Abstract
During the past 2 years, an atypical clinical outbreak, caused by a highly pathogenic porcine reproductive and respiratory syndrome virus (PRRSV) with a unique 30-amino-acid deletion in its Nsp2-coding region, was pandemic in China. In this study, we generated four full-length infectious cDNA clones: a clone of the highly virulent PRRSV strain JXwn06 (pWSK-JXwn), a clone of the low-virulence PRRSV strain HB-1/3.9 (pWSK-HB-1/3.9), a chimeric clone in which the Nsp2 region containing the 30-amino-acid deletion was replaced by the corresponding region of the low-virulence PRRSV strain HB-1/3.9 (pWSK-JXwn-HB1nsp2), and a mutated HB-1/3.9 clone with the same deletion in Nsp2 as JXwn06 (pWSK-HB1-ND30). We also investigated the pathogenicities of the rescued viruses (designated RvJXwn, RvJXwn-HB1nsp2, RvHB-1/3.9, and RvHB1-ND30, respectively) in specific-pathogen-free piglets in order to determine the role of the 30-amino-acid deletion in the virulence of the highly pathogenic PRRSV. All the rescued viruses could replicate stably in MARC-145 cells. Our findings indicated that RvJXwn-HB1nsp2 retained high virulence for piglets, like RvJXwn and the parental virus JXwn06, although the survival time of piglets infected with RvJXwn-HB1nsp2 was obviously prolonged. RvHB1-ND30 exhibited low virulence for piglets, like RvHB-1/3.9 and the parental virus HB-1/3.9. Therefore, we conclude that the 30-amino-acid deletion is not related to the virulence of the highly pathogenic PRRSV emerging in China.
Porcine reproductive and respiratory syndrome (PRRS) is one of the most economically devastating diseases for the pig industry worldwide (14, 30, 33). The disease was first reported in the late 1980s in the United States (19); similar clinical outbreaks occurred in Germany in 1990 and were widespread throughout Europe by 1991. The etiological agent of the disease—porcine reproductive and respiratory syndrome virus (PRRSV)—was identified in Europe and the United States in 1991 and 1992 (4, 7, 45). By now, PRRS has become a well-recognized global swine disease (1, 5, 17, 22, 41).
PRRSV is classified in the order Nidovirales, family Arteriviridae, genus Arterivirus, along with equine arteritis virus (EAV), lactate dehydrogenase-elevating virus of mice, and simian hemorrhagic fever virus (6). The virus contains a 5′-capped and 3′-polyadenylate single-strand positive-sense RNA genome of approximately 15 kb (8, 29, 37). The viral genomic RNA contains at least nine open reading frames (ORFs)—ORF1a, ORF1b, and ORFs 2 (2a and 2b) to 7—and 5′ and 3′ untranslated regions. ORF1a and ORF1b, occupying the 5′ terminus and three-fourths of the genome, encode the replicase polyprotein, which is considered to be autoproteolytically cleaved into at least 13 nonstructural proteins assumed to be associated with viral replication and transcription (9, 39, 42, 44). ORFs 2 to 7 encode viral structural proteins (3, 25, 27). Among the nonstructural proteins of PRRSV, Nsp2, the largest part of the cleavage product of the replicative protein, possesses a chymotrypsin-like cysteine protease domain (40). The Nsp2 protein of PRRSV has an organization similar to that of the Nsp2 counterpart of EAV and can be divided into three major domains: the conserved N-terminal cysteine proteinase domain, a hydrophobic transmembrane region at the C terminus, and a heterogeneous and variable region of unknown function, which is the spacer region between the former two domains (49). The biological functions of PRRSV Nsp2 are currently unclear, although EAV Nsp2 has been considered to assemble the replicative double-membrane vesicles in collaboration with Nsp3 (38).
A number of studies have indicated that PRRSV Nsp2 is able to tolerate amino acid deletion and foreign gene insertion (10, 11, 13, 15, 20 34, 41). It has been verified that an unparalleled large-scale, atypical PRRS outbreak in China in 2006 was caused by a highly virulent PRRSV strain with a unique 30-amino-acid (30-aa) deletion in its Nsp2-coding region (23, 41, 48). The question of whether the deletion in PRRSV Nsp2 is related to its virulence and pathogenicity is becoming an interesting one. The emergence of the highly virulent PRRSV strain and the subsequent pandemic have brought great economic loss to the swine industry in China. Analyzing the relationship between the 30-aa deletion of PRRSV and its virulence and pathogenicity is helpful for understanding the associated molecular mechanisms. In the present study, we constructed four infectious clones: a clone of the highly virulent PRRSV strain JXwn06, a clone of the low-virulence PRRSV strain HB-1/3.9, a chimeric clone in which the Nsp2 region containing the 30-aa deletion was replaced by the corresponding region of the low-virulence PRRSV strain HB-1/3.9, and a mutated HB-1/3.9 clone with the same deletion in Nsp2 as JXwn06. We rescued the viruses and simultaneously investigated the pathogenicities of the rescued viruses in specific-pathogen-free (SPF) piglets in comparison with the pathogenicity of the parental viruses, in order to reveal the role of the 30-aa deletion in the virulence and pathogenicity of the highly virulent PRRSV strain.
MATERIALS AND METHODS
Cells and viruses.
Cells of the MARC-145 line—a subclone of the African green monkey kidney epithelial cell line that is highly permissive for PRRSV replication—were maintained in Gibco Dulbecco's modified Eagle medium (DMEM) (Invitrogen Corporation, NY) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories Inc., South Logan, UT) at 37°C under 5% CO2. BHK-21 cells used for transfection of full-length cDNA were cultured by the same method as MARC-145 cells. Two PRRSV strains with different levels of virulence, JXwn06 and HB-1/3.9, were used for the study. Strain JXwn06 was isolated from an intensive pig farm with an atypical PRRS outbreak in the Jiangxi province of China in 2006. HB-1/3.9, adapted in MARC-145 cells, was derived from HB-1(sh)/2002, a low-virulence strain isolated in 2002 (13); its complete genomic sequence has been determined and deposited in GenBank (accession no. EU360130). There was no nucleotide deletion in the Nsp2-coding region of HB-1/3.9.
Pathogenicity analyses of JXwn06.
Healthy Large White-Dutch Landrace crossbred pigs, including 6-week-old nursery pigs and 12-week-old finishing pigs, were obtained from a pig farm that was negative for PRRSV and porcine circovirus type 2 (PCV2) infections. All animals were confirmed to be free of PRRSV and PCV2 infections by use of commercial enzyme-linked immunosorbent assay (ELISA) kits for the detection of antibodies against PRRSV (Idexx Labs Inc.) and PCV2 (Ingezim Circovirus IgG/IgM kit; Ingenasa, Spain) and by reverse transcription-PCR (RT-PCR) or PCR for viral nucleic acid detection. The animals were shown to be negative for classical swine fever virus, porcine parvovirus, pseudorabies virus, swine influenza virus, and Mycoplasma hyopneumoniae infections by serological methods and/or RT-PCR or PCR. The pigs were transported to biosafety level 3 animal facilities at the Chengdu Veterinary Biologic Factory 1 week prior to virus challenge. Ten nursery pigs were divided into an infection group and a control group, as were 10 finishing pigs. The animals in each group (n = 5) were raised separately in different isolation rooms, with individual ventilation. The animals received food and water ad libitum. Each pig in the infection groups was inoculated intranasally with 2 ml of viral culture, each milliliter containing 105 tissue culture infective doses (TCID50), from the fifth passage on MARC-145 cells. Each pig in the control groups was inoculated with 2 ml of DMEM. All pigs were clinically examined, and their rectal temperatures were measured daily until 21 days postinfection. The date and time of death of each animal was recorded.
Genomic sequencing of JXwn06.
