Abstract
Background
Polycystic ovary syndrome (PCOS), whose genetic basis is not completely well understood, is the most common endocrine disorder in women and it typically develops during adolescence. The aim of this study is to investigate the possible association between single nucleotide polymorphisms (SNPs) of FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 genes and PCOS in adolescent girls.
Methods
DNA samples from forty-four adolescent girls with PCOS and 50 healthy controls were analyzed by PCR-RFLP and direct DNA sequencing to determine the genotypic frequency of 17 different polymorphic loci on the FSHR (A307T, N680S), CYP17 (−34 T/C), CYP1A1 (T6235C), CAPN10 (44, 43, 19, 63), INSR (exon 17 C/T), SERPINE1 (4G/5G) genes. Genotyping of exon 12 (six polymorphisms) and intron 12 (one polymorphism) of INSR gene by direct DNA sequencing was performed for the first time in this study.
Results
No significant differences were observed in the genotype and allele distributions of above mentioned polymorphisms between cases and control groups.
Conclusion
Our data does not support an association between SNPs of FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 genes and susceptibility to PCOS or related traits in Turkish adolescent girls.
Keywords: Adolescent girl, Genetic polymorphisms, Polycystic ovary syndrome
Introduction
Polycystic ovary syndrome (PCOS) affects an estimated 5–10% of women of reproductive age and arises as a result of a genetically determined disorder of ovarian function at the onset of puberty [1]. The search for PCOS susceptibility genes has mainly focused on genes involved in sex hormones and regulators, insulin sensitivity, type 2 diabetes and cardiovascular disease, steroid metabolism and biosynthesis [2]. Although multiple genetic factors including mutations and polymorphisms to several genes have been associated with PCOS risk [3], the inheritance mode and the molecular genetic mechanisms underlying PCOS risk are not fully understood [4].
PCOS is characterized by endocrinological abnormalities; therefore, polymorphisms in genes encoding sex hormones or regulators of their activity have been investigated [5]. The follicle stimulating hormone receptor (FSHR) gene contains two important single nucleotide polymorphisms (SNPs) in exon 10, which are in linkage disequilibrium and change two amino acids at positions A307T and N680S. A307T, situated at the extracellular domain of FSHR, the site responsible for high affinity hormone binding [6], has been reported to affect hormone trafficking and signal transduction. Phosphorylation of the Ser and Thr residues within the intracellular regions of FSHR may influence the uncoupling from adenylyl cyclase [7]. As a result, amino acid alteration related to the corresponding SNPs might affect the post-translational modifications of the FSHR protein, hence the function of the receptor including FSH efficacy [8]. A few genetic studies have examined the association between FSHR gene polymorphisms and PCOS [9–11].
Several studies have investigated whether polymorphisms in enzymes involved in the biosynthesis and metabolism of sex steroids confer PCOS susceptibility [5]. A polymorphism has been found in the regulatory region of the 17α-hydroxylase (CYP17) gene, being a T to C substitution −34 base pairs (bp) from the translation initiation point in the 5′ promoter region of the gene that creates a new MspA1 restriction site [12]. The less common “C” allele also results in an additional Sp1-type (CCACC box) promoter site that is hypothesized to increase transcription of the gene [13] and thus lead to higher androgen levels. Although this base pair substitution is not the primary genetic defect in PCOS, it may aggravate the clinical picture of hyperandrogenemia, particularly when homozygosity exists [14]. On the other hand, in one of the previous studies, CYP17 gene was not associated with steroid hormone synthesis in PCOS [15]. Several genetic risk factors for PCOS have been studied [16, 17]. The CYP1A1 gene, located at 15q22–q24, comprises seven exons and six introns. A polymorphism in the CYP1A1 gene, which encodes the enzyme cytochrome P450 1A1, has been shown to have an association with PCOS [18, 19]. Furthermore, studies have indicated that a pentanucleotide repeat in the gene is associated with PCOS susceptibility [20, 21].
