Abstract
Previous studies have demonstrated the interaction between the Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA3C) and the metastatic suppressor Nm23-H1 both in vitro and in vivo (C. Subramanian, M. A. Cotter II, and E. S. Robertson, Nat. Med. 7:350-355, 2001). Importantly EBNA3C can reverse the ability of Nm23-H1 to suppress migration of human cells in vitro. EBNA3C contributes to EBV-associated human cancers by regulating transcription of a number of cellular and viral promoters as well as targeting and altering the transcription activities of the metastasis suppressor Nm23-H1. Furthermore, Necdin is a cellular protein which is highly induced in terminally differentiated cells; it contributes to the regulation of cell growth and is also known to interact with viral oncoproteins. In this report, we show that Nm23-H1 and EBNA3C can modulate the biological functions of Necdin in the context of EBV infection and transformation. The levels of Necdin were consistently lower in EBV-positive cells, and EBNA3C could change the subcellular localization of Necdin as well as rescue cells from the antiangiogenic and antiproliferative effects mediated by Necdin. We also show that Necdin directly interacts with Nm23-H1, resulting in modulation of the biochemical function of Nm23-H1 as well as the biological function of Necdin. Both EBNA3C and Nm23-H1 were able to rescue not only Necdin-mediated transcriptional repression of the downstream vascular endothelial growth factor promoter but also Necdin-mediated growth suppression and antiangiogenic effects on cancer cells. The majority of this response was mediated through amino acid residues 191 to 222 of Necdin, which are also known to be important for nuclear matrix targeting. These studies suggest a role for Necdin in the regulation of downstream cellular targets in a hypoxic environment in virus-associated human cancers.
Epstein-Barr virus (EBV) is a human gammaherpesvirus which predominantly targets B cells and epithelial cells and is associated with a number of human cancers, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, AIDS-associated and transplant-associated immunoblastic lymphoma, and, controversially, invasive breast carcinoma (9, 61). In vitro infection of B cells with EBV gives rise to lymphoblastoid cell lines (LCLs) which express a subset of 12 latent viral transcripts (61). These 12 transcripts encode six nuclear antigens (EBNAs), three latent membrane proteins, two early RNAs (32), and the BARF transcripts (61). Studies have shown that EBNA3C, one of the six nuclear antigens, interacts with the suppressor of cell migration and metastasis Nm23-H1 (69). This interaction between Nm23-H1 and EBNA3C has been shown to result in an increase in transcriptional activity on a targeted responsive promoter (70). Nm23-H1 tethered to DNA by a Gal4 DNA binding domain can activate transcription from a basal promoter at relatively low levels. However, when EBNA3C was introduced, the transactivation activity was shown to be substantially increased (70). These results suggest that Nm23-H1 may possess transcriptional regulatory activities which could function independent of its direct binding with DNA. This could possibly be through its interaction with EBNA3C (69). Interestingly, the presence of EBNA3C mediates the cellular translocation of Nm23-H1 from a mostly cytoplasmic to a predominantly nuclear signal. Moreover, EBNA3C can reverse the antimigratory effects of Nm23-H1 in vitro (69) as well as in vivo (23). The interaction between EBNA3C and Nm23-H1 has also been shown to be important in the regulation of COX-2, MMP-9, and alpha V integrin (13, 24, 26).
The Nm23 gene family is highly conserved among a wide variety of eukaryotic species. Eight Nm23 genes have been identified for humans: Nm23-H1 (62), Nm23-H2 (68), Nm23-H3 (79), Nm23-H4 (45), Nm23-H5 (47), Nm23-H6 (44, 76), Nm23-H7 (78), and Nm23-H8 (56). Members of the Nm23 gene family are structurally and functionally conserved, consisting of four to six identically folded subunits enclosing a large (25-Å) central cavity (84). Expression of Nm23 genes has been linked to suppression of tumor metastasis, differentiation, apoptosis, proliferation, and DNA mutation rate (14) and is also associated with nucleoside diphosphate (NDP) kinase activity (7, 49, 82), serine phosphorylation (8, 20, 37, 48), histidine protein kinase activities (81), and transcriptional stimulatory activities (2, 6, 59, 60). The human Nm23-H1 gene product is the best-characterized member of this family of proteins. It is 152 amino acids (aa) in length with leucine repeats and alpha-helical and basic domains (61). Nm23-H1 is 88% identical to Nm23-H2 and maps 4 kb apart at position q21.3 on chromosome 17 near the BRCA1 locus, which is known to be associated with early onset of familial breast and ovarian cancers (4). The Nm23-H1 gene product is more closely associated with the inhibition of metastasis and signal transduction, has an acidic pI, and is identical to the A subunit of NDP kinase (17). Importantly, in a number of human cancers there is an inverse relationship between Nm23-H1 expression and metastasis (14, 39). In neuroblastoma, mutations in the leucine zipper motif of Nm23-H1 directly correlates with increased metastatic potential (19, 31). Nm23-H1 is also known to regulate the activity of the oncoprotein Dbl1 (52, 53).
The mechanism by which Nm23-H1 regulates metastasis is not fully understood; however, the experimental data accumulated thus far strongly implicate Nm23-H1 in the regulation of metastasis in a diverse number of human cancers (38). Recently, Nm23-H1 has also been identified as a granzyme A-activated, apoptosis-inducing DNase which forms part of the endoplasmic reticulum-associated complex (SET complex) (16). The SET complex translocates to the nucleus of target cells within minutes of granzyme A loading or cytotoxic T-lymphocyte attack (16). Although the normal function of the SET complex is not known, its constituent proteins have been highly implicated in tumorigenesis. They have been linked to diverse functions, including chromatin modification, transcriptional activation of proto-oncogenes, DNA repair, and mRNA stability (10, 15, 35, 65, 66). The SET complex has also been suggested to be involved in repair and transcriptional activation in response to oxidative stress (16). All these observations indicate that translocation of Nm23-H1 from the cytoplasm to the nucleus may be an important event that results in modulation of expression of cellular genes that are important in tumorigenesis. The domain of EBNA3C that specifically binds to Nm23-H1 has been identified, and it lies within the region comprising aa 637 to 675 of EBNA3C, flanked by the proline- and glutamine-rich domains (68). A Blast analysis of this domain sequence shows homology to a cellular protein called Necdin.
The genetic inactivation of the Necdin gene is associated with a developmental disorder in humans, Prader-Willi syndrome (PWS), in which three other genes are inactivated (11, 18). The main features which characterize this syndrome are a delay in development; short stature; abnormalities in behavior; the onset of severe obesity in childhood; a consistent drive to eat, referred to as hyperphagia; inadequate ventilation; and reproductive system defects (11, 18). Necdin belongs to a family of proteins called the MAGE proteins, which are increasingly shown to have roles in a number of cellular processes, including cell cycle regulation and cell death (5). The most important feature in all MAGE family proteins is the presence of a large central region which is referred to as the MAGE homology domain (MHD), whereas their amino- and carboxy-terminal domains are typically shown to be unique for the individual family members (5). The localization of Necdin in differentiated neurons was previously demonstrated to show predominantly cytoplasmic signals, with a striking change in location to the nucleus during specific physiological changes (55). Necdin has also been shown to interact with DNA, which may enable it to regulate transcription of other genes involved in cellular differentiation and proliferation through direct binding with DNA or through its interaction with other transcription factors, including p53 and E2F1 (5, 40). In fact, Necdin has been shown to act as a cellular transcriptional repressor by directly binding to multiple guanosine clusters within the promoters of target genes (43). Importantly, in the context of cancer and tumor development, Necdin can associate with Hif-1α, a major transcription regulator in hypoxia, and may directly regulate its activity (46. Thus, the regulation of Hif-1α expression by Necdin may have important consequences, including downstream effects on cell growth and proliferation. Additional studies have shown that Necdin interacts with a number of other cellular proteins; however, the functional role of these interactions in the biology of PWS continues to be explored (11. In this report, we present evidence to suggest that the essential EBV latent protein EBNA3C can modulate Necdin-associated functions in association with the metastasis suppressor protein Nm23-H1.
MATERIALS AND METHODS
Constructs and cell lines.
The T-antigen-transformed human embryonic kidney cell line 293T (HEK293T) was obtained from Jon Aster (Department of Pathology, Brigham and Women's Hospital, Harvard Medical School). The HEK293T and HeLa cell lines used in transient-transfection experiments were maintained in Dulbecco's modified Eagle's medium supplemented with 5% bovine growth serum (BGS), 2 mM glutamine, 25 U/ml of penicillin-streptomycin, and 20 μg/ml of gentamicin. The EBV-negative BJAB cell line was obtained from Elliott Kieff (83). The BJAB cell line was maintained in RPMI 1640 medium (Invitrogen/Gibco-BRL, Rockville, MD) supplemented with 7% BGS, 2 mM glutamine, and 25 U/ml of penicillin-streptomycin (Gemini Bioproducts, Inc., Carlsbad, CA). The EBNA3C and Nm23-H1 expression constructs used in these experiments were as described earlier (51, 69). The EBNA3C mutant construct with aa 621 to 675 deleted (the Δ621-675 mutant) was generated by PCR amplification and deletion of aa 621 to 675 in the EBNA3C open reading frame and was cloned in frame in the expression vector pA3M Myc tagged at the carboxy terminus (3). The wild-type hypoxia response element (HRE) consisted of a trimerized 24-mer containing 18 bp of sequence from the PGK promoter, including the HRE (5-TGTCACGTCCTGCACGACTCTAGT [the HRE is underlined]), and its mutant (MHRE) had the ACG of the HIF-1 binding site mutated to CAT, abolishing binding, in the pGL3-Basic vector. These were provided by Craig B. Thompson (University of Pennsylvania School of Medicine, Philadelphia, PA). Plasmid pGL3-VEGF, containing the 47-bp HRE, was provided by Amit Maity (University of Pennsylvania School of Medicine).
Transfection.
BJAB, HEK293T, and HeLa cells were transfected by electroporation with a Bio-Rad Gene Pulser II electroporator. Ten million cells were harvested in the exponential phase, collected, washed in phosphate-buffered saline (PBS), and then resuspended in 400 μl of RPMI or Dulbecco's modified Eagle's medium with DNA for transfection. Resuspended cells were transferred to a 0.4-cm cuvette and electroporated at 975 μF and 220 V. The electroporated cells were then transferred to 10 ml of complete medium, followed by incubation at 37°C with 5% CO2. Transfection products were harvested after 24 h and assayed for activity.
Real-time quantitative PCR.
