Abstract
The maturation inhibitor bevirimat [3-O-(3′,3′dimethysuccinyl)betulinic acid; BVM; also known as PA-457 or DSB] potently inhibits human immunodeficiency virus type 1 (HIV-1) replication by blocking protease (PR)-mediated cleavage at the junction between capsid (CA) and spacer peptide 1 (SP1) in Gag. We previously isolated a panel of single-amino-acid substitutions that confer resistance to BVM in vitro (C. S. Adamson, S. D. Ablan, I. Boeras, R. Goila-Gaur, F. Soheilian, K. Nagashima, F. Li, K. Salzwedel, M. Sakalian, C. T. Wild, and E. O. Freed, J. Virol. 80:10957-10971, 2006). The BVM resistance mutations cluster at or near the CA-SP1 cleavage site. Because BVM likely will be used clinically in patients harboring viruses resistant to PR inhibitors (PIs), in this study we evaluated the interplay between a PI-resistant (PIR) PR and the BVM resistance mutations in Gag. As expected, the PIR mutations had no effect on inhibition by BVM; however, we observed general processing defects and a slight delay in viral replication in Jurkat T cells associated with the PIR mutations, even in the absence of compound. When combined, most BVM resistance and PIR mutations acted additively to impair viral replication, particularly in the presence of BVM. The BVM-resistant mutant SP1-A1V was an exception, as it supported robust replication in the context of either wild-type (WT) or PIR PR, even at high BVM concentrations. Significantly, the emergence of BVM resistance was delayed in the context of the PIR PR, and the SP1-A1V mutation was acquired most frequently with either WT or PIR PR. These results suggest that resistance to BVM is less likely to emerge in patients who have failed PIs than in patients who are PI naïve. We predict that the SP1-A1V substitution is the most likely to emerge in vivo, as this mutant replicates robustly independently of PR mutations or BVM. These findings offer insights into the effect of PIR mutations on the evolution of BVM resistance in PI-experienced patients.
The maturation of human immunodeficiency virus type 1 (HIV-1) virions is mediated by the virally encoded protease (PR). PR cleaves the HIV-1 Gag polyprotein precursor during or shortly after assembled virus particles are released from the plasma membrane of the infected cell. Gag is cleaved at five major sites in a stepwise cascade that generates the four mature Gag domains, matrix (MA or p17), capsid (CA or p24), nucleocapsid (NC or p7), and p6, and two spacer peptides, SP1 (or p2) and SP2 (or p1) (4, 12, 39). The rate of proteolytic cleavage at each site kinetically controls the processing cascade (11, 20, 29, 31, 38-40).
3-O-(3′,3′-dimethylsuccinyl)betulinic acid, known variously as bevirimat (BVM), PA-457, or DSB, is the first in a potentially new class of anti-HIV-1 drugs known as maturation inhibitors. BVM disrupts HIV-1 infectivity by specifically inhibiting a late step in the Gag processing cascade (5, 6, 33, 44), the cleavage of SP1 from the C terminus of CA (21, 49). The inhibition of CA-SP1 cleavage prevents proper maturation and potently inhibits virion infectivity (21, 40, 49). Under certain circumstances, BVM also has been shown to interfere with virus assembly and release (9, 15); however, this phenomenon occurs only at high, nonphysiological drug concentrations (9).
PR-mediated Gag processing prepares the virion for the infection of a new cell by inducing a morphological rearrangement within the virus particle. During maturation, CA reassembles into a conical condensed core containing the viral RNA in complex with NC and the viral enzymes reverse transcriptase (RT) and integrase (4, 12, 39). Disrupting processing at any of the individual Gag cleavage sites, or altering the order in which the sites are cleaved, results in the formation of aberrant particles that have significantly reduced infectivity (1, 16, 19, 21, 31, 39, 40, 49). The disruption of CA-SP1 cleavage, either by BVM treatment or mutation, leads to the generation of noninfectious particles that exhibit an electron-dense layer of Gag inside the viral membrane and fail to form conical cores (21, 40).
The mechanism by which BVM inhibits CA-SP1 processing is not fully defined. However, several lines of evidence suggest that BVM directly targets the CA-SP1 junction in Gag. All mutations reported thus far to confer resistance to BVM map to the CA-SP1 junction (3, 21, 22, 46, 49). HIV-2 and simian immunodeficiency virus from rhesus macaques (SIVmac) are naturally resistant to BVM (49); the amino acid sequence at the CA-SP1 junction of these lentiviruses diverges from that of HIV-1. Importantly, some sites of sequence divergence between HIV-1 and HIV-2/SIVmac occur at amino acid residues to which BVM resistance maps in HIV-1 (3, 49). Swapping residues between HIV-1 and SIVmac at the CA-SP1 junction renders SIVmac sensitive to BVM (47). The incorporation of BVM into immature HIV-1 particles has been demonstrated, and this incorporation is reduced by some of the reported BVM resistance mutations (46, 48). The observations that BVM does not inhibit the processing of monomeric Gag in solution (21), is specifically incorporated into immature particles (48), and requires immature virus assembly for activity (21, 32) suggest that BVM binds an undefined pocket in Gag that is formed upon Gag oligomerization. The disruption of a specific proteolytic cleavage event in the Gag processing cascade distinguishes the mechanism of action of BVM from that of clinically approved protease inhibitors (PIs), which directly target the catalytic activity of the enzyme (10, 37, 42, 43). The continued clinical development of maturation inhibitors such as BVM would add another much-needed class of anti-HIV-1 drugs to the arsenal currently available for the treatment of HIV/AIDS (5).
Phase II clinical trials are being conducted with BVM both in treatment-naïve patients and in patients who have failed their PI and/or RT inhibitor regimen due to the emergence of viruses resistant to these drugs (26, 33, 35). The fact that BVM likely will be used in PI inhibitor-experienced patients raises interesting and clinically significant questions about the emergence of resistance to BVM in the context of PR enzymes bearing PI-resistant (PIR) mutations. Resistance to PIs typically emerges via the stepwise acquisition of multiple mutations in PR; changes that confer resistance to the PIs emerge first, followed by substitutions in PR that compensate for viral fitness defects imposed by the primary resistance-conferring mutations (7, 28). For example, the HIV-1 isolate with PIR mutations L10R/M46I/L63P/V82T/I84V was isolated from HIV-1 patients failing treatment with the PI indinavir (variously known as IDV or MK-639) (8). These mutations confer resistance to six structurally diverse PIs and reportedly restore replicative fitness to wild-type (WT) levels (8, 27).
