Abstract
The human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) genomes encode several auxiliary proteins that have increasingly shown their importance in the virus-host relationship. One of these proteins, Vpx, is unique to the HIV-2/SIVsm lineage and is critical for viral replication in macrophages. The functional basis for this requirement, as well as the Vpx mode of action, has remained unexplained, and it is all the more enigmatic that HIV type 1 (HIV-1), which has no Vpx counterpart, can infect macrophages. Here, we underscore DCAF1 as a critical host effector of Vpx in its ability to mediate infection and long-term replication of HIV-2 in human macrophages. Vpx assembles with the CUL4A-DDB1 ubiquitin ligase through DCAF1 recruitment. Precluding Vpx present in the incoming virions from recruiting DCAF1 in target macrophages leads to a postentry block characterized by defective accumulation of HIV-2 reverse transcripts. In addition, Vpx from SIVsm functionally complements Vpx-defective HIV-2 in a DCAF1-binding-dependent manner. Altogether, our data point to a mechanism in which Vpx diverts the Cul4A-DDB1DCAF1 ligase to inactivate an evolutionarily conserved factor, which restricts macrophage infection by HIV-2 and closely related simian viruses.
Human immunodeficiency virus type 2 (HIV-2), the second causative agent of human AIDS (5), arose from cross-transmission of sooty mangabey simian immunodeficiency virus (SIVsm), a lentivirus that naturally infects sooty mangabeys without inducing overt disease (14). Although AIDS caused by HIV-2 is as lethal as that caused by HIV-1, most HIV-2 carriers remain asymptomatic much longer than HIV-1 carriers (36). On the other hand, cross-transmission of SIVsm to rhesus macaques gave rise to highly pathogenic viral strains, and infected animals develop a disease similar to human AIDS (14). Thus, the SIVsm lentiviral lineage exhibits markedly different pathogenicities in its original and recently acquired hosts. It is currently unknown whether cross-species transmission of primate lentiviruses leads to specific functional changes in viral proteins that might directly modulate pathogenesis in the new host species.
The HIV-1 and HIV-2 lentiviral lineages differ in the auxiliary proteins encoded by their genomes, which likely reflects different selective pressures in their adaptation to the host cell environment. Thus, the Vpx protein found in HIV-2 has no counterpart in HIV-1, whereas the genetically related Vpr protein is present in both lineages (47). Both Vpx and Vpr are actively packaged into the virions (1, 40), which suggests a role in early infection steps, i.e., prior to viral synthesis. Lack of Vpx strongly decreases the pathogenicity of SIVsmPBj in infected macaques (17). In vitro, Vpx is dispensable for infection of immortalized T cells but has been shown to be critical for primary macrophage infection (9, 23). In contrast, no cell culture system has so far revealed a strong requirement for Vpr in HIV infection, although Vpr-defective HIV-1 shows attenuated replication in human macrophages (2, 6, 16). Despite the absence of a clear defective phenotype of HIV lacking Vpr, this viral protein has so far attracted more attention than Vpx, presumably because of its intriguing ability to arrest the cell cycle at the G2/M transition (15, 21, 34, 35). We and others recently demonstrated that Vpr recruits DCAF1/VprBP, an adaptor of the CUL4A-DDB1 ubiquitin ligase complex (3, 8, 18, 26, 38, 46). A major role of ubiquitin ligases is the labeling of specific proteins for proteasome-mediated degradation, and the current hypothesis is that Vpr diverts the DCAF1 ubiquitin ligase to induce the degradation of a host protein required for entry into mitosis. We previously showed that Vpx from SIVsm also interacts with DCAF1, although unlike Vpr, it does not arrest the cell cycle (26). The ability to recruit DCAF1 might thus represent an ancient functional acquisition that predates the emergence of the genetically related but functionally distinct Vpr and Vpx genes. Here, we explore this hypothesis and address the potential role of Vpx-DCAF1 interaction in the context of HIV-2 replication in human macrophages.
MATERIALS AND METHODS
HIV-2 proviral clones and virus production.