Total RNAs from EBV-positive and -negative B-cell lines was collected using Trizol reagent (Invitrogen, Inc., Carlsbad, CA) following the manufacturer's instructions. cDNA was made with a Superscript II reverse transcription kit (Invitrogen, Inc., Carlsbad, CA) by following the manufacturer's instructions. The specific primers for Necdin were as follows: sense, 5′-GAGTTTGCCCTGGTCAAAGC-3′; antisense, 5′-CATGGGCATACGGTTGTTGAG-3′. PCR amplification yielded a 92-bp product. For β-actin, the primers were as follows: sense, 5′-GCTCGTCGTCGACAACGGCTC-3′; antisense, 5′-CAAACATGATCTGGGTCATCTTCTC-3′. PCR amplification yielded a product 352 bp in length. The cDNA was amplified using Sybr green real-time Mastermix (MJ Research, Inc., Waltham, MA), 1 mM concentrations of each primer, and 1 μl of the cDNA product in a total volume of 20 μl. Thirty-five cycles of 1 min at 94°C, 1 min at 56°C, and 30 s at 72°C, followed by 7 min at 72°C, were performed with an MJ Research Opticon II thermocycler (MJ Research, Inc., Waltham, MA). Each cycle was followed by two plate reads, with the first at 72°C and the second at 85°C. A melting curve analysis was performed to verify the specificity of the products, and the values for the relative quantitation were calculated by the ΔΔCT method. The experiment was performed in triplicate.
Preparation of GST fusion proteins for in vitro binding assays.
Necdin-hemagglutinin (Necdin-HA) was expressed in vitro by use of a coupled in vitro transcription/translation system (TNT) (Promega, Inc., Madison, WI) according to the manufacturer's instructions, by using [35S]methionine-cysteine (Perkin-Elmer, Inc., Boston, MA) in the TNT reaction mixture. Nm23-H1 glutathione S-transferase (GST) (34) fusion proteins (approximately 10 μg) expressed in Escherichia coli were used for in vitro bindings. In vitro-translated proteins were precleared with glutathione-Sepharose beads in binding buffer (1× PBS, 0.1% NP-40, 0.5 mM dithiothreitol, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml) for 30 min. Precleared labeled protein was then incubated with either GST or GST-Nm23H1 fusion proteins in binding buffer. Binding was performed overnight at 4°C with constant rotation, followed by collection of the beads by centrifugation. The beads were washed three times with 1 ml of binding buffer, followed by resuspension in sodium dodecyl sulfate (SDS) lysis buffer and resolution on SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The bound fraction was analyzed after drying the gel and exposing it to a PhosphorImager plate (Molecular Dynamics, Inc., Sunnyvale, CA).
Immunoprecipitation and Western blotting.
After 48 h of incubation, the transfected cells were lysed in cold radioimmunoprecipitation (IP) assay buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, aprotinin [1 μg/ml], and pepstatin [1 μg/ml]) on ice and homogenized. The lysates were precleared with protein A/G-Sepharose beads (Amersham Biosciences, Inc., Piscataway, NJ) and then incubated first with antibodies against Myc or HA (1 μg) in IP buffer (20 mM Tris-HCl [pH 8.0], 10% glycerol, 5 mM MgCl2, 0.1% Tween 20, 0.1 M KCl, 1× protease inhibitor cocktail [Amersham Biosciences, Inc., Piscataway, NJ], and 0.5 mM dithiothreitol) overnight at 4°C with constant rotation and then with protein A/G (50:50)-Sepharose beads for 1 h. Beads were washed three times with IP buffer and resuspended in 40 μl of 2× SDS Laemmli buffer. The sample was subjected to SDS-PAGE. Western blot analyses were performed using primary monoclonal antibodies against Myc (9E10) or HA (12CA5) and β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Detection and quantification were done using a Li-Cor Odyssey scanner (Li-Cor, Inc., Lincoln, NE).
Fractionation of nuclear and cytoplasm proteins.
Twenty million transfected cells were harvested and washed twice with ice-cold PBS, followed by resuspension of the cell pellet in hypotonic buffer A (10 mM HEPES-K+ [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 0.5 dithiothreitol) in the presence of protease inhibitor cocktail (PIC) (1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 10 μg of pepstatin A/ml, 10 μg of phenanthroline/ml, 16 μg of benzamidine/ml). Cells were pelleted by spinning them at 1,000 rpm for 5 min. The cells were lysed in ice-cold 0.5% NP-40-containing buffer A with PIC on ice for 10 min. The nuclei were pelleted by centrifugation at 3,000 rpm for 2 min at 4°C. The supernatant (cytoplasm protein) was harvested and frozen at −80°C for use. The pellet containing insoluble cytoplasmic membrane fraction was stored separately for use. The nuclear pellets were washed with buffer A (without NP-40), followed by resuspension in buffer C (20 mM HEPES-K+ [pH 7.9], 420 mM NaCl, 0.2 mM EDTA, 1.5 mM MgCl2, 0.5 dithiothreitol, 25% glycerol) with PIC. Nuclei were incubated on ice for 30 min and mixed periodically. Supernatants containing nuclear protein were collected by spinning at 14,500 rpm for 10 min at 4°C and then snap frozen for further use. The pellet containing the insoluble nuclear membrane fraction was stored separately for use. Antibodies to nuclear protein Sp1 (1C6; Santa Cruz, Inc., Santa Cruz, CA) and cytoplasm protein Hsp70 (BD Transduction Laboratory) were used as markers.
Immunofluorescence assays.
Transfected HeLa cells were overlaid on coverslips and allowed to grow for 24, 48, or 72 h. The cells were then washed with PBS, fixed with 1:1 methanol-acetone for 10 min at −20°C, dried, and rehydrated with PBS. For blocking, cells were incubated with PBS containing 3% bovine serum albumin and 1% glycine for 30 min. Cells were then incubated with appropriate antibodies (a 1:100 dilution of anti-Myc antibody and a 1:100 dilution of anti-EBNA3C antibody). Slides were washed three times in PBS and further incubated with a 1:1,000 dilution of goat anti-mouse or goat anti-human immunoglobulin fluorescein isothiocyanate-conjugated secondary antibodies. Slides were examined with an Olympus IX70 fluorescence microscope, and images were captured with a PixelFly digital camera (Cooke, Inc., Auburn Hills, MI).
Western blot analysis.
Protein was extracted from cultured cells, and the concentration was calculated. One hundred micrograms of cultured cell lysates was electrophoresed and transferred onto a nitrocellulose membrane. Equal loading of samples was confirmed with Ponceau-S staining of the membrane in all cases. The HA-tagged Necdin, Myc-tagged Nm23-H1, or Myc-tagged EBNA3C was analyzed as reported with the use of anti-HA or anti-Myc antibody (80).
Nm23 autophosphorylation assay.
GST fusion proteins of Nm23-H1 were incubated in the absence or presence of increasing amounts of immunoprecipitated Necdin. The reaction was initiated by the addition of 5 μCi of [γ-32P]ATP. After 5 min at 25°C, the reaction was stopped with Laemmli buffer; the samples were incubated for an additional 10 min and were subjected to 12% SDS-PAGE with and without boiling. The procedure without boiling was used to preserve the phosphorylation of Nm23 on the histidine residues. The gels were dried, exposed to a PhosphorImager plate (Molecular Dynamics, Inc., Sunnyvale, CA), and analyzed.
NDP kinase activity assay.
The NDP kinase assay was performed with TMD buffer (20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 1 mM dithiothreitol). In brief, 3 μg of GST-Nm23-H1 with increasing amounts of immunoprecipitated Necdin was incubated with 1 μM [γ-32P]ATP (0.2 mCi/ml) and 100 μM GDP in TMD buffer in a final volume of 20 μl for 10 min at room temperature. The reaction was stopped by the addition of 20 μl of 50 mM EDTA (pH 8.0). The reaction mixture (2 μl) was spotted onto 20- by 20-cm polyethyleneimine-cellulose thin-layer chromatography (TLC) plates (45), and reaction products were resolved by capillary action in 0.75 M KH2PO4, pH 3.65. The TLC plates were dried, exposed to a PhosphorImager plate (Molecular Dynamics, Inc., Sunnyvale, CA), and analyzed.
Luciferase promoter assay.
HEK293T cells were collected at 70% confluence; 10 million cells were resuspended, along with plasmid DNA, in 400 μl of medium. The cells were transfected by electroporation with a Bio-Rad Gene Pulser II at 210 V and 975 μF. Transfected cells were transferred to 100-mm plates in 10 ml of Dulbecco's modified Eagle's medium with 7% BGS. The plates were incubated at 37°C with 5% CO2 for 20 h. At 20 h, cells were harvested and washed once with PBS (Invitrogen-Gibco, Inc., Bethesda, MD). The cells were subsequently lysed with 300 μl of reporter lysis buffer (Promega, Inc., Madison, WI); 40 μl of the lysate was mixed with 25 μl of luciferase assay reagent. Luminescence was measured for 10 s with an Opticomp I luminometer (MGM Instruments, Inc., Hamden, CT). The lysates were also tested at various dilutions to ensure that luciferase activity was within the linear range of the assay. The results shown represent experiments performed in triplicate.
Matrigel assay.
293T cells were transfected with Necdin and vector control, EBNA3C, or Nm23-H1 expression constructs. The cells were allowed to grow for 48 h, after which the supernatant medium was harvested and stored at 4°C until used as conditioned medium (CM). Each well of a 24-well plate was coated with 150 μl of ice-cold Matrigel (BD Biosciences, San Jose, CA), which was incubated at 37°C for about 30 min. A total of 5 × 104 human umbilical vein endothelial cells (HUVECs) or HeLa cells were seeded on each well and grown in the 500 μl CM with 10% fetal bovine serum (FBS). After 16 h of incubation, the cells were photographed to detect the formation of tubes structure by HUVECs (tube formation assay) and the growth and invasiveness of HeLa cells.
RESULTS
Necdin transcript levels are reduced in EBV-positive lymphoblastoid cells.
Necdin is expressed predominantly in postmitotic neurons, although its expression levels vary among neuronal cell types, being the highest in the hypothalamus and brain stem (1, 77). Studies of the expression profiles of Necdin in different tissues have shown that although its expression is highest in brain and placenta, it is also expressed in all other tissues, including heart, brain, placenta, lung, liver, kidney, and muscle (36). To determine the effect of EBV on Necdin expression levels in cancer cells, we investigated whether or not there was any difference in the level of Necdin expression between known EBV-positive and EBV-negative cancer cells. These cell lines were assayed by real-time quantitative PCR to determine the level of Necdin mRNA. The values were normalized by β-actin mRNA levels for each cell line. The results of the real-time quantitative PCR for EBV-positive cell lines (LCL1 and LCL2) showed that relatively lower levels of Necdin mRNA were present in these lines than in the EBV-negative cell lines (Louckes, DG75, and BJAB) (Fig. 1a). The results indicated that Necdin mRNA expression levels in LCLs were approximately 10- to 100-fold lower than those in EBV-negative cells (Fig. 1a). We also tested the level of Necdin transcripts in Kaposi's sarcoma-associated herpesvirus (KSHV)-positive cells, which also showed slightly lower levels than those for KSHV/EBV-negative cells.
