Abstract
UL31 and UL34 of herpes simplex virus type 1 form a complex necessary for nucleocapsid budding at the inner nuclear membrane (INM). Previous examination by immunogold electron microscopy and electron tomography showed that pUL31, pUL34, and glycoproteins D and M are recruited to perinuclear virions and densely staining regions of the INM where nucleocapsids bud into the perinuclear space. We now show by quantitative immunogold electron microscopy coupled with analysis of variance that gD-specific immunoreactivity is significantly reduced at both the INM and outer nuclear membrane (ONM) of cells infected with a UL34 null virus. While the amount of gM associated with the nuclear membrane (NM) was only slightly (P = 0.027) reduced in cells infected with the UL34 null virus, enrichment of gM in the INM at the expense of that in the ONM was greatly dependent on UL34 (P < 0.0001). pUL34 also interacted directly or indirectly with immature forms of gD (species expected to reside in the endoplasmic reticulum or nuclear membrane) in lysates of infected cells and with the cytosolic tail of gD fused to glutathione S-transferase in rabbit reticulocyte lysates, suggesting a role for the pUL34/gD interaction in recruiting gD to the NM. The effects of UL34 on gD and gM localization were not a consequence of decreased total expression of gD and gM, as determined by flow cytometry. Separately, pUL31 was dispensable for targeting gD and gM to the two leaflets of the NM but was required for (i) the proper INM-versus-ONM ratio of gD and gM in infected cells and (ii) the presence of electron-dense regions in the INM, representing nucleocapsid budding sites. We conclude that in addition to their roles in nucleocapsid envelopment and lamina alteration, UL31 and UL34 play separate but related roles in recruiting appropriate components to nucleocapsid budding sites at the INM.
Herpesvirus virions comprise a nucleocapsid containing genomic viral DNA, a proteinaceous tegument layer surrounding the nucleocapsid, and a virion envelope surrounding the tegument. The envelope of extracellular herpes simplex virus (HSV) virions contains glycoproteins gB, gC, gD, gE, gI, gG, gH, gK, gL, and gM (23, 51).
As viewed by electron microscopy, nascent virions form as the nucleocapsid buds through densely staining regions of the nuclear membrane (NM) (21, 41). Electron tomograms of HSV perinuclear virions compared to those of extracellular virions infer that the former contain glycoproteins of considerably less glycosylation and a relatively sparse tegument layer compared to their counterparts in mature extracellular virions (6). The lower levels of glycosylation in HSV perinuclear virions are consistent with the fact that the lumen of the perinuclear space is continuous with that of the endoplasmic reticulum. Thus, the polysaccharide moieties of virion glycoproteins become fully processed as virions access Golgi enzymes during their egress to the extracellular space. Although the full proteome of the nascent perinuclear virion is unknown, immunogold studies have shown that they contain at least pUL31, pUL34, pUS3, gB, gC, gD, gH, gM, and the VP16 and pUL11 tegument proteins in addition to the proteins that comprise the viral capsid (4, 5, 15, 25, 37, 40, 47, 50, 55).
The UL31 and UL34 gene products of HSV-1 (pUL31 and pUL34, respectively) form a complex that localizes at the inner and outer NMs (INM and ONM, respectively) of infected cells (40). Both proteins are essential for nucleocapsid envelopment at the INM and become incorporated into nascent virions when nucleocapsids bud through the INM into the perinuclear space (39, 40, 42). The proteins and their essential role in nucleocapsid envelopment are conserved in all herpesvirus subfamilies (14, 20, 32, 45). pUL31 of HSV-1 is a mostly hydrophobic phosphoprotein that is held in close approximation to the nucleoplasmic face of the INM by interaction with pUL34, an integral membrane protein of type II orientation (33, 40, 46, 56). The first 248 amino acids of pUL34 are predicted to reside in the nucleoplasm or cytoplasm, depending on whether the protein localizes in the INM or ONM, respectively. This is followed by an approximately 22-amino acid transmembrane domain with up to 5 amino acids residing in the perinuclear space or lumen of the endoplasmic reticulum.
In the most prominent model of herpesvirion egress, the envelope of the perinuclear virion fuses with the ONM, releasing the deenveloped nucleocapsid into the cytoplasm, where it subsequently buds into cytoplasmic membranous organelles such as the Golgi or trans-Golgi network (34, 49). This model is supported by the observation that pUL31 and pUL34 are located in the perinuclear virion but not extracellular virions (18, 40). Thus, these proteins are lost from the virion upon fusion of the virion envelope with the ONM. Also supporting this egress model is the observation that deletion of both gB and gH causes virions to accumulate aberrantly in the perinuclear space (15). The involvement of gH and gB is potentially satisfying because these proteins comprise essential components of the machinery that mediates fusion of the virion envelope with the plasma or endosomal membranes during the initiation of infection (9, 12, 16, 44, 52). Moreover, expression of a combination of gB, gD, gH, and gL is sufficient to mediate fusion of cell membranes, whereas coexpression with gM or gK inhibits this fusion (3, 8, 11). Although the mechanism of fusion is unclear, gD is known to bind viral receptors on cell surfaces, and the structure of gB indicates features reminiscent of other viral fusion proteins (24, 35, 48). gD has been shown to interact with gB and gH at least transiently, suggesting that these interactions may be important for the fusion reaction (1, 2). Thus, fusion between the nascent and mature virion envelopes with target membranes may share mechanistic similarities.
On the other hand, it is likely that the two fusion events are mechanistically distinct because (i) single deletion of either gH or gB precludes viral entry and cell/cell fusion but does not cause nascent virions to accumulate in the perinuclear space (9, 16, 31, 43) and (ii) the activity of a viral kinase encoded by US3 is dispensable for entry but believed to promote fusion of the perinuclear virion and ONM (28, 40). Moreover, the lack of glycoproteins from the pseudorabies virus perinuclear virion suggests that fusion is mediated by an entirely different mechanism in this system (26).
The current study focuses on how glycoproteins are incorporated into the nascent virion. We show that optimal recruitment of gD to both leaflets of the NM and gM to the INM requires pUL34 and pUL31. We also show that immature gD interacts with pUL34, suggesting a mechanism by which pUL34 might recruit gD to the NM.
MATERIALS AND METHODS
Cells and viruses.
The UL34 and UL31 deletion viruses and the HSV-1(F) virus from which they were derived have been described previously (10, 13, 42). Viral stocks of the UL34 and UL31 null viruses were propagated on complementing cell lines derived from rabbit skin cells, and these cell lines were maintained as described previously (30, 39).
