Abstract
The adenovirus type 5 (Ad5) late region 4 (L4) 100-kDa nonstructural protein (L4-100K) mediates inhibition of cellular protein synthesis and selective translation of tripartite leader (TL)-containing viral late mRNAs via ribosome shunting. In addition, L4-100K has been implicated in the trimerization and nuclear localization of hexon protein. We previously proved that L4-100K is a substrate of the protein arginine methylation machinery, an emergent posttranslational modification system involved in a growing list of cellular processes, including transcriptional regulation, cell signaling, RNA processing, and DNA repair. As understood at present, L4-100K arginine methylation involves protein arginine methyltransferase 1 (PRMT1), which asymmetrically dimethylates arginines embedded in arginine-glycine-glycine (RGG) or glycine-arginine-rich (GAR) domains. To identify the methylated arginine residues and assess the role of L4-100K arginine methylation, we generated amino acid substitution mutations in the RGG and GAR motifs to examine their effects in Ad-infected and plasmid-transfected cells. Arginine-to-glycine exchanges in the RGG boxes significantly diminished L4-100K methylation in the course of an infection and substantially reduced virus growth, demonstrating that L4-100K methylation in RGG motifs is an important host cell function required for efficient Ad replication. Our data further indicate that PRMT1-catalyzed arginine methylation in the RGG boxes regulates the binding of L4-100K to hexon and promotes the capsid assembly of the structural protein as well as modulating TL-mRNA interaction. Furthermore, substitutions in GAR, but not RGG, regions affected L4-100K nuclear import, implying that the nuclear localization signal of L4-100K is located within the GAR sequence.
With the onset of the late phase, one of the first adenovirus type 5 (Ad5) late proteins translated, the late region 4 (L4) 100-kDa protein (L4-100K), starts to perform a number of functions that are essential for efficient completion of lytic virus infection. This Ad nonstructural late protein alters the cellular machinery in favor of translating large amounts of virus products, leading to their subsequent nuclear accumulation for capsid assembly. L4-100K achieves this by contributing to the transport and selective translation of late viral mRNAs (12, 13, 21), acting as a chaperone for hexon trimerization and being involved in its transport (8, 9, 24) and also playing a role in preventing apoptosis of the infected cell by interacting with granzyme B and inhibiting its activity (1). However, most of the mechanisms underlying these processes, and how L4-100K is regulated to accomplish these, remain unclear.
One of the most striking features of L4-100K is promoting viral mRNA translation through ribosome shunting and preventing cellular mRNA translation by eliminating the cap-dependent translation pathway (12, 13, 15, 21, 49-51). Underlying these processes is the interaction of L4-100K with both the tripartite leader (TL) sequence possessed by all the late viral transcripts and the scaffolding element of the cap-dependent translation initiation complex, eukaryotic initiation factor 4G (eIF4G) (13, 49). Recently it was shown that L4-100K is posttranslationally modified to confer selective binding to virus-specific mRNAs although it has a general RNA binding motif, and this modification was reported to be tyrosine phosphorylation (49). Tyrosine phosphorylation of L4-100K was considered essential for efficient ribosome shunting and late protein synthesis but not to be involved in eIF4G binding (49).
In the late phase, L4-100K interacts with hexon monomers, which are the major components of the Ad capsid, to form their trimeric structure (8, 9, 24). L4-100K not only acts as a chaperone for hexon trimerization but also assists their nuclear transport and capsid assembly (35). It was reported that in the cytoplasm L4-100K can associate both with trimeric and monomeric hexons but that in the nucleus it interacts only with trimers (24), suggesting that only the trimeric hexons can be transported in the nucleus.
As a multifunctional shuttling phosphoprotein, L4-100K might have more modification sites that allow regulation of its functions and interactions mentioned above. Indeed, our group showed the arginine methylation of L4-100K by protein arginine methyltransferase 1 (PRMT1) (29), meaning that L4-100K was the first Ad protein reported to be methylated. Arginine methylation of proteins is a posttranslational modification mediated by ubiquitously expressed PRMTs, which utilize S-adenosylmethionine to yield mono- or dimethylated arginine residues (3, 17, 22). This modification affects major processes in the cell, such as protein-protein interactions, regulation of transcription, RNA processing, DNA repair, nucleocytoplasmic shuttling of proteins, and protein stability (reviewed in references 2 and 3). Following the discovery of protein arginine methylation, many proteins were found to be modified by these enzymes, including fibrillarin, nucleolin, GTPase P, myelin basic proteins, myosin, heat shock proteins, and H3 histones (reviewed in references 11 and 31). Recent findings indicated the significance of methylation not only for cellular proteins but also for viral transcripts. The human immunodeficiency virus (HIV) transactivator protein was identified as the first HIV protein to contain methylated arginines (6). Inhibition of methylation was reported to increase HIV gene expression, and the authors suggested that increasing this modification might provide protection against HIV infection. Conversely, blocking of methylation was suggested as protection against hepatitis delta virus, since inhibition of this modification was found to prevent hepatitis delta virus replication (31). Also, herpes simplex virus RNA binding protein ICP27 was reported to be methylated in its RNA binding domain (33).
Interestingly, all substrates of type I PRMTs were found to be methylated at a common Arg-Gly-Gly (RGG) motif and/or in a Gly-Arg-rich (GAR) sequence. Since L4-100K possesses three RGG domains in its C terminus, we investigated whether this protein is methylated by PRMT1 at these arginine residues. We therefore constructed a mutant virus with three arginine residues replaced by glycines in the C-terminal RGG boxes of L4-100K and investigated the properties of this mutant protein in the course of an infection. This mutant exhibited a severely reduced L4-100K methylation pattern. We examined the effects of such inefficient methylation on subcellular localization, protein-protein/RNA interactions, and virus growth. We suggest that triple RGG boxes at the C terminus are the major but not the only target for this modification and that methylation of L4-100K in these boxes plays no role in subcellular localization but has regulatory effects on the functions of this protein, especially in modulating protein-protein/RNA interactions, and is necessary for efficient completion of the late phase to yield large amounts of virus particles.
MATERIALS AND METHODS
Cells and viruses.
A549 cells (DSMZ ACC 107), H1299 cells (34), and K16 cells (23) (kindly provided by A. Amalfitano) were maintained as monolayers in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS) and 100 units of penicillin and 100 μg of streptomycin/ml. Primary human hepatocytes, which were kindly provided by Thomas Weiss (University of Regensburg Hospital, Regensburg, Germany), were cultured in growth medium containing 125 mU/ml insulin, 60 ng/ml hydrocortisone, and 10 ng/ml glucagon.
H5pg4100 was used as the wild-type (wt) Ad5 in these studies (19, 27). Construction of the L4-100K mutant virus H5pm4151 is described below. The viruses were propagated in K16 monolayer cultures. Infections were performed in one-fifth of the normal culture volume in DMEM for 2 h at 37°C with gentle rocking every 10 min. The virus suspension was then replaced with normal growth medium. At 3 to 5 days postinfection, infected cells and supernatants were harvested and freeze-thawed four times to release progeny virions. Virus was harvested by sequential centrifugation in discontinuous and equilibrium cesium chloride gradients exactly as described previously (47). The titers of the viruses used in this study were determined by a fluorescent-focus assay. Confluent cells grown in 35-mm-diameter dishes were infected with serial 10-fold dilutions of virus in Ad infection medium (phosphate-buffered saline [PBS] supplemented with 2 mM MgCl2, 0.2 mM CaCl2, and 2% fetal calf serum). At 24 h after infection, cells were washed twice with PBS and fixed in ice-cold methanol for 15 min at −20°C, followed by two washes with TBS-BG buffer (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 0.05% Tween 20, 0.05% sodium azide, 5 mg/ml glycerol, and 5 mg/ml bovine serum albumin). Primary mouse monoclonal antibody (MAb) B6-8, specific for the viral E2A-72K protein, was added to the cells at a 1:10 dilution in TBS-BG, and the cells were incubated at room temperature for 2 h. The cells were washed twice in TBS-BG, and Alexa 488-goat anti-mouse immunoglobulin G (IgG) (Molecular Probes) diluted at 1:1,000 was added and left for 2 h at room temperature. The cells were washed twice with TBS-BG, and the titer (fluorescence-forming units [FFU]) was calculated on the basis of the average number of fluorescing cells per 35-mm-diameter dish determined with a Zeiss Axiovert inverted microscope.
