Abstract
The human immunodeficiency virus type 1 (HIV-1) envelope (Env) protein contains numerous N-linked carbohydrates that shield conserved peptide epitopes and promote trans infection by dendritic cells via binding to cell surface lectins. The potent and broadly neutralizing monoclonal antibody 2G12 binds a cluster of high-mannose-type oligosaccharides on the gp120 subunit of Env, revealing a conserved and highly exposed epitope on the glycan shield. To find an effective antigen for eliciting 2G12-like antibodies, we searched for endogenous yeast proteins that could bind to 2G12 in a panel of Saccharomyces cerevisiae glycosylation knockouts and discovered one protein that bound weakly in a Δpmr1 strain deficient in hyperglycosylation. 2G12 binding to this protein, identified as Pst1, was enhanced by adding the Δmnn1 deletion to the Δpmr1 background, ensuring the exposure of terminal α1,2-linked mannose residues on the D1 and D3 arms of high-mannose glycans. However, optimum 2G12 antigenicity was found when Pst1, a heavily N-glycosylated protein, was expressed with homogenous Man8GlcNAc2 structures in Δoch1 Δmnn1 Δmnn4 yeast. Surface plasmon resonance analysis of this form of Pst1 showed high affinity for 2G12, which translated into Pst1 efficiently inhibiting gp120 interactions with 2G12 and DC-SIGN and blocking 2G12-mediated neutralization of HIV-1 pseudoviruses. The high affinity of the yeast glycoprotein Pst1 for 2G12 highlights its potential as a novel antigen to induce 2G12-like antibodies.
The human immunodeficiency virus (HIV) has evolved numerous means to evade the humoral immune response, including a two-receptor mechanism for entry that recesses and protects highly conserved binding sites in the gp120 subunit of the viral envelope (Env) protein, trimerization of Env to further protect neutralizing epitopes readily exposed on the monomer, and rapid and continual mutation in the face of immune selective pressure (8, 9). Another highly effective defense mechanism is found in the extensive array of oligosaccharides covering gp120, with approximately 25 N-linked glycosylation sites per gp120 monomer (26). These glycans facilitate HIV type 1 (HIV-1) escape from immune surveillance by presenting immunologically “self” molecules with highly variable glycoforms that mask polypeptide epitopes along the “silent face” of gp120 (46, 49). Additionally, high-mannose-type N-linked glycans on gp120 have been implicated in inducing immunosuppressive responses from dendritic cells (DCs) (40), and in helping viral dissemination by binding to DCs through C-type lectins, such as DC-SIGN (DC-specific intercellular adhesion molecule 3-grabbing nonintegrin) (18, 33, 34). The high affinity of DC-SIGN for mannose structures on gp120 (29, 41), and evidence that DC-SIGN+ mucosal cells assist trans infection of permissive T cells, imply a key role for DC-SIGN in early HIV infection after sexual transmission (19).
The high-mannose-type glycans of gp120 also represent a vulnerability for HIV-1. Mannose-binding lectins, such as cyanovirin N (16), actinohivin (12), and human mannose-binding protein (17), can interact with gp120 and inhibit HIV-1 infection in vitro. More critically for vaccine studies, high-mannose glycans are also the target of 2G12, one of the few broadly neutralizing monoclonal antibodies (MAbs) isolated from HIV-1-infected patients (36, 37, 42). The potency of this MAb stems from its unique epitope on the exposed and relatively conserved “silent face” of gp120, comprised of a cluster of terminal Manα1,2-Man residues on the D1 and D3 arms of up to three high-mannose glycans (10, 11, 36, 37). 2G12 is thought to have a high affinity for these gp120 glycans due to a unique heavy chain variable (VH) domain-swapped configuration that forms a multivalent binding surface with a potential noncanonical binding site at the novel VH/VH interface in addition to the two conventional VH/light chain variable (VL) binding sites. This extended antigen binding surface is thought to allow 2G12 to interact with multiple clustered high-mannose glycans (11).
Due to the broadly neutralizing activity of 2G12, the high-mannose glycans on gp120 have aroused interest in the design of glycoantigens that recapitulate the 2G12 epitope. Several such antigens have been created by using flexible linkers to cross-link natural or chemically synthesized high-mannose glycans to various molecular scaffolds, each showing that multivalency of high-mannose glycans is the key to higher 2G12 affinity (2, 25, 27, 43-45). An alternative approach is to express heterologous glycoproteins with natural high-mannose glycans able to support 2G12 binding (28, 38). The yeast Saccharomyces cerevisiae expresses many proteins with high densities of N-linked glycans, and the enzymes involved in its N-glycosylation pathway are easily manipulated to produce glycans with various high-mannose structures (3, 31). We previously showed that an engineered strain lacking the OCH1, MNN1, and MNN4 genes for carbohydrate-processing enzymes expressed at least four highly glycosylated proteins that supported 2G12 binding and that immunization of rabbits with whole yeast cells from this strain elicited antibodies that cross-reacted with the glycans of gp120 (28). Here, we describe a second approach to modify the glycosylation machinery of S. cerevisiae and the subsequent discovery of Pst1, a yeast glycoprotein able to bind MAb 2G12. We show that Pst1 displays increased 2G12 binding as the dominant glycans on the protein become more similar to the glycans on the 2G12 epitope of gp120. This form of Pst1, containing strictly Man8GlcNAc2 glycans, displayed high affinity for 2G12 and effectively blocked the interaction of gp120 with 2G12 and DC-SIGN. This identifies Pst1 as a candidate molecular scaffold for an effective presentation of the 2G12 epitope and as a potential immunogen to induce mannose-specific antibodies.
MATERIALS AND METHODS
Materials.
2G12 and 2F5 were purchased from Polymum Scientific (Vienna, Austria). Antibodies against Pst1 (anti-Pst1) and clade B gp120 consensus sequence (anti-gp120) were made using the peptides DSLESITDSLNLQSLT and KIEPLGVAPTKAKRRVVQRE, respectively. Horseradish peroxidase (HRP)-labeled goat anti-rabbit immunoglobulin G (IgG) and goat anti-human IgG were purchased from Jackson ImmunoResearch (West Grove, PA). Homozygous diploid strains in the BY4743 S. cerevisiae background with single deletions in MNN1, PMR1, and MNS1 were obtained from Open Biosystems (Huntsville, AL), while the wild-type (WT) S. cerevisiae strain INVSc1 and single deletions in OCH1 and MNN9 were obtained from Invitrogen (Carlsbad, CA). HIV-1 gp120 glycoproteins from strains ADA, JR-FL, and Yu2 were expressed in 293T cells using recombinant vaccinia virus vectors and purified by Galanthus nivalis lectin-agarose (Vector Laboratories, Burlingame, CA) affinity chromatography (39). S. cerevisiae mannan, d-mannose, and α1-acid glycoprotein were purchased from Sigma-Aldrich (St. Louis, MO).
S. cerevisiae mating.
Yeast strains were mated, sporulated, and PCR verified as previously described (28). The confirmed Δmnn1 Δpmr1 double mutants were of two genotypes, MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 MNN1::kanmx4 PMR1::kanmx4 (labeled Δmnn1 Δpmr1-clone 1 and -clone 5), and MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 MNN1::kanmx4 PMR1::kanmx4 (labeled Δmnn1 Δpmr1-clone 4).
Protein purification and identification.
The culture supernatant from Δpmr1 Δmnn1 cells grown to log phase in yeast synthetic complete medium containing 2% galactose was concentrated and dialyzed against phosphate-buffered saline (PBS). Bio-Lyte 3/10 ampholytes (Bio-Rad, Hercules, CA) were added to 0.5% (wt/vol). The sample was separated by preparative isoelectric focusing (IEF) on a Rotofor Cell (Bio-Rad) operated according to the manufacturer's protocol. 2G12-positive fractions were determined by a standard Western blot in which samples were denatured by boiling them for 5 min at 100°C in 2× Laemmli sodium dodecyl sulfate (SDS) sample buffer (0.09 M Tris-HCl, pH 6.8, 20% glycerol, 2% SDS, 0.1 M dithiothreitol, 0.02% bromophenol blue).
Purified protein detected by 2G12 was excised from SDS-polyacrylamide gel electrophoresis (PAGE) gels and identified by Proteomic Research Services, Inc. (Ann Arbor, Michigan) using trypsin digestion followed by nanoliter liquid chromatography coupled to tandem mass spectrometry (LC/MS/MS), as previously described (28). The protein sequence of Pst1 (accession no. NP_010340) was used to search the HomoloGene database (release 50.1) of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/HomoloGene). For immunoprecipitation, culture supernatants from Δpmr1 and Δpmr1 Δmnn1 cells were precipitated with 2G12 as previously described (28), with Western blot analysis using anti-Pst1.
