Abstract
Binding of the human immunodeficiency virus (HIV) envelope glycoprotein (Env) to the cellular CD4 receptor and a chemokine coreceptor initiates a series of conformational changes in the Env subunits gp120 and gp41. Eventually, the trimeric gp41 folds into a six-helix bundle, thereby inducing fusion of the viral and cellular membranes. C peptides derived from the C-terminal heptad repeat (CHR) of gp41 are efficient entry inhibitors as they block the six-helix bundle formation. Previously, we developed a membrane-anchored C peptide (maC46) expressed from a retroviral vector that also shows high activity against virus strains resistant to enfuvirtide (T-20), an antiviral C peptide approved for clinical use. Here, we present a systematic analysis of mutations in Env that confer resistance of HIV type 1 (HIV-1) to maC46. We selected an HIV-1 BaL strain with 10-fold reduced sensitivity to maC46 (BaL_C46) by passaging virus for nearly 200 days in the presence of gradually increasing concentrations of maC46. In comparison to wild-type BaL, BaL_C46 had five mutations at highly conserved positions in Env, three in gp120, one in the N-terminal heptad-repeat (NHR), and one in the CHR of gp41. No mutations were found in the NHR domain around the GIV motif that are known to cause resistance to enfuvirtide. Instead, maC46 resistance was found to depend on complementary mutations in the NHR and CHR that considerably favor binding of the mutated NHR to the mutated CHR over binding to maC46. In addition, resistance was highly dependent on mutations in gp120 that accelerated entry. Taken together, resistance to maC46 did not develop readily and required multiple cooperating mutations at conserved positions of the viral envelope glycoproteins gp120 and gp41.
The entry process of the human immunodeficiency virus type 1 (HIV-1) has become a major target for new antiviral drugs. Viral entry is initiated by binding of the HIV-1 envelope glycoprotein subunit gp120 to the CD4 receptor and a chemokine coreceptor, generally CCR5 or CXCR4. Upon coreceptor binding, the viral transmembrane subunit gp41 undergoes conformational changes that eventually lead to the formation of the six-helix bundle (6HB) and membrane fusion. The 6HB is composed of a central trimeric coiled-coil structure formed by the N-terminal heptad repeat (NHR) domains of three gp41 molecules and the corresponding C-terminal heptad repeats (CHRs) that pack into the longitudinal grooves on the surface of the NHR coiled-coil in an antiparallel orientation (23). C-peptide fusion inhibitors (CFI) derived from the CHR of gp41 compete with the viral CHR for binding to the NHR trimer, thus blocking 6HB formation and viral entry (18).
T-20 (enfuvirtide) is the first clinically approved CFI with high antiviral activity and a low-toxicity profile. However, as with many anti-HIV-1 drugs, resistance can emerge rapidly (13). The majority of the resistance mutations are found in the NHR of gp41 among the amino acids 544 to 553 (32, 35) (numbering refers to gp160 of the HIV-1 HXB2 strain throughout the article). Most of these mutations cause resistance by reducing the affinity of the NHR target region to inhibitory C peptides (13). Additionally, viral entry kinetics were found to correlate with the baseline susceptibility of different HIV strains to CFI. Determinants for viral entry kinetics are found in gp41 as well as in gp120 (1, 14, 35). Here, the influence of coreceptor affinity on virus entry kinetics and CFI susceptibility has been studied extensively (28, 30, 31). Recently, a statistical approach was used that highlighted positions in gp120 that underwent mutations in patients under enfuvirtide treatment (38). However, to our knowledge, selected CFI resistance mutations outside of gp41 have never been confirmed experimentally.
Previously, we developed a retroviral vector expressing a membrane-anchored antiviral C peptide (maC46) that efficiently inhibits a broad range of different HIV-1 isolates. Enfuvirtide-resistant HIV-1 strains with mutations in the GIV motif of NHR were fully susceptible to maC46 (10). In the present study, we selected an HIV-1 variant with reduced sensitivity to maC46 by passaging an enfuvirtide-resistant BaL strain of HIV-1 on cells expressing increasing concentrations of maC46. Mutations in gp120 and gp41 were found to contribute to maC46 resistance.
MATERIALS AND METHODS
Retroviral vectors.
The vectors C46a (original name, M87o-hIgG2-Ineo), C46b (original name, M87/om-Ineo), C46b1 (original name, M87/omc-Ineo), C46c (original name, M87/omc) (15), and M87o (10) have been described previously. For a schematic overview of all vectors, see Fig. S1 in the supplemental material.
Cells and viruses.
The human embryonic kidney cell line 293T and the human astroglioma cell line U87 stably expressing CD4 and CXCR4 (U87.CD4.CXCR4) or CD4 and CCR5 (U87.CD4.CCR5) were maintained in Dulbecco's modified Eagle medium. The T-cell line PM-1, a subclone of HuT78 expressing CD4, CXCR4, and CCR5, was cultured in RPMI 1640 medium. All media were supplemented with 5% fetal calf serum, 2 mM glutamine, and 2 mM penicillin-streptomycin (complete media). The maC46-expressing cell lines were generated by transduction of PM-1 cells with retroviral vectors at low multiplicities of infection, as described below, to avoid multiple integrations. Cells transduced with vectors harboring a neomycin resistance gene were selected with G418 (0.8 mg/ml) for 10 days. Transduced cells without the resistance gene were enriched by fluorescence-activated cell sorting (FACS) for C-peptide expression to more than 95% purity using a FACSCalibur FACS sorter (BD, Heidelberg, Germany). The gamma-retroviral vectors C46a, C46b, C46c, C46b1, and M87o-dPRE express identical transgenes (C-peptide maC46) but differ in their expression levels. The diverse expression is achieved by differences in the vector backbone, codon optimization of the transgene, and the presence of cis elements (15). The viruses NL4-3, BaL, and BaL_V_K (original name, BaL/sel [19]) were propagated on PM-1 cells. Viral replication was monitored by p24 enzyme-linked immunosorbent assay (Innotest HIV antigen monoclonal antibody; Innogenetics, Gent, Belgium).
Peptides and coreceptor antagonist.
The peptides T-1249 (34) and soluble C46 were custom synthesized (Thermo Fisher Scientific, Ulm, Germany). The peptides constructed from the BaL_C46 gp41 wild-type NHR (NWT), NHR with the mutation A582T (NT582), a double mutant NHR (NV559T582), wild-type CHR (CWT/C46), and the CHR with the mutation N637K (CK637) (Table 1) were custom synthesized (Genscript, Piscataway, NJ, USA). T-20 (enfuvirtide) was obtained from Roche. TAK-779 (2) was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH.
TABLE 1.
Sequences of peptides used for the thermal denaturation studies
| Peptide | Sequencea |
|---|---|
| NWT | GIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARVLAVERYLRDQ |
| NT582 | GIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARVLTVERYLRDQ |
| NV559T582 | GIVQQQNNLLRAVEAQQHLLQLTVWGIKQLQARVLTVERYLRDQ |
| CWT/C46 | WMEWDREINNYTSIIYSLIEESQNQQEKNEQELL |
| CK637 | WMEWDREINKYTSIIYSLIEESQNQQEKNEQELL |
Deviations from the wild-type sequence are underlined.
Lentiviral and gamma-retroviral packaging.
