Abstract
A vital arm of the innate immune response to viral infection is the induction and subsequent antiviral effects of interferon (IFN). Rotavirus reduces type I IFN induction in infected cells by the degradation of IFN regulatory factors. Here, we show that the monkey rotavirus RRV and human rotavirus Wa also block gene expression induced by type I and II IFNs through a mechanism allowing signal transducer and activator of transcription 1 (STAT1) and STAT2 activation but preventing their nuclear accumulation. In infected cells, this may allow rotavirus to block the antiviral actions of IFN produced early in infection or by activated immune cells. As the intracellular expression of rotavirus nonstructural proteins NSP1, NSP3, and NSP4 individually did not inhibit IFN-stimulated gene expression, their involvement in this process is unlikely. RRV and Wa rotaviruses also prevented the tumor necrosis factor alpha-stimulated nuclear accumulation of NF-κB and NF-κB-driven gene expression. In addition, NF-κB was activated by rotavirus infection, confirming earlier findings by others. As NF-κB is important for the induction of IFN and other cytokines during viral infection, this suggests that rotavirus prevents cellular transcription as a means to evade host responses. To our knowledge, this is the first report of the use of this strategy by a double-stranded RNA virus.
The induction of the interferon (IFN) system of innate cellular defense is crucial for the control of viral infection. Following infection, the host recognition of viral components, including nucleic acids, results in the activation of IFN response factors (IRF), leading to the expression of the type I IFNs IFN-α and IFN-β (IFN-α/β). IFN-γ, a type II IFN, also can be produced by activated immune cells during infection. Secreted IFN-α/β and IFN-γ bind specific receptors on surrounding uninfected cells, triggering a cascade of signaling events involving Janus kinases (JAK) and signal transducer and activator of transcription (STAT) molecules. Once activated by phosphorylation, STAT molecules translocate to the nucleus and induce the expression of many IFN-stimulated genes (ISG) with antiviral properties to establish an antiviral state. The effectiveness of IFN in combating viral infection is highlighted by the fact that a multitude of viruses have evolved counterstrategies, such as blocking IFN-α/β and ISG expression and abrogating the antiviral actions of ISG (40).
The activation of NF-κB by viruses is important for innate immune responses to infection. NF-κB activation occurs downstream of retinoic acid-inducible gene I-like helicases, Toll-like receptors, and receptors for proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α), and it is required for the optimal expression of IFN-β (6, 46, 47). In virus-infected cells, the expression of other cytokines that are important for cellular immune responses, such as interleukin-8 (IL-8), also is largely dependent on the activation of NF-κB (36). There are five forms of NF-κB, namely, Rel (c-Rel), RelA (p65), RelB, NF-κB1 (p105/p50), and NF-κB2 (p100/p52), which are activated and translocated to the nucleus following the phosphorylation and subsequent degradation of the inhibitor of κB (IκB) (25). The phosphorylation and dimerization of NF-κB subunits also is important for their activity. Due to the key role of NF-κB in antiviral defenses, its action is the target of a number of viruses, such as human papillomavirus and African swine fever virus (39, 45).
Rotavirus, a member of the Reoviridae family, is the major cause of severe infantile gastroenteritis. Although sensitive to IFN, rotaviruses replicate well in cells that can produce IFN (5). For several rotavirus strains, including the monkey rotaviruses SA11 and RRV and bovine rotavirus B641, this is at least partly due to the prevention of IFN production in infected cells by rotavirus nonstructural protein 1 (NSP1) (2, 18, 26). This protein targets IRF-3 and IRF-7, transcription factors that are crucial for IFN-α/β gene expression, for proteasome-mediated degradation (2, 3). However, IFN still could be produced in certain cell types, such as myeloid dendritic cells (18), or early in infection before IRF-3 is efficiently targeted for degradation. Evidence also exists that NSP1 from certain rotavirus strains such as OSU is inefficient in degrading IRF-3 (26). IFN-γ production might occur in uninfected but activated immune cells. Supporting this, serum IFN levels are elevated in infected children, whose IFN-γ levels were negatively associated with symptoms (29). Interestingly, IFN treatment just before or after the infection of the human intestinal Caco-2 cell line has little effect on rotavirus replication (5), so rotavirus also might evade innate defenses by blocking IFN-mediated signaling. Rotavirus has been shown to induce the expression of various cellular genes in Caco-2 cells (15). In addition, rotavirus can activate NF-κB-dependent gene expression and induce the production of IL-8 and other cytokines in HT-29 cells and mouse intestine (9, 10, 42, 44). To date, no solid evidence exists that rotavirus possesses strategies to reduce the induction of cytokines other than type I IFN.
It is reported here that rotavirus efficiently blocks type I and II IFN-induced gene expression by preventing the nuclear accumulation of activated STAT1 and STAT2. We show that the nuclear accumulation of NF-κB and NF-κB-driven gene expression in response to TNF-α, and to rotavirus infection itself, also are prevented. These novel findings suggest that rotavirus employs multiple strategies to inhibit cytokine production and signaling and also dampen immune responses. This is the first report showing such an effect during infection by a double-stranded RNA virus.
MATERIALS AND METHODS
Cell lines, viruses, and infection.
Virus origin, propagation, titration, and purification have been described previously (11, 12, 28, 30, 33). For this study, purified RRV (monkey), partially purified Wa (human), and unpurified SA11 (monkey) and UK (bovine) rotavirus strains propagated in MA104 monkey kidney cells were used. Due to the technical difficulty of fully purifying Wa rotavirus to high titer, this virus was partially purified by omitting the glycerol gradient ultracentrifugation and final pelleting steps from the previously published protocol (28). The source and cultivation of MA104 and Caco-2 cell lines has been described previously (28, 33). 293T cells were obtained from the ATCC and maintained as described for MA104 cells (33). Cell passage numbers ranged from 40 to 95 (MA104), 40 to 90 (Caco-2), and 20 to 60 (293T). For cell infection, virus was activated at 37°C with porcine trypsin (10 μg/ml; Sigma, St. Louis, MO), diluted in serum-free medium, and adsorbed to cells for 1 h. The inoculum then was removed and replaced with serum-free medium for the remainder of the infection period.
Reporter gene assays.
