Abstract
Little is known about the transmission or tropism of the newly discovered human retrovirus, human T-cell lymphotropic virus type 3 (HTLV-3). Here, we examine the entry requirements of HTLV-3 using independently expressed Env proteins. We observed that HTLV-3 surface glycoprotein (SU) binds efficiently to both activated CD4+ and CD8+ T cells. This contrasts with both HTLV-1 SU, which primarily binds to activated CD4+ T cells, and HTLV-2 SU, which primarily binds to activated CD8+ T cells. Binding studies with heparan sulfate proteoglycans (HSPGs) and neuropilin-1 (NRP-1), two molecules important for HTLV-1 entry, revealed that these molecules also enhance HTLV-3 SU binding. However, unlike HTLV-1 SU, HTLV-3 SU can bind efficiently in the absence of both HSPGs and NRP-1. Studies of entry performed with HTLV-3 Env-pseudotyped viruses together with SU binding studies revealed that, for HTLV-1, glucose transporter 1 (GLUT-1) functions at a postbinding step during HTLV-3 Env-mediated entry. Further studies revealed that HTLV-3 SU binds efficiently to naïve CD4+ T cells, which do not bind either HTLV-1 or HTLV-2 SU and do not express detectable levels of HSPGs, NRP-1, and GLUT-1. These results indicate that the complex of receptor molecules used by HTLV-3 to bind to primary T lymphocytes differs from that of both HTLV-1 and HTLV-2.
The primate T-cell lymphotropic virus (PTLV) group of deltaretroviruses consists of three types of human T-cell lymphotropic viruses (HTLVs) (HTLV-1, HTLV-2, HTLV-3), their closely related simian T-cell lymphotropic viruses (STLVs) (STLV-1, STLV-2, STLV-3), an HTLV (HTLV-4) for which a simian counterpart has not been yet identified, and an STLV (STLV-5) originally described as a divergent STLV-1 (5-7, 30, 35, 37, 38, 45, 51, 53). HTLV-1 and HTLV-2, which have a 70% nucleotide homology, differ in both their pathobiology and tropism (reviewed in reference 13). While HTLV-1 causes a neurological disorder (tropical spastic paraparesis/HTLV-1-associated myelopathy) and a hematological disease (adult T-cell leukemia/lymphoma) (15, 42, 55), HTLV-2 is only rarely associated with tropical spastic paraparesis/HTLV-1-associated myelopathy-like disease and is not definitively linked to any lymphoproliferative disease (12, 20). In vivo, both HTLV-1 and HTLV-2 infect T cells. Although HTLV-1 is primarily found in CD4+ T cells, other cell types in the peripheral blood of infected individuals have been found to contain HTLV-1, including CD8+ T cells, dendritic cells, and B cells (19, 29, 33, 36, 46).
Binding and entry of retroviruses requires specific interactions between the Env glycoproteins on the virus and cell surface receptor complexes on target cells. For HTLV-1, three molecules have been identified as important for entry, as follows: heparan sulfate proteoglycans (HSPGs), neuropilin-1 (NRP-1), and glucose transporter 1 (GLUT-1) (16, 22, 26, 28, 29, 34, 39, 44). Recent studies support a model in which HSPG and NRP-1 function during the initial binding of HTLV-1 to target cells, and GLUT-1 functions at a postattachment stage, most likely to facilitate fusion (29, 34, 49). Efficient HTLV-2 binding and entry requires NRP-1 and GLUT-1 but not HSPGs (16, 26, 39, 49).
This difference in the molecules required for binding to target cells reflects differences in the T-cell tropisms of these two viruses. Activated CD4+ T cells express much higher levels of HSPGs than CD8+ T cells (26). In infected individuals, HTLV-1 is primarily found in CD4+ T cells, while HTLV-2 is primarily found in CD8+ T cells (21, 43, 46). In vitro, HTLV-1 preferentially transforms CD4+ T cells while HTLV-2 preferentially transforms CD8+ T cells, and this difference has been mapped to the Env proteins (54).
We and others have reported the discovery of HTLV-3 in two Cameroonese inhabitants (6, 7, 53). We recently uncovered the presence of a third HTLV-3 strain in a different population living several hundred kilometers away from the previously identified groups (5), suggesting that this virus may be common in central Africa. Since the HTLV-3 sequences were obtained by PCR amplification of DNA isolated from peripheral blood mononuclear cells (PBMCs) of infected individuals, little is known about its tropism and pathobiology in vivo. Based on the correlation between HSPG expression levels and viral tropisms of HTLV-1 and HTLV-2, we reasoned that knowledge about the HTLV-3 receptors might provide insight into the tropism of this virus. We therefore generated vectors expressing HTLV-3 Env proteins and used them to begin to characterize the receptor complex used by HTLV-3 to bind and enter cells.
MATERIALS AND METHODS
Amino acid sequence analyses.
All sequence alignments were performed using ClustalW software.
Phylogenetic analyses.
The 27 complete PTLV Env sequences available in GenBank were aligned with the DAMBE program (version 4.5.68) on the basis of a previous amino acid alignment created from the original sequences. The phylogeny was derived by maximum likelihood and maximum parsimony methods, performed in the PAUP program (version 4.0b10).
Isolation, separation, and culture of primary cells.
Leukopaks of peripheral blood from healthy donors were collected according to the NIH-approved Institutional Review Board protocols, PBMCs were separated, and CD8+ T cells were isolated from PBMCs using immunomagnetic bead technology (Miltenyi Biotec Inc.), activated, and cultured as previously described (25). Monocyte-derived dendritic cells were generated from monocytes by culturing them in medium containing transforming growth factor β, granulocyte-macrophage colony-stimulating factor, and interleukin-4 (IL-4), as recently described (29).
Cell culture and reagents.
MOLT4, SupT1, Raji, CHO-K1, CHO-K1-pgsA-745 (glycosaminoglycan-negative), HEK 293T/17, and U87 cells were obtained from the American Type Culture Collection. The B cell line 729B was a gift from P. Green (Ohio State University, Columbus, OH). The C5/U87 and C8/U87 cell lines were generated by limiting dilution cloning of U87. CHO-K1 and CHO-K1-pgsA-745 were maintained in Dulbecco's modified Eagle medium-Ham's F-12 medium (1:1 dilution); SupT1, Jurkat, MOLT4 and Raji in RPMI 1640; 729B in Iscove's modified Dulbecco's medium; and HEK 293T/17, C5/U87, and C8/U87 in Dulbecco's modified Eagle medium. All of the cell culture media were supplemented with l-glutamine (2 mM), penicillin (100 U/ml), streptomycin (100 ng/ml), and fetal calf serum (10%).
Plasmids and peptides.
The plasmid HTLV-3 Env/PCR2.1 was generated by PCR amplification of the Env region (nucleotides [nt] 5047 to 6787) of a plasmid encoding HTLV-3Pyl43 (8) as the template with the oligonucleotide primers 5′AAACCAAAGACCATCAACTCC3′ and 5′CCTGCATATCTACTTCCGGAGGC3′. The resultant DNA fragment was inserted into the PCR2.1 vector using the TA cloning kit (Invitrogen). To construct the plasmid (HTSU3-IgG/pCMV-Env) encoding full-length, soluble HTLV-3 surface glycoprotein (SU) (designated “HTLV-3 SU” throughout the article), a DNA fragment was generated using HTLV-3 Env/PCR2.1 as the template and the oligonucleotide primers 5′CCTCAAGCGAGCTGCATGCATGGGTAAGTCCGGTCTTTAT3′ and 5′GGTGCGGATCCACTAGTTCTAGATCGGCGCTTCCGGGGTC3′. The resultant DNA fragment was digested with SphI and SpeI and used to replace the SphI-SpeI fragment of HTSU-IgG/pCMV-Env (27).