Vector supernatants were produced by transient transfection with the calcium phosphate method. The day before transfection, 6 × 106 293T cells were plated on a 10-cm dish. On the day of transfection, medium was exchanged for complete Dulbecco's modified Eagle medium (gamma-retroviral vectors) or for complete medium without fetal calf serum (lentiviral vectors) supplemented with 25 μM chloroquine. For each transfection, 7.5 μg of lentiviral or gamma-retroviral transfer vector plasmid, 12.5 μg of Gag/Pol plasmid (murine leukemia virus Gag/Pol plasmid M57 [10] or lentiviral Gag/Pol plasmid pCMV-dR8.91 [24]), and 1 μg (lentiviral vectors) or 2 μg (gamma-retroviral vectors) of envelope plasmid were used. Lentiviral vectors were either packaged with HIV Env or with vesicular stomatitis virus G protein. Retroviral particles were packaged with gibbon ape leukemia virus Env. The medium was exchanged after 6 h. Supernatants containing the viral particles were collected 24 to 36 h after transfection, filtered (0.45-μm pore size) and stored at −80°C until usage. Lentiviral supernatants were concentrated by ultracentrifugation (L8-M, SW32 Ti rotor; Beckman, Krefeld, Germany) (2 h at 25,000 rpm at 4°C) before being stored.
Determination of cell surface expression level of membrane-anchored peptides by a quantitative flow cytometry assay.
Quantification of the antibody-binding sites (ABS) of the maC46-expressing cell lines was performed as described previously by Lee et al. (17) with minor modifications (15, 33). In short, the mean fluorescence intensity was converted into ABS using a Quantum Simply Cellular Microbead Kit (Bangs Laboratory, Fishers, IN). The kit contains five different bead populations, each with defined ABS. Beads and cells were stained in parallel with fluorescein isothiocyanate-conjugated 2F5 antibody (Polymun, Vienna, Austria), which recognizes the C peptide. Then the mean fluorescence was determined, and the fluorescence signal of the beads was used to generate a calibration curve, which was then used to calculate the ABS of each cell line.
HIV envelope gene cloning.
Cell-free virus supernatant (1 ml) was pelleted by ultracentrifugation (TL100; Beckman, Krefed, Germany) at 25,000 rpm for 2.5 h at 4°C. The virus pellet was resuspended in 140 μl of phosphate-buffered saline, and viral RNA was extracted (QiaAmp Viral RNA kit; Qiagen, Hilden, Germany). Viral RNA was reverse transcribed to cDNA using the primer N (5′-CTG CCA ATC AGG GAA GTA GCC TTG TGT-3′) with Superscript II (Invitrogen, Karlsruhe, Germany) as described by the manufacturer. The Env gene was amplified in a nested PCR with a High Fidelity Plus PCR system (Roche, Mannheim, Germany) in 50-μl reaction volumes using amplification buffer 2 provided in the High Fidelity Plus PCR system. The first round of PCR was performed with the primers A (5′-GGC TTA GGC ATC TCC TAT GGC AGG AAG AA-3′) and TNE3_fh (5′-CTT ATA GCA AAA TCC TTT CCA AGC CCT GTC TTA TTC TTC TAG G-3′). The second PCR was performed with the primers HIVkozakEnv6253f-(2) (5′-GCC ACC ATG AGA GTG ACG GAG ATC AGG-3′) and HIVnef9047rev (5′-CTG AGG TCT GAC TGG AAA ACC-3′). All reactions were run on a Biometra Thermal Cycler T-Personal Combi (10 cycles of 94°C for 10s, 59°C for 30s, and 68°C for 200 s, followed by 25 cycles of 94°C for 10s, 59°C for 30s, and 68°C for 180s, with a 20-s increment for each round, and a final step at 68°C for 10 min). The PCR products were purified by gel extraction (JETquick Gel Extraction Spin Kit; Genomed, Löhne, Germany). Fractions of the bulk PCR product were sequenced to identify mutations.
The PCR products were treated with Taq polymerase and a 0.3 mM concentration of each deoxynucleoside triphosphate for 10 min to generate A-overhangs at the 3′ ends of the PCR products. The PCR fragments were then cloned into the pCR-XL TOPO vector (Invitrogen, Karlsruhe, Germany). Then the Env gene was inserted into the pHCMV expression vector downstream of the rabbit beta-globin intron between the BamHI and PstI sites (11, 41).
maC46 sensitivity of Env constructs.
PM-1 cell lines expressing the maC46 peptide and the control cell line PM-1 were transduced with replication-incompetent lentiviral vectors harboring an enhanced green fluorescent protein (eGFP) marker gene. Five days later eGFP and transgene expression levels were determined by flow cytometry using a FACSCalibur machine (BD, Heidelberg, Germany). The maC46 peptide was detected by immunostaining with the monoclonal antibody 2F5 (Polymun, Vienna, Austria) and an antihuman secondary antibody conjugated with Cy5.5 or allophycocyanin. The percentage of cells that were doubly positive for C-peptide and eGFP expression was used to determine the titer of each vector on the respective cell line. The titer was normalized relative to the control cell line. For the dose-response assay, the reciprocal of the titer on the C-peptide-expressing cell line relative to the titer on the control cell line was used as a measure for the inhibitory capacity of each construct, as described previously (15, 33).
Entry kinetic assay.
PM-1 cells and replication-incompetent lentiviral vectors were mixed in ice-cold RPMI medium. A total of 104 cells were seeded per well into a 96-well plate and centrifuged for 1 h at 4°C. The temperature was then raised to 37°C (time point 0) to initiate viral entry. A completely inhibitory concentration of T-20 (2.5 μM) was added at the zero time point or later time points. For each time point triplicate samples were prepared. The plate was incubated at 37°C for 5 days, and the transduction efficacy in each well was determined by FACS. Relative transduction values were obtained by dividing the transduction efficacy at each time point by the transduction efficacy in the control wells where no inhibitor had been added.
Susceptibility to entry inhibitors.
Cells (2 × 104 U87.CD4.CCR5 or 1 × 104 to 2 × 104 PM-1 cells per well) were seeded in 96-well plates in replicates and mixed with lentiviral particles. Different entry inhibitors (T-20, C46, T-1249, and TAK-779) were added at increasing concentrations. The plate was centrifuged (1,000 × g) for 1 h at 31°C. Subsequently, cells were incubated at 37°C. If the lentiviral particles harbored an eGFP marker gene, transduction efficacy was determined by FACS after 5 days of incubation. If the lentiviral particles carried a luciferase marker gene, cells were incubated at 31°C for 3 days. Cells were then lysed, and the firefly luciferase activity was determined using a Luciferase Assay System (Promega, Mannheim, Germany).
Thermal denaturation studies.
N peptides (in 50 mM sodium formate, pH 3.5) and C peptides (in 20 mM sodium phosphate containing 0.2 M NaCl, pH 7.0) were mixed in 20 mM sodium phosphate (pH 7.0) containing 0.2 M NaCl. Circular dichroism temperature curves were collected on a Jasco J-810 spectrapolarimeter. Measurements were carried out in a 2-mm-path-length quartz cuvette; final peptide concentration was 25 μM (9). The signal at 222 nm was recorded between 20°C and 100°C. The heating speed was 1°C/min, and every 0.1°C a data point was collected. Temperatures at the transition midpoints (melting temperature [Tm]) were estimated from first-derivative plots of the curves.
RESULTS
Generation of maC46-resistant virus strains.