We previously identified a panel of six independent, single-amino-acid substitutions that confer resistance to BVM in vitro (Fig. 1) (3). A subset of these mutations was identified in other studies (21, 49). The BVM resistance mutations cluster at or near the CA-SP1 cleavage site (3, 21, 49). In this study, we evaluated the interplay between the L10R/M46I/L63P/V82T/I84V PIR mutations in PR and the BVM resistance mutations in the CA-SP1 region of Gag. We observed general processing defects associated with the PIR PR and a slight replication delay in Jurkat T cells. When combined, the BVM resistance and PIR mutations acted additively to negatively impact viral replication, particularly in the presence of BVM. As a result of this additive effect, the emergence of BVM resistance was delayed in the presence of the PIR mutations. The data presented in this study suggest that resistance to BVM is less likely to emerge in patients who have failed PIs than in patients who are PI naïve.
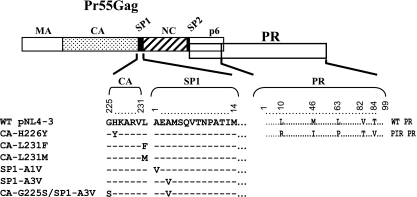
FIG. 1.
Representation of the BVM resistance and PIR PR mutations examined in this study. HIV-1 Gag and PR are represented at the top. The MA, CA, NC, and p6 domains and the SP1 and SP2 spacer peptides in Gag are indicated. The alignment shows each of the BVM resistance and PR mutations. (Adapted from reference 3.)
MATERIALS AND METHODS
BVM, cell culture, and transfections.
BVM was prepared as described previously (14) and used at the concentrations indicated. HeLa cells were maintained in Dulbecco's modified Eagle medium supplemented with 5% (vol/vol) fetal bovine serum (FBS). The Jurkat T-cell line was maintained in RPMI 1640 supplemented with 10% (vol/vol) FBS. All media were supplemented with l-glutamine (2 mM), penicillin, and streptomycin. HeLa cells were transfected with ExGen500 (Fermentas) or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Jurkat T cells were transfected using DEAE-dextran (18).
Generation of PIR HIV-1 derivatives.
A full-length HIV-1 molecular clone derivative of pNL4-3 (2) containing the PR mutations L10R/M46I/L63P/V82T/I84V, which confer resistance to multiple structurally diverse PIs (8), was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID). The PIR mutations were introduced into the previously described BVM-resistant pNL4-3 derivatives CA-H226Y, CA-L231M, CA-L231F, SP1-A1V, SP1-A3V, and CA-G225S/SP1-A3V (3, 21) by subcloning the 3,737-bp ApaI-EcoRI fragment containing the PIR changes (nucleotides 2006 to 5743) into each of the BVM-resistant clones, creating six pNL4-3 derivatives: CA-H226Y PIR, CA-L231M PIR, CA-L231F PIR, SP1-A1V PIR, SP1-A3V PIR, and CA-G225S/SP1-A3V PIR. The identities of all plasmids generated were confirmed by DNA sequencing.
Radioimmunoprecipitation analysis.
The methods used for the metabolic labeling of HeLa cells, the preparation of cell and virus lysates, and immunoprecipitation have been described in detail previously (13, 41). Briefly, media and solutions containing BVM at the indicated concentrations were prepared immediately before use and vortexed. BVM was maintained throughout the transfection and radioimmunoprecipitation procedures. HeLa cells were transfected with WT (2) or PIR (8) pNL4-3 clones and their derivatives encoding the BVM resistance mutations. Transfected HeLa cells were starved in Cys/Met-free medium for 30 min and then metabolically radiolabeled for 2 h with [35S]Cys/Met Pro mix (Amersham). Virions were pelleted by ultracentrifugation. Cell and virus lysates were immunoprecipitated with pooled immunoglobulin from HIV-1-infected patients (HIV-Ig) obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID. The radioimmunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and exposed to X-ray film and a phosphorimager plate (Fuji), and the bands were quantified by using Quantity One software (Bio-Rad).
Replication kinetics.
Jurkat T cells were transfected with WT and mutant pNL4-3 derivatives. BVM was added at the time of transfection at the indicated concentrations and was maintained throughout the course of the experiment. The Jurkat cells were split every 2 days, supernatant was collected at each time point, and viral replication was monitored by RT activity as previously described (13). Replication kinetics experiments were repeated independently four times. On the last repeat, cell pellets and virus supernatants were harvested on the days of peak RT activity. To verify that the BVM resistance and PIR mutations were maintained, genomic DNA was extracted from cells on the day of peak RT activity using a whole-blood DNA extraction kit (Qiagen). The entire Gag-PR coding region then was amplified by PCR using the forward and reverse primers NL516F (5′-TGC CCG TCT GTT GTG TGA CTC-3′) and NL2897R (5′-AAA ATA TGC ATC GCC CAC AT-3′). The resultant 2.3-kb PCR product was purified using the QIAquick PCR purification kit (Qiagen) and sequenced using the primers NL1410F (5′-GGA AGC TGC AGA ATG GGA TA-3′), NL1754F (5′-TGG TCC AAA ATG CGA ACC-3′), and NL2135F (5′-TTC AGA GCA GAC CAG AGC CAA-3′). Following the first round of replication kinetics, virus supernatants from WT, PIR, and SP1-A3V PIR at the day of peak RT activity were normalized for RT activity and used to infect fresh Jurkat T cells. The infected cells were cultured, and then genomic DNA was extracted, PCR amplified, and sequenced as described above to identify any secondary mutations.
Selection of BVM resistance in the context of WT and PIR PR in vitro.
BVM-resistant virus isolates were selected by the serial passage of Jurkat T cells transfected with WT pNL4-3 or the PIR derivative. Ten flasks for each plasmid were transfected in parallel, and cultures were maintained at 50 ng/ml BVM throughout the experiment. Virus replication during the selection process was monitored by RT activity as previously described (13). Cell pellets and virus supernatants were harvested on the days of peak RT activity. To identify mutations conferring BVM resistance, genomic DNA was extracted, PCR amplified, and sequenced as described above.
RESULTS
HIV-1 encoding PIR PR is sensitive to the HIV-1 maturation inhibitor BVM.
To examine the interplay between the evolution of drug resistance to BVM and mutations that confer resistance to clinically approved PR inhibitors, we obtained a derivative of the full-length HIV-1 molecular clone pNL4-3 containing the following PIR mutations: L10R/M46I/L63P/V82T/I84V (Fig. 1) (8). This clone was selected because it encodes a combination of mutations that confer resistance to multiple structurally diverse PR inhibitors and secondary mutations that compensate for defects in virus replication imposed by the PIR mutations (8, 27).