The fifth-passage viral cultures were harvested and used for genomic sequencing according to the method described previously (13). Briefly, by use of specific primers, 14 overlapped fragments covering the whole genome were amplified by RT-PCR. The 5′ region was amplified using a 5′ full RACE kit (TaKaRa, Dalian, China) according to the manufacturer's instructions. The amplicons were cloned into pEASY-Blunt vectors (Transgen, Beijing, China) and then submitted to Invitrogen (Beijing, China) for sequencing. Genomic analyses were conducted using DNAMAN (University of California) and DNAStar (Lasergene) software.
Construction of full-length cDNA clones for JXwn06 and HB-1/3.9, a chimeric full-length cDNA clone, and an HB-1/3.9 cDNA clone with a 30-aa deletion in Nsp2.
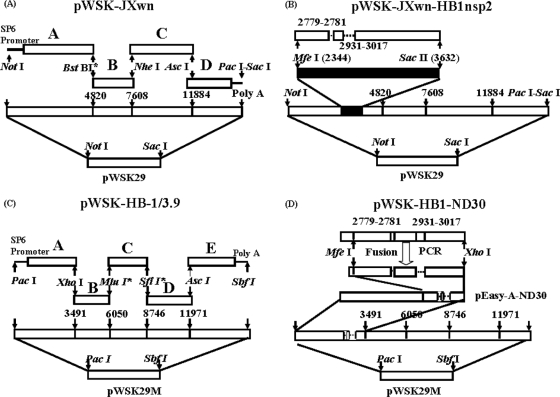
The strategy for the construction of the four full-length cDNA clones is illustrated in Fig. 1. For JXwn06, the MARC-145 cell culture supernatants were centrifuged first at 12,000 rpm for 30 min and then at 40,000 rpm for 4 h. Total RNAs were extracted from the pellet by using a QIAamp viral RNA kit (Qiagen), followed by reverse transcription with SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) and reverse primers of each fragment (Table 1). The cDNA was synthesized by incubation at 50 to 55°C for 2 h. The genome of JXwn06 was divided into four overlapping fragments spanning the appropriate restriction site. The G at position 4821 was mutated to C by PCR in order to create a BstBI restriction enzyme site, both for fragment A and B ligation and for a genetic marker. The SP6 polymerase promoter and one nontemplated G residue were added to the forward primer for fragment A (34). Amplicons were gel purified and subcloned into pEASY-Blunt vectors (Transgen, Beijing, China). Four fragments in subclone vectors were sequenced. Five point mutations, separately located in fragments B and C, were incidentally introduced by PCR. Return mutations were carried out by using the QuikChange Multi site-directed mutagenesis kit (Stratagene, La Jolla, CA) and designed mutagenic primers by following the manufacturer's suggestions in the instruction manual (Table 1). After the reaction was modified, fragments B and C were checked by sequencing again. A low-copy-number plasmid, pWSK29, was used to assemble the four overlapping fragments of JXwn06, as shown in Fig. 1A. Each fragment from D to A was excised from the subclone vector pEASY-Blunt by NotI and other specific restriction enzymes and was ligated into plasmid pWSK29 by T4 ligase (Promega, Madison, WI). After each ligation reaction, the product was transformed into Escherichia coli DH5α cells and grown overnight at 37°C in the presence of ampicillin. The full-length cDNA clone, plasmid pWSK-JXwn, was sequenced.
FIG. 1.
Strategy for the construction of full-length cDNA clones. (A) Capital letters (A, B, C, D) represent four overlapping fragments amplified from the JXwn06 genome according to the unique restriction enzyme cleavage sites in viral cDNA. A NotI enzyme cleavage site was added to the 5′ end of each fragment, while a 37-nt polyadenosine tail followed by PacI-SacI was added to the 3′ end of the D fragment by PCR mutagenesis. The BstBI site between fragments A and B was created by mutation to be the genetic marker as well as the ligation site. An SP6 promoter with one nontemplated G residue preceded the viral genome. The fragments were inserted into the low-copy-number vector pWSK29 in order from D to A. The completed full-length clone was named pWSK-JXwn. (B) An Nsp2 fragment of 1,378 nt covering the deletion region in JXwn06 was amplified from HB-1/3.9. Two suitable restriction enzyme sites (MfeI and SacII) in fragment A were used to exchange the corresponding fragment of Nsp2. The completed full-length chimeric clone was named pWSK-JXwn-HB1nsp2. (C) Five fragments amplified from HB-1/3.9 were inserted into the modified plasmid pWSK29M to construct the infectious clone. PacI was added to the 5′ end of each fragment, and MluI and SfiI were induced by PCR mutation to ligate the B and C fragments and the C and D fragments. This clone was named pWSK-HB-1/3.9. (D) A discontinuous 90-bp sequence in Nsp2 was deleted by integrating three overlapping segments using fusion PCR, and the product was then cloned into pEasy-A-ND30 using MfeI and XhoI. The modified fragment A, with a 90-bp deletion in the Nsp2 coding region, was finally assembled into the backbone of pWSK-HB-1/3.9 to construct clone pWSK-HB1-ND30.
TABLE 1.
Primers used in this study
| Primera | Positionb | Sequence (5′-3′)c | Use | |||
|---|---|---|---|---|---|---|
| JXwn06 | ||||||
| JAF (sp6) | 1-25 | GCGGCCGCGCGATTTAGGTGACACTATAGATGACGTATAGGTGTTGGCTCTATG (NotI) | Fragment clone | |||
| JAR | 4804-4826 | GGCTTCGAAATTTGCCTGATCTT (BstBI) | Fragment clone | |||
| JBF | 4801-4826 | GCGGCCGCCTAAAGATCAGGCAAATTTCGAAGC (NotI, BstBI) | Fragment clone | |||
| JBR | 8206-8184 | ATGGTTGTAGCCAAGACGGAGCG | Fragment clone | |||
| JCF | 7535-7559 | GCGGCCGCCCAAAGAACTGGAGAAACTGAAAAG (NotI) | Fragment clone | |||
| JCR | 12385-12409 | CCAAACCACTAATGCGAGACAATGT | Fragment clone | |||
| JDF | 11553-11577 | GCGGCCGCTACGCCTCATACATCCGAGTTCCTG (NotI) | Fragment clone | |||
| JDR | Poly(A) | GAGCTCGGCCGGCCTTAATTAA(T)37 (SacI, FseI, PacI) | Fragment clone | |||
| JBm5169 | 5151-5185 | GTGCATCTTACTCGCAATCGCTAGTTATGTTTGGG | Mutagenesis | |||
| JBm6657 | 6639-6671 | CTTTGCCGAGGGAAAGTTGAGGGAAGGGGTGTC | Mutagenesis | |||
| JBm7655 | 7643-7677 | CGGCTTAGTTGTTACTGAGACAGCGGTAAAAATAG | Mutagenesis | |||
| JCm9167 | 9152-9186 | CTCCATGCCAAACTACCACTGGTGGGTTGAACATC | Mutagenesis | |||
| JCm9965 | 9954-9985 | CGCAGCAGGTTTATCATCGGCCCACCCGGTGC | Mutagenesis | |||
| J3F | 2115-2137 | CCTCCGTGGCGCAACAAGTCTTG | Amplification of Nsp2 fragment of HB-1/3.9 | |||
| J4R | 4308-4329 | GGCGATCTCATTAGGAGCAGTT | ||||
| J5F | 4214-4234 | TGCTTAGGCTTGGCATTGTTG | Genetic marker detection | |||
| J5R | 5544-5564 | ACGGTGTTCAGTGAGGGCTTT | ||||
| DetectF | 2497-2517 | CTTAAAGACCAGATGGAGGAGG | Nsp2 detection | |||
| DetectR | 3156-3178 | CGATGATGGCTTGAGCTGAGTAT | ||||
| N312F | 14860-14881 | AGCTGTGCCAAATGCTGGGTAA | ORF7 detection | |||
| N312R | 15150-15171 | ATCATGCTGAGGGTGATGCTGT | ||||
| HB-1/3.9 | ||||||