FIG. 1.
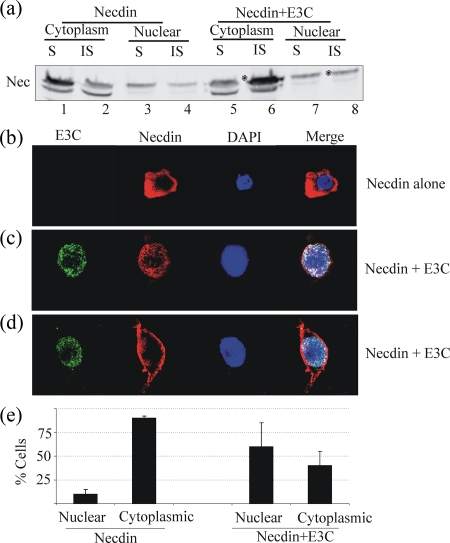
(a) Levels of Necdin are much lower in EBV- and KSHV-positive B cells than in control cells when compared at the mRNA level by quantitative real-time PCR. The specific primers for Necdin used were as follows: sense, 5′-GAGTTTGCCCTGGTCAAAGC-3′; antisense, 5′-CATGGGCATACGGTTGTTGAG-3′. PCR amplification yielded a 92-bp product. For β-actin, the primers were as follows: sense, 5′-GCTCGTCGTCGACAACGGCTC-3′; antisense, 5′-CAAACATGATCTGGGTCATCTTCTC-3′. A melting curve analysis was performed to verify the specificity of the products, and the values for the relative quantitation were calculated by the ΔΔCT method. (b) EBNA3C affects the localization of Necdin. Twenty million transfected cells were harvested and washed twice with ice-cold PBS, followed by resuspension of the cell pellet in hypotonic buffer A (10 mM HEPES-K+ [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 0.5 dithiothreitol) in the presence of PIC. Cells were pelleted by spinning them at 1,000 rpm for 5 min. The cells were lysed in ice-cold 0.5% NP-40-containing buffer A with PIC on ice for 10 min. The nuclei were pelleted by centrifugation at 3,000 rpm for 2 min at 4°C. The supernatant (cytoplasmic protein) was harvested and frozen at −80°C for use. The nuclear pellets were washed with buffer A (without NP-40), followed by resuspension in buffer C (20 mM HEPES-K+ [pH 7.9], 420 mM NaCl, 0.2 mM EDTA, 1.5 mM MgCl2, 0.5 dithiothreitol, 25% glycerol) with PIC. Nuclei were incubated on ice for 30 min and vortex mixed periodically. Supernatants containing nuclear protein were collected by spinning at 14,500 rpm for 10 min at 4°C and then snap frozen for further use. In the presence of EBNA3C, Necdin localization changes from primarily cytoplasmic to primarily noncytoplasmic. Soluble nuclear (N) and cytoplasmic (C) fractions were compared for cells coexpressing Necdin with EBNA3C (lanes 3 and 4) and cells expressing Necdin alone (lanes 5 and 6). Lanes 1 and 2 represent 293T cells as negative controls. (c) Expression levels of EBNA3C. Antibodies to nuclear protein Sp1 (1C6; Santa Cruz, Inc., Santa Cruz, CA) and cytoplasmic protein Hsp70 (BD Transduction Laboratory) were used as markers. (d and e) The localizations of HSP70, which is primarily cytoplasmic, and Sp1, which is nuclear, show the efficiency of cellular fractionation in nuclear and cytoplasmic fractions. Error bars indicate standard deviations.
EBNA3C induces change in localization of Necdin.
Previous studies have shown that EBNA3C can induce translocation of Nm23-H1 from a primarily cytoplasmic to a primarily nuclear localization (69). Necdin is also known to alter its subcellular distribution under the influence of other proteins, including the E2F1 transcription factor and p75NTR, the neurotropin receptor (27). Since Necdin did not show any direct interaction with EBNA3C in vitro (data not shown), we investigated whether the intracellular distribution of Necdin is altered in the presence of EBNA3C (Fig. 1b). 293T cells were transfected with expression vectors for HA-tagged Necdin and Myc-tagged EBNA3C. The majority of the signal was nuclear. Necdin was detected in the cytoplasm when it was expressed alone, but the majority was in the nucleus when coexpressed with EBNA3C (Fig. 1b). These patterns were similar to those for Nm23-H1 localization in cells expressing Nm23-H1 with and without EBNA3C (69). Thus, EBNA3C induced a change in the localization pattern of Necdin. However, the total expression of Necdin detected in both the cytoplasmic and nuclear fractions was somewhat lower when expressed with EBNA3C than when expressed alone (Fig. 1b, compare lanes 3 and 4 with lanes 5 and 6). We hypothesized that Necdin potentially interacts with membrane proteins which then relocalize to insoluble membrane fractions when expressed in the presence of EBNA3C. We then wanted to determine the presence of Necdin in insoluble cytoplasmic or nuclear membrane fractions. The results show that in the presence of EBNA3C, the majority of cytoplasmic Necdin was localized in the cytoplasmic membrane fraction compared to when Necdin was expressed alone, which showed that the majority of Necdin signal was in the soluble cytoplasmic fraction (Fig. 2a, compare lanes 1 and 2 with lanes 5 and 6). Moreover, the amounts of soluble Necdin were similar in the nuclear fraction and the cytoplasm when Necdin was expressed alone and with EBNA3C (Fig. 2a, compare lanes 1 and 3 with lanes 5 and 7). We then tested the localization of Necdin in the presence of EBNA3C by immunofluorescence. HEK-293T cells were transfected with expression vectors for HA-tagged Necdin with or without Myc-tagged EBNA3C and visualized by confocal laser microscopy (Fig. 2b, c, and d). When expressed alone, Necdin was distributed mainly to the cytoplasm (Fig. 2b). However, when coexpressed with EBNA3C, Necdin was localized to the nucleus as well (Fig. 2c) and to the periphery of the cytoplasm, possibly the cell membrane component (Fig. 2d). Quantification of the cells showing preferential nuclear and cytoplasmic redistribution patterns revealed that Necdin was localized mainly to the cytoplasm (90% of cells, with a cell count of 100) when alone and that its localization was altered to membranes and/or nucleus (60% of cells, with a cell count of 100) when coexpressed with EBNA3C (Fig. 1e). The localization of Necdin close to cellular membranes (30% of cells, with a cell count of 50) was seen mainly in the presence of EBNA3C.
FIG. 2.
(a) The majority of Necdin associates with the insoluble membrane fraction (asterisks) of cytoplasmic (compare lanes 2 and 6) or nuclear (compare lanes 4 and 8) extracts in the presence of EBNA3C. The cytoplasmic soluble fraction had a lower proportion of Necdin (lanes 1 and 5), whereas not much change was seen in the nuclear soluble fraction (lanes 3 and 7). S, soluble; IS, insoluble. (b) Immunofluorescence experiments were performed to visualize the localization of Necdin and EBNA3C. The subcellular localization of Necdin is altered in the presence of EBNA3C. Transfected HeLa cells were overlaid on coverslips and allowed to grow for 48 h. Cells were then washed with PBS, fixed with 1:1 methanol-acetone for 10 min at −20°C, dried, and rehydrated with PBS. For blocking, cells were incubated with PBS containing 3% bovine serum albumin and 1% glycine for 30 min. Cells were then incubated with appropriate antibodies (a 1:100 dilution of anti-Myc antibody and a 1:100 dilution of anti-HA antibody). Slides were washed three times in PBS and further incubated with a 1:1,000 dilution of goat anti-mouse or goat anti-human immunoglobulin fluorescein isothiocyanate-conjugated secondary antibodies. Slides were examined with a confocal microscope and images captured. The cells which did not show any Necdin signal in the nucleus were classified as cytoplasmic. Those cells which also showed a Necdin signal in nucleus were classified as those in which a change in localization had occurred. Necdin localizes primarily in the cytoplasm in most cells. (c and d) Necdin localization is altered in the presence of EBNA3C (c) and occurs in insoluble fractions probably complexing with membranous proteins (d). (d and e) Analysis of 100 cells for Necdin staining showed that changes in localization of Necdin to membranes and nucleus were seen in about 60% cells when they were coexpressing EBNA3C. Error bars indicate standard deviations.
Nm23-H1 interacts with Necdin in vitro.
Previous studies in our laboratory have shown that Nm23-H1 specifically interacts with a region of EBNA3C between aa 629 and 792, which encompasses the proline- and glutamine-rich region of EBNA3C (70). To determine other possible interacting partners of Nm23-H1, we analyzed the Nm23-H1-interacting sequence of EBNA3C by Blast analysis. The results showed that EBNA3C aa 648 to 662 had a high degree of homology to Necdin, with a 70% identity (data not shown). Since Necdin had a domain which showed homology to the Nm23-H1 binding domain of EBNA3C, we wanted to study whether Necdin can in fact interact with Nm23-H1. The interaction was first determined in the context of bacterially expressed human Nm23-H1. The results clearly showed that GST-Nm23-H1 beads precipitated in vitro-translated, 35S-labeled Necdin, while negligible signal was observed with GST control beads (Fig. 3a).
FIG. 3.
(a) Necdin binds to Nm23H1. The in vitro-translated Necdin protein was tested for binding with GST-tagged Nm23H1. Necdin showed binding with GST-Nm23H1 but not with control GST-2TK. In vitro-translated Necdin was precleared with glutathione-Sepharose beads in binding buffer (1× PBS, 0.1% NP-40, 0.5 mM dithiothreitol, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml) for 30 min. Precleared labeled protein were then incubated with either GST or GST-Nm23H1 fusion proteins in binding buffer. Binding was performed overnight at 4°C with constant rotation, followed by collection of the beads through centrifugation. The beads were washed three times with 1 ml of binding buffer, followed by resuspension in SDS lysis buffer and resolution by SDS-PAGE. The bound fraction was analyzed after drying the gel and exposing it to a PhosphorImager plate (Molecular Dynamics, Inc.). Necdin binds with GST-Nm23H1 (upper panel, lane 2) but not with GST control (upper panel, lane 3). Lane 1 (upper panel) shows the Necdin input control, whereas the middle and lower panels show GST protein loading controls. (b) Necdin binds to Nm23H1 in vivo. After 48 h of incubation, the transfected cells were lysed in cold IP assay buffer on ice and homogenized. The lysates were precleared with protein A/G-Sepharose beads (Amersham Biosciences, Inc., Piscataway, NJ) and then incubated first with antibodies against Myc or HA (1 μg) in IP buffer overnight at 4°C with constant rotation and then with protein A/G (50/50)-Sepharose beads for 1 h. The beads were washed three times with IP buffer and resuspended in 40 μl of 2× SDS Laemmli buffer. The sample was subjected to SDS-PAGE. Western blot analyses were performed using primary monoclonal antibodies against Myc (9E10) or HA (12CA5) and β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Detection and quantification were done using a Li-Cor Odyssey scanner (Li-Cor, Inc., Lincoln, NE). Co-IP experiments were carried out using HA-tagged Necdin and Myc-tagged Nm23H1 with anti-Myc and anti-HA antibodies. Nm23H1-Myc was immunoprecipitated with Myc antibody (left panel) and then tested for Necdin co-IP by blotting with HA antibody. The results show that Necdin coimmunoprecipitated with Nm23-H1 (right panel). In each panel, lane 1 shows the input (20%), lane 2 is preclear, and lane 3 is IP or co-IP. (c) Results of the reverse experiment using HA antibody for immunoprecipitating Necdin-HA and testing co-IP of Nm23-H1. The results show that Nm23-H1 coimmunoprecipitated with Necdin (right panel). Error bars indicate standard deviations.