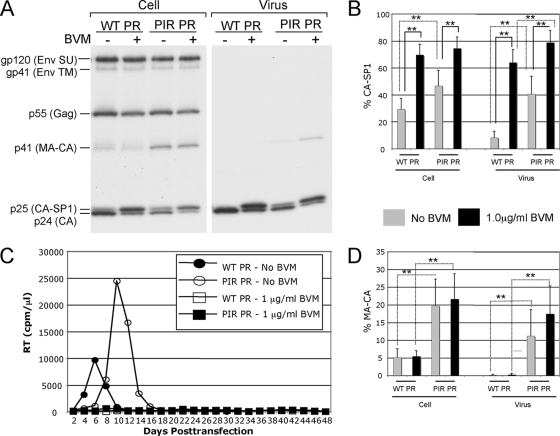
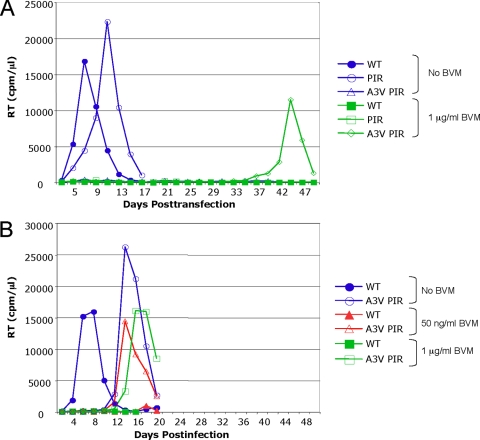
Potent BVM activity against HIV-1 isolates containing PIR PR has been demonstrated previously (21, 49). To verify the sensitivity of the PIR clone to the action of BVM, we conducted a quantitative biochemical CA-SP1 processing assay (Fig. 2A and B ). HeLa cells transfected with WT pNL4-3 or pNL4-3/PIR were cultured either without BVM or with 1 μg/ml BVM. The cells were metabolically labeled with [35S]Met/Cys, and cell- and virion-associated proteins were immunoprecipitated with HIV-Ig. CA-SP1 cleavage was detected (Fig. 2A) and quantified (Fig. 2B). The BVM treatment of PIR-transfected cells resulted in the marked and statistically significant accumulation of unprocessed CA-SP1 in both cell- and virus-associated fractions, which was equivalent to or greater than the levels observed for the BVM-treated WT. This accumulation of CA-SP1 indicates that the PIR clone is sensitive to the action of BVM. The sensitivity of the PIR clone to BVM was confirmed in virus replication assays in which detectable virus spread in Jurkat T cells transfected with either WT or PIR molecular clones was completely blocked at 1 μg/ml BVM for 48 days in culture (Fig. 2C).
FIG. 2.
PIR mutations induce general defects in Gag processing and a modest delay in virus replication. (A) Biochemical characterization of Gag expression and proteolytic processing. Transfected HeLa cells were cultured with 0 or 1 μg/ml BVM and were metabolically labeled with [35S]Met/Cys. Released virions were pelleted by ultracentrifugation. Cell and virus lysates were immunoprecipitated with HIV-Ig and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and fluorography. SU, surface; TM, transmembrane. (B) Phosphorimager analysis of radioimmunoprecipitation assays to quantify the percentage of proteolytic processing intermediate CA-SP1 relative to total CA-SP1 plus CA: % CA-SP1 = [CA-SP1/(CA-SP1 + CA)] × 100. Error bars indicate standard deviations (n ≥ 8). (C) Replication kinetics in Jurkat T cells of WT and PIR molecular clones. Cultures were maintained in 0 or 1 μg/ml BVM. Cells were split every 2 days, and supernatants were reserved at each time point for RT analysis. The experiment shown is representative of eight independent experiments in which the delay between the replication of the WT and PIR clones cultured without BVM ranged from 0 to 12 days, with an average delay of 5 days. Detectable virus replication was not observed in the presence of 1 μg/ml BVM. (D) Phosphorimager analysis of radioimmunoprecipitation assays to quantify the percentage of proteolytic processing intermediate MA-CA relative to total Gag: cellular % MA-CA = [MA-CA/(Pr55 Gag + MA-CA + CA-SP1 + CA)] × 100, and virion % MA-CA = [MA-CA/(MA-CA + CA-SP1 + CA)] × 100. Error bars indicate standard deviations (n ≥ 8). One-way ANOVA was performed using jmp software to test differences between means. Statistically significant differences between pairs of means are indicated with a solid line for BVM-treated versus non-BVM-treated samples with the same PR and a dashed line for samples encoding WT or PIR PR but with the same inhibitor treatment. **, P < 0.01; *, P < 0.05.
PIR mutations are associated with general processing defects and a modest replication delay in Jurkat T cells.
The CA-SP1 processing assay, performed to confirm PIR sensitivity to BVM, demonstrated a general proteolytic processing defect associated with the PIR mutations (Fig. 2A). This defect was characterized by the accumulation of the Gag processing intermediates CA-SP1 and MA-CA (p41) in both the cell- and virion-associated fractions (Fig. 2A, B, and D). The PIR-associated increase in MA-CA levels was statistically significant (P < 0.01 by analysis of variance [ANOVA]) for both cell- and virus-associated fractions. This general proteolytic processing defect likely accounts for the slight delay in virus replication observed for the PIR clone, relative to that of the WT, in the absence of BVM (Fig. 2C). This slight replication delay in Jurkat T cells, which was observed in multiple independent experiments (data not shown and figures cited below), is in contrast to a previous report of WT replication kinetics for the PIR clone in non-Jurkat target cells (27).
General Gag processing defects are induced by PIR mutations independently of BVM resistance mutations surrounding the CA-SP1 cleavage site.
We and others previously have identified a panel of single-amino-acid substitutions that independently confer resistance to BVM (3, 21, 49). All BVM resistance mutations identified thus far are located in the vicinity of the CA-SP1 cleavage site. To understand the interplay between PIR mutations in PR and BVM resistance mutations surrounding the CA-SP1 cleavage site, we examined whether the BVM resistance mutations affected Gag processing efficiency in the context of the PIR PR. To address this question, we introduced the PIR mutations into each of the pNL4-3 clones encoding the following BVM resistance mutations: CA-H226Y, CA-L231M, CA-L231F, SP1-A1V, and SP1-A3V (Fig. 1). We also included a double mutant, CA-G225S/SP1-A3V, containing the CA-G225S mutation, which compensates for the replication defect imposed by SP1-A3V (3).