The retroviral vector M87o expresses the membrane-anchored CFI peptide maC46 (46 amino acids) as a fusion protein, with an N-terminal signal peptide to mediate translocation into the endoplasmic reticulum, followed by the C46 sequence, a flexible linker derived from human immunoglobulin G2 (IgG2) and the membrane anchor from human CD34 (Fig. 1). To generate drug-resistant virus strains against low-molecular-weight antiviral agents, the virus is generally passaged in the presence of increasing concentrations of the drug, starting with a suboptimal dose. For the generation of a maC46-resistant virus strain, we had to mimic this strategy as the vector M87o, which expresses high levels of maC46 (>105 maC46 surface peptides per cell [15]), suppresses viral replication completely (10, 19). Therefore, PM-1 T-helper cell lines were transduced with retroviral vectors that contain less efficient cis elements and support lower levels of maC46 expression (see Fig. S1 in the supplemental material). The lowest-expressing PM-1 line had approximately 103 maC46 surface peptides per cell (PM/C46a cells). Passaging was initiated with the CXCR4-tropic HIV-1 virus strain NL4-3. In addition, the CCR5-tropic virus BaL_V_K, a virus that had been selected for resistance to the shorter membrane-anchored C-peptide maC36, was used (19). Both virus strains could be passaged on cells with up to 7.8 × 103 maC46 molecules per cell (PM/C46b cells). However, despite repeated passaging at this level, no NL4-3 strain could be generated that productively replicated on cell lines expressing higher maC46 levels. In addition, the envelope of this virus was cloned and showed no mutations relative to the wild-type NL4-3 Env sequence, and the sensitivity of the thus passaged NL4-3 strain to maC46 was not altered in a single-round infection dose-response assay (data not shown).
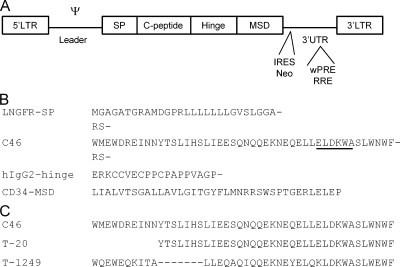
FIG. 1.
Design of the retroviral vectors expressing membrane-anchored C peptide (maC46) and C-peptide sequences. (A) The retroviral vectors are flanked by long terminal repeats (LTR) and encode the HIV entry inhibitor maC46 consisting of the following modules: a signal peptide (SP), the antiviral C-peptide maC46, a hinge/linker domain, and a membrane-spanning domain (MSD). The leader region contains the packaging signal (Ψ). Some vectors coexpress a neomycin resistance gene (Neo) via a polio internal ribosome entry site (IRES) as a marker, while others carry RNA elements like the Rev-responsive element (RRE) or the woodchuck hepatitis posttranscriptional regulatory element (wPRE) in the 3′ untranslated region (3′ UTR). (B) Amino acid sequence of the transgene. The maC46 transgene consists of the N-terminal signal peptide of the low-affinity nerve growth factor receptor (LNGFR-SP), the inhibitory C peptide (C46), the linker domain of the human IgG2 (hIgG2), and the membrane-spanning domain (MSD) of the human CD34. The epitope of the human monoclonal antibody 2F5 is underlined within the C peptide. Restriction sites flanking the C peptide encode the amino acids RS. (C) The amino acid sequences of the soluble C peptides C46, T-20, and T-1249 (34).
In contrast, the BaL_V_K strain could be passaged at higher levels of cell surface maC46 and replicated on cells expressing 3 × 104 surface maC46 peptides per cell (PM/C46c cells). This selected virus was called BaL_C46 (Table 2).
TABLE 2.
Schematic overview of the selection process for the BaL_V_K-strain
| Passage no. | Cell line | C-peptide expression (no. of ABS/cell) | Selection time (day no.)
|
|
|---|---|---|---|---|
| Start of selection round | End of selection round | |||
| 1 | PM/C46a | 800 | 0 | 49 |
| 2 | PM/C46a | 800 | 49 | 61 |
| 3 | PM/C46b | 7,800 | 61 | 92 |
| 4 | PM/C46c | 32,000 | 92 | 136 |
| 5 | PM/C46c | 32,000 | 136 | 159 |
| 6 | PM/C46c | 32,000 | 159 | 196 |
Susceptibility of BaL_C46 to maC46.
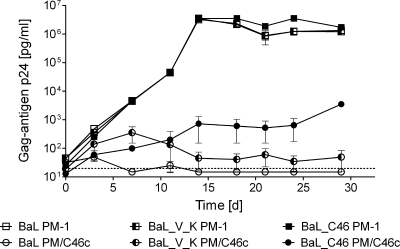
Next, the sensitivity of the selected virus (BaL_C46) to maC46 was compared to the wild-type BaL and the maC36-resistant BaL_V_K. PM-1 and PM/C46c cells, the latter expressing approximately 3 × 104 surface maC46 peptides per cell, were infected with wild-type BaL, BaL_V_K, and BaL_C46. The infectious doses were chosen to produce similar replication kinetics on native PM-1 cells (control) for all three virus strains (Fig. 2). On the maC46-expressing cell line PM/C46c, the replication of the wild-type BaL virus was completely suppressed. In the presence of maC46, BaL_V_K retained a residual replication capacity (p24 concentration of ∼50 pg/ml), which was slightly above the detection limit. The virus BaL_C46 replicated at 10-fold higher levels (p24 concentration of ∼600 pg/ml) than BaL_V_K on PM/C46c cells (Fig. 2). However, replication of BaL_C46 was still suppressed by around 3 orders of magnitude in PM/C46c cells in comparison to the replication on wild-type PM-1 cells. Interestingly, at similar levels of p24 production, BaL_C46 was much more cytopathic than BaL_V_K or BaL, showing strong syncytia formation, while BaL_V_K infection produced only few syncytia (see Fig. S2 in the supplemental material).
FIG. 2.
The selected virus isolate BaL_C46 has a reduced sensitivity to maC46. Wild-type PM-1 cells and PM-1 cells expressing maC46 (PM/C46c) were infected with the HIV variants BaL, BaL_V_K, or BaL_C46. Error bars represent the standard deviations of triplicates. To follow the course of virus replication, the p24 concentration in the supernatant was measured by enzyme-linked immunosorbent assay. d, days.
Mapping of resistance mutations.
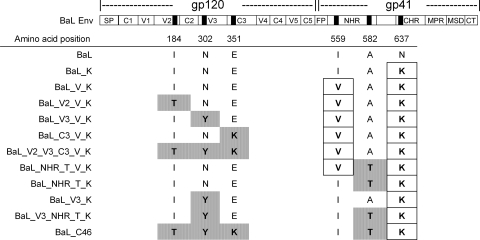
Viral RNA was isolated from cell culture supernatants, and the complete HIV envelope genes were amplified by reverse transcription-PCR. The bulk PCR products of BaL, BaL_V_K, and BaL_C46 Env were sequenced and compared (Fig. 3). Relative to wild-type BaL, BaL_V_K only had the two published mutations in the gp41 transmembrane protein (19): I559V in the NHR and N637K in the CHR. No mutations in gp120 were found. A more complex pattern of mutations was found in the Env of BaL_C46. Here, the V559 in NHR had reverted back to the wild-type amino acid configuration, I559, present in the BaL strain. Another exchange was found further downstream in the NHR of BaL_C46: alanine at position 582 was mutated to threonine (A582T). The N637K mutation in the CHR, already present in BaL_V_K, was also found in BaL_C46. Finally, three additional mutations had emerged in gp120 of BaL_C46: I184T in gp120 V2, N302Y in gp120 V3, and E351K in gp120 C3 (Fig. 3).
FIG. 3.