The plasmid encoding HTLV-3 SU and TM (CMV-ENV-LTR-III; designated “HTLV-3 Env” throughout) was constructed by digesting HTLV-3 Env/PCR2.1 with BamHI and PsiI, generating a fragment that included the entire HTLV-3 Env coding sequences. The plasmid CMV-ENV-LTR was digested with BamHI and PsiI to remove the homologous fragment of the HTLV-1 provirus envelope region, and the fragment was replaced with the HTLV-3 sequence. All four constructs were confirmed by being sequenced.
The peptides homologous to the NRP-1 ligand VEGF165 were obtained as follows. Tuftsin (TU) (TKPR) was obtained from Sigma. TU analog (TU-A; TKPPR) and the control for TU-A (L5D; LTRKD), generated from a variant of VEGF165 (VEGF165b) that does not bind NRP-1, were obtained from Eurogentec.
SU binding assay.
The soluble fusion proteins carrying HTLV-1, HTLV-2, and HTLV-3 SU as well as a negative control (avian retrovirus subgroup A avian leukosis and sarcoma virus [ALSV-A] SU) protein were obtained following transfection of HEK 293T/17 cells, as previously described (28). The amount of SU protein was standardized using a rabbit immunoglobulin G (IgG) enzyme-linked immunosorbent assay (ZeptoMetrix), and 200 ng of each SU protein was used, unless otherwise noted. Specific binding of soluble HTLV SU proteins to target cells was performed, as previously described (28).
Removal of cell surface HSPGs.
Heparan sulfate (HS) lyase (heparitinase III) was obtained from Seikagaku Corp. (Tokyo, Japan) and used as previously described (28). Briefly, MOLT4 cells (106) were spun down, resuspended in 200 μl of HS lyase buffer, and then incubated for 2 h at 37°C with 10 mU HS lyase or in buffer only. Two hours later, the cells were washed twice, resuspended in 400 μl of phosphate-buffered saline, and analyzed by flow cytometry.
Cell surface expression of HSPGs, NRP-1, and GLUT-1.
The levels of HSPGs and NRP-1 expression were determined using the monoclonal antibody clone F58-10E4 (Seikagaku Corp.) and clone AD5-17F6 (Miltenyi Biotec), respectively. The level of GLUT-1 expression was determined using two antibodies which recognize extracellular domains of human GLUT-1: a mouse anti-human monoclonal antibody from R&D Systems (MAB1418) and a rabbit anti-human polyclonal from Alpha Diagnostics (GT14-A), as previously described (28). All cells were fixed in 4% paraformaldehyde after being stained, with the exception of red blood cells, which were fixed in 1% paraformaldehyde. The level of cell surface expression was determined as previously described (26).
Retroviral vector transduction.
Pseudotyped retroviral vectors were generated and used to transduce the target cells as previously described (27). Briefly, target cells were incubated with 10-fold dilutions of supernatant containing the pseudotyped viruses, and negative control cultures (cells transduced with murine leukemia virus [MLV] vectors with no Env) were included in each experiment. Four days later, cells were analyzed by flow cytometry for the expression of enhanced green fluorescent protein to determine the percentage of transduced cells. Relative titers were determined from the well with the lowest percentage that was at least 5% positive using the following formula: (% positive − % positive in negative control) × (number of cells in well on day of transduction). Titers were corrected to 1 ml.
Statistical analysis.
Data were analyzed by one-way analysis of variance followed by Tukey's test. A P value of <0.05 was considered statistically significant. All results are presented as means ± standard errors of the means (SEM).
RESULTS
HTLV-3 SU sequence displays similarities with those of HTLV-1 and HTLV-2.
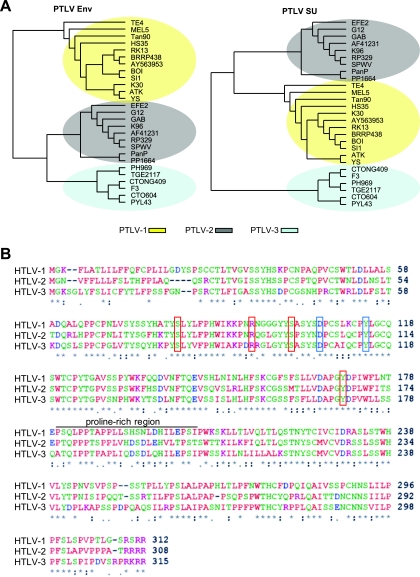
Previous phylogenetic studies conducted on concatenated viral sequences (6,812 bp) revealed that the HTLV-3 genome is as divergent from that of HTLV-1 as it is from that of HTLV-2 (6, 17). To determine whether this was also true for the HTLV Env sequences, we performed a series of phylogenetic analyses on both the complete Env genes of PTLVs and the portion encoding the SU. These results revealed that the sequences of both the HTLV-3 Env and the HTLV-3 SU proteins fit into a clade that is distinct from those of HTLV-1 and HTLV-2 (see Pyl43) (Fig. 1A). Alignment of the SU amino acid consensus sequences of HTLV-1, HTLV-2, and HTLV-3 revealed that, despite being grouped in a cluster different from that of both HTLV-1 and HTLV-2, the HTLV-3 SU sequence displays some similarities with those of the other two SU proteins (Fig. 1B). For example, the alignment revealed that the structure of HTLV-3 SU would be predicted to consist of two domains linked by a short, proline-rich (amino acids 180 to 205) “hinge” region. This has previously been shown to be the structure of a number of retroviral SU proteins (1-3, 10, 14, 18, 56), including HTLV-1, HTLV-2, and bovine leukemia virus deltaretroviruses (24, 31, 32). Moreover, amino acids previously shown to be important for HTLV-1 SU functions are also conserved in HTLV-3 SU. These include residues involved in the interaction with GLUT-1 (D106 and Y114) (39) and residues important for HTLV-1 Env-mediated fusion and infection (S81, R94, S101, Y170) (11, 47) (Fig. 1B).
FIG. 1.
HTLV-3 Env and HTLV-3 SU belong to different phylogenetic clades than the HTLV-1 and HTLV-2 Env and SU but display a similar domain organization. (A) Phylogenetic trees resulting from the analysis of 27 sequences of either the entire env gene (1,479 nt) (left) or the portion that encodes the SU (951 nt) (right). The sequences were aligned using ClustalW (in DAMBE program). The trees shown were generated in PAUP using the maximum likelihood method. (B) The SU amino acid consensus sequences of HTLV-1, HTLV-2, and HTLV-3 were aligned using ClustalW. An asterisk indicates identical residues, a colon indicates conserved substitutions, and a period indicates semiconserved substitutions. Red boxes, residue involved in Env-mediated fusion and infection (S81, R94, S101, Y170); blue boxes, residues involved in GLUT-1 interactions (D106 and Y114).
HTLV-3 SU binds to activated primary CD4+ T cells and CD4+ T-cell lines.
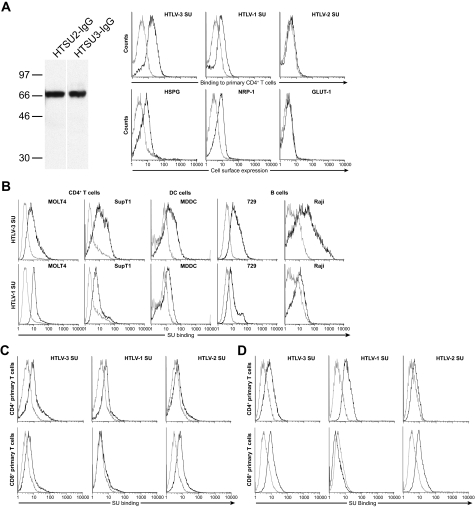
It was previously shown that the level of binding of soluble forms of HTLV-1 and HTLV-2 SU proteins to T cells parallels the level of binding and internalization of HTLV virions (26, 28, 41, 49). We therefore generated a construct encoding a soluble form of the HTLV-3 SU protein using an approach that we previously used to generate soluble forms of the HTLV-1 and HTLV-2 SU proteins (26, 28). Western blot analysis confirmed that the soluble HTLV-3 SU protein generated from this construct had the correct size and was recognized by anti-HTLV SU antibodies (Fig. 2A, left).
