Abstract
The broadly neutralizing antibody immunoglobulin G1 (IgG1) b12 binds to a conformationally conserved surface on the outer domain of the human immunodeficiency virus type 1 (HIV-1) gp120 envelope (Env) glycoprotein. To develop outer domain proteins (ODs) that could be recognized selectively by CD4-binding-site (CD4-BS) antibodies, membrane-anchored ODs were generated from an HIV-1 clade B virus, TA1 R3A, which was highly sensitive to neutralization by the IgG1 b12 antibody. A 231-residue fragment of gp120 (residues 252 to 482) linked to transmembrane regions from CD4 showed b12 binding comparable to that of the native Env spike as measured by flow cytometry. Truncation of the β20-β21 hairpin (residues 422 to 436 to Gly-Gly) improved overall protein expression. Replacement of the immunodominant central 20 amino acids of the V3 loop (residues 302 to 323) with a basic hexapeptide (NTRGRR) increased b12 reactivity further. Surface calculations indicated that the ratio of b12 epitope to exposed immunogenic surface in the optimized OD increased to over 30%. This OD variant [OD(GSL)(Δβ20-21)(hCD4-TM)] was recognized by b12 and another CD4-BS-reactive antibody, b13, but not by eight other CD4-BS antibodies with limited neutralization potency. Furthermore, optimized membrane-anchored OD selectively absorbed neutralizing activity from complex antisera and b12. Structurally designed membrane-anchored ODs represent candidate immunogens to elicit or to allow the detection of broadly neutralizing antibodies to the conserved site of CD4 binding on HIV-1 gp120.
The human immunodeficiency virus type 1 (HIV-1) envelope is composed of surface gp120 and transmembrane gp41. Initial attempts to develop HIV vaccines through the induction of antibodies focused on recombinant gp120 glycoproteins. Two phase III clinical trials conducted in the United States and Thailand showed no protection from a gp120-based subunit vaccine against HIV infection, nor did the vaccine delay HIV-1 disease progression (11, 25). In addition, a phase II trial completed in Thailand with a live recombinant HIV-1 canarypox vaccine (vCP1452) in combination with a gp120 subunit protein did not stimulate a markedly improved immune response (28). The lack of efficacy was likely to be related to its failure to elicit broadly neutralizing antibodies (4, 10, 33).
Several broadly neutralizing human monoclonal antibodies (MAbs) have been derived from infected individuals, including immunoglobulin G1 (IgG1) b12, 2G12, 2F5 and 4E10, which are directed against CD4-binding-site (BS), carbohydrate, and membrane-proximal regions of HIV Env (reviewed in reference 9). Among the most potent, the b12 antibody occludes the site of CD4 binding on gp120 and prevents virus attachment to CD4 on target cells (39). Other CD4-BS antibodies recognize epitopes on monomeric gp120 that overlap with b12 but lack the ability of b12 to neutralize primary HIV-1 isolates (5). An understanding of the specificity of b12 binding, neutralization, and protection should aid in the development of immunogens that induce neutralizing antibodies of a similar specificity.
The structure of the b12-gp120 complex (39) shows that b12 binds to a conformationally conserved surface, which is centered around the CD4-binding loop on the outer domain of gp120. In the CD4-bound conformation of gp120, the CD4-binding loop or β15-strand makes antiparallel intermolecular hydrogen bonds to the C″ strand of CD4 (14). Overall, the outer domain of gp120 comprises 82% of the gp120 contact surface with b12, while most of the contacts outside of the outer domain have marginal importance (39). One exception, however, are contacts to the loop connecting the outer domain with the α5-helix of the inner domain (39), which appear to be important.
Because it represents the smallest structural unit containing the b12 epitope, and therefore maximizes the b12-immunogenic surface relative to the overall surface, an outer domain-only immunogen with high b12 affinity represents an attractive immunogen. An outer domain construct (named OD1) was previously derived from HIV-1 strain YU2 gp120 and found to bind 2G12 and a number of anti-V3 antibodies (36); however, b12 binding to this construct was difficult to detect by enzyme-linked immunosorbent assay, probably due to an enhanced off rate (36, 39). A large, relatively flat interface exists between the inner and outer domains of gp120 in both CD4-bound and b12-bound conformations. We reasoned that the removal of the inner domain might partially destabilize it and decided to replace the inner domain with another polar surface, the cell membrane. We expressed outer domain proteins (ODs) in various membrane-anchored forms and tested their abilities to bind b12. An HIV-1 clade B R5 and X4 dual-tropic virus, R3A, was selected as a prototype (20). Laboratory-adapted virus strain R3A TA1 contains a truncated V1/V2 and a truncated V3 (named 9,9), maintains CCR5 tropism, and is highly sensitive to b12 neutralization (15, 23). We used available atomic-level structures to model an R3A gp120 core and to design truncations of flexible, potentially immunodominant structures, which emanate from OD, including the β20-β21 hairpin and the V3 loop. Thus, by using structure-based design to modify the OD form of R3A TA1, we attempted to remove strain-specific determinants, to enhance cell-surface expression, and to increase specific b12 binding compared to other native forms.
MATERIALS AND METHODS
Cell lines, media, and antibodies.