For IFN-responsive reporter assays, MA104 and Caco-2 cells at 50 to 70% confluence in 24-well trays were cotransfected using Transit LT-1 (Mirus Bio, Madison, WI), with either 0.5 μg of the IFN-α/β-responsive luciferase reporter plasmid p(9-27)4thΔ(−39)Lucter (ISRE-Luc) or the IFN-γ-responsive luciferase reporter p(IRF-1*GAS)6tkΔ(−39)Lucter (GAS-Luc) (31), along with 0.5 μg of the β-galactosidase expression vector pCMV-β (Clontech, Palo Alto, CA). All cell transfections and treatments were performed at 37°C. Following infection with RRV or Wa rotavirus for the indicated times, transfected cells were treated for the indicated times with or without IFN-α (500 U/ml; Calbiochem, La Jolla, CA) or IFN-γ (50 ng/ml; BD Pharmingen, San Diego, CA). Luciferase expression was measured using the Luciferase assay system (Promega, Madison, WI) in a Topcount NXT scintillation and luminescence counter (Packard Bell, Wijchen, The Netherlands) and normalized for β-galactosidase levels measured with the β-galactosidase enzyme assay (Promega).
For NF-κB-responsive reporter assays, cells were cotransfected with 0.4 μg of pNF-κB-Luc (Clontech), consisting of the firefly luciferase gene under the control of a promoter containing multiple NF-κB binding sites, and 0.1 μg of pRL-TK (Promega), comprising the Renilla luciferase gene under the control of the herpes simplex virus thymidine kinase promoter. Following rotavirus infection for the indicated times, cells were either left untreated or treated with TNF-α (20 ng/ml; eBioscience, San Diego, CA) for 4 h. Firefly and Renilla luciferase levels were measured using the dual-luciferase reporter assay system (Promega) in the Topcount NXT instrument. For each sample, the expression of firefly luciferase was normalized for Renilla luciferase levels.
Real-time PCR.
Following MA104 cell infection for 4 or 8 h and then IFN-α treatment (500 U/ml) for an additional 4 h, total cellular RNA was extracted as reported previously (28). cDNA was generated by reverse transcription using random hexamers with Multiscribe reverse transcriptase (Applied Biosystems, Foster City, CA). Real-time PCRs were carried out using ABI TaqMan PCR master mix with gene expression assays for MxA (unique identifier Hs00182073_ml), ISG56 (unique identifier Hs00356631_g1), and 18S rRNA (unique identifier Hs99999901_s1). Data were collected and analyzed as previously described (28) or using an MX3000P real-time PCR system and MxPro software (Stratagene, La Jolla, CA). Relative gene expression was calculated using the comparative threshold cycle method, employing 18S rRNA as the reference.
Western blotting.
Confluent MA104 cell monolayers were mock or RRV infected for the indicated times and then left untreated or treated with IFN-α (500 U/ml), IFN-γ (50 ng/ml), or TNF-α (20 ng/ml) for 30 min. Cell lysis and Western blotting were performed as described previously (28). Blots were probed with rabbit polyclonal antibodies directed to STAT1, phospho-STAT1 (Tyr701), STAT2, phospho-p65 (Ser536), and β-actin (Cell Signaling Technology, Beverly, MA); phospho-STAT2 (Tyr689) (Upstate/Millipore, Billerica, MA); and p65 and IRF-3 (Santa Cruz Biotechnology, Santa Cruz, CA). The anti-FLAG M2 mouse monoclonal antibody was from Sigma.
Cell staining and confocal microscopy.
Caco-2 cells were seeded onto glass coverslips that had been coated overnight at 4°C with human placental type I collagen, which was used according to the manufacturer's protocol (50 μg/ml; Sigma). MA104 cells were grown on uncoated coverslips. Cells were infected with RRV, Wa, SA11, or UK rotavirus for 6 h, left untreated or treated with IFN-α, IFN-γ, or TNF-α for 30 min, and fixed with 3.7% (wt/vol) formaldehyde (Sigma) in phosphate-buffered saline (PBS) for 10 min, followed by permeabilization with acetone-methanol (1:1, vol/vol) at −20°C for 15 min. Fixed cells were blocked in 3% (wt/vol) bovine serum albumin (Sigma) in PBS for 30 min and reacted at 20°C for 1 h with a combination of polyclonal rabbit antibody (2 μg/ml; Santa Cruz) to STAT1 (SC-345), STAT2 (SC-476), or p65 (SC-109) and mouse monoclonal antibody RVA to rotavirus VP6 (14). Cells washed with PBS were incubated for 30 min with a combination of Alexa Fluor 488-conjugated anti-mouse immunoglobulin G and Alexa Fluor 594-conjugated anti-rabbit immunoglobulin G (10 μg/ml each; Invitrogen, Carlsbad, CA). Coverslips washed in PBS were mounted on microscope slides with Prolong Gold containing 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Images were obtained with an LSM 510 META confocal microscope (Carl Zeiss, Göttingen, Germany) at 630× magnification with an optical slice thickness of 1.2 to 1.6 μm.
Cloning of rotavirus NSP cDNA and functional assays.
Viral double-stranded RNA was extracted from RRV as previously described (20) and was used as a template for cDNA synthesis using the Superscript first-strand synthesis system (Invitrogen) and gene-specific primers. Following amplification by PCR using Phusion polymerase (Finnzymes; Keilaranta, Espoo, Finland), cDNA was inserted into pCMV-3Tag-6 (Stratagene) using standard cloning techniques to yield plasmids expressing proteins fused to three N-terminal copies of the FLAG epitope (DYKDDDDK). Plasmid integrity was confirmed by DNA sequencing. The predicted amino acid sequence of each of the encoded NSPs was identical to the GenBank sequences (accession numbers AY117048, AY065842, and L41247; Entrez Database, NCBI, NIH), except for the substitution of Val for Leu at position 50 of NSP1. To assess the potential role of each rotavirus NSP in the inhibition of IFN signaling, 0.7 μg of empty pCMV-3Tag-6 or plasmids expressing RRV NSP1, NSP3, and NSP4 were cotransfected with 0.2 μg of ISRE-Luc and 0.1 μg pCMV-β into 293T cells in 24-well trays. As a positive control, a plasmid expressing the parainfluenza virus 5 (PIV5) V protein also was included (17). At 24 h after transfection, cells were left untreated or treated with IFN-α for 7 h and assayed as described above for luciferase and β-galactosidase levels. Cell lysates from transfected cells, without IFN treatment, were assayed by Western blotting as described above for levels of FLAG-tagged protein, IRF-3, and β-actin.
RESULTS
Rotavirus infection blocked expression of IFN-dependent reporter genes.