Glycan analyses.
N-linked glycans on purified Pst1 were analyzed at the Glycotechnology Core Resource, University of California, San Diego, by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS. Deglycosylation of purified Pst1 expressed in Δpmr1 Δmnn1 cells for Western blotting was conducted using endoglycosidase H (Endo H), jack bean mannosidase, and Aspergillus saitoi mannosidase (Glyko Inc., San Leandro, CA) as previously described (28).
Cloning and protein expression.
A plasmid containing the coding sequence of Pst1 without the glycosylphosphatidylinositol (GPI) anchor signal sequence was constructed in two steps. First, a pair of overlapping PCR primers was used to amplify a StrepII and eight-His tag. The PCR product was digested with EcoRI and XbaI (New England Biolabs, Ipswich, MA) and inserted into a similarly digested pYES2/CT vector (Invitrogen). Second, the coding sequence of Pst1 from nucleotides 1 to 1194 (GenBank accession number NC_001136) (23) was PCR cloned from total S. cerevisiae cDNA and inserted into pYES2/CT downstream of the StrepII/His tag by digestion with BamHI and EcoRI, with the final construct confirmed by DNA sequencing. Diploid Δoch1 Δmnn1 Δmnn4 yeast, engineered as described previously (28), was transformed using the quick lithium acetate method (1). Pst1 expression was induced by transfer of a yeast culture from -ura/glucose (0.67% yeast nitrogen base without amino acids, 0.2% ura dropout mix, 2% glucose) to −uracil/galactose (with 2% galactose) at an optical density at 600 nm of 0.4 for 72 h. Pst1 protein was purified by preparative IEF as described above. The concentrations of purified Pst1 and gp120 glycoproteins were measured by the bicinchoninic acid protein assay (Pierce Biotechnology, Rockford, IL), using α1-acid glycoprotein as a standard.
For DC-SIGN, the entire extracellular domain of human DC-SIGN (GenBank accession number AF290886) (5) was PCR cloned into pET100/D-TOPO (Invitrogen), which contains an N-terminal six-His tag. Escherichia coli BL21(DE3) bacteria (Novagen, Madison, WI) were transformed with the vector, and protein expression was induced for 3 h with 1 mM isopropyl β-d-thiogalactopyranoside. DC-SIGN was purified using nickel-nitrilotriacetic acid-agarose resin (Qiagen, Venlo, The Netherlands) as previously described (41). Imidazole was removed from the eluate fractions, and 4 mM CaCl2 was added for purification on S. cerevisiae mannan-agarose (Sigma) preequilibrated with 30 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 4 mM CaCl2. DC-SIGN was eluted with 30 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 25 mM EDTA and dialyzed to replace EDTA with 4 mM CaCl2.
ELISA.
For enzyme-linked immunosorbent assays (ELISAs), Costar high-binding enzyme immunoassay 96-well plates (Corning Inc., Corning, NY) were used. All steps were performed at room temperature in a 100-μl working volume. Standard ELISAs were conducted as previously described (28), with some variations. Briefly, microwell plates were coated with 100 ng of Pst1 or gp120 protein per duplicate well overnight. The wells were blocked with blocking buffer (1× PBS, pH 7.2, 5% Carnation nonfat dried milk, 0.02% thimerosal) for 2 h. After the plates were rinsed, a serial dilution of 2G12 (starting at 4 μg/ml) in blocking buffer was incubated with the antigens for 2 h. 2G12 binding was detected using HRP-labeled goat anti-human IgG at 1:10,000 in blocking buffer for 1 h, followed by the addition of 3,3′5,5′-tetramethylbenzydine solution.
For 2G12 inhibition ELISAs, wells were coated with 1 μg/ml of gp120 from strain JR-FL of HIV-1 and blocked as described above. Twofold serial dilutions of the indicated inhibitors were titrated outside the plate in 1× PBS, 2.5% Carnation nonfat dried milk, 0.01% thimerosal and added to the wells in duplicate. MAb 2G12 diluted in blocking buffer was used to quickly overlay each well, for a final dilution of 0.5 μg/ml. The plates were incubated for 1 h with mild shaking. 2G12 binding was detected using HRP-labeled goat anti-human IgG followed by 3,3′5,5′-tetramethylbenzydine as described above. Endo H digestion of Pst1 for inhibition studies was conducted using native conditions and the manufacturer's recommended buffer (EMD Chemicals, San Diego, CA), and digestion was verified by SDS-PAGE.
DC-SIGN inhibition ELISA was conducted as previously described (41), with modifications. Wells were coated with 5 μg/ml of DC-SIGN in 30 mM Tris-HCl, pH 8.3, 30 mM NaHCO3, 3 mM CaCl2 overnight. The wells were blocked with DC-blocking buffer (30 mM Tris-HCl, pH 8.3, 3 mM CaCl2, 5% Carnation nonfat dried milk, 0.02% thimerosal) for 2 h. In a separate plate, twofold serial dilutions of Pst1, S. cerevisiae mannan, d-mannose, and bovine serum albumin (BSA) were titrated in duplicate using DC-blocking buffer, and JR-FL gp120 was added with the final concentration at 1 μg/ml. The gp120/inhibitor mixture was added over the DC-SIGN-coated plates and incubated for 1 h with mild shaking. gp120 binding was detected with anti-gp120, followed by goat anti-rabbit IgG-HRP, with thorough washes after each incubation with DC wash buffer (30 mM Tris-HCl, pH 8.3, 3 mM CaCl2, 0.05% Tween 20, 0.02% thimerosal).
Surface plasmon resonance (SPR) measurement.
Experiments to test Pst1 binding to 2G12 were performed on a BIAcore T100 instrument (BIAcore, Piscataway, NJ) at 25°C. Immobilization of 2G12 was conducted as previously described (25). Briefly, two flow cells with a CM5 chip (BIAcore) were activated by a mixture of N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide and N-hydroxysuccinimide. A reference cell was fully blocked with 1.0 M ethanolamine-HCl, pH 8.5, while a sample cell was injected with 20 μg/ml 2G12 in 10 mM sodium acetate, pH 5.0, until the number of response units (RU) reached the target level of 10,000 RU (actually immobilized, 9,257 RU) and then blocked by 1.0 M ethanolamine-HCl, pH 8.5. Pst1 and gp120 from strain Yu2 were tested on the 2G12-immobilized CM5 chip in serial twofold dilutions from 800 nM to 12.5 nM. In each cycle, the glycoprotein samples were injected over both flow cells at 30 μl/min for 120 s for binding association, and then dissociation was monitored for 300 s. For regeneration, 3.5 M MgCl2 was injected at 60 μl/min for 30 s after each cycle. The binding curves were obtained by subtraction of the reference cell curve from the 2G12 cell curve. The data were processed by BIAcore evaluation software following a steady-state fitting method, using the following equation: Y = Rmax × X/(KD + X), where Y is the binding response unit, Rmax is the maximal response unit, X is the sample concentration, and KD is the equilibrium constant. For measurement of the polypeptide KD, the concentration of Pst1 was adjusted by multiplying by a factor of 0.6 to account for the net polypeptide in the standard used to measure the glycoprotein concentration, α1-acid-glycoprotein.
Virus neutralization assays.
HIV-1 enveloped pseudoviruses were generated by cotransfection of 293T cells with pNL4-3.Luc.E−R− (Nathaniel Landau, Salk Institute for Biological Studies, La Jolla, CA) and the gp160 Env-expressing plasmids pcDNA3-SF162 and pSV7D-HxB. For the neutralization assay, 10,000 TZM-b1 cells (NIH AIDS Research and Reference Reagent Program; contributed by J. Kappes, X. Wu, and Tranzyme Inc.) were seeded in 96-well plates (Costar 96-well assay plate; Corning) in 100 μl of Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum and cultured overnight. The next day, in a separate plate, 50 μl of pseudovirus suspensions containing 2 × 105 relative light units was added to 25 μl of twofold serially diluted protein inhibitors (Pst1 or Endo H-digested Pst1) or DMEM (mock) in duplicate. Then, 25 μl of MAb 2G12 or 2F5 diluted in DMEM was added over the pseudovirus/inhibitor mixture to a final concentration of 0.2 μg/ml and 1.5 μg/ml, respectively, and incubated for 1 h at 37°C. After incubation, medium from the plated cells was replaced with 100 μl of the pseudovirus mixture, incubated for 3 h at 37°C in a CO2 incubator, and then replaced with 100 μl of fresh DMEM. After 72 h, the cells were lysed using the Bright-Glo Luciferase Assay System (Promega, Madison, WI), and luciferase activity was measured using a Veritas Microplate Luminometer (Turner Biosystems, Sunnyvale, CA). Neutralization activity was measured as the percent reduction in relative light units of sample wells compared to virus control wells (VC) after subtraction of background signal in cell control wells (CC) using the following formula: ([VC − sample]/[VC − CC]) × 100.