Necdin and Nm23-H1 can associate with one another and form a complex in human cells.
To assess whether Necdin associates with Nm23-H1 in vivo, we then tested the interaction between Necdin and Nm23-H1 in cells by using co-IP assays for HEK-293T cells that were transiently transfected with HA-tagged Necdin and Myc-tagged Nm23-H1 expression constructs. Whole-cell extracts of the transfected 293T cells were precipitated with anti-Myc or anti-HA antibody, and the precipitates were analyzed by Western blotting with anti-HA and anti-Myc antibodies, respectively. As shown in Fig. 3b, Necdin associated in a complex with Nm23-H1 in human cells. Importantly, results of the reverse IP analysis with anti-HA antibody and subsequent Western blotting with anti-Myc antibody showed that Nm23-H1 can form a complex with Necdin (Fig. 3c). Almost 10% of the total Necdin was coimmunoprecipitated with Nm23-H1, suggesting a strong association in human cells.
Necdin increases NDP kinase activity but decreases autophosphorylation activity of Nm23-H1.
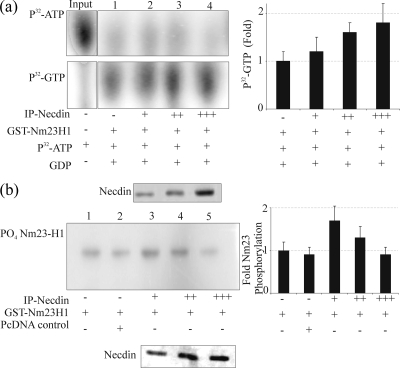
Biochemical studies have shown that Nm23-H1 has several types of enzymatic activity, including autophosphorylation and NDP kinase activities. To determine whether the interaction of Nm23-H1 with Necdin affected its biochemical properties, we examined the NDP kinase (Fig. 4a) and autophosphorylation (Fig. 4b) activities of Nm23-H1 in the presence of increasing amounts of Necdin. It is known that the NDP kinase activity of Nm23-H1 can transfer γ-phosphate from all of nucleoside triphosphates (NTPs) to form corresponding NDPs (74). To determine whether there was an effect of Necdin on the NDP kinase activity of Nm23-H1, [α-32P]ATP and GDP were incubated with GST-Nm23-H1 alone or with a mixture of GST-Nm23-H1 and immunoprecipitated Necdin. As expected, GST-Nm23-H1 alone quickly led to conversion of the GDP to GTP by 5 min. Moreover, the addition of Necdin further increased the amounts of GTP formed by transfer of γ-phosphate from [α-32P]ATP to GDP (Fig. 4a). Necdin has no known NDP kinase activity; therefore, an increase in NDP kinase activity as indicated by increased generation of labeled GTP is most likely a result of modulation of the Nm23-H1 NDP kinase activity as a result of its interaction with Necdin.
FIG. 4.
(a) The NDP kinase activity of Nm23H1 is increased in the presence of Necdin. Three micrograms of GST-Nm23-H1 with increasing amounts of immunoprecipitated Necdin was incubated with 1 μM [γ-32P]ATP (0.2 mCi/ml) and 100 μM GDP in TMD buffer in a final volume of 20 μl for 10 min at room temperature. The reaction was stopped by the addition of 20 μl of 50 mM EDTA (pH 8.0). The reaction mixture (2 μl) was spotted onto 20- by 20-cm polyethyleneimine-cellulose TLC plates (45), and reaction products were resolved by capillary action in 0.75 M KH2PO4, pH 3.65. The TLC plates were dried, exposed to PhosphorImager plates (Molecular Dynamics, Inc.), and analyzed. The top panel shows [γ-32P]ATP. The amount of [γ-32P]ATP used in each reaction is shown in the input lane. Lanes 1 to 4 (upper panel) show the unused [γ-32P]ATP, and lanes 1 to 4 in lower panel show the amount of [γ-32P]GTP synthesized. The right panel shows the quantification of [γ-32P]GTP as averages from three separate experiments. (b) Autophosphorylation activity of Nm23H1 is reduced in the presence of Necdin. A GST fusion protein of Nm23-H1 was incubated in the absence or presence of increasing amounts of immunoprecipitated Necdin. The reaction was initiated by the addition of 5 μCi of [γ-32P]ATP. After 5 min at 25°C, the reaction was stopped with Laemmli buffer; the samples were incubated for an additional 10 min and were subjected to 12% SDS-PAGE with and without boiling. The procedure without boiling was used to preserve the phosphorylation of Nm23 on the histidine residues. The gels were dried, exposed to PhosphorImager plates, and analyzed. In the left panel, lanes 1 and 2 represent the controls, and lanes 3, 4, and 5 show levels of Nm23-H1 autophosphorylation. The quantitation of autophosphorylation levels is shown in the right panel. Error bars indicate standard deviations.
The second important catalytic feature of Nm23-H1 is its ability to undergo autophosphorylation on histidine and serine residues. These effects occur when ATP is used as the main donor of phosphate (30, 74). To determine the effect of Necdin on Nm23-H1 autophosphorylation, we incubated the GST fusion Nm23-H1 protein with immunoprecipitated Necdin in the presence of [γ-32P]ATP and analyzed the phosphorylation by SDS-PAGE. Care was taken to maintain the phosphohistidine in place by not boiling the sample. In the absence of Necdin, the autophosphorylation of Nm23-H1 was detected after incubation with [γ-32P]ATP (Fig. 4b). Interestingly, the addition of Necdin markedly increased autophosphorylation of Nm23-H1. However, in the presence of increasing amounts of Necdin, the autophosphorylation of Nm23-H1 was inhibited in a dose-dependent manner (Fig. 4b). These data suggest that the interaction of Nm23-H1 with Necdin may lead to enhanced autophosphorylation but that further increases in Necdin will eventually lead to a suppression of this activity, likely due to its competition with kinase activity through sequestration of the active site of phosphorylation.
EBNA3C and Nm23-H1 can modulate Necdin-mediated regulation of downstream cellular targets.
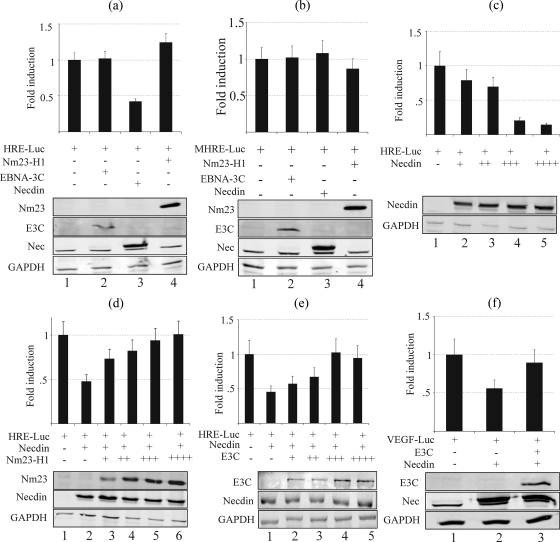
Angiogenesis is required for invasive tumor growth and metastasis and contributes to the control of cancer progression (64). Tumors secrete various angiogenic factors, including vascular endothelial growth factor (VEGF), which are important for angiogenesis (64). Hypoxia-inducible factor alpha (HIF-1α), which is a transcription factor regulated by oxygen concentrations, is directly involved in upregulation of VEGF through upregulation of its promoter. HIF-1α dimerizes with HIF-1β, and the dimer binds to HREs within promoters of target genes, such as VEGF (21). Necdin has previously been shown to interact with HIF-1α and downregulate its stability (46). To determine whether interaction of Necdin with Nm23-H1 or changes in its localization pattern in the presence of EBNA3C play any role in Necdin-mediated regulation of HIF-1α and its downstream targets, we tested the effect of Necdin on HRE promoter activity by using luciferase promoter assays. Our results show that Necdin downregulates HRE promoter activity, whereas EBNA3C and Nm23-H1 did not have any significant direct effect on the activity of HRE promoter (Fig. 5a). Importantly, Necdin did not affect the activity of the MHRE promoter, suggesting that the effect of Necdin on the HRE element is specific (Fig. 5b). The downregulation of HRE by Necdin was also dose dependent (Fig. 5c). Moreover, both Nm23-H1 and EBNA3C were able to rescue the HRE-dependent transcription from Necdin-mediated downregulation in a dose-dependent manner (Fig. 5d and e). Further analysis using the specific VEGF promoter demonstrated that EBNA3C was able to rescue its activity from Necdin-mediated downregulation (Fig. 5f).
FIG. 5.
Necdin downregulates the HRE sequence containing promoter activity, whereas EBNA3C and Nm23-H1 rescue it from Necdin-mediated downregulation. In all experiments, 10 million 293T cells were transfected with 5.0 μg of pGL3-HRE reporter construct (except in panel b, where 5.0 μg of pGL3-MHRE was used) along with other expression constructs as explained below. All the results are expressed as means ± standard errors of the means from three separate experiments performed in duplicate or triplicate. (a) Five micrograms of pA3M-Nm23-H1, pA3M-EBNA3C, pA3HA-Necdin, or a vector control was transfected. The results show that Necdin downregulates HRE promoter activity (lane 3), whereas EBNA3C and Nm23-H1 (lanes 2 and 4) do not have any significant direct effect on the activity of HRE promoter. (b) Five micrograms of Nm23-H1, EBNA3C, Necdin, or a vector control was transfected. The results show that Necdin did not induce any effect on MHRE promoter activity (lane 3), suggesting that the effect of Necdin on HRE is specific. (c) Cells were transfected with 0, 2.5, 5.0, 7.5, or 10.0 μg of pA3HA-Necdin. The results show that the downregulation of HRE by Necdin is dose dependent (lanes 2 to 5). (d) Five micrograms of pA3HA-Necdin, along with 0, 0.1, 0.2, 0.3, or 0.5 μg of pA3M-Nm23-H1, was transfected. The results show that Nm23-H1 was able to rescue the HRE-dependent transcription from Necdin-mediated downregulation in a dose-dependent manner (lanes 3 to 6). (e) Five micrograms of pA3HA-Necdin, plus 0, 0.1, 0.2, 0.3, or 0.5 μg of pA3M-EBNA3C, was transfected. The results show that EBNA3C was able to rescue the HRE-dependent transcription from Necdin-mediated downregulation in a dose-dependent manner (lanes 2 to 6). (f) Five micrograms of pA3HA-Necdin with or without 5.0 μg EBNA3C was transfected. The results confirmed that EBNA3C is able to rescue activity of the VEGF promoter from Necdin-mediated downregulation (lane 3).