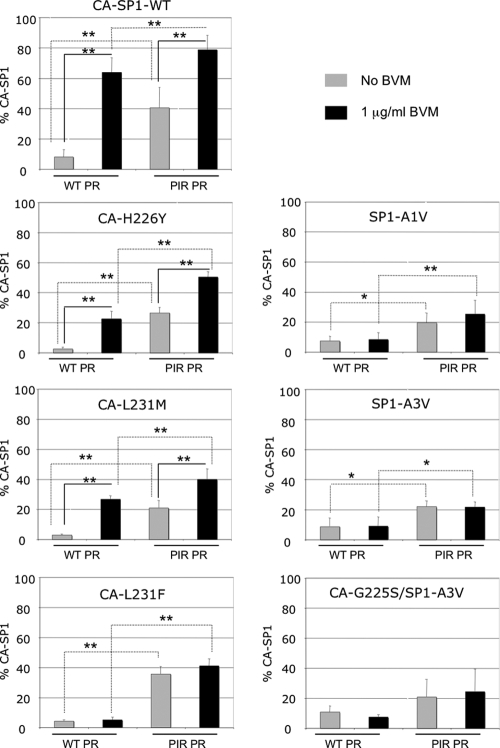
To determine whether the general processing defects observed for PIR PR also were observed in the context of the BVM-resistant mutants, we compared the levels of MA-CA in virions bearing either WT or PIR PR (see Fig. S1 in the supplemental material). The results indicated that the levels of MA-CA in each of the BVM-resistant clones in the context of PIR PR were elevated relative to those in the WT PR series. A statistically significant increase was observed for all BVM-resistant clones, with the exception of the CA-L231M WT PR versus PIR PR when cultured without BVM. We next examined CA-SP1 levels in the absence of BVM to evaluate whether the BVM resistance mutations affected the ability of PIR PR to process the CA-SP1 cleavage site (Fig. 3). The levels of virion-associated CA-SP1 clearly were elevated for each PIR PR clone compared to that of the WT PR clones in the absence of BVM. This was demonstrated by a statistically significant increase in CA-SP1 levels in all BVM-resistant clones except CA-G225S/SP1-A3V. These results demonstrate that the PIR mutations lead to general processing defects, as determined by the accumulation of MA-CA and CA-SP1, independently of BVM resistance mutations.
FIG. 3.
Biochemical quantification of virion-associated CA-SP1 in BVM-resistant clones expressing either WT or PIR PR. Viral lysates were prepared, and the percent CA-SP1 was calculated as indicated in the Fig. 2 legend. Error bars indicate standard deviations (n ≥ 3). ANOVA was performed as described in the Fig. 2 legend. **, P < 0.01; *, P < 0.05.
BVM resistance mutations reduce BVM-induced CA-SP1 accumulation in the context of either WT or PIR PR.
Resistance to BVM is characterized biochemically by complete or nearly complete CA-SP1 processing, even in the presence of the compound (3, 21, 49). To determine whether our previously characterized BVM resistance mutations still confer BVM resistance in the context of the PIR PR, we analyzed the effect of BVM treatment on CA-SP1 levels (Fig. 3). For the CA-L231F, SP1-A1V, SP1-A3V, and CA-G225S/SP1-A3V mutants, BVM treatment did not increase the levels of CA-SP1 in the context of either WT or PIR PR. However, because of the general processing defects noted in the context of PIR PR, the overall CA-SP1 levels were elevated compared to those in the WT PR series independently of BVM treatment. This increase in the percent CA-SP1 accumulation was statistically significant in all BVM-resistant mutants, with the exception of CA-G225S/SP1-A3V. In samples with mutations CA-H226Y and CA-L231M, a statistically significant accumulation of unprocessed CA-SP1 occurred upon BVM treatment in the context of either WT or PIR PR. Elevated levels of CA-SP1 also were noted for CA-L231F PIR. These observations indicate that these CA mutations confer partial resistance to BVM, which is consistent with previous reports (3, 46). A combination of this partial resistance to BVM and the general elevation of CA-SP1 levels in the context of the PIR PR resulted in the high-level accumulation of CA-SP1 in BVM-treated PIR CA-H226Y, CA-L231M, and CA-L231F virions. However, levels of CA-SP1 for these mutants did not reach those observed for the WT CA-SP1 clones in the presence of BVM (Fig. 3).
The BVM resistance and PIR mutations act additively to decrease viral replication capacity.
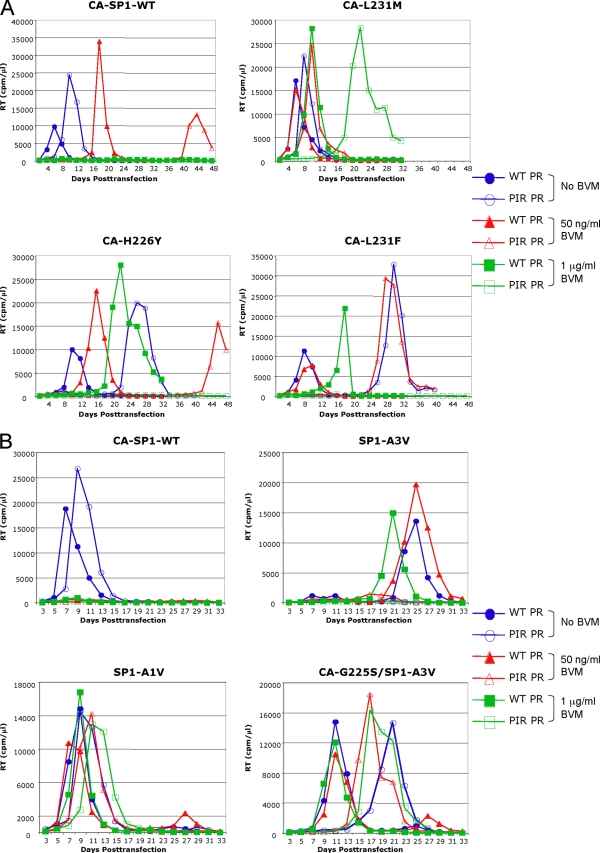
The biochemical analyses presented above demonstrate that the PIR PR mutations are associated with general Gag processing defects. The replication of the PIR clone in Jurkat T cells is slightly delayed, most likely as a consequence of these processing defects (Fig. 2A). Higher levels of unprocessed CA-SP1 also were observed upon treatment with BVM when the CA mutations were coupled with those in PR. To investigate the consequences for virus replication of combining the BVM resistance and PIR mutations, each molecular clone was transfected into Jurkat T cells, and cultures were passaged without BVM or in the presence of suboptimal (50 ng/ml) or inhibitory (1 μg/ml) concentrations of the compound. The BVM resistance mutations originally were selected at the suboptimal BVM concentration (3, 21), and the replication capacity of these mutants in the context of WT PR previously has been evaluated at these BVM concentrations (3). The replication assays were divided into two groups: a CA mutant set (H226Y, L231M, and L231F) (Fig. 4A) and an SP1 mutant set (A1V, A3V, and G225S/A3V) (Fig. 4B). Similar results were obtained in four independently repeated experiments (data not shown).
FIG. 4.
PIR and BVM resistance mutations synergize to delay virus replication. Jurkat T cells were transfected with the indicated BVM-resistant CA (A) or SP1 (B) mutants expressing either WT PR (closed symbols) or PIR PR (open symbols) and were cultured without BVM (blue circle) or with 50 ng/ml (red triangle) or 1 μg/ml (green square) BVM. Cells were split every 2 days, and supernatants were reserved at each time point for RT analysis. Results shown are representative of four independent experiments.