The increased risk of the type 2 diabetes mellitus (T2DM) and cardiovascular disease in women with PCOS has led to numerous association studies in genes related to these diseases [5]. The gene encoding calpain-10 (CAPN10) consists of 15 exons and it is located on chromosome 2q37.3 and encodes a ubiquitously expressed member of the calpain cysteine protease family. It was identified as a T2DM susceptibility locus by Horikawa et al [22] and has been shown to be related to proinsulin processing, insulin secretion and insulin resistance [23]. Potential associations between five different polymorphisms and PCOS have been investigated [5, 24–27]. Insulin receptor gene (INSR) comprises 22 exons spanning 120 kilobases on chromosome 19p13.3–p13.2 [28]. The region of exons 17–21 encodes the tyrosine kinase domain of the receptor, which is necessary for insulin signal transduction. C/T single nucleotide polymorphism at exon 17 in tyrosine kinase domain of INSR has been associated with PCOS most possibly by the resultant effects on the autophosphorylation of the INSR function in some women with PCOS [29–31]. On the other hand, Lee et al investigated a total of nine polymorphisms in this region [32]. Plasminogen activator inhibitor-1 (PAI-1), a member of the serine protease inhibitor (SERPINE1) family, is involved in blood coagulation. It is located on chromosome 7q21.3–q22 and has been implicated in cardiovascular disease. A common diallelic polymorphism located at −675 bases from the transcription start site of PAI-1 is the best characterized genetic determinant. The 4G/5G polymorphism in the SERPINE1 gene has been linked with PCOS susceptibility [33], but no association was found in other studies [34–36].
We aim to elucidate, for the first time in adolescent girls in Turkish population, the putative functional significance of FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 gene polymorphisms in the development of the PCOS. In addition, this study aims to perform direct DNA sequencing method in order to investigate polymorphic variants in exon 12 and intron 12 of INSR gene.
Materials and methods
Subjects
The study was conducted within Gazi University, Faculty of Medicine, Department of Medical Biology and Genetics. Cases and controls were recruited from Gazi University, Faculty of Medicine, Department of Pediatric Endocrinology between 2006 and 2008. A total of 94 unrelated Turkish adolescent girls were studied. Forty-four were patients with PCOS and fifty were non-PCOS with normal menstrual cycles without signs of clinical or biochemical hyperandrogenism. Clinical characteristics of the subjects were obtained from medical records. All cases and controls were of Caucasian origin and were matched according to age. The diagnosis of PCOS was based on the Rotterdam 2003 critera [37]. None of the subjects had been taking any medications known to affect hormone, lipid or carbohydrate metabolism for at least 3 months before entering the study. Clinical and biochemical characteristics of adolescent girls with PCOS and healthy controls are listed in Table 1. The study was approved by the Committee of Ethics of the Gazi University and written informed consents were obtained from of all participants and their parents.
Table 1.
Clinical and biochemical characteristics in adolescent girls with polycystic ovary syndrome and healthy controls
| PCOS (n = 44) | Controls (n = 50) | P | |
|---|---|---|---|
| Age (years) | 14.5 ± 1.3 | 14.0 ± 3.3 | 0.248a |
| BMI (kg/m2) | 25.0 ± 5.5 | 20.7 ± 4.2 | <0.001a |
| Ferriman-Gallwey score | 13.1 ± 4.3 | 1.3 ± 1.1 | <0.001b |
| Acanthosis nigricans (%) | 8 (18.2%) | 0 | 0.002c |
| Total testosterone (ng/mL) | 0.9 ± 0.3 | 0.5 ± 0.2 | <0.001b |
| FSH (mIU/mL) | 5.9 ± 1.8 | 4.6 ± 1.3 | <0.001b |
| LH (mIU/mL) | 12.7 ± 9.5 | 4.4 ± 2.1 | <0.001b |
| Prolactin (ng/mL) | 16.9 ± 11.9 | 13.8 ± 12.0 | <0.001b |
| TSH (μIU/mL) | 2.6 ± 1.2 | 1.6 ± 1.2 | <0.001b |
| 17-OHP (ng/dL) | 1.7 ± 1.1 | 1.5 ± 0.6 | 0.936b |
| Fasting blood glucose (mg/dL) | 94.4 ± 29.3 | 90.1 ± 6.4 | 0.452b |
| Fasting insulin (μIU/mL) | 15.2 ± 9.1 | 12.8 ± 6.5 | 0.386b |
BMI, body mass index; FSH, follicle stimulating hormone; LH, luteinizing hormone; TSH, thyroid stimulating hormone; 17-OHP, 17-OH-progestrone
aStudent’s t test., bMann Whitney U test., cFisher’s Exact test.