To measure virus growth, infected cells were harvested at 48 h and 72 h postinfection and lysed by four cycles of freeze-thawing. The cell lysates were serially diluted in Ad infection medium for infection of K16 cells, and virus yield was determined by quantitative E2A-72K immunofluorescence staining at 24 h after infection as described above. Viral DNA replication was determined by quantitative PCR exactly as described previously (42). PCR products were analyzed on a 1% agarose gel and quantified using the ChemiDoc system and QuantityOne software (Bio-Rad).
Construction of mutant virus H5pm4151.
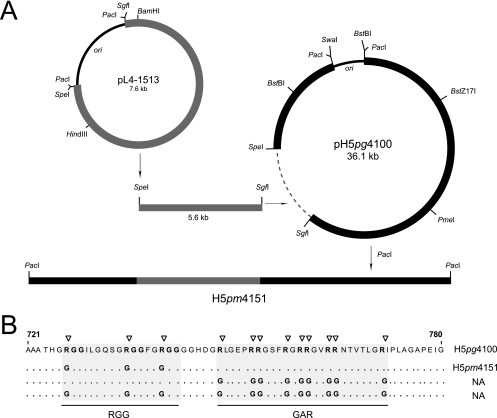
To generate the Ad5 mutant carrying defined amino acid changes in the RGG boxes of L4-100K (Fig. 1B), point mutations were first introduced into the L4-100K gene at nucleotides 26239, 26266, and 26281 (nucleotide numbering is according to the published Ad5 sequence from GenBank, accession no. AY339865) in pL4-1513 by site-directed mutagenesis with oligonucleotide primers 1266 and 1267 (Table 1), resulting in pL4-1599. This plasmid carries three arginine-to-glycine substitutions at amino acid positions 727, 736, and 741 (Fig. 1B). The mutations in the plasmid were verified by DNA sequencing. Finally, the 5.6-kb SpeI/SgfI fragment from pH 5pg4100 was replaced with the corresponding fragment from plasmid pL4-1599 to generate Ad plasmid pH 5pm4151. The recombinant plasmid was partially sequenced to confirm the mutations in the L4-100K open reading frame.
FIG. 1.
(A) Generation of L4-100K mutant virus H5pm4151. Point mutations were introduced into the L4-100K gene in pL4-1513 by site-directed mutagenesis. The L4 plasmid and bacmid pH5pg4100 were linearized with SgfI plus SpeI, and the 5.6-kb SgfI/SpeI fragment from pH 5pg4100 was replaced with the corresponding fragment from the mutated pL4 construct. The viral genome was released by PacI digestion and used for transfection of the L4-100K-complementing cell line K16. Viruses were then isolated and propagated using K16 cells. (B) The positions of the amino acid changes within the L4-100K mutants are marked by triangles. Numbers refer to amino acid residues in the wt L4-100K protein from H5pg4100. NA, not applicable.
TABLE 1.
Primers
| Primer | Sequence |
|---|---|
| 1266 | 5′-GCCACCCACGGAGGAGGAGGAATACTGGGACAGTCAGGCGGAGGAGGTTTTGGAGGAGGAGGAGGAGGACATGATGG-3′ |
| 1267 | 5′-CCATCATGTCCTCCTCCTCCTCCTCCAAAACCTCCTCCGCCTGACTGTCCCAGTATTCCTCCTCCTCCGTGGGTGGC-3′ |
| 1330 | 5′-GGACATGATGGAGGACTGGGAGAGCCTGGAGGAGGAAGCTTCGGAGGTCGAAGAGG-3′ |
| 1331 | 5′-CCTCTTCGACCTCCGAAGCTTCCTCCTCCAGGCTCTCCCAGTCCTCCATCATGTCC-3′ |
| 1332 | 5′-GCTTCGGAGGTGGAGGAGGTGTCGGAGGAAACACCGTCACCCTCGGTGGCATTCCCCTCGC-3′ |
| 1333 | 5′-GCGAGGGGAATGCCACCGAGGGTGACGGTGTTTCCTCCGACACCTCCTCCACCTCCGAAGC-3′ |
| 1222 | 5′-CCAACGTGGAGGCCTGCAACGCGGTCTCCTACGCTGGAGCTTTGCACGAAAACCGC-3′ |
| 1223 | 5′-GCGGTTTTCGTGCAAAGCTCCAGCGTAGGAGACCGCGTTGCAGGCCTCCACGTTGG-3′ |
| 1161 | 5′-AAGCTAGCCCTGCAAACATCA-3′ |
| 1162 | 5′-CCCAAGCTACCAGTGGCAGTA-3′ |
| 1204 | 5′-CATCCTGGGCTACACTGA-3′ |
| 1205 | 5′-TTGACAAAGTGGTCGTTG-3′ |
| 1369 | 5′-ACTCTCTTCCGCATCGCTGTC-3′ |
| 1370 | 5′-GCGACTGTGACTGGTTAGACG-3′ |
For the generation of mutant virus H5pm4151, the viral genome was released from the recombinant plasmids by PacI digestion. Five micrograms of viral DNA was used to transfect 3 × 105 complementing K16 cells by the calcium phosphate procedure (18). After 5 days, cells were harvested and viruses were released by three cycles of freezing and thawing. The viruses were propagated in K16 monolayer cells and purified as described above. Viral DNA was isolated from viral particles as described previously (44) and analyzed by HindIII restriction endonuclease digestion. In addition, the viral DNA was partially sequenced to verify the presence of the inserted mutations.
Antibodies, protein analysis, and inhibitors.
Primary antibodies specific for Ad proteins used in this study included L4-100K rat MAb 6B10 (29), E2A-72K (DBP) mouse MAb B6-8 (40), E1B-55K mouse MAb 2A6 (43), Ad5 hexon polyclonal rabbit antibody (Abcam), and Ad5 rabbit polyclonal serum L133 (27). Primary antibodies specific for cellular proteins included anti-PRMT1 rabbit polyclonal antibody (Upstate); anti-β-actin (Sigma); anti-SAF-A rabbit polyclonal antibody (22); anti-eIF4G rabbit polyclonal antibody (7); Asym-24 rabbit polyclonal antibody (Upstate), which recognizes asymmetrically dimethylated arginines; and mouse MAb Ab412 (Abcam), which recognizes methylated/asymmetrically dimethylated arginines.
Secondary antibodies conjugated to horseradish peroxidase for detection of proteins by immunoblotting were donkey anti-rabbit IgG, sheep anti-mouse IgG, and goat anti-rat IgG (Amersham Biosciences). Fluorescent secondary antibodies were affinity-purified fluorescein isothiocyanate- or Texas red-conjugated donkey anti-mouse, anti-rat, and anti-rabbit IgGs (Dianova). These were used at a 1:100 dilution in all immunofluorescence experiments.
For the analysis of proteins, total cell extracts were prepared from mock- or Ad-infected cells at the indicated times and washed two times in ice-cold PBS. The cells were resuspended in NP-40 (Nonidet P-40) 1% lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 1% NP-40) or in RIPA light (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0,1% sodium dodecyl sulfate [SDS], and 0,1% Triton X-100) supplemented with 1 mM dithiothreitol (DTT) and a protease inhibitor cocktail (Roche). After 1 h on ice, the lysates were sonicated and the insoluble debris was pelleted at 15,000 × g at 4°C.