The replication of the CA mutants in the context of WT PR was largely consistent with our previous analysis (3). The L231M and L231F mutants replicated with essentially WT kinetics in the absence of BVM, and their replication was not significantly affected by 50 ng/ml BVM (Fig. 4A). The replication of H226Y was only slightly delayed relative to that of WT in the absence of BVM, while a more significant delay was seen in the presence of 50 ng/ml BVM (Fig. 4A). A several-day delay was reproducibly observed for all of these mutants at 1 μg/ml BVM. This replication delay at 1 μg/ml BVM is indicative of a residual level of BVM sensitivity and is consistent with the increased accumulation of unprocessed CA-SP1 upon BVM treatment (Fig. 3) (3). The replication of CA-H226Y and CA-L231F in the context of PIR PR was considerably delayed compared to the replication of the WT PR series. Long delays in replication were observed without BVM and at 50 ng/ml, and no detectable replication was observed at 1 μg/ml BVM. The replication of CA-L231M PIR was more robust, as replication without BVM and at 50 ng/ml BVM was only slightly delayed relative to that of the WT PR series. However, in contrast to CA-L231M with WT PR, the replication of CA-L231M with PIR PR at 1 μg/ml BVM was reproducibly and significantly delayed (Fig. 4A and data not shown).
The replication of viruses containing SP1 mutations (A1V, A3V, and CA-G225S/SP1-A3V) in the context of WT PR also was largely consistent with our previous analysis (3). The SP1-A1V WT PR mutant replicated with WT kinetics in the absence of BVM, and its replication was not significantly affected even by high BVM concentrations (Fig. 4B). The replication of SP1-A1V PIR also was robust under all conditions tested, and the observed delays essentially matched those observed between WT and PIR PR in the context of WT Gag.
The replication of SP1-A3V with WT PR was markedly delayed and exhibited a degree of drug dependence at 1 μg/ml BVM (3). The replication of SP1-A3V PIR PR typically was not detected in the presence or absence of BVM (Fig. 4B) but was observed in one experiment (Fig. 5A). In this instance, virus replication in 1 μg/ml BVM peaked on day 44. As a number of secondary mutations are associated with the replication of the SP1-A3V mutant (3), the virus from day 44 was repassaged to select for secondary mutations (Fig. 5B). Upon repassage, the virus replicated much earlier under all conditions tested (0 ng/ml, 50 ng/ml, and 1 μg/ml BVM). Genomic DNA was extracted from cells harvested at the RT peak, and the Gag and PR coding regions were amplified by PCR and sequenced. Two secondary mutations were identified within the population: the previously observed SP1-V7I (3) and a mutation distal to the CA-SP1 junction, CA-N203T. The SP1-A3V mutation and all of the PIR PR mutations were maintained. We also examined the previously characterized CA-G225S secondary mutation, which reverses the replication delay imposed by SP1-A3V (3). The CA-G225S mutation also compensated for the delay imposed by SP1-A3V in the context of PIR PR as replication occurred at all BVM concentrations, albeit with a moderate delay compared to that of the WT PR context (Fig. 4B).
FIG. 5.
Replication kinetics of the SP1-A3V mutant coupled with the PIR changes. Jurkat T cells were transfected with WT pNL4-3 (closed symbols), pNL4-3/PIR, or pNL4-3/A3V PIR. (A) Cultures were maintained either without BVM (blue symbols) or with 1 μg/ml BVM (green symbols). (B) Cultures were maintained in no BVM (blue circle), 50 ng/ml (red triangle) BVM, or 1 μg/ml (green square) BVM. Cells were split every 2 days, and supernatants were reserved at each time point for RT analysis. Virus from the SP1-A3V PIR flask (A) was harvested on day 44 and used to infect fresh Jurkat T cells (B). DNA was extracted at peak RT activity, and the Gag-PR coding region was PCR amplified and sequenced.
The maintenance of all five PIR PR mutations upon the repassage of A3V PIR PR confirms the previously reported stability of the PIR mutations (27). Indeed, in the final repeat of the virus replication experiments, we confirmed that all PIR and BVM resistance mutations still were present in all samples with detectable viral replication (data not shown). Only one additional secondary mutation, CA-V230I, was identified. This mutation arose in the CA-L231M PIR PR virus that arose 30 days posttransfection when cultured at 1 μg/ml BVM. The additional characterization of the secondary changes identified here, along with several others associated with SP1-A3V (3 and C. S. Adamson and E. O. Freed, unpublished data), will be reported elsewhere.
Overall, when combined, the BVM resistance and PIR mutations acted additively to negatively impact viral replicative capacity, particularly in the presence of BVM. For example, the combination of a mutation that impaired virus replication (e.g., SP1-A3V) with PIR PR essentially abolished virus replication. The combination of mutations that induced various degrees of BVM resistance (e.g., CA-H226Y or L231F) with the PIR mutations delayed virus replication in the absence of BVM and at low BVM concentrations and abolished replication at high concentrations of the compound. However, for a virus that replicated robustly even at a high BVM concentration, such as SP1-A1V, combination with PIR had a minimal impact on virus replication kinetics.
Emergence of BVM resistance is delayed in the context of PIR PR.
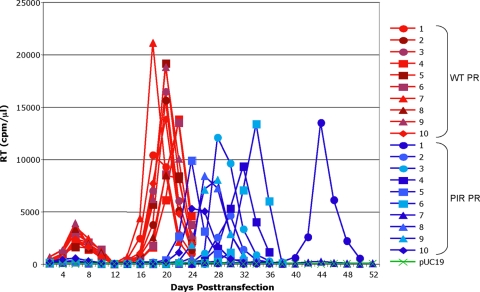
Replication assays showed that the combination of the BVM resistance and PIR mutations generally impaired viral fitness, particularly in the presence of BVM. We therefore hypothesized that the PIR mutations would delay or prevent the emergence of BVM resistance and would limit the number of distinct BVM resistance mutations that emerge during virus propagation in BVM. Resistance in Jurkat T cells typically emerges 3 to 4 weeks posttransfection with WT pNL4-3 at 50 ng/ml BVM (3, 21) (e.g., Fig. 4A). In contrast, in a parallel culture transfected with pNL4-3/PIR, virus replication did not peak until 44 days posttransfection (Fig. 4A). To determine which BVM resistance mutation(s) arose in the context of PIR PR in this culture, genomic DNA was extracted from cells harvested at peak RT activity, and the PCR-amplified Gag-PR region was sequenced. This analysis revealed the acquisition of the previously characterized mutation SP1-A1V. All five PIR PR mutations were maintained, and no other changes in Gag or PR were observed.