The pGL-AN plasmid, which carries a replication-competent HIV-2 provirus from the GH-1 isolate, as well as its derivatives defective in the Vpr and/or the Vpx genes (48), were kindly provided by A. Adachi. Env-defective viruses were obtained by creating a frameshift mutation at the unique NsiI site as previously described (48). To create a HIV-2 proviral clone encoding the Vpx Q76 mutant, the 400-bp SacI fragment of pGL-AN that overlaps the 3′ and 5′ sequences of the vpx and vpr genes was subcloned into PUC19 prior to site-directed mutagenesis, and the mutagenized fragment was cloned back into the HIV-2 backbone. Replicative viruses were produced from 293T cells transiently transfected with proviral clones by using SuperFect reagent (Qiagen). For the production of vesicular stomatitis virus G protein (VSV-G)-pseudotyped single-cycle viruses, pCMV-G encoding VSV-G (pMD2 VSV-G) was cotransfected in a 1:1 ratio with the HIV-2 proviral constructs using either Superfect or calcium-phosphate coprecipitation. In some experiments, Vpx-defective HIV-2 viruses were trans-complemented with wild-type (WT) or mutant Vpx protein expressed from cotransfected expression plasmids. Cell culture supernatants were harvested 48-hour posttransfection, clarified by centrifugation at 1,500 rpm for 5 min, and filtered (0.45 μm) before storage at −80°C. When appropriate, the viruses were partially purified and concentrated by ultracentrifugation over a 20% sucrose cushion. Replication-competent viruses were titrated on HeLa P4P cells (49) by quantification of HIV-2 capsid (CA) p27 in cell supernatants using an enzyme-linked immunosorbent assay (ELISA) (ZeptoMetrix Corporation). Env-pseudotyped virions were titrated on P4P cells by CA quantification in cell lysates 72 h posttransduction.
Mammalian expression vectors.
The pAS1B vector, which provides an N-terminal hemagglutinin (HA) epitope coding sequence, was used to express the different Vpx proteins. pAS1B-Vpx-SIV has been previously described (39). HIV-2 Vpx was PCR amplified from GL-AN prior to being cloned into pAS1B. The Q76R mutation in HIV-2 Vpx and the K77A mutation in SIV Vpx were introduced by site-directed mutagenesis. Plasmids encoding DDB1-HA and FLAG-DCAF1 have been previously described (27, 42).
MDMs and CD4 T lymphocytes.
Human monocytes were isolated from buffy coats of healthy seronegative donors (Centre de Transfusion Sanguine Ile-de-France, Rungis, and Hôpital de la Pitié-Salpêtrière, Paris, France), and differentiated into macrophages as described previously (7). Briefly, monocytes were separated from peripheral blood mononuclear cells by adherence and then detached and cultured for 7 to 11 days in hydrophobic Teflon dishes (Lumox; D Dutcher) in monocyte-derived macrophage (MDM) medium (RPMI 1640 supplemented with 200 mM l-glutamine, 100 U penicillin, 100 μg streptomycin, 10 mM HEPES, 10 mM sodium pyruvate, 50 μM β-mercaptoethanol, 1% minimum essential medium vitamins, and 1% nonessential amino acids) containing 15% human AB serum. The MDMs were washed and diluted in MDM medium containing 10% heat-inactivated fetal calf serum prior to infection experiments. CD4 T lymphocytes were isolated from nonadherent peripheral blood leukocytes by positive selection with antibody-coated magnetic beads (Milteny Biotech, France) following the manufacturer's instructions and were activated for 3 days with phytohemagglutinin in the presence of interleukin-2 in RPMI 1640 medium, 10% fetal calf serum, 100 U/ml penicillin/streptomycin (RPMI).
MDMs and CD4 T-cell infection.
MDMs and activated CD4 T cells were infected using a spinoculation protocol (30). After infection, the cells were washed in phosphate-buffered saline. Infected MDMs were then cultured in MDM medium (105 MDMs/well in 96-well plates or 0.5 × 106 MDMs/well in 12-well plates) and CD4 T cells in RPMI. Replication-competent viruses were used at a multiplicity of infection of 10−2. The supernatants were harvested every 3 or 4 days for quantification of Gag CA p27. For single-round infections, MDMs were infected with VSV-G-pseudotyped viruses at equivalent multiplicities of infection, and intracellular CA was measured in MDM lysates 72 to 96 h postinfection.
Real-time PCR quantification of HIV-2 cDNA in infected MDMs.
MDMs were infected with DNase-treated VSV-G-pseudotyped viruses as described previously (24), and total cellular DNA was extracted at different times after infection using a DNeasy kit (Qiagen). Quantitative real-time PCR analysis of late reverse transcription (RT) products (U5/5′-end noncoding region) and of two-long-terminal-repeat (LTR) circles were carried out using the primers and probes described by Fujita et al. (10) with an ABI Prism 7000 sequence detection system. The copy number of late RT products was determined with reference to a standard curve generated by serial dilution of DNA from HIV-2-infected P4P cells in which the viral-DNA copy number was quantified in comparison with cloned HIV-2 DNA. The levels of two-LTR circles were determined with reference to standard curves obtained by concurrent amplification of serial dilutions of DNA from HIV-2-infected P4P cells and were expressed in arbitrary units. The albumin gene was used as a reference gene for normalization, as described previously (7).
siRNA transfection.