FIG. 2.
HTLV-3 SU binds to CD4+ and CD8+ T cells, dendritic cells, and B cells. (A) HTLV-3 SU binding to CD4+ T cells. (Left) HEK 293T/17 cells were transfected with either the plasmid encoding the HTLV-3 SU protein fused to rabbit IgG (HTSU3-IgG/pCMV-Env) or, as a control, a similar construct encoding the HTLV-2 SU fusion protein (HTSU2-IgG/pCMV-Env) (26). Cells were lysed, and Western blot analysis was performed as previously described (28) with the following modifications: the immunoblots were incubated overnight at 4°C with an antibody directed against HTLV SU (ZeptoMetrix). Numbers on the left indicate the positions of the molecular mass markers (in kDa). (Right) CD4+ T cells were isolated and activated for 24 h with phytohemagglutinin and IL-2. (Top) Cells were incubated with 200 ng of either the soluble form of HTLV-1 SU, HTLV-2 SU, HTLV-3 SU, or a similar fusion protein containing the SU protein from the avian retrovirus ALSV-A as a negative control. The amount of binding was determined by flow cytometry. In this and all subsequent SU binding studies: black line, HTLV SU; gray line, ALSV-A SU. (Bottom) Studies performed in parallel to determine the cell surface levels of components of the HTLV-1 receptor complex on the CD4+ T cells used in the SU binding studies. Cells were incubated with antibodies specific for HSPGs, NRP-1, or GLUT-1, and analysis was performed as described in Materials and Methods. Black line, binding of anti-HSPG antibody, anti-NRP-1 antibody, or anti-GLUT-1 antibody; gray line, isotype controls. (B) CD4+ T-cell lines (MOLT4, SupT1), dendritic cells (monocyte-derived dendritic cells [MDDC]), and B cell lines (729, Raji) were incubated with SU proteins, and the level of binding to SU was determined as described above. (Top) HTLV-3 SU binding. (Bottom) HTLV-1 SU binding. (C and D) CD4+ and CD8+ T cells were activated for either 2 days (C) or 4 days (D) and exposed to 100 ng of SU proteins, and the level of binding was determined. Flow cytometry results are representative of six (A, right), three (B and D) and four (C) independent experiments.
Although HTLV-3 proviral sequences have been found in the PBMCs of infected individuals, nothing is known about which cell types are infected with HTLV-3. We initially examined the binding of HTLV-3 SU to several cell types that can be infected by HTLV-1. Experiments performed with HTLV-3 SU revealed that it bound at high levels to primary activated CD4+ T cells, as did HTLV-1 SU but not HTLV-2 (Fig. 2A, right and top). Flow analyses performed in parallel confirmed that, as previously reported (4, 29, 48, 49), the primary activated CD4+ T cells expressed high levels of HSPGs and NRP-1, but not GLUT-1, on the cell surface (Fig. 2A, right and bottom). HTLV-3 SU also bound at high levels to CD4+ T-cell lines (Fig. 2B, top two panels on the left). Further studies revealed that HTLV-3 SU also efficiently binds to two other cell types (dendritic cells and B cells) that can be infected by HTLV-1 (Fig. 2B, top right three panels). The level of HTLV-3 SU binding to all these cells was higher than that observed for HTLV-1 SU (Fig. 2B, compare top and bottom rows).
HTLV-3 SU binds efficiently to both activated CD4+ and CD8+ T cells.
Comparison of the abilities of HTLV-1, HTLV-2, and HTLV-3 SUs to bind to primary CD4+ and CD8+ T lymphocytes activated for 2 (Fig. 2C) or 4 (Fig. 2D) days revealed that the pattern of HTLV-3 SU binding was unique. Like the results shown in Fig. 2A, HTLV-1 and HTLV-3 SU both bound efficiently to activated CD4+ T cells (Fig. 2C and D, top, left and middle). However, HTLV-3 SU was also able to bind to activated CD8+ T cells (Fig. 2C and D, bottom and left). In contrast, and like previous reports (26), HTLV-2 SU binding was markedly higher on CD8+ T cells than on CD4+ T cells (Fig. 2C and D, right).
Role of HSPGs in HTLV-3 SU binding to T cells.
We next examined whether HTLV-3 SU binding to cells involves any of the molecules previously shown to be required for the binding of HTLV-1 and/or HTLV-2 SU. HSPGs have been shown to be required for efficient binding to and infection of target cells by HTLV-1. The levels of binding of soluble HTLV-1 SU and HTLV-1 virions, the titers of HTLV-1 Env-pseudotyped viruses, and infection of primary CD4+ T cells and dendritic cells are dramatically reduced in cells lacking HSPGs (26, 28, 29, 44, 49). In contrast, HTLV-2 does not require HSPGs for efficient binding and entry (26).
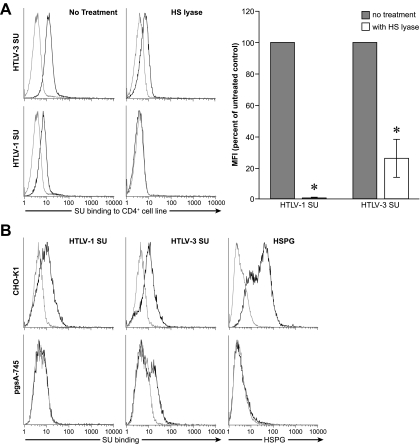
As a first step to investigate whether HSPGs play a role in HTLV-3 Env binding, we examined the ability of HTLV-3 SU to bind to a CD4+ T-cell line (MOLT4) from which HSPGs had been enzymatically removed by treatment with HS lyase. Although removal of HSPGs lowered the level of binding, HTLV-3 SU could still bind to these cells (Fig. 3A). This was in contrast to the level of HTLV-1 SU binding, which, as expected from previous studies (26, 28, 44), was consistently low or undetectable on the cells lacking HSPGs (Fig. 3A, left and bottom). The notion that HSPGs play a less important role in HTLV-3 SU binding than in HTLV-1 SU binding is supported by the studies of binding to primary T cells shown above. As we have previously reported, primary CD4+ T cells activated for 4 days express higher levels of HSPGs than the cells activated for 2 days (data not shown) and bind higher levels of HTLV-1 SU (compare top middle panels of Fig. 2D [4 days] and Fig. 2C [2 days]). In contrast, the binding of HTLV-3 SU was not significantly higher on the cells that expressed higher levels of HSPGs (compare top left panels of Fig. 2D and C).
FIG. 3.
Contribution of HSPGs to HTLV-3 SU binding. (A) MOLT4 cells were incubated for 2 h in HS lyase buffer in the presence or absence of HS lyase, and the levels of binding to HTLV-1 SU and HTLV-3 SU were determined. Staining with anti-HSPG antibodies (not shown) was performed following the incubation to verify that the level of cell surface HSPGs was reduced to below the level of detection. (Left) Histogram of one representative experiment. (Right) Graph representing an average of three independent experiments. Mean fluorescent intensity (MFI) was determined by subtracting the MFI of the negative control (ASLV-A SU) from the MFI of the HTLV SU. MFI obtained in the absence of HS lyase was normalized to 100%, and the average value from three independent experiments was determined; error bars denote SEM. Statistical significance was determined by analysis of variance followed by Tukey's test. An asterisk denotes significant differences from controls (binding in the absence of HS lyase). The P values were as follows: no treatment with HTLV-1 SU versus HS lyase treatment, <0.001; no treatment with HTLV-3 SU versus HS lyase treatment, <0.001. (B) CHO-K1 and HSPG-negative CHO-K1 mutant (CHO-K1-pgsA-745) cells were incubated with HTLV-1 and HTLV-3 SU, and the levels of SU binding (left) and anti-HSPG antibody staining (right) were determined. The MFIs for the SU binding were as follows: CHO-K1/SU-1, 2.2; pgsA-745/SU-1, 0.0; CHOK1/SU-3, 4.5; pgsA-745/SU-3, 2.6. For the left two panels: black line, HTLV SU; gray line, ALSV-A SU. For the right panel: black line, binding of anti-HSPG antibody; gray line, isotype control.