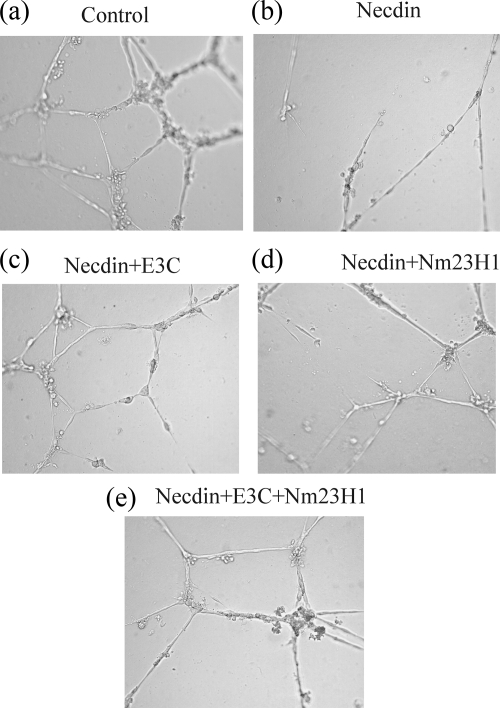
EBNA3C and Nm23-H1 rescue cells from Necdin-mediated antiangiogenic and anti-invasive effects.
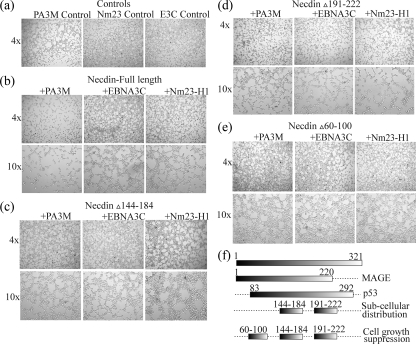
Necdin has been previously shown to have antiangiogenic activities (46). We tested the effect of Nm23-H1 and EBNA3C on the antiangiogenic activity of Necdin by using a tube formation assay with HUVECs on Matrigel (Fig. 6). CM taken from cells transfected with Necdin alone and Necdin with EBNA3C or Nm23-H1 was tested for activity. As expected, Necdin inhibited the formation of tube structures compared to results for the vector control (Fig. 6a and b). CM harvested from cells expressing EBNA3C with Necdin and from cells expressing EBNA3C together with Nm23-H1 and Necdin did not show an antiangiogenic effect (Fig. 6c and e). This result suggests that EBNA3C can rescue the cells from Necdin-mediated antiangiogenic effects on HUVECs. However, Nm23-H1 alone may be able to only partially rescue the cells from Necdin-mediated antiangiogenic effects, as evidenced by partial or negligible inhibition of tube formation (Fig. 6d). We also studied the role of EBNA3C and Nm23-H1 in Necdin-mediated effects on invasive and growth characteristics of HeLa cells in Matrigel (Fig. 7). EBNA3C or Nm23-H1 alone did not have any effects on the growth characteristics of HeLa cells (Fig. 7a). However, both EBNA3C and Nm23-H1 were able to rescue the cells from Necdin-mediated growth inhibition of HeLa cells (Fig. 7b). The domains of Necdin important for different biological functions have been mapped previously (71). The domains consisting of aa 144 to 184 and 191 to 222 are important for subcellular distribution for Necdin as well as for cell growth suppression, whereas the domain consisting of aa 60 to 100 is important for subcellular distribution (71). We analyzed the domains to determine their role in EBNA3C or Nm23-H1 regulation of Necdin-mediated growth inhibition. The Necdin Δ144-184 and Δ60-100 mutants did not show any suppression of HeLa cell growth, suggesting a role of these domains in Necdin-mediated growth inhibition (Fig. 7c and e). However, the Necdin Δ191-222 mutant did show partial inhibition of HeLa cell growth, and, more importantly, neither EBNA3C nor Nm23-H1 was able to rescue the cells from the Necdin Δ191-222 mutant-mediated growth suppression (Fig. 7d). This result suggests that aa 191 to 222 of Necdin are important for EBNA3C- and Nm23-H1-modulated regulation of Necdin-mediated functions.
FIG. 6.
EBNA3C and Nm23-H1 can rescue HUVECs from antiangiogenic activity of Necdin. 293T cells were transfected with Necdin and vector control, EBNA3C, or Nm23-H1 expression constructs. The cells were allowed to grow for 48 h, after which the supernatant medium was harvested and stored at 4°C until used as CM. Each well of a 24-well plate was coated with 150 μl of ice-cold Matrigel (BD Biosciences), which was incubated at 37°C for about 30 min. A total of 5 × 104 HUVECs were seeded on each well and grown in 500 μl of CM with 10% FBS. After 16 h of incubation, the cells were photographed to detect the formation of tubes structure by HUVECs. Tube structures were formed with the vector control (a) but not with Necdin (b). CM harvested from cells expressing EBNA3C with Necdin and from cells expressing EBNA3C together with Nm23-H1 and Necdin did not show an antiangiogenic effect (c and e), but medium from cells expressing Nm23-H1 with Necdin did show some antiangiogenic activity (d).
FIG. 7.
EBNA3C and Nm23-H1 rescue HeLa cells from Necdin-mediated anti-invasive effects. 293T cells were transfected with Necdin and vector control, EBNA3C, or Nm23-H1 expression constructs. The cells were allowed to grow for 48 h, after which the supernatant medium was harvested and stored at 4°C until used as CM. Each well of a 24-well plate was coated with 150 μl of ice-cold Matrigel (BD Biosciences), which was incubated 37°C for about 30 min. A total of 5 × 104 HeLa cells were seeded on each well and grown in the 500 μl CM with 10% FBS. After 16 h of incubation, the cells were photographed to detect the growth properties of HeLa cells. Necdin suppresses growth of HeLa cells (a, left panel), whereas Nm23-H1 (center panel) and EBNA3C (right panel) do not have any effect. Both EBNA3C and Nm23-H1 were able to rescue the cells from Necdin-mediated growth inhibition of HeLa cells (b). Necdin Δ144-184 (c) and Necdin Δ60-100 (e) did not show any suppression of HeLa cell growth, suggesting a role of these domains in Necdin-mediated growth inhibition. However, Necdin Δ191-222 did show partial inhibition of HeLa cell growth, and, more importantly, neither EBNA3C nor Nm23-H1 was able to rescue the cells from Necdin Δ191-222-mediated growth suppression (d).
DISCUSSION
Earlier studies have revealed that the expression of the MAGE family member Necdin is induced in terminally differentiated cells, suggesting a role in cell differentiation and cellular pathways linked to differentiation (41). Indeed, Necdin was discovered because its expression was markedly increased when cancer cells were induced to differentiate (41). Necdin is recognized as a basic nuclear DNA binding protein that can associate with other transcription factors to regulate the activity of a number of cellular genes (40). However, the Necdin gene is more commonly associated with the neurogenetic disorder PWS, in which this gene is inactivated or deleted (36). Necdin is located in the PWS chromosomal region and expressed exclusively from the paternally inherited allele, whereas the maternal allele is imprinted (22).
Recently, it has been reported that EBV-positive transformed lymphocytes show a higher level of methylation within the CpG sites of the Necdin promoter than do primary lymphocytes (29). This suggests that the EBV latent antigens may be involved in regulation of Necdin-mediated functions possibly related to cell cycle regulation and apoptosis. EBV is also known to be associated with a number of human lymphoid and epithelial tumors. Moreover, the essential EBV antigen EBNA3C has been shown to interact with the known metastasis suppressor Nm23-H1 (70). This interaction between EBNA3C and Nm23-H1 is of interest, as it presents a potential scenario where the interaction of this essential viral oncoprotein and the known cellular metastasis suppressor (Nm23-H1), a regulator of cell proliferation, can lead to reversion of cell migration in vitro as well as affect cell proliferation and metastasis in EBV-positive tumors (69). The binding domain for Nm23-H1 has been mapped to aa 637 to 675 on EBNA3C (70). This Nm23-H1 binding domain on EBNA3C shows a high degree of sequence homology with Necdin, which is also known to act as a transcriptional factor (43). Necdin is localized in the cytoplasm of fully differentiated neurons and is translocated to the perinuclear compartments, possibly associating with heterochromatin under certain conditions where it can perform its transcriptional activities (55). We therefore tested the possibility that EBNA3C might promote angiogenesis and growth of cancer cells by modulation of Necdin-regulated functions.
Our results clearly show that the expression of Necdin in EBV-positive cells is significantly lower than that in EBV-negative cells, suggesting a role for EBV in modulation of Necdin expression. EBNA3C is one of the critical EBV proteins that are expressed in type III latency during EBV infection. Type III latency is commonly associated with AIDS-associated lymphomas and posttransplant lymphoproliferative disorders (75). In at least one study, two cell lines derived from EBV-positive gastric carcinoma tissue have been shown to express a type III latency program, indicating that EBNA3C may also be expressed in tumors of epithelial origin (50). In addition, EBV transformation of B lymphocytes requires a subset of six latent proteins, including EBNA3C. Previous studies have reported that EBV-positive LCLs show significantly increased methylation of CpG sites within the Necdin promoter compared to circulating B lymphocytes (29). DNA methylation in the promoters of imprinted target genes such as Necdin is one of the strategies for regulation of their allelic and tissue-specific expression. Therefore, it is probable that a much-reduced level of Necdin in EBV-positive cells compared to EBV-negative cells may be the result of increased methylation and epigenetic silencing of the Necdin promoter, which is mediated by the recruitment of methyl transferases by EBV latent antigens, including EBNA3C, to these sites.