Cell surface DC-SIGN inhibition assay.
For DC-SIGN inhibition, 1 μg of JR-FL gp120 and the indicated amounts of protein inhibitor (mannan, BSA, or Pst1) were mixed together in 100 μl binding buffer (1× Tris-buffered saline, 2 mM CaCl2, 2.5% FBS, 0.05% sodium azide) and allowed to bind THP-1 cells stably expressing DC-SIGN (4, 18). After 1 h of incubation, the cells were washed twice in binding buffer. Then, 10 μg of anti-gp120 was added to the cells for 30 min on ice, followed by 10 μg of R-phycoerythrin-conjugated anti-rabbit (Invitrogen) for 30 min on ice in the dark. After being washed, the cells were analyzed immediately by fluorescence-activated cell sorting (FACS) on a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) machine. Data were analyzed using FlowJo 8.7.3 software (Tree Star Inc., Ashland, OR).
RESULTS
Discovery of an endogenous yeast glycoprotein that cross-reacts with 2G12.
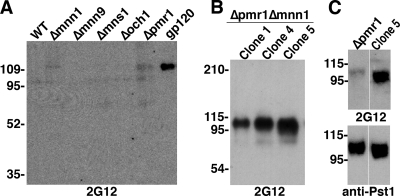
In an earlier study, we found that a triple-mutant (TM) yeast strain lacking the carbohydrate-processing enzymes Och1, Mnn1, and Mnn4 produced N-linked glycans that were strictly Man8GlcNAc2. This led to the discovery of at least four highly glycosylated yeast proteins that bound 2G12 (28). Immunization with whole TM yeast resulted in antibodies that cross-reacted with the glycans of gp120 but could not neutralize the virus. Given these promising results, we screened additional S. cerevisiae mutants with knockouts in genes known to affect normal yeast N glycosylation (Fig. 1), including genes important for cleaving core Man9GlcNAc2 to Man8GlcNAc2 in the endoplasmic reticulum (ER) (MNS1), hypermannosylation in the Golgi apparatus (MNN9 and OCH1), capping of Manα1,2-Man residues with terminal α1,3-linked Man (MNN1), and ion-dependent protein sorting and processing in the Golgi apparatus (PMR1) (13, 35). Cell lysates from yeast strains with individual deletions in these genes, along with wild-type (WT) S. cerevisiae, were separated by SDS-PAGE and probed with 2G12. One protein at ∼110 kDa in the cell lysate of the Δpmr1 yeast was weakly detected by 2G12 (Fig. 2A).
FIG. 1.
Critical proteins involved in S. cerevisiae N-linked glycosylation. In the ER, Mns1 cleaves mannose from core Man9GlcNAc2 on nascent polypeptides to form Man8GlcNAc2. In the Golgi apparatus, S. cerevisiae adds mannose residues to this oligosaccharide to form core-type and polymannose-type glycans with up to 200 mannoses per structure. Mnn1 adds terminal α1,3-linked mannose caps (open boxes), Och1 adds the initial α1,6-linked mannose of the polymannose side chain (dark-gray boxes), and Mnn9 elongates the α1,6-linked backbone (light-gray box). The deletion of PMR1 results in aberrant glycosylation with severely truncated polymannose side chains so that the N-glycans are similar to those found in the Δmnn9 mutant (35), like yeast core-type N glycosylation (curved arrow). Throughout processing in the Golgi apparatus, the ER-secreted Man8GlcNAc2 remains intact (trapezoidal boxes). Phosphomannose residues were omitted for simplicity.
FIG. 2.
2G12 detection of a yeast protein in Δpmr1-containing mutants. (A) Equal amounts of yeast cell lysates from single-mutant and WT yeast were screened for 2G12 binding by Western blotting, with 50 ng of 293-expressed ADA gp120 as a control. (B) Cell lysate samples from the three Δpmr1 Δmnn1 clones were tested for 2G12 binding by Western blotting. (C) Culture medium samples from Δpmr1 and Δpmr1 Δmnn1-clone 5 (Clone 5) were blotted with 2G12 (top) and an antibody specific to the unknown protein, later identified as Pst1 (bottom). The migration positions of molecular mass markers, in kilodaltons, are indicated to the left of the blots.
Binding of 2G12 to HIV-1 gp120 is dependent upon terminal α1,2-linked mannose residues, which are normally capped with α1,3-Man in yeast by Mnn1 (Fig. 1). Pmr1 is a Ca2+/Mn2+ P-type ATPase in the Golgi apparatus, and the Δpmr1 mutation is known to affect the function of Mn2+-dependent mannosyltransferases in the Golgi apparatus, like Mnn1, likely resulting in the partial exposure of 2G12-binding residues in this mutant (15, 35). To enhance 2G12 binding to the unknown protein detected in Δpmr1 yeast, we created Δpmr1 Δmnn1 mutants to ensure that potential α1,3-linked mannose residues are not added to the core Man8GlcNAc2 (data not shown). Three separate Δpmr1 Δmnn1 clones were selected, and proteins in the cell lysate were tested against 2G12. As expected, stronger signals were seen at ∼110 kDa in all three clones (Fig. 2B) than in the previous lysate blot with Δpmr1 cells. As further proof, a side-by-side comparison of Δpmr1 and Δpmr1 Δmnn1 culture media showed that 2G12 bound more strongly to the unknown protein in the Δpmr1 Δmnn1 strain than in the Δpmr1 mutant, despite similar amounts of protein in both samples (Fig. 2C), later identified as Pst1 (see below). Additionally, the migration of the 2G12-binding protein in the Δpmr1 Δmnn1 strain was slightly faster than in the single mutant, due to the loss of terminal α1,3-Man residues. Together, these results show that stronger 2G12 reactivity to the 110-kDa protein is likely due to the loss of α1,3-Man residues added by Mnn1 that cap the known binding site for 2G12, terminal Manα1,2-Man residues on the D1 and D3 arms of Man8GlcNAc2.
Identification of the yeast protein that cross-reacts with 2G12.
The unknown 2G12-binding protein from the culture supernatant of Δpmr1 Δmnn1 yeast was purified using preparative IEF, which demonstrated a high isoelectric point (pI), between 9 and 10 (data not shown). After IEF separation, the protein was identified by nano-LC/MS/MS as Pst1 (Fig. 3A), a GPI-anchored yeast cell wall protein important for cell wall integrity (32). The mature protein contains 400 amino acids, with the pI predicted to be 9.25, similar to the experimental pI, and the molecular mass predicted to be 41.3 kDa. The disparity between the predicted and experimental molecular masses in Δpmr1 Δmnn1 cells (∼110 kDa) is due to heavy glycosylation of the 15 potential N-linked glycosylation sites and numerous potential O-linked glycosylation sites (Fig. 3A). Further analysis using the HomoloGene database of NCBI revealed that out of the 18 completely sequenced eukaryotic genomes, Pst1 is specific to yeast, with homologues found only in Schizosaccharomyces pombe and Kluyveromyces lactis (data not shown).
FIG. 3.
Identification and verification of the yeast glycoprotein that cross-reacts with MAb 2G12. (A) The amino acid sequence of the glycoprotein Pst1, isolated from Δpmr1 Δmnn1 yeast and identified by nano-LC/MS/MS, is depicted. The detected peptides are underlined, with each being identified at least twice; the boldface letters at the N and C termini depict the signaling peptide and GPI anchor signal, respectively; and the highlighted letters are potential N-linked glycosylation sites. The amino acid sequence was obtained from GenBank under the accession number NP_010340 (23). (B) Pst1 was precipitated from Δpmr1 and Δpmr1 Δmnn1 culture media by 2G12 and detected by Western blotting with anti-Pst1. Starting material (S), flowthrough (F), wash (W), and eluate (E) samples are indicated.