The location of the mutations in the BaL envelope gene selected with maC36 and maC46. Black bars indicate the positions of mutations that were identified after maC36 selection (boxed) (19) and maC46 selection (gray shading). The amino acid numbering refers to gp160 of the HIV-1 HXB2 strain. The envelope protein has been subdivided into specific regions. In the gp120 subunit, the signal peptide (SP) and constant (C1 to C5) and variable (V1 to V5) regions are marked, while in the gp41 subunit the fusion peptide (FP), NHR, CHR, membrane proximal region (MPR), membrane-spanning domain (MSD), and cytoplasmatic tail (CT) are highlighted.
Comparison with 406 reference sequences of the HIV-1 B subtype revealed that the mutations found in the BaL_C46 Env affected highly conserved amino acid positions; i.e., the mutant amino acids are very rarely present at the respective positions in the reference strains. An exception was the reversion of the rare valine found in the BaL_V_K strain (19) to the more common isoleucine at position 559 (V559I) in BaL_C46, the latter corresponding to the amino acid configuration of wild-type BaL and most other HIV-1 clade B strains (see Table S1 in the supplemental material).
Mutations in the glycoprotein are responsible for the maC46 resistance.
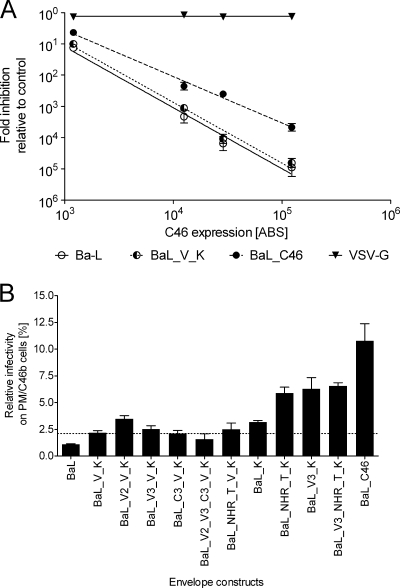
To elucidate the contribution of the identified mutations in Env to maC46 resistance, the envelope genes of BaL, BaL_V_K, and BaL_C46 were cloned into expression plasmids and were used to package replication-incompetent lentiviral vectors. These vectors were then used in a single-round infection assay on PM-1 cells expressing different concentrations of maC46 (Fig. 4A). This dose-response assay has been described by us in detail elsewhere (15). BaL_C46 Env-mediated virus entry was approximately 10-fold less susceptible to maC46 inhibition than BaL wild-type Env-mediated entry. The BaL_V_K Env conferred a slightly reduced susceptibility to maC46 relative to the BaL Env. These results indicate that mutations in BaL_C46 Env are responsible for the maC46 resistance of BaL_C46.
FIG. 4.
Mutations in Env determine maC46 resistance phenotype. Replication-incompetent lentiviral vectors pseudotyped with different envelope glycoproteins were titrated on native PM-1 cells or PM-1 cells expressing maC46. GFP expression in the target cells was determined by FACS 5 days after transduction. Titers achieved on maC46-expressing cells were normalized relative to the titer on native PM-1 cells. Error bars represent the standard deviation of triplicates. (A) PM-1 cells were transduced with the retroviral vectors C46a, C46b1, C46c, and M87o to generate cell lines with different maC46 expression levels. The surface expression of maC46 was quantified by FACS (15) and is given as the number of ABS per cell. Lentiviral vectors were pseudotyped with the envelope glycoproteins of BaL, BaL_V_K, BaL_C46, and vesicular stomatitis virus G protein (control). For the respective vector, the reciprocal of the normalized titers was defined as the relative inhibition of each maC46 cell line. (B) The lentiviral vectors were pseudotyped with the indicated mutant envelope glycoproteins and were used for a single-round infection assay. The dotted line marks the transduction efficacy reached with the BaL_V_K Env and serves as an optical orientation.
maC46 resistance is determined by mutations in gp120 and gp41.
The individual contribution of each amino acid mutation to the maC46 resistance phenotype of BaL_C46 was determined. Different combinations of mutations were introduced into the Env derived from the parental BaL_V_K virus isolate, and the resulting Env genes were cloned into an expression plasmid (Fig. 3). Replication-incompetent lentiviral vectors were then pseudotyped with these Env mutants, and single-round infection assays on maC46-expressing cells or control cells were performed to determine their susceptibility to maC46 inhibition (Fig. 4B).
The N637K mutation in the gp41 CHR, already present in the initial BaL_V_K isolate used for selection in this study, reduced susceptibility to maC46 slightly relative to the wild-type BaL strain (as described previously by Lohrengel et al. [19]) (Fig. 4B, compare BaL and BaL_K). In contrast, the second mutation in the BaL_V_K isolate, I559V (in gp41 NHR), obviously did not contribute to maC46 resistance and was found reverted to the wild-type isoleucine at this position in BaL_C46 (compare constructs BaL_K and BaL_V_K and BaL). The N637K mutation (CHR) in BaL_K was then combined either with the gp41 NHR mutation A582T (NHR_T) or the gp120 V3 mutation N302Y. In each case, susceptibility to maC46 was further decreased, indicating that mutations in gp41 NHR and gp120 both contribute to maC46 resistance (compare construct pairs BaL_K/BaL_NHR_T_K and BaL_K/BaL_V3_K). However, combining the two mutations A582T (NHR_T) and N302Y (V3) with the N637K (CHR) mutation did not further enhance maC46 resistance (compare constructs BaL_NHR_T_K, BaL_V3_K, and BaL_V3_NHR_T_K). Finally, the triple-mutant (BaL_V3_NHR_T_K) was found to be still less resistant to maC46 than the selected BaL_C46. This finding shows that the additional two gp120 mutations (in V2 and C3) are required to achieve the full resistance phenotype of BaL_C46.
Interestingly, the reversion of valine to isoleucine at position 559 was a prerequisite for all other resistance mutations to take effect. The construct pairs BaL_NHR_T_V_K/BaL_NHR_T_K and BaL_V3_V_K/BaL_V3_K each differ only at amino acid 559 (mutant V or wild-type I). However, only the constructs that have an isoleucine at position 559 exhibit significant maC46 resistance while a valine at position 559 seems to counteract the other maC46 resistance mutations. Taken together, the resistance phenotype of BaL_C46 is caused by a combination of mutations in gp120 (as in constructs BaL_V3_K and BaL_C46) and mutations in gp41 (as in construct BaL_NHR_T_K).
6HB stability.
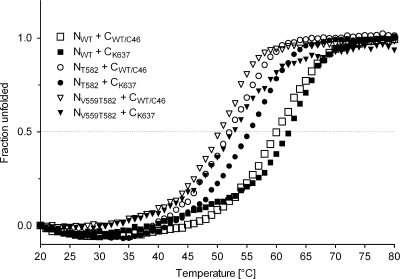
Resistance to CFI is frequently associated with mutations in the gp41 NHR that reduce the binding affinity of the antiviral CFI peptides to their target domains (6, 25, 32). Other resistance mutations are found in the CHR, where they increase the affinity of the CHR to the NHR, thereby outcompeting the binding of the CFI (3, 25). To analyze the effect of amino acid substitutions in the NHR and CHR on 6HB stability in vitro, mixtures of synthetic N and C peptides (Table 1) were used for thermal denaturing studies (Fig. 5). The peptides corresponded to the mutated NHR and CHR of BaL_C46, i.e., NT582 and CK637, respectively, as well as the wild-type NHR and CHR, NWT and CWT/C46, respectively. Of note, maC46 contains the wild-type C46 sequence.