P values < 0.001 and P = 0.002 are shown in bold.
Biochemical and hormonal measures
After an overnight fast, venous blood samples were obtained for genetic study and for hormonal profile, glucose and insulin assays on days 3 to 5 from menstrual cycling patients. In the case of amenorrheic patients, blood samples were obtained after a progesterone-induced menstruation. Transabdominal pelvic sonography was performed on all of the subjects with PCOS. Plasma glucose was measured with hexokinase method. Plasma concentrations of total testosterone, FSH, LH, prolactin, TSH and insulin were also measured with chemiluminessence assays using an autoanalyser (Abbott Laboratories, Chicago, IL, USA). Plasma concentration of 17-OHP was analyzed using commercially available radioimmunoassay kits (Diagnostic System Laboratories, Webster, TX, USA).
Polymerase Chain Reaction- Restriction Fragment Length Polymorphism (PCR-RFLP) genotyping and DNA Sequencing
Analyzed FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 polymorphisms used to detect the base changes are shown in Table 2 (Methodical Nomenclature recommended by Human Genome Variation Society -www.hgvs.org-).
Table 2.
Polymorphisms in the FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 genes and the methods of their genotyping
| Analyzed polymorphisms | |||
|---|---|---|---|
| Gene | Common nomenclature used in paper (alleles) | Methodical nomenclature | db SNP |
| FSHR | A307T | g.997 A/G | rs6165 |
| N680S | g.2117 A/G | rs6166 | |
| CYP17 | -34 T/C promoter region | -34 T/C | rs743572 |
| CYP1A1 | T6235C | g.6235 T/C | rs4646903 |
| CAPN10 | UCSNP44 | g.4841 T/C | rs2975760 |
| UCSNP43 | g.4852 G/A | rs3792267 | |
| UCSNP19 | g.7920 ins/del32bp | rs3842570 | |
| UCSNP63 | g.16378 C/T | rs5030952 | |
| INSR | Exon17 C/T | g.3364 T/C | rs1799817 |
| SERPINE1 | 4G/5G | -675 ins/delG | rs1799889 |
Each PCR was carried out in a total volume 50 µL consisting of 2.5 µL extracted DNA, 50 pmol/µL each primer, 100 µM dNTP, 1 U/µL unit Taq DNA polymerase and 2 mM MgCl2. Primer sets, annealing temperatures and restriction enzymes used for the PCR-RFLP assay are shown in Table 3. PCR conditions, except the annealing temperatures, were set as provided in the References column in Table 3 [29, 38–45]. Product and allele sizes are also given in Table 3. The PCR and restriction enzyme products were separated by electrophoresis in a 2.0% and 4.0% agarose gel respectively, and subsequently stained with ethidium bromide. In addition, all gels were reread blindly by three persons without any change, and 15% of the analyses were randomly repeated.
Table 3.