To verify that Pst1 is indeed recognized by 2G12, the culture supernatant from Δpmr1 Δmnn1 yeast, as well as Δpmr1 yeast, was immunoprecipitated by 2G12 and detected by Western blotting with anti-Pst1 (Fig. 3B). A band of approximately 110 kDa was recognized in both clones, with the signal being stronger in the Δpmr1 Δmnn1 yeast, suggesting that 2G12 bound more protein. This result confirmed that the identity of the 2G12-binding protein found in both mutants is Pst1 and demonstrated that the glycoprotein can bind to 2G12 under native conditions. Additionally, we conducted two-dimensional separation of proteins in the culture supernatant from Δpmr1 Δmnn1 yeast, and both 2G12 and anti-Pst1 detected a protein spot at pH 9 to 10 and ∼110 kDa (data not shown). This further corroborates that Pst1 can cross-react with 2G12.
Analysis of the glycans on Pst1.
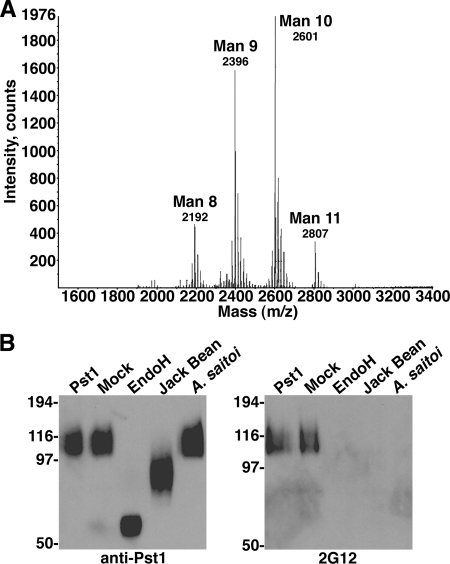
Due to stronger signals seen in 2G12 assays using Δpmr1 Δmnn1 than using Δpmr1 yeast, we analyzed the N-linked glycans released from Δpmr1 Δmnn1-expressed Pst1 (Δpmr1 Δmnn1-Pst1). MALDI-TOF MS showed that the predominant glycans were Man9-10GlcNAc2 (Fig. 4A). This result confirmed the loss of the hypermannosylated outer chain on N-glycans on Δpmr1 Δmnn1-Pst1 (Fig. 1), as WT-expressed Pst1 migrated to over 200 kDa (data not shown).
FIG. 4.
Analysis of Pst1 glycans. (A) MALDI-TOF mass spectrogram of N-linked glycans released from purified Δpmr1 Δmnn1-Pst1. (B) Δpmr1 Δmnn1-Pst1 digested with Endo H or with jack bean or A. saitoi mannosidase was analyzed by Western blotting using anti-Pst1 (left) or 2G12 (right), with mock-digested and untreated Pst1 used as controls.
To characterize the 2G12 epitope on Δpmr1 Δmnn1-Pst1, the purified protein was digested by a variety of glycosidases. Endo H cleaves high-mannose glycans at the β1,4 linkage connecting the two GlcNAc residues; jack bean mannosidase cleaves terminal α1,6-, α1,3-, and α1,2-Man residues; and A. saitoi mannosidase cleaves only terminal α1,2-Man residues (22, 36). Digestion of Δpmr1 Δmnn1-Pst1 with Endo H and with jack bean and A. saitoi mannosidases resulted in the expected relative masses as detected by anti-Pst1 (Fig. 4B, left). Probing these digested proteins with 2G12 showed that it could detect only undigested and mock-digested Pst1 (Fig. 4B, right). This result shows that 2G12 cross-reactivity to Δpmr1 Δmnn1-Pst1 is glycan dependent, with terminal Manα1,2-Man residues on high-mannose glycans being critical for binding, similar to previous results with gp120 (36, 37). The data also indicate that the vital Manα1,2-Man residues are exposed on the D1 and D3 arms of the high-mannose glycans released from Δpmr1 Δmnn1-Pst1, as terminal α1,3-Man would prevent digestion by A. saitoi mannosidase. Thus, the loss of hypermannosylation and two terminal α1,3-Man residues by deletion of PMR1 and MNN1, respectively, clearly increased 2G12 reactivity to Pst1. The Man9-10GlcNAc glycans on Δpmr1 Δmnn1-Pst1 likely represent a core structure of Man8GlcNAc2, with one or two mannose residues added as a defunct side chain, similar to yeast core-type glycans (illustrated in Fig. 1, top right) without terminal α1,3-Man added by Mnn1.
Improvement of MAb 2G12 cross-reactivity to Pst1.
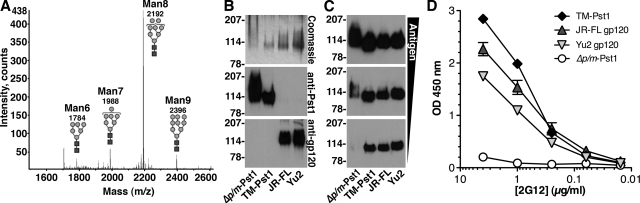
While 2G12 clearly recognized Δpmr1 Δmnn1-Pst1, the dominant glycans produced in this mutant, core Man8GlcNAc2 with one or two mannose residues added on a defunct side chain, are structurally different from the core Man9GlcNAc2 found on HIV-1 gp120 and may not be optimal for 2G12 binding. Recently, we created an S. cerevisiae Δoch1 Δmnn1 Δmnn4 strain (called TM yeast) that produced proteins with strictly core Man8GlcNAc2 glycans and induced mannose-specific antibodies in rabbits that bound gp120 (28). Considering that Man8GlcNAc2 is the preferred substrate for 2G12 (11, 37), this strain should express Pst1 with the ideal glycans for 2G12 binding. Unfortunately, endogenous Pst1 expression could not be detected in TM yeast (data not shown). Therefore, we cloned the Pst1 gene into the pYES2/CT yeast expression vector and transformed TM yeast. Pst1 expression was induced with galactose via the GAL1 promoter and purified by preparative IEF as described above. MALDI-TOF analysis of N-linked glycans from TM-expressed Pst1 (TM-Pst1) showed that Man8GlcNAc2 was the predominant glycan, verifying the production of Man8GlcNAc2 glycans on exogenous proteins expressed in TM yeast (Fig. 5A). SDS-PAGE staining and Western analysis showed that TM-Pst1 migrated noticeably faster than Δpmr1 Δmnn1-Pst1, representing the change in glycan structure from Man9-10GlcNAc2 on Δpmr1 Δmnn1-Pst1 to Man8GlcNAc2 on TM-Pst1 (Fig. 5B). Additionally, TM-Pst1 showed greater homogeneity in SDS-PAGE than Δpmr1 Δmnn1-Pst1, suggesting a more uniform glycan structure on the former.
FIG. 5.
Analysis of 2G12 reactivity between Pst1 and gp120. (A) MALDI-TOF mass spectrogram of N-linked glycans released from purified TM-Pst1. (B) A comparison of purified Δpmr1 Δmnn1-Pst1, TM-Pst1, JR-FL gp120, and Yu2 gp120 is shown with 500 ng of each protein stained by Coomassie blue (top) or 250 ng detected by Western blotting with anti-Pst1 (middle) and anti-gp120 (bottom). (C) Western blot comparing 2G12 (1 μg/ml) binding to purified Δpmr1 Δmnn1-Pst1, TM-Pst1, JR-FL gp120, and Yu2 gp120 loaded at 1 μg (top), 0.5 μg (middle), and 0.25 μg (bottom). (D) Comparison of 2G12 binding to 100 ng of purified Δpmr1 Δmnn1-Pst1 (Δp/m-Pst1), TM-Pst1, JR-FL gp120, and Yu2 gp120 proteins by ELISA. The values are the averages ± standard errors of the means (error bars) of three independent experiments. The gp120 proteins were produced in 293T cells. OD, optical density.