HSPGs are just one of several different types of glycosylaminoglycans (GAGs) used by viruses to facilitate binding to target cells. HTLV-1 SU appears only to be able to use HSPGs since other GAGs do not enhance HTLV-1 SU binding (28 and data not shown). One possible explanation for the binding of HTLV-3 SU to cells that lack HSPGs is that HTLV-3 can use GAGs other than HSPGs to enhance binding. To address this, we examined the ability of HTLV-3 to bind to the cell line pgsA-745, a derivative of CHO-K1 cells that does not express any GAGs. Flow analysis revealed that, although the level was reduced, HTLV-3 SU could bind to a portion of the cells in the absence of GAGs (Fig. 3B, middle and bottom). The level of HTLV-3 SU binding varied among experiments: sometimes the binding was higher than that shown in Fig. 3B, while in other experiments, binding was just slightly above the control (data not shown). In contrast, HTLV-1 SU binding to the cells lacking GAGs was never observed (Fig. 3B, left and bottom). The right panels demonstrate the lack of HSPG expression in the pgsA-745 cells. All together, these data indicate that HTLV-3 SU binding involves interaction with molecules other than GAG and suggest that those molecules are expressed differently at different times in the cell cycle. Taken together, these results show that, while HSPGs can enhance HTLV-3 SU binding, HTLV-3 SU can bind to cells in the absence of HSPGs and other GAGs.
Role of NRP1 in HTLV-3 SU binding.
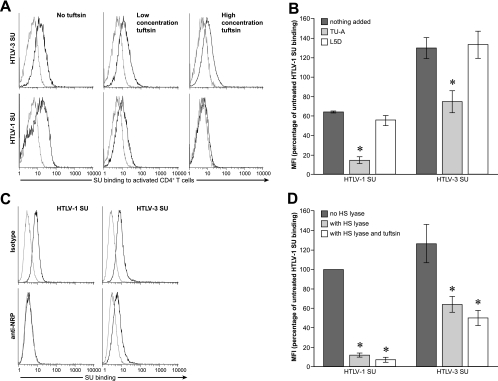
We next examined whether two other molecules involved in HTLV entry, NRP-1 and GLUT-1, contributed to the non-HSPG binding by HTLV-3 SU. NRP-1 has been shown to directly bind to HTLV-1 SU (16, 34). Moreover, the level of NRP-1 on target cells parallels the titers of HTLV-1 and HTLV-2 Env-pseudotyped virions, and blocking interactions with NRP-1 with its ligand VEGF-A165 decreases HTLV-1 binding, entry, and infection (16, 34). Peptides homologous to the region of VEGF-A165 that bind directly to NRP-1 (50, 52) have recently been shown to block HTLV-1 SU binding (34). To determine whether blocking NRP-1 interactions also decreased HTLV-3 SU binding, activated primary CD4+ T cells were incubated with an NRP-1-binding peptide (tuftsin [TU]) prior to and during incubation with HTLV-1 and HTLV-3 SU proteins. Blocking interactions with NRP-1 decreased the level of HTLV-3 SU binding to primary CD4+ cells (Fig. 4A). However, under conditions where the HTLV-1 SU binding was almost completely blocked, HTLV-3 SU still bound at significant levels (Fig. 4A, right, top and bottom). The effect of blocking NRP-1 interactions was also examined by incubating the cells with a second peptide that binds to NRP-1 (TU-A) or, as a negative control, a peptide of the same length (L5D) corresponding to the same region of a variant of VEGF165 (VEGF165b) which does not bind to NRP-1 (Fig. 4B). The TU-A peptide reduced the level of HTLV-3 SU binding to activated primary CD4+ T cells by about 50%, while the control peptide did not significantly block binding. In contrast, and consistent with what has been recently reported (34), the TU-A had a more significant effect on the levels of HTLV-1 SU: binding was reduced by more than 87%. The fact that the control peptide also modestly reduced HTLV-1 SU binding suggests that the peptides, which have a positive charge, may be interacting with the negatively charged HSPGs.
FIG. 4.
Contribution of NRP-1 to HTLV-3 SU binding. (A and B) Primary activated CD4+ T cells were incubated with peptides homologous to an NRP-1-binding domain of the NRP-1 ligand VEGF165, and the levels of HTLV-1 and HTLV-3 SU binding were determined. (A) Cells were incubated with either 5 μg/ml (low concentration TU), 50 μg/ml of the 4-mer peptide TU (TKPR) (high concentration TU), or left untreated (no TU). The MFIs for the SU binding were as follows: no TU/SU-1, 5.2; 5 μg/ml TU/SU-1, 4.0; 50 μg/ml TU/SU-1, 0.7; no TU/SU-3, 8.3; 5 μg/ml TU/SU-3, 7.5; 50 μg/ml TU/SU-3, 4.9. (B) Cells were incubated with 50 μg/ml of either the 5-mer peptide TU-A (TKPPR) or a 5-mer control peptide (L5D; LTRKD) or left untreated. The MFIs for SU binding were determined and normalized to the MFI of HTLV-1 SU binding in the absence of peptides (100%), and the average value from three independent experiments was determined. Statistical significance was determined as above; an asterisk denotes significant differences from controls. P values were as follows: HTLV-1 SU with no peptide versus TU-A, <0.05; HTLV-1 SU with no peptide versus L5D, >0.05; HTLV-3 SU with no peptide versus TU-A, <0.01; HTLV-3 SU with no peptide versus L5D, >0.05. (C) The CD4+ T-cell line SupT1 was incubated for 1 h with 10 μg/ml of either an antibody directed against an extracellular portion of NRP-1 or an isotype control (mouse IgG1) and then exposed to the SU proteins. The MFIs for the SU binding were as follows: isotype/SU-1, 3.5; anti-NRP-1/SU-1, 0.2 (5.7% of isotype); isotype/SU-3, 5.5; anti-NRP-1/SU-3, 2.69 (48.5% of isotype). (D) CHO-K1 cells were treated with HS lyase, and binding assays were performed as in panel A, except that a portion of the cells treated with HS lyase were also incubated with 20 μg/ml TU. Shown is the MFI for SU binding from three independent experiments, normalized as described above. An asterisk denotes significant differences from binding of the same SU to untreated cells. P values were as follows: no treatment with HTLV-1 SU versus HS lyase treatment, <0.001; no treatment with HTLV-1 SU versus HS lyase and TU treatment, <0.001; no treatment with HTLV-3 SU versus HS lyase treatment, <0.01; no treatment with HTLV-3 SU versus HS lyase and TU treatment, <0.001. HTLV-3 SU bound at significant levels to cells that had been enzymatically treated to remove HSPGs (light gray bar). HTLV-3 SU also bound to cells when NRP-1 interactions were blocked and HSPGs were removed (white bar).
The effect of blocking NRP-1 interactions was also examined using an antibody directed against NRP-1. The comparative effects on HTLV-1 and HTLV-3 SU binding paralleled what were observed when peptides were used to block binding to NRP-1. Exposure of a CD4+ T-cell line (SupT1) to an anti-NRP-1 antibody reduced HTLV-1 SU binding by 85 to 98%, while the level of HTLV-3 SU binding was reduced by 30 to 45% (Fig. 4C and data not shown).