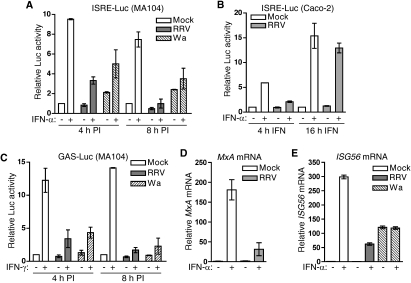
The effect of rotavirus infection on ISG expression was examined in permissive MA104 and Caco-2 epithelial cells. Following the mock infection of ISRE-Luc/pCMV-β-transfected cells for 4 h, treatment with IFN-α for 4 h produced a 9.5-fold increase in luciferase levels (Fig. 1A). In contrast, the infection of identically transfected cells with RRV or Wa for 4 h at a multiplicity of infection (MOI) of 5 or 10, respectively, decreased IFN-α-stimulated luciferase expression by 73 and 53%, respectively, compared to that of stimulated, mock-infected cells (Fig. 1A). Unstimulated RRV- and mock-infected cells showed similar low levels of luciferase expression, while unstimulated Wa-infected cells showed a 2.1-fold increase in luciferase levels. RRV infection for 8 h prior to IFN-α addition completely abolished IFN-α-stimulated luciferase expression, while Wa infection at this time reduced luciferase expression by 61% (Fig. 1A). In unstimulated cells at 8 h postinfection (PI), luciferase levels were reduced by 50% by RRV but were increased 2.4-fold by Wa. These data indicate that although both viruses can reduce IFN-stimulated gene expression in MA104 cells, RRV is more effective than Wa. In addition, Wa induced a low level of IFN-stimulated gene expression. The RRV infection of Caco-2 cells (MOI, 10) for 8 h, followed by 4 h of stimulation with IFN-α, also reduced IFN-α-stimulated luciferase expression by 78% compared to that of mock-infected cells (Fig. 1B), showing that the ability of rotavirus to block IFN-induced gene expression is maintained in an intestinal cell line. When the length of IFN-α stimulus was increased to 16 h in Caco-2 cells, the block to luciferase expression caused by RRV essentially was lost. At this time, untreated RRV-infected cells showed no alteration in luciferase levels.
FIG. 1.
Effect of rotavirus infection on IFN-induced gene expression. (A) MA104 cells were cotransfected with the ISRE-Luc and pCMV-β constructs and mock infected or infected with RRV (MOI, 5) or Wa (MOI, 10) for 4 or 8 h, followed by incubation for 4 h in the presence or absence of IFN-α. Data are provided as the means ± ranges of luciferase (Luc) activity normalized for β-galactosidase levels from two independent experiments. (B) Normalized luciferase activity in Caco-2 cells cotransfected as described for panel A and mock infected or infected with RRV at an MOI of 10 for 8 h, followed by incubation for 4 or 16 h in the presence or absence of IFN-α. Data represent the means and ranges of two independent experiments. (C) MA104 cells were cotransfected with the GAS-Luc and pCMV-β constructs and infected as described for panel A, followed by incubation for 4 h in the presence or absence of IFN-γ. Data are provided as the means ± ranges of luciferase activity normalized for β-galactosidase levels from two independent experiments. (D) MA104 cells were mock infected or infected with RRV at an MOI of 5 for 4 h, followed by incubation for 4 h in the presence or absence of IFN-α. Total cellular RNA was extracted and analyzed for MxA mRNA and 18S rRNA expression by real-time PCR. The means ± standard deviations of MxA mRNA levels, normalized for 18S rRNA levels, are provided from triplicate analyses. (E) MA104 cells were mock infected or infected with RRV (MOI, 5) or Wa (MOI, 10) for 8 h, followed by incubation for 4 h in the presence or absence of IFN-α, and analyzed for ISG56 mRNA and 18S rRNA levels by real-time PCR. Data represent the means ± ranges of ISG56 mRNA levels, normalized for 18S rRNA levels, from duplicate analyses.
The effect of rotavirus infection on IFN-γ-stimulated gene expression also was examined. Infection with RRV or Wa for 4 h in GAS-Luc/pCMV-β-transfected MA104 cells reduced the level of luciferase expression stimulated through 4 h of treatment with IFN-γ by 79 and 70%, respectively, compared to those of controls (Fig. 1C). At 8 h after infection with RRV or Wa, IFN-γ-stimulated luciferase expression was reduced by 95 and 90%, respectively. In the absence of stimulation, infection with Wa or RRV had no effect on luciferase expression. These results clearly show that RRV is able to efficiently block both IFN-α- and IFN-γ-dependent gene expression and strongly suggest that rotavirus uses this as a means to evade innate immune responses, particularly early in the replication cycle.
Rotavirus infection reduced the levels of endogenous ISG mRNA.
To determine if the block to ISG expression following rotavirus infection is at the level of transcription or translation, the effect of infection on the IFN-stimulated transcription of the endogenous ISGs MxA and ISG56 was assessed. Following the mock infection of MA104 cells for 4 h, IFN-α treatment for 4 h produced a 181-fold increase in MxA mRNA levels (Fig. 1D). In line with the effects on reporter gene expression, this increase was reduced by 83% in RRV-infected cells. After the mock infection of MA104 cells for 8 h, 4 h of IFN-α stimulation induced a 300-fold increase in ISG56 mRNA levels (Fig. 1E). RRV or Wa infection reduced this response by 79 and 60%, respectively. In line with ISRE reporter assays, unstimulated RRV-infected cells showed no alteration in ISG56 mRNA levels compared to those of uninfected cells, while ISG56 mRNA increased 120-fold in unstimulated Wa-infected cells. Furthermore, the stimulation of Wa-infected cells did not change ISG56 mRNA levels compared to those of unstimulated infected cells. Overall, these data demonstrate that the block to IFN-dependent gene expression is at the level of transcription or mRNA stability, and it occurs for multiple endogenous ISGs.
Rotavirus infection did not cause STAT degradation or interfere with IFN-induced STAT activation.
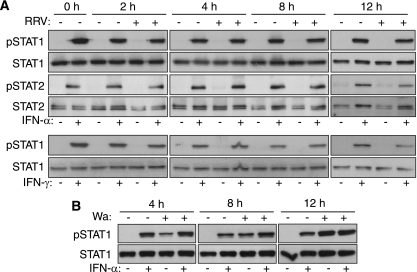
Many viruses block ISG expression by interfering with STAT signaling. For PIV5, this is achieved by targeting STAT1 for proteasome-mediated degradation (17), while West Nile virus prevents STAT1 and STAT2 activation without degradation (32). The ability of RRV and Wa to affect STAT molecules similarly was examined by Western blotting for activated (phosphorylated) and total levels of cellular STAT1 and STAT2. Irrespective of IFN-α stimulation, total STAT1 and STAT2 levels remained constant in uninfected cells (0 h) and mock- or RRV-infected cells at 2 to 12 h PI (Fig. 2A). Bands corresponding to activated STAT1 and STAT2 were detected following the IFN-α stimulation of uninfected cells. Similar levels of activated STAT1 and STAT2 were observed following the IFN-α stimulation of mock- or RRV-infected cells at 2 to 12 h PI. The IFN-γ treatment of MA104 cells also produced STAT1 activation that was unaffected by RRV infection (Fig. 2A). In unstimulated MA104 cells, infection with Wa rotavirus for 4 h led to the activation of STAT1 (Fig. 2B). The level of activated STAT1 in these infected cells increased at 8 and 12 h PI. Particularly in cells stimulated at 8 h after infection, it was evident that Wa infection synergized with IFN-α to increase STAT1 activation. The RRV infection of Caco-2 cells had no effect on IFN-α-stimulated STAT1 and STAT2 activation (data not shown). These findings indicate that the rotavirus-mediated block to ISG expression is not due to STAT1 or STAT2 degradation or the inhibition of their activation.
