Abstract
Herpes simplex virus 1 (HSV-1) nucleocapsids exit the nucleus by budding into the inner nuclear membrane, where they exist briefly as primary enveloped virions. These virus particles subsequently fuse their envelopes with the outer nuclear membrane, permitting nucleocapsids to then enter the cytoplasm and complete assembly. We have developed a method to isolate primary enveloped virions from HSV-1-infected cells and subjected the primary enveloped virion preparation to MALDI-MS/MS (matrix-assisted laser desorption ionization-tandem mass spectrometry) analyses. We identified most capsid proteins, a tegument protein (VP22), a glycoprotein (gD), and a cellular protein (annexin A2) in the primary enveloped virion preparation. We determined that annexin A2 does not play an essential role in infection under our experimental conditions. Elucidating the structure and biochemical properties of this unique virus assembly intermediate will provide new insights into HSV-1 biology.
Herpes simplex virus 1 (HSV-1) is a large virus containing a double-stranded DNA genome enclosed within an icosahedral capsid, which is in turn surrounded by an amorphous layer of proteins, termed the tegument. The outermost structural component is an envelope composed of a lipid bilayer and derived from a host cell organelle.
HSV-1 has a complicated life cycle, which involves interaction with many host cell organelles and disruption of multiple host cell processes. During the initial stages of infection, viral capsids travel on microtubules and dock at the nuclear pore, whereupon the viral genome is injected into the nucleus (38) and DNA replication and transcription occur. Several viral proteins, including pUL6, pUL15, pUL25, pUL28, and pUL33, are required for cleavage and packaging of the newly replicated viral DNA into preassembled procapsids in the nucleus (25, 34). The newly assembled nucleocapsids then travel on actin cables toward the inner nuclear membrane (6). Three viral proteins (pUL31, pUL34, and pUs3) and the cellular protein kinase C work in coordination to rearrange the nuclear lamina to allow capsids access to the inner nuclear membrane (26, 29, 33). The functions of pUL31, pUL34, and pUs3 seem to be conserved among several other herpesviruses, such as human cytomegalovirus virus (HCMV) and pseudorabies virus (PrV) (18, 23), and the interaction between pUL34 and pUL31 which occurs during nuclear egress is essential for HSV-1 virions to complete assembly (30, 32). Finally, in a step unique to herpesviruses, the capsid buds into the inner nuclear membrane to exist transiently as a primary enveloped virion in the perinuclear space. The nuclear-membrane-derived envelope is then lost by fusion with the outer nuclear membrane (36), and the nucleocapsids are delivered into the cytoplasm. Once in the cytoplasm, they acquire a second and final envelope, most likely derived from endosomes or the trans-Golgi network (2, 11, 28, 39-41).
It has been shown that primary enveloped virions of HSV-1 lying between the two nuclear membranes appear to be morphologically distinct from mature virions (10). The differences are also apparent between primary enveloped virions and mature virions for other members of the herpesvirus family, including PrV, equine herpesvirus, and gallid infectious laryngotracheitis virus (10). The structure of the perinuclear HSV-1 particle is poorly defined. However, several components of the primary enveloped virion have been elucidated using immunogold electron microscopy (EM) and immunofluorescent microscopy (8, 24, 31). In PrV, pUL31 and pUL34 can form vesicles in the perinuclear space when stably expressed in RK-13 cells, indicating that they may compose the essential budding machinery (16), while mutant HSV-1 particles lacking the envelope glycoproteins gB and gH accumulate in the perinuclear space, indicating that these proteins either work together or redundantly to promote fusion with the outer nuclear membrane (5).
The use of proteomic approaches in the study of viruses has gained momentum in recent years (21). There have been extensive proteomics studies for various members of the herpesvirus family, such as HCMV, Epstein-Barr virus, and Kaposi's sarcoma-associated herpesvirus, among others (1, 14, 42). However, it was not until recently that these types of large-scale proteomic analyses were carried out for PrV and HSV-1 (19, 37). The aim of this study was to isolate the HSV-1 primary enveloped virion and perform whole-particle proteomic analysis to elucidate its components. In order to attempt this type of analysis of the primary enveloped virion, we first isolated nuclear envelopes from infected cells and then harvested primary enveloped virions from the nuclear membranes. After extensive testing of the efficacy of the preparation, limited proteomic analysis using MALDI-MS/MS (matrix-assisted laser desorption ionization-tandem mass spectrometry) was initiated. We found that the primary enveloped virion preparation contains known viral proteins as well as the cellular protein annexin A2, one of a large family of proteins that bind calcium and associate tightly with membranes (9). Isolation of the primary enveloped virions provides a new approach to studying the events of nuclear egress and virion assembly.
MATERIALS AND METHODS
Cells and viruses.
COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% penicillin-streptomycin (PS) and 10% fetal bovine serum (FBS). Vero cells were grown in DMEM supplemented with 1% PS and 10% newborn calf serum. LnCAP cells were grown in RPMI 1640 with 1% PS and 10% FBS. The HSV-1 strain SC16 was grown and titers determined on Vero cells. Mutant viruses Hr81-2 (ΔUL15), KΔUL36, and SC16ΔgH were grown and titers determined on their complementing cell lines, M3, HS30, and M6, respectively.
Isolation of nuclear membranes.
