Johnson, M. Buller, J. Liu, B. Gardner, G. Downing, P. S. Blum, T. Kemp, R. Nichols, and R. Weltzin. 2004. ACAM2000 clonal Vero cell culture vaccinia virus (New York City Board of Health strain)—a second-generation smallpox vaccine for biological defense. Int. J. Infect. Dis. 8(Suppl. 2)S31-S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 969345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicolson, C., D. Major, J. M. Wood, and J. S. Robertson. 2005. Generation of influenza vaccine viruses on Vero cells by reverse genetics: an H5N1 candidate vaccine strain produced under a quality system. Vaccine 232943-2952. [DOI] [PubMed] [Google Scholar]
- 33.Ober, B. T., P. Bruhl, M. Schmidt, V. Wieser, W. Gritschenberger, S. Coulibaly, H. Savidis-Dacho, M. Gerencer, and F. G. Falkner. 2002. Immunogenicity and safety of defective vaccinia virus Lister: comparison with modified vaccinia virus Ankara. J. Virol. 767713-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obst, R., H. M. van Santen, R. Melamed, A. O. Kamphorst, C. Benoist, and D. Mathis. 2007. Sustained antigen presentation can promote an immunogenic T cell response, like dendritic cell activation. Proc. Natl. Acad. Sci. USA 10415460-15465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orr, N., M. Forman, H. Marcus, S. Lustig, N. Paran, I. Grotto, E. Klement, Y. Yehezkelli, G. Robin, S. Reuveny, A. Shafferman, and D. Cohen. 2004. Clinical and immune responses after revaccination of Israeli adults with the Lister strain of vaccinia virus. J. Infect. Dis. 1901295-1302. [DOI] [PubMed] [Google Scholar]
- 36.Oseroff, C., F. Kos, H. H. Bui, B. Peters, V. Pasquetto, J. Glenn, T. Palmore, J. Sidney, D. C. Tscharke, J. R. Bennink, S. Southwood, H. M. Grey, J. W. Yewdell, and A. Sette. 2005. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc. Natl. Acad. Sci. USA 10213980-13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palese, P., and A. Garcia-Sastre. 2002. Influenza vaccines: present and future. J. Clin. Investig. 1109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plotkin, S. A., A. Murdin, and E. Vidor. 1999. Inactivated polio vaccine, p. 345-363. In S. A. Plotkin and W. A. Orenstein (ed.), Vaccines, 3rd ed. W. B. Saunders, Philadelphia, PA.
- 39.Ramirez, J. C., M. M. Gherardi, D. Rodriguez, and M. Esteban. 2000. Attenuated modified vaccinia virus Ankara can be used as an immunizing agent under conditions of preexisting immunity to the vector. J. Virol. 747651-7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 41.Rubinstein, S., and M. Schoofs. 2007. Canceled vaccine may have boosted HIBV risk. Wall Street J. http://online.wsj.com/article/SB119445916050785399.html.
- 42.Sheets, R. 2000. History and characterization of the Vero cell line. Report prepared for the Vaccines and Related Biological Products Advisory Committee Meeting. FDA, Rockville, MD. http://www.fda.gov/ohrms/dockets/AC/00/backgrd/3616b1a.pdf.
- 43.Sutter, G., and B. Moss. 1992. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. USA 8910847-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutter, G., L. S. Wyatt, P. L. Foley, J. R. Bennink, and B. Moss. 1994. A recombinant vector derived from the host range-restricted and highly attenuated MVA strain of vaccinia virus stimulates protective immunity in mice to influenza virus. Vaccine 121032-1040. [DOI] [PubMed] [Google Scholar]
- 45.Tscharke, D. C., W. P. Woo, I. G. Sakala, J. Sidney, A. Sette, D. J. Moss, J. R. Bennink, G. Karupiah, and J. W. Yewdell. 2006. Poxvirus CD8+ T-cell determinants and cross-reactivity in BALB/c mice. J. Virol. 806318-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veits, J., A. Romer-Oberdorfer, D. Helferich, M. Durban, Y. Suezer, G. Sutter, and T. C. Mettenleiter. 2008. Protective efficacy of several vaccines against highly pathogenic H5N1 avian influenza virus under experimental conditions. Vaccine 261688-1696. [DOI] [PubMed] [Google Scholar]
- 47.Wyatt, L. S., P. L. Earl, J. Vogt, L. A. Eller, D. Chandran, J. Liu, H. L. Robinson, and B. Moss. 2008. Correlation of immunogenicities and in vitro expression levels of recombinant modified vaccinia virus Ankara HIV vaccines. Vaccine 26486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wyatt, L. S., S. T. Shors, B. R. Murphy, and B. Moss. 1996. Development of a replication-deficient recombinant vaccinia virus vaccine effective against parainfluenza virus 3 infection in an animal model. Vaccine 141451-1458. [DOI] [PubMed] [Google Scholar]
- 49.Yamada, S., Y. Suzuki, T. Suzuki, M. Q. Le, C. A. Nidom, Y. Sakai-Tagawa, Y. Muramoto, M. Ito, M. Kiso, T. Horimoto, K. Shinya, T. Sawada, M. Kiso, T. Usui, T. Murata, Y. Lin, A. Hay, L. F. Haire, D. J. Stevens, R. J. Russell, S. J. Gamblin, J. J. Skehel, and Y. Kawaoka. 2006. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444378-382. [DOI] [PubMed] [Google Scholar]
- 50.Yap, K. L., and G. L. Ada. 1978. The recovery of mice from influenza A virus infection: adoptive transfer of immunity with influenza virus-specific cytotoxic T lymphocytes recognizing a common virion antigen. Scand. J. Immunol. 8413-420. [DOI] [PubMed] [Google Scholar]
- 51.Yuen, L., and B. Moss. 1987. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc. Natl. Acad. Sci. USA 846417-6421. [DOI] [PMC free article] [PubMed] [Google Scholar]