FIG. 2.
Rotavirus infection did not affect STAT1 and STAT2 protein levels or the IFN-induced activation of STAT1 and STAT2. (A) MA104 cells were left untreated (0 h), mock infected or infected with RRV (MOI, 5) for 2, 4, 8, or 12 h, and then incubated for 30 min in the presence or absence of IFN-α or IFN-γ. Cell lysates were analyzed by Western blotting for total levels of STAT1 and STAT2 and for levels of activated (tyrosine-phosphorylated) STAT1 and STAT2 (pSTAT1 and pSTAT2). (B) MA104 cells were mock infected or infected with Wa (MOI, 10) for 4 or 8 h, treated with IFN-α for 30 min, and analyzed for levels of STAT1 and pSTAT1.
Rotavirus infection blocked IFN-stimulated STAT1 and STAT2 nuclear accumulation.
The observed integrity of IFN-induced STAT activation suggests that IFN-activated STAT1 and STAT2 are prevented from accumulating in the nucleus of rotavirus-infected cells. This possibility was assessed by determining the localization of immunofluorescently stained STAT1 and STAT2 in infected cells before and after IFN treatment. Infection at low MOI and costaining for viral antigen allowed the assessment of STAT localization in infected and uninfected cells in the same sample.
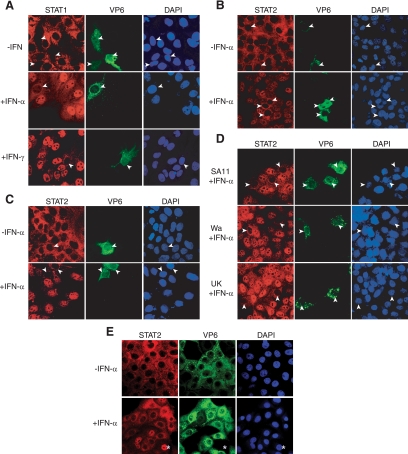
As expected, STAT1 and STAT2 localized predominantly to the cytoplasm and were largely excluded from the nucleus in uninfected and unstimulated MA104 cells, as identified by DAPI staining (Fig. 3A and B). RRV infection of unstimulated cells did not affect STAT1 or STAT2 localization. IFN-α stimulated nuclear accumulation in 99 (STAT1) and 98% (STAT2) of uninfected cells. In striking contrast, IFN-α induced nuclear accumulation in only 6 (STAT1) and 2% (STAT2) of RRV-infected cells, as identified by VP6 staining (Fig. 3A and B). IFN-γ stimulated STAT1 nuclear accumulation in 99% of uninfected cells but in only 7% of RRV-infected cells (Fig. 3A). Importantly, the RRV infection of Caco-2 cells also prevented the IFN-α-mediated nuclear accumulation of STAT2 and STAT1 (Fig. 3C and data not shown). IFN-α-stimulated STAT2 nuclear accumulation also was blocked in ≥95% of MA104 cells infected with SA11, Wa, or UK rotavirus (Fig. 3D). In line with reporter assays, the infection of MA104 cells with RRV at an MOI of 5 inhibited IFN-α-stimulated STAT2 nuclear accumulation in nearly all cells viewed, apart from the few that remained uninfected. STAT2 remained in the cytoplasm in unstimulated cells (Fig. 3E). Taken together, these findings demonstrate that the prevention of STAT1 and STAT2 nuclear accumulation can account for the RRV-mediated block to IFN-α- and IFN-γ-stimulated gene expression, which occurs at the transcriptional level. The ability of SA11, Wa and UK rotaviruses to block STAT nuclear accumulation indicates that this property is highly conserved among rotaviruses.
FIG. 3.
Rotavirus inhibition of IFN-induced STAT1 and STAT2 nuclear accumulation. (A) MA104 cells were infected with RRV at an MOI of 0.05 for 6 h, followed by incubation for 30 min in the presence or absence of IFN-α or IFN-γ. Cells were fixed, permeabilized, and stained with antibodies to STAT1 and rotavirus VP6, as described in Materials and Methods. (B) MA104 cells infected, treated with IFN-α, fixed, and permeabilized as described for panel A were stained for STAT2, VP6, and nuclear DNA. (C) Caco-2 cells were infected, treated, and stained as described for panel B. (D) MA104 cells were infected with SA11, Wa, or UK rotavirus at an MOI of 0.1 for 6 h and then treated and stained as described for panel B. (E) MA104 cells were infected with RRV at an MOI of 5 and treated as described for panel A. Nuclear DNA was stained with DAPI, and images were obtained using confocal microscopy. Arrowheads indicate nuclei of infected cells. Asterisks indicate a representative uninfected cell.
Rotavirus infection blocked expression of an NF-κB-driven reporter gene without reducing NF-κB activation.
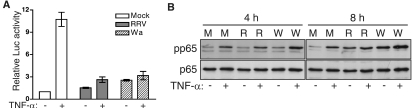
To examine whether rotavirus could interfere with signaling pathways that are distinct from the IFN pathway, an NF-κB-driven luciferase reporter system was employed with exogenous TNF-α as the stimulus. In cells that had been mock infected for 8 h, the addition of TNF-α for 4 h resulted in a 10.7-fold increase in luciferase expression (Fig. 4A). In cells infected with RRV (MOI, 5) or Wa (MOI, 10), the level of TNF-α-stimulated luciferase expression, compared to that of mock-infected cells, was reduced by 83 and 78%, respectively. Interestingly, a small increase in luciferase expression was observed in RRV-infected (1.5-fold) and Wa-infected (2.5-fold) cells without TNF-α stimulation, suggesting that rotavirus can activate NF-κB during the infection of MA104 cells. This was confirmed by the Western blot analysis of infected cells (Fig. 4B). In cells mock infected for 4 h, stimulation with TNF-α led to an increase in levels of phosphorylated p65 (pp65) compared to those of unstimulated cells. After 4 h of RRV infection, a similar increase in TNF-α-stimulated pp65 levels was observed. Both unstimulated and stimulated Wa-infected cells showed a small increase in pp65 above that seen in mock- and RRV-infected cells. Total levels of p65 were unaltered by infection or stimulation by TNF-α at this time. Levels of unstimulated and TNF-α-stimulated pp65 were similar at 4 and 8 h after mock inoculation. However, in the absence of stimulation, RRV infection increased pp65 to a level similar to that of stimulated, mock-infected cells. The TNF-α stimulation of RRV-infected cells further enhanced the pp65 level. Wa infection enhanced pp65 levels in both unstimulated and stimulated cells to an even higher degree than RRV. Again, total levels of p65 were unchanged by infection with RRV or Wa. Taken together with the reporter assay data, these results suggest that although rotavirus can robustly activate NF-κB during infection, the ability of this activated transcription factor to enhance gene expression is efficiently blocked in MA104 cells.