To examine the efficiency with which 2G12 detected purified Δpmr1 Δmnn1- and TM-Pst1, we loaded equivalent amounts of both, along with purified mammalian-cell-expressed JR-FL and Yu2 gp120, and detected the proteins by Western blotting with 2G12. As the amount of each protein was reduced, the staining intensity of Δpmr1 Δmnn1-Pst1 decreased more substantially than that of the other proteins, so that 2G12 signal was absent at 0.25 μg for Δpmr1 Δmnn1-Pst1 (Fig. 5C). In addition, we conducted an ELISA with equivalent amounts of TM-Pst1, Δpmr1 Δmnn1-Pst1, and JR-FL and Yu2 gp120 captured to wells and detected with serial dilutions of 2G12. We found that 2G12 bound only weakly to Δpmr1 Δmnn1-Pst1 at the highest 2G12 concentration tested (4 μg/ml) (Fig. 5D). In contrast, 2G12 strongly detected TM-Pst1, with reactivity comparable to that of the two gp120 proteins. These results show that TM-Pst1 has stronger binding to 2G12 than Δpmr1 Δmnn1-Pst1, especially under the nondenaturing conditions of ELISA, likely due to the presence of homogenous Man8GlcNAc2 glycans on TM-Pst1.
Analysis of Pst1 affinity for 2G12.
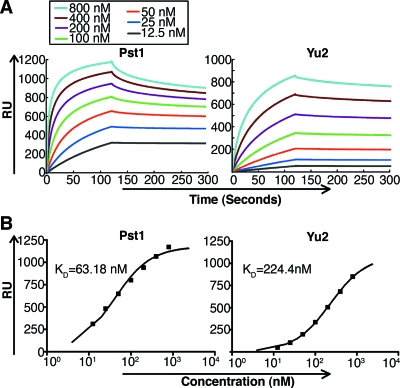
TM-Pst1 was characterized by using SPR to measure its binding affinity for immobilized 2G12. Concentration-dependent binding showed that TM-Pst1 bound quickly and strongly to 2G12, with rapid association especially at high concentrations (more than 200 nM), to reach a steady state of binding (Fig. 6A, left). The dissociation rate was extremely low, so that the bound TM-Pst1 required a high concentration of regeneration reagent (3.5 M MgCl2) to be released from 2G12, perhaps due to the multiple high-mannose glycans on the surface of TM-Pst1. Real-time binding curves did not fit a simple 1:1 Langmuir model, indicating rather complex kinetics that do not provide precise kon (association rate constant) and koff (dissociation rate constant) values. Nonetheless, steady-state fitting models show that TM-Pst1 has a KD of 63 nM (Fig. 6B, left). This KD value was calculated for the whole protein (polypeptide and glycans) using the apparent molecular mass seen by SDS-PAGE (Fig. 5C). Due to the heterogeneous nature of glycoproteins, it is difficult to determine the true molecular mass. For reference, we also calculated the KD based on a calibrated concentration of the polypeptide portion of mature Pst1, obtaining a KD of approximately 56 nM, similar to that calculated with the whole protein. For comparison, we also measured the affinity of Yu2 gp120 under the same concentrations and conditions as Pst1 (Fig. 6A, right). The same steady-state fitting equation calculated a KD of 224 nM for Yu2 (Fig. 6B, right). Together, these data show that TM-Pst1 bound to 2G12 more quickly and with a lower KD value than did Yu2 gp120 produced in human 293T cells, perhaps due to a large number (up to 15) of homogenous Man8GlcNAc2 glycans on the TM-Pst1 protein. This is in contrast to a mixture of complex-, hybrid-, and high-mannose-type glycans on the surface of gp120.
FIG. 6.
TM-Pst1 binding to 2G12 by SPR. (A) Real-time SPR RU of purified TM-Pst1 (left) and Yu2 gp120 (right) from 12.5 nM to 800 nM on a 2G12-immobilized sensor chip. (B) Steady-state fitting of TM-Pst1 (left) and Yu2 gp120 (right) with the indicated KD values.
Pst1 inhibition of MAb 2G12 binding to gp120.
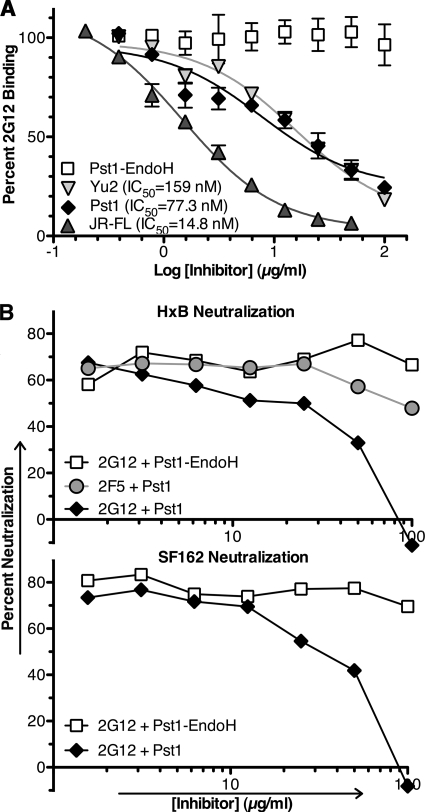
We next evaluated TM-Pst1 for the ability to competitively inhibit 2G12 binding to immobilized JR-FL gp120 in ELISA. Immobilized JR-FL gp120 was detected with 2G12 in the presence of decreasing concentrations of TM-Pst1, Endo H-digested TM-Pst1, Yu2 gp120, and JR-FL gp120. In this format, TM-Pst1 inhibited binding of 2G12 to JR-FL gp120 with a molar 50% inhibitory concentration (IC50) of 77 nM (Fig. 7A). This inhibition was specifically attributed to the Man8GlcNAc2 glycans found on TM-Pst1, as Endo H-digested Pst1 showed no inhibition of 2G12 interaction with gp120. Based upon molar IC50s, TM-Pst1 showed twofold-stronger inhibition of 2G12 binding to JR-FL gp120 than did gp120 from a heterologous strain (Yu2), while the homologous gp120 was fivefold better than TM-Pst1 (Fig. 7A). As a reference, we calculated a molar IC50 for d-mannose at 143 mM (data not shown). To our knowledge, this calculated molar IC50 for TM-Pst1 represents the lowest value measured for inhibition of 2G12 binding to immobilized gp120 by a glycoantigen to date.
FIG. 7.
TM-Pst1 inhibition of 2G12-gp120 interaction. (A) The percent 2G12 binding to 100 ng of immobilized JR-FL gp120 was analyzed by ELISA in the presence of TM-Pst1, Pst1-Endo H, Yu2 gp120, and JR-FL gp120. There was no preincubation of inhibitors with 2G12, and 100% represents 2G12 binding in the absence of inhibitor. The values are the averages ± standard errors of the means (error bars) of three independent experiments. Molar IC50s were calculated by nonlinear regression using GraphPad Prism 5.0, using the apparent molecular masses of TM-Pst1, Yu2, and JR-FL gp120 at 100 kDa (Fig. 5B). (B) (Top) Neutralization activity of MAb 2G12 against HxB-pseudotyped virus in the presence of Pst1-Endo H and TM-Pst1, with MAb 2F5 as a control. The results are representative of three separate experiments. (Bottom) Neutralization activity of 2G12 assessed against SF162-pseudotyped virus in the presence of Pst1-Endo H and TM-Pst1. The results are representative of two separate experiments.
We also evaluated TM-Pst1 for its ability to block 2G12 neutralization of HIV-1 pseudoviruses. To do this, we incubated an IC80 of 2G12 with virus pseudotypes bearing either the HxB or the SF162 Env protein in the presence of various concentrations of TM-Pst1 or Endo H-digested Pst1. Under these conditions, increasing concentrations of TM-Pst1 decreased 2G12-mediated neutralization of both viruses, with complete inhibition achieved by 100 μg/ml (representing 1 μM TM-Pst1) (Fig. 7B). In contrast, Endo H-digested-Pst1 did not inhibit 2G12 neutralization of either strain, again indicating that the 2G12-Pst1 interaction is glycan dependent. Additionally, Pst1 did not inhibit the ability of the gp41-specific MAb 2F5 to neutralize HxB pseudoviruses.
Pst1 inhibition of gp120 interactions with DC-SIGN.