As the acquisition of resistance is essentially a stochastic process, we repeated the selection experiment in multiple flasks to determine whether the emergence of BVM resistance in the context of PIR is consistently delayed and to identify the BVM resistance mutations that arise. Ten flasks each were transfected with WT pNL4-3 or pNL4-3/PIR and then cultured in 50 ng/ml BVM (Fig. 6). Viral replication was consistently delayed by the PIR mutations. Virus emerged in 9 out of 10 PIR-transfected flasks, with delay in RT peaks ranging from 2 to 22 days after the latest RT peak in WT-transfected flasks. In the one remaining PIR-transfected flask, detectable virus replication did not occur during 90 days in culture (data not shown). Sequencing revealed that the SP1-A1V mutation emerged with approximately equal frequency in the context of WT and PIR PR (Table 1). Of the 10 flasks transfected with WT PR plasmid, the SP1-A1V mutation arose 90% of the time. One flask did not contain SP1-A1V but instead acquired a previously unidentified mutation, SP1-M4V. Another flask contained a mixture of SP1-A1V and CA-L231W mutations. Although CA-L231W has not been observed previously, BVM-resistant mutants L231M and L231F map to the same residue. For the set of 10 flasks transfected with PIR PR, the SP1-A1V mutation arose 80% of the time. The only other mutation to arise in this series of flasks was SP1-A3V. The PIR PR mutations were maintained in all nine flasks in which virus emerged, and no other mutations in PR were detected. In a repeat of the selection experiment, virus emerged in 8 of a set of 10 flasks transfected with the WT PR clone but just 1 flask of the set of 10 PIR PR-transfected flasks (data not shown). These data demonstrate that the acquisition of BVM resistance is either delayed or prevented in the context of PIR PR.
FIG. 6.
PIR changes in PR delay the development of BVM resistance. Ten Jurkat flasks each were transfected with WT pNL4-3 (red) or pNL4-3/PIR (blue) and cultured in parallel in the presence of 50 ng/ml BVM. Cells were split every 2 days, and supernatants were reserved at each time point for RT analysis. DNA was extracted at peak RT activity, and the Gag-PR coding region was PCR amplified and sequenced.
TABLE 1.
Mutations acquired at the CA-SP1 junction in Jurkat T cells transfected with WT pNL4-3 or pNL4-3/PIR and cultured in 50 ng/ml BVM
| Virus no. | Day of peak RT | Mutation(s) acquired |
|---|---|---|
| WT PR | ||
| 1 | 18 | SP1-A1V |
| 2 | 20 | SP1-A1V |
| 3 | 20 | SP1-A1V |
| 4 | 22 | SP1-A1V, CA-L231W |
| 5 | 20 | SP1-A1V |
| 6 | 22 | SP1-A1V |
| 7 | 18 | SP1-A1V |
| 8 | 20 | SP1-M4V |
| 9 | 20 | SP1-A1V |
| 10 | 20 | SP1-A1V |
| PIR PR | ||
| 1 | 44 | SP1-A1V |
| 2 | 30 | SP1-A3V |
| 3 | 30 | SP1-A1V |
| 4 | 32 | SP1-A1V |
| 5 | 24 | SP1-A1V |
| 6 | 34 | SP1-A1V |
| 7 | No replication | No replication |
| 8 | 26 | SP1-A1V |
| 9 | 28 | SP1-A1V |
| 10 | 24 | SP1-A1V |
DISCUSSION
Many novel antiretroviral drugs, including BVM, likely will be used clinically in patients harboring viruses resistant to RT inhibitors and PIs. Because BVM targets PR-mediated processing at the CA-SP1 cleavage site, and mutations that confer BVM resistance map to this region of Gag, in this study we sought to examine the interplay between the evolution of resistance to BVM and PIR mutations in PR. We demonstrate that the emergence of BVM resistance is delayed in the context of the PIR PR, which induces a general Gag processing defect independently of BVM resistance mutations. The SP1-A1V mutation arises most frequently in clones expressing either WT or PIR PR. Combining our previously characterized panel of BVM resistance changes with PIR mutations led in most cases to severely inhibited virus replication, particularly in the presence of BVM. However, in the case of SP1-A1V, we observed no significant effect of the PIR mutations on virus replication, even at high BVM concentrations.
Mutations in PR could influence the emergence of BVM resistance primarily by two mechanisms: (i) the ability of the mutant PR to cleave at a CA-SP1 junction containing BVM resistance changes could be altered, and/or (ii) the PR mutations could reduce the overall rate of virus replication, thereby limiting the ability of BVM resistance to emerge. In the latter instance, the mechanism is not due to the nature of the PR mutations per se but rather their overall deleterious effect on replication, therefore a similar phenomenon might also be observed for other mutants that display a reduced replication capacity. We chose the L10R/M46I/L63P/V82T/I84V PIR clone for this study because it has been reported to replicate with essentially WT kinetics (27). However, we repeatedly observed a slight delay in the replication of this clone relative to that of the WT, most likely due to the above-mentioned general Gag processing defects. Despite this slight replication defect, we observed that the PIR mutations were very stable during long-term culture in Jurkat T cells; in no case did we observe the loss of any of these PIR mutations or the acquisition of other mutations in PR. The stability of the PIR mutations upon long-term culture obviated the need for the inclusion of PIs in the BVM selection experiments. The overall reduction in the replication capacity of the PIR clone likely contributed to the delayed emergence of BVM resistance in the context of PIR PR. To the extent that PIR mutations circulating in patients reduce replication fitness, these findings may be relevant to the potential emergence of BVM resistance in vivo.
In both this and our previous study (3), we have noted the frequent emergence of the SP1-A1V mutation. This can be explained by the robust replication kinetics of SP1-A1V in the context of either WT or PIR PR even at high BVM concentrations. In contrast, the combination of the other BVM resistance mutations with PIR PR significantly decreased viral replication capacity, particularly in the presence of BVM. For example, the replication of viral clones containing the CA mutations (H226Y, L231M, and L231F) in combination with the PIR changes was significantly delayed or abolished in the presence of BVM. This is likely due to high CA-SP1 levels resulting from the general Gag processing defects induced by PIR mutations, combined with only a partial degree of BVM resistance being conferred by the CA mutations (3, 46, 49). Interestingly, a CA-V230I mutation emerged upon the passage of CA-L231M with PIR PR at 1 μg/ml BVM. This secondary mutation may compensate for the replication delay induced by the PIR changes. It is well documented that mutations in Gag cleavage sites can accumulate during the acquisition of resistance to PIs (17, 23, 25, 30, 45). However, these mutations are rarely seen at the CA-SP1 site (23, 25). The residues to which BVM resistance map are highly conserved (3), and to date only CA-L231M has been reported in the context of one PI-experienced patient sample (24). However, the CA-V230I mutation has been reported in ∼10% of both PI-experienced and treatment-naïve patients (23, 24, 34). It also is possible that CA-V230I confers a degree of BVM resistance. Residue CA-230 occurs at position P2 of the PR recognition site, and the insertion of the P2 and P1 residues from SIVmac, which is BVM resistant, into HIV-1 resulted in the transfer of BVM resistance to HIV-1 (47). This study identified two additional novel mutations that may confer BVM resistance: CA-L231W and SP1-M4V.