The procedure for isolation of nuclear membranes was derived from a previously published method (15). COS-7 cells were infected at a multiplicity of infection (MOI) of 10 for 18 h and incubated at 37°C. Cells were washed, harvested, and pelleted in sucrose buffer A (10 mM Tris [pH 7.4], 0.32 M sucrose, 3 mM MgCl2). Pelleted cells were resuspended in fresh sucrose buffer A and 60 μl protease inhibitor cocktail (AEBSF [4-(2-aminoethyl) benzenesulfonyl fluoride], pepstatin A, E-64, bestatin, leupeptin, aprotinin) per 6.0 × 107 cells in accordance with the manufacturer's recommendations (Sigma-Aldrich, St. Louis, MO) and then manually disrupted using a 25 5/8-gauge needle. A portion of the lysed cells was set aside for subsequent studies and was termed total cell extract (TCE). The lysed cells in sucrose buffer A were then subjected to centrifugation for 10 min at 700 × g in order to pellet the nuclei. The crude nuclear pellet was resuspended in sucrose buffer B (10 mM Tris [pH 7.4], 2.4 M sucrose, 3 mM MgCl2) and then ultracentrifuged at 50,000 × g for 1 h at 4°C. The purified nuclear pellet was resuspended in sucrose buffer C (10 mM Tris [pH 7.4], 0.25 M sucrose, 3 mM MgCl2) and centrifuged at 700 × g for 10 min for a final wash. The washed nuclear pellet was resuspended in 1 mM DNase I in 0.1 mM MgCl2 and digestion buffer (10 mM Tris [pH 8.0], 5 mM β-mercaptoethanol, 0.3 M sucrose). Incubation with DNase I and digestion buffer proceeded at room temperature for 20 min, and then nuclear membranes were ultracentrifuged at 40,000 × g for 15 min. The digestion incubation was repeated for 1 h at room temperature, and the membranes were ultracentrifuged again for 15 min. The infected nuclear membrane pellet was then resuspended in sucrose buffer C. Throughout the procedure, all the supernatants from each centrifugation step were collected and pooled. A small volume (250 μl) of those pooled supernatants was subjected to trichloroacetic acid precipitation and then resuspended in Tris-EDTA, pH 8.0. This fraction was termed the intermediate fraction (INT). At the end of the procedure, there were thus three fractions: TCE, INT, and infected nuclear membrane preparation (NMP).
Isolation of primary enveloped virions.
The resuspended NMP was sonicated to break apart the envelopes and release the perinuclear HSV-1 particles. The material was then incubated with 50 μg/ml proteinase K on ice for 1 h, and the proteinase K was quenched with 1 mM phenylmethylsulfonyl fluoride. Finally, the preparation was overlaid on a 30% sucrose cushion (10 mM Tris [pH 7.4], 30% sucrose [wt/vol]) and ultracentrifuged at 40,000 × g for 1 h at 4°C. The sucrose cushion was discarded, and the remaining pellet of primary enveloped virions was resuspended in sucrose buffer C. This fraction was termed the perinuclear HSV-1 preparation (pnHSV-1).
Isolation of extracellular virions.
Medium was collected from cells which had been infected at an MOI of 10 with the SC16 wild-type strain of HSV-1 and was cleared by centrifugation at 1,000 × g for 15 min. The cleared medium was then ultracentrifuged at 100,000 × g for 3 h at 4°C in a Beckman SW28 rotor. For Fig. 3C, pelleted extracellular virions were resuspended in medium without serum, overlaid on a 30% sucrose cushion (10 mM Tris [pH 7.4], 30% sucrose [wt/vol]), and ultracentrifuged at 40,000 × g for 1 h at 4°C. The final pellet was resuspended in an isotonic sucrose solution (10 mM Tris [pH 7.4], 0.25 M sucrose, and 3 mM MgCl2).
FIG. 3.
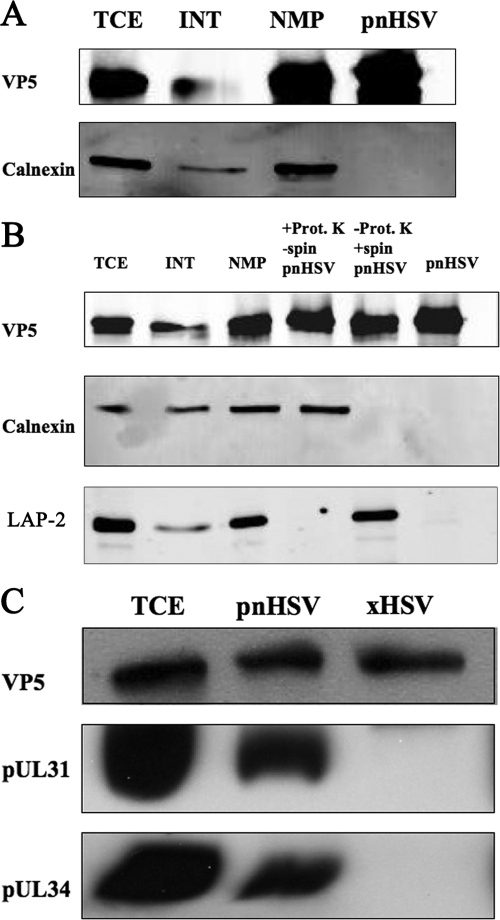
Isolation of primary enveloped virions from infected nuclear membranes. (A) Western blot of SC16-infected TCE, INT, NMP, and pnHSV-1. Fractions were probed for the viral capsid protein VP5 and the nuclear membrane marker and endoplasmic reticulum marker calnexin. (B) Western blot of TCE, INT, NMP, pnHSV-1, and two variations of the pnHSV-1: (i) pnHSV plus proteinase K without centrifugation (−spin) and (ii) pnHSV without proteinase K and with centrifugation (+spin). Blots were probed for VP5, calnexin, and LAP-2, a nuclear membrane marker, as indicated at left. (C) TCE, pnHSV, and xHSV-1 were subjected to Western blotting. The Western blots were probed for viral capsid protein VP5 and for pUL31 and pUL34, markers of primary enveloped virions.
For Fig. 5A, a modification of a recently published method was employed (19). After centrifugation for 3 h at 100,000 × g in a SW28 rotor, the pellet was resuspended in STE buffer (10 mM Tris [pH 7.4], 50 mM NaCl, 5 mM EDTA) and sonicated to break up viral aggregates. The mixture was filtered through a 0.8-μm filter, then overlaid on a 10% Ficoll 400 cushion in 10 mM Tris [pH 7.4], and centrifuged at 26,000 × g for 2 h at 4°C in a SW41 rotor.
FIG. 5.