293F and 293T human embryonic kidney cell lines were purchased from Invitrogen (Carlsbad, CA) and were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Sigma, St. Louis, MO). Human IgG1 isotype control was obtained from Alexis Biochemicals (San Diego, CA). MAbs b12 and b13 were produced in 293F cells and purified with a protein I affinity column. CD4 was obtained from the NIH AIDS Research and Reference Reagent Program. 17b, 15e, and F91 were kindly provided by James Robinson (12, 14, 21, 24, 29). b3, b6, and b11 were generated as described previously (1, 3, 24, 27, 29). m14 and m18 were prepared by Dimiter S. Dimitrov (29, 37, 38). F105 was made by Marshall R. Posner (24, 26, 29, 31), and 2G12 was kindly provided by Hermann Katinger (39). fluorescein isothiocyanate-conjugated Affinipure goat anti-human IgG(H+L) was purchased from Jackson ImmunoResearch (West Grove, PA). Human sera and plasma samples were described previously (17).
Outer domain genes.
gp145ΔCFIΔV12(9,9) was derived from the HIV-1 R3A TA1 envelope (15, 23) with cytoplasmic, cleavage-site, fusion peptide, interhelical-domain, and V1V2 deletions, as previously described (7, 34). The outer domain construct OD(9,9)(hCD4-TM) was made by PCR of amino acids 252 to 482 (252PVVST……WRSE482) (numbering based on HXBc2) from R3A-gp145ΔCFIΔV12(9,9). This fragment was fused to the mouse interleukin-2 (IL-2) signal peptide sequence (MYSMQLASCVTLTLVLLVNSGPR) and the human CD4 transmembrane domain (GSGSLIVLGGVAGLLLFIGLGI). V3 derivatives of the same construct and β20-21 replacement with Gly-Gly were made using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The corresponding D368R mutants were also generated using the QuikChange mutagenesis kit. All envelope constructs were expressed under the cytomegalovirus (CMV) promoter in the CMV/R vector (34).
Structure-based OD design and surface area calculations.
An R3A gp120 was modeled from the structure of the gp120 core with V3 (Protein Data Bank accession number 2B4C) (13) using SwissModel (http://swissmodel.expasy.org/SWISS-MODEL.html) in alignment mode, while the target sequence of the R3A core with V3 was aligned to the JR-FL sequence. Variants of OD containing the primary binding site for b12 were visualized with the program XFIT as provided in the XtalView package (19). Truncations of the β20-β21 hairpin were designed, which retained OD-hydrogen bonding while removing areas of structural differences between the CD4- and b12-bound structures of core gp120 (Protein Data Bank accession numbers 2NXY and 2NY7) (39). Truncations of the V3 loop were designed to retain hydrogen bonding at the V3 base while providing variation in the connecting loop. Variants included Ser-Gly minimal linkers, basic and acidic linkers, and linkers containing N-linked glycosylation. Surface areas were calculated with the program GRASP (22).
Surface staining of ODs.
293T cells were transfected with plasmids expressing OD constructs using the ProFection mammalian calcium phosphate transfection system (Promega, Madison, WI) according to the manufacturer's instructions. Twenty-four hours after transfection, cells were collected with phosphate-buffered saline (PBS) containing 2 mM EDTA and washed with PBS. Cells were stained with the indicated antibodies (5 μg/ml), human serum, or plasma samples (1:100 dilutions) for 30 min on ice. Cells were then washed twice with PBS and incubated with fluorescein isothiocyanate-conjugated Affinipure goat anti-human IgG(H+L) (5 μg/ml) for 30 min on ice and then washed with PBS and resuspended in PBS with 0.5% paraformaldehyde. Samples were run on a BD FACSCalibur (see Fig. 1 and 5) or BD LSR-II (see Fig. 2 to 4 and 6) flow cytometer using FACSDiva software (BD Biosciences, San Jose, CA) and analyzed with FlowJo 8.6.1 software (Tree Star, Ashland, OR).
FIG. 1.
R3A gp145ΔCFIΔV12(9,9) can bind b12, and binding is eliminated by the D368R mutation. (A) Schematic representation of functional domains and mutations in the HIV-1 R3A Env glycoprotein gp145ΔCFIΔV12(9,9). Deletions include the V1-V2 loop, the tip of the V3 loop, the gp120/gp41 cleavage site and fusion domain, and the region between the two heptad repeats. (B) MAb b12 (gray lines) or 2G12 (solid black lines) or human control IgG (dashed lines) binding to 293T cells transfected with a CMV/R vector expressing gp145ΔCFIΔV12(9,9) or gp145ΔCFIΔV12(9,9)(D368R) was analyzed by flow cytometry.
FIG. 5.
Specificity of OD binding to selected neutralizing human antisera. Binding of MAb b12 (solid lines) and human sera or plasma from HIV-positive patients (Pt.) (solid lines) to 293T cells transfected with OD(GSL)(Δβ20-21)(hCD4-TM), gp145ΔCFIΔV12(9,9), or their D368R mutations was analyzed by flow cytometry. Human control IgG or normal human sera (dashed lines) were used as controls. SM, serum; PL, plasma.
FIG. 2.