The additive effect of combining an already replication-impaired virus, such as SP1-A3V, with PIR PR essentially abolished virus replication. We previously reported that the SP1-A3V mutant in the context of WT PR exhibits a degree of BVM dependence at 1 μg/ml BVM and generally requires a secondary mutation to facilitate replication at low BVM concentrations or in the absence of the compound (3). In this study, we observed two mutations that may rescue the replication of the SP1-A3V PIR clone, SP1-V7I and a mutation distal to the CA-SP1 cleavage site, CA-N203T. The significance of the distal CA-N203T mutation has not been established; however, it is unlikely to confer BVM resistance, as all previously identified BVM resistance mutations cluster to the CA-SP1 junction. It is plausible that the CA-N203T mutation compensates for defects imposed by SP1-A3V and/or the PIR mutations; however, this remains to be determined. SP1-V7I previously has been identified during the propagation of SP1-A3V in the context of WT PR (3), therefore its emergence is unlikely to be a direct consequence of the PIR mutations. We previously detected a number of secondary mutations associated with SP1-A3V ([3] and Adamson and Freed, unpublished). The characterization of these secondary mutations will be reported elsewhere, but they are of interest because they map to nonconserved residues at the periphery of the CA-SP1 junction, and polymorphisms at these amino acid positions appear to be correlated with variable clinical responses to BVM (K. Salzwedel, unpublished data).
This study demonstrates that the emergence of BVM resistance is delayed or prevented in the context of a PIR PR compared to that in the context of WT PR. The reduced replication fitness of mutants bearing substitutions in both the CA-SP1 region and in PR likely is due to a slight replication impairment of the PIR clone and because the majority of BVM resistance mutations act additively with PIR changes to inhibit virus replication. Understanding the nature of BVM resistance arising in the context of both WT and PIR PR elucidates the interplay between these two sets of mutations and may help to predict the types of mutations that are likely to arise in vivo. We anticipate that the SP1-A1V substitution is the most likely to emerge in vivo, as this mutant replicates robustly independently of PR mutations or BVM. Because an Ala at SP1 residue 1 is highly conserved among HIV-1 isolates, it might be predicted that a fitness cost would be associated with the replication of this virus in vivo. However, we observe that SP1-A1V can replicate efficiently in primary lymphocytes and macrophages (C. S. Adamson, S. D. Ablan, and E. O. Freed, unpublished data), and this mutant confers resistance to BVM and replicates with no fitness impairment in SCID-hu Thy/Liv mice (36). The SP1-A1V mutation also has been observed in virus from 2 of 46 patients participating in the phase IIb clinical trials (K. Salzwedel, unpublished data). Ongoing clinical trials with BVM ultimately will allow comparisons to be made between the evolution of BVM resistance in vitro and in vivo.
Supplementary Material
Acknowledgments
We thank members of the Freed laboratory for critical reviews of the manuscript. We thank M. Fivash for assistance with statistical analysis. HIV-Ig and the PIR plasmid construct were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by the Intramural AIDS Targeted Antiviral Program.
Footnotes
Published ahead of print on 11 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 722072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamson, C. S., S. D. Ablan, I. Boeras, R. Goila-Gaur, F. Soheilian, K. Nagashima, F. Li, K. Salzwedel, M. Sakalian, C. T. Wild, and E. O. Freed. 2006. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (bevirimat). J. Virol. 8010957-10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adamson, C. S., and E. O. Freed. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 55347-387. [DOI] [PubMed] [Google Scholar]
- 5.Adamson, C. S., and E. O. Freed. 2008. Recent progress in antiretrovirals—lessons from resistance. Drug Discov. Today 13424-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aiken, C., and C. H. Chen. 2005. Betulinic acid derivatives as HIV-1 antivirals. Trends Mol. Med. 1131-36. [DOI] [PubMed] [Google Scholar]
- 7.Condra, J. H. 1998. Resistance to HIV protease inhibitors. Haemophilia 4610-615. [DOI] [PubMed] [Google Scholar]
- 8.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374569-571. [DOI] [PubMed] [Google Scholar]
- 9.DaFonseca, S., A. Blommaert, P. Coric, S. S. Hong, S. Bouaziz, and P. Boulanger. 2007. The 3-O-(3′,3′-dimethylsuccinyl) derivative of betulinic acid (DSB) inhibits the assembly of virus-like particles in HIV-1 Gag precursor-expressing cells. Antivir. Ther. 121185-1203. [PubMed] [Google Scholar]
- 10.Emini, E. A., and H. Y. Fan. 1997. Immunological and pharmacological approaches to the control of retroviral infections. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 11.Erickson-Viitanen, S., J. Manfredi, P. Viitanen, D. E. Tribe, R. Tritch, C. A. Hutchison III, D. D. Loeb, and R. Swanstrom. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retrovir. 5577-591. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 2511-15. [DOI] [PubMed] [Google Scholar]
- 13.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 682503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujioka, T., Y. Kashiwada, R. E. Kilkuskie, L. M. Cosentino, L. M. Ballas, J. B. Jiang, W. P. Janzen, I. S. Chen, and K. H. Lee. 1994. Anti-AIDS agents. 11. Betulinic acid and platanic acid as anti-HIV principles from Syzigium claviflorum, and the anti-HIV activity of structurally related triterpenoids. J. Nat. Prod. 57243-247. [DOI] [PubMed] [Google Scholar]
- 15.Kanamoto, T., Y. Kashiwada, K. Kanbara, K. Gotoh, M. Yoshimori, T. Goto, K. Sano, and H. Nakashima. 2001. Anti-human immunodeficiency virus activity of YK-FH312 (a betulinic acid derivative), a novel compound blocking viral maturation. Antimicrob. Agents Chemother. 451225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan, A. H., J. A. Zack, M. Knigge, D. A. Paul, D. J. Kempf, D. W. Norbeck, and R. Swanstrom. 1993. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of non-infectious particles. J. Virol. 674050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaufmann, G. R., K. Suzuki, P. Cunningham, M. Mukaide, M. Kondo, M. Imai, J. Zaunders, and D. A. Cooper. 2001. Impact of HIV type 1 protease, reverse transcriptase, cleavage site, and p6 mutations on the virological response to quadruple therapy with saquinavir, ritonavir, and two nucleoside analogs. AIDS Res. Hum. Retrovir. 17487-497. [DOI] [PubMed] [Google Scholar]