Control short interfering RNA (siRNA) and DCAF1-specific siRNAs have been previously described (26). MDMs were plated at a density of 0.5 × 106/well in 12-well plates or at 105/well in 96-well plates prior to siRNA transfection using InterferIN (PolyPlus Transfection). A final concentration of 100 nM siRNA was routinely used. MDMs were infected 72 h post-siRNA transfection. The knockdown efficiency was assessed by Western blot (WB) analysis at the time of infection.
Immunoprecipitation procedures and WB analyses.
Expression constructs (3 μg) were transfected into HeLa cells grown in 10-cm dishes by the Lipofectamine-reagent plus method (Invitrogen). The cells were lysed 48 h posttransfection in 300 μl SD buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Triton X-100) supplemented with an anti-protease cocktail (Sigma). Following clarification, the cell lysates were rocked for 2 h at 4°C with beads coated with either anti-FLAG M2 (Sigma) or anti-HA 3F10 (Roche). The beads were washed twice in SD buffer, and bound proteins were eluted using 3× FlAG peptide (Sigma) for anti-FLAG immunoprecipitation and direct boiling in Laemmli buffer for anti-HA precipitation. For WB analyses, the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto polyvinylidene difluoride membranes, and revealed by a chemiluminescence procedure (enhanced chemiluminescence [GE Healthcare] or CDP-star [Applied Biosystems]). The following antibodies were used: rabbit anti-VprBP/DCAF1 from Shanghai Genomics Inc., mouse anti-DDB1 and mouse anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) from AbCam, mouse anti-FLAG M2 from Sigma, and mouse anti-HA from Covance. The mouse monoclonal antibodies specific for the HIV-2/SIV capsid and Vpx protein were provided by the MRC AIDS directed program, National Institute for Biological Standards and Control, Hertfordshire, United Kingdom.
RESULTS
The ability of Vpx to recruit DCAF1 is conserved between HIV-2 and its parental SIVsm primate lentivirus.
We first examined the abilities of Vpx proteins from two distinct lentiviral origins to form a complex with DCAF1 in human cells. Vpx from HIV-2 (GH-1 isolate) was retained in amounts comparable to those of Vpx from SIVsm (PBj isolate) in DCAF1 immunoprecipitates (Fig. 1A, compare lanes 2 and 4), indicating similar affinities of the two viral proteins for human DCAF1. In addition, both Vpx proteins failed to bind DCAF1 when they carried the Q76R mutation (Fig. 1A, lanes 3 and 5), as expected due to the phenotype of the homologous substitution in HIV-1 Vpr (26). HIV-1 Vpr uses the adaptor function of DCAF1 to assemble with the CUL4A-DDB1 ubiquitin ligase and presumably to divert its activity (3, 8, 18, 26, 38, 46). A similar scenario may occur for HIV-2 Vpx, since endogenous DDB1 coimmunoprecipitated with WT Vpx, but not with the Vpx Q76R mutant (Fig. 1B, compare lanes 2 and 4). In addition, the endogenous pool of DCAF1 sufficiently mediated the association of WT Vpx with DDB1 (Fig. 1B, compare lanes 2 and 3).
FIG. 1.
The HIV-2 Vpx protein recruits DDB1 through DCAF1 binding in a Q76 residue-dependent manner. Cell lysates were prepared from HeLa cells expressing the indicated HA-tagged Vpx proteins along with FLAG-tagged DCAF1, where indicated. Crude cell lysates (inputs) and immunoprecipitates (IP) were analyzed by WB to reveal the transfected tagged proteins (A and B) and endogenous DDB1 (B).
Inactivation of the DCAF1-binding property of Vpx phenocopies the absence of Vpx and causes a severe HIV-2 growth defect in macrophages, but not in CD4 T cells.
When placed in the context of the HIV-2 provirus, the Q76R substitution crippled the virus as severely as Vpx deletion in its ability to replicate in MDMs (hereafter referred to as macrophages). Virtually no p27 viral CA was detected in cell supernatants over an 18-day period for both viruses (Fig. 2A, left). In contrast, both mutant viruses were as competent as the WT virus for replication in primary CD4-positive T cells (Fig. 2A, right). Efficient encapsidation of Vpx Q76R excluded the mutant mimicking lack of Vpx because of a mere packaging defect (Fig. 2B). Altogether, these data show that defective DCAF1 binding by Vpx correlates with impaired HIV-2 replication in macrophages.
FIG. 2.
The Q76R mutation in Vpx strongly impairs HIV-2 replication in human primary macrophages. (A) The Q76R mutation impairs HIV-2 replication in macrophages to an extent similar to that with the Vpx deletion. Macrophages (left) or CD4 T cells (right) were infected with the indicated HIV-2 strains, and the CA contents of the culture supernatants were monitored over time by ELISA. Shown is a representative experiment out of four performed with macrophages from different donors. The results are expressed as means of the CA levels from triplicate samples ± standard deviations. (B) The Q76R Vpx mutant is efficiently encapsidated into virions. The culture supernatants of 293T cells were harvested 48 h after transfection with the indicated HIV-2 proviral plasmids and were ultracentrifuged through a 20% sucrose cushion. After lysis in sodium dodecyl sulfate, the pelleted virions were analyzed by WB using anti-Vpx and anti-CA antibodies. Δ, lack of Vpx.