A novel virus bearing an influenza hemagglutinin (HA) epitopic tag at the C terminus of pUL34 was constructed using en passant mutagenesis of a bacterial artificial chromosome (BAC) containing the entire HSV-1(F) genome as previously described, with some modifications (53, 54). Briefly, a DNA amplicon containing a kanamycin (Kan) resistance cassette and Sce1 site and encoding HA fused to the C terminus of pUL34 was flanked by sequences homologous to UL34 and was PCR amplified from a pEPkan-S plasmid using the primer pairs listed in Table 1. The resulting amplicon was electroporated into recombination-competent GS1783 Escherichia coli, harboring an HSV-1(F) BAC and an Sce1 endonuclease gene integrated into its chromosome under the control of an arabinose-inducible promoter (this strain was a kind gift of Greg Smith, Northwestern University). Following screening for Kan resistance and genotypic confirmation by restriction fragment length polymorphism, the Kanr gene was removed by arabinose-induced SceI expression, resulting in Red recombination between the homologous sequences within the PCR primers. Fusion of the HA tag to the C terminus of pUL34 was verified through restriction fragment length polymorphism and DNA sequencing of Kans clones. Rabbit skin cells were transfected with the BAC and a plasmid encoding the FLP recombination target recombinase to remove the BAC sequences as described previously (30). Virus within viral plaques was amplified into viral stocks using Vero cells, and the phenotype (nuclear rim staining of the tagged pUL34) was verified by indirect immunofluorescence using HA-specific antibody (data not shown). Growth curves of the resulting recombinant virus revealed growth kinetics similar to that of wild-type HSV-1(F) (not shown).
TABLE 1.
Primers for constructing UL34-HA recombinant virus
| Gene and direction | Sequencea |
|---|---|
| UL34 | |
| Forward | 5′-GGCCGCTATTTGGTGGGTGGTTGGTGCTGGCGCGCGCCTATACCCATACGACGTCCCAGACTACGCGTAAAAAAGTAGGGATAACAGGGT-3′ |
| Reverse | 5′-GGAACGCACTGGCGATTAGGGCGGCGGTGCGTCCTTTTTTACGCGTAGTCTGGGACGTCGTATGGGTATAGGCGCGCCAGTGTTACAACC-3′ |
HA insertion sequences are in italics, UL34 homologous sequences are underlined, and pEPKanS template priming sequences are bold.
Immunogold electron microscopy.
Electron microscopy, photography, and immunogold staining of gD in plastic-embedded sections were performed essentially as described previously (40) except that (i) LRWhite plastic was polymerized at 50°C without polymerization accelerator, (ii) some thin sections were probed with gD-specific mouse monoclonal antibody (a kind gift from Gary Cohen and Roselyn Eisenberg) diluted 1:50 in phosphate-buffered saline (PBS) supplemented with 0.5% Tween 20 and 1% fish gelatin, and (iii) these sections were reacted with goat anti-mouse immunoglobulin G (IgG) conjugated with 12-nm colloidal gold (catalog no. 115-205-146; Jackson ImmunoResearch). Statistical analyses were performed using the JMP statistical package.
Conventional electron microscopy.
At 18 hours after mock infection or infection with 5.0 PFU/cell of the indicated viruses, Hep2 cells were fixed with 2.5% glutaraldehyde (Electron Microscopy Sciences) in 0.1 M sodium cacodylate buffer, pH 7.4, for 30 min at room temperature and then 90 min at 4°C. After three rinses for 5 min each with the same buffer, cells were treated with 4% osmium tetroxide for 1 h at room temperature, rinsed again with 0.1 M sodium cacodylate buffer, and subsequently dehydrated with a graduated series of ethanol concentrations (10%, 30%, 50%, 70%, and 100%), followed by increasing concentrations of acetone (50% and then two incubations with 100%). This was followed by stepwise infiltration with Epon-Araldite resin (Electron Microscopy Sciences) over the course of 48 h at room temperature. Samples were dispensed into Beem capsules, and the resin was polymerized at 65°C for 18 h. Thin sections (60- to 90-nm thick) were collected on 300-mesh copper grids (Ted Pella, Inc., Redding, CA). Thin sections were counterstained with 2% aqueous uranyl acetate for 20 min and then with Reynolds lead citrate for 7 min. Stained grids were viewed in a Philips 201 transmission electron microscope. Conventionally rendered negatives of electron microscopic images were scanned by using a Microtek ScanMaker 5 and ScanWizard Pro PPC 1.02 software. Positive images were rendered from digitized negatives with Adobe Photoshop software.
Flow cytometry.
Hep2 cells were infected with HSV-1(F) or the UL31 or UL34 null viruses at 3 PFU/cell. At 16 h postinfection, cells were removed from the culture dishes by trypsinization, rinsed two times with cold PBS, fixed with 3% paraformaldehyde in PBS for 15 min, and rinsed three times in excess PBS. Autofluorescence was quenched by treatment with 50 mM NH4Cl in PBS for 15 min, followed by another PBS rinse. Cells were permeabilized with 0.1% Triton X-100 in PBS for 2 min, followed by three rinses in PBS. Blocking of nonspecific immunoreactivity was done by incubation in PBS supplemented with 10% pooled human sera and 10% goat serum for 15 min.
In one experiment, the infected cells were reacted with gD-specific primary mouse monoclonal antibody diluted 1:100 in PBS supplemented with 0.1% Tween 20. The cells were then reacted sequentially for 30 min with rabbit anti-ICP8 antibody diluted 1:200 (a gift from Bill Ruyechan), fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (diluted 1:100), and Cy5-conjugated anti-rabbit conjugate diluted 1:100. The cells were washed in excess cold PBS supplemented with 0.2% Tween 20 between each antibody reaction.
In the other experiment, infected cells were reacted with rabbit anti-gM antiserum diluted 1:200 and were reacted sequentially for 30 min with a mouse monoclonal antibody directed against ICP4 (diluted 1:100), FITC-conjugated anti-rabbit IgG (diluted 1:100), and Cy5-conjugated anti-mouse IgG (diluted 1:100). Between each reaction, the cells were washed three times in PBS-0.2% Tween 20.
Cells from both experiments were analyzed separately on a FACSCalibur flow cytometer using CellQuest software (BD Biosciences). Data analysis was performed using CellQuest v3.3 software.
GST pull down from HSV-infected cell lysates.