FIG. 4.
Effect of rotavirus infection on NF-κB signaling and activation. (A) MA104 cells cotransfected with the NF-κB-Luc and pRL-TK constructs were mock infected or infected with RRV (MOI, 5) or Wa (MOI, 10) for 8 h, followed by incubation for 4 h in the presence or absence of TNF-α. Data show the means ± ranges of firefly luciferase (Luc) activity normalized for Renilla luciferase levels from two independent experiments. (B) MA104 cells were mock infected (M) or infected with RRV (R) or Wa (W) as described for panel A for 4 or 8 h, followed by incubation for 30 min in the presence or absence of TNF-α. Cell lysates were analyzed by Western blotting for levels of activated (serine-phosphorylated) p65 (pp65) and total p65.
Rotavirus infection blocked the nuclear accumulation of NF-κB.
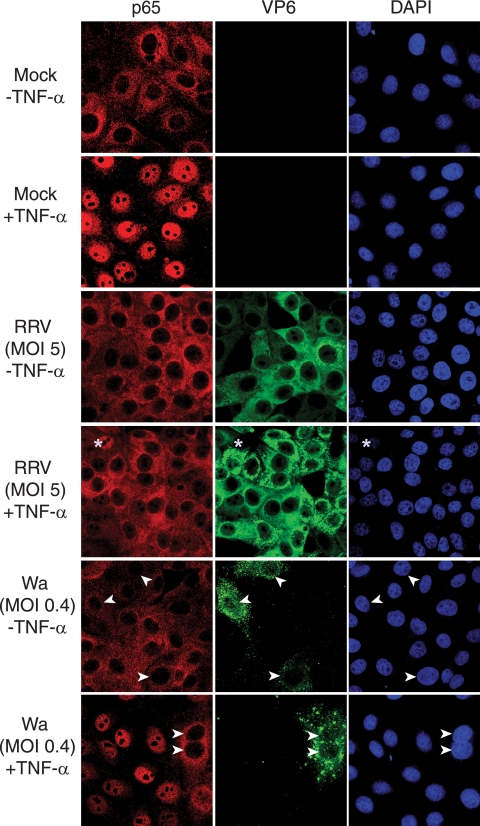
The mechanism by which rotavirus can reduce NF-κB-dependent gene expression was examined by assessing the nuclear localization of p65 in infected MA104 cells by confocal microscopy (Fig. 5). In unstimulated mock-infected cells, p65 was localized primarily to the cytoplasm, whereas TNF-α stimulation led to p65 nuclear accumulation in 93% of cells. In unstimulated cells infected with RRV for 6 h at an MOI of 5, p65 again was largely absent from the nucleus. However, TNF-α treatment induced the nuclear accumulation of p65 in only 6% of cells expressing rotavirus antigen at this MOI, while p65 was observed in the nucleus of the few cells that remained uninfected. In contrast, the stimulation of cells infected with RRV at an MOI of 0.2 resulted in p65 nuclear accumulation in 84% of infected cells (data not shown). Interestingly, cell cultures infected with Wa for 6 h at an MOI of 0.4 and stimulated with TNF-α showed nuclear accumulation of p65 in only 5% of infected cells (Fig. 5). It therefore appears that at 6 h PI with RRV or Wa, transcriptional enhancement by p65 is blocked through the prevention of its nuclear accumulation. However, RRV is less efficient at producing this effect, as it requires a higher MOI than Wa.
FIG. 5.
Rotavirus infection inhibited the TNF-α-induced nuclear accumulation of p65. (A) MA104 cells mock infected or infected with RRV or Wa at the indicated MOI for 6 h were incubated for 30 min in the presence or absence of TNF-α. Fixed and permeabilized cells were stained with antibodies to p65 and rotavirus VP6, as described in the text. Nuclear DNA was stained with DAPI. Images were obtained using confocal microscopy, as described in Materials and Methods. Arrowheads indicate nuclei of Wa-infected cells. Asterisks indicate one representative uninfected cell in the RRV-inoculated cell population.
Intracellular expression of RRV NSP1, NSP3, or NSP4 did not inhibit IFN-α- or TNF-α-stimulated reporter gene expression.
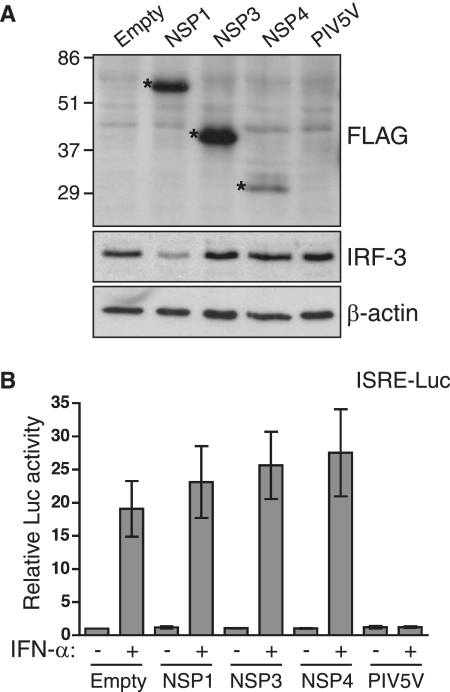
Viral NSPs often are responsible for antagonizing IFN signaling. Therefore, plasmids expressing FLAG-tagged NSP1, NSP3, and NSP4 were constructed to allow the testing of these proteins for their effect on IFN-stimulated reporter gene expression. The expression of the RRV NSPs in 293T cells was confirmed by Western blotting using anti-FLAG antibodies (Fig. 6A), with protein sizes approximating predicted values (FLAG-NSP1, 57 kDa; FLAG-NSP3, 38 kDa; and FLAG-NSP4, 32 kDa). The functional integrity of NSP1 also was confirmed by a blot showing the degradation of IRF-3 in 293T cells expressing NSP1 (Fig. 6B). Lower IRF-3 levels were not observed in cells transfected with the other constructs, and the equal loading of the blot was confirmed by reprobing for β-actin. In 293T cells cotransfected with empty vector and ISRE-Luc/pCMV-β, IFN stimulation increased luciferase levels by 19-fold (Fig. 6A). Transfection with constructs expressing NSP1, NSP3, and NSP4 had no impact on IFN-stimulated luciferase levels. In contrast, under identical conditions the expression of a well-characterized inhibitor of IFN signaling, the PIV5 V protein, completely abolished IFN-stimulated luciferase expression. It also was found that NSP1, NSP3, or NSP4 did not reduce TNF-α-induced reporter gene expression (data not shown). These findings show that under the conditions used, the expression of RRV NSP1, NSP3, or NSP4 could not account for the block to IFN-α-stimulated or NF-κB-driven gene expression observed during RRV infection.