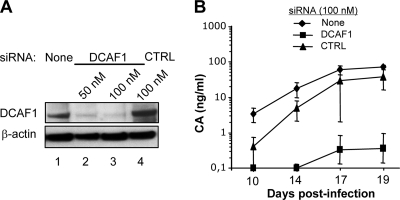
DCAF1 silencing strongly decreases the permissivity of macrophages to WT HIV-2 replication.
We next investigated the effect of siRNA-mediated depletion of DCAF1 on WT HIV-2 replication in macrophages. Under conditions in which DCAF1 expression was efficiently knocked down at the time of infection (Fig. 3A), HIV-2 replication was strongly compromised (Fig. 3B). HIV-2 CA was undetectable in DCAF1-depleted macrophage supernatants until day 14 postinfection (p.i.), and at later time points, the CA levels were more than 2 orders of magnitude less than in the control cultures (Fig. 3B). The residual HIV-2 replication observed likely results from the impossibility of achieving and/or maintaining complete DCAF1 silencing during long-term macrophage culture. The deleterious effect of DCAF1 depletion on HIV-2 replication was reproduced with macrophages from three different donors (data not shown). Since both Vpr and Vpx recruit DCAF1, silencing of DCAF1 might have interfered with Vpr rather than Vpx function. However, in agreement with previous studies of HIV-2 and SIVsm/mac (9, 32, 48), HIV-2 replication in macrophages was only mildly affected by Vpr deletion compared to Vpx deletion (see Fig. S1 in the supplemental material). A recent large-scale siRNA screen underscored DCAF1 as a positive factor in the life cycle of Vpr-defective HIV-1 in HeLa cells (4). The possibility that DCAF1 was required for efficient HIV-2 infection regardless of any dependence on auxiliary proteins had to be considered. We observed that depletion of DCAF1 in HeLa cells did not affect infection by either HIV-2 or SIVsmPBj (data not shown). We therefore conclude that the dependence of HIV-2 on host DCAF1 for efficient infection is specific for macrophages and mimics its dependence on the Vpx auxiliary protein in these cells.
FIG. 3.
DCAF1 knockdown inhibits HIV-2 replication in macrophages. Macrophages were mock treated or transfected with either DCAF1-specific or control (CTRL) siRNA at the indicated concentrations. (A) DCAF1 was detected by immunoblot analysis of cell lysates prepared 72 h posttransfection. (B) Macrophages were transfected with the indicated siRNAs 72 h prior to infection with WT HIV-2, and viral CA in culture supernatants was monitored over time as described in the legend to Fig. 2.
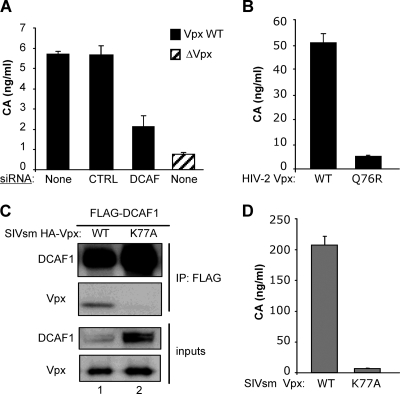
Single-round infection of macrophages by HIV-2 requires interaction between DCAF1 from target cells and Vpx from incoming viral particles.
The presence of Vpx in HIV-2 virions suggests that Vpx-DCAF1 interaction exerts its function during early postentry steps of the viral cycle. To address this hypothesis, we conducted single-round infections of macrophages using Envelope (Env)-defective HIV-2 pseudotyped with VSV-G Env. The amount of intracellular viral capsid newly synthesized from integrated proviral DNA was used as a marker of viral transduction. Silencing of DCAF1 in the target macrophages, as well as Vpx deletion, strongly reduced macrophage transduction by the HIV-2 particles (Fig. 4A). The decrease in viral transduction directly depended on the efficiency of DCAF1 silencing. These data suggested that the active partners are DCAF1 from the target macrophage and Vpx from the incoming virions. To further examine this hypothesis, we produced VSV-G-pseudotyped particles made from Vpx-defective (ΔVpx) HIV-2 provirus trans-complemented with either WT or mutant Vpx (see Fig. S2 in the supplemental material). The infectious potential of these viral particles was compared in macrophages. WT Vpx, but not its Q76R mutant counterpart, promoted efficient HIV-2 transduction of macrophages (Fig. 4B).