Approximately 4 × 108 Hep2 cells were infected with 5.0 PFU/cell of wild-type HSV-1(F). Sixteen hours later, the cells were lysed in 30 ml modified RIPA buffer {50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% 3-[(3-cholamidopropyl)-diamethylammonio]-1-propanesulfonate [CHAPS], 0.5% Na-deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA} containing 1× complete protease inhibitor cocktail (Roche) and phosphatase inhibitors (10 mM NaF, 10 mM Na3VO4) with gentle rocking for 2 h at 4°C. The lysates were clarified by centrifugation for 20 min at 10,000 × g at 4°C and were precleared by the reaction mixture with excess glutathione-Sepharose beads (GE) for 2 h at 4°C. Glutathione S-transferase (GST) fused to pUL34 (GST-pUL34; 300 μg) was prepared as described previously (36) except that the affinity-purified protein was cross-linked to glutathione-Sepharose beads with 5 mM bis(sulfosuccinimidyl) suberate (Pierce Chemical) according to the manufacturer's protocol. GST similarly cross-linked to glutathione-Sepharose beads served as a control. After overnight incubation of the GST- or GST-pUL34-laden Sepharose beads with the precleared cellular lysates at 4°C, the beads were washed two times with ice-cold RIPA buffer and then three times with 0.5% Tween 20 in PBS (sodium phosphate buffer, pH 7.4, supplemented with 150 mM NaCl). Bound proteins were eluted in SDS-polyacrylamide gel electrophoresis sample buffer (10 mM Tris-HCl [pH 8.0], 10 mM β-mercaptoethanol, 20% glycerol, 5% SDS, and trace amounts of bromophenol blue) by immersion in a boiling water bath for 10 min. Eluted proteins were electrophoretically separated on a 10% SDS-polyacrylamide gel and visualized by Sypro-Ruby staining. Bands that by inspection were more heavily or uniquely present in the elution from GST-pUL34 beads as opposed to from GST beads were excised and submitted to a central mass spectrometry facility at the Biotechnology Resource Center, Cornell University, where the incorporated proteins were digested by trypsin and the masses of peptides were determined by liquid chromatography-mass spectrometry and subsequently identified by comparison with an NCBI virus database using Mascot software (Matrix Science).
Immunoprecipitation and immunoblotting.
Approximately 2 × 107 Hep2 cells were infected with 5.0 PFU of the pUL34-HA recombinant HSV-1 per cell and were lysed at 16 h postinfection in 1.5 ml RIPA buffer (50 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1% CHAPS, 0.25% Na-deoxycholate, 1 mM EDTA) containing 1× complete protease inhibitor cocktail (Roche) and phosphatase inhibitor (10 mM NaF, 10 mM Na3VO4) with gentle rocking for 3 h. This and all subsequent steps were performed on ice or at 4°C. The lysates were clarified by centrifugation for 15 min at 14,000 × g, and the supernatants were incubated with 2 μg rabbit anti-HA antibody (sc-805; Santa Cruz Biotechnology) or nonimmune rabbit serum. After overnight incubation, the insoluble material was removed by centrifugation for 15 min at 14,000 × g. Twenty microliters of a slurry of protein A/G Plus-agarose beads (Santa Cruz Biotechnology) was then added to the supernatants and incubated for another 2 h. The beads were washed three times with RIPA buffer and boiled in SDS-polyacrylamide gel electrophoresis sample buffer (10 mM Tris-HCl [pH 8.0], 10 mM β-mercaptoethanol, 20% glycerol, 5% SDS, and trace amounts of bromophenol blue) and subjected to electrophoresis in a 12% polyacrylamide gel in the presence of 0.1% SDS. Resolved protein samples were transferred to nitrocellulose sheets for immunoblotting.
Nitrocellulose sheets bearing proteins of interest were blocked in 5% nonfat milk plus 0.2% Tween 20 for at least 2 h. The membrane was probed with a mouse monoclonal antibody directed against gD (antibody DL-6; a gift of Roselyn Eisenberg and Gary Cohen), followed by polyclonal pUL34 chicken antibody (a gift from Richard Roller) as needed. Primary antibody was detected by horseradish peroxidase-conjugated bovine anti-mouse and anti-chicken secondary antibodies, respectively (Santa Cruz Biotechnology). All bound immunoglobulins were visualized by enhanced chemiluminescence (Pierce), followed by exposure to X-ray film.
gDtail-GST pull down of pUL34 expressed in vitro.
The cytosolic tail region (amino acids 359 to 394) of gD (gDtail) was amplified by PCR from a full-length cDNA construct (kindly provided by Gary Cohen and Roselyn Eisenberg, University of Pennsylvania) using an upstream primer containing an EcoRI site (5′-ATATGAATTCGGAATTGTGTACTGGATGCG) and a downstream primer containing a XhoI site (5′-ATATCTCGAGCTAGTAAAACAAGGGCTGGTGCGA) (restriction sites in italics). The PCR product was cloned as an EcoRI/XhoI fragment into the vector pGEX4T-1 in frame with the GST gene. This plasmid was designated pJB650.
The construct described above was used to transform a chemically competent BL21(DE3) codon plus E. coli. For production of gDtail-GST, 2 ml of fresh stationary-phase culture was inoculated into 200 ml of Luria broth supplemented with ampicillin and grown at 37°C to an optical density at 600 nm of 0.6, when protein expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside. Incubation was continued at 30°C for 3 h. Bacteria were pelleted and lysed as previously described by Frangioni and Neel (17) except that one tablet of complete EDTA-free protease inhibitor cocktail (Roche) was added during bacterial lysis with lysozyme and Sarkosyl. The final supernatant contained 1.5% Sarkosyl and 4% Triton-X in cold STE buffer (50 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The mixture was incubated overnight at 4°C with glutathione-Sepharose 4B beads (Amersham Biosciences). The beads were then pelleted and washed extensively with cold sterile PBS.
A plasmid containing full-length UL34 (pJB234) has been described previously (39). Full-length UL34 was expressed and radiolabeled with [35S]methionine using the Promega TNT rabbit reticulocyte transcription/translation coupled system programmed with pJB234 in the presence of canine microsomal membranes according to the manufacturer's protocol. Ten microliters of the UL34 protein reaction mixture was either electrophoretically separated on an SDS-8% polyacrylamide gel or incubated overnight at 4°C with 20 μg of gDtail-GST fusion protein or 20 μg of GST bound to glutathione-Sepharose 4B beads in cold PBS. The beads were then washed four times with an excess volume of cold PBS. Bound proteins were eluted by being boiled in 2× SDS loading buffer and electrophoretically separated on an SDS-8% polyacrylamide gel. The gel was soaked for 30 min in 20% sodium salicylate and dried. Fluorography was performed with CL-XPosure film (Thermo Scientific) exposed overnight at −80°C.
RESULTS
PUL34 interacts with immature gD.