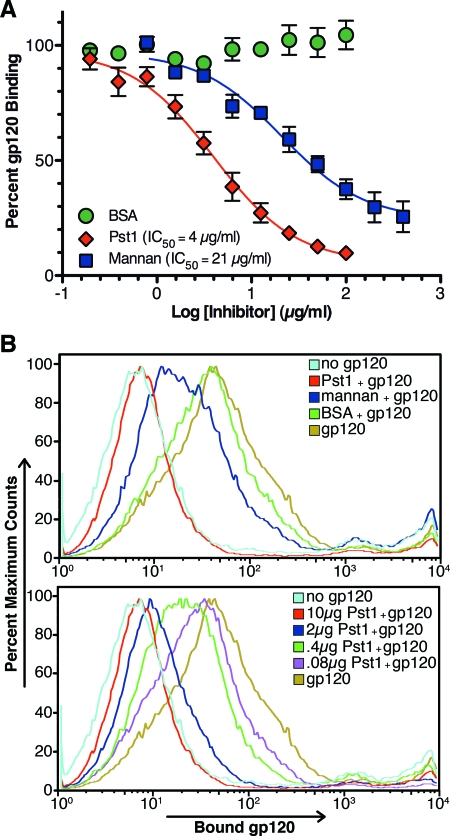
DC-SIGN binds HIV-1 gp120 through high-mannose glycans (20, 29). Therefore, TM-Pst1 was tested for inhibition of DC-SIGN binding to gp120. Competition ELISA was conducted to test solid-phase DC-SIGN binding to gp120 in the presence of inhibitors, as this format showed relatively strong and moderate inhibition of DC-SIGN-gp120 interaction by mannan and 2G12, respectively, without background inhibition by the V3 loop MAb 447-52D (6). Using this format, TM-Pst1 inhibited gp120 binding to recombinant monomeric DC-SIGN with an IC50 of 4 μg/ml (molar IC50 = 40 nM), while mannan had an IC50 of 21 μg/ml (due to the extreme heterogeneity of mannan, the molar IC50 was not calculated) (Fig. 8A). In contrast, d-mannose had a molar IC50 of 34.2 mM (data not shown), and BSA showed no inhibition of gp120 binding to DC-SIGN (Fig. 8A).
FIG. 8.
TM-Pst1 inhibition of DC-SIGN-gp120 interaction. (A) Graded concentrations of TM-Pst1, mannan, and BSA were evaluated for direct inhibition of soluble JR-FL gp120 binding to immobilized DC-SIGN by ELISA. The results are shown relative to 100% gp120 binding, calculated in the absence of inhibitor. The values are the averages ± standard errors of the means (error bars) of three independent experiments for Pst1 and mannan and two for BSA. IC50s were calculated by nonlinear regression using GraphPad Prism 5. (B) (Top) Inhibition of JRFL gp120 (1 μg) binding to THP-DC-SIGN by 10 μg of mannan, BSA, or TM-Pst1. (Bottom) Dose-dependent inhibition of JR-FL gp120 (1 μg) binding to THP-DC-SIGN cells by TM-Pst1. As controls, cells were incubated with only antibodies (no gp120) or gp120 without inhibitors (gp120). The results are representative of two independent experiments.
TM-Pst1 was also investigated for inhibition of gp120 binding to native DC-SIGN tetramers on the surfaces of mammalian cells. THP-1 cells stably transfected with DC-SIGN (THP-DC-SIGN) were incubated with JR-FL gp120 in the presence of TM-Pst1, mannan, or BSA. After the cells were washed, the amount of gp120 bound to DC-SIGN was measured by FACS analysis. When 10 μg/ml of each inhibitor was tested, TM-Pst1 showed almost complete inhibition of gp120 binding to THP-DC-SIGN cells, while mannan showed moderate inhibition and BSA was ineffective (Fig. 8B, top). Dose-dependent inhibition of gp120 binding to THP-DC-SIGN cells by TM-Pst1 was also shown (Fig. 8B, bottom), with approximately 50% inhibition occurring at 4 μg/ml (40 nM), a result consistent with the IC50 calculated in the DC-SIGN competition ELISA. In contrast, mannan could not obtain 50% inhibition even at the highest concentration tested, 100 μg/ml (data not shown). Altogether, the ability of TM-Pst1 to prevent gp120 interaction with solid-phase DC-SIGN and THP-DC-SIGN cells highlights this protein as an interesting immunogen to potentially elicit antibodies against mannose clusters on gp120 that can block the capture of intact virus by DCs at mucosal surfaces.
DISCUSSION
The 2G12 epitope consists of several closely spaced, high-mannose carbohydrate structures with D1 and D3 arms presenting clusters of terminally exposed Manα1,2-Man moieties (11, 36, 37, 42). Several features of the 2G12 epitope, and the glycan shield in general, make it a viable target for vaccine design. First, since high-mannose glycans are rare and/or sparsely distributed on mammalian glycoproteins, antibodies such as 2G12 should exhibit limited autoreactivity (7, 24). Second, glycans are surface exposed on the mature Env trimer (49) and not recessed like the CD4 binding site or transiently available like CD4-induced epitopes (8, 9). Third, high-mannose glycans form clusters on gp120, the majority of which are found on the most conserved glycosylation sites (26, 50), suggesting a need to maintain these glycans to protect highly neutralizing peptide epitopes (21, 46). Finally, the large number of high-mannose glycans on gp120 raises the possibility that clusters of mannose glycans other than the 2G12 epitope may be viable antibody targets (26, 30).
Since various immunization strategies using HIV-1 Env have failed to elicit 2G12-like antibodies, attempts have been made to produce the 2G12 epitope in different contexts. Synthetic glycoantigens, often created using chemical linkers to conjugate synthetic or enzyme-cleaved high-mannose glycans to molecular scaffolds, have consistently supported 2G12 binding and shown that multivalency of high-mannose glycans can markedly improve 2G12 affinity (2, 25, 27, 43-45). However, these glycoantigens have not yet approached the affinity seen in 2G12 binding to gp120, usually showing IC50s in the micromolar to 200 nM range for blocking 2G12 binding to immobilized gp120, while the homologous gp120 shows an IC50 in the low nanomolar range (2, 27, 44, 45). Additionally, these glycoantigens have not, to the best of our knowledge, elicited antibodies capable of cross-reacting with gp120.
An alternative approach is to recapitulate the 2G12 epitope using natural high-mannose glycans on heterologous glycoproteins. We previously found that when S. cerevisiae was modified to produce Man8GlcNAc2 due to deletion of specific carbohydrate-processing genes, several highly glycosylated yeast cell wall proteins supported 2G12 binding (28). Similarly, treatment of human 293T cells with kifunensine, an alpha-mannosidase inhibitor, resulted in binding of 2G12 to the human protein CEACAM1 (38). By manipulating the glycosylation machinery of S. cerevisiae and subsequently identifying the yeast proteins that support 2G12 binding, we have begun to characterize the properties needed for this genetic-scaffold approach. Pst1, like the other four yeast proteins we have identified, is very heavily glycosylated and presents closely arrayed N-linked glycans, particularly in the area from amino acids 190 to 310 (Fig. 3). In total, there are 15 potential N-linked glycosylation sites in the 400 amino acids of the mature protein, giving a calculated N-linked site density of 3.8%, which is only slightly less than that found on HIV-1 Env. Such a high density of N-linked glycosylation sites is a distinguishing feature of yeast cell wall proteins, as analysis of the S. cerevisiae genome reveals numerous proteins with densities of greater than 3.0% (data not shown), providing a large number of potential scaffolds for 2G12 binding. In fact, it was recently discovered that the pathogenic yeasts Candida albicans and Candida tropicalis can bind to 2G12 with high affinity (14). A similar analysis of the human genome revealed that such glycan densities are rare, reemphasizing the limited autoreactivity expected in targeting dense clusters of high-mannose glycans.
The density of N-linked sites alone does not account for 2G12 binding to Pst1, as the structure of the high-mannose glycans has a strong effect on 2G12 binding. By taking advantage of the power and ease of genetic manipulation of yeast, we were able to analyze 2G12 binding to Pst1 containing a variety of high-mannose structures. In Δmnn1 yeast, 2G12 did not bind Pst1 or any other protein, showing that polymannose-type glycans containing numerous exposed Manα1,2-Man residues on both the core Man8GlcNAc2 and the extended side chain could not cross-react with 2G12. This is consistent with the idea that an optimal spatial configuration of high-mannose glycans is required for the 2G12 epitope, since the large extended side chains might keep core Man8GlcNAc2 structures too far apart to support 2G12 binding, while the tightly packed Manα1,2-Manα1,2-Man trisaccharides on the side chain (structures similar to the D1 arm of Man8GlcNAc2) might be too close. In Δpmr1 yeast, Pst1 showed weak 2G12 binding. We reasoned that the Δpmr1 mutation's effect on the normal balance of Mn2+ in the Golgi apparatus inhibited the function of the manganese-dependent Mnn1 to some degree, leading to the exposure of some Manα1,2-Man residues needed for 2G12 reactivity (15, 35). In Δpmr1 Δmnn1 yeast, the reactivity of 2G12 to Pst1 was markedly improved, showing that the loss of both hypermannosylation and terminal Manα1,3-Man residues were sufficient for consistent 2G12 binding. Indeed, Δpmr1 Δmnn1-Pst1 showed properties similar to those of the 2G12 epitope on gp120, since enzymatic removal of all high-mannose glycans and specific removal of terminal α1,2-Man destroyed reactivity. However, 2G12 bound with even higher affinity to Pst1 produced in TM yeast, which expressed Pst1 with mostly Man8GlcNAc2. The only difference between Δpmr1 Δmnn1- and TM-Pst1 is that the majority of high-mannose glycans in Δpmr1 Δmnn1-Pst1 are structurally distinct from those found on gp120, such that they represent Man8GlcNAc2 with one or two mannose residues added as a defunct side chain. Thus, the difference in 2G12 reactivity between the two proteins can most likely be attributed to the interference of the additional one or two side chain mannose residues in optimal clustering of the core Man8GlcNAc2 glycans on Δpmr1 Δmnn1-Pst1.