R3A outer domain construct containing the human CD4 transmembrane recognizes b12. (A) Structure of core gp120 in CD4-bound and b12-bound contexts. The gp120 core structure is shown in ribbon diagram format with the surface corresponding to the b12 epitope highlighted in blue. (B) Domain separation. Residues 252 to 482, which correspond roughly to the outer domain of gp120 with the α5-helix, are depicted separate from the rest of the inner domain. (C) Outer domain with the α5-helix, rotated 90° from that shown in B, with the surface colored blue for b12 epitope and otherwise red for the outer domain and gray for the α5-helix. (D) Same as C but colored according to the chemistry of the underlying amino acids (red, negatively charged; blue, positively charged; white, apolar). (E) Outer domain with the α5-helix, with transmembrane linker shown with membrane and N-linked glycan. C-term, C terminus. (F) Schematic representation of the outer domain construct OD(9,9)(hCD4-TM). The construct is from R3A gp145ΔCFIΔV12(9,9) residues 252 to 482 (based on the amino acid residue numbers of HXB2) with an IL-2 leader sequence and the hCD4 transmembrane domain. The β20-β21 loop deletion construct is shown below. (G) MAb b12 (blue) or 2G12 (green) or human control IgG (red) binding to 293T cells transfected with OD(9,9)(hCD4-TM) or OD(9,9)(Δβ20-21)(hCD4-TM) was analyzed by flow cytometry.
FIG. 4.
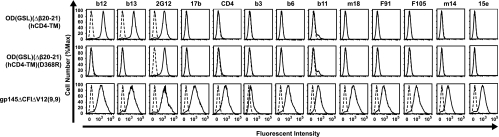
Specific binding of ODs to CD4-BS MAbs. MAb (solid lines) or human control IgG (dashed lines) binding to 293T cells transfected with OD(GSL)(Δβ20-21)(hCD4-TM), its D368R mutation, and R3A gp145ΔCFIΔV12(9,9) was analyzed by flow cytometry.
FIG. 6.
Neutralization of HIVJRFL clade B virus by leukapheresis sera from an HIV-positive patient, Zeptometrix 1642, and absorption by wild-type but not mutant OD-TM. (A) Zeptometrix 1642 antisera (left), b13 antibody (middle), and b12 antibody (right) were absorbed with 293F cells transfected with OD(GSL)(Δβ20-21)(hCD4-TM) (gray lines) or 293F cells transfected with OD(GSL)(Δβ20-21)(hCD4-TM)(D368R) (solid black lines). After absorption, residual binding was analyzed by flow cytometry for 293T cells expressing wild-type OD(GSL)(Δβ20-21)(hCD4-TM). Normal human serum (dashed lines) was used as a negative control. (B) Neutralizations of absorbed Zeptometrix 1642 antisera against HIV-1 clade B virus JRFL (left) and absorbed MAb b12 against HIV-1 clade B virus HxBc2 (right) were analyzed. Percent neutralization is shown for antisera absorbed with 293F cells transfected with OD(GSL)(Δβ20-21)(hCD4-TM) (diamonds) or with 293F cells transfected with OD(GSL)(Δβ20-21)(hCD4-TM)(D368R) (squares).
Human plasma absorption and neutralization assay.
293F cells were transfected with OD(GSL)(Δβ20-21)(hCD4-TM) or OD(GSL)(Δβ20-21)(hCD4-TM)(D368R) plasmids using 293fectin (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Cells were harvested 36 to 48 h after transfection, washed with PBS, and incubated with human plasma Zeptometrix 1642 (5 × 107cells/50 μl plasma in 500 μl PBS) or MAb b12 (1 × 109cells/10 μg MAb in 1000 μl PBS) on ice for 30 min. The absorbed supernatants were harvested and filtered for cell surface staining or neutralization assays.
Neutralization was performed using a pseudovirus-based single-cycle infectivity assay using TZM-bl cells as described previously (17). The results are reported as the 50% inhibitory concentration (the plasma dilution producing 50% virus neutralization) in Table 1. In the absorbed plasma neutralization assay, dilutions of absorbed plasma or MAb were used, and the percent neutralization was calculated compared to that of controls with no plasma added.
TABLE 1.