Primers, annealing temperatures, product sizes, restriction enzymes, and allele sizes
| Gene | Polymorphism | Primer Sequence | Annealing temperature (0C) | Product size (bp) | Restriction enzyme* | Allele size | Reference |
|---|---|---|---|---|---|---|---|
| FSHR | A307T | F: 5'-CCTGCACAAAGACAGTGATG-3' | 55 | 577 | AhdI | Ala:403 + 174 | 38 |
| rs6165 | R: 5'-TGGCAAAGACAGTGAAAAAG-3' | Thr:403 + 143 + 31 | |||||
| N680S | F 5'CCCAAATTTATAGGACAG 3' | 50 | 114 | BsrI | Asn:114 | 39 | |
| rs6166 | R 5'GAGGGACAAGTATGTAAGTG 3' | Ser:86 + 28 | |||||
| CYP17 | rs743572 | F 5’-GCCCAGATACCATTCGCACT -3’ | 55 | 368 | MspA1 | A1:368 | 40 |
| R 5’-TAAGCAGCAAGAGAGCCACG-3’ | A2:305 + 63 | ||||||
| CYP1A1 | T6235C | F 5’ CAGTGAAGAGGTGTAGCCGCT-3’ | 55 | 297 | MspI | T:297 | 41 |
| rs4646903 | R 5’-AGGCAGGTGGATCACTTGAG-3’ | C:162 + 135 | |||||
| UCSNP44 | F 5’-GCAGGGCGCTCACGCTTGCCG-3’ | 60 | 166 | BstFNI (isoschizomer of AccII) | T:166 | 42 | |
| rs2975760 | R 5’-GCATGGCCCCCTCTCTGATTC-3’ | C:145 + 21 | |||||
| UCSNP43 | F 5'- GCAGGGTTGGAGCTTGAGAG -3' | 55 | 175 | FauNDI (isoschizomer of NdeI) | G:175 | 43 | |
| CAPN10 | rs3792267 | R 5'-AAGTCAAGGCTTAGCCTCACCTTCATA-3' | A:148 + 27 | ||||
| UCSNP19 | F 5'- CAGTTTGGTTCTCTTCAGCG-3' | 60 | Del:146 | - | - | 43 | |
| rs3842570 | R 5'-GCAGGGTCTAAGCAGCAGC-3' | Ins:178 | |||||
| UCSNP63 | F 5’-AAGGGGGGCCAGGGCCTGACGGGGGTGGCG-R 5’-AGCACTCCCAGCTCTCGATC-3’ | 55 | 189 | HhaI | C:159 + 30 | 44 | |
| rs5030952 | R 5’- TCAGGAAAGCCAGCCCATGTC −3’ | T:189 | |||||
| INSR | Exon17 C/T | F 5’- CCAAGGATGCTGTGTAGATAAG −3’ | 55 | 317 | PmlI | T:317 | 29 |
| rs1799817 | R 5’-CCAACAGAGGACTCTTGGTCT-3’ | C:274 + 43 | |||||
| SERPINEI | 4G/5G | F 5’-CACAGAGAGAGTCTGGCCACGT-3’ | 55 | 99 | BslI | 4G:99 | 45 |
| (-675 ins/delG) | R 5’-CCAACAGAGGACTCTTGGTCT-3’ | 5G:77 + 22 | |||||
| rs1799817 |
*Temperatures of restriction enzymes: AhdI, BsrI, MspA1, MspI, FauNDI, HhaI, PmlI at 37°C; BstFNI at 60°C; BslI at 55°C
We amplified exon 12 of INSR gene with primers (F: 5'-TGATGGTGATGGTGTCATCATA-3' and R: 5'-TGTCCTTGGTCAGCCTTGATGT-3') as proposed earlier by Seino et al [46]. The PCR products (379 bp) of INSR gene exon 12 were subjected to direct sequencing with the forward primer by using BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA). Products were analyzed by using ABI PRISM® 310 (Applied Biosystems, Foster City, CA, USA). The conditions of sequencing were the same as described by Seino et al [46], except that our annealing temperature was 60°C.