FIG. 4.
The recruitment of DCAF1 by incoming Vpx is critical for efficient macrophage transduction by HIV-2. (A) VSV-G-pseudotyped HIV-2 strains were produced by cotransfection of 293T cells with Env-defective proviral DNA and a plasmid encoding the VSV-G envelope. Macrophages were mock treated or transfected with either DCAF1-specific or control (CTRL) siRNA 72 h prior to transduction with the indicated HIV-2 particles. The intracellular CA content was measured by ELISA 72 h postinfection. Shown is a representative experiment out of four performed with macrophages from different donors. The bars represent mean values calculated from triplicate cell lysates, and the corresponding standard deviations are indicated. (B) The VSV-G-pseudotyped HIV-2 particles were produced as for panel A using a Vpx-defective proviral DNA, along with a construct expressing the indicated HIV-2 Vpx proteins. The content in intracellular CA was measured 96 h posttransduction. (C) Vpx WT and Vpx K77A from SIVsmPBj were expressed as HA-tagged proteins and compared for their abilities to interact with FLAG-DCAF1 using coimmunoprecipitation as described in the legend to Fig. 1B. (D) Macrophages were transduced with VSV-G-pseudotyped HIV-2 particles complemented with the indicated SIVsmPbj Vpx proteins, and the content in intracellular CA was measured as for panel B. Similar results were obtained using macrophages from four different donors.
Vpx from SIVsm functionally complements Vpx-defective HIV-2 in a DCAF1-binding-dependent manner.
Ubiquitination of SIVsm Vpx has recently been suggested to control its activity based on the functional defect resulting from lysine substitutions (41). We noticed that the K77 residue in SIVsm Vpx, which is conserved in HIV-2 Vpx, lies adjacent to the Q76 residue that is critical for binding to DCAF1. This raised the possibility that a defect in DCAF1 binding, rather than in ubiquitination, was responsible for the loss of function shown by Vpx with lysine replaced. In agreement with this hypothesis, the SIVsm Vpx K77A mutant failed both to coimmunoprecipitate with DCAF1 (Fig. 4C) and to promote macrophage transduction by Vpx-defective HIV-2 (Fig. 4D), in contrast to the WT SIV Vpx.
Knockdown of host DCAF1 and lack of Vpx result in decreased accumulation of HIV-2 reverse transcripts in infected macrophages.
To further characterize the defect in HIV-2 replication caused by the absence of host DCAF1 and of functional Vpx in incoming virions, we used real-time PCR to quantify HIV-2 RT intermediates. As discussed above, macrophages were subjected to single-round infections with VSV-G-pseudotyped HIV-2 particles. DCAF1 knockdown led to a strong decrease in the accumulation of two-LTR circles (Fig. 5A, left), as well as a marked reduction in late RT products (Fig. 5A, right). This suggested a defect occurring at the RT step, i.e., prior to nuclear import of viral cDNA. Residual DNA carryover in the viral preparations, despite DNase treatment, precluded confident analysis of early reverse transcripts. Lack of Vpx affected the same replication steps as DCAF1 silencing and caused a gross defect in the accumulation of both two-LTR circles and late RT products (Fig. 5B).
FIG. 5.
Both Vpx and DCAF1 are required in the preintegration steps of the HIV-2 life cycle in macrophages. (A) Decreased amounts of two-LTR circles and late RT products in DCAF1-depleted macrophages. Macrophages were transfected with DCAF1-specific or control (CTRL) siRNAs 72 h prior to infection with VSV-G-pseudotyped HIV-2 particles. The accumulation of two-LTR circles was monitored over time using real-time PCR (left). The products were quantified by comparison to a standard curve generated by serial dilutions of DNA from HIV-2-infected HeLa P4P cells, and the results are expressed as arbitrary units. Levels of late RT products were assessed by real-time PCR 72 h postinfection, when accumulation was maximal (right). All PCR experiments were performed on duplicate samples of cell DNA. (B) Defective HIV-2 RT in the absence of Vpx. Macrophages were infected with the indicated VSV-G-pseudotyped HIV-2 virions. DNA was extracted and analyzed as described for panel A. Similar results were obtained using macrophages from four different donors.
DISCUSSION
Our work underscores DCAF1 as a critical host effector of Vpx in its ability to impose macrophage permissivity to HIV-2 infection. Active packaging of Vpx, which ensures its delivery into the cells by incoming virions, has long been taken as a clue to a role in the early steps of viral replication in macrophages. In agreement with this prediction, we show that susceptibility of macrophages to HIV-2 infection requires the presence of both DCAF1-binding-proficient Vpx from the incoming virions and DCAF1 from the target cell. In addition, our data show that preventing incoming Vpx from encountering DCAF1 impairs the accumulation of HIV-2 reverse transcripts, which in turn precludes completion of the subsequent steps of the viral life cycle. While this work was in progress, a report focusing on Vpx from SIVmac reached similar conclusions (43). Whether the RT process itself is inhibited or newly synthesized viral cDNA undergoes abnormal degradation requires further study. In any event, our observations, together with other recent reports (10, 41, 43), challenge the previous view of a purely mechanical role of Vpx in the transport of the viral preintegration complex to the nucleus in nondividing cells (9, 29, 31).