We next examined the effect of blocking both HSPG and NRP-1 interactions on HTLV-3 SU binding. As reported above in Fig. 3, HTLV-3 SU bound at significant levels to cells that had been enzymatically treated to remove HSPGs (Fig. 4D). HTLV-3 SU also bound to cells when NRP-1 interactions were blocked and HSPGs were removed, confirming that HTLV-3 can bind efficiently to cells when interactions with both HSPGs and NRP1 are blocked. In contrast, the combination of TU and HS lyase almost completely abolished HTLV-1 SU binding to the cells.
GLUT-1 is not involved in HTLV-3 SU binding.
Cell surface levels of GLUT-1 do not correlate with the level of binding of HTLV-1 SU or that of HTLV-1 virions (26, 49). However, the level of GLUT-1 does correlate with the binding of a truncated form of HTLV-1 SU lacking the C terminus, the titers of HTLV-1 Env-pseudotyped viruses, and cell-cell transmission of HTLV-1, suggesting that this molecule functions after the initial binding to the cell (31, 39, 49). In contrast, GLUT-1 appears to be the major determinant for the level of binding of HTLV-2 SU (26).
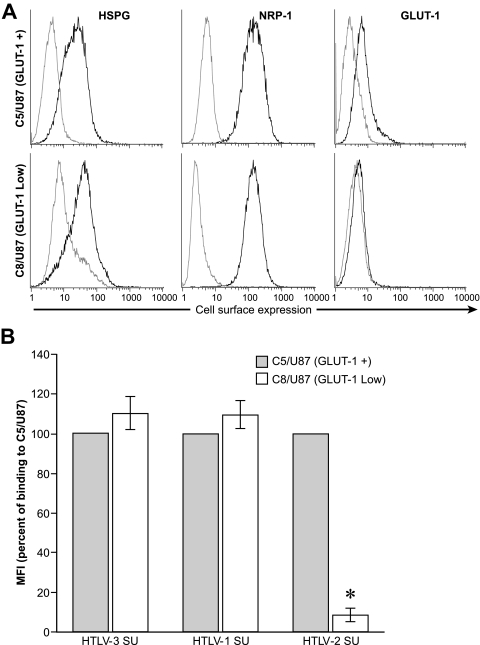
To examine whether GLUT-1 is involved in the binding of HTLV-3 SU, we generated a pair of cell lines (C5/U87 and C8/U87) that differ in their levels of cell surface GLUT-1. These cells were cloned from the U87 cell line, which had previously been shown to express low but detectable levels of GLUT-1 on the cell surface (23). C5/U87 expresses significant levels of cell surface GLUT-1, while the level of GLUT-1 on C8/U87 cells is very low (Fig. 5A, right). Both the C5/U87 and C8/U87 clones express high levels of HSPG and NRP-1 (Fig. 5A, left and middle). SU binding studies revealed that, as expected from previous studies, HTLV-2 SU binding was significantly lower on C8/U87, which expresses low levels of GLUT-1, than on C5/U87 (Fig. 5B). In contrast, the levels of binding of both HTLV-3 and HTLV-1 SU were not reduced on the cells expressing lower levels of GLUT-1. These results suggest that, as for HTLV-1, GLUT-1 does not play a significant role in the initial binding of HTLV-3 to the target cell.
FIG. 5.
Contribution of GLUT-1 to HTLV-3 SU binding. (A) The cell surface levels of HSPG, NRP-1, and GLUT-1 on U87 clones C5 (GLUT-1+) and C8 (GLUT-1Low) were determined by flow cytometry, as in Fig. 2A. Black line, specific antibodies; gray line, isotype control. The result shown here is representative of four independent experiments. (B) Levels of binding of HTLV SU to C5/U87 and C8/U87. Data shown were calculated as for Fig. 4. An asterisk denotes significant differences from binding of the same SU to the C5/U87 cells. P values were as follows: HTLV-3 SU C5/U87 versus C8/U87, >0.05; HTLV-1 SU C5/U87 versus C8/U87, >0.05; HTLV-2 SU C5/U87 versus C8/U87, <0.001.
Role of HSPGs, NRP-1, and GLUT-1 in HTLV-3 Env-mediated entry.
We next wanted to examine whether HSPGs, NRP-1, and/or GLUT-1 play a role in HTLV-3 Env-mediated entry at a step after the initial binding to the cell surface. To do this, we generated a vector expressing both the HTLV-3 SU and TM proteins and used this construct to pseudotype retroviral vectors. The titers of such vectors reflect the ability of the Env proteins to facilitate both the initial binding and subsequent virus-cell fusion and entry.
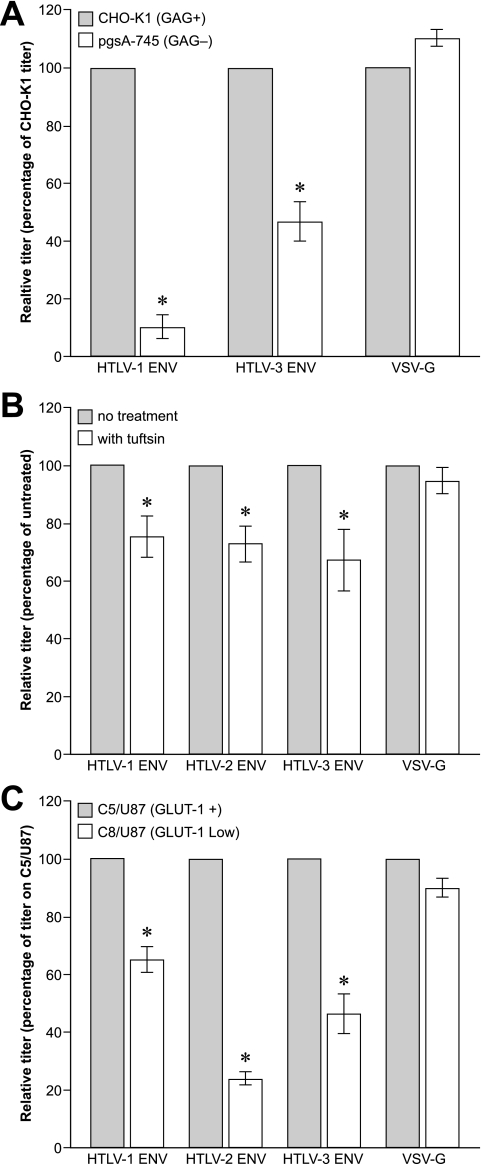
Previous studies have shown that, as for SU binding, the titers of HTLV-1 Env-pseudotyped viruses are dramatically lowered when the level of HSPGs on target cells is reduced (28, 44). To investigate whether HSPGs were also important for efficient HTLV-3 Env-mediated entry, the titers of MLV-based virions pseudotyped with either HTLV-1 Env, HTLV-3 Env, or vesicular stomatitis virus G protein (VSV-G) on CHO-K1 and pgsA-745 cells were determined. As expected, the titer of HTLV-1 Env-pseudotyped virions was dramatically lower on the GAG-negative pgsA-745 cells than on CHO-K1 cells (Fig. 6A). The absence of GAG also significantly reduced the titer of the HTLV-3 Env-pseudotyped virus, but the effect was less dramatic than for the HTLV-1 Env-pseudotyped virus (Fig. 6A, middle). The effect observed appeared to involve specific interactions between the Env proteins and the target cells, since entry of VSV-G-pseudotyped virus, which has previously been shown to be independent of GAG, was not affected by this treatment (Fig. 6A, right). Thus, as for SU binding, GAGs enhance, but are not required for, HTLV-3 Env-mediated binding and entry into target cells.
FIG. 6.