Statistical analysis
Differences between the means of two continuous variables were evaluated by Student t-test, Mann Whitney U test and Fisher’s exact test. Using the χ2 test, polymorphisms were tested for deviation from Hardy-Weinberg equilibrium (HWE). The relative association between patients and controls for genotype and allele frequencies was assessed by Pearson’s χ2 test. The corresponding odds ratios (ORs) and confidence intervals (95% CIs) were calculated with SPSS version 15.0. Multivariate unconditional logistic regression analysis with adjustment for clinical and biochemical characteristics and genotype distribution was performed to calculate adjusted ORs and 95% CIs. Strong association (significance) was assumed at P < 0.01. From P = 0.01 to 0.05, a weak but still significant association was considered.
Results
Clinical and laboratory variables
The clinical and biochemical characteristics of adolescent girls enrolled in this study are summarized in Table 1. PCOS girls presented a higher frequency of acanthosis nigricans and increased values of Ferriman-Gallwey score. Furthermore, total testosterone, FSH, LH, prolactin, TSH levels and body mass index were significantly higher in the PCOS patients when compared to controls. However, there were no significant differences between patients and controls in terms of 17-OHP, fasting blood glucose, fasting insulin levels and mean age.
Genotype and allele frequencies
The genotype and allele frequencies of the polymorphisms in the control and PCOS groups were consistent with the HWE equilibrium distribution (P > 0.05) except for INSR exon 17 C/T polymorphism (P < 0.05).
Genotype distribution of genes encoding sex hormones and hormone regulators are shown in Table 4. The genotype frequencies of the SNP A307T and N680S were not different between the cases and controls (P > 0.05).
Table 4.
Genotype distribution of genes encoding sex hormones and hormone regulators in adolescent girls with polycystic ovary syndrome and healthy controls
| Gene | Genotype | Cases (n = 44) | Controls (n = 50) | P | OR (95% CIs);P |
|---|---|---|---|---|---|
| FSHR | A307T (rs6165) | 0.804 | |||
| Thr/Thr | 16 (36%) | 16 (32%) | 1a | ||
| Ala/Thr | 19 (43%) | 25 (50%) | 0.76 (0.31-1.90); 0.556 | ||
| Ala/Ala | 9 (21%) | 9 (18%) | 1.00 (0.32-3.17); 1.000 | ||
| Ala/Thr + Ala/Ala | 28 (64%) | 34 (68%) | 0.82 (0.35-1.94); 0.656 | ||
| Allele frequency | 0.895 | ||||
| Ala | 51 (58%) | 57 (57%) | 1a | ||
| Thr | 37 (42%) | 43 (43%) | 0.96 (0.54-1.72); 0.895 | ||
| N680S (rs6166) | 0.638 | ||||
| Asn/Asn | 13 (30%) | 14 (28%) | 1a | ||
| Ser/Asn | 20 (45%) | 27 (54%) | 0.80 (0.31-2.07); 0.641 | ||
| Ser/Ser | 11 (25%) | 9 (18%) | 1.32 (0.41-4.20); 0.642 | ||
| Ser/Asn + Ser/Ser | 31 (70%) | 36 (72%) | 0.93 (0.38-2.27); 0.869 | ||
| Allele frequency | 0.708 | ||||
| Asn | 46 (52%) | 55 (55%) | 1a | ||
| Ser | 42 (48%) | 45 (45%) | 1.12 (0.63-1.98); 0.708 |
OR, odds ratio; CIs, confidence intervals.
aReference genotype/allele.
Genotype distribution of genes encoding enzymes involved in metabolism and biosynthesis are shown in Table 5. In terms of the CYP17 −34 T/C polymorphism, proportionally, individuals carrying TC + CC variants of the gene were more frequently observed in patients than in controls. However, the genotype and allele frequencies were not different between the cases and controls (P > 0.05). The frequency for CYP1A1 TT genotype was higher among controls when compared with adolescent girls with PCOS. On the other hand, the frequency for CYP1A1 CT genotype in adolescent girls with PCOS was higher than that in the controls. The CYP1A1 CC genotypes were not detected in cases or controls. Although no statistically significant difference was observed in genotype and allele distribution between the cases and controls in terms of CYP1A1 polymorphism (P > 0.05), high genotype frequency of TT homozygotes were observed in both groups.