For immunoblotting, equal amounts of total protein were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Schleicher & Schuell). Membranes were incubated overnight in PBS-0.1% Tween 20 containing 5% nonfat dry milk and then for 2 h in PBS-1% nonfat dry milk containing the appropriate primary antibody. Membranes were washed three times in PBS-0.1% Tween 20, incubated with a secondary antibody linked to horseradish peroxidase (Amersham) in PBS-0.1% Tween 20, and washed three times in PBS-0.1% Tween 20. The bands were visualized by enhanced chemiluminescence as recommended by the manufacturer (Pierce) on X-ray films (CL-XPosure [Pierce] or Kodak X-Omat AR). Autoradiograms were scanned and cropped using Adobe Photoshop CS2, and figures were prepared using Adobe Illustrator CS2 software.
For immunoprecipitations, 500 μg of protein samples was precleared with either protein A- or G-Sepharose and incubated overnight with MAbs bound to protein A/G-Sepharose. The immunocomplexes were washed five times with RIPA light lysis buffer, separated by SDS-PAGE, and analyzed by immunoblotting.
For the inhibition of total methylation reactions in the cell, AdOx (Adenosine, periodate oxidized [Sigma]) was added to the replacement medium after infection. For time course analysis, the medium was replaced every 24 h with fresh AdOx containing medium. Leptomycin B (LMB) was purchased from Biomol and used to block the CRM1-dependent nuclear export pathway.
Cell fractionation.
Nucleocytoplasmic fractionation was performed using the Sigma CelLytic nuclear extraction kit (Sigma) according to the manufacturer's protocol. Cells were collected 36 h after mock or virus infection and washed with PBS once. Cell pellets were incubated in the hypotonic lysis buffer for 15 min on ice. Samples were centrifuged at 450 × g, and the supernatant was removed. Resuspended cells were then passed through a 27-gauge needle seven times. After centrifugation of the lysate at 11,000 × g for 20 min at 4°C, the supernatant was collected as the cytoplasmic fraction. Pellets were further incubated with the extraction buffer for 30 min at 4°C with agitation. Samples were then centrifuged at 21,000 × g for 5 min, and the supernatant was collected as the nuclear fraction.
Immunoprecipitation of RNA.
RNA was coimmunoprecipitated from infected and noninfected cells by using rat 6B10 MAb as described previously (16, 50). A549 cells (3 × 106) were collected 48 h after infection, washed with cold PBS, and resuspended in two pellet volumes of polysome lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM HEPES (pH 7.0), and 0.5% Nonidet P-40 supplemented with 1 mM DTT, 100 units/ml RNase OUT, 0.2 mM phenylmethylsulfonyl fluoride, 1 mg/ml pepstatin A, 5 mg/ml bestatin, and 20 mg/ml leupeptin (16). Resuspended cells were incubated on ice for 5 min and then centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was used for the further immunoprecipitation procedures. Protein G-Sepharose beads were swollen in NT2 buffer containing 50 mM Tris-Cl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, and 0.5% Nonidet P-40 supplemented with 5% bovine serum albumin (16) for 1 h at 4°C and further incubated with MAb 6B10 for 1 h at 4°C. Antibody-coupled beads were then washed twice with ice-cold NT2 buffer and resuspended in NT2 buffer supplemented with 100 units/ml RNase OUT and 1 mM DTT. Half of the cell lysate containing mRNPs was incubated with the antibody-coupled beads overnight at 4°C. Beads were then washed six times with ice-cold NT2 buffer. Washed beads were resuspended in 100 μl NT2 buffer supplemented with 0.1% SDS and 30 μg proteinase K and incubated for 30 min at 55°C. The immunoprecipitated mRNA was isolated by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. Isolated RNA was quantified by real-time PCR as described below.
Quantitative real-time PCR.
A549 cells (3 × 106) were infected with H5pg4100 and H5pm4151. At the indicated times, total RNA from cells was purified using an RNeasy Mini Kit (Qiagen) or precipitated as described above and reverse transcribed using SuperScriptIII First Strand (Invitrogen) plus oligo(dT) primers (Invitrogen). Real-time PCR was performed with a Rotor-Gene 6000 instrument (Corbett Life Sciences, Sydney, Australia) with Sybr green PCR master mix (Applied Biosystems). The PCR conditions were 10 min at 95°C and 55 cycles of 10 s at 95°C, 30 s at 60°C, and 20 s at 72°C. The average threshold cycle value was determined from triplicate reactions, and levels of the TL and fiber mRNAs relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were calculated as described previously (20). For the quantification of coimmunoprecipitated mRNA, threshold cycle values of these reactions were normalized with total TL and fiber RNA. Primers 1161, 1162, 1204, 1205, 1369, and 1370 for the quantitative real-time PCR are listed in Table 1.
Plasmids and transient transfections.
All plasmids used in this study express Ad2 wt and mutant L4-100K proteins under the control of the cytomegalovirus major immediate-early promoter from vector pcDNA3 (Invitrogen). Plasmid pTL-flag-100K (generously provided by R. Schneider, New York University of Medicine) produces Flag-tagged wt L4-100K translated from the TL (50). Plasmids pTL-100K-RGG, pTL-100K-GAR, pTL-100K-RGG.GAR, pTL-100K-RGG3, and pTL-100K-NES were derived from pTL-flag L4-100K by site-directed mutagenesis with oligonucleotide primers 1266, 1267, 1330, 1331, 1332, 1333, 1472, 1473, 1222, and 1223 (Table 1) and directly expressed L4-100K containing amino acid changes in the RGG boxes, GAR region, both segments, and the nuclear export signal (NES) (see Fig. 5). The mutations were confirmed by DNA sequencing. H1299 cells (2.5 × 106) seeded in 10-cm-diameter dishes were transfected with 10 μg of plasmids by polyethylenimine (PEI) (25 kDa, linear; Polysciences) transfection. Fifty micrograms of PEI was added to the DNA dissolved in 600 μl DMEM and kept at room temperature for 10 min. The DNA-PEI mixture was added to the cells seeded in DMEM without antibiotics. At 6 hours posttransfection, the medium was replaced with normal growth medium.
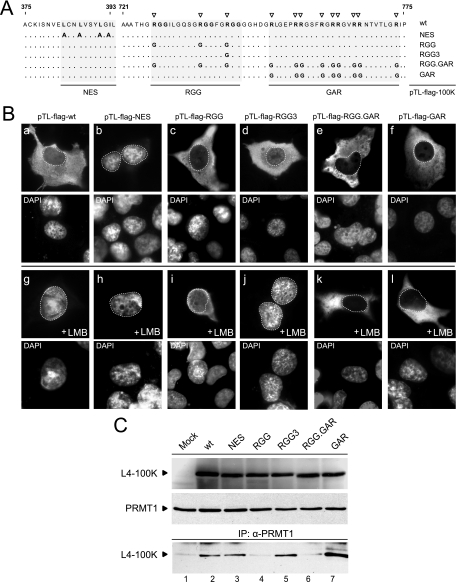
FIG. 5.
Effect of amino acid changes on subcellular distribution of L4-100K mutant proteins. (A) Amino acid substitution mutations in L4-100K wt and mutant plasmids. The leucine residues in the L4-100K NES (pTL-100K-NES) known to be critical for CRM1-mediated nuclear transport (13) are indicated in bold. Numbers refer to amino acids residues in the wt L4-100K protein from pTL-100K. Amino acid changes in L4 proteins from pTL-100K-RGG, pTL-100K-RGG3, pTL-100K-RGG.GAR, and pTL-100K-GAR are indicated below. (B) Subcellular distribution of wt and mutant L4 proteins in the absence or presence of the CRM1 inhibitor LMB. H1299 cells were transfected with the indicated plasmids. At 36 h after transfection, LMB was added to the medium (+LMB). Three hours later, cells were fixed and steady-state localization of wt and mutant L4-100K was examined by indirect immunofluorescence using MAb 6B10. In all panels nuclei are visualized by DAPI staining and are indicated by a dotted line. Magnification, ×7,600. (C) H1299 cells were transfected with L4-100K wt and mutant plasmids and harvested at 36 h after transfection. Fifty-microgram aliquots from total cell lysates were separated by SDS-10% PAGE, and proteins were visualized with 6B10 and anti-PRMT1 antibody (upper two panels). Alternatively, 500-μg aliquots were used for immunoprecipitation with anti-PRMT1 antibody, and coprecipitated L4-100K proteins were visualized with MAb 6B10.