Neutralization (50% inhibitory concentration) by antiserum from HIV-positive patient serum Zeptometrix 1642 against HIV-1 isolates of diverse clade origin
| Virus | Source | Clade | Location | Zeptometrix 1642 dilution | HIV-positive IgG concn (μg/ml) |
|---|---|---|---|---|---|
| MN | TCLA | B | United States | 3,717 | 7.1 |
| SF162.LS | B | United States | 1,295 | 4.6 | |
| Bal.26 | B | United States | 426 | 40.1 | |
| SS1196.1 | ccPBMC | B | United States | 158 | 161 |
| SHIV-89.6P | B | United States | 307 | 281 | |
| SHIV-SF162.P3 | B | United States | 333 | 436 | |
| 6535.3 | ccPBMC | B | United States | 485 | 142 |
| QH0692.42 | ccPBMC | B | Trinidad | 114 | 1,571 |
| SC422661.8 | Plasma | B | Trinidad | 184 | 303 |
| PVO.4 | ccPBMC | B | Italy | 54 | 1,203 |
| TRO.11 | ccPBMC | B | Italy | 98 | 583 |
| AC10.0.29 | ccPBMC | B | United States | 309 | 1,961 |
| RHPA4259.7 | Plasma | B | United States | 80 | 242 |
| THRO4156.18 | Plasma | B | United States | 307 | 845 |
| REJO4541.67 | Plasma | B | United States | 524 | 262 |
| TRJO4551.58 | Plasma | B | United States | 34 | 1,151 |
| WITO4160.33 | Plasma | B | United States | 1,080 | 399 |
| CAAN5342.A2 | Plasma | B | United States | 152 | 817 |
| Du123.6 | ccPBMC | C | South Africa | 98 | 998 |
| Du156.12 | ccPBMC | C | South Africa | 575 | 348 |
| CAP45.2.00.G3 | ccPBMC | C | South Africa | 370 | 952 |
| CAP210.2.00.E8 | Plasma | C | South Africa | 157 | 890 |
| ZM197 M.PB7 | ucPBMC | C | Zambia | 441 | 201 |
| ZM233 M.PB6 | ucPBMC | C | Zambia | 263 | 454 |
| ZM249 M.PL1 | Plasma | C | Zambia | 105 | 502 |
| ZM135 M.PL10a | Plasma | C | Zambia | 448 | 460 |
| Q23.17 | ucPBMC | A | Kenya | 154 | 429 |
| Q168.a2 | ucPBMC | A | Kenya | 113 | 931 |
| Q461.e2 | ucPBMC | A | Kenya | 108 | >2,500 |
| Q769.d22 | ucPBMC | A | Kenya | 139 | 415 |
| T250-4 | CRF02_AG | West Africa | 100 | 942 | |
| T251-18 | CRF02_AG | West Africa | 112 | 2,316 | |
| T278-50 | CRF02_AG | West Africa | 79 | 2,056 | |
| T255-34 | CRF02_AG | West Africa | 129 | 1,116 | |
| CH115.12 | ccPBMC | CRF07_BC | Beijing, People's Republic of China | 127 | 1,817 |
| CH070.1 | ccPBMC | CRF07_BC | Beijing, People's Republic of China | 89 | 1,492 |
| CH111.8 | ccPBMC | CRF07_BC | Beijing, People's Republic of China | 100 | 976 |
| CH038.12 | ccPBMC | CRF07_BC | Beijing, People's Republic of China | 149 | 801 |
aTCLA, T-cell line adapted; ccPBMC, cocultured peripheral blood mononuclear cells; ucPBMC, uncultured peripheral blood mononuclear cells.
Serum or plasma samples from patients 1, 20, 45, US21, and Zeptometrix 1642 all displayed moderate to potent neutralizing activity against diverse strains of HIV-1. Sera/plasma from patients 1, 20, 45 and US21 were derived from clinically symptomatic HIV-1-infected subjects who were not on antiretroviral therapy. Zeptometrix 1642 was a gift from David Montefiori (Duke University) and was initially obtained from the Zeptometrix Corporation (Buffalo, NY). There was no clinical information available on this plasma. Sera/plasma samples from patients 1, 20, and 45 were provided by Mark Connors, NIAID (17, 18).
RESULTS
Binding of transmembrane forms of the OD glycoprotein to the b12 antibody.
To evaluate the reactivity of R3A TA1 envelope derivatives, several deletion mutations, including the elimination of the V1 and V2 regions, truncation of V3 leaving 9 amino acids on each side of the stem V3(9,9), and deletion of the cleavage, fusion, and interhelical (ΔCFI) domains, were prepared in a gp145 form of this Env to generate gp145ΔCFIΔV12(9,9) (Fig. 1A) (7, 15, 23). When expressed on transfected 293T cells, this glycoprotein was able to bind to the broadly neutralizing MAbs b12 and 2G12 (Fig. 1B). A negative control vector was created with a mutation (D368R) in the β15-strand, a portion of gp120 which is critical for its interaction with CD4 (14). gp145ΔCFIΔV12(9,9) (D368R) showed no binding to b12, whereas 2G12 binding was unaffected (Fig. 1B) (2G12 recognizes a cluster of α1-2 mannose residues on the silent face of the outer domain of gp120 [29] and whose binding is independent of b12). A membrane-anchored OD variant was generated from R3A TA1 gp145ΔCFIΔV12(9,9) by expressing gp120 residues 252 to 482 with the IL-2 signal peptide sequence and human CD4 transmembrane domain [OD(9,9)(hCD4-TM)] (Fig. 2A to D and F). This variant of gp120 replaces the inner domain of gp120 with the cell membrane (Fig. 2E and F). Residues 252 and 482 are spatially close in unliganded, CD4-bound, and b12-bound forms of gp120, and charged residues at position 252 and 482 were retained to define the membrane boundary. In addition, the truncation and membrane anchor were designed to place the membrane into a position similar to one that the native viral spike is expected to occupy. OD(9,9)(hCD4-TM) exhibited b12 and 2G12 binding when expressed on 293T cells by flow cytometry similarly to the gp145ΔCFI version (Fig. 2G, left, versus Fig. 1B, middle).