Table 5.
Genotype distribution of genes encoding enzymes involved in metabolism and biosynthesis in adolescent girls with polycystic ovary syndrome and healthy controls
| Gene | Genotype | Cases (n = 44) | Controls (n = 50) | P | OR (95% CIs);P |
|---|---|---|---|---|---|
| CYP17 | -34 T/C (rs743572) | 0.383 | |||
| TT | 15 (34%) | 20 (40%) | 1a | ||
| TC | 19 (43%) | 24 (48%) | 1.06 (0.43-2.60); 0.906 | ||
| CC | 10 (23%) | 6 (12%) | 2.22 (0.66-7.48); 0.193 | ||
| TC + CC | 29 (66%) | 30 (60%) | 1.29 (0.56-2.99); 0.554 | ||
| Allele frequency | 0.245 | ||||
| T | 49 (55.7%) | 64 (64%) | 1a | ||
| C | 39 (44.3%) | 36 (36%) | 1.42 (0.79-2.54); 0.245 | ||
| CYP1A1 | T6235C (rs4646903) | 0.188 | |||
| TT | 26 (59%) | 36 (72%) | 1a | ||
| CT | 18 (41%) | 14 (28%) | 1.78 (0.75-4.21); 0.188 | ||
| CC | 0 (0%) | 0 (0%) | NC | ||
| Allele frequency | 0.240 | ||||
| T | 70 (79.54%) | 86 (86%) | 1a | ||
| C | 18 (20.45%) | 14 (14%) | 1.58 (0.73-3.40); 0.240 |
OR, odds ratio; CIs, confidence intervals.
aReference genotype/allele.
NC, Not Calculated
The genotype distribution and the relative allele frequencies for the CAPN10, INSR, SERPINE1 genes are shown in Table 6. No significant differences were observed for genotype distribution (P > 0.05) and relative allele frequencies (P > 0.05) between cases and controls for the UCSNP44, 43, 19 and 63 polymorphisms of the CAPN10 gene. The relative frequencies of “C” allele for UCSNP44, “A” allele for UCSNP43, “T” allele for UCSNP63 polymorphisms were found quite less than those of their wild-type alleles in cases and controls. The frequency of “Del” allele for UCSNP19 polymorphism was 45.4% in cases and 44% in controls. We also did not find any statistically significant differences in the genotypes and allele frequencies between the cases and controls for exon 17 C/T, exon 12 G/A and exon 12 G/C variants in the INSR gene. For exon 12 G/A, two GA genotypes were observed (one in cases and one in controls) in all 94 subjects, whereas no AA genotype was observed. Similarly, for exon 12 G/C, two GC genotypes were observed (one in cases and one in controls) in all 94 subjects, whereas no CC genotype was observed. Moreover, for exon 17 C/T, no CC genotype was observed in cases or controls. Lastly, genotype and allele frequencies of SERPINE1 gene were not significantly different between the cases and controls.
Table 6.