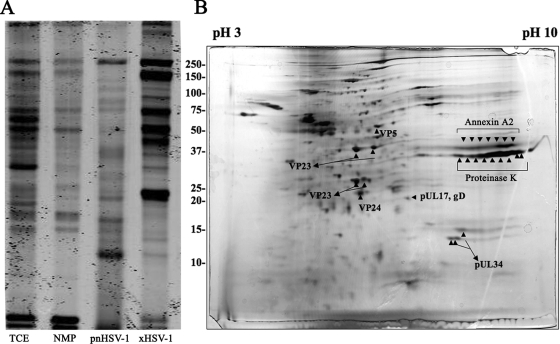
Analysis of primary enveloped virion protein profile. (A) Coomassie-stained 10% SDS-PAGE gel shows the protein profile of TCE, NMP, pnHSV-1, and xHSV-1. (B) Silver-stained 2D gel displays the pnHSV-1 fraction protein profile. Arrowheads denote the spots picked that corresponded with the proteins identified and listed in Table 1.
Analysis of viral DNA.
DNA was prepared and subjected to Southern blot analysis as previously described (4). A DNA probe which hybridizes with the BamHI SQ junction fragment was used for Southern blot analysis (kindly provided by Sandra K. Weller).
Western blotting.
Fractions were boiled in Laemmli buffer and loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane or an Immobilon-FL transfer membrane (Millipore, Billerica, MA). The membrane was blocked in 4% milk in either Tris-buffered or phosphate-buffered saline supplemented with Tween and then incubated with the appropriate antibody overnight at 4°C. The PVDF membrane was then incubated with peroxidase-conjugated secondary antibody for 1 h. Alternatively, the Immobilon-FL membranes were incubated with fluorescent-conjugated secondary antibodies for 1 h in the dark. PVDF membranes were developed using chemiluminescence, while the Immobilon-FL membranes were imaged with an Odyssey infrared imaging system and software (Licor Biosciences, Lincoln, NE). Primary antibodies used include mouse anti-VP5, rabbit anti-gH, rabbit anti-pUL31, chicken anti-pUL34, mouse anti-Rab 5, mouse anti-α-tubulin, mouse anti-β-Cop, mouse anti-PCNA, mouse anti-LAP-2, rabbit anticalnexin, and mouse anti-annexin A2; secondary antibodies include peroxidase-conjugated anti-mouse immunoglobulin G (IgG), anti-rabbit IgG, anti-chicken IgG, and Alex Fluor 680-conjugated goat anti-mouse IgG and IRDye800-conjugated goat anti-rabbit IgG.
Determination of PFU production.
Viral extracts were prepared by infecting cells at an MOI of 0.01 and harvesting the infected cells after 1 h at 37°C or 72 h at 37°C, when the infection had progressed to full cytopathic effect (CPE). Confluent monolayers of Vero cells were infected in duplicate with serial dilutions of viral extracts for 1 h at 37°C. Then cells were overlaid with DMEM-PS-FBS containing carboxymethyl cellulose and incubated for 72 h. Cells were then fixed in 10% formyl saline and stained with 0.1% toluidine blue to visualize plaques.
2D gel electrophoresis.
The primary enveloped virion fraction was subjected to two-dimensional (2D) gel electrophoresis. Fifty micrograms of the fraction was applied to a 13-cm Immobiline dry gel strip, pH 3-10L (GE Healthcare Biosciences AB, Uppsala, Sweden). The strip was focused in an IPGphor isoelectric focusing system (GE Healthcare Biosciences AB). The proteins were diffused into the strip using active rehydration, and isoelectric focusing occurred overnight with a total voltage-hours of approximately 15 kV·h. The isoelectric focusing gel strips were rehydrated, equilibrated, and stained as previously described (20). The strip was overlaid on a 13% SDS-PAGE gel and was run until the dye front reached the bottom of the gel. The gel was silver stained as previously described (35).
MALDI-MS/MS analysis.
The most intensely stained 36 spots from the 2D gel were picked, destained, and subjected to in-gel trypsin digestion as previously described (13). The reaction was stopped with 0.1% trifluoroacetic acid. The peptides in 50 mM ammonium bicarbonate (pH 8.5) were pipetted onto a C18 ZipTip (Millipore, Billerica, MA) and eluted with α-cyano-4-hydroxycinnamic acid in 50% (vol/vol) acetonitrile-water plus 0.1% trifluoroacetic acid (Agilent Technologies, Santa Clara, CA) directly onto the MALDI-MS/MS target plate. Spotted peptides were analyzed with a 4700 proteomics analyzer (Applied Biosystems, Foster City, CA). The MALDI-TOF/TOF (MALDI-tandem time-of-flight) mass spectrometer was operated in reflector mode with an accelerating voltage of 20 kV and using delayed extraction. An in-house copy of the MASCOT search engine, version 1.8 (Matrix Science Ltd., United Kingdom), using the NCBInr database was employed to identify peptides. Carbamidomethylation was set as a fixed modification for cysteine residues; mass tolerance was 100 ppm for MS data and 300 ppm for MS/MS data. Proteins were considered identified if the MOWSE (molecular weight search) score was significant (P > 0.05). Spectra corresponding to unique peptides were manually validated.
EM.
The INT, NMP, and primary enveloped virion preparations were subjected to EM. Fractions were fixed in 2.5% glutaraldehyde-0.1 M sodium cacodylate for 45 min, rinsed in 0.1 M sodium cacodylate, postfixed in 1% osmium tetroxide-0.1 M sodium cacodylate, stained in 1% uranyl acetate, dehydrated, embedded, and sectioned. Images were examined under a JEOL CX100 or JEOL 1200EX transmission electron microscope.
Immunogold EM.