To date, the only role ascribed to DCAF1 is as a substrate-specific adaptor of the Cul4A-DDB1 ubiquitin ligase (25). Except for Merlin, a cytoskeleton-associated protein (20), the endogenous substrates of the Cul4A-DDB1DCAF1 complex are so far unknown. The fact that DCAF1 depletion mimics the absence of Vpx argues against Vpx-mediated inhibition of endogenous DCAF1 activity. Conversely, Vpx function is unlikely to come down to a mere stimulation of DCAF1 activity, given that both Vpx and Vpr bind to DCAF1 but lack functional redundancy. We therefore favor the hypothesis of a DCAF1 gain of function upon Vpx binding, which relieves the constitutive refractoriness of human macrophages to HIV-2 infection. Interestingly, a recent study showed that resistance of macrophages to infection by Vpx-deficient SIVsm is dominant over the permissive state of other cell types and pinpointed DDB1 as a host effector of Vpx function (41). Taken together with our finding that Vpx assembles with DDB1 through DCAF1 binding, a picture is now emerging: Vpx diverts the ubiquitin ligase activity of the CUL4A-DDB1DCAF1 complex in order to inactivate a macrophage-specific restriction factor that targets an early postentry step of the viral life cycle (Fig. 6).
FIG. 6.
Presumed mode of action of HIV-2 Vpx in macrophage infection. Vpx connects the DCAF1 adaptor of the CUL4A-DDB1 ubiquitin (Ub) ligase to a macrophage-specific restriction factor (RF), which targets the HIV-2/SIVsm lentiviruses. As a result, RF is polyubiquitinated and is subsequently degraded by the proteasome.
What predictions can be made regarding this restriction factor? First, it is conserved across several primate species based on our finding that Vpx from SIVsm can substitute for HIV-2 Vpx and on the reciprocal observations that HIV-2 Vpx can complement Vpx-defective SIVmac (43). In contrast, species specificity is expected for the factor targeted by Vpr, which induces cell cycle arrest (33, 44), and we suspect that Vpr and Vpx evolved under distinct selective pressures. Moreover, evolutionary conservation of the Vpx-targeted factor contrasts with the positive selection that occurred for the Trim5 alpha restriction factor (28, 37, 45), despite the two factors targeting similar viral replication steps. Second, the restriction factor counteracted by Vpx senses a component specific for the HIV-2/SIVsm type of viral particles, given the well-known susceptibility of macrophages to HIV-1 infection. However, the recently described helper function of SIV Vpx in macrophage infection by HIV-1 suggests that the virus does not fully escape the restriction (41).
The possibility that cells other than macrophages express a similar restriction factor remains to be clarified. Conflicting data have been reported on the role of Vpx in HIV-2 infection of lymphoid cells using T-cell lines or peripheral blood mononuclear cells (13, 19, 22, 23, 48). However, we have not observed a dependence on Vpx in primary CD4 T-lymphocyte infection by HIV-2. Monocyte-derived dendritic cells have been reported to be refractory to infection by SIVmac in the absence of Vpx, due to an impaired RT step (12). A recent attempt to characterize the underlying mechanism surprisingly pointed to a DCAF1-independent action of Vpx in dendritic cells, as well as monocytic THP1 cells differentiated with phorbol ester (11). However, differentiated THP1 cells may not fully recapitulate the physiological properties of the primary macrophages used in our own study. Whether Vpx counteracts an innate antiviral defense in human macrophages, as strongly suggested by our study, while complementing the lack of cellular factors in other cell types requires further investigation. In any event, Vpx has won its spurs and should now be viewed as a valuable probe of cellular mechanisms that modulate susceptibility to primate lentivirus infection.
Supplementary Material
Acknowledgments
We are grateful to Françoise Barré-Sinoussi for her continuous support. We thank Akio Adachi for providing replication-competent HIV-2 proviral clones. We thank K. Kent and K. Krohn, as well as the NIBSC Centre for AIDS Reagents, for providing antibodies.
D.A., A.B., E.L.R., and M.M. receive support from the Ministère de l'Education Nationale, the Agence Nationale de la Recherche sur le SIDA et les Hépatites Virales (ANRS), the Mairie de Paris, and Sidaction and Fondation de France, respectively. This work was supported by the ANRS, by Sidaction and Fondation de France, and by the Mairie de Paris.