Indirect immunofluorescence.
A549 and H1299 cells were grown on glass coverslips and infected or transfected as described above. At the indicated times, cells were fixed with ice-cold methanol for 15 min at −20°C and stained with antibodies and 4′-6-diamidino-2-phenylindole (DAPI) as described previously (16). Coverslips were mounted in Glow medium (Energene), and digital images were acquired on a DMRB fluorescence microscope (Leica) with a charge-coupled device camera (Leica). These images were cropped and decoded with Photoshop CS2 (Adobe Systems) and assembled with Illustrator CS2 (Adobe Systems). Nuclei were identified by DAPI staining, and nuclear borders were digitally added as dotted lines.
RESULTS
Construction of L4-100K mutant virus H5pm4151.
Figure 1A outlines our strategy for constructing the Ad5 L4-100K mutant virus H5pm4151. This involved two recombinant plasmids containing Ad5 L4 from nucleotide 21438 to 27081 (pL4-1513) or the Ad5 wt genome lacking a portion of E3 (pH5pg4100) (19). Plasmid pL4-1513 was used to introduce mutations into the L4-100K gene by site-directed mutagenesis, while plasmid pH5pg4100 served as the DNA backbone to recombine the mutations into the Ad5 chromosome by direct cloning. To generate L4-100K mutant virus, infectious viral DNA was released from the recombinant plasmid by PacI digestion and transfected into the L4-100K-complementing cell line K16 (23). Viral progenies were isolated and amplified, and viral DNA was analyzed by restriction digestion and DNA sequencing. The amino acid exchanges introduced into the RGG boxes of the Ad5 L4-100K polypeptide are summarized in Fig. 1B.
Using the same approach, we also tried to generate an L4-100K mutant virus carrying arginine-to-glycine substitutions in the GAR region and a double mutant containing identical amino acid exchanges in both the RGG boxes and the GAR region (Fig. 1B, NA). Interestingly, neither mutant virus could be reproducibly grown on K16 cells after transfecting the viral DNA. The reason for these results is unclear, but they may be due to severe defects in the late functions of L4-100K that K16 cells, which typically express low levels of the wt L4-100K (23), are unable to complement.
Amino acid substitutions in the RGG boxes reduce arginine methylation of L4-100K during infection.
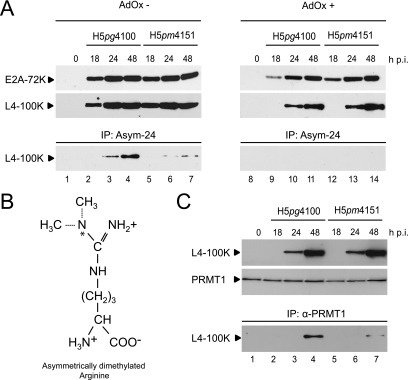
To test whether H5pm4151 produces a stable protein, we determined the steady-state concentration of L4-100K by immunoblot analysis using total cell lysates from infected A549 cells. In parallel we assessed the effect of amino acid exchanges on L4-100K arginine methylation in the absence or presence of the methylation inhibitor AdOx. Representative samples of these analyses are shown in Fig. 2. In the absence of AdOx, H5pm4151 accumulated L4-100K to levels comparable to those for wt H5pg4100 in the course of the infection (Fig. 2A). A similar result was obtained in the presence of the arginine methylation inhibitor, although expression of the viral protein was delayed under such conditions (Fig. 2A). This delay in viral protein production was also seen when extracts were analyzed for the expression of E2A-72K, an early Ad protein necessary for viral DNA replication. This confirms that arginine methylation is a critical cellular function required for efficient viral early and late gene expression, as previously observed (25, 29). Next, total cell lysates were subjected to immunoprecipitation using polyclonal antibody Asym-24, which reacts with asymmetrically dimethylated arginines (Fig. 2B). wt and mutant L4-100K proteins were then detected by immunoblotting (Fig. 2A) using the anti-L4-100K MAb 6B10. In line with our previous work (29) and a recent report (25), a substantial amount of L4-100K present in wt-infected cells was found to be methylated on arginine residues (Fig. 2A, lanes 3 and 4). In contrast, no L4-100K was detected in the immunoprecipitates from wt-infected cell extracts in the presence of AdOx (Fig. 2A, lanes 9 to 11), verifying the specificity of the polyclonal antibody Asym-24. Also as anticipated, the amount of methylated L4-100K was greatly decreased in H5pm4151-infected cells (Fig. 2A, lanes 5 to 7), demonstrating that the amino acid substitutions in the RGG boxes substantially reduce, but do not fully eliminate L4-100K arginine methylation. Consistent with this, no methylated form of the mutant protein was immunoprecipitated with Asym-24 in the presence of AdOx (Fig. 2A, lanes 12 to 14). This result is further supported by the observation that the amino acid exchanges in the RGG boxes greatly reduce binding of this Ad5 protein to cellular PRMT1 (Fig. 2C), which binds its targets through arginine residues clustered within in the context of RGG boxes or GAR regions (5, 39). Taken together, these data show that Ad5 L4-100K is methylated by PRMT1 at arginine residues mainly in the RGG boxes and hint at a possible partial methylation in the following GAR region.
FIG. 2.
Protein steady-state levels and L4-100K methylation in A549 cells infected with H5pg4100 and H5pm4151 viruses. (A) Cells were infected in the absence (AdOx−) or presence (AdOx+) of AdOx and harvested at the indicated times after infection (h p.i.). Total cell extracts were prepared from noninfected (0) and infected cells, and 25-μg aliquots of lysates were separated by SDS-10% PAGE and analyzed by immunoblotting using L4-100K rat MAb 6B10 or mouse MAb B6-8, recognizing the E2A-72K protein. The same lysates were used for immunoprecipitation (IP) with MAb Asym-24, detecting asymmetrically dimethylated arginine residues (B). The immunocomplexes were separated by SDS-10% PAGE and analyzed by immunoblotting using MAb 6B10. (C) Binding of PRMT1 to L4-100K in infected cells. Whole lysates were prepared as described above and used for immunoblotting with MAb 6B10 (upper panel) and immunoprecipitation with anti-PRMT1 MAb.
Arginine methylation of RGG motifs in L4-100K is required for maximal virus growth.
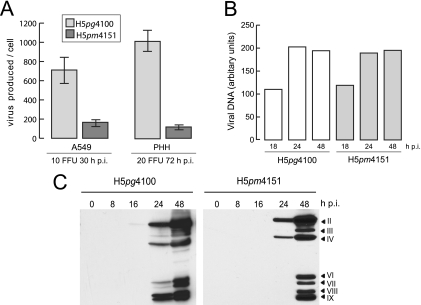
To evaluate the contribution of arginine methylation in the RGG boxes of L4-100K to overall virus growth properties, total virus yields were determined in infected A549 cells and primary human hepatocytes (Fig. 3A). Arginine substitutions in L4-100K resulted in a 6- to 10-fold reduction in virus growth, depending on the cell type. This defect could not be attributed to reductions in early viral protein synthesis or Ad DNA replication, since H5pm4151 accumulated E2A-72K (Fig. 2A) and viral DNA comparably to wt H5pg4100 (Fig. 3B). Rather, the decrease in virus production was directly related to levels of late viral proteins, since H5pm4151 showed substantially reduced hexon, penton, and particularly fiber protein expression (Fig. 3C). These data show that methylation of arginine residues in the context of RGG boxes contributes to efficient late protein production and maximal virus growth in infected A549 cells and primary human hepatocytes.
FIG. 3.