FIG. 5.
Mutations in gp41 disrupt the maC46 binding site. Thermal denaturing studies of equimolar mixtures of the N and C peptides were performed by circular dichroism measurements at 222 nm as a function of temperature. The dotted line serves as an optical orientation to ease the comparison of the melting temperatures.
NWT and CWT/C46 form highly stable complexes (Tm = 60.5°C). This observation is consistent with the strong inhibitory activity of maC46 against wild-type BaL. The resistance mutations in gp41 of BaL_C46 have two effects. First, the mutation A582T in the NHR disrupts the C46 binding domain, leading to a considerable decrease in stability of the complex NT582/CWT/C46 (Tm = 52.5°C) relative to NWT/CWT/C46. Second, this loss of stability is partially alleviated by the additional mutation in the CHR, and the complex of NT582/CK637 is again more stable (Tm = 56.5°C) than the NT582/CWT/C46 complex. Thus, the mutated NHR in BaL_C46 has a higher affinity to the mutated CHR in the BaL_C46 gp41 than to the wild-type sequence found in maC46 (3, 25). This combination of both mutations in the BaL_C46 gp41, A582T (NHR) and N637K (CHR), has created a gp41 protein that can discriminate between the viral CHR and the inhibitory peptide in maC46, leading to a reduced susceptibility to maC46. However, this resistance phenotype develops at the cost of a reduced stability of the mutated 6HB for the BaL_C46 gp41 relative to wild-type BaL gp41.
As we observed that a valine at position 559 in the NHR hindered the manifestation of the resistance phenotype (Fig. 4), we analyzed the influence of V559 on 6HB stability. Only low-stability complexes formed after combining the peptide NV559T582 with CWT/C46 (Tm= 50.5°C) or CK637 (Tm = 52.5°C), indicating that V559 considerably hampers 6HB formation. This may be the reason why this valine was reverted to an isoleucine in Bal_C46.
Entry kinetics.
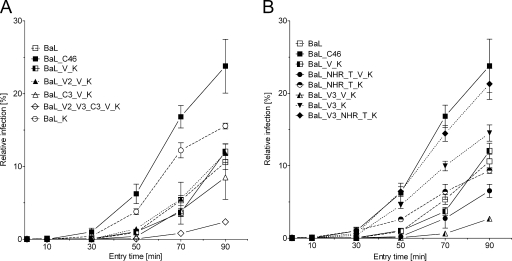
During the entry process, the inhibitory effect of C peptides is restricted to a relatively small window of opportunity after the virus has bound to its cellular receptors. Only in this phase are CFI able to compete with the natural ligand (CHR) for binding to the NHR and thus inhibit fusion (22, 23, 36). Accordingly, several studies have shown that an accelerated viral entry is associated with a reduced susceptibility to CFI (13, 28, 30, 31, 37). Therefore, we analyzed whether the mutations found in BaL_C46 Env affect entry kinetics.
PM-1 target cells were incubated with an eGFP encoding a lentiviral vector pseudotyped with the different envelope variants. The entry process was stopped with excess concentrations of T-20 at different time points, and the level of completed virus entry was quantified by FACS analysis of eGFP expression (Fig. 6). Relative to wild-type BaL and BaL_V_K, entry of BaL_C46 was considerably accelerated. It is interesting that the entry kinetics correlated with the observed degree of maC46 resistance for the different Env constructs (see Fig. S3 in the supplemental material). BaL_C46 was most rapid to enter the cell and was also the most resistant Env variant (Fig. 4). The constructs BaL_NHR_T_K, BaL_V3_K, BaL_V3_NHR_T_K, which showed a less pronounced acceleration of entry kinetics (especially during the early time points at 30 and 50 min), also had an intermediate level of resistance. Of note, these constructs all carry the wild-type isoleucine residue at position 559 in the NHR. All other constructs, which have a valine residue in the same position, showed slow virus entry and were highly susceptible to maC46 (BaL_V3_V_K and BaL_V2_V3_C3_V_K). The entry kinetics of these constructs was especially slow during early time points. In summary, the accelerated entry kinetics of BaL_C46 seemed to be a major cause for the observed maC46 resistance. Combinations of mutations in gp120 (especially N302Y in V3) and in gp41 of BaL_C46 were responsible for the enhancement of entry kinetics, and again the reversion V559I is the basis for all other mutations to show effect.
FIG. 6.
The mutations of BaL_C46 Env enhance entry kinetics. PM-1 target cells were challenged with replication-incompetent lentiviral vectors. The viral particles were pseudotyped with the indicated Env constructs and encoded an eGFP marker gene. The viral entry process was stopped with fully inhibitory concentrations of enfuvirtide/T-20 at the indicated time points. The amount of completed virus entry was determined by FACS analysis of eGFP expression 5 days later. The level of infection is given relative to the level achieved without addition of T-20. The results of a single representative experiment have been divided on two panels for better clarity. Error bars represent the standard deviations of triplicates.
Sensitivity toward coreceptor antagonists.
Several studies have found that coreceptor usage can influence the sensitivity to CFI although others have not seen such a connection (12). In the present study, the coreceptor usage was not altered in the selected virus strains. With regard to the BaL wild-type, entry mediated by the BaL_V_K and BaL_C46 envelopes was dependent on the presence of CCR5. The coreceptor CXCR4 did not support entry for these envelope variants although it is also expressed on the PM-1 cell line used for selection (see Fig. S4 in the supplemental material).
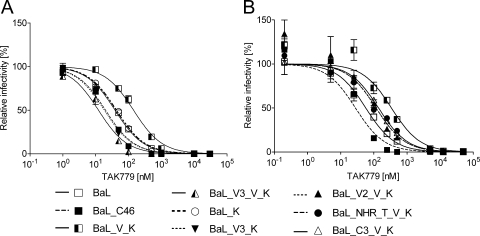
Interestingly, sensitivity to TAK-779, a CCR5 antagonist (2), varied markedly among the BaL envelope variants as tested in a single-round infection assay (Fig. 7). Enhanced affinity of gp120 to the coreceptor has been found to reduce sensitivity to TAK-779. In turn, several studies found that an enhanced affinity to CCR5 coincides with a reduced sensitivity to CFI (28, 30) although others could not confirm such a correlation (31). The increase of coreceptor affinity is proposed to accelerate the entry process, thus reducing the time span during which CFI can bind (30). Relative to wild-type BaL, sensitivity of BaL_V_K to TAK-779 inhibition was markedly reduced, which is usually considered as a sign of increased coreceptor affinity (Fig. 7). Such an increase in coreceptor affinity, in turn, would have been predicted to accelerate entry. However, the BaL_V_K and wild-type BaL had similar entry kinetics (Fig. 5). In contrast, the sensitivity of BaL_C46 to TAK-779 was even slightly higher than observed for BaL, which is indicative of a reduced coreceptor affinity, although entry kinetics of BaL_C46 were strongly accelerated. Analysis of the single amino acid mutations showed that V559 is primarily responsible for the low sensitivity of BaL_V_K toward TAK-779 inhibition. Reversion of V559 to isoleucine increased the sensitivity to levels of wild-type BaL (compare constructs BaL_V_K and BaL_K).
FIG. 7.