A primary enveloped virion preparation was fixed in 4% paraformaldehyde, 0.05% glutaraldehyde, and 0.1 M sodium cacodylate, rinsed, blocked with 0.05 M glycine, and embedded in 10% gelatin. Gelatin cubes were then infiltrated with 2.3 M sucrose, frozen in liquid nitrogen on stubs, and sectioned. Sections on nickel grids were rinsed, glycine blocked, blocked with 1% BSA, and then incubated with primary antibody in 1% BSA. They were then rinsed, incubated with 10-nm-gold-conjugated goat anti-mouse IgG, rinsed, contrast stained with 2% aqueous uranyl acetate, embedded in 0.75% methyl cellulose, and observed in a JEOL 1200 EX transmission microscope.
RESULTS
Preparation of infected nuclear membranes is devoid of contaminant cellular markers.
The method used to prepare HSV-1-infected nuclear membranes was derived from a protocol developed to study rat liver nuclei (15). COS-7 cells, infected at an MOI of 10, were harvested 18 h postinfection into a sucrose buffer and manually disrupted using a 25 5/8-gauge needle, yielding a TCE. Nuclei were pelleted and subjected to serial washes. Pure nuclei were then subjected to two rounds of DNase I digestion at pH 8.0 to rid the preparation of chromatin and to release the nuclear envelopes, resulting in the NMP. Supernatants obtained at each stage were pooled, resulting in a combined INT.
The NMP was subjected to several tests to determine its purity. First, we used Western blotting to test for the presence of classical cell compartment markers. These markers include Rab5 (early endosomes), α-tubulin (cytoplasm), β-COP (Golgi apparatus), proliferating cell nuclear antigen (nucleoplasm), and lamin-associated protein 2 (LAP-2), an integral membrane protein found only in nuclear membranes. Figure 1 is a representative blot in which all of these compartment markers were present in the TCE and INT fractions, as expected. However, all markers were absent from the NMP fraction, with the exception of LAP-2, the nuclear membrane marker. This demonstrates that soluble and membrane-associated components of the cytoplasm and nucleoplasm had been removed, leaving only the nuclear envelope.
FIG. 1.
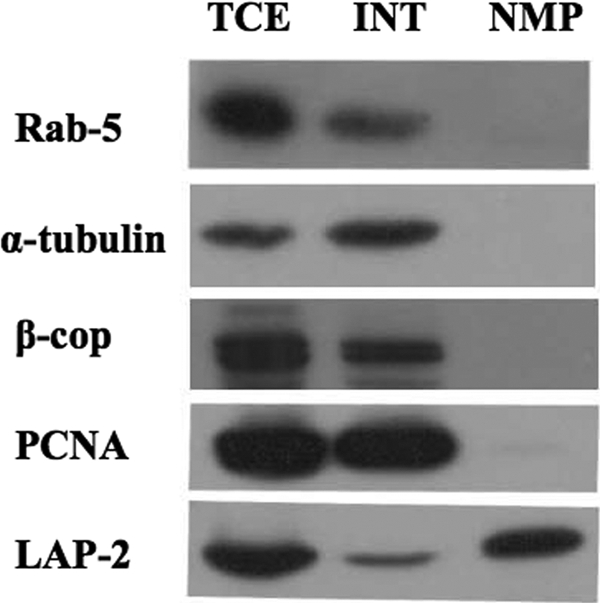
Isolation of infected nuclear membranes. COS-7 cells were infected at an MOI of 10 and infected cells harvested 18 h postinfection. TCE, INT, and NMP were collected and subjected to Western blot to test for exclusion of cell compartment markers. The blots were probed with antibodies against a variety of cellular antigens, as indicated on the left.
Mature HSV particles were excluded from the infected NMP.
Since the goal of this study was to analyze perinuclear HSV-1 particles, we needed to ensure that mature HSV-1 virions were excluded from the preparation. We employed several HSV-1 mutants and three mixing experiments to demonstrate this.
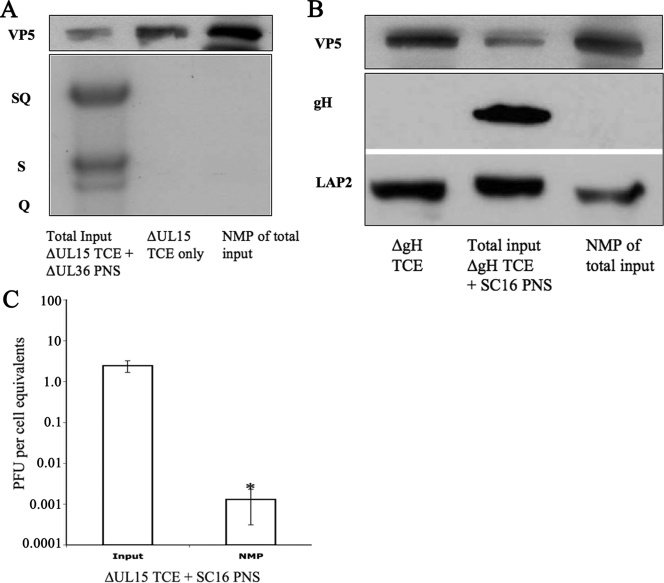
The first experiment employed an HSV-1 ΔUL15 mutant and a KΔUL36 HSV-1 mutant virus. The former has an insertion in the UL15 open reading frame which results in the inability of this strain to cleave and package viral DNA (44). The resulting phenotype is an accumulation of B capsids (44). The latter mutant lacks a portion of the large UL36 open reading frame, which results in disruption of secondary envelopment and accumulation of mature packaged C capsids in the cytoplasm (17). The postnuclear supernatant (PNS) of COS-7 cells infected with KΔUL36 HSV-1 was mixed with a TCE of COS-7 cells infected with ΔUL15 HSV-1. In the resulting mixture, the only packaged DNA is present in mature, cytoplasmic capsids. Nuclear membranes were then prepared from this mixture using the protocol we have developed. Three fractions were subjected to Western and Southern blotting: (i) total input mixture consisting of KΔUL36 HSV-1 PNS and ΔUL15 HSV-1 TCE, (ii) ΔUL15 HSV-1 TCE only, and (iii) NMP from total input (Fig. 2A). The major capsid protein VP5 was present in all three mixtures as expected (Fig. 2A, top), indicating that there are comparable numbers of viral capsids in all three mixtures. In the Southern blot (Fig. 2A, bottom), total DNA was subjected to digestion with the restriction endonuclease BamHI and then probed for the SQ restriction fragment within the HSV-1 genome. The SQ band represents noncleaved, immature viral DNA, and the S and Q bands represent terminal cleavage fragments, which indicate a mature, packaged genome. Although the starting extract clearly contained packaged HSV-1 DNA (Fig. 2A, leftmost lane), the NMP obtained from processing the total input mixture shows complete exclusion of viral genomes, which would be present if there were any contaminating mature HSV-1 nucleocapsids (Fig. 2A, rightmost lane).