Effects of amino acid substitutions in L4-100K on virus growth, viral DNA replication, and late protein synthesis. (A) Virus yield. A549 cells and primary human hepatocytes (PHH) were infected with wt H5pg4100 or H5pm4151 at the indicated multiplicities. Viral particles were harvested at the indicated time points post infection (h p.i.), and virus yield was determined by quantitative E2A-72K immunofluorescence staining on K16 cells. The results represent the averages from three independent experiments. Error bars indicate the standard errors of the means. (B) Viral DNA accumulation. A549 cells were infected with H5pg4100 and H5pm4151 viruses at a multiplicity of 10 FFU per cell. Total nuclear DNA was isolated at the indicated times after infection and subjected to semiquantitative PCR. PCR products were analyzed and quantified using the ChemiDoc system and QuantityOne software (Bio-Rad). The results shown represent the averages from two independent experiments. (C) Viral late protein synthesis. A549 cells were infected with the wt or mutant virus at a multiplicity of 10 FFU per cell. Total cell extracts were prepared at the indicated times postinfection. Proteins (40-μg samples) were separated by SDS-10% PAGE, transferred to nitrocellulose membranes, and probed with the anti-Ad5 rabbit polyclonal serum L133. Bands corresponding to viral late proteins hexon (II), penton (III), fiber (IV), and minor capsid proteins (VI, VII, VIII, and IX) are indicated on the right.
The phenotype observed with the mutant virus H5pm4151 could be attributed to a number of possible defects in L4-100K, including altered subcellular localization, RNA binding, or protein-protein interactions. We therefore compared the L4-100K RGG mutant and the wt protein for nucleocytoplasmic shuttling activity and binding to TL-containing mRNA (TL-mRNA), eIF4G, and hexon.
Effect of amino acid substitutions in the RGG motifs on the subcellular distribution of L4-100K during infection.
Previous work has shown that L4-100K actively shuttles between the nuclear and cytoplasmic compartments in plasmid-transfected cells (13). Nucleocytoplasmic export of L4-100K is mediated by a leucine-rich NES (see Fig. 5A) of the HIV type 1 Rev type and can be blocked by point mutations within the L4-100K NES. Also, fragment deletion analysis has shown that the carboxy-terminal region of L4-100K (amino acids 727 to 807) contains a potential nuclear localization signal (13), raising the possibility that nuclear import of L4-100K is modulated by methylation of arginine residues within the RGG boxes and/or GAR region. Consistent with this observation, a recent study showed that the replacement of arginine and the glycine residues in the third RGG box (RGG3) by alanines in a plasmid construct eliminates import of L4-100K into the nucleus (25).
We therefore determined the subcellular distribution of the wt and mutant proteins in virus-infected A549 cells (Fig. 4). In the course of infection, staining for wt L4-100K was seen mostly in the cytoplasm at earlier time points (18 h) (Fig. 4A, panel a) in the late phase, while most of the protein exhibited a dominant nuclear fluorescence as the infection proceeded (48 h) (Fig. 4A, panel c). A similar subcellular distribution was observed for the mutant protein in H5pm4151-infected cells. Comparable to the wt product, the L4-100K mutant was mostly cytoplasmic early after expression and accumulated in the nuclei of the infected cells during the late phase (Fig. 4A, panels b and d). We note that no substantial differences in the localizations of wt L4-100K and its variants were observed when cells were fixed with paraformaldehyde (data not shown).
FIG. 4.
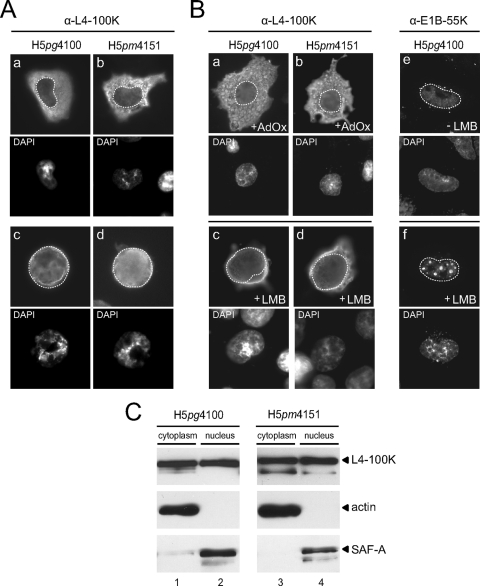
Subcellular localization of wt and mutant L4-100K during productive infection. A549 cells were infected with H5pg4100 and H5pm4151 viruses at a multiplicity of 10 FFU per cell. (A) Infected cells were fixed at 18 h (a and b) and 48 h (c and d) after infection and stained with MAb 6B10. (B) At 36 h postinfection, cells were fixed and the subcellular localization of L4-100K was examined by immunofluorescence using MAb 6B10. Methylation inhibitor AdOx (100 μM) was added at 18 h postinfection to the infection medium (a and b). Infected cells were kept in the presence of LMB (20 nM) for 3 h before fixation (c and d). Right panels, cells were stained with MAb 2A6 to detect E1B-55K localization in the absence (e) or presence (f) of LMB. In all panels nuclei are visualized by DAPI staining and are indicated by a dotted line. Magnification, ×7,600. (C) At 36 h postinfection, cells were collected and fractionated, and 5-μg aliquots of cytoplasmic and 2-μg aliquots of nuclear lysates were separated by SDS-10% PAGE and analyzed by Western blotting using L4-100K rat MAb 6B10. Antiactin and anti-SAF-A antibodies were used as cytoplasmic and nuclear markers, respectively.
To test the ability of L4-100K to enter the nucleus in a hypomethylated environment, we determined its steady-state localization in the presence of the methylation inhibitor AdOx by indirect immunofluorescence. As before, the wt and mutant L4-100K proteins could be detected in both the cytoplasm and the nuclei of infected cells (Fig. 4B, panels a and b). To further test the nuclear localization of L4-100K, we took advantage of the fact that the majority of cytoplasmic L4-100K is relocalized to the nucleus by mutational inactivation of its NES (13) or in the presence of the CRM1 inhibitor LMB. Thus, if the amino acid exchanges in the RGG boxes interfere with nuclear import, the mutant protein should not be able to accumulate in the nucleus in LMB-treated cells. As control, we determined the subcellular distribution of E1B-55K, which continuously shuttles between the nuclear and cytoplasmic compartments through the CRM1 pathway (28). Treatment of H5pg4100-infected cells with LMB caused the nuclear accumulation of wt L4-100K, and notably also of mutant L4-100K, in H5pm4151-infected cells (Fig. 4B, panels c and d). As expected (27, 28), a large portion of E1B-55K was relocalized from the cytoplasm to the nucleus upon LMB treatment, where it accumulated in dot-like structures (Fig. 4B, panels e and f). Consistent with immunofluorescence data, nucleocytoplasmic fractionation of H5pg4100- and H5pm4151-infected cells showed that similar amounts of L4-100K are present in both cytoplasmic and nuclear fractions regardless of the methylation status of the RGG boxes (Fig. 4C).
To further assess the role of arginine methylation in L4-100K nuclear localization, we introduced mutations into plasmid pTL-flag-100K (Fig. 5A) containing Flag-tagged, wt L4-100K cDNA including the TL. As in the L4-100K virus mutant H5pm4151, arginine-to-glycine substitutions were introduced into the RGG boxes. In addition, the leucine residues in the L4-100K NES were changed to alanines, and the arginines in the RGG3 box, the GAR, and the RGG plus GAR regions were converted to glycines. H1299 cells were transfected, and steady-state localization of wt and mutant proteins was determined by indirect immunofluorescence in the absence or presence of LMB (Fig. 5B). Consistent with a previous report (13), mutational inactivation of the NES resulted in predominant nuclear staining of L4-100K in 92% of the cells examined (n > 50) in the absence or presence of LMB (Fig. 5B, panels b and h). Also, in a high percentage (86%) of cells (n > 50), wt L4-100K relocalized from the cytoplasm to the nucleus upon LMB treatment (Fig. 5B, panels a and g). An identical result was obtained when cells producing the RGG and RGG3 mutants were treated with LMB (Fig. 5B, panels c, d, i, and j). Interestingly, however, no substantial increase in the nuclear fluorescence was seen in all LMB-treated cells expressing L4-100K variants carrying amino acid exchanges in the GAR region (Fig. 5B, panels f and l) or the RGG plus GAR region (Fig. 5B, panels e and k). The observed differences were not attributable to changes in steady-state concentrations, since the L4-100K variants accumulated to levels comparable to that of wt L4-100K (Fig. 5C). On the other hand, wt and mutant proteins differed in their abilities to interact with PRMT1 (Fig. 5C). As predicted, L4-100K levels were significantly reduced in the coprecipitates containing the RGG and RGG.GAR mutant proteins, indicating that both variants are substantially less methylated than the wt and the other L4-100K mutants. Altogether these results show that neither the arginines in the RGG boxes nor their methylation contributes to L4-100K nuclear import; instead, the arginine residues in the GAR region play a critical role in this process, consistent with the previous work (13). Identification of the nuclear import signal in the GAR region may also explain why we were unable to grow the corresponding GAR and RGG.GAR virus mutants from the linearized viral DNAs (Fig. 1B).