The β20-β21 region of gp120 differs significantly among unliganded, b12-bound, and CD4-bound conformations of gp120 (8, 14, 39). In the CD4-bound conformation, β20-β21 forms half of a well-ordered four-stranded sheet, named the bridging sheet, which contains elements critical for both CD4 and CCR5 binding (14). Meanwhile, in unliganded and b12-bound conformations, β20-β21 forms a flexible hairpin (8, 39). A truncation of β20-β21 between residues 424 and 432 was previously observed to increase b12 binding in strains derived from HIV-1 clade B virus IIIB and SHIV-89.6 (2). To determine whether an analogous truncation in an OD expression vector could similarly increase b12 reactivity, a Gly-Gly dipeptide replacement of β20-β21 residues between residues 422 and 436 was introduced into OD-transmembrane (OD-TM) vectors. While the introduction of this deletion increased reactivity with b12, binding to the control 2G12 MAb also improved (Fig. 2B, right), suggesting that the β20-β21 truncation improved the overall protein expression of OD and did not selectively enhance the exposure of the site of b12 binding.
Selective modification of the V3 base after V3 truncation increased b12 reactivity with OD-TMs.
The structure of a V3-containing gp120 core reveals that the V3 is exposed and flexible in the CD4-bound state, with an accordion-like stem, a conserved fixed base, and a β-hairpin tip (13). OD(9,9)(hCD4-TM) has a V3 loop tip deletion and only retains 9 amino acids on each base and the partial stem of V3 and is linked with Gly-Val-Gly (15). To diminish the length of the stem and to preserve the distance between residues on opposite sides of the base, we deleted the V3 loop-stem closer to the base and replaced it with one of several sequences, including a Gly-Ser linker (GGSGSG [9c]), an acidic/negatively charged linker (ENGEGE [AN]), a basic/positively charged linker (KNGKGK [BP]), a glycan-shielded linker (GGNGSG [GS]), or a native glycosylation site adjacent to a shorter/charged loop-stem (NTRGRR [GSL]) (Fig. 3A and B). The OD-TM glycoproteins with either Gly-Ser, acidic/negative, or glycan-shielded linkers showed no increase in b12 and 2G12 binding compared to the OD(9,9)(hCD4-TM) glycoprotein [9c, AN, or GS versus 9,9 OD(hCD4-TM)] (Fig. 3C). In contrast, the basic/positive or glycosylation site adjacent to a shorter/charged loop linker showed significantly increased binding to both b12 and 2G12, with b12 recognition being even greater than 2G12 binding (BP and GSL) (Fig. 3C). Thus, these V3 linkers improved the relative exposure and reactivity of the CD4-BS determinant recognized by b12.
FIG. 3.
Differential charges at the base of V3 affect b12 recognition. (A) Structure of gp120 core with V3 and V3 sequences for the different V3 loop mutations: original V3 from a R3A TA1 strain (9,9), V3 deletion with a Gly-Ser link (9c), V3 deletion with acidic/negative charge (AN), V3 deletion with basic/positive charge (BP), V3 deletion with glycan shield (GS), and V3 deletion with native glycan and a shorter/charged loop-stem (GSL). WT, wild type. (B) Models of V3 with the surface colored according to the chemistry of the underlying amino acids (red, negatively charged; blue, positively charged; white, apolar). (C) MAb b12 (blue) or 2G12 (green) or human control IgG (red) binding to 293T cells transfected with OD(hCD4-TM) with different V3 mutations was analyzed by flow cytometry.
Binding of CD4-BS antibodies to OD-TMs.
An increasing number of CD4-BS antibodies have been identified and characterized, some of which neutralize diverse viruses and others of which are weakly neutralizing or restricted in their breadth (1, 3, 6, 12, 21, 24, 26, 27, 30-32, 37, 38). We tested OD(GSL)(Δβ20-21)(hCD4-TM), which showed increased binding to b12. This OD-TM did not bind to CD4, CD4-induced antibodies (17b), or most nonpotent neutralizing CD4-BS antibodies, while these MAbs were able to bind to a gp145 Env (b3, b6, b11, m14, m18, F91, F105, and 15e) (Fig. 4, top versus bottom). The OD-TM did bind to neutralizing CD4-BS antibody b12 as well as to one of the nonpotent CD4-BS antibodies (b13) (Fig. 4, top). No binding, however, to a D368R mutation that abolishes the interaction of CD4 with Env was observed by flow cytometry (b12, b13, and wild type versus D386R) (Fig. 4, middle).
We also tested the binding of OD(GSL)(Δβ20-21)(hCD4-TM) to five complex antisera (Fig. 5). One specimen (from patient 1) was derived from a subject with broadly neutralizing CD4-BS antibodies to HIV-1 (17). This antiserum along with a second antiserum (from patient 20) failed to react with OD(GSL)(Δβ20-21)(hCD4-TM). Another antiserum (from patient 45) showed marginal reactivity, whereas a fourth (US21) showed moderate reactivity; however, with both of these sera, similar reactivity to a D368R mutant was observed. In contrast, antisera from another subject (Zeptometrix 1642) with neutralizing activity specifically bound to this OD-TM but not to the D368R mutant (Fig. 5). This specificity was not apparent using a transmembrane form containing a nearly complete Env that included the complete gp120 with a truncated V3 region [gp145ΔCFIΔV12(9,9); wild type versus D368R] (Fig. 5), suggesting that the OD-TM eliminated irrelevant specificities and allowed the detection of specific CD4-BS reactivity in complex antisera.