FIG. 2.
Testing NMP. (A) Exclusion of cytoplasmic viral DNA from NMP. Three mixtures were subjected to Southern blot probing for packaged viral DNA and Western blotting against VP5, the major capsid protein: the total input mixture, ΔUL15-infected TCE only, and NMP from the total input mixture. For the Southern blot, total DNA was subjected to digestion by endonuclease BamHI. The SQ band represents noncleaved immature viral genomes, and the S and Q bands represent cleaved and packaged viral DNA. (B) Exclusion of cytosolic virions from NMP. Three mixtures were subjected to Western blot: ΔgH-infected TCE, total input mixture, and NMP of the total input mixture. The blots were probed with antibodies as indicated on the left. (C) Exclusion of infectious virions from NMP. PNS from SC16-infected COS-7 cells was mixed with TCE from ΔUL15-infected cells, and then the mixture was subjected to NMP. Error bars represent standard deviations. P = 0.04 for the NMP; n = 3.
A second mixing experiment was performed using a ΔgH HSV-1 mutant virus (7) and a wild-type strain of HSV-1, SC16. TCE from ΔgH HSV-1-infected COS-7 cells was mixed with PNS from SC16-infected COS-7 cells. In this mixture, perinuclear virions lack gH and some cytoplasmic virions contain gH. Nuclear membranes were prepared from this mixture. Three fractions were subjected to Western blotting: (i) TCE of ΔgH HSV-1-infected COS-7 cells, (ii) total input mixture consisting of ΔgH HSV-1 TCE and PNS from wild type SC16-infected COS-7 cells, and (iii) an NMP from this total input mixture. While the viral marker VP5 and cellular marker LAP-2 were present in all three lanes, gH was present only in the total input lane (Fig. 2B, middle). We conclude that the NMP includes nucleocapsids but has lost all detectable gH-containing mature virions.
The final mixing experiment employed ΔUL15 HSV-1 and a wild-type strain of HSV-1, SC16. TCE from ΔUL15 HSV-1 COS-7-infected cells and PNS from SC16 COS-7-infected cells were mixed. Infectious particles will be present only in the nonnuclear, cytoplasmic portion of the mixture. Nuclear membranes were then isolated from the mixture, and PFU titers were determined from cell equivalent amounts of the NMP and the total input mixture. The titer of the NMP was found to be three logs lower than the titer of the total input mixture on a per-cell basis (Fig. 2C). The level of contamination with infectious cytoplasmic particles can therefore be estimated to be 0.05% or less. It can thus be concluded that the NMP excludes mature cytoplasmic virions.
Perinuclear virions are harvested from infected nuclear membranes.
Once we had confirmed the efficacy of our NMP, we proceeded to harvest primary enveloped virions from nuclear membranes. The NMP was resuspended in an isotonic sucrose solution (10 mM Tris [pH 7.4], 1 mM MgCl2, 0.25 M sucrose), and the membranes were disrupted using sonication. Then the preparation was treated with 50 μg/ml proteinase K for 1 h on ice to digest any remaining polypeptides external to the perinuclear particles. Finally, the mixture was overlaid on a 30% sucrose cushion (10 mM Tris [pH 7.4], 30% sucrose [wt/vol]) and ultracentrifuged for 1 h. We termed our new preparation the pnHSV-1 and subjected it to Western blotting along with the infected TCE, INT, and NMP fractions. We probed for the major capsid protein VP5 and the cellular marker calnexin, an integral membrane protein found in the endoplasmic reticulum which is contiguous with the nuclear membranes (Fig. 3A). The new fraction, pnHSV-1, contains only the viral marker and excludes the cellular marker, while TCE, INT, and NMP contain both proteins (Fig. 3A).
To further test for the efficiency of exclusion of nuclear membrane markers, we subjected TCE, INT, NMP, pnHSV-1, and two variations of the pnHSV-1 fraction to Western blot. The first variation was a resuspended NMP pellet that had been sonicated and treated with proteinase K but not subjected to ultracentrifugation over the 30% sucrose cushion (Fig. 3B). The second variation was resuspended NMP pellet that had been sonicated and then subjected to ultracentrifugation over the 30% sucrose cushion but had not been subjected to proteinase K treatment (Fig. 3B). All fractions showed the presence of VP5, the major capsid protein. However, while both calnexin and LAP-2 were present as expected in the TCE, INT, and NMP fractions, there were differences among pnHSV-1 and the two pnHSV-1 variations. Calnexin was present in the first variation (pnHSV plus proteinase K without centrifugation) but not in the second variation (pnHSV without proteinase K and with centrifugation) or the pnHSV-1 fraction proper. LAP-2 was present in the second variation but absent from the first variation and the pnHSV-1 fraction proper. It can be concluded that depending on the protein, both proteinase K treatment and centrifugation through a 30% sucrose cushion are critical for separation of primary enveloped virions from proteins present in the NMP.
If the pnHSV-1 fraction contains primary enveloped virions, then known components of the primary enveloped particle should be present. Two viral proteins, pUL34 and pUL31, are present only in primary enveloped virions and not extracellular virions (31). Three preparations were subjected to Western blotting: infected TCE, pnHSV-1, and extracellular mature virions (xHSV-1). The proteins VP5, pUL31, and pUL34 were present in TCE and pnHSV-1, while the xHSV-1 fraction contained only VP5 (Fig. 3C). Therefore, we have demonstrated that our pnHSV-1 fraction contains known components of primary enveloped virions.