Mutations in the RGG boxes of L4-100K reduce interactions with TL RNAs.
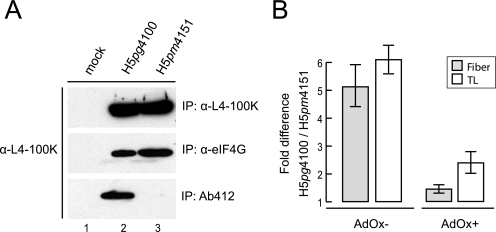
We next tested whether methylation of arginine residues in the RGG boxes might affect the known interactions of L4-100K with TL-containing late viral mRNAs and eIF4G (13, 49, 50). Binding of wt and mutant L4-100K to eIF4G was tested by combined immunoprecipitation and immunoblotting experiments using the appropriate antibodies (Fig. 6A). Results from these assays demonstrated that amino acid substitutions in the L4-100K RGG boxes do not affect eIF4G binding, although the same mutations significantly reduced amounts of arginine methylated L4-100K to undetectable levels (Fig. 6A). To monitor the mutations’ effects on interactions with TL-mRNAs, wt and mutant proteins were immunoprecipitated and the associated RNAs isolated from the immune complexes were identified by quantitative reverse transcription-PCR with specific primers for TL, fiber, and cellular GAPDH mRNAs (Fig. 6B). wt L4-100K reproducibly associated with TL-mRNAs sixfold more efficiently than the mutant L4-100K RGG protein and with fiber mRNA around fivefold more efficiently in three independent experiments. No measurable interaction was observed with wt and mutant L4-100K in these assays with GAPDH mRNAs, excluding the possibility that the arginine residues in wt L4-100K contribute to nonspecific RNA interactions (46; also data not shown). To determine whether efficient binding to TL-mRNAs is dependent on the methylation of arginines in the RGG boxes, we repeated the experiments in the presence of AdOx (Fig. 6B). Inhibition of global arginine methylation significantly reduced but did not completely abolish the ability of the wt protein to bind to TL-RNAs. This indicates that arginine methylation in the RGG boxes likely contributes to efficient binding to TL-mRNAs.
FIG. 6.
Binding of wt and mutant L4-100K to eIF4G and viral late mRNAs. (A) A549 cells were infected with H5pg4100 and H5pm4151 viruses at a multiplicity of 10 FFU per cell. At 48 h after infection, total cell lysates were prepared and subjected to immunoprecipitation (IP) with L4-100K antibody, eIF4G antibody, or Ab412 (recognizing methylated arginines). The immunocomplexes were separated by SDS-10% PAGE and analyzed by immunoblotting using MAb 6B10. (B) RNA was purified from immunoprecipitates of total cell lysates obtained from infected cells grown in the absence (AdOx−) or presence (AdOx+) of AdOx. RNA was reverse transcribed and subjected to quantitative real-time PCR as described in the text. L5 fiber and TL RNA levels are shown relative to the wt values. The results represent the averages from three independent experiments. Error bars indicate the standard errors of the means.
Arginine methylation of L4-100K may regulate hexon biogenesis.
In addition to its important role in blocking host cell protein synthesis by stimulating cap-independent translation of viral late mRNAs, L4-100K further contributes to efficient virus growth by facilitating trimerization and nuclear accumulation of hexons by acting as a chaperone for hexon trimerization in the cytoplasm and promoting their nuclear import (9, 24).
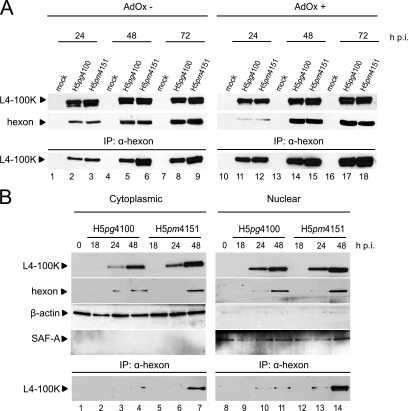
To monitor the effect of mutations on L4-100K binding to hexon we performed combined immunoprecipitation and immunoblotting assays (Fig. 7). As predicted, wt L4-100K coprecipitated with hexon during the late phase of infection (Fig. 7A, lanes 2, 5, and 8). Unexpectedly, we noticed a positive effect on hexon binding with the L4-100K RGG mutant. We reproducibly observed a four- to sixfold increase in binding of the mutant protein to hexon, in particular at later time points (48 and 72 h) of infection (Fig. 7A, lanes 6 and 9). Significantly, a similar binding pattern was evident when we monitored the interaction of both wt and mutant L4-100K with hexon in the presence of the arginine methylation inhibitor AdOx (Fig. 7A, lanes 14, 15, 17, and 18), confirming that PRMT1-mediated methylation of arginines in the RGG boxes regulates hexon binding. This was further supported by coimmunoprecipitation experiments using fractionated cell lysates (Fig. 7B). Again, in these assays substantially more L4-100K RGG mutant protein than the wt product precipitated with hexon in both the cytoplasmic (Fig. 7B, lanes 3, 4, 6, and 7) and the nuclear (Fig. 7B, lanes 10, 11, 13, and 14) fractions. Also, it appeared that more of the L4-100K RGG mutant protein was found in a complex with hexon in the nuclear fraction than in the cytoplasmic fraction (Fig. 7B, compare lanes 7 and 14). One possible explanation for this result is that arginine methylation in the RGG boxes of L4-100K negatively regulates the interaction with hexon, presumably favoring dissociation of the complex in the nucleus.
FIG. 7.
Arginine methylation in RGG boxes modulates hexon binding to L4-100K. (A) A549 cells were infected with H5pg4100 and H5pm4151 at a multiplicity of 10 FFU per cell in the absence or presence of AdOx (AdOx− and AdOx+, respectively) and harvested at the indicated time points after infection (h p.i.). Total cell extracts were prepared from noninfected (mock) and infected cells, and 50-μg aliquots of lysates were separated by SDS-10% PAGE and analyzed by Western blotting using L4-100K rat MAb 6B10 or rabbit MAb recognizing the hexon protein. The same lysates were used for immunoprecipitation (IP) with antihexon MAb. The immunocomplexes were separated by SDS-10% PAGE and analyzed by immunoblotting using MAb 6B10. (B) Total cell extracts were prepared and subjected to nucleocytoplasmic fractionation. Aliquots of cell extracts corresponding to cytoplasmic (5 μg) and nuclear (2 μg) fractions were separated by SDS-10% PAGE and analyzed by immunoblotting using 6B10 and antihexon MAb. The same extracts were subjected to immunoprecipitation with MAb hexon and visualized by SDS-10% PAGE and immunoblotting with MAb 6B10. Antiactin and anti-SAF-A antibodies were used as cytoplasmic and nuclear markers, respectively.
DISCUSSION
In this report, we investigated the consequences of L4-100K methylation throughout the infection cycle, particularly focusing on its late-phase functions and interactions. Our results confirmed that methylated L4-100K is required during Ad infection to ensure sufficient late protein synthesis and virus yield.