To identify pUL34-interacting partners, GST-pUL34 or GST was bound and cross-linked separately to glutathione-Sepharose beads. Proteins within clarified lysates of HSV-1(F)-infected Hep2 cells that bound the Sepharose beads were eluted in SDS and electrophoretically separated in a denaturing SDS-polyacrylamide gel. Bands overrepresented or unique to eluates of the GST-pUL34 beads were excised, and the masses of tryptic peptides derived from proteins within the bands were determined by mass spectrometry and identified by comparison with the NCBI virus databases.
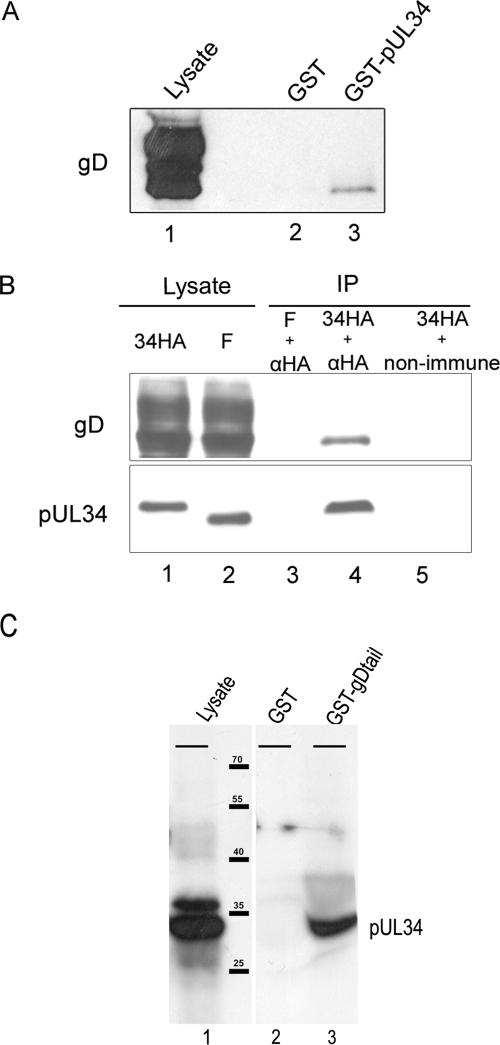
A single peptide (SVLLNAPSEAPQIVR) that corresponded to the predicted peptide of gD amino acids 93 to 107 was identified in the GST-pUL34 pull down. To verify the putative gD/pUL34 interaction, mock-infected Hep2 cell lysates, lysates of Hep2 cells infected with HSV-1(F), and the eluates from the GST and GST-pUL34 pull-down reactions were electrophoretically separated, transferred to nitrocellulose membranes, and probed with an antibody directed against gD. As shown in Fig. 1A, whereas gD was not detected in mock-infected cells, a broad band indicative of both mature and immature forms of gD reacted with the gD-specific antibody in lysates of cells infected with HSV-1(F). More importantly for the purposes of this report, gD was detected in material eluted from the GST-pUL34-containing Sepharose beads but was absent from eluates from beads containing GST. Moreover, the migration of gD eluted from GST-pUL34 was faster than that of most gD species detected in the HSV-1(F) lysate. These observations suggested that pUL34 interacted preferentially with immature forms of gD over mature forms. This was consistent with the fact that pUL34 is located primarily within the NM and perinuclear space of HSV-1(F)-infected cells (40).
FIG. 1.
Interaction between gD and pUL34. (A) gD immunoblot of GST pull down. Lysates of cells infected with HSV-1(F) (lane 1) were reacted with GST (lane 2) or GST fused to pUL34 (lane 3). After the beads were washed, bound proteins were eluted in denaturing buffer, electrophoretically separated, transferred to nitrocellulose membranes, and probed with gD-specific antibody. (B) Coimmunoprecipitation (IP) of gD- and HA-tagged pUL34. Cells were infected with HSV-1(F) (F) or a recombinant virus bearing a HA tag fused to the C terminus of pUL34 (34HA). Cellular lysates (lanes 1 and 2) were reacted with anti-HA antibody (lanes 3 and 4) or nonimmune antibody (lane 5). Antibody-antigen complexes were purified, electrophoretically separated, and subjected to immunoblotting with mouse monoclonal anti-gD (top) or pUL34-specific anti-IgY (bottom). (C) Interaction of pUL34 and gDtail in rabbit reticulocyte lysates. [35S]methionine-labeled pUL34 was expressed in a transcription/translation-coupled rabbit reticulocyte lysate expression system in the presence of pancreatic microsomes, and 5 μl was either electro- phoretically separated (lane 1) or reacted with Sepharose beads bearing GST (lane 2) or GST fused to gD amino acids 359 to 394 (lane 3). After beads with bound proteins were washed, proteins bound to beads were eluted and electrophoretically separated on a denaturing polyacrylamide gel. The gel was then dried and subjected to fluorography. The migration positions and sizes of protein standards are indicated in thousands. The bars at the top of each lane indicate the origin of the resolving gel.
To confirm the putative interaction between gD and pUL34, a novel virus was constructed using BAC technology as described in Materials and Methods. This mutant viral genome encoded a HA epitopic tag fused to the C terminus of pUL34, where it was expected to reside within the perinuclear space of infected cells. The virus bearing the tag replicated normally, indicating that the tag did not interfere with viral replication (data not shown). Cells were infected with 5.0 PFU of the pUL34-HA-tagged virus and were lysed at 16 h after infection. The lysates were then reacted with preimmune antibody or a monoclonal antibody directed against the HA tag, and immune complexes were purified on protein A/G-containing agarose beads. Bound immune complexes were then eluted in SDS-containing buffer, electrophoretically separated on an SDS-polyacrylamide gel, transferred to nitrocellulose membranes, and subjected to immunoblotting with gD- and pUL34-specific antibodies.
Immunoblot analyses of the lysates of HSV-1(F)-infected cells and cells infected with the recombinant virus revealed that the presence of DNA encoding the HA tag slowed the migration of pUL34, thus indicating that the tag was fused to pUL34. Probing material immunoprecipitated with the HA-specific antibody with a pUL34-specific antibody revealed that HA-tagged pUL34 was readily immunoprecipitated from lysates of cells infected with the recombinant virus but not from lysates of HSV-1(F)-infected cells. Most important for the purposes of this study, the immunoprecipitations also contained gD, as revealed by immunoblotting with the gD-specific antibody. The gD that was immunoprecipitated migrated faster than most gD species in the lysates, suggesting that underglycosylated gD preferentially interacted with pUL34. In a control reaction, gD was not immunoprecipitated by the HA antibody from lysates of cells infected with HSV-1(F). These studies lend further support to the conclusion that immature gD and pUL34 interact in infected cells.