To determine whether these antibodies in Zeptometrix 1642 mediated HIV neutralization, we evaluated its neutralization activity against HIV-1. Zeptometrix 1642 antisera showed neutralization of HIV-1 strains from diverse clades (Table 1), and it thus showed increased breadth compared to those of most sera (16). This activity was nearly completely removed by OD-TM but not the D368R mutant [OD(GSL)(Δβ20-21)(hCD4-TM) versus D368R] (Fig. 6A and B, left) when assayed against JRFL, a CD4-BS antibody-sensitive virus. Finally, the CD4-BS reactivity of OD-TM was confirmed with b12, whose neutralization activity was removed by the wild type but not the D368R mutant (Fig. 6A and B, right).
DISCUSSION
The development of immunogens that elicit broadly neutralizing antibodies remains a high priority for the development of an effective AIDS vaccine. Among the targets of neutralization is the highly conserved CD4-BS. MAb b12 binds primarily to the surface of gp120 utilized in the initial contact with the CD4 receptor. This site is recessed on the viral spike, is surrounded by both N-linked glycans and immunodominant variable loops, and represents only one among many potential B-cell epitopes within gp160.
One strategy for optimizing the presentation of this site is “minimization,” to remove all immunogenic Env surfaces other than the target site. Such minimization would have utility both to define antibodies to this region in complex antisera and to develop new prototypes as potential immunogens. However, it has been a challenge to isolate the site of b12 binding independent of the rest of the HIV Env, particularly the inner domain. In this report, we show that an expression vector encoding the OD and α5-helix of Env linked to a transmembrane domain retains b12-binding activity and interacts specifically with antisera with broadly neutralizing CD4-BS antibodies. Absorption of one of those antisera selectively removed neutralizing activity against several viruses. These data suggest that it is possible to generate simplified proteins that can eliminate multiple specificities detected by anti-HIV-1 sera and to focus recognition on the CD4-BS.
Previous studies have shown that a soluble outer domain protein, OD1, is recognized by 2G12 and a number of V3 antibodies, but recognition by b12 was hampered by an enhanced off rate (36, 39). Laboratory-adapted virus strain R3A TA1 was chosen for further modification because it was highly sensitive to b12 neutralization and lacked most of the variable loops. It is likely that similar results can be obtained by removing analogous Env regions in other strains, and while a soluble OD based on R3A TA1 was similar to previously described ODs (36; and data not shown), here, the addition of a cellular transmembrane domain significantly improved b12 reactivity as determined by flow cytometry. This feature was observed with a variety of membrane anchors and was retained upon modification of the V3 region. It is important to remove the V3 from candidate immunogens because of its strong but strain-specific antigenicity.
The OD-TM described here was generated after attempting to develop secreted versions of OD. Secreted OD formed aggregates when expressed in 293 cells and failed to interact with IgG1 b12, similar to another soluble OD described previously (36). We reasoned that the loss of reactivity of soluble OD was due in part to the loss of appropriate folding, a result of inner domain removal. We further reasoned that an alternative surface might stabilize OD and that, possibly, a cell membrane might provide a suitable surface. Thus, appropriately anchoring OD to a membrane might stabilize it sufficiently to retain a conformation recognized with high affinity by b12. Other fusion protein approaches had failed to confer this property, although it remains possible that secreted ODs could be generated with heterologous domains using independent mutations.
The reasons for the improved reactivity of membrane-anchored OD are not fully known but include the possibility that membrane anchoring allows protein contacts with the cell surface that stabilize exposed regions that would otherwise interact with the inner domain in the nontruncated Env (Fig. 2A to E). In this context, the membrane may also occlude ligand binding to the anchored OD in a manner similar to that of the native viral spike. Finally, we note that cell surface avidity, present in the membrane-anchored context but not the soluble context, should reduce the off rate, thereby stabilizing b12 interactions.
Interestingly, the V3 loop modification mutants tested here showed that both the length and charge of V3 affect b12 binding. This result is somewhat unexpected since the V3 region emanating from the core is distal from both the site of inner domain truncation and the site of b12 recognition. To eliminate the V3 immunodominance, we both truncated V3 and made modifications to reduce its immunogenicity. In some cases, modifications affected both 2G12 and b12 binding, indicating an effect on OD-TM expression, but in others, b12 recognition was specifically enhanced, suggesting that its exposure was increased relative to those of other epitopes on the OD-TM.
The deletion of the β20-β21 hairpin has been suggested to reduce steric hindrance around the CD4-BS, to expand the CD4-binding pocket, and to increase b12 accessibility (2). Our data showed that the deletion of the β20-β21 hairpin improved b12 and 2G12 binding. These data suggest that the β20-β21 loop is not necessary for b12 binding to the membrane-anchored OD proteins, although the increased binding to 2G12 suggests that this deletion improves overall protein expression.