Roles of HSPGs, NRP-1, and GLUT-1 in HTLV-3 Env-mediated entry. (A) CHO-K1 and CHO-K1-pgsA-745 cells were incubated overnight with MLV-based retroviral vectors pseudotyped with either HTLV-1 Env, HTLV-3 Env, or VSV-G and harvested 4 days later, and the titers were determined as described in Materials and Methods. Titers obtained on CHO-K1 were normalized to 100, and the relative titer was determined using the following formula: (titer on CHO-K1/titer on CHO-K1-pgsA-745) × 100. P values were as follows: HTLV-1 Env CHOK-1 versus CHO-K1-pgsA-745, <0.001; HTLV-3 Env CHOK-1 versus CHO-K1-pgsA-745, <0.001; VSV-G CHOK-1 versus CHO-K1-pgsA-745, >0.05. (B) 293 cells were incubated in either the presence or absence of 8 μg/ml of TU. Thirty minutes later, equal volumes of media containing either HTLV-1 Env, HTLV-2 Env, HTLV-3 Env, or VSV-G-pseudotyped vectors were added, so that the final concentration of TU was 4 μg/ml. Six hours later, the cells were washed, and the titers were determined 4 days later. P values were as follows: no treatment with HTLV-1 Env versus TU, <0.05; no treatment with HTLV-2 Env versus TU, <0.05; no treatment with HTLV-3 Env versus TU, <0.01; no treatment with VSV-G versus TU, >0.05. (C) C5/U87 and C8/U87 were incubated overnight with pseudotyped vectors, and the titers were determined as described above. Shown are means and SEM of four (A) or three (B and C) independent experiments. P values were as follows: HTLV-1 Env C5/U87 versus C8/U87, <0.001; HTLV-2 Env C5/U87 versus C8/U87, <0.05; HTLV-3 Env C5/U87 versus C8/U87, <0.01; VSV-G C5/U87 versus C8/U87, >0.05.
We next investigated the role of NRP-1 and GLUT-1 in HTLV-3 Env-mediated entry. In the first series of experiments, decreasing NRP-1 interactions by the addition of TU significantly decreased the titers of all three HTLV Env-pseudotyped viruses, suggesting that NRP-1 is involved in the entry of all three viruses (Fig. 6B).
Similar results were obtained when the role of GLUT-1 in entry was examined. The titers of retroviral vectors pseudotyped with all three HTLV Env proteins were significantly lower on the cells expressing lower levels of cell surface GLUT-1 (Fig. 6C). The vectors pseudotyped with HTLV-2 Env had the lowest titers on U87/C8, consistent with observations that, for HTLV-2, GLUT-1 plays a role in binding as well as in postbinding events (26). These results combined with those of Fig. 4 and 5 indicate that, as for HTLV-1, GLUT-1 is involved in HTLV-3 Env-mediated fusion at a step after the initial binding to the cell.
HTLV-3 SU binds efficiently to naïve CD4+ cells in the absence of known components of the HTLV receptor complex.
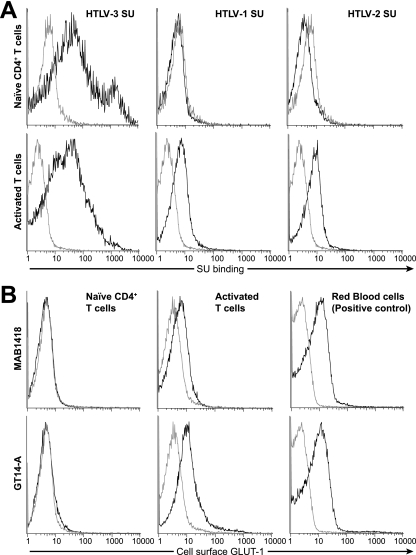
The results presented in Fig. 3, 4, and 5 strongly suggest that HTLV-3 SU binding to cells can also involve molecules other than HSPGs, NRP-1, and GLUT-1. Studies performed prior to the characterization of the HTLV receptor complexes demonstrated that freshly isolated quiescent CD4+ T cells do not bind either HTLV-1 or HTLV-2 SU (40, 41). More recent studies have revealed that naïve unactivated CD4+ T cells do not express HSPGs or NRP-1 and express low or undetectable amounts of GLUT-1 (4, 26, 28, 29, 48, 49). To determine whether HTLV-3 SU binding can occur in the absence of the HSPGs, NRP-1, and GLUT-1, naïve (CD45RA+) CD4+ T cells were isolated. Activated (CD45RO+) T cells were also isolated and used as a positive control. Twenty-four hours later, the level of HTLV SU binding was determined. We observed that, unlike HTLV-1 and HTLV-2 SU, HTLV-3 SU could bind efficiently to quiescent CD4+ T cells (Fig. 7A, top, compare left panel with middle and right panels). As expected (26, 28) (Fig. 2), all three HTLV SU proteins bound to the activated T cells (Fig. 7A, bottom). Analysis of the level of cell surface expression of GLUT-1 on these cells performed in parallel using the monoclonal antibody MAB1418 confirmed that the level of GLUT-1 on the naïve cells was below the level of detectability (Fig. 7B, top and left). As expected, GLUT-1 was expressed on activated T cells and on red blood cells, which express high levels of GLUT-1 (Fig. 7B, top, middle and right). Similar results were obtained with experiments performed in parallel with a polyclonal antibody directed against GLUT-1 (GT14-A) (Fig. 7B, bottom), confirming that the naïve cells used in these experiments did not express detectable levels of GLUT-1 on the cell surface.
FIG. 7.
HTLV-3 SU binds to naïve CD4+ T cells. Naïve CD4+ T cells, activated T cells (both CD4+ and CD8+), and red blood cells were isolated from adult PBMCs. The naïve cells were left under the conditions to keep them unactivated, and the red blood cells were left as a pellet, both at 4°C. The T cells were stimulated by culturing them at 37°C in RPMI media containing 10% sera, phytohemagglutinin, and IL-2. The levels of SU binding and GLUT-1 expression were determined 24 h later. (A) SU binding on naïve CD4+ T cells (top) and activated T cells (bottom). Note that the level of HTLV-2 SU binding for the naïve cells is below that of the negative control (ASLV-A SU). Shown is a representative result of three independent experiments. (B) Cell surface expression of GLUT-1 on naïve CD4+ T cells (left), activated T cells (middle), or red blood cells (right), which express high levels of GLUT-1. The level of GLUT-1 was determined using two independently derived antibodies that recognize extracellular domains of human GLUT-1: a mouse anti-human monoclonal antibody from R&D Systems (top) and a rabbit anti-human polyclonal from Alpha Diagnostics (bottom).
These observations confirm that HTLV-3 SU can bind efficiently to naïve CD4+ T cells in the absence of detectable levels of HSPGs, NRP-1, and GLUT-1, indicating that a different, currently unidentified molecule is sufficient to support efficient binding of HTLV-3 SU to T cells.
DISCUSSION
We present here the initial characterization of the molecules used by HTLV-3 to bind and enter T cells. Sequence comparisons revealed that HTLV-3 Env proteins have an overall organization similar to those of HTLV-1 and HTLV-2 and that residues in the Env proteins that are important for HTLV-1 and HTLV-2 infectivity are conserved in HTLV-3.
Recent studies have determined that HSPGs, NRP-1, and GLUT-1 are involved in the entry of HTLV-1. We report here that, as for HTLV-1, the cell surface levels of both HSPGs and NRP-1 can influence HTLV-3 SU binding. However, in sharp contrast to HTLV-1 SU, HTLV-3 SU can bind to cell lines that do not express HSPG or any other GAG and when interactions with both HSPGs and NRP-1 are blocked. Consistent with these observations, HTLV-3 SU bind efficiently to primary CD8+ T cells, which express low or undetectable levels of HSPGs (26). Further examination of HTLV-3 Env interactions also revealed that the level of cell surface GLUT-1 correlates with Env-mediated entry but not with SU binding. This observation indicates that, as for HTLV-1 (26, 49), GLUT-1 facilitates entry at a step after the initial attachment to the target cell.
These results strongly suggest that HTLV-3 SU can bind to target cells using molecules other than those involved in HTLV-1 and HTLV-2 binding. This hypothesis was confirmed by our studies with freshly isolated primary T cells. HTLV-3 SU bound efficiently to naïve CD4+ T cells, which do not bind HTLV-1 or HTLV-2 SU and which do not express detectable levels of HSPGs, NRP-1, or GLUT-1. These results also suggest that, as is the case for certain other viruses including HIV-1, HTLV-3 may use different receptor complexes to infect different cell types. This notion is supported by a previous report of GLUT-1-independent HTLV-1 Env-mediated fusion and entry into a glioblastoma/astroglioma cell line (23).