Footnotes
Published ahead of print on 4 March 2009.
Supplemental material for this study may be found at http://jvi.asm.org/.
REFERENCES
- 1.Accola, M. A., A. A. Bukovsky, M. S. Jones, and H. G. Gottlinger. 1999. A conserved dileucine-containing motif in p6gag governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIVmac and SIVagm. J. Virol. 739992-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balliet, J. W., D. L. Kolson, G. Eiger, F. M. Kim, K. A. McGann, A. Srinivasan, and R. Collman. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200623-631. [DOI] [PubMed] [Google Scholar]
- 3.Belzile, J. P., G. Duisit, N. Rougeau, J. Mercier, A. Finzi, and E. A. Cohen. 2007. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 3e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319921-926. [DOI] [PubMed] [Google Scholar]
- 5.Clavel, F., D. Guetard, F. Brun-Vezinet, S. Chamaret, M. A. Rey, M. O. Santos-Ferreira, A. G. Laurent, C. Dauguet, C. Katlama, C. Rouzioux, et al. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233343-346. [DOI] [PubMed] [Google Scholar]
- 6.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206935-944. [DOI] [PubMed] [Google Scholar]
- 7.David, A., A. Saez-Cirion, P. Versmisse, O. Malbec, B. Iannascoli, F. Herschke, M. Lucas, F. Barre-Sinoussi, J. F. Mouscadet, M. Daeron, and G. Pancino. 2006. The engagement of activating FcγRs inhibits primate lentivirus replication in human macrophages. J. Immunol. 1776291-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeHart, J. L., E. S. Zimmerman, O. Ardon, C. M. Monteiro-Filho, E. R. Arganaraz, and V. Planelles. 2007. HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol. J. 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher, T. M., III, B. Brichacek, N. Sharova, M. A. Newman, G. Stivahtis, P. M. Sharp, M. Emerman, B. H. Hahn, and M. Stevenson. 1996. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM). EMBO J. 156155-6165. [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, M., M. Otsuka, M. Miyoshi, B. Khamsri, M. Nomaguchi, and A. Adachi. 2008. Vpx is critical for reverse transcription of the human immunodeficiency virus type 2 genome in macrophages. J. Virol. 827752-7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goujon, C., V. Arfi, T. Pertel, J. Luban, J. Lienard, D. Rigal, J. L. Darlix, and A. Cimarelli. 2008. Characterization of simian immunodeficiency virus SIVsm/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J. Virol. 8212335-12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goujon, C., L. Riviere, L. Jarrosson-Wuilleme, J. Bernaud, D. Rigal, J. L. Darlix, and A. Cimarelli. 2007. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guyader, M., M. Emerman, L. Montagnier, and K. Peden. 1989. VPX mutants of HIV-2 are infectious in established cell lines but display a severe defect in peripheral blood lymphocytes. EMBO J. 81169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287607-614. [DOI] [PubMed] [Google Scholar]
- 15.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 696705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 917311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch, V. M., M. E. Sharkey, C. R. Brown, B. Brichacek, S. Goldstein, J. Wakefield, R. Byrum, W. R. Elkins, B. H. Hahn, J. D. Lifson, and M. Stevenson. 1998. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat. Med. 41401-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrecka, K., M. Gierszewska, S. Srivastava, L. Kozaczkiewicz, S. K. Swanson, L. Florens, M. P. Washburn, and J. Skowronski. 2007. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. USA 10411778-11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, W., N. Vander Heyden, and L. Ratner. 1989. Analysis of the function of viral protein X (VPX) of HIV-2. Virology 173624-630. [DOI] [PubMed] [Google Scholar]
- 20.Huang, J., and J. Chen. 2008. VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene 274056-4064. [DOI] [PubMed] [Google Scholar]
- 21.Jowett, J. B., V. Planelles, B. Poon, N. P. Shah, M. L. Chen, and I. S. Chen. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 696304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kappes, J. C., J. A. Conway, S. W. Lee, G. M. Shaw, and B. H. Hahn. 1991. Human immunodeficiency virus type 2 vpx protein augments viral infectivity. Virology 184197-209. [DOI] [PubMed] [Google Scholar]
- 23.Kawamura, M., H. Sakai, and A. Adachi. 1994. Human immunodeficiency virus Vpx is required for the early phase of replication in peripheral blood mononuclear cells. Microbiol. Immunol. 38871-878. [DOI] [PubMed] [Google Scholar]
- 24.Koh, K. B., M. Fujita, and A. Adachi. 2000. Elimination of HIV-1 plasmid DNA from virus samples obtained from transfection by calcium-phosphate co-precipitation. J. Virol. Methods 9099-102. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J., and P. Zhou. 2007. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 26775-780. [DOI] [PubMed] [Google Scholar]