Genotype distribution of genes encoding proteins involved in type 2 diabetes and cardiovacular disease in adolescent girls with polycystic ovary syndrome and healthy controls
| Gene | Genotype | Cases (n = 44) | Controls (n = 50) | P | OR (95% CIs);P |
|---|---|---|---|---|---|
| CAPN10 (Calpain-10) | UCSNP44 (rs2975760) | 0.535 | |||
| TT | 33 (75%) | 34 (68%) | 1a | ||
| TC | 11 (25%) | 15 (30%) | 0.76 (0.30-1.88); 0.547 | ||
| CC | 0 (0%) | 1 (2%) | NC | ||
| TC + CC | 11 (25%) | 16 (32%) | 0.71 (0.29-1.75); 0.454 | ||
| Allele frequency | 0.387 | ||||
| T | 77 (88%) | 83 (83%) | 1a | ||
| C | 11 (12%) | 17 (17%) | 0.70 (0.31-1.58); 0.387 | ||
| UCSNP43 (rs3792267) | 0.771 | ||||
| GG | 29 (66%) | 33 (66%) | 1a | ||
| GA | 13 (29.5%) | 16 (32%) | 0.93 (0.38-2.24); 0.862 | ||
| AA | 2 (4.5%) | 1 (2%) | 2.28 (0.20-26.42); 0.602 | ||
| GA + AA | 15 (34%) | 17 (34%) | 1.00 (0.43-2.36); 0.993 | ||
| Allele frequency | 0.817 | ||||
| G | 71 (80.7%) | 82 (82%) | 1a | ||
| A | 17 (19.3%) | 18 (18%) | 1.09 (0.52-2.28); 0.817 | ||
| UCSNP19 (rs3842570) | 0.314 | ||||
| Ins/Ins | 10 (22.8%) | 16 (32%) | 1a | ||
| Del/Ins | 28 (63.6%) | 24 (48%) | 1.87 (0.72-4.88); 0.200 | ||
| Del/Del | 6 (13.6%) | 10 (20%) | 0.96 (0.27-3.47); 0.950 | ||
| Del/Ins + Del/Del | 34 (77.2%) | 34 (68%) | 1.79 (0.72-4.47); 0.211 | ||
| Allele frequency | 0.841 | ||||
| Ins | 48 (54.6%) | 56 (56%) | 1a | ||
| Del | 40 (45.4%) | 44 (44%) | 1.06 (0.60-1.89); 0.841 | ||
| UCSNP63 (rs5030952) | 0.160 | ||||
| CC | 37 (84.1%) | 36 (72%) | 1a | ||
| CT | 7 (15.9%) | 14 (28%) | 0.49 (0.18-1.35); 0.160 | ||
| TT | 0 (0%) | 0 (0%) | NC | ||
| Allele frequency | 0.189 | ||||
| C | 81 (92.1%) | 86 (86%) | 1 a | ||
| T | 7 (7.9%) | 14 (14%) | 0.53 (0.20-1.38); 0.189 | ||
| INSR (insulin receptor) | Exon17 C/T (rs1799817) | 0.437 | |||
| TT | 22 (50%) | 21 (42%) | 1a | ||
| CT | 22 (50%) | 29 (58%) | 0.72 (0.32-1.64); 0.437 | ||
| CC | 0 (0%) | 0 (0%) | NC | ||
| Allele frequency | 0.538 | ||||
| T | 66 (75%) | 71 (71%) | 1a | ||
| C | 22 (25%) | 29 (29%) | 0.82 (0.43-1.56); 0.538 | ||
| Exon12 G/A (rs2229434) | 1.000 | ||||
| GG | 43 (97.7%) | 49 (98%) | 1 a | ||
| GA | 1 (2.3%) | 1 (2%) | 1.14 (0.07-18.78); 1.000 | ||
| AA | 0 (0%) | 0 (0%) | NC | ||
| Allele frequency | 1.000 | ||||
| G | 87 (98.9%) | 99 (99%) | 1a | ||
| A | 1 (1.1%) | 1 (1%) | 1.14 (0.07-18.47); 1.000 | ||
| Exon12 G/C (rs2229430) | 1.000 | ||||
| GG | 43 (97.7%) | 49 (98%) | 1a | ||
| GC | 1 (2.3%) | 1 (2%) | 1.14 (0.07-18.47); 1.000 | ||
| CC | 0 (0%) | 0 (0%) | NC | ||
| Allele frequency | 1.000 | ||||
| G | 87 (98.9%) | 99 (99%) | 1a | ||
| C | 1 (1.1%) | 1 (1%) | 1.14 (0.07-18.47); 1.000 | ||
| SERPINE1 (Plasminogen activator inhibitor-1, PAI-1) | 4G/5G (-675 ins/delG) (rs1799817) | 0.589 | |||
| 4G/4G | 7 (16%) | 12 (24%) | 1a | ||
| 4G/5G | 24 (54.5%) | 26 (52%) | 1.58 (0.54-4.68); 0.405 | ||
| 5G/5G | 13 (29.5%) | 12 (24%) | 1.86 (0.55-6.28); 0.317 | ||
| 4G/5G + 5G/5G | 37 (84%) | 38 (76%) | 1.70 (0.59-4.71); 0.330 | ||
| Allele frequency | 0.350 | ||||
| 4G | 38 (43.2%) | 50 (50%) | 1a | ||
| 5G | 50 (56.8%) | 50 (50%) | 1.32 (0.74-2.34); 0.350 |
OR, odds ratio; CIs, confidence intervals.