| HAF (sp6) | 1-25 | GCCTTAATTAAGGCCATACGATTTAGGTGACACTATAGAAATGACGTA TAGGTGTTGGCTCTATG (PacI) | Fragment clone | |||
| HAR | 3588-3610 | TGCGTGGAACATCCTCAGTGGCT | Fragment clone | |||
| HBF | 3464-3488 | GCCTTAATTAAGGCGGTTTGAGTTTCTCCCAAAGATGAT (PacI) | Fragment clone | |||
| HBR | 6043-6066 | CTGGCCTGAGGGACGCGTGACAAT (MluI) | Fragment clone | |||
| HCF | 6044-6066 | GCCTTAATTAAGGCTTGTCACGCGTCCCTCAGGCCAG (PacI, MluI) | Fragment clone | |||
| HCR | 8729-8751 | CGGCCCGATGGGCCAACGCAATG (SfiI) | Fragment clone | |||
| HDF | 8735-8755 | GCCTTAATTAAGGCGTTGGCCCATCGGGCCGCGTT (PacI, SfiI) | Fragment clone | |||
| HDR | 12037-12061 | AGGCCTAAAGTTGGTTCAATGACAG | Fragment clone | |||
| HEF | 11925-11949 | GCCTTAATTAAGGCACTGGGAATGGTGAGGACTGGGAGG (PacI) | Fragment clone | |||
| HER | polyA | AGGCCCTGCAGGGCC(T)39 (SbfI) | Fragment clone | |||
| Seg1F | 2231-2249 | CGCATCAGACAACCGAACA | Nsp2 deletion | |||
| Seg1R | 2760-2778, 2782-2795 | GGACCACCGACAGTATCTTCCGAACCGTTAGGG | Nsp2 deletion | |||
| Seg2F | 2767-2778, 2782-2799 | GGTTCGGAAGATACTGTCGGTGGTCCCCTC | Nsp2 deletion | |||
| Seg2R | 2912-2930, 3018-3029 | AGCGTTGTTGTCACAGTTCTACGCGGTGCAG | Nsp2 deletion | |||
| Seg3F | 2917-2930, 3018-3036 | CCGCGTAGAACTGTGACAACAACGCTGACGCAC | Nsp2 deletion | |||
| Seg3R | 3589-3607 | GTGGAACATCCTCAGTGGC | Nsp2 deletion | |||
| H5F | 5701-5721 | GACGGGAAAATCAAGTGCGTA | Genetic marker detection | |||
| H5R | 6993-7013 | ACGGTATCGGCAAAAGCCTCA | ||||
F denotes a forward PCR primer; R denotes reverse transcription or a reverse PCR primer.
Numbers refer to nucleotide positions within the genome of JXwn06 (GenBank accession no: EF641008) or HB-1/3.9 (GenBank accession no: EU360130), as indicated.
Restriction sites introduced by PCR are shown in boldface and specified in parentheses at the end of the sequence. The sequence of the SP6 promoter (underlined), followed by a nongenomic G (italicized), is added in the JAF and HAF primers. The mutated nucleotides (boldface italics) in primers JAR, JBF, HBR, HCF, HCR, and HDF were used to create BstBI, MluI, and SfiI restriction enzyme sites, both for fragment ligation and as genetic markers. Mutated nucleotides in site-directed mutagenesis primers (JBm5169, JBm6657, JBm7655, JCm9167, and JCm9965) are italicized. Each site-directed mutagenesis primer was 5′ phosphorylated for maximum mutagenesis efficiency.
For the chimeric full-length cDNA clone, the partial Nsp2 fragment (1,378 nucleotides [nt]) of HB-1/3.9, covering the deletion region in JXwn06, was amplified by RT-PCR using primers W3F and W4R (Table 1). Two suitable restriction enzyme sites (MfeI and SacII) in fragment A, within the full-length cDNA clone of JXwn06, were used to exchange the corresponding fragment of Nsp2. After a sequencing check, fragment A, with a partial Nsp2 fragment of HB-1/3.9, was inserted back into the pWSK-JXwn backbone (Fig. 1B). The chimeric full-length cDNA clone was designated pWSK-JXwn-HB1nsp2.
The construction of a full-length cDNA clone for HB-1/3.9 was similar to that for JXwn06. The genome of HB-1/3.9 was divided into five fragments to be amplified with the primers in Table 1. To render the low-copy-number plasmid pWSK29 suitable for HB-1/3.9 construction, a fragment with KpnI, PacI, SbfI, and SacI enzyme sites was inserted into it (Fig. 1C). This modified plasmid was designated pWSK29M. The full-length cDNA clone, plasmid pWSK-HB1/3.9, was sequenced.
For the HB-1/3.9 cDNA clone with a 30-aa deletion in Nsp2, three overlapping segments around the two deletion regions were generated by PCR. Second-round PCR was performed to generate the deletion fragment by using the upstream primer from segment 1 and the downstream primer from segment 3 (Table 1), with the three overlapping segments as templates. A 1,287-bp fragment of the PCR product was digested with restriction enzymes MfeI (2,344 nt in the HB-1/3.9 genome) and XhoI (3,491 nt in the HB-1/3.9 genome) and was then ligated into the shuttle plasmid pEASY-Blunt-Fragment A, which had been digested by the same enzymes. The positive clone was sequenced and designated pEB-A-nsp2del30. Finally, pEB-A-nsp2del30 was inserted back into the pWSK-HB1 backbone (Fig. 1D) and designated pWSK-HB1-ND30.
In vitro transcription and transfection.
The full-length cDNA clone of JXwn06 and the chimeric full-length cDNA clone were separately linearized by cleavage with restriction enzyme PacI. Correspondingly, pWSK-HB1 and pWSK-HB1-ND30 were linearized by restriction enzyme SbfI. The capped RNAs were transcribed with SP6 RNA polymerase by using an mMessage high-yield capped RNA transcription kit (Ambion, Austin, TX) according to the manufacturer's instructions. The transcribed RNAs were individually treated with DNase I to remove the input plasmid and were purified with a MEGAclear kit (Ambion, Austin, TX) according to the protocol recommended by the manufacturer. The RNAs were quantified by spectrophotometry and then transfected into BHK-21 cells by using DMRIE-C reagent (Invitrogen) according to the manufacturer's instructions. The cell culture supernatant was obtained at 24 h posttransfection and was then serially passaged on MARC-145 cells. The cytopathic effect (CPE) was observed daily. The rescued viruses were confirmed by an immunofluorescence assay using two monoclonal antibodies: (i) SDOW17, which is specific for the N protein of PRRSV, and (ii) E3G11, prepared in our laboratory, which is specific for PRRSV Nsp2 (46).
Detection of the rescued viruses.
To differentiate the rescued viruses from the parental viruses, viral RNAs were extracted from cell culture supernatants of the infected cells by using a QIAamp viral RNA kit (Qiagen). RT-PCR was performed with selected primer pairs: J5F+/J5R (for JXwn06, RvJXwn, and RvJXwn-HB1nsp2), H5F+/H5R (for HB-1/3.9, RvHB-1/3.9, and RvHB1-ND30), and DetectF+/DetectR (Table 1). The products amplified with J5F+/J5R and H5F+/H5R were digested by BstBI and MluI (New England Bio, Beijing, China), respectively, to check the genetic markers, and the products amplified with DetectF+/DetectR were sequenced to check for Nsp2 substitution or deletion.
Genomic sequencing of the rescued viruses.
The third-passage viral cultures of the rescued viruses were harvested and used for genomic sequencing by the method described above.