The selected maC46-resistant envelope has an increased sensitivity to the coreceptor antagonist TAK-779. U87.CD4.CCR5 cells were challenged with replication-incompetent lentiviral vectors. The viral particles were pseudotyped with the indicated Env constructs and encoded a luciferase marker gene. The single-round infection assay was carried out in the presence of increasing concentrations of TAK-779. The transduction efficacy was determined by luciferase activity 3 days later. Nonlinear regression (variable slope) was performed with GraphPad Prism (version 5). Top and bottom constraints were set to equal 100% and 0%, respectively. The results of two representative experiments have been divided on two panels for better clarity. Error bars represent the standard deviations of triplicates.
The higher TAK-779 sensitivity of BaL_C46 relative to wild-type BaL is primarily caused by the N302Y mutation in V3. All constructs that carry this mutation show TAK-779 sensitivities similar to BaL_C46 (compare constructs BaL_V3_V_K and BaL_V3_K) (Fig. 7). The effect of the other mutations was less pronounced. In conclusion, previous studies showed that an enhanced sensitivity to TAK-779 is associated with a reduced coreceptor binding affinity, slower viral entry, and higher sensitivity to CFI. In contrast, we found that a mutation in the V3 loop that enhanced sensitivity to TAK-779, which is indicative of a reduced coreceptor affinity, accelerated the fusion process and reduced the sensitivity to maC46. Thus, the effect of the V3 loop mutations on the sensitivity to CFI does not strictly correlate with the effect on coreceptor affinity (31).
Susceptibility to soluble C-peptide fusion inhibitors.
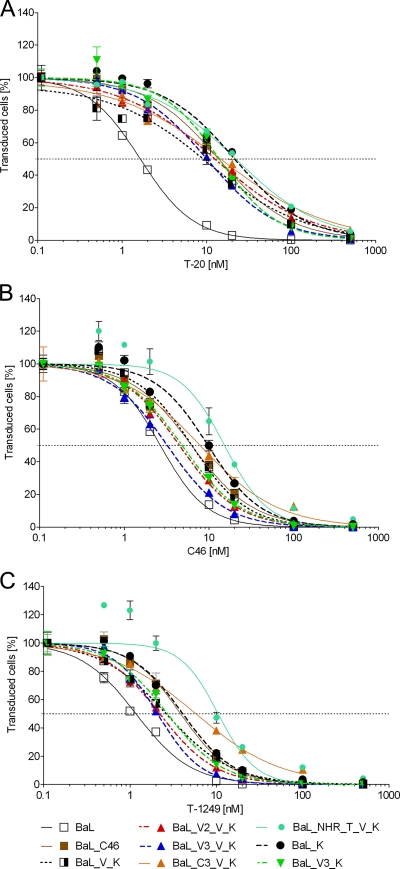
Although variations in the V3 region of gp120 that alter coreceptor affinity have been found to influence baseline susceptibility to CFI (7), gp120 is generally not analyzed in fusion inhibitor-resistant HIV-1 strains. In addition, the A582T (NHR) is not a mutation generally found to be associated with resistance to CFI. We therefore determined whether the maC46 resistance mutations also conferred a reduced sensitivity to the soluble CFI T-20 (enfuvirtide), T-1249, and soluble C46 (sequences are given in Fig. 1C) in a single-round infection assay (Fig. 8). Relative to enfuvirtide, the peptides T-1249 and C46 are elongated at the N terminus, interact with a conserved groove at the C terminus of the NHR coiled-coil, and thus have been found to inhibit enfuvirtide-resistant HIV strains.
FIG. 8.
Selected resistance mutations to maC46 reduce the susceptibility to soluble CFI. PM-1 cells were challenged with replication-incompetent lentiviral vectors. The viral particles were pseudotyped with the indicated Env constructs and encoded an eGFP marker gene. The amino acid substitutions within the BaL Env mutants are given in Fig. 3. The single-round infection assay was carried out in the presence of increasing concentrations of T-20 (A), C46 (B), or T-1249 (C) (Fig. 1). Five days later, eGFP expression was determined by FACS. Error bars represent the standard deviations of triplicates. Nonlinear regression (variable slope) was performed with GraphPad Prism (version 5). Top and bottom constraints were set to equal 100% and 0%, respectively.
All Env variants that carried the mutation N637K in the CHR showed an approximately 10-fold reduced sensitivity to T-20 compared to wild-type BaL. The other mutations in the BaL_C46 envelope did not seem to affect enfuvirtide sensitivity significantly. For T-1249 and C46 the data were nonhomogeneous. Wild-type BaL showed the highest degree of susceptibility while the level of resistance was comparable for BaL_V_K and BaL_C46. In contrast to the resistance against the membrane-anchored C peptide, the resistance toward the soluble C46 peptide and T-1249 did not correlate with the entry kinetics of the tested constructs (see Fig. S3 in the supplemental material). For soluble C peptides, mutations in the target site (NHR), which reduce affinity, seem to be more important than mutations which accelerate entry kinetics (Fig. 5). Interestingly, the strongest resistance against soluble C46 and T-1249 was observed for the construct BaL_NHR_T_V_K, which showed only a small loss of susceptibility to maC46. Thus, determinants of resistance differ between soluble and membrane-anchored CFI.
DISCUSSION
In a previous study, the membrane-anchored C peptide, maC46, was found to efficiently inhibit HIV-1 entry. maC46 was also fully active against HIV strains resistant to the shorter C peptide T-20/C36 (enfuvirtide) (10). After prolonged passaging of a T-20-resistant virus (BaL_V_K), we succeeded in selecting an HIV strain, BaL_C46, with a 10-fold reduced susceptibility to maC46. Compared to BaL, BaL_C46 had five point mutations located in gp120 and in gp41. The mutations with the highest impact on maC46 resistance were N302Y (gp120 V3), A582T (gp41 NHR), V559I (the revertant to wt BaL sequence, gp41 NHR) and N637K (gp41 CHR; already present in the parental virus BaL_V_K).
Two types of resistance mechanisms to C-peptides have been described: first, changes that alter the C-peptide binding site (direct effects) and, second, alterations that influence the kinetics of 6HB formation (indirect effects). Alterations in the C-peptide binding site located in the NHR of gp41 have been found during in vitro selection and/or in clinical trials with the soluble C-peptides T-20, C34, and T-1249 (21, 25, 32, 40). These mutations are found primarily at amino acid positions 544 to 553 around the GIV motif in the NHR and have been shown to reduce the affinity of the C peptides to the NHR (25, 32, 39).
In the present study, we have identified a new resistance mutation (A582T) in the NHR. Thermal denaturing studies using synthetic C peptides showed that this amino acid substitution considerably decreases the stability of the 6HB with the wild-type C46 sequence found in maC46 although the mutation in A582T in the NHR does not make direct contact with the CHR, according to the published crystal structure (5). The mutation N637K found in the CHR of BaL_C46, which has also been found by others in CFI-resistant viruses (3, 25), partially restored 6HB stability. The combination of NHR (A582T) and CHR (N637K) is thus expected to form a more stable 6HB during viral entry than NHR (A582) and maC46. As a result, at the expense of 6HB stability, NHR and CHR in BaL_C46 were mutated to escape inhibition by CFI by favoring 6HB formation with the mutated viral CHR over the complex containing the CFI maC46.
This effect may be even more pronounced in vivo as the N673K mutation deletes a potential N-glycosylation site in the CHR, which we found to be glycosylated in maC46 (data not shown). The lack of glycosylation in the CHR (N637K) may even further enhance the binding affinity between the two gp41 heptad repeats and thus stabilize the 6HB. In accordance with such an effect, we previously found that the antiviral activity of recombinant soluble C peptides, produced in cell culture and thus glycosylated on N637, increased up to sevenfold after deglycosylation in vitro (8).