- 26.Le Rouzic, E., N. Belaidouni, E. Estrabaud, M. Morel, J. C. Rain, C. Transy, and F. Margottin-Goguet. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6182-188. [DOI] [PubMed] [Google Scholar]
- 27.Le Rouzic, E., M. Morel, D. Ayinde, N. Belaidouni, J. Letienne, C. Transy, and F. Margottin-Goguet. 2008. Assembly with the Cul4A-DDB1DCAF1 ubiquitin ligase protects HIV-1 Vpr from proteasomal degradation. J. Biol. Chem. 28321686-21692. [DOI] [PubMed] [Google Scholar]
- 28.Li, X., B. Gold, C. O'Huigin, F. Diaz-Griffero, B. Song, Z. Si, Y. Li, W. Yuan, M. Stremlau, C. Mische, H. Javanbakht, M. Scally, C. Winkler, M. Dean, and J. Sodroski. 2007. Unique features of TRIM5α among closely related human TRIM family members. Virology 360419-433. [DOI] [PubMed] [Google Scholar]
- 29.Mahalingam, S., B. Van Tine, M. L. Santiago, F. Gao, G. M. Shaw, and B. H. Hahn. 2001. Functional analysis of the simian immunodeficiency virus Vpx protein: identification of packaging determinants and a novel nuclear targeting domain. J. Virol. 75362-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 7410074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pancio, H. A., N. Vander Heyden, and L. Ratner. 2000. The C-terminal proline-rich tail of human immunodeficiency virus type 2 Vpx is necessary for nuclear localization of the viral preintegration complex in nondividing cells. J. Virol. 746162-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, I. W., and J. Sodroski. 1995. Functional analysis of the vpx, vpr, and nef genes of simian immunodeficiency virus. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 8335-344. [PubMed] [Google Scholar]
- 33.Planelles, V., J. B. Jowett, Q. X. Li, Y. Xie, B. Hahn, and I. S. Chen. 1996. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J. Virol. 702516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Re, F., D. Braaten, E. K. Franke, and J. Luban. 1995. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J. Virol. 696859-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogel, M. E., L. I. Wu, and M. Emerman. 1995. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 69882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowland-Jones, S. L., and H. C. Whittle. 2007. Out of Africa: what can we learn from HIV-2 about protective immunity to HIV-1? Nat. Immunol. 8329-331. [DOI] [PubMed] [Google Scholar]
- 37.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 1022832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrofelbauer, B., Y. Hakata, and N. R. Landau. 2007. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. USA 1044130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selig, L., S. Benichou, M. E. Rogel, L. I. Wu, M. A. Vodicka, J. Sire, R. Benarous, and M. Emerman. 1997. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J. Virol. 714842-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selig, L., J. C. Pages, V. Tanchou, S. Preveral, C. Berlioz-Torrent, L. X. Liu, L. Erdtmann, J. Darlix, R. Benarous, and S. Benichou. 1999. Interaction with the p6 domain of the gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J. Virol. 73592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharova, N., Y. Wu, X. Zhu, R. Stranska, R. Kaushik, M. Sharkey, and M. Stevenson. 2008. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS. Pathog. 4e1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sitterlin, D., F. Bergametti, and C. Transy. 2000. UVDDB p127-binding modulates activities and intracellular distribution of hepatitis B virus X protein. Oncogene 194417-4426. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava, S., S. K. Swanson, N. Manel, L. Florens, M. P. Washburn, and J. Skowronski. 2008. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS. Pathog. 4e1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stivahtis, G. L., M. A. Soares, M. A. Vodicka, B. H. Hahn, and M. Emerman. 1997. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J. Virol. 714331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427848-853. [DOI] [PubMed] [Google Scholar]
- 46.Tan, L., E. Ehrlich, and X. F. Yu. 2007. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J. Virol. 8110822-10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tristem, M., C. Marshall, A. Karpas, and F. Hill. 1992. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 113405-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ueno, F., H. Shiota, M. Miyaura, A. Yoshida, A. Sakurai, J. Tatsuki, A. H. Koyama, H. Akari, A. Adachi, and M. Fujita. 2003. Vpx and Vpr proteins of HIV-2 up-regulate the viral infectivity by a distinct mechanism in lymphocytic cells. Microbes Infect. 5387-395. [DOI] [PubMed] [Google Scholar]
- 49.Verrier, F. C., P. Charneau, R. Altmeyer, S. Laurent, A. M. Borman, and M. Girard. 1997. Antibodies to several conformation-dependent epitopes of gp120/gp41 inhibit CCR-5-dependent cell-to-cell fusion mediated by the native envelope glycoprotein of a primary macrophage-tropic HIV-1 isolate. Proc. Natl. Acad. Sci. USA 949326-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.