In vitro stability and growth kinetics of the rescued viruses.
To assess the replication stability of the rescued viruses, the cloned viruses were serially passaged (F1 to F12) in MARC-145 cells, and the CPE and the presence of genetic markers in the viruses were observed. Virus titers in cell cultures for each of three passages were determined by a microtitration infectivity assay and recorded as TCID50 per milliliter by using the Reed-Muench method. Briefly, cells were prepared in 96-well plates and inoculated with virus suspensions (100 μl/well), which were prepared by serial 10-fold dilution. After absorption for 1 h at 37°C, the liquids in the wells were removed, and DMEM with 5% FBS was added to the wells. Plates were incubated for an additional 72 to 96 h; virus titers were determined by the presence of a visible CPE.
For the in vitro growth kinetics of the rescued virus, MARC-145 cell monolayers in T-25 flasks were infected with individual rescued viruses or with the parental virus at a multiplicity of infection of 0.01, washed with 0.01 M phosphate-buffered saline (pH 7.4) after adsorption for 1 h, and incubated at 37°C under a humid 5% CO2 atmosphere in DMEM supplemented with 5% FBS. The virus-infected supernatants were obtained at various time points after inoculation. Viral titers were determined by a microtitration infectivity assay and were expressed as TCID50 per milliliter.
Pathogenicity analyses of the rescued viruses.
Thirty-five 6-week-old SPF Landrace piglets were obtained from the Beijing Center for SPF Swine Breeding and Management. The animals were randomly allotted to seven groups (five piglets per group). Each group was housed separately in a different isolation room, with individual ventilation. Each piglet was intranasally inoculated with 2 ml of virus containing 105 TCID50 (JXwn06, HB-1/3.9, RvJXwn, RvJXwn-HB1nsp2, RvHB-1/3.9, or RvHB1-ND30). The piglets in the control group were mock inoculated with the same dosage of MARC-145 cell culture supernatant. The animals were observed daily for clinical signs, and rectal temperatures were measured every day for 21 days postinoculation (dpi). Serum and nasal swab samples were also collected. Serum samples from the challenged animals were used for the titration of viral loads by a microtitration infectivity assay and for the detection of antibodies specific for PRRSV N protein by using an Idexx Herdchek PRRS 2XR ELISA kit. Nasal swabs were used for the detection of viral RNA by RT-PCR amplifying a 312-bp PRRSV ORF7 fragment in order to analyze the excretion of virus by the inoculated pigs.
Statistical analysis.
Data were expressed as means ± standard deviations. The significance of the variability among the animal trials was determined by one-way or two-way analysis of variance using GraphPad Prism (version 4.0) software. Differences were considered statistically significant at a P value of <0.05.
Nucleotide sequence accession number.
The genomic sequence of JXwn06 has been deposited in GenBank under accession no. EF641008.
RESULTS
The JXwn06 isolate exhibited high virulence for pigs.
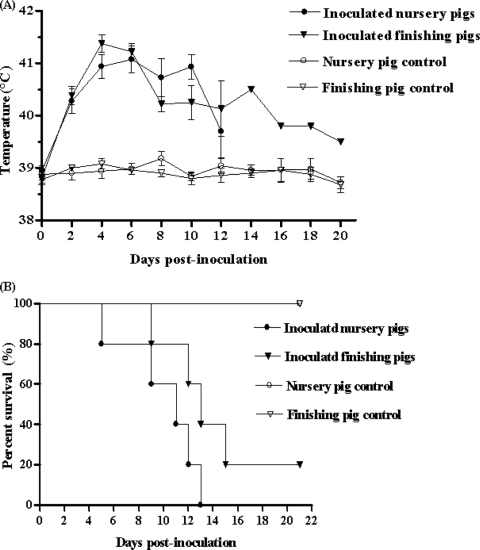
On day 2 postinoculation, all the inoculated pigs started to develop elevated body temperatures (>40°C) that lasted for 3 to 5 days (Fig. 2A) and marked clinical presentations, including depression, anorexia, and lethargy, rubefaction on the skin and in the ears, respiratory distress, shivering, and diarrhea. Five 6-week-old nursery pigs died within 5 to 13 days; four of five 12-week-old finishing pigs died within 9 to 15 days, and one survived throughout the animal experiment period (Fig. 2B). These data indicate that JXwn06 is a highly virulent PRRSV isolate with fatal pathogenicity for pigs.
FIG. 2.
Analyses of the pathogenicity of JXwn06. Shown are rectal temperature measurements (A) and survival rates (B) for each group of pigs (n = 5) inoculated with JXwn06. Body temperatures shown are means ± standard deviations (error bars), except for the data from day 14 on for the one inoculated finishing pig that survived.
The JXwn06 isolate had a characteristic 30-aa deletion in its Nsp2-coding region.
The sequences of RT-PCR-generated overlapping cDNA fragments from JXwn06 were assembled into full-length consecutive sequences of 15,320 nt, excluding the poly(A) tail, with a 189-nt 5′ noncoding region and a 150-nt 3′ untranslated region. The genomic sequence of JXwn06 shared 89.7% nucleotide identity with the North American PRRSV prototype VR-2332, 99.4 to 99.6% identity with highly pathogenic PRRSV isolates (JXA1, HuN4, and JX143) described recently in China, and 97.2% identity with HB-1(sh)/2002 (an earlier strain), as well as 97.4% identity with the cell-adapted virus HB-1/3.9. Amino acid alignment revealed that JXwn06 had the same unique deletion of 30 discontinuous amino acids (located at aa 481 and aa 533 to 561) in its Nsp2-coding region as JXA1, HuN4, JX143, and SY0608, in contrast to the sequences of VR-2332 and the earlier Chinese isolates CH1a, HB-1(sh)/2002, and HB-1/3.9 (Fig. 3).
FIG. 3.
Alignment of amino acids in the Nsp2 deletion region of JXwn06 with the sequences of VR-2332 (GenBank accession no. U87392); the earlier Chinese isolates CH-1a (GenBank accession no. AY032626), HB-1(sh)/2002 (GenBank accession no. AY150312), and HB-1/3.9 (GenBank accession no. EU360130); and the highly pathogenic isolates JXA1 (GenBank accession no. EF112445), JX143 (GenBank accession no. EU708726), HuN4 (GenBank accession no. EF635006), and SY0608 (GenBank accession no. EU144079). Dots indicate conserved residues; dashes indicate deleted amino acids at positions 481 and 533 to 561. The deleted amino acid positions were determined based on the genome of VR-2332.
Viable viruses could be rescued from the four full-length cDNA clones.