FIG. 6.
Effect of RRV NSP expression on IFN-induced gene expression. 293T cells were cotransfected with ISRE-Luc, pCMV-β, and empty vector (empty) or constructs expressing FLAG-tagged RRV NSP1, NSP3, NSP4, or untagged PIV5 V protein. At 24 h posttransfection, cells were incubated for 7 h in the presence or absence of IFN-α. (A) Cells (without IFN treatment) were lysed and analyzed by Western blotting with antibodies to the FLAG epitope, IRF-3, and β-actin. Asterisks indicate bands corresponding to FLAG-tagged NSPs. The positions of molecular mass markers are shown in kilodaltons. (B) Cells were analyzed for luciferase (Luc) and β-galactosidase expression. Data show the means ± ranges of luciferase activity normalized for β-galactosidase levels from two independent experiments.
DISCUSSION
This study clearly shows that rotavirus infection can block the nuclear accumulation of STAT1, STAT2, and NF-κB and reduce their ability to act as transcriptional enhancers. It is likely that this rotavirus function facilitates the virus evasion of the antiviral effects of IFN and reduces the expression of cytokines that are required for effective innate and adaptive immune responses.
In reporter assays, rotavirus strains RRV and Wa were effective in reducing IFN-responsive gene expression. It is known that RRV, but not Wa, can block cellular translation, so translational inhibition cannot account for the reductions we observed in reporter protein levels (13, 43). This suggests that rotavirus reduces mRNA abundance, a conclusion supported by the reduced levels of transcripts from the IFN-inducible genes MxA and ISG56 that we observed in rotavirus-infected cells following IFN stimulation. Interestingly, Wa rotavirus also induced a low level of ISG expression in the absence of exogenous IFN, suggesting the induction of IFN production by the virus. Supporting IFN induction by Wa, our data show that STAT1 activation occurs in Wa-infected cells in the absence of added IFN. It may be that Wa, in contrast to RRV, cannot efficiently degrade IRF-3, although this needs to be formally tested. Another possible explanation is that the partially purified Wa used, in contrast to the purified RRV, contains replication-defective particles that might induce IFN production. In any case, it is apparent that the rotavirus-induced block to ISG expression does not result from interference with the activation of STAT molecules or upstream components of the IFN pathway.
In our studies, NF-κB-dependent gene expression also was blocked by rotavirus infection. This effect was similar to the rotavirus-mediated block to IFN-induced gene expression, as p65 activation by TNF-α was not affected but its nuclear accumulation was inhibited. NF-κB was found to be activated during the RRV and Wa infection of MA104 cells, supporting previous studies showing NF-κB activation, DNA binding, and the NF-κB-dependent expression of IL-8 in infected HT-29 cells (10, 42). It is reasonable to propose that rotavirus infection also blocks NF-κB nuclear accumulation in HT-29 cells. The efficient production of IL-8 by rotavirus-infected HT-29 cells in the face of a probable reduction in nuclear NF-κB levels requires explanation. It is feasible that any effect on nuclear accumulation could be preceded by a rapid burst of IL-8 production that is inhibited once virus replication is fully established. Interestingly, evidence exists showing the activation of STAT1 and STAT2 early during the rotavirus infection of HT-29 cells but not of MA104 or Caco-2 cells (42 and T. T. Truong, G. Holloway, and B. S. Coulson, unpublished data). This suggests that these infected HT-29 cells also produce type I IFN, despite efficient IRF-3 degradation. Indeed, the transient production of IFN-α mRNA was detected in RRV-infected HT-29 cells, and IFN-β mRNA has been found in rotavirus-infected mouse intestine (42). The production of cytokines in rotavirus-infected cells would depend on the relative timing of the response of the cell to infection and the rotavirus disruption of gene expression, both of which may vary between cell types. In contrast to our findings in MA104 cells, NF-κB has been shown translocate to the nucleus at 6 h after the RRV infection of mouse embryonic fibroblasts (18). It is possible that NF-κB is activated in mouse embryonic fibroblasts before RRV can inhibit its nuclear accumulation, or that RRV does not inhibit the nuclear accumulation of NF-κB in cells of mouse origin.
NF-κB is required for optimal IFN-β production. Therefore, in addition to the degradation of IRF-3, the reduction of NF-κB nuclear accumulation may be a strategy used by rotavirus to reduce IFN expression (2, 47). A proportion of the antiviral effect of IFN requires NF-κB (19), so this part of the IFN signaling cascade also might be blocked in rotavirus-infected cells. As Wa is more efficient than RRV in blocking NF-κB nuclear accumulation, based on the MOI required, it may be that different rotavirus strains have evolved to preferentially target distinct pathways of the host response. It has been demonstrated that TNF-α can synergize with type I and II IFN to produce an elevated antiviral state (4, 22). As TNF levels are elevated in the serum and stools of rotavirus-infected children (1, 29), blocking NF-κB nuclear accumulation also might prevent the antiviral action of TNF-α in infected cells.
When the profile of cellular gene expression during the RRV infection of Caco-2 cells was analyzed by microarray, only 0.5% of genes were upregulated by >4-fold (15). Few of the many genes known to be responsive to IFN stimulation were upregulated, suggesting that either IFN production did not occur efficiently or IFN signaling was blocked (16). In a recent study of expression profiles in human PIV type 1-infected cells, mutant viruses that were highly attenuated by the loss of a functional C protein induced a far greater transcriptional response than wild-type virus (8). IFN-responsive genes and those containing transcription factor binding sites for NF-κB were upregulated, suggesting that the wild-type virus was largely restricting innate cellular responses. It can be hypothesized that a mutant rotavirus lacking the ability to antagonize signaling through IFN and NF-κB also would induce a much greater transcriptional response in infected cells than wild-type rotavirus.