EM shows expected differences in mature and pnHSV-1 virion morphology.
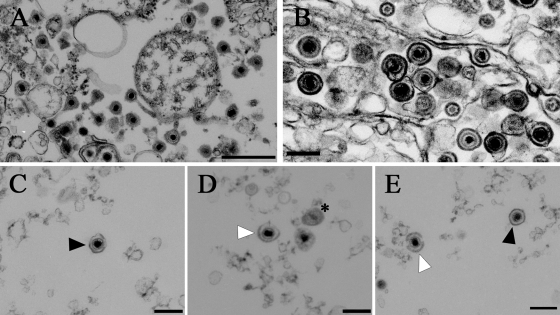
Three fractions were sectioned and prepared for EM: INT, NMP, and pnHSV-1. The INT fraction shows an abundance of membranous organelles and mature HSV-1 virions (Fig. 4A). The NMP fraction contains mainly membranous material, viral capsids, and primary enveloped virions (Fig. 4B). Figure 4C, D, and E show three representative pictures of particles that we observed in the pnHSV-1 fraction, including intact primary enveloped virions (Fig. 4C and E), partially intact primary enveloped virions (Fig. 4D and E), and occasional free capsids (Fig. 4D). We conclude that the INT fraction contains mature HSV-1 virions and other cellular cytoplasmic contents. The pnHSV-1 preparation excludes most cytoplasmic and nucleoplasmic contents and includes mostly primary enveloped virions.
FIG. 4.
Ultrastructural analysis of INT, NMP, and pnHSV fractions. (A) The INT contains mature virions and membranous structures. (B) The NMP shows primary enveloped virions, membranes, partially enveloped capsids, and naked capsids. (C to E) pnHSV-1. Images at the ultrastructural level show both intact (black arrowheads) and partially damaged (open arrowheads) primary enveloped virions. Also visible are both immature and mature HSV-1 capsids (asterisk). Bars, 500 nm (A) and 200 nm (B to E).
Proteomic analysis of the pnHSV-1 preparation.
Ultrastructural analysis reveals that primary enveloped virions are morphologically distinct from mature HSV-1 particles, suggesting a different protein composition (10). Four fractions were loaded onto a 10% SDS-PAGE gel: mature virions isolated from medium (xHSV-1), primary enveloped virions (pnHSV-1), an NMP, and TCE. The gel was stained with Coomassie brilliant blue in order to observe the protein profile of primary enveloped virions (pnHSV-1) and to compare this preparation to the other tested fractions. The protein profile of pnHSV-1 is unique in comparison to TCE, NMP, and mature HSV-1 virions (Fig. 5A).
We then attempted proteomic analysis of the pnHSV-1 preparation by initially employing a 13% 1D SDS-PAGE gel to separate the proteins. The 1D gel was stained with Coomassie brilliant blue, and bands were excised and then subjected to in-gel trypsin digestion. The trypsin-digested peptides were subjected to MALDI-MS/MS analysis, and three proteins were identified using the MASCOT search engine (27): VP5, VP19c, and VP22 (Table 1).
TABLE 1.
Proteins identified from MALDI-MS/MS analysis in a pnHSV-1 preparation
| Gene | Protein | Mol mass (kDa) | No. of unique peptides | % coverage |
|---|---|---|---|---|
| UL17 | DNA packaging protein | 75 | 1 | 2 |
| UL18 | VP23 | 35 | 13 | 48 |
| UL19 | VP5a | 150 | 32 | 36 |
| UL26 | VP24 | 67 | 3 | 14 |
| UL34 | pUL34 | 30 | 5 | 33 |
| UL38 | VP19cb | 50 | 9 | 28 |
| UL49 | VP22b | 32 | 9 | 49 |
| US6 | gD | 44 | 4 | 23 |
| Anxa-2 | Annexin A2 | 36 | 9 | 26 |
| Proteinase K | 29 | 6 | 43 |
The protein was identified from both 1D and 2D gels, but the mass spectrometry data correspond only to the band excised from the 1D gel.
Protein excised from the 1D gel before MALDI-MS/MS (gel not shown). All other proteins were excised from the 2D gel before MALDI-MS/MS analysis.
In order to enhance separation of proteins and to visualize the protein profile of pnHSV-1, we used 2D gel electrophoresis. Fifty micrograms of the pnHSV-1 fraction was subjected to isoelectric focusing on a 13-cm immobilized pH gradient gel strip. The strip was then overlaid on a 13% SDS-PAGE gel in order to separate the isoelectrically focused proteins by size. The gel was then silver stained (Fig. 5), and the 36 most intensely stained spots (as determined by eye) were excised and subjected to in-gel trypsin digestion.
Trypsin-digested peptides next underwent MALDI-MS/MS analysis, and proteins were identified by using the MASCOT search engine (27). A short list of identified proteins was generated (Table 1) from this limited proteomic analysis, including capsid proteins VP5 and VP23, VP24, the DNA packaging protein pUL17, pUL34, and glycoprotein D. Additionally, proteinase K was identified, as it was used during pnHSV-1 preparation to remove any possible contaminating proteins outside the pnHSV-1 particle. Finally, a cellular protein, annexin A2 was identified. Interestingly, the location of annexin A2 on the 2D gel corresponded to an intensely stained series of spots, indicating that it was abundant and present in multiple forms in the pnHSV-1 (Fig. 5B).
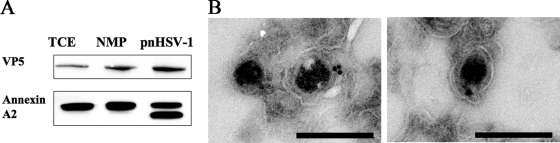
Annexin A2 is packaged into primary enveloped virions.