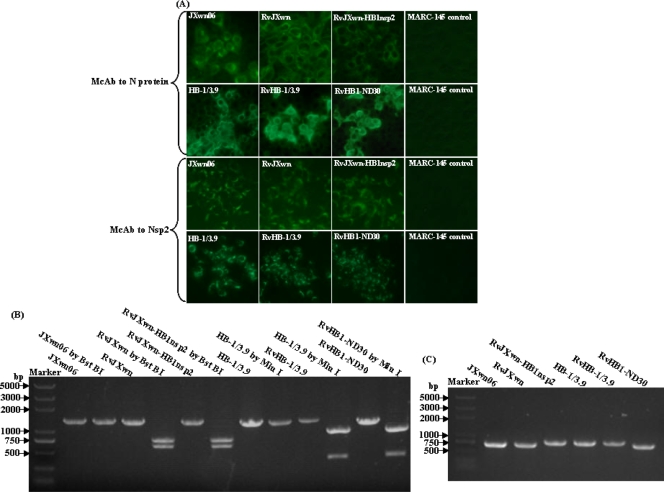
After the MARC-145 cell monolayers were inoculated with the supernatants obtained from BHK-21cells transfected with the transcribed capped RNAs at 24 h posttransfection and the virus collected from the infected MARC-145 supernatants was serially passaged, a typical PRRSV CPE could be observed at the first passage for all full-length cDNA clones. The four rescued viruses were individually designated RvJXwn, RvJXwn-HB1nsp2, RvHB-1/3.9, and RvHB1-ND30. Identification by an indirect immunofluorescence assay, genetic marker checks, and partial Nsp2 sequencing for the rescued viruses were carried out at each of three passages. MARC-145 cells infected with the rescued viruses could be stained by monoclonal antibodies specific for the Nsp2 and N proteins of PRRSV (Fig. 4A). By RT-PCR with primer pair J5F-J5R or H5F-H5R and restriction enzyme digestion, the 1,350-bp fragment amplified from the rescued viruses RvJXwn, and RvJXwn-HB1nsp2 could be cleaved into two fragments (approximately 600 bp and 750 bp) by BstBI; a 1,312-bp fragment from RvHB-1/3.9 and RvHB1-ND30 could be cleaved into two fragments (approximately 300 bp and 1,000 bp) (Fig. 4B). RT-PCR and sequencing showed that a fragment of the expected size for the Nsp2 deletion region (682 bp for RvJXw and RvHB1-ND30; 772 bp for RvHB-1/3.9 and RvJXwn-HB1nsp2) could be amplified from the rescued viruses (Fig. 4C). The genomes of the third-passage cultures of the four rescued viruses were sequenced. It was shown that, compared with the parental viruses JXwn06 and HB-1/3.9, only two nucleotides in RvJXwn, one in RvJXwn-HB1nsp2, and one in RvHB-1/3.9 changed. The nucleotide C at positions 10004 and 10059 in the Nsp10 gene changed to T in RvJXwn, resulting in amino acid changes from L to F; the nucleotide G at position 9949 in the Nsp10 gene was replaced by C in RvJXwn-HB1nsp2, resulting in an amino acid change from G to R; and the nucleotide T at position 12938 changed to C, resulting in a silent mutation in RvHB-1/3.9. These results indicate that the constructed full-length cDNA clones are infectious and that viable viruses are recovered.
FIG. 4.
Identification and differentiation of the rescued viruses. (A) MARC-145 cells infected with third-passage cultures of the viruses were fixed at 48 h postinoculation and examined by immunofluorescence assays using monoclonal antibodies against the N protein (SDOW17) and the Nsp2 protein (E3G11) of PRRSV. McAb, monoclonal antibody. (B) Genetic markers of the rescued viruses were detected by RT-PCR. (C) Partial Nsp2 fragments of the rescued viruses were examined by RT-PCR.
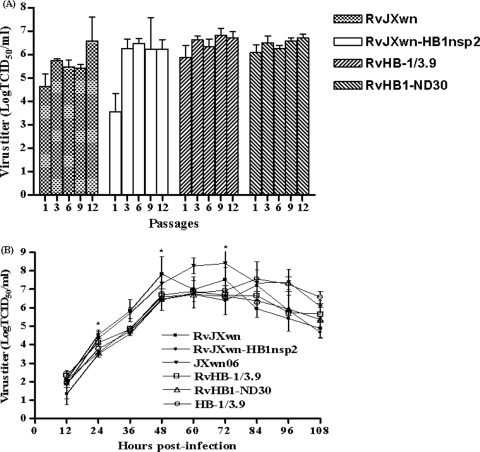
All the rescued viruses could replicate stably, and RvJXwn-HB1nsp2 displayed slower replication in vitro.
The rescued viruses were subjected to serial passage in MARC-145 cells. The virus titers in passages 1, 3, 6, 9, and 12 were determined. As shown in Fig. 5A, the four rescued viruses could replicate stably in vitro. When the growth characteristics of the rescued viruses were compared with those of their parental viruses by one-step growth kinetics assays, it was shown that RvJXwn and RvHB-1/3.9 had growth curves similar to those of their parental viruses until they reached peak titers; subsequently, they exhibited titers lower than those of their parental viruses. However, it was found that RvJXwn-HB1nsp2 had a 12-h delay in reaching the same titer as RvJXwn (Fig. 5B). At all time points before virus titers reached a peak at 84 h postinfection, RvJXwn-HB1nsp2 had a lower titer, with significant differences from RvJXwn and JXwn06 at 24 h, 48 h, and 72 h postinfection (P < 0.05), indicating that the replication rate of RvJXwn-HB1nsp2 on MARC-145 cells was lower than those of RvJXwn and JXwn06.
FIG. 5.
Stability and growth kinetics of the rescued viruses. (A) The virus titers of the rescued viruses at passages 1, 3, 6, 9, and 12 were determined by microtitration infectivity assays. (B) The growth curves of the four rescued viruses were drawn by assaying the viral titers of the supernatants obtained from 12 h to108 h postinfection by using microtitration infectivity assays. Data are means ± standard deviations (error bars) from three independent trials. Asterisks indicate a significant difference in the viral titer between RvJXwn-HB1nsp2 and RvJXwn or between RvJXwn-HB1nsp2 and JXwn06 (P < 0.05).
Both RvJXwn and RvJXwn-HB1nsp2 presented high-virulence for piglets, while RvHB-1/3.9 and RvHB1-ND30 exhibited low-virulence, like HB-1/3.9.
The pathogenicities of RvJXwn, RvJXwn-HB1nsp2, RvHB-1/3.9, RvHB1-ND30, JXwn06, and HB-1/3.9 were analyzed and compared by an inoculation experiment performed with 6-week-old SPF piglets. Rectal temperature measurements indicated that the body temperatures of piglets inoculated with JXwn06, RvJXwn, or RvJXwn-HB1nsp2 started to rise at 24 h postinoculation and reached more than 40°C at 4 dpi. The peak in these three groups was 41.4°C (Fig. 6A), whereas the body temperatures of pigs inoculated with RvHB-1/3.9, RvHB1-ND30, or HB-1/3.9 were significantly lower than those of the groups inoculated with JXwn06, RvJXwn, or RvJXwn-HB1nsp2. No body temperature reaction was observed in the control group during the experiment period.
FIG. 6.
Analyses of the pathogenicities of the rescued viruses for pigs. (A) The rectal temperatures of the inoculated pigs were measured. Data are means ± standard deviations (error bars), except for the temperatures of the one RvJXwn-inoculated pig that survived at 10 and 12 dpi. (B) The survival of the inoculated pigs was recorded.
Observation of clinical symptoms showed that piglets inoculated with RvJXwn or RvJXwn-HB1nsp2 showed signs of depression, anorexia, lethargy, rubefaction, respiratory distress, and shivering at 2 dpi, like piglets inoculated with JXwn06. In contrast, piglets infected with RvHB-1/3.9, RvHB1-ND30, or HB-1/3.9 had no obvious clinical presentations, except for occasional depression and anorexia when their body temperatures rose.
Five piglets infected with JXwn06 died within 5 to 10 dpi; five piglets infected with RvJXwn died within 7 to 14 days. Compared with these two groups, the survival of the piglets infected with RvJXwn-HB1nsp2 was obviously prolonged; five piglets died within 10 to 17 days (Fig. 6B), suggesting that replacement of the Nsp2 deletion region might lower the virulence of highly pathogenic PRRSV to some extent. However, RvJXwn-HB1nsp2 retained high virulence for pigs. All the animals in the groups infected with RvHB-1/3.9, RvHB1-ND30, or HB-1/3.9, as well as those in the control group, survived throughout the experiment period. These data suggest that the 30-aa deletion in the Nsp2 region had no obvious direct relationship with the virulence of highly pathogenic PRRSV.
RvJXwn-HB1nsp2 exhibited lower rates of replication and excretion in infected piglets, but no significant differences were observed between RvHB-1/3.9 and RvHB1-ND30.