To determine whether the interaction between pUL34 and gD required other viral proteins, a GST fusion protein bearing gDtail was purified from Escherichia coli on Sepharose beads and reacted with full-length pUL34 labeled with [35S]methionine in a rabbit reticulocyte lysate. As a control, GST was reacted with radiolabeled pUL34 in parallel. After beads with bound proteins were washed extensively, proteins bound to the beads were eluted, electrophoretically separated, and subjected to fluorography. As shown in Fig. 1C, GST fused to gDtail pulled down pUL34 expressed in the rabbit reticulocyte lysate, whereas GST did not pull down radiolabeled pUL34. These data indicate that gDtail can interact with pUL34 in the absence of other viral proteins.
pUL31 and pUL34 promote gD localization at the NM.
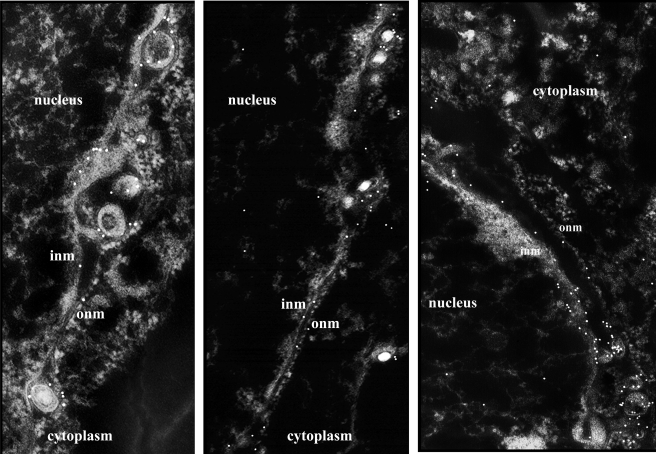
As a first step to determine the significance of the interaction between pUL34 and immature gD, we tested whether gD recruitment to the NM was dependent on pUL34 and pUL34's interacting partner pUL31. Cells were therefore infected with HSV-1(F) or mutant viruses lacking UL31 or UL34. At 12 to 14 h after infection, the cells were fixed and embedded in LRWhite, and thin sections (20- to 40-nm thick) were reacted with monoclonal antibody directed against gD, followed by a reaction with anti-mouse IgG conjugated to 12-nm colloidal gold beads. Examples of such reactions in cells infected with HSV-1(F) are shown in Fig. 2. As noted previously, both gM and gD colocalized with both leaflets of the NM and with virions located between these leaflets. Examination of cells infected with the UL31 and UL34 deletion viruses indicated that gD was at least occasionally detectable at the INM of cells infected with all three viruses (not shown). However, our initial impression was that less gD-specific signal was present in the INM of cells infected with the pUL31 and pUL34 null viruses. To ascertain whether this was the case, the number of gD-specific gold beads in individual leaflets of the NM was determined in cells infected with the various viruses. The results are presented in Tables 2 and 3 and are summarized as follows. (i) Analysis of variance of the amount of gD-specific immunoreactivity at both leaflets of the NM of cells infected with the UL34 deletion virus was significantly reduced relative to the amount of immunoreactivity associated with the NM of cells infected with HSV-1(F) or the UL31 deletion mutant (P = 0.0004 and P = 0.0126, respectively). (ii) The ratio of gD-specific immunoreactivity in the INM versus ONM of cells infected with HSV-1(F) was approximately 1.0 (mean, 1.15 ± 0.72). With the caveat that there were significantly fewer beads associated with the NM of cells infected with the UL34 deletion virus, statistically this ratio was not significantly different from the ratio of gD at the INM versus ONM of cells infected with the UL34 deletion mutant (Table 3). (iii) The total amount of gD immunoreactivity at the NM was not significantly different in cells infected with the UL31 deletion virus from that in cells infected with HSV-1(F). (iv) The ratio of gD at the INM versus ONM in cells infected with the UL31 deletion virus was decreased, but given the variability of immunostaining from section to section, this difference was not significantly different from that in cells infected with HSV-1(F) (P = 0.125) (Table 3).
FIG. 2.
Example of gD and gM immunogold electron microscopy. Cells were infected with HSV-1(F), embedded in plastic, sectioned, and reacted with either gD-specific (left and middle) or gM-specific (right) antibodies. After sections with bound immunoglobulin were washed extensively, bound immunoglobulin was revealed by a reaction with anti-mouse (left and middle) or anti-rabbit (right) immunoglobulins conjugated to 12-nm gold beads. Negatives of the transmission electron micrographs are shown to better illustrate the presence of gold beads associated with various structures. As a size standard, viral capsids are approximately 125 nm in diameter. Data from many sections are summarized and analyzed in Tables 2 and 3.
TABLE 2.
Amount of gD-specific immunoreactivity associated with the NM (sum of both leaflets) of cells infected with various viruses
| Virus | Total NM examined (μM) | Total no. of beads counted | Mean (gD-specific beads per μM NM) | SE | 95% CI |
|---|---|---|---|---|---|
| HSV-1(F) | 160 | 1,190 | 7.44 | 1.12 | 5.13-9.75 |
| UL34 null | 259 | 435 | 1.68 | 0.88 | 0-3.5a |
| UL31 null | 83 | 502 | 6.06 | 1.55 | 2.86-9.3 |
The difference in means from HSV-1(F)-infected cells and UL31 deletion virus-infected cells is statistically significant (P values of 0.0004 and 0.0216, respectively).
TABLE 3.
Comparison of means of gD localization in INM versus ONM in cells infected with various viruses
| Virus | Mean ratio of gM INM/ONM | SD | SEM | 95% CI | P value [difference from HSV-1(F)]a |
|---|---|---|---|---|---|
| HSV-1(F) | 1.14 | 0.727 | 0.247 | 0.630-1.65 | ND |
| UL34 null | 1.27 | 0.194 | 0.195 | 0.541-1.055 | 0.671 |
| UL31 null | 0.47 | 0.285 | 0.343 | 0-1.17 | 0.125 |
Determined by using Student's t test. ND, not determined.
Together, these data indicate that UL34 is necessary for accumulation of gD at both leaflets of the NM.
Neither pUL34 nor pUL31 affects expression of gD in infected cells.