Annexin A2 has previously been shown to be a component of extracellular herpesvirus particles (19, 22, 43). However, its presence in the HSV-1 perinuclear particle had not previously been demonstrated. Western blot and immunogold EM analysis further validated our mass spectrometry data. First, three fractions, TCE, NMP, and pnHSV-1, were subjected to Western blotting to test for the presence of annexin A2, and it was found to be present in all three fractions, with two bands in the pnHSV-1 fraction (Fig. 6A). Next, we subjected the pnHSV-1 preparation to immunogold EM by probing for annexin A2. Figure 6B shows two examples of immunogold labeling on primary enveloped virions. We can conclude that the cellular protein annexin A2 is packaged into primary enveloped virions.
FIG. 6.
Annexin A2 is a component of the primary enveloped virion. (A) Three fractions, TCE, NMP, and pnHSV-1, were subjected to Western blotting and probed with antibodies against VP5 and annexin A2. (B) The pnHSV-1 preparation was subjected to immunogold EM labeling for annexin A2. The images show two examples of annexin A2 labeling within the primary enveloped virion. Bars, 200 nm.
Testing the role of annexin A2 in HSV-1 replication.
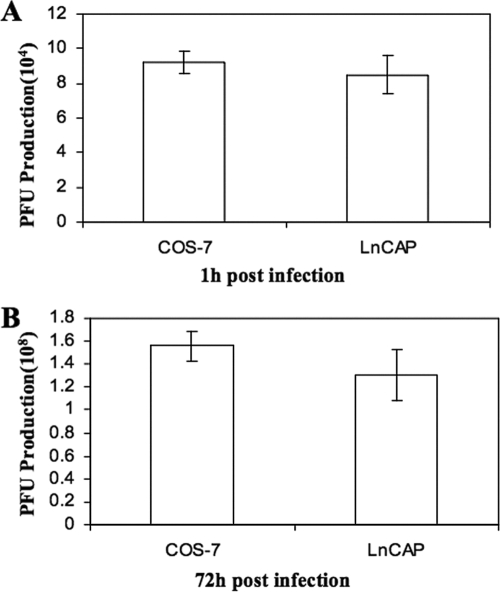
Despite its presence in both perinuclear and mature HSV-1 particles, the role of annexin A2 in viral maturation had not been tested. There is a prostate cancer cell line, LnCAP, in which annexin A2 expression has been lost (3). We exploited this cell line to test whether lack of annexin A2 affected HSV-1 replication. COS-7 cells and LnCAP cells were infected at an MOI of 0.01, and cells were harvested after 1 h of infection at 37°C or after 72 h, when the infection had proceeded to full CPE. The extent of viral replication was determined by titration on Vero cells, and the results are shown in Fig. 7. There was no apparent difference in the yields of HSV-1 from COS-7 or LnCAP cells, suggesting that under these conditions annexin A2 does not play an important role in total viral yield.
FIG. 7.
Viral titers in the presence and absence of annexin A2. (A) Yields from COS-7 and LnCAP cells that were infected with SC16 at an MOI of 0.01 and harvested after 1 h. (B) Yields from COS-7 and LnCAP cells that were infected with SC16 at an MOI of 0.01 and harvested at 72 h at full CPE. Titers were determined on Vero cells. Error bars represent standard deviations; n = 4.
DISCUSSION
The purpose of this study was to isolate primary enveloped virions in order to gain insight into their protein composition. We were able to isolate nuclear envelopes from infected cells and then harvest primary enveloped virions from them. The purity of the nuclear envelope preparation was demonstrated by exclusion of numerous cellular compartment markers and inclusion of the nuclear membrane protein LAP-2. Furthermore, three independent mixing experiments demonstrated the efficiency with which mature HSV-1 particles were removed during the NMP isolation. From the data presented in Fig. 2C, we conclude that the NMP excludes 99.95% of mature HSV-1 particles present in the starting cell extract. Additionally, after harvesting primary enveloped virions (pnHSV-1) from the NMP, we were able to exclude calnexin, an endoplasmic reticulum marker that is integrally associated with the nuclear membranes. This was the case despite the fact that the number of cell equivalents of pnHSV-1 and NMP fractions loaded were more than 50 and 30 times (respectively) the number of TCE cell equivalents loaded (Fig. 3A). Finally, it is known that the gene products of UL34 and UL31 are present only in the primary enveloped virion (31) and are essential for primary enveloped virion morphogenesis (8, 30, 32). Our pnHSV-1 preparation included both of those gene products as well as the major capsid protein VP5, in comparison to our extracellular virion preparation, which contained only VP5 (Fig. 3C).
The biochemical approach of isolating primary enveloped HSV-1 virions complements the analysis of this assembly intermediate by electron and fluorescent microscopy. After confirming the efficiency of the pnHSV-1 preparation, the next step was to subject the pnHSV-1 fraction to whole-particle analysis. We first observed the protein profile of the pnHSV-1 fraction and compared it to the profiles of mature virions (xHSV-1), an NMP, and TCE. The Coomassie-stained gel shown in Fig. 5A demonstrates that each fraction has its own unique protein profile. At this point it was clear that a proteomics study was a natural next step in attempting whole-particle analysis of the pnHSV-1 preparation: a protein population that is relatively small and prepared in the absence of detergents lends itself ideally to mass spectrometry studies. The protein profile of pnHSV-1 was visualized using 2D gel electrophoresis, and there were approximately 140 distinct protein spots stained on the gel. We took a limited approach by subjecting only the most intense spots (as determined by eye), which numbered 36, to MALDI-TOF/TOF. We were able to identify several known proteins, including most of the capsid proteins and pUL34, a component of primary enveloped virions (8, 31). We were also able to identify gD, which is considered a component of primary enveloped virions (36, 39), and the tegument protein VP22. Finally, we were able to identify a cellular protein, annexin A2. Annexin A2 corresponded to a series of intensely stained spots on the 2D gel, indicating that it is rather abundant in this fraction (Fig. 5B). Annexin A2 has previously been shown to be a minor component of extracellular HSV-1 virions (19) as well as other members of the herpesviruses, PrV and HCMV (22, 43).