In this study, requirements for binding and entry were examined using a soluble form of the SU protein and Env-pseudotyped viruses, respectively. For other retroviruses, studies using this approach have proven useful for identifying molecules required for the initial steps in infection. However, the presence of receptors on cells is not necessarily sufficient for generating a productive infection. Indeed, it has been shown that, while HTLV-1 and HTLV-2 Env-pseudotyped viruses can transduce a wide variety of cells, only a small number of cell types (including T cells, dendritic cells, and B cells) can be productively infected by these viruses (19, 29, 33, 36, 46). Until recently, similar studies of HTLV-3 tropism were not possible due to the lack of productively infected cell lines and the lack of an infectious HTLV-3 clone. We have recently generated such an infectious clone of HTLV-3 (8), as well as one of the closely related STLV-3 (9). These tools will allow us to expand the current study by characterizing additional steps in the viral life cycle required for productive HTLV-3 infection.
Acknowledgments
We thank Claudine Pique for her helpful comments and Sara Reynolds for her technical help.
This work was supported by fellowships from le Ministère de la Recherche and from La Fondation pour la Recherche Médicale to S.A.C. R.M. was supported by INSERM and is now supported by ENS Lyon. The financial support from the Programme Interdisciplinaire CNRS, Maladies Infectieuses Émergentes, and from the Virus Cancer Prevention Association is also acknowledged. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Footnotes
Published ahead of print on 11 March 2009.
REFERENCES
- 1.Battini, J. L., J. M. Heard, and O. Danos. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 661468-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini, J. L., J. E. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 961385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boomer, S., M. Eiden, C. C. Burns, and J. Overbaugh. 1997. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J. Virol. 718116-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruder, D., M. Probst-Kepper, A. M. Westendorf, R. Geffers, S. Beissert, K. Loser, H. von Boehmer, J. Buer, and W. Hansen. 2004. Neuropilin-1: a surface marker of regulatory T cells. Eur. J. Immunol. 34623-630. [DOI] [PubMed] [Google Scholar]
- 5.Calattini, S., E. Betsem, S. Bassot, S. A. Chevalier, R. Mahieux, A. Froment, and A. Gessain. 1999. A new HTLV-3 strain in a Pygmy from Cameroon with a peculiar HTLV serology. J. Infect. Dis. 199561-564. [DOI] [PubMed] [Google Scholar]
- 6.Calattini, S., S. A. Chevalier, R. Duprez, P. Afonso, A. Froment, A. Gessain, and R. Mahieux. 2006. Human T-cell lymphotropic virus type 3: complete nucleotide sequence and characterization of the human tax3 protein. J. Virol. 809876-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calattini, S., S. A. Chevalier, R. Duprez, S. Bassot, A. Froment, R. Mahieux, and A. Gessain. 2005. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in Central Africa. Retrovirology 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevalier, S. A., N. L. Ko, S. Calattini, A. Mallet, M. C. Prevost, K. Kehn, J. N. Brady, F. Kashanchi, A. Gessain, and R. Mahieux. 2008. Construction and characterization of a human T-cell lymphotropic virus type 3 infectious molecular clone. J. Virol. 826747-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevalier, S. A., M. Walic, S. Calattini, A. Mallet, M. C. Prevost, A. Gessain, and R. Mahieux. 2007. Construction and characterization of a full-length infectious simian T-cell lymphotropic virus type 3 molecular clone. J. Virol. 816276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey, R. A., Y. Zuo, and J. M. Cunningham. 1999. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J. Virol. 733758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delamarre, L., A. R. Rosenberg, C. Pique, D. Pham, and M. C. Dokhelar. 1997. A novel human T-leukemia virus type 1 cell-to-cell transmission assay permits definition of SU glycoprotein amino acids important for infectivity. J. Virol. 71259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dooneief, G., R. Marlink, K. Bell, K. Marder, B. Renjifo, Y. Stern, and R. Mayeux. 1996. Neurologic consequences of HTLV-II infection in injection-drug users. Neurology 461556-1560. [DOI] [PubMed] [Google Scholar]
- 13.Feuer, G., and P. L. Green. 2005. Comparative biology of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2. Oncogene 245996-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gemeniano, M., O. Mpanju, D. R. Salomon, M. V. Eiden, and C. A. Wilson. 2006. The infectivity and host range of the ecotropic porcine endogenous retrovirus, PERV-C, is modulated by residues in the C-terminal region of its surface envelope protein. Virology 346108-117. [DOI] [PubMed] [Google Scholar]
- 15.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de The. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii407-410. [DOI] [PubMed] [Google Scholar]
- 16.Ghez, D., Y. Lepelletier, S. Lambert, J. M. Fourneau, V. Blot, S. Janvier, B. Arnulf, P. M. van Endert, N. Heveker, C. Pique, and O. Hermine. 2006. Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. J. Virol. 806844-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goubau, P., M. Van Brussel, A. M. Vandamme, H. F. Liu, and J. Desmyter. 1994. A primate T-lymphotropic virus, PTLV-L, different from human T-lymphotropic viruses types I and II, in a wild-caught baboon (Papio hamadryas). Proc. Natl. Acad. Sci. USA 912848-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heard, J. M., and O. Danos. 1991. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J. Virol. 654026-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hishizawa, M., K. Imada, T. Kitawaki, M. Ueda, N. Kadowaki, and T. Uchiyama. 2004. Depletion and impaired interferon-alpha-producing capacity of blood plasmacytoid dendritic cells in human T-cell leukaemia virus type I-infected individuals. Br. J. Haematol. 125568-575. [DOI] [PubMed] [Google Scholar]
- 20.Hjelle, B., O. Appenzeller, R. Mills, S. Alexander, N. Torrez-Martinez, R. Jahnke, and G. Ross. 1992. Chronic neurodegenerative disease associated with HTLV-II infection. Lancet 339645-646. [DOI] [PubMed] [Google Scholar]
- 21.Ijichi, S., M. B. Ramundo, H. Takahashi, and W. W. Hall. 1992. In vivo cellular tropism of human T cell leukemia virus type II (HTLV-II). J. Exp. Med. 176293-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jassal, S. R., R. G. Pohler, and D. W. Brighty. 2001. Human T-cell leukemia virus type 1 receptor expression among syncytium-resistant cell lines revealed by a novel surface glycoprotein-immunoadhesin. J. Virol. 758317-8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, Q., L. Agrawal, Z. Vanhorn-Ali, and G. Alkhatib. 2006. GLUT-1-independent infection of the glioblastoma/astroglioma U87 cells by the human T cell leukemia virus type 1. Virology 35399-110. [DOI] [PubMed] [Google Scholar]
- 24.Johnston, E. R., L. M. Albritton, and K. Radke. 2002. Envelope proteins containing single amino acid substitutions support a structural model of the receptor-binding domain of bovine leukemia virus surface protein. J. Virol. 7610861-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, K. S., S. Akel, C. Petrow-Sadowski, Y. Huang, D. C. Bertolette, and F. W. Ruscetti. 2005. Induction of human T cell leukemia virus type I receptors on quiescent naive T lymphocytes by TGF-beta. J. Immunol. 1744262-4270. [DOI] [PubMed] [Google Scholar]