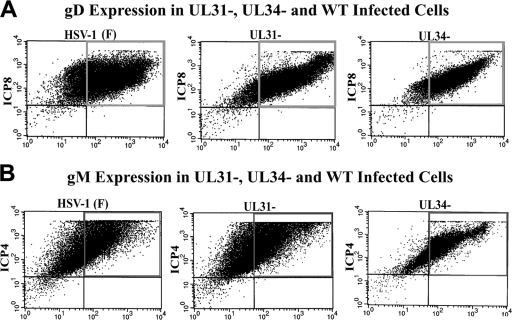
To determine whether the lower levels of gD at the INM of cells infected with the UL34 deletion virus were a consequence of lower overall expression of gD, cells were infected with HSV-1(F) or the UL34 deletion virus, and the level of expression of gD was determined by indirect immunofluorescence and quantified by flow cytometry. Because pUL31 affects gD localization at the nuclear rim, cells were also infected with the UL31 null virus and immunostained similarly. Infected cells were identified by immunostaining with a rabbit antibody to a second viral protein, ICP8. As shown in Fig. 3A, although a population of cells expressing high levels of ICP8 and relatively low levels of gD was unique to the HSV-1(F) infection, there was little difference in the numbers of cells expressing large amounts of gD in infections with the different viruses.
FIG. 3.
Flow cytometry of gD and gM immunostaining of Hep2 cells infected with wild-type and mutant viruses. (A) Monolayers of Hep2 cells were infected with the indicated viruses, and the cells were removed from the substrate by trypsinization and then fixed, permeabilized in paraformaldehyde and Triton X-100, and reacted sequentially with mouse monoclonal antibody to gD, rabbit anti-ICP8, FITC-conjugated anti-mouse, and Cy5-conjugated anti-rabbit immunoglobulins. The cells were analyzed on a FACSCalibur flow cytometer, and the levels of ICP8-specific (y axis) and gD-specific (x axis) antibody are shown for each cell. (B) Hep2 cells were infected as described for panel A, except that the permeabilized cells were immunostained with rabbit antibody to gM (x axis) and mouse monoclonal antibody to ICP4 (y axis), followed by a reaction with FITC-conjugated anti-rabbit and Cy5-conjugated anti-mouse immunoglobulins.
Thus, the UL34-dependent increase in gD at the NM was not explainable by a defect in gD expression by the UL34 deletion virus. This conclusion is consistent with the results of others, showing virtually no defect in synthesis of viral proteins by the UL34 null virus (42).
gM recruitment to the INM requires UL31 and UL34.
To determine whether the UL34 and UL31 effects on glycoprotein recruitment to the NM were specific for gD, we also examined the localization of gM in cells infected with HSV-1(F) and the UL31 and UL34 null viruses by using immunogold electron microscopy. The data are presented in Tables 4 and 5 and are summarized as follows. (i) Upon summation of the numbers of gold beads in both leaflets of the NM, there was a slight decrease in the amount of gM immunoreactivity associated with the NM of cells infected with HSV-1(F) compared to that of cells infected with the UL34 null virus (P = 0.027) (Table 4). (ii) The mean ratio of gM-specific immunoreactivity at the INM versus ONM in cells infected with HSV-1(F) was approximately 1.9 (95% confidence interval [CI], 1.62 to 2.20). (iii) The relative amount of gM-specific immunoreactivity at the INM versus ONM of cells infected with both the UL31 and UL34 deletion viruses was significantly reduced (P < 0.0001). Specifically, the mean ratio of INM/ONM in cells infected with the UL31 deletion virus was 0.55 (95% CI, 0.28 to 0.81), whereas the mean of this ratio was 0.80 (95% CI, 0.54 to 1.10) in cells infected with the UL34 null virus.
TABLE 4.
Amount of gM-specific immunoreactivity associated with the NM (sum of both leaflets) of cells infected with various viruses
| Virus | Total NM examined (μM) | Total no. of beads counted | Mean (gD-specific beads per μM NM) | SE | 95% CI |
|---|---|---|---|---|---|
| HSV-1(F) | 203 | 1,307 | 9.15 | 1.77 | 5.36-12.93 |
| UL34 null | 188 | 435 | 3.38 | 1.55 | 0.08-6.68a |
| UL31 null | 172 | 502 | 4.62 | 1.62 | 1.17-8.07 |
Difference in means from HSV-1(F)-infected cells is statistically significant (P = 0.027).
TABLE 5.
Comparison of means of gM localization in INM versus ONM in cells infected with various viruses
| Virus | Mean ratio of gM INM/ONM | SD | SEM | 95% CI | P value [difference from HSV-1(F)]a |
|---|---|---|---|---|---|
| HSV-1(F) | 1.910 | 0.254 | 0.138 | 1.615-2.204 | ND |
| UL34 null | 0.798 | 0.314 | 0.120 | 0.541-1.055 | <0.0001 |
| UL31 null | 0.545 | 0.339 | 0.126 | 0.277-0.814 | <0.0001 |
Determined by using Student's t test.
We conclude that both UL31 and UL34 are necessary for proper recruitment of gM to the INM.
To ensure that the defects in accumulation of gM at the nuclear rim were not a consequence of poor expression of gM in cells infected with the UL31 and UL34 deletion viruses, flow cytometry was performed as indicated above except that infected cells were identified by immunostaining with a mouse monoclonal antibody directed against ICP4 and a rabbit antibody directed against gM. As shown in Fig. 3B, similar numbers of cells infected with the three viruses expressed large amounts of gM.
The presence of dense-staining budding sites in the INM requires pUL31.
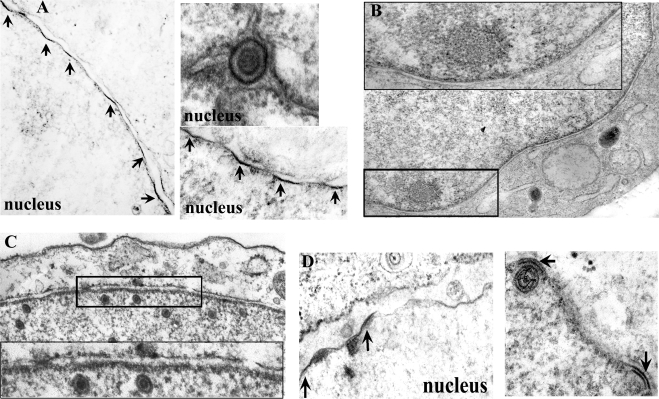
During the course of these studies, it became apparent that densely staining regions of the INM (termed INM budding sites for purposes of discussion) were not apparent in cells infected with the UL31 deletion mutant. To quantify this difference, cells were infected with HSV-1(F) or the UL31 and UL34 deletion viruses, and the presence or absence of INM budding sites was scored as a function of the amount of linear distance of INM examined. The length of individual INM budding sites was also determined. To ensure that only infected cells were examined, sections lacking intranuclear viral capsids were excluded from the analysis. Typical results are shown in Fig. 4 and were quantified as shown in Table 6.