Both 2G12 and b12 are conformational antibodies (5, 29, 39). Our results showed that without the inner domain, the outer domain of gp120 itself can form a stable structure that allows those antibodies to become accessible. The optimized ODs developed here specifically recognize b12 but not most other nonpotent neutralizing CD4-BS antibodies. Moreover, OD(GSL)(Δβ20-21)(hCD4-TM) can absorb the HIV-1-neutralizing antibody from the human sample Zeptometrix 1642. Zeptometrix 1642 serum has reasonable breadth and potency, although its activity is lower than that of patient 1 sera described previously (17, 18). Other HIV-positive patient sera have been well characterized, including sera from patients 1 and 45 (18). Both samples contain CD4-BS antibodies, but their relationship to b12, b13, and Zeptometrix 1642 is unknown, and they might be expected to differ since these sera show broader reactivity across divergent clades than b12. A possible explanation for the lack of binding of patient 1 serum with the membrane-anchored OD is that it may require an interaction with an additional region, such as the inner domain or bridging sheet, not present in the outer domain. It is also unclear whether the activity of patient 1 sera can be explained by a single antibody or a combination of CD4-BS antibodies directed against diverse epitopes. The identification of other broadly neutralizing MAbs to the CD4-BS from such subjects will be instructive in this regard.
Efforts to use OD-TM as a vaccine immunogen are currently in progress. Structure-based calculations show that the proportion of b12-reactive surface relative to the total exposed surface on the optimized OD is significantly higher than that in core gp120 or OD1 contexts (Table 2 and Fig. 7). Because of its transmembrane nature, gene-based vaccination is required to elicit immune responses. Nonetheless, the findings reported here suggest that the membrane-anchored OD represents an alternative form of the Env glycoprotein that can detect neutralizing antibodies directed to the site of CD4 binding and may contribute to the rational design of an AIDS vaccine.
TABLE 2.
Surface area calculations
| Immunogen | Surface area (Å2)
|
Relative b12 surface (b12/exposed) (%) | ||||
|---|---|---|---|---|---|---|
| Total surface | b12 epitope | Masked by glycana | Occluded by membraneb | Total exposed protein surface | ||
| gp120 core | 14,627 | 746 | 7,017 | 0 | 7,610 | 0.098 |
| OD1 | 10,675 | 739 | 5,330 | 0 | 5,345 | 0.138 |
| OD-TM | 8,304 | 697 | 4,527 | 1,510 | 2,267 | 0.307 |
Glycan masking calculated with a 7-Å radius around each N-linked site of glycosylation (this radius results in full coverage of the gp120 silent face) (35).
Membrane occlusion calculated by using a 4-Å radius of the inner domain in the core context.
FIG. 7.
b12 epitope relative surface area on core gp120, OD1, and membrane-anchored OD. The b12 surface area was calculated in the context of three immunogens: core gp120 (14), OD1 (36), and the membrane-anchored OD developed here, OD(GSL)(Δβ20-21)(hCD4-TM). A model of the immunogen, with the b12 surface shown in blue, and expected locations of glycan and membrane are shown. The relative surface area of the b12 epitope is shown, after immunogenic masking by glycan and membrane components of the immunogens is taken into account (Table 2). C-term, C terminus.
Acknowledgments
This research was supported by the Intramural Research Program, Vaccine Research Center, NIAID, NIH.
We thank Dimiter Dimitrov for m6, m14, and m18; Marshall Posner for F105; James Robinson for 17b, F91, F105, and 15e; Ati Tislerics for assistance with manuscript preparation; the NIH Fellows Editorial Board for editorial assistance; Michael Cichanowski, Brenda Hartman, and Jonathan Stuckey for assistance with figures; and members of the Structural Biology Section and Virology Laboratory for helpful advice, discussions, and comments on the manuscript.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Barbas, C. F., III, E. Bjorling, F. Chiodi, N. Dunlop, D. Cababa, T. M. Jones, S. L. Zebedee, M. A. Persson, P. L. Nara, and E. Norrby. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. USA 899339-9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkower, I., C. Patel, Y. Ni, K. Virnik, Z. Xiang, and A. Spadaccini. 2008. Targeted deletion in the beta20-beta21 loop of HIV envelope glycoprotein gp120 exposes the CD4 binding site for antibody binding. Virology 377330-338. [DOI] [PubMed] [Google Scholar]
- 3.Bublil, E. M., S. Yeger-Azuz, and J. M. Gershoni. 2006. Computational prediction of the cross-reactive neutralizing epitope corresponding to the monclonal [sic] antibody b12 specific for HIV-1 gp120. FASEB J. 201762-1774. [DOI] [PubMed] [Google Scholar]
- 4.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5233-236. [DOI] [PubMed] [Google Scholar]
- 5.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 2661024-1027. [DOI] [PubMed] [Google Scholar]
- 6.Cavacini, L. A., C. L. Emes, J. Power, A. Buchbinder, S. Zolla-Pazner, and M. R. Posner. 1993. Human monoclonal antibodies to the V3 loop of HIV-1 gp120 mediate variable and distinct effects on binding and viral neutralization by a human monoclonal antibody to the CD4 binding site. J. Acquir. Immune Defic. Syndr. 6353-358. [PubMed] [Google Scholar]