Pst1 protein containing mostly natural Man8GlcNAc2 showed high affinity for 2G12 and a low molar IC50 to inhibit gp120-2G12 interactions. Several factors could be involved in the high-affinity binding of 2G12 to the TM-Pst1 glycoprotein. First, the large number and high density of Man8GlcNAc2 glycans on the protein may be crucial, as it was previously shown that increasing the number and density of synthetic Man9 or Man4 on oligodendrons directly correlated with an increase in 2G12 binding (45). Second, the Pst1 polypeptide contains several areas where three or more neighboring N glycans could potentially form a cluster able to bind 2G12, allowing the possibility of more than one 2G12 epitope, similar to gp120 expressed with homogenous core Man9GlcNAc2 in 293T cells with kifunensine (38). Last, unlike synthetic constructs, the Man8GlcNAc2 structures of TM-Pst1 are biologically linked to the protein through two GlcNAc residues. Naturally linked core Man9GlcNAc2 and Man8GlcNAc2 likely limit the conformations of glycan clusters in relation to the polypeptide backbone due to the rigid nature of the GlcNAc residues and a relative conservation of topology (47, 48), allowing 2G12 to bind at a higher affinity.
To summarize, Pst1 produced in TM yeast bound 2G12 with a KD of approximately 63 nM. Consistent with this high affinity, TM-Pst1 was able to inhibit 2G12 binding to immobilized gp120 and did so more efficiently than a heterologous gp120 from the same subtype. Additionally, purified TM-Pst1 inhibited 2G12 neutralization of HIV-1 pseudoviruses, highlighting the protein's ability to block 2G12 binding to functional, trimeric Env protein. Finally, this protein strongly inhibited the interaction between gp120 and solid-phase DC-SIGN and between gp120 and cell surface DC-SIGN, as demonstrated by ELISA and FACS, respectively. Together, the high binding affinity of TM-Pst1 for 2G12 and DC-SIGN is fairly consistent between these experiments, as the calculated molar IC50 and KD values were all in the 40 to 80 nM range. These results suggest that targeting high-mannose structures in vaccine design may elicit antibodies that provide a dual function: neutralization of HIV by blocking gp120 binding to its receptors and prevention of HIV interaction with DCs via C-type lectins like DC-SIGN.
Altogether, we have discovered and analyzed a heterologous yeast protein capable of supporting strong 2G12 binding and inhibition of gp120 interaction with DC-SIGN. While we have successfully produced a protein containing large numbers of Man8GlcNAc2 glycans, the manner in which they are clustered on the protein's surface may not be optimal. By identifying the positions of the N-glycans on TM-Pst1 to which 2G12 binds, it may be possible to further enhance binding via a mutagenesis approach. Additionally, it will be important to perform immunization studies with purified TM-Pst1. Our previous immunizations with whole TM yeast (which did not express detectable levels of Pst1) elicited anti-mannose antibodies that cross-reacted with gp120, although they did not neutralize the virus (28). We detected these antibodies despite the extremely large number of other immunogenic epitopes exposed on the surfaces of whole yeast cells. By utilizing an iterative process in which 2G12-binding yeast proteins, such as Pst1, are identified, purified, used as immunogens, and modified to improve immunogenicity, we hope to induce mannose-specific antibodies capable of neutralizing HIV-1. This approach has potential for discovering a cheap and easily produced antigen to target the HIV-1 glycan shield.
Acknowledgments
We thank Regina Guo and Bingfen Liu for their excellent technical help and Natasha Naidu (University of California, San Diego) for her glycan analysis.
This work was supported by National Institutes of Health grants AI058724 (to Y.G.) and AI067111 (to L.-X.W.). Work in R. W. Doms' laboratory was supported by the International AIDS Vaccine Initiative's Neutralizing Antibody Consortium and by NIH T32 AI007632 to C.A.-G.
Yu Geng, president and chief executive officer of ProSci Inc., acknowledges a potential financial conflict of interest.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Amberg, D., D. Burke, and J. Strathern. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 2.Astronomo, R. D., H. K. Lee, C. N. Scanlan, R. Pantophlet, C. Y. Huang, I. A. Wilson, O. Blixt, R. A. Dwek, C. H. Wong, and D. R. Burton. 2008. A glycoconjugate antigen based on the recognition motif of a broadly neutralizing human immunodeficiency virus antibody, 2G12, is immunogenic but elicits antibodies unable to bind to the self glycans of gp120. J. Virol. 826359-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballou, C. E. 1990. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 185440-470. [DOI] [PubMed] [Google Scholar]
- 4.Baribaud, F., S. Pohlmann, G. Leslie, F. Mortari, and R. W. Doms. 2002. Quantitative expression and virus transmission analysis of DC-SIGN on monocyte-derived dendritic cells. J. Virol. 769135-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashirova, A. A., T. B. Geijtenbeek, G. C. van Duijnhoven, S. J. van Vliet, J. B. Eilering, M. P. Martin, L. Wu, T. D. Martin, N. Viebig, P. A. Knolle, V. N. KewalRamani, Y. van Kooyk, and M. Carrington. 2001. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 193671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley, J. M., S. Ngo-Abdalla, P. Moore, M. Bobardt, U. Chatterji, P. Gallay, D. R. Burton, I. A. Wilson, J. H. Elder, and A. de Parseval. 2006. Inhibition of HIV Env binding to cellular receptors by monoclonal antibody 2G12 as probed by Fc-tagged gp120. Retrovirology 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braibant, M., S. Brunet, D. Costagliola, C. Rouzioux, H. Agut, H. Katinger, B. Autran, and F. Barin. 2006. Antibodies to conserved epitopes of the HIV-1 envelope in sera from long-term non-progressors: prevalence and association with neutralizing activity. AIDS 201923-1930. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5233-236. [DOI] [PubMed] [Google Scholar]
- 9.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA 10214943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calarese, D. A., H. K. Lee, C. Y. Huang, M. D. Best, R. D. Astronomo, R. L. Stanfield, H. Katinger, D. R. Burton, C. H. Wong, and I. A. Wilson. 2005. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. USA 10213372-13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 3002065-2071. [DOI] [PubMed] [Google Scholar]
- 12.Chiba, H., J. Inokoshi, H. Nakashima, S. Omura, and H. Tanaka. 2004. Actinohivin, a novel anti-human immunodeficiency virus protein from an actinomycete, inhibits viral entry to cells by binding high-mannose type sugar chains of gp120. Biochem. Biophys. Res. Commun. 316203-210. [DOI] [PubMed] [Google Scholar]
- 13.Dean, N. 1999. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426309-322. [DOI] [PubMed] [Google Scholar]
- 14.Dunlop, D. C., A. Ulrich, B. J. Appelmelk, D. R. Burton, R. A. Dwek, N. Zitzmann, and C. N. Scanlan. 2008. Antigenic mimicry of the HIV envelope by AIDS-associated pathogens. AIDS 222214-2217. [DOI] [PubMed] [Google Scholar]
- 15.Durr, G., J. Strayle, R. Plemper, S. Elbs, S. K. Klee, P. Catty, D. H. Wolf, and H. K. Rudolph. 1998. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol. Biol. Cell 91149-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esser, M. T., T. Mori, I. Mondor, Q. J. Sattentau, B. Dey, E. A. Berger, M. R. Boyd, and J. D. Lifson. 1999. Cyanovirin-N binds to gp120 to interfere with CD4-dependent human immunodeficiency virus type 1 virion binding, fusion, and infectivity but does not affect the CD4 binding site on gp120 or soluble CD4-induced conformational changes in gp120. J. Virol. 734360-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezekowitz, R. A., M. Kuhlman, J. E. Groopman, and R. A. Byrn. 1989. A human serum mannose-binding protein inhibits in vitro infection by the human immunodeficiency virus. J. Exp. Med. 169185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100587-597. [DOI] [PubMed] [Google Scholar]