FIG. 4.
Conventional electron microscopy of Hep2 cells mock infected or infected with various viruses. Cells were mock infected or infected at a multiplicity of infection of 5.0 PFU/cell, and 18 h later they were fixed, embedded, sectioned, and stained with uranyl acetate and osmium tetroxide. (A) Cells infected with HSV-1(F). Arrows indicate densely staining portions of INM. The top right panel shows a perinuclear virion exhibiting dense staining of the virion envelope. (B) Mock-infected cells. The inset shown at the bottom of this panel is illustrated at a higher magnification at the top. (C) Cells infected with the UL31 null virus. The inset at the top of the panel is magnified at the bottom of the panel. Electron densities at the INM are not apparent. (D) Cells infected with UL34 null virus. Densely staining areas of the NM are indicated by arrows.
TABLE 6.
Electron densities in INM of cells infected with various viruses
| Virus | Total INM examined (nm) | Total dense areas (nm) | Mean length of dense areas (nm) | Densely staining INM (%) |
|---|---|---|---|---|
| HSV-1(F) | 29,597 | 4,063 | 123 | 14 |
| UL34 null | 10,061 | 1,642 | 137 | 16 |
| UL31 null | 24,048 | 0 | 0 |
The results indicated that budding sites were common in cells infected with HSV-1(F), with approximately 14% of the almost 30,000 nm of INM examined staining densely in distinct patches. Each INM budding site was of average length, 123 nm, but the range varied considerably, from 15 to 229 nm in length. Similarly, cells infected with the UL34 null virus contained ample INM budding sites. Thus, of the approximately 10,000 nm of INM examined, the total distance incorporated into densely staining INM budding sites was approximately 16%, having an average length of each site of approximately 137 nm and a length range of 44 to 311 nm. The average length was not statistically different from those observed in HSV-1(F)-infected cells. In contrast to these results, no densely staining sites were observed in cells infected with the UL31 deletion mutant, despite examination of over 24,000 nm of INM from cells known to be infected as determined by the presence of intranuclear capsids. We conclude from these data that UL31 is required for the accumulation of densely staining budding sites in the INM of infected cells.
DISCUSSION
A longstanding observation has been that nucleocapsids bud from densely staining regions of the INM as viewed in thin sections stained with uranyl acetate and osmium tetroxide. As viewed by electron tomography of freeze-substituted material, incorporation of these densely staining regions of INM into nascent virions results in a virion envelope that appears to be approximately twice as thick as other regions of the INM (6). The biochemical nature of these densely staining sites is unknown, but the sites may reflect accumulations of viral tegument proteins and glycoproteins that comprise nucleocapsid docking and budding sites at the INM. Supporting this possibility is the observation that at least the tegument proteins pUL11 and VP16 and the glycoproteins M, C, D, and B of perinuclear virions are recruited to the INM of infected cells (4, 5, 19, 25, 37, 55). The presence of these proteins in perinuclear virions argues for their accumulation at budding sites prior to or at the time of envelopment. In contrast, pseudorabies virus glycoproteins have not been detected in perinuclear virions, suggesting that the electron densities seen in the INM of cells infected with that virus comprise components other than glycoproteins (21, 26).
Neither the lack of pUL34 nor the associated defect in recruitment of at least gD and gM to the INM of cells infected with the HSV UL34 deletion mutant affected the morphology of densely staining INM budding sites, at least as viewed by transmission electron microscopy. On the other hand, the presence of INM budding sites was dependent on UL31. Densely staining regions were not observed in sections not stained with osmium tetroxide (such as in sections prepared for immunogold electron microscopy). While the electron densities might simply reflect the physical presence of pUL31 at the INM, the propensity of INM budding sites to stain densely with OsO4 may also reflect biochemical changes other than recruitment of proteins. For example, OsO4 preferentially stains the double bonds of unsaturated fatty acids (22), suggesting that subsets of unsaturated fatty acids may be preferentially recruited to INM budding sites. Although recruitment of subsets of lipids in the milieu of the INM is unprecedented, there are many examples in which specific types of lipids are recruited to virion budding sites, including the preferential involvement of cholesterol-rich lipid rafts in budding of many enveloped viruses from the plasma membrane, including influenza viruses and retroviruses (38). In the case of retroviruses, the lipid environment in lipid rafts promotes budding by concentrating Gag and glycoproteins at virion budding sites (7, 29). Similarly, the pUL31 and pUL34 homologs in pseudorabies viruses are sufficient to bud from the INM to form virion-like densely staining particles (27). The absence of densely staining regions of the INM in cells infected with a UL31 deletion mutant suggests that if the dense staining is a consequence of lipid recruitment to budding sites, UL31 must be necessary for this recruitment. Although such a role for UL31 is speculative, it is theoretically consistent with its mostly hydrophobic composition and its maintenance at the nucleoplasmic face of the INM by interaction with pUL34. Thus, pUL31 may act in a fashion similar to the Gag protein of retroviruses and the matrix proteins of other viruses to orchestrate assembly of the budding machinery.
These data also support the conclusion that UL31 and UL34 play important roles in the recruitment of proteins to INM budding sites to help orchestrate their incorporation into virions. Why might recruitment of glycoproteins represent an important function of UL31 and UL34? We suggest that pUL31/pUL34 orchestrates assembly of INM budding sites for the success of later steps in the egress pathway. For example, fusion of the perinuclear virion envelope with the ONM should require some type of fusion machinery within the envelope of the perinuclear virion or ONM. Although it is unknown which proteins comprise the fusion machinery, gH and gB are likely involved because excess numbers of virions are observed in the perinuclear space of cells infected with a mutant lacking both proteins (15). In further indirect support of this hypothesis, fusion of neighboring cells can be mediated by coexpression of gD, gH/gL, and gB, suggesting that at least in the context of the plasma membrane, these proteins comprise a competent fusion apparatus (8). gM, in contrast, decreases cell-cell fusion when coexpressed in vitro, suggesting a potential regulatory role (11). The pUL34-dependent recruitment of gD to the INM is of particular interest because gD has been shown to interact with both gH and gB on or about the time of fusion (1, 2). The current studies suggest that one function of pUL31/pUL34 is to recruit at least gD and gM and possibly other virus glycoproteins to the INM for their eventual incorporation into perinuclear virions. Clearly, further studies are warranted to test whether pUL34 interacts directly or indirectly with the fusogenic proteins gH and gB and whether it affects recruitment of these proteins to the INM.
It should also be noted that fusion at the nuclear and plasma membranes is likely to involve different mechanisms. For example, virions do not accumulate aberrantly in the perinuclear space of cells infected with viruses lacking genes encoding single glycoproteins, such as gH, gD or gB, whereas absence of any one of these proteins completely abrogates viral entry and fusion of plasma membranes in the in vitro fusion assay.