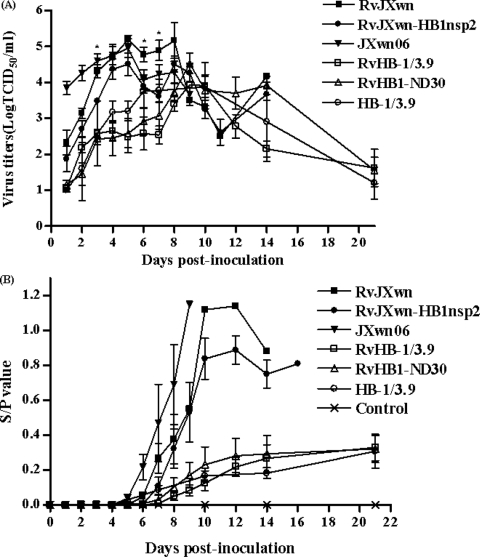
The virus loads in serum samples of infected piglets were assayed by a microtitration infectivity assay. As shown in Fig. 7A, the data revealed that virus titers in the sera of pigs inoculated with RvJXwn-HB1nsp2 were lower than those for pigs inoculated with RvJXwn or JXwn06 within 1 to 7 dpi (with significant differences compared with RvJXwn at 3, 6, and 7 dpi [P < 0.05]) but significantly higher than those for RvHB-1/3.9-, RvHB1-ND30-, or HB-1/3.9-infected pigs within 1 to 14 dpi. The levels of antibody specific for PRRSV N protein in infected pigs were examined by ELISA. The results indicated that serum antibody seroconverted on day 7 postinoculation in JXwn06-infected pigs, on day 8 postinoculation in RvJXwn-infected pigs, and on day 9 postinoculation for RvJXwn-HB1nsp2-infected pigs; RvHB1-ND30- and HB-1/3.9-infected piglets seroconverted on day 11 postinoculation, 3 days earlier than RvHB-1/3.9-infected piglets (Fig. 7B), suggesting that the humoral immune response induced by RvJXwn-HB1nsp2 infection was generated later than that induced by RvJXwn or JXwn06. Nasal swab samples were examined by RT-PCR in order to analyze the excretion of virus by the virus-infected pigs. The results showed that RvJXwn-HB1nsp2-infected pigs started to excrete virus later than pigs infected with JXwn06 or RvJXwn but that there was no difference among pigs infected with HB-1/3.9, RvHB-1/3.9, or RvHB1-ND30 (Table 2). These findings suggest that the replication ability of RvJXwn-HB1nsp2 was weaker than those of RvJXwn and the parental virus, JXwn06, in vivo.
FIG. 7.
Viral loads and antibody kinetics in the sera of pigs inoculated with the rescued viruses. (A) Virus titers were determined by a microtitration infectivity assay. Data are means ± standard deviations (error bars), except for the data representing the one pig inoculated with RvJXwn that survived at 14 dpi. Asterisks indicate significant differences in viral load between RvJXwn-HB1nsp2 and RvJXwn and between RvJXwn-HB1nsp2 and JXwn06 (P < 0.05). (B) An antibody specific for PRRSV N protein was detected using an Idexx Herdchek PRRS 2XR ELISA kit, and the level of antibody was expressed as a sample value/positive value (S/P) ratio. A ratio of ≥0.4 was regarded as indicating seroconversion.
TABLE 2.
Detection by RT-PCRa of viral RNA in nasal swab samples collected from inoculated piglets
| Day postinoculation | No. of piglets inoculated with the following virus in whose samples viral RNA was detected/total no.b:
|
||||||
|---|---|---|---|---|---|---|---|
| JXwn06 | RvJXwn | RvJXwn-HB1nsp2 | RvHB-1/3.9 | RvHB1-ND30 | HB-1/3.9 | Control | |
| 0 | 0/5a | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| 1 | 0/5 | 5/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| 2 | 4/5 | 5/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| 3 | 5/5 | 5/5 | 3/5 | 2/5 | 2/5 | 4/5 | 0/5 |
| 4 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 0/5 |
| 5 | 4/4 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 0/5 |
| 6 | 4/4 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 0/5 |
| 7 | 3/3 | 4/4 | 5/5 | 5/5 | 5/5 | 5/5 | 0/5 |
| 8 | 3/3 | 2/2 | 5/5 | 5/5 | 5/5 | ND | ND |
| 9 | 1/1 | 1/1 | 5/5 | 5/5 | 5/5 | ND | ND |
| 10 | — | 1/1 | 4/4 | 5/5 | 5/5 | 5/5 | 0/5 |
| 11 | — | 1/1 | 4/4 | ND | ND | ND | ND |
| 12 | — | 1/1 | 2/3 | 5/5 | 5/5 | ND | ND |
| 14 | — | — | 2/2 | 3/5 | 4/5 | 4/5 | 0/5 |
| 21 | — | — | — | 1/5 | 2/5 | 3/5 | 0/5 |
RT-PCR used the primers specific to the ORF7 gene of PRRSV listed in Table 1.
—, no samples; ND, not detected.
DISCUSSION
Since May 2006, atypical PRRS (called porcine high fever syndrome in China) has been pandemic in major pig-producing areas of China. Studies have confirmed that the causative agent of this clinical outbreak was a highly pathogenic PRRSV strain, with a genomic marker consisting of a 30-aa deletion in Nsp2 (23, 41, 48). First, we determined, through an animal inoculation trial, that the PRRSV isolate JXwn06 was a highly pathogenic virus. Complete sequence determination showed that the virus had the same genomic characteristics as the isolates described previously (23, 41, 48), namely, a noncontiguous 30-aa deletion in its Nsp2. In recent years, reverse genetic operation methods have been extensively applied to study the genomic functions of PRRSV (11, 21, 26, 28, 31, 32, 34, 43). A new study indicated that the rescued virus from an infectious cDNA clone of a highly pathogenic PRRSV variant was highly virulent for pigs (24). In our study, a viable virus recovered from an infectious cDNA clone of JXwn06 induced typical clinical symptoms identical to those observed in the field, displaying high pathogenicity for pigs, like the parent virus. Our present data unambiguously point to the same conclusion: highly virulent PRRSV is the etiological agent of atypical PRRS in China. Our data also provide further etiological evidence for the clinical outbreak in China.
It was necessary to determine whether the 30-aa deletion in the Nsp2 of highly pathogenic PRRSV is related to its virulence, although the virus has been confirmed to be the causative agent of the emerging disease. This point is valuable for elucidating the molecular mechanism associated with the virulence and pathogenicity of PRRSV. Therefore, in this study, we rescued two mutant viruses: (i) RvJXwn-HB1nsp2, from a full-length infectious cDNA clone where the Nsp2 region containing the 30-aa deletion of a high-virulence PRRSV strain (JXwn06) was replaced with a corresponding 1,378-nt region from a low-virulence PRRSV strain (HB-1/3.9), and (ii) RvHB1-ND30, from a cDNA clone of HB-1/3.9 with a 30-aa deletion in Nsp2 corresponding to that in JXwn06. We systematically analyzed the pathogenicities of these mutant viruses for pigs together with those of the parental viruses and the rescued viruses from full-length infectious cDNA clones of JXwn06 and HB-1/3.9. This animal inoculation experiment indicates that RvJXwn-HB1nsp2 retains fatal pathogenicity for piglets. The five piglets inoculated all died, although the survival of the infected animals was prolonged compared with that of animals infected with the parental virus JXwn06 or the rescued virus RvJXwn. In contrast, RvHB1-ND30 maintained its low virulence, similar to that of the parental virus. These findings suggest that the 30-aa deletion in Nsp2 is not related to virulence. Interestingly, the replacement of the Nsp2 deletion region of JXwn06 with a corresponding region of a low-virulence PRRSV strain could make the rescued virus grow less vigorously. Meanwhile, our data based on analysis of growth characteristics in vitro and in vivo showed that (i) the replication rate of RvJXwn-HB1nsp2 on MARC-145 cells was lower than those of JXwn06 and RvJXwn; (ii) viral loads in the sera of pigs inoculated with RvJXwn-HB1nsp2 were significantly lower than those in pigs inoculated with JXwn06 or RvJXwn; (iii) the humoral immune response induced by RvJXwn-HB1nsp2 infection was generated later than those induced by RvJXwn and JXwn06; (iv) RvJXwn-HB1nsp2-infected pigs started to excrete virus later than pigs infected with JXwn06 or RvJXwn, indicating a lower replication rate of RvJXwn-HB1nsp2 on MARC-145 cells and in the host; and (v) RvHB1-ND30 showed no significant differences from the parental virus HB-1/3.9 in the features mentioned above. Previous studies have noted that infection of susceptible pigs with highly virulent isolates of PRRSV resulted in longer periods of viremia, increased severity of clinical signs, increased mortality, and significantly higher viral loads in blood and tissues than infection with mildly virulent or cell culture-adapted viruses (18). Hence, our findings could not exclude the possibility that aside from the deleted 30 aa, the rest of the substituted fragment in HB-1/3.9 Nsp2 might contribute to the virulence and replication rate alterations in RvJXwn-HB1nsp2.