The mechanism used by rotaviruses to inhibit STAT and NF-κB nuclear accumulation is unknown. STAT1, STAT2, and p65 remain intact and can be activated efficiently in infected cells, so it is possible that rotavirus mediates the sequestration of these activated proteins in the cytoplasm. This strategy is used by measles virus for STAT (37), although the viral sequestration of NF-κB has not been reported to date. Another plausible explanation is that rotavirus interferes with components of the cellular machinery required for STAT and NF-κB nuclear import or export. Proteins expressed by Ebola virus and the severe acute respiratory syndrome coronavirus affect STAT1 in this way by interacting with karyopherins, proteins involved in nuclear import (24, 41). At least for STAT1, DNA binding is required for its retention in the nucleus following translocation (35). It may be that rotavirus blocks STAT nuclear accumulation by disrupting STAT/DNA binding. The nuclear import of both STAT and NF-κB has been reported to require interactions between nuclear localization signals within these proteins and importin molecules that mediate nuclear entry via the nuclear pore complex (21, 34). Interference with importins by rotavirus could affect the nuclear import of STAT1/STAT2 and NF-κB during infection. Alternatively, the interaction of rotavirus with proteins that make up the nuclear pore complex, known as nucleoporins, could affect the nuclear import of multiple factors. In this manner, poliovirus has the ability to induce the relocalization of many cellular proteins (27). It is possible that rotavirus causes a general block to nuclear accessibility and global transcription; however, this remains to be tested. Although RRV efficiently blocked IFN-induced gene expression in Caco-2 cells, the effect was greatly reduced when IFN treatment was sustained for long periods. It is conceivable that the rotavirus-induced block to gene expression is more important early in infection, or that a sustained block to cellular transcription would be detrimental to either virus production or cell survival and thus is avoided by rotavirus.
The antagonism of IFN signaling by other viruses often is mediated by nonstructural proteins, such as the V proteins of paramyxoviruses (17, 38). This might be the case for rotaviruses, although the involvement of structural proteins also is possible. The expression of tagged RRV NSP1, NSP3, and NSP4 did not inhibit IFN-α- or TNF-α-induced gene expression in our assays. In the case of NSP1, the expression of a functional protein was confirmed by its ability to reduce IRF-3 levels. It remains possible that the inhibition of IFN or TNF-α signaling requires higher levels of NSP expression than IRF-3 degradation. The presence of multiple FLAG tags also may differentially affect NSP1 functions and affect the functions of NSP3 and NSP4.
Recent evidence shows that IFN-α and IFN-γ signaling is required for the resolution of viral replication and extraintestinal pathology in mice infected with certain strains of rotavirus but not others (23, 48). Taken together with our current data, this suggests that the control of rotavirus replication and spread in vivo depends on overcoming the rotavirus-mediated blockade of both IFN induction and signaling. Although the degree to which the IFN system is antagonized probably varies between different virus strain and host combinations, it is likely that the inhibition of STAT and p65 nuclear accumulation will be of importance, particularly in light of the conservation of these functions between human and animal rotaviruses.
The demonstrated ability of rotavirus to reduce cellular gene expression requiring STAT and NF-κB nuclear accumulation significantly enhances our understanding of the immune evasion strategies of rotavirus. As the ability to evade host defenses likely is a major determinant of virulence, the manipulation of a viral gene responsible for blocking STAT or NF-κB signaling could be used in the design of a rationally attenuated, second-generation rotavirus vaccine. Drugs targeting a viral protein responsible for the block to gene expression could enhance innate and adaptive immunity against rotaviruses. Peptide drugs based on a novel rotavirus antagonist of IFN or NF-κB also could be used to treat diseases involving dysregulated IFN production, such as systemic lupus erythematosus (7) or uncontrolled inflammation.
ADDENDUM IN PROOF
Recently, Graff and coworkers reported the inhibition of NF-κB signaling by NSP1 of bovine and porcine rotavirus strains (J. W. Graff, K. Ettayebi, and M. E. Hardy, PLoS Pathog. 5:e1000280, 2009).
Acknowledgments
We thank Steven Goodbourn and Richard Randall for permission to use the ISRE and GAS luciferase reporter constructs and the PIV5 V protein expression vector and Alexander Khromykh for their provision. We are grateful to Fiona Fleming for rotavirus propagation and purification.
This work was supported by project grants 350252 and 509006 from the National Health and Medical Research Council of Australia (NHMRC) and NHMRC Senior Research Fellowship 350253 (B.S.C.).
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Azim, T., S. M. Ahmad, E. K. Sefat, M. S. Sarker, L. E. Unicomb, S. De, J. D. Hamadani, M. A. Salam, M. A. Wahed, and M. J. Albert. 1999. Immune response of children who develop persistent diarrhea following rotavirus infection. Clin. Diagn. Lab. Immunol. 6690-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barro, M., and J. T. Patton. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. USA 1024114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barro, M., and J. T. Patton. 2007. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J. Virol. 814473-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartee, E., M. R. Mohamed, M. C. Lopez, H. Baker, and G. McFadden. 2009. The addition of tumor necrosis factor plus β-interferon induces a novel synergistic antiviral state against poxviruses in primary human fibroblasts. J. Virol. 83498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bass, D. M. 1997. Interferon-γ and interleukin 1, but not interferon-α, inhibit rotavirus entry into human intestinal cell lines. Gastroenterology 11381-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baud, V., and M. Karin. 2001. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11372-377. [DOI] [PubMed] [Google Scholar]
- 7.Blanco, P., A. K. Palucka, M. Gill, V. Pascual, and J. Banchereau. 2001. Induction of dendritic cell differentiation by IFN-α in systemic lupus erythematosus. Science 2941540-1543. [DOI] [PubMed] [Google Scholar]
- 8.Boonyaratanakornkit, J. B., E. J. Bartlett, E. Amaro-Carambot, P. L. Collins, B. R. Murphy, and A. C. Schmidt. 2009. The C proteins of human parainfluenza virus type I (HPIV1) control the transcription of a broad array of cellular genes that would otherwise respond to HPIV1 infection. J. Virol. 831892-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casola, A., M. K. Estes, S. E. Crawford, P. L. Ogra, P. B. Ernst, R. P. Garofalo, and S. E. Crowe. 1998. Rotavirus infection of cultured intestinal epithelial cells induces secretion of CXC and CC chemokines. Gastroenterology 114947-955. [DOI] [PubMed] [Google Scholar]