However, the mutations within gp41 that influenced 6HB stability were not found to be sufficient to cause the full maC46 resistance phenotype observed in BaL_C46. In this study, we found that three additional amino acid substitutions in gp120 (especially in V3) were necessary to achieve the full extent of resistance observed. These gp120 mutations accelerate entry kinetics, thereby presumably evading inhibition by maC46 (Fig. 5; see also Fig. S3 in the supplemental material). An accelerated entry is known to reduce the sensitivity of HIV to C-peptide inhibition as the time of target site exposure is minimized (1, 30). The margins of this kinetic window are governed by the speed at which Env undergoes its conformational changes and passes through the different transitional states until the entry process is completed. It is noteworthy that the time frame during which maC46 can inhibit the entry process is shorter than for soluble C peptides. While soluble peptides can already access the target region within gp41 after binding of Env to CD4, membrane-anchored C peptides are active only after the subsequent coreceptor engagement (22). Therefore, accelerated entry is expected to have a greater impact on inhibition by maC peptides than on the antiviral activity of soluble CFI.
The overall kinetics of 6HB formation can be accelerated in a direct manner by increasing the affinity of the heptad repeats for one another. We could show that this is the primary mode of action for the mutation N637K in the CHR as discussed above. However, in combination with the A582T mutation in the NHR, the accelerating effect of N637K on entry kinetics is almost completely abolished. Thus, although the combination of A582T and N637K reduces sensitivity to maC46 by disfavoring the incorporation of maC46 into the 6HB, the mutated 6HB is less stable than the wild-type form and thus slows down viral entry. This slowdown is compensated by the mutations in gp120 found in BaL_C46.
The acceleration of entry kinetics in BaL_C46 depended primarily on the mutation in the V3 region of gp120 in conjunction with the mutated gp41. The mutation N302Y in V3 probably influences entry kinetics in an indirect manner. The V3 domain is associated with coreceptor affinity and usage (16, 30). The mutation N302Y caused a reduction in coreceptor affinity (compare constructs BaL_NHR_T_K and BaL_V3_NHR_T_K) (Fig. 5). Therefore, it seems that the N302Y mutation may destabilize coreceptor binding, thereby enhancing the progression through subsequent transition states during entry. Similarly, Reeves et al. (31) have identified a mutation (P438A) in the coreceptor binding site of gp120 that causes a strong increase in TAK-779 sensitivity (i.e., reduced coreceptor affinity) while supporting rapid entry kinetics. In this case, the mutations were found to destabilize the Env-coreceptor complex after CD4 engagement (31). Interestingly, the N302Y (N300Y in JR-CSF) mutation in V3 has been described previously by Platt and coworkers (29) in an HIV-1JR-CSF strain selected for the ability to use the low-affinity coreceptor CCR5(Y14N). This virus showed reduced sensitivity to T-20. These authors also argued that the effect was most likely due to an acceleration of viral entry (29).
A striking observation in this study was that the mutations N302Y and A582T increased maC46 resistance and entry kinetics only when combined with the revertant V559I in NHR. In a previous study, V559 was found to increase resistance to T-20 and maC36 but did not contribute to maC46 resistance. Instead, it was reported to reduce viral fitness (19). According to the 6HB crystal structure, the amino acid at position 559 of each NHR faces toward the center of the inner coiled-coil and is in very tight contact with the residues at position 559 of the neighboring two NHR helices (5). A valine at this position is extremely rare (see Table S1 in the supplemental material). It can be assumed that valine destabilizes the central coiled-coil due to steric hindrance. Such hindrance probably hampers 6HB formation and leads to the generally observed low entry kinetics of constructs that carry V559 and the previously observed reduced viral fitness of the mutated virus. Furthermore, the central coiled-coil formed by the NHR helices is responsible for the trimerization of Env. Mutations in this region have been reported to influence coreceptor binding and infectivity of HIV (27). This conclusion is in agreement with our finding that the V559 massively increases coreceptor affinity in BaL_V_K.
Surprisingly, when Env constructs were analyzed for resistance against soluble C peptides, no correlation between entry kinetics and C-peptide resistance could be found. The requirement of gp120 mutations for maC46 resistance, which was not observed for resistance to soluble CFI, could be explained by the different modes of binding to the target site for soluble and membrane-bound C peptides. Peptides are not rigid structures but are better represented as a manifold of low-energy conformations and conformational substrates (energy landscape) (4, 20, 26). The binding of a flexible soluble peptide to its target is governed purely by random molecular diffusion, and it can therefore be assumed that upon its first encounter with the target region, only a small segment of each peptide will be aligned correctly. After this first interaction, binding will progress in discrete steps during which each section of the peptide has to be properly adjusted relative to the target site until perfect association has been achieved. Therefore, even minor alterations within the target region, which hamper the initial contact, will lead to the complete abrogation of binding. On the other hand, due to its membrane-bound presentation, maC46 may depend less on random association processes as it is already aligned correctly relative to the target structure with fewer degrees of freedom and has therefore a more favorable energy landscape to allow binding. Additionally, in contrast to soluble antiviral peptides, the membrane-bound expression of a C peptide may lead to a higher effective local concentration. Therefore, compared to soluble CFI, a mere reduction of the binding site affinity may not be sufficient to outcompete maC46 and ensure 6HB formation between the viral CHR and NHR domains.
Taken together, these results suggest that, in contrast to soluble CFI, resistance to the elongated membrane-anchored C46 does not readily develop and is relatively weak. A complex combination of mutations in gp120 and gp41 associated with different resistance mechanisms must be acquired by HIV-1 to evade entry inhibition by maC46.
Supplementary Material
Acknowledgments
We thank Tefik Merovci and Patricia Schult-Dietrich for excellent technical assistance.
The study was sponsored by the EC STREP project PoxGene, LSHP-CT-2005-018680, the project TreatID funded by the German Federal Ministry for Research BMBF, and the ANRS.