- 19.Gurney, K. B., J. Elliott, H. Nassanian, C. Song, E. Soilleux, I. McGowan, P. A. Anton, and B. Lee. 2005. Binding and transfer of human immunodeficiency virus by DC-SIGN+ cells in human rectal mucosa. J. Virol. 795762-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong, P. W., K. B. Flummerfelt, A. de Parseval, K. Gurney, J. H. Elder, and B. Lee. 2002. Human immunodeficiency virus envelope (gp120) binding to DC-SIGN and primary dendritic cells is carbohydrate dependent but does not involve 2G12 or cyanovirin binding sites: implications for structural analyses of gp120-DC-SIGN binding. J. Virol. 7612855-12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, Q., N. Mahmood, and R. J. Shattock. 2007. High-mannose-specific deglycosylation of HIV-1 gp120 induced by resistance to cyanovirin-N and the impact on antibody neutralization. Virology 368145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichishima, E., M. Arai, Y. Shigematsu, H. Kumagai, and R. Sumida-Tanaka. 1981. Purification of an acidic alpha-d-mannosidase from Aspergillus saitoi and specific cleavage of 1,2-alpha-d-mannosidic linkage in yeast mannan. Biochim. Biophys. Acta 65845-53. [DOI] [PubMed] [Google Scholar]
- 23.Jacq, C., J. Alt-Morbe, B. Andre, W. Arnold, A. Bahr, J. P. Ballesta, M. Bargues, L. Baron, A. Becker, N. Biteau, H. Blocker, C. Blugeon, J. Boskovic, P. Brandt, M. Bruckner, M. J. Buitrago, F. Coster, T. Delaveau, F. del Rey, B. Dujon, L. G. Eide, J. M. Garcia-Cantalejo, A. Goffeau, A. Gomez-Peris, P. Zaccaria, et al. 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome IV. Nature 38775-78. [PubMed] [Google Scholar]
- 24.Joos, B., A. Trkola, H. Kuster, L. Aceto, M. Fischer, G. Stiegler, C. Armbruster, B. Vcelar, H. Katinger, and H. F. Gunthard. 2006. Long-term multiple-dose pharmacokinetics of human monoclonal antibodies (MAbs) against human immunodeficiency virus type 1 envelope gp120 (MAb 2G12) and gp41 (MAbs 4E10 and 2F5). Antimicrob. Agents Chemother. 501773-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krauss, I. J., J. G. Joyce, A. C. Finnefrock, H. C. Song, V. Y. Dudkin, X. Geng, J. D. Warren, M. Chastain, J. W. Shiver, and S. J. Danishefsky. 2007. Fully synthetic carbohydrate HIV antigens designed on the logic of the 2G12 antibody. J. Am. Chem. Soc. 12911042-11044. [DOI] [PubMed] [Google Scholar]
- 26.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 26510373-10382. [PubMed] [Google Scholar]
- 27.Li, H., and L. X. Wang. 2004. Design and synthesis of a template-assembled oligomannose cluster as an epitope mimic for human HIV-neutralizing antibody 2G12. Org. Biomol. Chem. 2483-488. [DOI] [PubMed] [Google Scholar]
- 28.Luallen, R. J., J. Lin, H. Fu, K. K. Cai, C. Agrawal, I. Mboudjeka, F. H. Lee, D. Montefiori, D. F. Smith, R. W. Doms, and Y. Geng. 2008. An engineered Saccharomyces cerevisiae strain binds the broadly neutralizing human immunodeficiency virus type 1 antibody 2G12 and elicits mannose-specific gp120-binding antibodies. J. Virol. 826447-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell, D. A., A. J. Fadden, and K. Drickamer. 2001. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 27628939-28945. [DOI] [PubMed] [Google Scholar]
- 30.Mizuochi, T., M. W. Spellman, M. Larkin, J. Solomon, L. J. Basa, and T. Feizi. 1988. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochem. J. 254599-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakanishi-Shindo, Y., K. Nakayama, A. Tanaka, Y. Toda, and Y. Jigami. 1993. Structure of the N-linked oligosaccharides that show the complete loss of alpha-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 26826338-26345. [PubMed] [Google Scholar]
- 32.Pardo, M., L. Monteoliva, P. Vazquez, R. Martinez, G. Molero, C. Nombela, and C. Gil. 2004. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 1504157-4170. [DOI] [PubMed] [Google Scholar]
- 33.Pohlmann, S., F. Baribaud, B. Lee, G. J. Leslie, M. D. Sanchez, K. Hiebenthal-Millow, J. Munch, F. Kirchhoff, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 754664-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pohlmann, S., E. J. Soilleux, F. Baribaud, G. J. Leslie, L. S. Morris, J. Trowsdale, B. Lee, N. Coleman, and R. W. Doms. 2001. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. USA 982670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudolph, H. K., A. Antebi, G. R. Fink, C. M. Buckley, T. E. Dorman, J. LeVitre, L. S. Davidow, J. I. Mao, and D. T. Moir. 1989. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell 58133-145. [DOI] [PubMed] [Google Scholar]
- 36.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 767293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J. Virol. 767306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scanlan, C. N., G. E. Ritchie, K. Baruah, M. Crispin, D. J. Harvey, B. B. Singer, L. Lucka, M. R. Wormald, P. Wentworth, Jr., N. Zitzmann, P. M. Rudd, D. R. Burton, and R. A. Dwek. 2007. Inhibition of mammalian glycan biosynthesis produces non-self antigens for a broadly neutralising, HIV-1 specific antibody. J. Mol. Biol. 37216-22. [DOI] [PubMed] [Google Scholar]
- 39.Selvarajah, S., B. Puffer, R. Pantophlet, M. Law, R. W. Doms, and D. R. Burton. 2005. Comparing antigenicity and immunogenicity of engineered gp120. J. Virol. 7912148-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan, M., P. J. Klasse, K. Banerjee, A. K. Dey, S. P. Iyer, R. Dionisio, D. Charles, L. Campbell-Gardener, W. C. Olson, R. W. Sanders, and J. P. Moore. 2007. HIV-1 gp120 mannoses induce immunosuppressive responses from dendritic cells. PLoS Pathog. 3e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su, S. V., P. Hong, S. Baik, O. A. Negrete, K. B. Gurney, and B. Lee. 2004. DC-SIGN binds to HIV-1 glycoprotein 120 in a distinct but overlapping fashion compared with ICAM-2 and ICAM-3. J. Biol. Chem. 27919122-19132. [DOI] [PubMed] [Google Scholar]
- 42.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 701100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, J., H. Li, G. Zou, and L. X. Wang. 2007. Novel template-assembled oligosaccharide clusters as epitope mimics for HIV-neutralizing antibody 2G12. Design, synthesis, and antibody binding study. Org. Biomol. Chem. 51529-1540. [DOI] [PubMed] [Google Scholar]
- 44.Wang, L. X., J. Ni, S. Singh, and H. Li. 2004. Binding of high-mannose-type oligosaccharides and synthetic oligomannose clusters to human antibody 2G12: implications for HIV-1 vaccine design. Chem. Biol. 11127-134. [DOI] [PubMed] [Google Scholar]
- 45.Wang, S. K., P. H. Liang, R. D. Astronomo, T. L. Hsu, S. L. Hsieh, D. R. Burton, and C. H. Wong. 2008. Targeting the carbohydrates on HIV-1: interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc. Natl. Acad. Sci. USA 1053690-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 47.Woods, R. J., C. J. Edge, and R. A. Dwek. 1994. Protein surface oligosaccharides and protein function. Nat. Struct. Biol. 1499-501. [DOI] [PubMed] [Google Scholar]
- 48.Woods, R. J., A. Pathiaseril, M. R. Wormald, C. J. Edge, and R. A. Dwek. 1998. The high degree of internal flexibility observed for an oligomannose oligosaccharide does not alter the overall topology of the molecule. Eur. J. Biochem. 258372-386. [DOI] [PubMed] [Google Scholar]
- 49.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393705-711. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, X., C. Borchers, R. J. Bienstock, and K. B. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 3911194-11204. [DOI] [PubMed] [Google Scholar]