Annexin A2 was an interesting candidate for follow-up analysis because it is the first example of a cellular protein associated with the primary enveloped virion and also because of its abundance in the fraction. A Western blot showed the presence of annexin A2 in three fractions (TCE, NMP, and pnHSV-1), with two bands present in the pnHSV-1 lane (Fig. 6A). We are attempting to determine the origin of the lower-molecular-weight form of annexin A2 in the pnHSV-1 fraction. Furthermore, we demonstrated using immunogold EM that annexin A2 is indeed packaged into primary enveloped virions (Fig. 6B).
Annexin A2 is known to be an organizer of membrane domains and to interact with the actin cytoskeleton (9). In fact, annexin A2 is known to be involved in the “rocketing” of endosomes through the cytoplasm on filamentous actin (12). This is interesting considering that HSV-1 nucleocapsids are thought to travel on actin cables to the nuclear periphery (6). Annexin A2 could facilitate intranuclear nucleocapsid movement, help to organize the budding machinery at the inner nuclear membrane, or both. Nevertheless, viral titer was unaffected when HSV-1 was grown on LnCAP cells, a prostate cancer cell line lacking annexin A2 (3). Therefore, despite its abundance in the fraction, it does not play an essential role in replication under our conditions.
In closing, we have isolated nuclear envelopes from HSV-1-infected cells and harvested primary enveloped virions from our NMP. We have performed proteomic analysis and identified both viral and cellular proteins in the pnHSV-1 fraction. Shotgun proteomic studies will be forthcoming, which should reveal a more complete list of pnHSV-1 fraction components.
Acknowledgments
We thank the following for kindly providing us with mutant virus strains and complementing cell lines: Preshant Desai for KΔUL36, Anthony Minson for SC16ΔgH, and Sandra Weller for Hr81-2 (ΔUL15). We also thank Richard Roller and Joel Baines for providing the pUL31 and pUL34 antibodies. We thank the Analytical Imaging Facility (AIF) and the Laboratory for Macromolecular Analysis and Proteomics (LMAP) at Albert Einstein College of Medicine for their expertise and guidance in the EM and proteomics studies, respectively. We thank Lily Huang for her expert technical assistance.
Footnotes
Published ahead of print on 11 March 2009.
REFERENCES
- 1.Bechtel, J. T., R. C. Winant, and D. Ganem. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 794952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 704311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chetcuti, A., S. H. Margan, P. Russell, S. Mann, D. S. Millar, S. J. Clark, J. Rogers, D. J. Handelsman, and Q. Dong. 2001. Loss of annexin II heavy and light chains in prostate cancer and its precursors. Cancer Res. 616331-6334. [PubMed] [Google Scholar]
- 4.Church, G. A., and D. W. Wilson. 1997. Study of herpes simplex virus maturation during a synchronous wave of assembly. J. Virol. 713603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA 10410187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forest, T., S. Barnard, and J. D. Baines. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7429-431. [DOI] [PubMed] [Google Scholar]
- 7.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerke, V., and S. E. Moss. 2002. Annexins: from structure to function. Physiol. Rev. 82331-371. [DOI] [PubMed] [Google Scholar]
- 10.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 753675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 751236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes, M. J., U. Rescher, V. Gerke, and S. E. Moss. 2004. Annexin-actin interactions. Traffic 5571-576. [DOI] [PubMed] [Google Scholar]
- 13.Hellman, U., C. Wernstedt, J. Gonez, and C. H. Heldin. 1995. Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224451-455. [DOI] [PubMed] [Google Scholar]
- 14.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 10116286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kay, R. R., D. Fraser, and I. R. Johnston. 1972. A method for the rapid isolation of nuclear membranes from rat liver. Characterisation of the membrane preparation and its associated DNA polymerase. Eur. J. Biochem. 30145-154. [DOI] [PubMed] [Google Scholar]
- 16.Klupp, B. G., H. Granzow, W. Fuchs, G. M. Keil, S. Finke, and T. C. Mettenleiter. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. USA 1047241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klupp, B. G., H. Granzow, G. M. Keil, and T. C. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 806235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 822363-2371. [DOI] [PubMed] [Google Scholar]
- 19.Loret, S., G. Guay, and R. Lippe. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 828605-8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo, Q., L. Siconolfi-Baez, P. Annamaneni, M. T. Bielawski, P. M. Novikoff, and R. H. Angeletti. 2007. Altered protein expression at early-stage rat hepatic neoplasia. Am. J. Physiol. Gastrointest. Liver Physiol. 292G1272-G1282. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell, K. L., and L. Frappier. 2007. Viral proteomics. Microbiol. Mol. Biol. Rev. 71398-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 801332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297854-857. [DOI] [PubMed] [Google Scholar]
- 24.Naldinho-Souto, R., H. Browne, and T. Minson. 2006. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J. Virol. 802582-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 7510923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 203551-3567. [DOI] [PubMed] [Google Scholar]
- 28.Remillard-Labrosse, G., G. Guay, and R. Lippe. 2006. Reconstitution of herpes simplex virus type 1 nuclear capsid egress in vitro. J. Virol. 809741-9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 785564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 758803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 768939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 36.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 755697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skiba, M., T. C. Mettenleiter, and A. Karger. 2008. Quantitative whole-cell proteome analysis of pseudorabies virus-infected cells. J. Virol. 829689-9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1361007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torrisi, M. R., C. Di Lazzaro, A. Pavan, L. Pereira, and G. Campadelli-Fiume. 1992. Herpes simplex virus envelopment and maturation studied by fracture label. J. Virol. 66554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turcotte, S., J. Letellier, and R. Lippe. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 798847-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Genderen, I. L., R. Brandimarti, M. R. Torrisi, G. Campadelli, and G. van Meer. 1994. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology 200831-836. [DOI] [PubMed] [Google Scholar]
- 42.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 7810960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright, J. F., A. Kurosky, E. L. Pryzdial, and S. Wasi. 1995. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 694784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, D., A. K. Sheaffer, D. J. Tenney, and S. K. Weller. 1997. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J. Virol. 712656-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]