L4-100K contains three RGG boxes followed by GAR sequences in its C terminus. To investigate whether these RGG boxes are the target domain for methylation, we designed a virus mutant encoding a L4-100K protein with arginine-to-glycine exchanges in all of the RGG boxes. Indeed, the mutant virus proved defective in L4-100K methylation compared to the wt virus, pointing to the RGG boxes being the major methylation target, although the protein was slightly methylated at later time points, hinting at other methylated arginine residues. Since methylation of arginines in an RGG cluster was shown to alter the confirmation of a protein (14) and affect its destination (3, 4, 36), protein interactions, and DNA (5) or RNA (33, 41) binding, we analyzed the properties of this mutant in more detail.
First, we investigated the subcellular distribution of the mutant protein. Surprisingly, we observed no obvious difference in the localization or appearance of mutant L4-100K compared to wt protein, either in A549 or in other tested cell lines such as H1299 and primary human hepatocytes. Furthermore, addition of a general methylation inhibitor (AdOx) did not affect the nucleocytoplasmic shuttling of L4-100K, although the efficiency of lytic infection decreased. These results were in contrast to a recent report claiming that methylation of the arginine in the third RGG box is required for the nuclear import of L4-100K (25). These different observations may be a result of different assay setups. The authors converted the RGG boxes into AAA sequences in plasmid constructs and transfected cells that were infected with wt virus. However, we used a virus mutant with only arginine-to-glycine exchanges in three RGG boxes. Since alanine also contains a methyl group, we wanted to replace the arginines with either lysines or glycines. In the viral context, to avoid affecting the L4-33K and -22K coding sequence, we opted for glycines rather than lysines. None of our virus mutants or plasmid constructs (including the RGG3 mutant) showed any defect in nuclear import, indicating that neither the arginines in the RGG boxes nor their methylation is involved in this process.
We further investigated possible residual methylation of the arginines in the downstream GAR region to assess their impact on nuclear localization. Examples of several methylated arginine residues in an RG cluster have been reported, some of which are partially methylated (32, 39, 45). To analyze this effect, we intended to construct a mutant virus that expresses L4-100K with no possible methylated arginine residues, including GAR motifs in the downstream sequence, as well as a GAR-only mutant. However, these constructs could not be grown as viruses in the helper cell line used here. This indicated how incompetent these mutants are compared to the RGG box mutant, which was successfully produced. Therefore, we generated RGG.GAR and GAR mutants using a pTL-flag100K plasmid, and indeed transfection studies revealed methylation differences compared to the wt and the RGG mutant. Coimmunoprecipitation assays with Asym-24 resulted in no precipitated RGG.GAR 100K protein, and while slight methylation of RGG 100K was observed, it was much less than that of the wt protein (data not shown). This result confirmed the partial methylation of arginine residues not in the RGG box domain but in the GAR region of the downstream sequence. Interestingly, these GAR and RGG.GAR mutants were located exclusively in the cytoplasm and apparently were unable to enter the nucleus. Since the nuclear localization signal of L4-100K is located in the carboxy terminus (13), we suggest that the GAR domain, not the RGG boxes, is responsible for nuclear import.
Next we analyzed the RNA and protein interactions of the mutant virus. Contributing to selective translation of Ad late mRNAs, L4-100K can bind both cellular and viral mRNAs in addition to the eIF4G scaffold protein of the translation initiation complex (12, 13, 49, 50). One of the most important characteristics of RNA binding proteins (such as hnRNPs) is arginine methylation in GAR RNA binding domains (30). Several examples demonstrated that arginine methylation acts as a direct signal for RNA interaction in either a positive or a negative way (3, 32, 33). In our experiments we detected up to a sixfold reduction in TL-mRNA binding activity of the L4-100K mutant protein, which was surprising in the light of a previous report demonstrating phosphorylation as the factor deciding L4-100K's TL-mRNA binding specificity (49). Thus, the reduction that we observed in TL-mRNA binding of the L4-100K RGG mutant may hint at a possible interplay between two posttranslational modifications, since one of the two phosphorylated tyrosine residues (Y365 and Y682) is in close proximity to the first RGG box (R727) and may be modulated by methylation of this arginine, as there is evidence of a masking effect of adjacent posttranslational modifications (3).
Another very important function of L4-100K involves interacting with hexon monomers to assist their maturation into trimers (24, 37). Although it was shown that hexon protein was precipitated in large amounts with L4-100K-specific antibodies (38, 41), the hexon-interacting domain of L4-100K has not been identified yet. Therefore, we asked whether the methylation of RGG boxes could be a signal for this vital interaction. In our investigations, we unexpectedly found improved binding of hexon to the L4-100K RGG mutant, although late protein synthesis efficiency was lower than that of the wt virus. This increase was unique to hexon interaction, since eIF4G binding was not affected by the inserted mutations. If L4-100K is sufficient for hexon trimerization and contributes to transporting trimeric hexons into the nucleus, the L4-100K interaction with hexon molecules should be transient so that L4-100K can translocate back to the cytoplasm for its other tasks. Dissociation of the complex was thought to occur only if the trimers are efficiently integrated into the capsid (24, 37). In efficient Ad infection, these maturation and integration steps should proceed rapidly. The tested H5pm4151 mutant might show increased L4-100K binding to hexon due to prolonged interactions caused by an interfering factor or the loss of some control mechanism. Such a delay in hexon maturation and capsid assembly could account in part for the reduced virus yield that we observed here. There is also evidence in the literature showing that arginine methylation can be a negative regulator of protein-protein interactions (3, 39). Thus, blocking the methylation of L4-100K may alter the tuning of L4-100K-hexon interactions, leading to inefficient hexon trimerization, transport, or capsid assembly which could further affect other L4-100K tasks such as TL-mRNA binding. This hypothesis raises a number of questions, including whether this cascade of events requires a reversible arginine methylation. Previously only deiminases were known to convert methylarginine to citrulline by demethylation (2). Recently, another enzyme, called JMJD6, was identified as a histone arginine demethylase located in the nucleus (10). To date, it remains unknown whether this arginine demethylase has other substrates, but it is tempting to speculate that this mechanism is also involved in the regulation of L4-100K-hexon interactions.
Several studies have demonstrated and discussed the role of L4-100K in the nuclear import of hexons (8, 9, 24). In addition, pVI protein has also been implicated in this process (26, 48). However some of these data are contradictory, particularly about the transport of hexon trimers favoring either L4-100K or pVI. It will be of great interest to study the triple interaction of L4-100K, hexon, and pVI and the effect of L4-100K arginine methylation on this process.
In summary, arginine methylation of L4-100K in triple RGG boxes was found not to regulate the subcellular localization of L4-100K or selective translation of viral late mRNAs via eIF4G binding. However, it did contribute to efficient Ad late protein synthesis by modulating protein-protein/RNA interactions or regulating intracellular trafficking and possibly regulating TL-mRNA binding, hexon biogenesis, and virus assembly, topics that should be further investigated.