- 18.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 724116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kräusslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 693407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kräusslich, H. G., H. Schneider, G. Zybarth, C. A. Carter, and E. Wimmer. 1988. Processing of in vitro-synthesized gag precursor proteins of human immunodeficiency virus (HIV) type 1 by HIV proteinase generated in Escherichia coli. J. Virol. 624393-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, F., R. Goila-Gaur, K. Salzwedel, N. R. Kilgore, M. Reddick, C. Matallana, A. Castillo, D. Zoumplis, D. E. Martin, J. M. Orenstein, G. P. Allaway, E. O. Freed, and C. T. Wild. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. USA 10013555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, F., D. Zoumplis, C. Matallana, N. R. Kilgore, M. Reddick, A. S. Yunus, C. S. Adamson, K. Salzwedel, D. E. Martin, G. P. Allaway, E. O. Freed, and C. T. Wild. 2006. Determinants of activity of the HIV-1 maturation inhibitor PA-457. Virology 356217-224. [DOI] [PubMed] [Google Scholar]
- 23.Malet, I., B. Roquebert, C. Dalban, M. Wirden, B. Amellal, R. Agher, A. Simon, C. Katlama, D. Costagliola, V. Calvez, and A. G. Marcelin. 2007. Association of Gag cleavage sites to protease mutations and to virological response in HIV-1 treated patients. J. Infect. 54367-374. [DOI] [PubMed] [Google Scholar]
- 24.Malet, I., M. Wirden, A. Derache, A. Simon, C. Katlama, V. Calvez, and A. G. Marcelin. 2007. Primary genotypic resistance of HIV-1 to the maturation inhibitor PA-457 in protease inhibitor-experienced patients. AIDS 21871-873. [DOI] [PubMed] [Google Scholar]
- 25.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 727632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, D. E., R. Blum, J. Wilton, J. Doto, H. Galbraith, G. L. Burgess, P. C. Smith, and C. Ballow. 2007. Safety and pharmacokinetics of bevirimat (PA-457), a novel inhibitor of human immunodeficiency virus maturation, in healthy volunteers. Antimicrob. Agents Chemother. 513063-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 733744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menéndez-Arias, L., M. A. Martinez, M. E. Quinones-Mateu, and J. Martinez-Picado. 2003. Fitness variations and their impact on the evolution of antiretroviral drug resistance. Curr. Drug Targets Infect. Disord. 3355-371. [DOI] [PubMed] [Google Scholar]
- 29.Mervis, R. J., N. Ahmad, E. P. Lillehoj, M. G. Raum, F. H. Salazar, H. W. Chan, and S. Venkatesan. 1988. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J. Virol. 623993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myint, L., M. Matsuda, Z. Matsuda, Y. Yokomaku, T. Chiba, A. Okano, K. Yamada, and W. Sugiura. 2004. Gag non-cleavage site mutations contribute to full recovery of viral fitness in protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 688017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakalian, M., C. P. McMurtrey, F. J. Deeg, C. W. Maloy, F. Li, C. T. Wild, and K. Salzwedel. 2006. 3-O-(3′,3′-dimethysuccinyl) betulinic acid inhibits maturation of the human immunodeficiency virus type 1 gag precursor assembled in vitro. J. Virol. 805716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salzwedel, K., D. E. Martin, and M. Sakalian. 2007. Maturation inhibitors: a new therapeutic class targets the virus structure. AIDS Rev. 9162-172. [PubMed] [Google Scholar]
- 34.Shelburne, S. A., III, R. J. Hamill, M. C. Rodriguez-Barradas, S. B. Greenberg, R. L. Atmar, D. W. Musher, J. C. Gathe, Jr., F. Visnegarwala, and B. W. Trautner. 2002. Immune reconstitution inflammatory syndrome: emergence of a unique syndrome during highly active antiretroviral therapy. Medicine (Baltimore) 81213-227. [DOI] [PubMed] [Google Scholar]
- 35.Smith, P. F., A. Ogundele, A. Forrest, J. Wilton, K. Salzwedel, J. Doto, G. P. Allaway, and D. E. Martin. 2007. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-o-(3′,3′-dimethylsuccinyl)betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob. Agents Chemother. 513574-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoddart, C. A., P. Joshi, B. Sloan, J. C. Bare, P. C. Smith, G. P. Allaway, C. T. Wild, and D. E. Martin. 2007. Potent activity of the HIV-1 maturation inhibitor bevirimat in SCID-hu Thy/Liv mice. PLoS ONE 2e1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Temesgen, Z., D. Warnke, and M. J. Kasten. 2006. Current status of antiretroviral therapy. Expert Opin. Pharmacother. 71541-1554. [DOI] [PubMed] [Google Scholar]
- 38.Tritch, R. J., Y. E. Cheng, F. H. Yin, and S. Erickson-Viitanen. 1991. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 gag polyprotein. J. Virol. 65922-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 21495-131. [DOI] [PubMed] [Google Scholar]
- 40.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 722846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willey, R. L., J. S. Bonifacino, B. J. Potts, M. A. Martin, and R. D. Klausner. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA 859580-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wlodawer, A., and J. W. Erickson. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62543-585. [DOI] [PubMed] [Google Scholar]
- 43.Wlodawer, A., and J. Vondrasek. 1998. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu. Rev. Biophys. Biomol. Struct. 27249-284. [DOI] [PubMed] [Google Scholar]
- 44.Yu, D., C. T. Wild, D. E. Martin, S. L. Morris-Natschke, C. H. Chen, G. P. Allaway, and K. H. Lee. 2005. The discovery of a class of novel HIV-1 maturation inhibitors and their potential in the therapy of HIV. Expert Opin. Investig. Drugs 14681-693. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 716662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, J., C. H. Chen, and C. Aiken. 2006. Human immunodeficiency virus type 1 resistance to the small molecule maturation inhibitor 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid is conferred by a variety of single amino acid substitutions at the CA-SP1 cleavage site in Gag. J. Virol. 8012095-12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, J., C. H. Chen, and C. Aiken. 2004. The sequence of the CA-SP1 junction accounts for the differential sensitivity of HIV-1 and SIV to the small molecule maturation inhibitor 3-O-{3′,3′-dimethylsuccinyl}-betulinic acid. Retrovirology 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, J., L. Huang, D. L. Hachey, C. H. Chen, and C. Aiken. 2005. Inhibition of HIV-1 maturation via drug association with the viral Gag protein in immature HIV-1 particles. J. Biol. Chem. 28042149-42155. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, J., X. Yuan, D. Dismuke, B. M. Forshey, C. Lundquist, K. H. Lee, C. Aiken, and C. H. Chen. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 78922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.