Nsp2 is recognized as the most variable region in the genome of PRRSV. Since the first natural Nsp2 deletion isolate, HB-2(sh)/2002, was identified as containing a unique 12-aa deletion at positions 470 to 481, a number of mutations, insertions, and deletions in Nsp2 have been documented (10, 13, 16, 36, 41). Meanwhile, Nsp2 has become one of the vital regions for monitoring the evolution of PRRSV and for molecular epidemiology research on PRRSV (2, 10, 12, 13, 16, 35, 47). Recently, mutations in the Nsp2-coding region resulting in a 403-aa manual deletion and expression of a foreign protein were reported (11, 15, 20). Therefore, further investigations are required, particularly on the role of Nsp2 in the cell and tissue tropism, replication and growth, and variation and pathogenicity of PRRSV, as well as in the difference in virulence among various strains. The virulence of PRRSV is considered to be associated with multiple factors; it is not easy to identify the unique determinant region in the genome of PRRSV (43). However, searching any genomic region or gene related to virulence is an essential step toward making a contribution to the understanding of the pathogenicity of PRRSV.
Taken as a whole, our findings indicate that the 30-aa deletion in the Nsp2-coding region of the highly virulent PRRSV emerging in China is not related to its virulence. Therefore, the Nsp2 deletion can be used as a marker to distinguish the highly pathogenic Chinese virus from North American strains but cannot be utilized as a definition of highly pathogenic strains of PRRSV in general. Further exploration of the virulence-determining region or point within the genome of PRRSV is essential.
Acknowledgments
This work was supported by the National Natural Science Funds for Distinguished Young Scholars (30825031) from the National Natural Science Foundation of China, by a National Key Basic Research Plan grant (2005CB523204), and by the National Key Technology R&D Program of China (grant 2006BAD06A03) from the Chinese Ministry of Science and Technology.
We thank Ying Fang at South Dakota State University for support and suggestions on the construction of infectious cDNA clones of PRRSV.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Albina, E. 1997. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet. Microbiol. 55309-316. [DOI] [PubMed] [Google Scholar]
- 2.Allende, R., T. L. Lewis, Z. Lu, D. L. Rock, G. F. Kutish, A. Ali, A. R. Doster, and F. A. Osorio. 1999. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 80307-315. [DOI] [PubMed] [Google Scholar]
- 3.Bautista, E. M., J. J. Meulenberg, C. S. Choi, and T. W. Molitor. 1996. Structural polypeptides of the American (VR-2332) strain of porcine reproductive and respiratory syndrome virus. Arch. Virol. 1411357-1365. [DOI] [PubMed] [Google Scholar]
- 4.Benfield, D. A., E. Nelson, J. E. Collins, L. Harris, S. M. Goyal, D. Robison, W. T. Christianson, R. B. Morrison, D. E. Gorcyca, and D. W. Chladek. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Investig. 4127-133. [DOI] [PubMed] [Google Scholar]
- 5.Bøtner, A., J. Nielsen, and V. Bille-Hansen. 1994. Isolation of porcine reproductive and respiratory syndrome (PRRS) virus in a Danish swine herd and experimental infection of pregnant gilts with the virus. Vet. Microbiol. 40351-360. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142629-633. [PubMed] [Google Scholar]
- 7.Collins, J. E., D. A. Benfield, W. T. Christianson, L. Harris, J. C. Hennings, D. P. Shaw, S. M. Goyal, S. McCullough, R. B. Morrison, H. S. Joo, et al. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Investig. 4117-126. [DOI] [PubMed] [Google Scholar]
- 8.Conzelmann, K. K., N. Visser, P. van Woensel, and H. J. Tiel. 1993. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the Arterivirus group. Virology 193329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Boon, J. A., K. S. Faaberg, J. J. Meulenberg, A. L. Wassenaar, P. G. Plagemann, A. E. Gorbalenya, and E. J. Snijder. 1995. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J. Virol. 694500-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, Y., D. Y. Kim, S. Ropp, P. Steen, J. Christopher-Hennings, E. A. Nelson, and R. R. Rowland. 2004. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 100229-235. [DOI] [PubMed] [Google Scholar]
- 11.Fang, Y., R. R. Rowland, M. Roof, J. K. Lunney, J. Christopher-Hennings, and E. A. Nelson. 2006. A full-length cDNA infectious clone of North American type 1 porcine reproductive and respiratory syndrome virus: expression of green fluorescent protein in the Nsp2 region. J. Virol. 8011447-11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, Y., P. Schneider, W. P. Zhang, K. S. Faaberg, E. A. Nelson, and R. R. Rowland. 2007. Diversity and evolution of a newly emerged North American Type 1 porcine arterivirus: analysis of isolates collected between 1999 and 2004. Arch. Virol. 1521009-1017. [DOI] [PubMed] [Google Scholar]
- 13.Gao, Z. Q., X. Guo, and H. C. Yang. 2004. Genomic characterization of two Chinese isolates of porcine respiratory and reproductive syndrome virus. Arch. Virol. 1491341-1351. [DOI] [PubMed] [Google Scholar]
- 14.Garner, M. G., I. F. Whan, G. P. Gard, and D. Phillips. 2001. The expected economic impact of selected exotic diseases on the pig industry of Australia. Rev. Sci. Tech. 20671-685. [DOI] [PubMed] [Google Scholar]
- 15.Han, J., G. Liu, Y. Wang, and K. S. Faaberg. 2007. Identification of nonessential regions of the nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J. Virol. 819878-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han, J., Y. Wang, and K. S. Faaberg. 2006. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 122175-182. [DOI] [PubMed] [Google Scholar]
- 17.Hopper, S. A., M. E. White, and N. Twiddy. 1992. An outbreak of blue-eared pig disease (porcine reproductive and respiratory syndrome) in four pig herds in Great Britain. Vet. Rec. 131140-144. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, W., M. Roof, E. Vaughn, J. Christopher-Hennings, C. R. Johnson, and M. P. Murtaugh. 2004. Pathogenic and humoral immune responses to porcine reproductive and respiratory syndrome virus (PRRSV) are related to viral load in acute infection. Vet. Immunol. Immunopathol. 102233-247. [DOI] [PubMed] [Google Scholar]
- 19.Keffaber, K. 1989. Reproductive failure of unknown etiology. Am. Assoc. Swine Pract. Newsl. 19. [Google Scholar]
- 20.Kim, D. Y., J. G. Calvert, K. O. Chang, K. Horlen, M. Kerrigan, and R. R. Rowland. 2007. Expression and stability of foreign tags inserted into nsp2 of porcine reproductive and respiratory syndrome virus (PRRSV). Virus Res. 128106-114. [DOI] [PubMed] [Google Scholar]