Footnotes
Published ahead of print on 11 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abrahamyan, L. G., S. R. Mkrtchyan, J. Binley, M. Lu, G. B. Melikyan, and F. S. Cohen. 2005. The cytoplasmic tail slows the folding of human immunodeficiency virus type 1 Env from a late prebundle configuration into the six-helix bundle. J. Virol. 79106-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 965698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, C. E., R. W. Sanders, Y. Deng, S. Jurriaans, J. M. Lange, M. Lu, and B. Berkhout. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J. Virol. 7812428-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogan, A. A., and K. S. Thorn. 1998. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 2801-9. [DOI] [PubMed] [Google Scholar]
- 5.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89263-273. [DOI] [PubMed] [Google Scholar]
- 6.Chinnadurai, R., D. Rajan, J. Munch, and F. Kirchhoff. 2007. Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J. Virol. 816563-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 748358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dervillez, X., A. Huther, J. Schuhmacher, C. Griesinger, J. H. Cohen, D. von Laer, and U. Dietrich. 2006. Stable expression of soluble therapeutic peptides in eukaryotic cells by multimerisation: application to the HIV-1 fusion inhibitory peptide C46. Chem. Med. Chem. 1330-339. [DOI] [PubMed] [Google Scholar]
- 9.Desmezieres, E., N. Gupta, R. Vassell, Y. He, K. Peden, L. Sirota, Z. Yang, P. Wingfield, and C. D. Weiss. 2005. Human immunodeficiency virus (HIV) gp41 escape mutants: cross-resistance to peptide inhibitors of HIV fusion and altered receptor activation of gp120. J. Virol. 794774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egelhofer, M., G. Brandenburg, H. Martinius, P. Schult-Dietrich, G. Melikyan, R. Kunert, C. Baum, I. Choi, A. Alexandrov, and D. von Laer. 2004. Inhibition of human immunodeficiency virus type 1 entry in cells expressing gp41-derived peptides. J. Virol. 78568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giroglou, T., J. Cinatl, Jr., H. Rabenau, C. Drosten, H. Schwalbe, H. W. Doerr, and D. von Laer. 2004. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J. Virol. 789007-9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg, M., N. Cammack, M. Salgo, and L. Smiley. 2004. HIV fusion and its inhibition in antiretroviral therapy. Rev. Med. Virol. 14321-337. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg, M. L., and N. Cammack. 2004. Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 54333-340. [DOI] [PubMed] [Google Scholar]
- 14.Heil, M. L., J. M. Decker, J. N. Sfakianos, G. M. Shaw, E. Hunter, and C. A. Derdeyn. 2004. Determinants of human immunodeficiency virus type 1 baseline susceptibility to the fusion inhibitors enfuvirtide and T-649 reside outside the peptide interaction site. J. Virol. 787582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann, F. G., H. Martinius, M. Egelhofer, T. Giroglou, T. Tonn, S. D. Roth, R. Zahn, P. Schult-Dietrich, A. Alexandrov, U. Dietrich, C. Baum, and D. von Laer. 6 February 2009. Protein scaffold and expression level determine antiviral activity of membrane-anchored antiviral peptides. Hum. Gene Ther. doi: 10.1089/hum.2006.158. [DOI] [PubMed]
- 16.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 25371-74. [DOI] [PubMed] [Google Scholar]
- 17.Lee, B., M. Sharron, L. J. Montaner, D. Weissman, and R. W. Doms. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 965215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, S., W. Jing, B. Cheung, H. Lu, J. Sun, X. Yan, J. Niu, J. Farmar, S. Wu, and S. Jiang. 2007. HIV gp41 C-terminal heptad repeat contains multifunctional domains. Relation to mechanisms of action of anti-HIV peptides. J. Biol. Chem. 2829612-9620. [DOI] [PubMed] [Google Scholar]
- 19.Lohrengel, S., F. Hermann, I. Hagmann, H. Oberwinkler, L. Scrivano, C. Hoffmann, D. von Laer, and M. T. Dittmar. 2005. Determinants of human immunodeficiency virus type 1 resistance to membrane-anchored gp41-derived peptides. J. Virol. 7910237-10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCammon, J. A. 1998. Theory of biomolecular recognition. Curr. Opin. Struct. Biol. 8245-249. [DOI] [PubMed] [Google Scholar]
- 21.Melby, T., R. Demasi, N. Cammack, G. D. Miralles, and M. L. Greenberg. 2007. Evolution of genotypic and phenotypic resistance during chronic treatment with the fusion inhibitor T-1249. AIDS Res. Hum. Retrovir. 231366-1373. [DOI] [PubMed] [Google Scholar]
- 22.Melikyan, G. B., M. Egelhofer, and D. von Laer. 2006. Membrane-anchored inhibitory peptides capture human immunodeficiency virus type 1 gp41 conformations that engage the target membrane prior to fusion. J. Virol. 803249-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naldini, L., U. Blömer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272263-267. [DOI] [PubMed] [Google Scholar]
- 25.Nameki, D., E. Kodama, M. Ikeuchi, N. Mabuchi, A. Otaka, H. Tamamura, M. Ohno, N. Fujii, and M. Matsuoka. 2005. Mutations conferring resistance to human immunodeficiency virus type 1 fusion inhibitors are restricted by gp41 and Rev-responsive element functions. J. Virol. 79764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nevo, R., V. Brumfeld, R. Kapon, P. Hinterdorfer, and Z. Reich. 2005. Direct measurement of protein energy landscape roughness. EMBO Rep. 6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, E. J., and G. V. Quinnan, Jr. 1999. Both neutralization resistance and high infectivity phenotypes are caused by mutations of interacting residues in the human immunodeficiency virus type 1 gp41 leucine zipper and the gp120 receptor- and coreceptor-binding domains. J. Virol. 735707-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platt, E. J., J. P. Durnin, and D. Kabat. 2005. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 794347-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platt, E. J., D. M. Shea, P. P. Rose, and D. Kabat. 2005. Variants of human immunodeficiency virus type 1 that efficiently use CCR5 lacking the tyrosine-sulfated amino terminus have adaptive mutations in gp120, including loss of a functional N-glycan. J. Virol. 794357-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 9916249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeves, J. D., J. L. Miamidian, M. J. Biscone, F. H. Lee, N. Ahmad, T. C. Pierson, and R. W. Doms. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 785476-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schambach, A., B. Schiedlmeier, K. Kuhlcke, M. Verstegen, G. P. Margison, Z. Li, K. Kamino, J. Bohne, A. Alexandrov, F. G. Hermann, D. von Laer, and C. Baum. 2006. Towards hematopoietic stem cell-mediated protection against infection with human immunodeficiency virus. Gene Ther. 131037-1047. [DOI] [PubMed] [Google Scholar]
- 34.Schneider, S. E., B. L. Bray, C. J. Mader, P. E. Friedrich, M. W. Anderson, T. S. Taylor, N. Boshernitzan, T. E. Niemi, B. C. Fulcher, S. R. Whight, J. M. White, R. J. Greene, L. E. Stoltenberg, and M. Lichty. 2005. Development of HIV fusion inhibitors. J. Pept. Sci. 11744-753. [DOI] [PubMed] [Google Scholar]
- 35.Sista, P. R., T. Melby, D. Davison, L. Jin, S. Mosier, M. Mink, E. L. Nelson, R. DeMasi, N. Cammack, M. P. Salgo, T. J. Matthews, and M. L. Greenberg. 2004. Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. AIDS 181787-1794. [DOI] [PubMed] [Google Scholar]
- 36.Steger, H. K., and M. J. Root. 2006. Kinetic dependence to HIV-1 entry inhibition. J. Biol. Chem. 28125813-25821. [DOI] [PubMed] [Google Scholar]
- 37.Sterjovski, J., M. J. Churchill, A. Ellett, L. R. Gray, M. J. Roche, R. L. Dunfee, D. F. Purcell, N. Saksena, B. Wang, S. Sonza, S. L. Wesselingh, I. Karlsson, E. M. Fenyo, D. Gabuzda, A. L. Cunningham, and P. R. Gorry. 2007. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su, C., T. Melby, R. DeMasi, P. Ravindran, and G. Heilek-Snyder. 2006. Genotypic changes in human immunodeficiency virus type 1 envelope glycoproteins on treatment with the fusion inhibitor enfuvirtide and their influence on changes in drug susceptibility in vitro. J. Clin. Virol. 36249-257. [DOI] [PubMed] [Google Scholar]
- 39.Trivedi, V. D., S. F. Cheng, C. W. Wu, R. Karthikeyan, C. J. Chen, and D. K. Chang. 2003. The LLSGIV stretch of the N-terminal region of HIV-1 gp41 is critical for binding to a model peptide, T20. Protein Eng. 16311-317. [DOI] [PubMed] [Google Scholar]
- 40.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yee, J.-K., A. Miyanohara, P. LaPorte, K. Bouic, J. C. Burns, and T. Friedmann. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. USA 919564-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.