- 7.Chakrabarti, B. K., W. P. Kong, B.-Y. Wu, Z.-Y. Yang, J. Friborg, Jr., X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 765357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433834-841. [DOI] [PubMed] [Google Scholar]
- 9.Douek, D. C., P. D. Kwong, and G. J. Nabel. 2006. The rational design of an AIDS vaccine. Cell 124677-681. [DOI] [PubMed] [Google Scholar]
- 10.Fauci, A. S., M. I. Johnston, C. W. Dieffenbach, D. R. Burton, S. M. Hammer, J. A. Hoxie, M. Martin, J. Overbaugh, D. I. Watkins, A. Mahmoud, and W. C. Greene. 2008. HIV vaccine research: the way forward. Science 321530-532. [DOI] [PubMed] [Google Scholar]
- 11.Flynn, N. M., D. N. Forthal, C. D. Harro, F. N. Judson, K. H. Mayer, and M. F. Para. 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191654-665. [DOI] [PubMed] [Google Scholar]
- 12.Ho, D. D., J. A. McKeating, X. L. Li, T. Moudgil, E. S. Daar, N. C. Sun, and J. E. Robinson. 1991. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J. Virol. 65489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 3101025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laakso, M. M., F. H. Lee, B. Haggarty, C. Agrawal, K. M. Nolan, M. Biscone, J. Romano, A. P. Jordan, G. J. Leslie, E. G. Meissner, L. Su, J. A. Hoxie, and R. W. Doms. 2007. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 3e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 7910108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 131032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y., K. Svehla, M. K. Louder, D. Wycuff, S. Phogat, M. Tang, S. A. Migueles, X. Wu, A. Phogat, G. M. Shaw, M. Connors, J. Hoxie, J. R. Mascola, and R. Wyatt. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 831045-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125156-165. [DOI] [PubMed] [Google Scholar]
- 20.Meissner, E. G., K. M. Duus, F. Gao, X. F. Yu, and L. Su. 2004. Characterization of a thymus-tropic HIV-1 isolate from a rapid progressor: role of the envelope. Virology 32874-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 701863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11281-296. [DOI] [PubMed] [Google Scholar]
- 23.Nolan, K. M., A. P. Jordan, and J. A. Hoxie. 2008. Effects of partial deletions within the human immunodeficiency virus type 1 V3 loop on coreceptor tropism and sensitivity to entry inhibitors. J. Virol. 82664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantophlet, R., S. E. Ollmann, P. Poignard, P. W. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitisuttithum, P., P. Gilbert, M. Gurwith, W. Heyward, M. Martin, F. van Griensven, D. Hu, J. W. Tappero, and K. Choopanya. 2006. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 1941661-1671. [DOI] [PubMed] [Google Scholar]
- 26.Posner, M. R., L. A. Cavacini, C. L. Emes, J. Power, and R. Byrn. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. 67-14. [PubMed] [Google Scholar]
- 27.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 684821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell, N. D., B. S. Graham, M. C. Keefer, M. J. McElrath, S. G. Self, K. J. Weinhold, D. C. Montefiori, G. Ferrari, H. Horton, G. D. Tomaras, S. Gurunathan, L. Baglyos, S. E. Frey, M. J. Mulligan, C. D. Harro, S. P. Buchbinder, L. R. Baden, W. A. Blattner, B. A. Koblin, and L. Corey. 2007. Phase 2 study of an HIV-1 canarypox vaccine (vCP1452) alone and in combination with rgp120: negative results fail to trigger a phase 3 correlates trial. J. Acquir. Immune Defic. Syndr. 44203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scanlan, C. N., R. Pantophlet, M. R. Wormald, S. E. Ollmann, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 767306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 673978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, J. Li, and J. Sodroski. 1991. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J. Virol. 655007-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 701100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, B. D., and D. R. Burton. 2008. Toward an AIDS vaccine. Science 320760-764. [DOI] [PubMed] [Google Scholar]
- 34.Wu, L., Z. Y. Yang, L. Xu, B. Welcher, S. Winfrey, Y. Shao, J. R. Mascola, and G. J. Nabel. 2006. Cross-clade recognition and neutralization by the V3 region from clade C human immunodeficiency virus-1 envelope. Vaccine 244995-5002. [DOI] [PubMed] [Google Scholar]
- 35.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393705-711. [DOI] [PubMed] [Google Scholar]
- 36.Yang, X., V. Tomov, S. Kurteva, L. Wang, X. Ren, M. K. Gorny, S. Zolla-Pazner, and J. Sodroski. 2004. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J. Virol. 7812975-12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, M. Y., Y. Shu, S. Phogat, X. Xiao, F. Cham, P. Bouma, A. Choudhary, Y. R. Feng, I. Sanz, S. Rybak, C. C. Broder, G. V. Quinnan, T. Evans, and D. S. Dimitrov. 2003. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods 28317-25. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, M. Y., X. Xiao, I. A. Sidorov, V. Choudhry, F. Cham, P. F. Zhang, P. Bouma, M. Zwick, A. Choudhary, D. C. Montefiori, C. C. Broder, D. R. Burton, G. V. Quinnan, Jr., and D. S. Dimitrov. 2004. Identification and characterization of a new cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody. J. Virol. 789233-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]