- 26.Jones, K. S., K. Fugo, C. Petrow-Sadowski, Y. Huang, D. C. Bertolette, I. Lisinski, S. W. Cushman, S. Jacobson, and F. W. Ruscetti. 2006. Human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 use different receptor complexes to enter T cells. J. Virol. 808291-8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, K. S., M. Nath, C. Petrow-Sadowski, A. C. Baines, M. Dambach, Y. Huang, and F. W. Ruscetti. 2002. Similar regulation of cell surface human T-cell leukemia virus type 1 (HTLV-1) surface binding proteins in cells highly and poorly transduced by HTLV-1-pseudotyped virions. J. Virol. 7612723-12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, K. S., C. Petrow-Sadowski, D. C. Bertolette, Y. Huang, and F. W. Ruscetti. 2005. Heparan sulfate proteoglycans mediate attachment and entry of human T-cell leukemia virus type 1 virions into CD4+ T cells. J. Virol. 7912692-12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, K. S., C. Petrow-Sadowski, Y. K. Huang, D. C. Bertolette, and F. W. Ruscetti. 2008. Cell-free HTLV-1 infects dendritic cells leading to transmission and transformation of CD4(+) T cells. Nat. Med. 14429-436. [DOI] [PubMed] [Google Scholar]
- 30.Kalyanaraman, V. S., M. G. Sarngadharan, M. Robert-Guroff, I. Miyoshi, D. Golde, and R. C. Gallo. 1982. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science 218571-573. [DOI] [PubMed] [Google Scholar]
- 31.Kim, F. J., N. Manel, E. N. Garrido, C. Valle, M. Sitbon, and J. L. Battini. 2004. HTLV-1 and -2 envelope SU subdomains and critical determinants in receptor binding. Retrovirology 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, F. J., I. Seiliez, C. Denesvre, D. Lavillette, F. L. Cosset, and M. Sitbon. 2000. Definition of an amino-terminal domain of the human T-cell leukemia virus type 1 envelope surface unit that extends the fusogenic range of an ecotropic murine leukemia virus. J. Biol. Chem. 27523417-23420. [DOI] [PubMed] [Google Scholar]
- 33.Koyanagi, Y., Y. Itoyama, N. Nakamura, K. Takamatsu, J. Kira, T. Iwamasa, I. Goto, and N. Yamamoto. 1993. In vivo infection of human T-cell leukemia virus type I in non-T cells. Virology 19625-33. [DOI] [PubMed] [Google Scholar]
- 34.Lambert, S., M. Bouttier, R. Vassy, M. Seigneuret, C. Petrow-Sadowski, S. Janvier, N. Heveker, F. W. Ruscetti, G. Perret, K. S. Jones, and C. Pique. 6 March 2009. HTLV-1 uses HSPG and neuropilin 1 for entry by molecular mimicry of VEGF165. Blood [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 35.Liegeois, F., B. Lafay, W. M. Switzer, S. Locatelli, E. Mpoudi-Ngole, S. Loul, W. Heneine, E. Delaporte, and M. Peeters. 2008. Identification and molecular characterization of new STLV-1 and STLV-3 strains in wild-caught nonhuman primates in Cameroon. Virology 371405-417. [DOI] [PubMed] [Google Scholar]
- 36.Macatonia, S. E., J. K. Cruickshank, P. Rudge, and S. C. Knight. 1992. Dendritic cells from patients with tropical spastic paraparesis are infected with HTLV-1 and stimulate autologous lymphocyte proliferation. AIDS Res. Hum. Retrovir. 81699-1706. [DOI] [PubMed] [Google Scholar]
- 37.Mahieux, R., and A. Gessain. 2008. The human HTLV-3 and HTLV-4 retroviruses: new members of the HTLV family. Pathol. Biol. (Paris) 57161-166. [DOI] [PubMed] [Google Scholar]
- 38.Mahieux, R., J. Pecon-Slattery, and A. Gessain. 1997. Molecular characterization and phylogenetic analyses of a new, highly divergent simian T-cell lymphotropic virus type 1 (STLV-1marc1) in Macaca arctoides. J. Virol. 716253-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manel, N., F. J. Kim, S. Kinet, N. Taylor, M. Sitbon, and J. L. Battini. 2003. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell 115449-459. [DOI] [PubMed] [Google Scholar]
- 40.Manel, N., S. Kinet, J. L. Battini, F. J. Kim, N. Taylor, and M. Sitbon. 2003. The HTLV receptor is an early T-cell activation marker whose expression requires de novo protein synthesis. Blood 1011913-1918. [DOI] [PubMed] [Google Scholar]
- 41.Nath, M. D., F. W. Ruscetti, C. Petrow-Sadowski, and K. S. Jones. 2003. Regulation of the cell-surface expression of an HTLV-I binding protein in human T cells during immune activation. Blood 1013085-3092. [DOI] [PubMed] [Google Scholar]
- 42.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i1031-1032. [DOI] [PubMed] [Google Scholar]
- 43.Persaud, D., J. L. Munoz, S. L. Tarsis, E. S. Parks, and W. P. Parks. 1995. Time course and cytokine dependence of human T-cell lymphotropic virus type 1 T-lymphocyte transformation as revealed by a microtiter infectivity assay. J. Virol. 696297-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinon, J. D., P. J. Klasse, S. R. Jassal, S. Welson, J. Weber, D. W. Brighty, and Q. J. Sattentau. 2003. Human T-cell leukemia virus type 1 envelope glycoprotein gp46 interacts with cell surface heparan sulfate proteoglycans. J. Virol. 779922-9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 777415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson, J. H., A. J. Edwards, J. K. Cruickshank, P. Rudge, and A. G. Dalgleish. 1990. In vivo cellular tropism of human T-cell leukemia virus type 1. J. Virol. 645682-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenberg, A. R., L. Delamarre, A. Preira, and M. C. Dokhelar. 1998. Analysis of functional conservation in the surface and transmembrane glycoprotein subunits of human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2. J. Virol. 727609-7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarris, M., K. G. Andersen, F. Randow, L. Mayr, and A. G. Betz. 2008. Neuropilin-1 expression on regulatory T cells enhances their interactions with dendritic cells during antigen recognition. Immunity 28402-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takenouchi, N., K. S. Jones, I. Lisinski, K. Fugo, K. Yao, S. W. Cushman, F. W. Ruscetti, and S. Jacobson. 2007. GLUT1 is not the primary binding receptor but is associated with cell-to-cell transmission of human T-cell leukemia virus type 1. J. Virol. 811506-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vander Kooi, C. W., M. A. Jusino, B. Perman, D. B. Neau, H. D. Bellamy, and D. J. Leahy. 2007. Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Natl. Acad. Sci. USA 1046152-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Dooren, S., L. Meertens, P. Lemey, A. Gessain, and A. M. Vandamme. 2005. Full-genome analysis of a highly divergent simian T-cell lymphotropic virus type 1 strain in Macaca arctoides. J. Gen. Virol. 861953-1959. [DOI] [PubMed] [Google Scholar]
- 52.von Wronski, M. A., N. Raju, R. Pillai, N. J. Bogdan, E. R. Marinelli, P. Nanjappan, K. Ramalingam, T. Arunachalam, S. Eaton, K. E. Linder, F. Yan, S. Pochon, M. F. Tweedle, and A. D. Nunn. 2006. Tuftsin binds neuropilin-1 through a sequence similar to that encoded by exon 8 of vascular endothelial growth factor. J. Biol. Chem. 2815702-5710. [DOI] [PubMed] [Google Scholar]
- 53.Wolfe, N. D., W. Heneine, J. K. Carr, A. D. Garcia, V. Shanmugam, U. Tamoufe, J. N. Torimiro, A. T. Prosser, M. Lebreton, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. M. Switzer. 2005. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc. Natl. Acad. Sci. USA 1027994-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie, L., and P. L. Green. 2005. Envelope is a major viral determinant of the distinct in vitro cellular transformation tropism of human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2. J. Virol. 7914536-14545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 792031-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Y., J. C. Rassa, M. E. deObaldia, L. M. Albritton, and S. R. Ross. 2003. Identification of the receptor binding domain of the mouse mammary tumor virus envelope protein. J. Virol. 7710468-10478. [DOI] [PMC free article] [PubMed] [Google Scholar]