Previous studies show that endogenous Necdin is localized predominantly to the cytoplasm and is translocated to the nucleus under specific conditions; there, it may then exert its transcriptional functions (55). These studies clearly show that the majority of Necdin is localized to the cytoplasm and that its localization is strikingly altered in the presence of the EBV latent antigen EBNA3C. Importantly, Necdin can also function as a transcriptional factor which can specifically bind to multiple guanosine clusters (43). Necdin has also been shown to bind directly with DNA sequences containing GN boxes, but it did not show any interaction with those containing GC boxes (43). An examination of this interaction on the endogenous c-myc P1 promoter did show that Necdin bound to its GN box and furthermore also repressed its Sp1-dependent transcriptional activities (43). Necdin is also known to interact with several transcriptional factors, including p53 (72), E2F1 (27), Hif-1α (46), and Sp family members (43). Thus, EBNA3C mediated alteration in localization of Necdin resulting in its sequestration in complexes associated with heterochromatin is likely to be important for modulation of Necdin-mediated functions by affecting Necdin function as a transcriptional factor and its downstream activities.
As with all MAGE proteins, Necdin has a central homologous domain with the melanoma-associated antigen (57) protein family members, which are expressed in different tumor cell types (12). The MAGE family of proteins has been known to play critical roles in various cellular processes, including cell cycle progression and apoptosis, and in a number of disease conditions, such as neurogenetic disorders, gastrointestinal carcinoma, esophageal carcinoma, and pulmonary carcinoma (5, 85). Other members of the MAGE family of proteins, such as Dlxin1, have been reported to be associated with membrane proteins, such as Ror1, which is a member of the mammalian Ror family of receptor tyrosine kinases (42). In a previous study, Ror2 was found to modulate its downstream Msx2 functions by sequestering Msx2 to membrane compartments (42). Our studies have clearly shown that the presence of EBNA3C results not only in the relocation of Necdin but also in the sequestration of Necdin into membrane or perinuclear compartments, probably through interaction with membrane- or heterochromatin-specific proteins. Therefore, EBNA3C may modulate Necdin-mediated functions by regulating its subcellular localization. Necdin shows sequence homology to the Nm23-H1 binding domain of EBNA3C. Nm23-H1 is also a transcriptional factor whose subcellular localization is modulated by EBNA3C (70). Our results clearly show that Necdin interacts with Nm23-H1. Moreover, this interaction results in modulation of the biological functions of both Nm23-H1 and Necdin. It would be interesting to determine the level of association of EBNA3C with Nm23-H1 in complex with Necdin and whether EBNA3C can compete with Necdin for binding to Nm23-H1 and so alter its ability to modulate cellular processes. These studies are currently ongoing in our laboratory.
Among the various biochemical activities of Nm23-H1, its NDP kinase activity is required for the energy transfer necessary to produce NTP pools at the expense of ATP (82). Nm23-H1 removes the terminal phosphate from NTP to autophosphorylate its own histidine 118 and then transfers the phosphate to NDP to recreate NTP (82). The most important role for NDP kinase is to provide NTPs for nucleic acid synthesis and to the cytosol (28). Numerous reports suggest that NDP kinase activity may also provide phosphorylation energy to cytosolic structures, such as the translation apparatus (63, 67), G proteins, microtubules (7, 58), and chaperones (33). Our results show that Necdin appears to increase the NDP kinase activity of Nm23-H1. This is indeed surprising, because Necdin is known to have antigrowth/antiproliferative functions, whereas Nm23-H1 NDP kinase activity is important for providing NTPs for nucleic acid synthesis and hence promotes cell growth. Moreover, the NDP kinase activity of Nm23-H1 is also responsible for biosynthesis of GTPs, which are important for signal transduction. It is possible that Nedcdin-Nm23-H1 interaction may also involve other interacting partners in the cell. In EBV-infected cells, the latent antigen EBNA3C, which is known to play a role in the translocation of Nm23-H1, may also play a role in altering the localization of Necdin. In this context, it becomes even more important to study the significance of changes in the localization of Necdin in the presence of EBNA3C in terms of their effect on biological activities of Necdin.
The autophosphorylation activity of Nm23-H1 has been directly related to the potential of Nm23-H1 to suppress metastasis by regulation of other signaling pathways (37). Our results show that at lower concentrations, Necdin upregulates the autophosphorylation activity of Nm23-H1. However, when the level of Necdin was increased, the results was a reduction of the autophosphorylation activity of Nm23-H1 in a dose-dependent manner to levels equal to or lower than basal levels. Thus, regulation of Necdin subcellular localization and its alterations may play a critical role in determining the functional consequences of Necdin-Nm23-H1 interaction. It is also important to note that Necdin levels are comparatively lower in EBV-positive cancer cells than in EBV-negative ones. These low Necdin levels can be important in the differential regulation of Nm23-H1 autophosphorylation activity and may also provide EBV-positive cancer cells with a growth advantage over EBV-negative cancer cells. Previous studies have shown that other proteins, including Rad, which is the prototypic member of a new class of Ras-related GTPases, interact with Nm23-H1 and are known to regulate Nm23-H1 by enhancing its NDP kinase activity and decreasing its autophosphorylation (86). Therefore, the biological consequences of EBNA3C interaction with Nm23-H1 in EBV-positive cells, such as suppression of its antimetastasis activity and promotion of tumorigenicity, may in part be explained by its regulation of the association of Necdin and Nm23-H1.
The most critical biological consequences of Necdin are its negative effect on the growth of cells and its antiangiogenic activity. The regulation of the angiogenic switch is very critical as the tumors increase in size and metastatic potential. As the tumors become neovascularized, the “angiogenic switch” can be regulated by perturbing the local balance of proangiogenic and antiangiogenic factors (25). VEGF is one of the angiogenic factors secreted by tumors that play major roles in this process (64). The interaction between Necdin and HIF-1α is known to contribute to the downregulation of the HIF-1α protein levels, resulting in a decrease in VEGF expression (46). This is due mainly to a decrease in the transcriptional activity of HIF-1α, which is known to act as a transcriptional regulator by targeting HRE sequences in promoters of downstream target genes (46). Our results confirm that Necdin downregulates the VEGF promoter containing HRE cis-acting elements. Nm23-H1 was able to rescue the HRE-responsive promoter from Necdin-mediated downregulation. This suggests that direct interaction of Nm23-H1 with Necdin also results in modulation of Necdin-mediated functions. Increased expression of Nm23-H1 in cancer cells is commonly associated with lower metastasis potential. Nm23-H1, however, has also been shown to increase the growth rate of cancer cells, resulting in high tumorigenicity (23). It is probable that rescuing the transcriptional activity of Necdin downstream genes could be one of the ways to achieve this goal. Interestingly, EBNA3C was also able to rescue the HRE-responsive cis-acting promoter from Necdin-mediated downregulation. This clearly provides EBV-positive cancer cells with a growth advantage by tilting the balance in favor of angiogenesis factors.
We also show that EBNA3C and (less so) Nm23-H1 were able to rescue the cells from Necdin-mediated growth inhibition as well as from its antiangiogenic effects. Since Nm23-H1 alone was able to only partially rescue the cells from the antiangiogenic effect of Necdin on HUVECs, a change in localization of Necdin is most likely the major strategy for modulation of its antiangiogenic activity, as a complete rescue was seen only with EBNA3C. The interaction with Nm23-H1 may also play a minor role. However, Nm23-H1 was able to rescue the cells from Necdin-mediated HeLa cell growth inhibition as efficiently as EBNA3C. This strongly suggests a mechanism independent of the subcellular localization changes of Necdin.
The amino-terminal region of Necdin (aa 1 to 100) is highly acidic and proline rich and is less conserved between mouse and human Necdin sequences (54). The rest of the region (aa 101 to 325), including the MHD, is highly conserved between mouse and human. The MHD is a 160- to 170-aa motif of shared sequence similarity among the MAGE family of gene products, which includes Necdin (5). The domains consisting of aa 60 to 100, aa 144 to 184, and aa 191 to 222 of Necdin have been shown to be important for the related biological functions of Necdin (71). Our results clearly show that Necdin aa 60 to 100 and 144 to 184 are critical for Necdin-mediated suppression of HeLa cell growth. More importantly, we show that both EBNA3C and Nm23-H1 were unable to rescue the cells from cell growth suppression mediated by the Necdin protein with residues 191 to 222 deleted. This shows that aa 191 to 222 of Necdin, although not important for Necdin-mediated growth suppression, may be critical for modulation of Necdin-mediated functions by EBNA3C and Nm23-H1. Our data therefore suggest that different domains of Necdin could be involved in interaction with upstream regulatory molecules and downstream effector molecules.
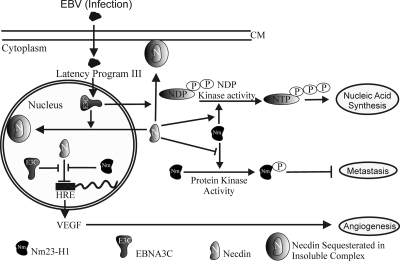
It is interesting that Necdin has also been shown to interact with simian virus 40 large T antigen and the adenovirus protein E1A (73). The interactions of Necdin with viral oncoproteins may have critical consequences in the context of the numerous functions of these proteins, which include activities related to cell growth, proliferation, and apoptosis (73). The modulation of subcellular localization of Necdin by viral proteins such as EBNA3C is likely to be another mechanism by which transforming viruses can regulate Necdin-mediated functions, including its interaction with other cellular proteins, such as Nm23-H1. We present a model (Fig. 8) for this interaction in which the presence of EBNA3C results in a change in the localization patterns of Necdin and Nm23-H1. This results in translocation of Nm23-H1 and Necdin from mainly cytoplasmic to mainly nuclear localization. The majority of Necdin is also sequestered in membranes bound to multifunctional complexes both in the cytoplasm and in the nucleus, which may contribute to its transcriptional repressive activities. This makes Necdin unavailable for regulating its functions, including effects on downstream promoters, growth, and angiogenesis. Nm23-H1 also interacts directly with Necdin, affecting its functions (Fig. 8). Moreover, the interaction of Necdin with Nm23-H1 modulates NDP kinase activity as well as its protein kinase activity (Fig. 8).
FIG. 8.
Schematic model for interaction of EBNA3C with Nm23-H1 and Necdin, resulting in regulation of Necdin- and Nm23-H1-mediated functions. The presence of EBNA3C results in a change in localization of Necdin and Nm23-H1 to the nucleus and membranes. Nm23-H1 and EBNA3C rescues Necdin-mediated downregulation of the downstream promoter. Interaction of Necdin with Nm23-H1 modulates NDP kinase activity as well as protein kinase activity.
Further information about the interactions between the growth suppressor protein Necdin and cell cycle regulatory machinery as well as their modulation by EBV latent antigens will lead to a better understanding of the molecular mechanisms underlying EBV-mediated B-cell transformation.