Acknowledgments
K16 cells were kindly provided by A. Amalfitano. We thank Robert J. Schneider for plasmid pTL-flag, Richard E. Lloyd for anti-eIF4G antibody, and Frank O. Fackelmayer for anti-SAF-A antibody. We thank Emre Koyuncu and Avril Arthur-Goettig (Bioxpress) for critical reviews of the manuscript.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Andrade, F., L. A. Casciola-Rosen, and A. Rosen. 2003. A novel domain in adenovirus L4-100K is required for stable binding and efficient inhibition of human granzyme B: possible interaction with a species-specific exosite. Mol. Cell. Biol. 236315-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford, M. T. 2007. Arginine methylation at a glance. J. Cell Sci. 1204243-4246. [DOI] [PubMed] [Google Scholar]
- 3.Bedford, M. T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18263-272. [DOI] [PubMed] [Google Scholar]
- 4.Boisvert, F. M., M. J. Hendzel, J. Y. Masson, and S. Richard. 2005. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle 4981-989. [DOI] [PubMed] [Google Scholar]
- 5.Boisvert, F. M., A. Rhie, S. Richard, and A. J. Doherty. 2005. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 41834-1841. [DOI] [PubMed] [Google Scholar]
- 6.Boulanger, M. C., C. Liang, R. S. Russell, R. Lin, M. T. Bedford, M. A. Wainberg, and S. Richard. 2005. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 79124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2005. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J. Biol. Chem. 28018610-18622. [DOI] [PubMed] [Google Scholar]
- 8.Cepko, C. L., and P. A. Sharp. 1983. Analysis of Ad5 hexon and 100K ts mutants using conformation-specific monoclonal antibodies. Virology 129137-154. [DOI] [PubMed] [Google Scholar]
- 9.Cepko, C. L., and P. A. Sharp. 1982. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell 31407-415. [DOI] [PubMed] [Google Scholar]
- 10.Chang, B., Y. Chen, Y. Zhao, and R. K. Bruick. 2007. JMJD6 is a histone arginine demethylase. Science 318444-447. [DOI] [PubMed] [Google Scholar]
- 11.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 2842174-2177. [DOI] [PubMed] [Google Scholar]
- 12.Cuesta, R., Q. Xi, and R. J. Schneider. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 193465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuesta, R., Q. Xi, and R. J. Schneider. 2004. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J. Virol. 787707-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dery, U., Y. Coulombe, A. Rodrigue, A. Stasiak, S. Richard, and J. Y. Masson. 2008. A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol. Cell. Biol. 283058-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolph, P. J., V. Racaniello, A. Villamarin, F. Palladino, and R. J. Schneider. 1988. The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J. Virol. 622059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eystathioy, T., E. K. Chan, S. A. Tenenbaum, J. D. Keene, K. Griffith, and M. J. Fritzler. 2002. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 131338-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fackelmayer, F. O. 2005. Protein arginine methyltransferases: guardians of the Arg? Trends Biochem. Sci. 30666-671. [DOI] [PubMed] [Google Scholar]
- 18.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52456-467. [DOI] [PubMed] [Google Scholar]
- 19.Groitl, P., and T. Dobner. 2007. Construction of adenovirus type 5 early region 1 and 4 virus mutants. Methods Mol. Med. 13029-39. [DOI] [PubMed] [Google Scholar]
- 20.Hartl, B., T. Zeller, P. Blanchette, E. Kremmer, and T. Dobner. 2008. Adenovirus type 5 early region 1B 55-kDa oncoprotein can promote cell transformation by a mechanism independent from blocking p53-activated transcription. Oncogene 273673-3684. [DOI] [PubMed] [Google Scholar]
- 21.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 642732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrmann, F., J. Lee, M. T. Bedford, and F. O. Fackelmayer. 2005. Dynamics of human protein arginine methyltransferase 1 (PRMT1) in vivo. J. Biol. Chem. 28038005-38010. [DOI] [PubMed] [Google Scholar]
- 23.Hodges, B. L., H. K. Evans, R. S. Everett, E. Y. Ding, D. Serra, and A. Amalfitano. 2001. Adenovirus vectors with the 100K gene deleted and their potential for multiple gene therapy applications. J. Virol. 755913-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong, S. S., E. Szolajska, G. Schoehn, L. Franqueville, S. Myhre, L. Lindholm, R. W. Ruigrok, P. Boulanger, and J. Chroboczek. 2005. The 100K-chaperone protein from adenovirus serotype 2 (subgroup C) assists in trimerization and nuclear localization of hexons from subgroups C and B adenoviruses. J. Mol. Biol. 352125-138. [DOI] [PubMed] [Google Scholar]
- 25.Iacovides, D. C., C. C. O'Shea, J. Oses-Prieto, A. Burlingame, and F. McCormick. 2007. Critical role for arginine methylation in adenovirus-infected cells. J. Virol. 8113209-13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kauffman, R. S., and H. S. Ginsberg. 1976. Characterization of a temperature-sensitive, hexon transport mutant of type 5 adenovirus. J. Virol. 19643-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kindsmuller, K., P. Groitl, B. Hartl, P. Blanchette, J. Hauber, and T. Dobner. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. USA 1046684-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krätzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55k oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19850-857. [DOI] [PubMed] [Google Scholar]
- 29.Kzhyshkowska, J., E. Kremmer, M. Hofmann, H. Wolf, and T. Dobner. 2004. Protein arginine methylation during lytic adenovirus infection. Biochem. J. 383259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kzhyshkowska, J., H. Schutt, M. Liss, E. Kremmer, R. Stauber, H. Wolf, and T. Dobner. 2001. Heterogeneous nuclear ribonucleoprotein E1B-AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem. J. 358305-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y. J., M. R. Stallcup, and M. M. Lai. 2004. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 7813325-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, Q., and G. Dreyfuss. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 152800-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 707445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsudomi, T., S. M. Steinberg, M. M. Nau, D. Carbone, D. D'Amico, H. K. Bodner, H. K. Oie, R. I. Linnoila, J. L. Mulshine, J. D. Minna, and A. F. Gazdar. 1992. p53 gene mutations in non-small-lung cell cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7171-180. [PubMed] [Google Scholar]
- 35.Morin, N., and P. Boulanger. 1986. Hexon trimerization occurring in an assembly-defective, 100K temperature-sensitive mutant of adenovirus 2. Virology 15211-31. [DOI] [PubMed] [Google Scholar]
- 36.Nichols, R. C., X. W. Wang, J. Tang, B. J. Hamilton, F. A. High, H. R. Herschman, and W. F. Rigby. 2000. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 256522-532. [DOI] [PubMed] [Google Scholar]
- 37.Oosterom-Dragon, E. A., and H. S. Ginsberg. 1981. Characterization of two temperature-sensitive mutants of type 5 adenovirus with mutations in the 100,000-dalton protein gene. J. Virol. 40491-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oosterom-Dragon, E. A., and H. S. Ginsberg. 1980. Purification and preliminary immunological characterization of the type 5 adenovirus nonstructural 100,000-dalton protein. J. Virol. 331203-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostareck-Lederer, A., D. H. Ostareck, K. P. Rucknagel, A. Schierhorn, B. Moritz, S. Huttelmaier, N. Flach, L. Handoko, and E. Wahle. 2006. Asymmetric arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein-arginine methyltransferase 1 inhibits its interaction with c-Src. J. Biol. Chem. 28111115-11125. [DOI] [PubMed] [Google Scholar]
- 40.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128480-484. [DOI] [PubMed] [Google Scholar]
- 41.Riley, D., and S. J. Flint. 1993. RNA-binding properties of a translational activator, the adenovirus L4 100-kilodalton protein. J. Virol. 673586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubenwolf, S., H. Schütt, M. Nevels, H. Wolf, and T. Dobner. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 711115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120510-517. [DOI] [PubMed] [Google Scholar]
- 44.Schmid, S. I., and P. Hearing. 1999. Adenovirus DNA packaging. Construction and analysis of viral mutants, p. 47-59. In W. S. Wold (ed.), Adenovirus methods and protocols, vol. 21. Humana Press Inc., Totowa, NJ. [Google Scholar]
- 45.Smith, J. J., K. P. Rucknagel, A. Schierhorn, J. Tang, A. Nemeth, M. Linder, H. R. Herschman, and E. Wahle. 1999. Unusual sites of arginine methylation in poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J. Biol. Chem. 27413229-13234. [DOI] [PubMed] [Google Scholar]
- 46.Tao, J., and A. D. Frankel. 1992. Specific binding of arginine to TAR RNA. Proc. Natl. Acad. Sci. USA 892723-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tollefson, A. E., T. W. Hermiston, and W. S. Wold. 1999. Preparation and titration of CsCl-banded adenovirus stocks, p. 1-9. In W. S. Wold (ed.), Adenovirus methods and protocols, vol. 21. Humana Press Inc., Totowa, NJ. [Google Scholar]
- 48.Wodrich, H., T. Guan, G. Cingolani, D. Von Seggern, G. Nemerow, and L. Gerace. 2003. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 226245-6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xi, Q., R. Cuesta, and R. J. Schneider. 2005. Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100K to viral mRNAs. J. Virol. 795676-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xi, Q., R. Cuesta, and R. J. Schneider. 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 181997-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yueh, A., and R. J. Schneider. 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 14414-421. [PMC free article] [PubMed] [Google Scholar]