- 10.Casola, A., R. P. Garofalo, S. E. Crawford, M. K. Estes, F. Mercurio, S. E. Crowe, and A. R. Brasier. 2002. Interleukin-8 gene regulation in intestinal epithelial cells infected with rotavirus: role of viral-induced IκB kinase activation. Virology 2988-19. [DOI] [PubMed] [Google Scholar]
- 11.Coulson, B. S., K. J. Fowler, R. F. Bishop, and R. G. Cotton. 1985. Neutralizing monoclonal antibodies to human rotavirus and indications of antigenic drift among strains from neonates. J. Virol. 5414-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coulson, B. S., and C. Kirkwood. 1991. Relation of VP7 amino acid sequence to monoclonal antibody neutralization of rotavirus and rotavirus monotype. J. Virol. 655968-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulson, B. S., J. M. Tursi, W. J. McAdam, and R. F. Bishop. 1986. Derivation of neutralizing monoclonal antibodies to human rotaviruses and evidence that an immunodominant neutralization site is shared between serotypes 1 and 3. Virology 154302-312. [DOI] [PubMed] [Google Scholar]
- 14.Coulson, B. S., L. E. Unicomb, G. A. Pitson, and R. F. Bishop. 1987. Simple and specific enzyme immunoassay using monoclonal antibodies for serotyping human rotaviruses. J. Clin. Microbiol. 25509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuadras, M. A., D. A. Feigelstock, S. An, and H. B. Greenberg. 2002. Gene expression pattern in Caco-2 cells following rotavirus infection. J. Virol. 764467-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69912-920. [PubMed] [Google Scholar]
- 17.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 739928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douagi, I., G. M. McInerney, A. S. Hidmark, V. Miriallis, K. Johansen, L. Svensson, and G. B. Karlsson Hedestam. 2007. Role of interferon regulatory factor 3 in type I interferon responses in rotavirus-infected dendritic cells and fibroblasts. J. Virol. 812758-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du, Z., L. Wei, A. Murti, S. R. Pfeffer, M. Fan, C. H. Yang, and L. M. Pfeffer. 2007. Non-conventional signal transduction by type 1 interferons: the NF-κB pathway. J. Cell Biochem. 1021087-1094. [DOI] [PubMed] [Google Scholar]
- 20.Dyall-Smith, M. L., and I. H. Holmes. 1984. Sequence homology between human and animal rotavirus serotype-specific glycoproteins. Nucleic Acids Res. 123973-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagerlund, R., L. Kinnunen, M. Kohler, I. Julkunen, and K. Melen. 2005. NF-κB is transported into the nucleus by importin α3 and importin α4. J. Biol. Chem. 28015942-15951. [DOI] [PubMed] [Google Scholar]
- 22.Feduchi, E., M. A. Alonso, and L. Carrasco. 1989. Human γ-interferon and tumor necrosis factor exert a synergistic blockade on the replication of herpes simplex virus. J. Virol. 631354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng, N., B. Kim, M. Fenaux, H. Nguyen, P. Vo, M. B. Omary, and H. B. Greenberg. 2008. Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J. Virol. 827578-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 819812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109S81-S96. [DOI] [PubMed] [Google Scholar]
- 26.Graff, J. W., J. Ewen, K. Ettayebi, and M. E. Hardy. 2007. Zinc-binding domain of rotavirus NSP1 is required for proteasome-dependent degradation of IRF3 and autoregulatory NSP1 stability. J. Gen. Virol. 88613-620. [DOI] [PubMed] [Google Scholar]
- 27.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holloway, G., and B. S. Coulson. 2006. Rotavirus activates JNK and p38 signaling pathways in intestinal cells, leading to AP-1-driven transcriptional responses and enhanced virus replication. J. Virol. 8010624-10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, B., L. Snipes-Magaldi, P. Dennehy, H. Keyserling, R. C. Holman, J. Bresee, J. Gentsch, and R. I. Glass. 2003. Cytokines as mediators for or effectors against rotavirus disease in children. Clin. Diagn. Lab. Immunol. 10995-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolly, C. L., B. M. Beisner, and I. H. Holmes. 2000. Rotavirus infection of MA104 cells is inhibited by ricinus lectin and separately expressed single binding domains. Virology 27589-97. [DOI] [PubMed] [Google Scholar]
- 31.King, P., and S. Goodbourn. 1994. The β-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 26930609-30615. [PubMed] [Google Scholar]
- 32.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 791934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Londrigan, S. L., M. J. Hewish, M. J. Thomson, G. M. Sanders, H. Mustafa, and B. S. Coulson. 2000. Growth of rotaviruses in continuous human and monkey cell lines that vary in their expression of integrins. J. Gen. Virol. 812203-2213. [DOI] [PubMed] [Google Scholar]
- 34.Melen, K., R. Fagerlund, J. Franke, M. Kohler, L. Kinnunen, and I. Julkunen. 2003. Importin-α nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 27828193-28200. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, T., A. Marg, P. Lemke, B. Wiesner, and U. Vinkemeier. 2003. DNA binding controls inactivation and nuclear accumulation of the transcription factor STAT1. Genes Dev. 171992-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mogensen, T. H., and S. R. Paludan. 2001. Molecular pathways in virus-induced cytokine production. Microbiol. Mol. Biol. Rev. 65131-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 777635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283230-239. [DOI] [PubMed] [Google Scholar]
- 39.Powell, P. P., L. K. Dixon, and R. M. Parkhouse. 1996. An IκB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 708527-8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 891-47. [DOI] [PubMed] [Google Scholar]
- 41.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 805156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rollo, E. E., K. P. Kumar, N. C. Reich, J. Cohen, J. Angel, H. B. Greenberg, R. Sheth, J. Anderson, B. Oh, S. J. Hempson, E. R. Mackow, and R. D. Shaw. 1999. The epithelial cell response to rotavirus infection. J. Immunol. 1634442-4452. [PubMed] [Google Scholar]
- 43.Sato, T., H. Suzuki, S. Kitaoka, T. Konno, and N. Ishida. 1986. Patterns of polypeptide synthesis in human rotavirus infected cells. Arch. Virol. 9029-40. [DOI] [PubMed] [Google Scholar]
- 44.Sheth, R., J. Anderson, T. Sato, B. Oh, S. J. Hempson, E. Rollo, E. R. Mackow, and R. D. Shaw. 1996. Rotavirus stimulates IL-8 secretion from cultured epithelial cells. Virology 221251-259. [DOI] [PubMed] [Google Scholar]
- 45.Spitkovsky, D., S. P. Hehner, T. G. Hofmann, A. Moller, and M. L. Schmitz. 2002. The human papillomavirus oncoprotein E7 attenuates NF-κB activation by targeting the IκB kinase complex. J. Biol. Chem. 27725576-25582. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi, O., and S. Akira. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 2017-22. [DOI] [PubMed] [Google Scholar]
- 47.Thanos, D., and T. Maniatis. 1992. The high mobility group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell 71777-789. [DOI] [PubMed] [Google Scholar]
- 48.Vancott, J. L., M. M. McNeal, A. H. Choi, and R. L. Ward. 2003. The role of interferons in rotavirus infections and protection. J. Interferon Cytokine Res. 23163-170. [DOI] [PubMed] [Google Scholar]