Acknowledgments
This work was supported by grants from the Leukemia and Lymphoma Society of America and Public Health Service grants from the NCI (CA108461, CA137894, and CA091792). E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
We are thankful to Rachel Wevrick (University of Alberta) for providing the Necdin expression construct. We are also grateful to Amit Maity and Craig B. Thompson for generously providing reagents.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Aizawa, T., K. Maruyama, H. Kondo, and K. Yoshikawa. 1992. Expression of Necdin, an embryonal carcinoma-derived nuclear protein, in developing mouse brain. Brain Res. Dev. Brain Res. 68265-274. [DOI] [PubMed] [Google Scholar]
- 2.Arcinas, M., and L. M. Boxer. 1994. Differential protein binding to the c-myc promoter during differentiation of hematopoietic cell lines. Oncogene 92699-2706. [PubMed] [Google Scholar]
- 3.Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J. Biol. Chem. 27211336-11343. [DOI] [PubMed] [Google Scholar]
- 4.Backer, J. M., C. E. Mendola, I. Kovesdi, J. L. Fairhurst, B. O'Hara, R. L. Eddy, Jr., T. B. Shows, S. Mathew, V. V. Murty, and R. S. Chaganti. 1993. Chromosomal localization and nucleoside diphosphate kinase activity of human metastasis-suppressor genes NM23-1 and NM23-2. Oncogene 8497-502. [PubMed] [Google Scholar]
- 5.Barker, P. A., and A. Salehi. 2002. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci. Res. 67705-712. [DOI] [PubMed] [Google Scholar]
- 6.Berberich, S. J., and E. H. Postel. 1995. PuF/NM23-H2/NDPK-B transactivates a human c-myc promoter-CAT gene via a functional nuclease hypersensitive element. Oncogene 102343-2347. [PubMed] [Google Scholar]
- 7.Biggs, J., E. Hersperger, P. S. Steeg, L. A. Liotta, and A. Shearn. 1990. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell 63933-940. [DOI] [PubMed] [Google Scholar]
- 8.Bominaar, A. A., A. D. Tepper, and M. Veron. 1994. Autophosphorylation of nucleoside diphosphate kinase on non-histidine residues. FEBS Lett. 3535-8. [DOI] [PubMed] [Google Scholar]
- 9.Bonnet, M., J. M. Guinebretiere, E. Kremmer, V. Grunewald, E. Benhamou, G. Contesso, and I. Joab. 1999. Detection of Epstein-Barr virus in invasive breast cancers. J. Natl. Cancer Inst. 911376-1381. [DOI] [PubMed] [Google Scholar]
- 10.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 1511-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush, J. R., and R. Wevrick. 2008. The Prader-Willi syndrome protein necdin interacts with the E1A-like inhibitor of differentiation EID-1 and promotes myoblast differentiation. Differentiation 76994-1005. [DOI] [PubMed] [Google Scholar]
- 12.Chibuk, T. K., J. M. Bischof, and R. Wevrick. 2001. A Necdin/MAGE-like gene in the chromosome 15 autism susceptibility region: expression, imprinting, and mapping of the human and mouse orthologues. BMC Genet. 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhuri, T., S. C. Verma, K. Lan, and E. S. Robertson. 2006. Expression of alpha V integrin is modulated by Epstein-Barr virus nuclear antigen 3C and the metastasis suppressor Nm23-H1 through interaction with the GATA-1 and Sp1 transcription factors. Virology 35158-72. [DOI] [PubMed] [Google Scholar]
- 14.de la Rosa, A., R. L. Williams, and P. S. Steeg. 1995. Nm23/nucleoside diphosphate kinase: toward a structural and biochemical understanding of its biological functions. Bioessays 1753-62. [DOI] [PubMed] [Google Scholar]
- 15.Demple, B., T. Herman, and D. S. Chen. 1991. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl. Acad. Sci. USA 8811450-11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan, Z., P. J. Beresford, D. Y. Oh, D. Zhang, and J. Lieberman. 2003. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112659-672. [DOI] [PubMed] [Google Scholar]
- 17.Gilles, A. M., E. Presecan, A. Vonica, and I. Lascu. 1991. Nucleoside diphosphate kinase from human erythrocytes. Structural characterization of the two polypeptide chains responsible for heterogeneity of the hexameric enzyme. J. Biol. Chem. 2668784-8789. [PubMed] [Google Scholar]
- 18.Gunay-Aygun, M., S. Schwartz, S. Heeger, M. A. O'Riordan, and S. B. Cassidy. 2001. The changing purpose of Prader-Willi syndrome clinical diagnostic criteria and proposed revised criteria. Pediatrics 108E92. [DOI] [PubMed] [Google Scholar]
- 19.Hailat, N., D. R. Keim, R. F. Melhem, X. X. Zhu, C. Eckerskorn, G. M. Brodeur, C. P. Reynolds, R. C. Seeger, F. Lottspeich, J. R. Strahler, et al. 1991. High levels of p19/nm23 protein in neuroblastoma are associated with advanced stage disease and with N-myc gene amplification. J. Clin. Investig. 88341-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemmerich, S., and I. Pecht. 1992. Oligomeric structure and autophosphorylation of nucleoside diphosphate kinase from rat mucosal mast cells. Biochemistry 314580-4587. [DOI] [PubMed] [Google Scholar]
- 21.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 957987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jay, P., C. Rougeulle, A. Massacrier, A. Moncla, M. G. Mattei, P. Malzac, N. Roeckel, S. Taviaux, J. L. Lefranc, P. Cau, P. Berta, M. Lalande, and F. Muscatelli. 1997. The human Necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat. Genet. 17357-361. [DOI] [PubMed] [Google Scholar]
- 23.Kaul, R., M. Murakami, T. Choudhuri, and E. S. Robertson. 2007. Epstein-Barr virus latent nuclear antigens can induce metastasis in a nude mouse model. J. Virol. 8110352-10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaul, R., S. C. Verma, M. Murakami, K. Lan, T. Choudhuri, and E. S. Robertson. 2006. Epstein-Barr virus protein can upregulate cyclo-oxygenase-2 expression through association with the suppressor of metastasis Nm23-H1. J. Virol. 801321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerbel, R., and J. Folkman. 2002. Clinical translation of angiogenesis inhibitors. Nat. Rev. Cancer 2727-739. [DOI] [PubMed] [Google Scholar]
- 26.Kuppers, D. A., K. Lan, J. S. Knight, and E. S. Robertson. 2005. Regulation of matrix metalloproteinase 9 expression by Epstein-Barr virus nuclear antigen 3C and the suppressor of metastasis Nm23-H1. J. Virol. 799714-9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuwako, K., H. Taniura, and K. Yoshikawa. 2004. Necdin-related MAGE proteins differentially interact with the E2F1 transcription factor and the p75 neurotrophin receptor. J. Biol. Chem. 2791703-1712. [DOI] [PubMed] [Google Scholar]
- 28.Lacombe, M. L., L. Milon, A. Munier, J. G. Mehus, and D. O. Lambeth. 2000. The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 32247-258. [DOI] [PubMed] [Google Scholar]
- 29.Lau, J. C., M. L. Hanel, and R. Wevrick. 2004. Tissue-specific and imprinted epigenetic modifications of the human NDN gene. Nucleic Acids Res. 323376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leone, A., U. Flatow, C. R. King, M. A. Sandeen, I. M. Margulies, L. A. Liotta, and P. S. Steeg. 1991. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell 6525-35. [DOI] [PubMed] [Google Scholar]
- 31.Leone, A., R. C. Seeger, C. M. Hong, Y. Y. Hu, M. J. Arboleda, G. M. Brodeur, D. Stram, D. J. Slamon, and P. S. Steeg. 1993. Evidence for nm23 RNA overexpression, DNA amplification and mutation in aggressive childhood neuroblastomas. Oncogene 8855-865. [PubMed] [Google Scholar]
- 32.Lerner, M. R., N. C. Andrews, G. Miller, and J. A. Steitz. 1981. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 78805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung, S. M., and L. E. Hightower. 1997. A 16-kDa protein functions as a new regulatory protein for Hsc70 molecular chaperone and is identified as a member of the Nm23/nucleoside diphosphate kinase family. J. Biol. Chem. 2722607-2614. [DOI] [PubMed] [Google Scholar]
- 34.Livingstone, J. I., W. Yasui, E. Tahara, and C. Wastell. 1995. Are Japanese and European gastric cancer the same biological entity? An immunohistochemical study. Br. J. Cancer 72976-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma, D., Z. Xing, B. Liu, N. G. Pedigo, S. G. Zimmer, Z. Bai, E. H. Postel, and D. M. Kaetzel. 2002. NM23-H1 and NM23-H2 repress transcriptional activities of nuclease-hypersensitive elements in the platelet-derived growth factor-A promoter. J. Biol. Chem. 2771560-1567. [DOI] [PubMed] [Google Scholar]
- 36.MacDonald, H. R., and R. Wevrick. 1997. The Necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum. Mol. Genet. 61873-1878. [DOI] [PubMed] [Google Scholar]
- 37.MacDonald, N. J., A. De la Rosa, M. A. Benedict, J. M. Freije, H. Krutsch, and P. S. Steeg. 1993. A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J. Biol. Chem. 26825780-25789. [PubMed] [Google Scholar]
- 38.MacDonald, N. J., A. de la Rosa, and P. S. Steeg. 1995. The potential roles of nm23 in cancer metastasis and cellular differentiation. Eur. J. Cancer. 31A1096-1100. [DOI] [PubMed] [Google Scholar]
- 39.Martin, K. K., and G. J. Pilkington. 1998. Nm23: an invasion suppressor gene in CNS tumours? Anticancer Res. 18919-926. [PubMed] [Google Scholar]
- 40.Maruyama, E. 1996. Biochemical characterization of mouse brain necdin. Biochem. J. 314895-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruyama, K., M. Usami, T. Aizawa, and K. Yoshikawa. 1991. A novel brain-specific mRNA encoding nuclear protein (Necdin) expressed in neurally differentiated embryonal carcinoma cells. Biochem. Biophys. Res. Commun. 178291-296. [DOI] [PubMed] [Google Scholar]
- 42.Matsuda, T., H. Suzuki, I. Oishi, S. Kani, Y. Kuroda, T. Komori, A. Sasaki, K. Watanabe, and Y. Minami. 2003. The receptor tyrosine kinase Ror2 associates with the melanoma-associated antigen (MAGE) family protein Dlxin-1 and regulates its intracellular distribution. J. Biol. Chem. 27829057-29064. [DOI] [PubMed] [Google Scholar]