In summary, the current study shows that UL31 and UL34 mediate recruitment of virion-destined glycoproteins gD and gM to the INM, where primary virion envelopment occurs. Proper assembly of INM budding sites is likely important to ensure the correct proteome of the nascent virion and for successful execution of later steps in the egress pathway.
Acknowledgments
We thank Gary Cohen and Roseyln Eisenberg for the gD-specific antibody, Bill Ruyechan for the ICP8-specific antibody, Greg Smith for the GS1783 strain of E. coli, and Bernard Roizman and Richard Roller for the UL31 and UL34 deletion viruses, respectively.
These studies were supported by Public Health Service grant AI52341 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 11 March 2009.
REFERENCES
- 1.Atanasiu, D., J. C. Whitbeck, T. M. Cairns, B. Reilly, G. H. Cohen, and R. J. Eisenberg. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. USA 10418718-18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 8111532-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avitabile, E., G. Lombardi, and G. Campadelli-Fiume. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J. Virol. 776836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines, J. D., E. G. Wills, J. Pennington, R. J. Jacob, and B. Roizman. 2007. Glycoprotein M of herpes simplex virus type 1 is incorporated into virions during budding at the inner nuclear membrane. J. Virol. 81800-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines, J. D., C. E. Hsieh, E. Wills, C. Mannella, and M. Marko. 2007. Electron tomography of nascent herpes simplex virions. J. Virol. 812726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya, J., A. Repik, and P. R. Clapham. 2006. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. J. Virol. 805292-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 821419-1422. [DOI] [PubMed] [Google Scholar]
- 9.Cai, W., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 622596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype of infected cells. J. Virol. 718307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crump, C. M., B. Bruun, S. Bell, L. E. Pomeranz, T. Minson, and H. M. Browne. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 853517-3527. [DOI] [PubMed] [Google Scholar]
- 12.Davis-Poynter, N., S. Bell, T. Minson, and H. Browne. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J. Virol. 687586-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2357-364. [DOI] [PubMed] [Google Scholar]
- 14.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H. J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 793703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA 10410187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frangioni, J. V., and B. G. Neel. 1993. Solubilization and purification of enzymatically active glutathione-S-transferase (pGEX) fusion proteins. Anal. Biochem. 210179-187. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, R., K. Ghosh, L. Rasile, and H. P. Ghosh. 1994. Membrane anchoring domain of herpes simplex virus glycoprotein gB is sufficient for nuclear envelope localization. J. Virol. 682272-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granato, M., R. Feederle, A. Farina, R. Gonnella, R. Santarelli, B. Hub, A. Faggioni, and H. J. Delecluse. 2008. Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 824042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 753675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes, T. L., F. T. Lindgren, and J. W. Gofman. 1963. A quantitative determination of the osmium tetroxide-lipoprotein interaction. J. Cell Biol. 19251-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heine, J. W., P. G. Spear, and B. Roizman. 1972. Proteins specified by herpes simplex virus. VI. Viral proteins in the plasma membrane. J. Virol. 9431-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313217-220. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, H. L., and B. Norrild. 1998. Herpes simplex virus type 1-infected human embryonic lung cells studied by optimized immunogold cryosection electron microscopy. J. Histochem. Cytochem. 46487-496. [DOI] [PubMed] [Google Scholar]
- 26.Klupp, B., J. Altenschmidt, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2008. Glycoproteins required for entry are not necessary for egress of pseudorabies virus. J. Virol. 826299-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupp, B. G., H. Granzow, W. Fuchs, G. M. Keil, S. Finke, and T. C. Mettenleiter. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. USA 1047241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 822363-2371. [DOI] [PubMed] [Google Scholar]
- 29.Leung, K., J. O. Kim, L. Ganesh, J. Kabat, O. Schwartz, and G. J. Nabel. 2008. HIV-1 assembly: viral glycoproteins segregate quantally to lipid rafts that associate individually with HIV-1 capsids and virions. Cell Host Microbe 3285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang, L., M. Tanaka, Y. Kawaguchi, and J. D. Baines. 2004. Cell lines that support replication of a novel herpes simplex 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 32968-76. [DOI] [PubMed] [Google Scholar]
- 31.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 621486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lötzerich, M., Z. Ruzsics, and U. H. Koszinowski. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 8073-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 691531-1574. [DOI] [PubMed] [Google Scholar]
- 34.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9423-429. [DOI] [PubMed] [Google Scholar]
- 35.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87427-436. [DOI] [PubMed] [Google Scholar]
- 36.Mou, F., T. Forest, and J. D. Baines. 2007. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 816459-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naldinho-Souto, R., H. Browne, and T. Minson. 2006. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J. Virol. 802582-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nayak, D. P., E. K.-W. Hui, and S. Barman. 2004. Assembly and budding of influenza virus. Virus Res. 106147-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds, A. E., B. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 758803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the HSV-1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 768939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roizman, B., and D. Furlong. 1974. The replication of herpesviruses, p. 229-403. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology. Plenum Press, New York, NY.
- 42.Roller, R., Y. Zhou, R. Schnetzer, J. Ferguson, and D. Desalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 672285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarmiento, M., M. Haffey, and P. G. Spear. 1979. Membrane proteins specified by herpes simplex virus type 1. III. Role of glycoprotein VP7(B2) in virion infectivity. J. Virol. 291149-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnee, M., Z. Ruzsics, A. Bubeck, and U. H. Koszinowski. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 8011658-11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiba, C., T. Daikoku, F. Goshima, H. Takakuwa, Y. Yamauchi, O. Koiwai, and Y. Nishiyama. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 812397-2405. [DOI] [PubMed] [Google Scholar]
- 47.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 755697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell. Microbiol. 6401-410. [DOI] [PubMed] [Google Scholar]
- 49.Stackpole, C. W. 1969. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J. Virol. 475-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stannard, L. M., S. Himmelhoch, and S. Wynchank. 1996. Intra-nuclear localization of two envelope proteins, gB and gD, of herpes simplex virus. Arch. Virol. 141505-524. [DOI] [PubMed] [Google Scholar]
- 51.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, NY.
- 52.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. USA 1042903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 771382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step RED-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40191-197. [DOI] [PubMed] [Google Scholar]
- 55.Torrisi, M. R., C. Di Lazzaro, A. Pavan, L. Pereira, and G. Campadelli-Fiume. 1992. Herpes simplex virus envelopment and maturation studies by fracture label. J. Virol. 66554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 741355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]