- 21.Kroese, M. V., J. C. Zevenhoven-Dobbe, J. N. Bos-de Ruijter, B. P. Peeters, J. J. Meulenberg, L. A. Cornelissen, and E. J. Snijder. 2008. The nsp1α and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 89494-499. [DOI] [PubMed] [Google Scholar]
- 22.Kuwahara, H., T. Nunoya, M. Tajima, A. Kato, and T. Samejima. 1994. An outbreak of porcine reproductive and respiratory syndrome in Japan. J. Vet. Med. Sci. 56901-909. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y., X. Wang, K. Bo, X. Wang, B. Tang, B. Yang, W. Jiang, and P. Jiang. 2007. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet. J. 174577-584. [DOI] [PubMed] [Google Scholar]
- 24.Lv, J., J. Zhang, Z. Sun, W. Liu, and S. Yuan. 2008. An infectious cDNA clone of a highly pathogenic porcine reproductive and respiratory syndrome virus variant associated with porcine high fever syndrome. J. Gen. Virol. 892075-2079. [DOI] [PubMed] [Google Scholar]
- 25.Meng, X. J., P. S. Paul, and P. G. Halbur. 1994. Molecular cloning and nucleotide sequence of the 3′-terminus genomic RNA of the porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 751795-1801. [DOI] [PubMed] [Google Scholar]
- 26.Meulenberg, J. J., J. N. Bos-de Ruijter, G. Wensvoort, and R. J. Moormann. 1998. An infectious cDNA clone of porcine reproductive and respiratory syndrome virus. Adv. Exp. Med. Biol. 440199-206. [DOI] [PubMed] [Google Scholar]
- 27.Meulenberg, J. J. M., A. Petersen-den Besten, E. P. De Kluyver, R. J. M. Moormann, W. M. M. Schaaper, and G. Wensvoort. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meulenberg, J. J. M., J. N. A. Bos-de Ruijter, R. van de Graaf, G. Wensvoort, and R. J. M. Moormann. 1998. Infectious transcripts from cloned genome-length cDNA of porcine reproductive and respiratory syndrome virus. J. Virol. 72380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meulenberg, J. J. M., M. M. Hulst, E. J. de Meijer, P. L. J. M. Moonen, A. den Besten, E. P. de Kluyver, G. Wensvoort, and R. J. M. Moormann. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 19262-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann, E. J., J. B. Kliebenstein, C. D. Johnson, J. W. Mabry, E. J. Bush, A. H. Seitzinger, A. L. Green, and J. J. Zimmerman. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 227385-392. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen, H. S., G. Liu, J. Nielsen, M. B. Oleksiewicz, A. Botner, T. Storgaard, and K. S. Faaberg. 2003. Generation of an infectious clone of VR-2332, a highly virulent North American-type isolate of porcine reproductive and respiratory syndrome virus. J. Virol. 773702-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei, Y., D. C. Hodgins, C. Lee, J. G. Calvert, S. K. Welch, R. Jolie, M. Keith, and D. Yoo. 2008. Functional mapping of the porcine reproductive and respiratory syndrome virus capsid protein nuclear localization signal and its pathogenic association. Virus Res. 135107-114. [DOI] [PubMed] [Google Scholar]
- 33.Pejsak, Z., T. Stadejek, and I. Markowska-Daniel. 1997. Clinical signs and economic losses caused by porcine reproductive and respiratory syndrome virus in a large breeding farm. Vet. Microbiol. 55317-322. [DOI] [PubMed] [Google Scholar]
- 34.Ran, Z. G., X. Y. Chen, X. Guo, X. N. Ge, K. J. Yoon, and H. C. Yang. 2008. Recovery of viable porcine reproductive and respiratory syndrome virus from an infectious clone containing a partial deletion within the Nsp2-encoding region. Arch. Virol. 153899-907. [DOI] [PubMed] [Google Scholar]
- 35.Ropp, S. L., C. E. Wees, Y. Fang, E. A. Nelson, K. D. Rossow, M. Bien, B. Arndt, S. Preszler, P. Steen, J. Christopher-Hennings, J. E. Collins, D. A. Benfield, and K. S. Faaberg. 2004. Characterization of emerging European-like porcine reproductive and respiratory syndrome virus isolates in the United States. J. Virol. 783684-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen, S., J. Kwang, W. Liu, and D. X. Liu. 2000. Determination of the complete nucleotide sequence of a vaccine strain of porcine reproductive and respiratory syndrome virus and identification of the Nsp2 gene with a unique insertion. Arch. Virol. 145871-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79961-979. [DOI] [PubMed] [Google Scholar]
- 38.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82985-994. [DOI] [PubMed] [Google Scholar]
- 39.Snijder, E. J., A. L. Wassenaar, and W. J. Spaan. 1994. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 685755-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snijder, E. J., A. L. Wassenaar, W. J. Spaan, and A. E. Gorbalenya. 1995. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 27016671-16676. [DOI] [PubMed] [Google Scholar]
- 41.Tian, K., X. Yu, T. Zhao, Y. Feng, Z. Cao, C. Wang, Y. Hu, X. Chen, D. Hu, X. Tian, D. Liu, S. Zhang, X. Deng, Y. Ding, L. Yang, Y. Zhang, H. Xiao, M. Qiao, B. Wang, L. Hou, X. Wang, X. Yang, L. Kang, M. Sun, P. Jin, S. Wang, Y. Kitamura, J. Yan, and G. F. Gao. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Dinten, L. C., A. L. Wassenaar, A. E. Gorbalenya, W. J. Spaan, and E. J. Snijder. 1996. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 706625-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Y., Y. Liang, J. Han, K. M. Burkhart, E. M. Vaughn, M. B. Roof, and K. S. Faaberg. 2008. Attenuation of porcine reproductive and respiratory syndrome virus strain MN184 using chimeric construction with vaccine sequence. Virology 371418-429. [DOI] [PubMed] [Google Scholar]
- 44.Wassenaar, A. L., W. J. Spaan, A. E. Gorbalenya, and E. J. Snijder. 1997. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J. Virol. 719313-9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wensvoort, G., C. Terpstra, J. M. Pol, E. A. ter Laak, M. Bloemraad, E. P. de Kluyver, C. Kragten, L. van Buiten, A. den Besten, F. Wagenaar, et al. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet. Q. 13121-130. [DOI] [PubMed] [Google Scholar]
- 46.Yan, Y., X. Guo, X. Ge, Y. Chen, Z. Cha, and H. Yang. 2007. Monoclonal antibody and porcine antisera recognized B-cell epitopes of Nsp2 protein of a Chinese strain of porcine reproductive and respiratory syndrome virus. Virus Res. 126207-215. [DOI] [PubMed] [Google Scholar]
- 47.Yoshii, M., T. Okinaga, A. Miyazaki, K. Kato, H. Ikeda, and H. Tsunemitsu. 2008. Genetic polymorphism of the nsp2 gene in North American type porcine reproductive and respiratory syndrome virus. Arch. Virol. 1531323-1334. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, Y. J., X. F. Hao, Z. J. Tian, G. Z. Tong, D. Yoo, T. Q. An, T. Zhou, G. X. Li, H. J. Qiu, T. C. Wei, and X. F. Yuan. 2008. Highly virulent porcine reproductive and respiratory syndrome virus emerged in China. Transbound. Emerg. Dis. 55152-164. [DOI] [PubMed] [Google Scholar]
- 49.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81853-879. [DOI] [PubMed] [Google Scholar]