- 43.Matsumoto, K., H. Taniura, T. Uetsuki, and K. Yoshikawa. 2001. Necdin acts as a transcriptional repressor that interacts with multiple guanosine clusters. Gene 272173-179. [DOI] [PubMed] [Google Scholar]
- 44.Mehus, J. G., P. Deloukas, and D. O. Lambeth. 1999. NME6: a new member of the nm23/nucleoside diphosphate kinase gene family located on human chromosome 3p21.3. Hum Genet. 104454-459. [DOI] [PubMed] [Google Scholar]
- 45.Milon, L., M. F. Rousseau-Merck, A. Munier, M. Erent, I. Lascu, J. Capeau, and M. L. Lacombe. 1997. nm23-H4, a new member of the family of human nm23/nucleoside diphosphate kinase genes localised on chromosome 16p13. Hum. Genet. 99550-557. [DOI] [PubMed] [Google Scholar]
- 46.Moon, H. E., M. Y. Ahn, J. A. Park, K. J. Min, Y. W. Kwon, and K. W. Kim. 2005. Negative regulation of hypoxia inducible factor-1alpha by necdin. FEBS Lett. 5793797-3801. [DOI] [PubMed] [Google Scholar]
- 47.Munier, A., C. Feral, L. Milon, V. P. Pinon, G. Gyapay, J. Capeau, G. Guellaen, and M. L. Lacombe. 1998. A new human nm23 homologue (nm23-H5) specifically expressed in testis germinal cells. FEBS Lett. 434289-294. [DOI] [PubMed] [Google Scholar]
- 48.Munoz-Dorado, J., S. Inouye, and M. Inouye. 1993. Eukaryotic-like protein serine/threonine kinases in Myxococcus xanthus, a developmental bacterium exhibiting social behavior. J. Cell Biochem. 5129-33. [DOI] [PubMed] [Google Scholar]
- 49.Munoz-Dorado, J., S. Inouye, and M. Inouye. 1990. Nucleoside diphosphate kinase from Myxococcus xanthus. II. Biochemical characterization. J. Biol. Chem. 2652707-2712. [PubMed] [Google Scholar]
- 50.Murakami, M., Y. Hoshikawa, Y. Satoh, H. Ito, M. Tajima, K. Okinaga, Y. Miyazawa, T. Kurata, and T. Sairenji. 2000. Tumorigenesis of Epstein-Barr virus-positive epithelial cell lines derived from gastric tissues in the SCID mouse. Virology 27720-26. [DOI] [PubMed] [Google Scholar]
- 51.Murakami, M., K. Lan, C. Subramanian, and E. S. Robertson. 2005. Epstein-Barr virus nuclear antigen 1 interacts with Nm23-H1 in lymphoblastoid cell lines and inhibits its ability to suppress cell migration. J. Virol. 791559-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murakami, M., P. I. Meneses, J. S. Knight, K. Lan, R. Kaul, S. C. Verma, and E. S. Robertson. 2008. Nm23-H1 modulates the activity of the guanine exchange factor Dbl-1. Int. J. Cancer 123500-510. [DOI] [PubMed] [Google Scholar]
- 53.Murakami, M., P. I. Meneses, K. Lan, and E. S. Robertson. 2008. The suppressor of metastasis Nm23-H1 interacts with the Cdc42 Rho family member and the pleckstrin homology domain of oncoprotein Dbl-1 to suppress cell migration. Cancer Biol. Ther. 7677-688. [DOI] [PubMed] [Google Scholar]
- 54.Nakada, Y., H. Taniura, T. Uetsuki, J. Inazawa, and K. Yoshikawa. 1998. The human chromosomal gene for necdin, a neuronal growth suppressor, in the Prader-Willi syndrome deletion region. Gene 21365-72. [DOI] [PubMed] [Google Scholar]
- 55.Niinobe, M., K. Koyama, and K. Yoshikawa. 2000. Cellular and subcellular localization of Necdin in fetal and adult mouse brain. Dev. Neurosci. 22310-319. [DOI] [PubMed] [Google Scholar]
- 56.Padma, P., A. Hozumi, K. Ogawa, and K. Inaba. 2001. Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene 275177-183. [DOI] [PubMed] [Google Scholar]
- 57.Pal, S., S. N. Vishwanath, H. Erdjument-Bromage, P. Tempst, and S. Sif. 2004. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 249630-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pinon, V. P., G. Millot, A. Munier, J. Vassy, G. Linares-Cruz, J. Capeau, F. Calvo, and M. L. Lacombe. 1999. Cytoskeletal association of the A and B nucleoside diphosphate kinases of interphasic but not mitotic human carcinoma cell lines: specific nuclear localization of the B subunit. Exp. Cell Res. 246355-367. [DOI] [PubMed] [Google Scholar]
- 59.Postel, E. H., S. J. Berberich, S. J. Flint, and C. A. Ferrone. 1993. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science 261478-480. [DOI] [PubMed] [Google Scholar]
- 60.Postel, E. H., and C. A. Ferrone. 1994. Nucleoside diphosphate kinase enzyme activity of NM23-H2/PuF is not required for its DNA binding and in vitro transcriptional functions. J. Biol. Chem. 2698627-8630. [PubMed] [Google Scholar]
- 61.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 62.Rosengard, A. M., H. C. Krutzsch, A. Shearn, J. R. Biggs, E. Barker, I. M. Margulies, C. R. King, L. A. Liotta, and P. S. Steeg. 1989. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature 342177-180. [DOI] [PubMed] [Google Scholar]
- 63.Sastre-Garau, X., L. Ovtracht, I. Lascu, M. L. Lacombe, M. Veron, K. Bourdache, and J. P. Thiery. 1992. Ultrastructural immunocytochemical localization of diphosphate kinase/Nm23 in human cancer cells. Bull. Cancer 79465-470. [PubMed] [Google Scholar]
- 64.Semenza, G. L. 2002. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol. Med. 8S62-S67. [DOI] [PubMed] [Google Scholar]
- 65.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104119-130. [DOI] [PubMed] [Google Scholar]
- 66.Shikama, N., H. M. Chan, M. Krstic-Demonacos, L. Smith, C. W. Lee, W. Cairns, and N. B. La Thangue. 2000. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 208933-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sonnemann, J., and R. Mutzel. 1995. Cytosolic nucleoside diphosphate kinase associated with the translation apparatus may provide GTP for protein synthesis. Biochem. Biophys. Res. Commun. 209490-496. [DOI] [PubMed] [Google Scholar]
- 68.Stahl, J. A., A. Leone, A. M. Rosengard, L. Porter, C. R. King, and P. S. Steeg. 1991. Identification of a second human nm23 gene, nm23-H2. Cancer Res. 51445-449. [PubMed] [Google Scholar]
- 69.Subramanian, C., M. A. Cotter II, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med. 7350-355. [DOI] [PubMed] [Google Scholar]
- 70.Subramanian, C., and E. S. Robertson. 2002. The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J. Virol. 768702-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taniura, H., M. Kobayashi, and K. Yoshikawa. 2005. Functional domains of Necdin for protein-protein interaction, nuclear matrix targeting, and cell growth suppression. J. Cell Biochem. 94804-815. [DOI] [PubMed] [Google Scholar]
- 72.Taniura, H., K. Matsumoto, and K. Yoshikawa. 1999. Physical and functional interactions of neuronal growth suppressor Necdin with p53. J. Biol. Chem. 27416242-16248. [DOI] [PubMed] [Google Scholar]
- 73.Taniura, H., N. Taniguchi, M. Hara, and K. Yoshikawa. 1998. Necdin, a postmitotic neuron-specific growth suppressor, interacts with viral transforming proteins and cellular transcription factor E2F1. J. Biol. Chem. 273720-728. [DOI] [PubMed] [Google Scholar]
- 74.Tepper, A. D., H. Dammann, A. A. Bominaar, and M. Veron. 1994. Investigation of the active site and the conformational stability of nucleoside diphosphate kinase by site-directed mutagenesis. J. Biol. Chem. 26932175-32180. [PubMed] [Google Scholar]
- 75.Thompson, M. P., and R. Kurzrock. 2004. Epstein-Barr virus and cancer. Clin. Cancer Res. 10803-821. [DOI] [PubMed] [Google Scholar]
- 76.Tsuiki, H., M. Nitta, A. Furuya, N. Hanai, T. Fujiwara, M. Inagaki, M. Kochi, Y. Ushio, H. Saya, and H. Nakamura. 1999. A novel human nucleoside diphosphate (NDP) kinase, Nm23-H6, localizes in mitochondria and affects cytokinesis. J. Cell Biochem. 76254-269. [DOI] [PubMed] [Google Scholar]
- 77.Uetsuki, T., K. Takagi, H. Sugiura, and K. Yoshikawa. 1996. Structure and expression of the mouse Necdin gene. Identification of a postmitotic neuron-restrictive core promoter. J. Biol. Chem. 271918-924. [DOI] [PubMed] [Google Scholar]
- 78.van Golen, K. L., S. Risin, A. Staroselsky, D. Berger, M. A. Tainsky, S. Pathak, and J. E. Price. 1996. Predominance of the metastatic phenotype in hybrids formed by fusion of mouse and human melanoma clones. Clin. Exp. Metastasis 1495-106. [DOI] [PubMed] [Google Scholar]
- 79.Venturelli, D., R. Martinez, P. Melotti, I. Casella, C. Peschle, C. Cucco, G. Spampinato, Z. Darzynkiewicz, and B. Calabretta. 1995. Overexpression of DR-nm23, a protein encoded by a member of the nm23 gene family, inhibits granulocyte differentiation and induces apoptosis in 32Dc13 myeloid cells. Proc. Natl. Acad. Sci. USA 927435-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verma, S. C., and E. S. Robertson. 2003. ORF73 of herpesvirus Saimiri strain C488 tethers the viral genome to metaphase chromosomes and binds to cis-acting DNA sequences in the terminal repeats. J. Virol. 7712494-12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wagner, P. D., and N. D. Vu. 1995. Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J. Biol. Chem. 27021758-21764. [DOI] [PubMed] [Google Scholar]
- 82.Wallet, V., R. Mutzel, H. Troll, O. Barzu, B. Wurster, M. Veron, and M. L. Lacombe. 1990. Dictyostelium nucleoside diphosphate kinase highly homologous to Nm23 and Awd proteins involved in mammalian tumor metastasis and Drosophila development. J. Natl. Cancer Inst. 821199-1202. [DOI] [PubMed] [Google Scholar]
- 83.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 642309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Webb, P. A., O. Perisic, C. E. Mendola, J. M. Backer, and R. L. Williams. 1995. The crystal structure of a human nucleoside diphosphate kinase, NM23-H2. J. Mol. Biol. 251574-587. [DOI] [PubMed] [Google Scholar]
- 85.Xiao, J., and H. S. Chen. 2004. Biological functions of melanoma-associated antigens. World J. Gastroenterol. 101849-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu, J., Y. H. Tseng, J. D. Kantor, C. J. Rhodes, B. R. Zetter, J. S. Moyers, and C. R. Kahn. 1999. Interaction of the Ras-related protein associated with diabetes rad and the putative tumor metastasis suppressor NM23 provides a novel mechanism of GTPase regulation. Proc. Natl. Acad. Sci. USA 9614911-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]