aReference genotype/allele.
NC, Not Calculated
There were no significant differences in clinical and biochemical characteristics of our cases with respect to the polymorphic variants of the 17 different loci (data not shown).
DNA sequence analysis
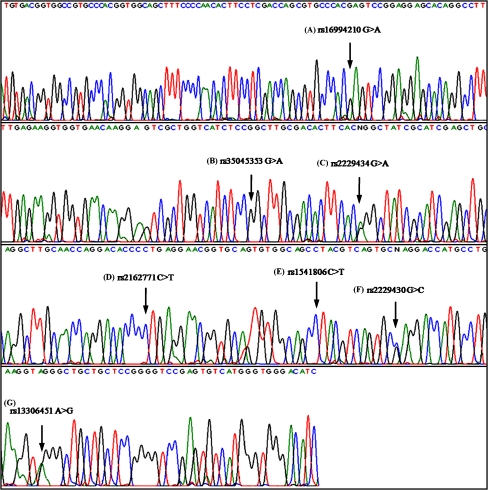
We screened exon 12 and intron 12 of the INSR gene for mutations in all 94 genotyped subjects. This is the first study to detect polymorphic variants rs16994210 G>A, rs35045353 G>A, rs2229434 G>A, rs2162771 C>T, rs1541806 C>T, rs2229430 G>C in exon 12 and rs13306451 A>G in intron 12 of INSR gene by using direct DNA sequencing method (Fig. 1). Among these, rs2229434 G>A and rs2229430 G>C are the silent mutations observed in both patients and controls. Frequencies of all seven polymorphic variants are similar in patients and controls, suggesting that these genetic variations are not major factors in the occurrence of PCOS in the Turkish adolescent girls.
Fig. 1.
Direct sequencing results for exon 12 of INSR gene. (A) Homozygous GG genotype corresponds to rs16994210 G>A; (B) homozygous GG genotype corresponds to rs35045353 G>A; (C) heterozygous GA genotype corresponds to rs2229434 G>A; (D) homozygous CC genotype corresponds to rs2162771 C>T; (E) homozygous CC genotype corresponds to rs1541806 C>T; (F) heterozygous GC genotype corresponds to rs2229430 G>C and (G) for intron 12 of the gene, homozygous AA genotype corresponds to rs13306451 A>G
Discussion
Genetic studies contribute to our understanding of molecular mechanisms of disease pathogenesis through characterization of natural variants or polymorphisms in the DNA sequence among individuals. We aimed, by investigating the SNPs of FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 genes, at identifying genetic factors which might have an impact on the etiology of the PCOS in adolescent girls.
First of all, due to the limited number of studies on adolescent girls with PCOS, we had to compare our results for all 17 polymorphisms investigated with those of similar studies on adults with PCOS. In addition, when we evaluated the clinical and biochemical characteristics of the adolescent girls with PCOS and controls, we observed that BMI, Ferriman-Gallwey score, percentages of acanthosis nigricans, levels of total testosterone, FSH, LH, prolactin, TSH were higher in PCOS girls (P < 0.01). These findings confirm the well-known associations between PCOS and these variants [1, 47].
Two known polymorphisms, Thr307Ala and Ser680Asn of FSHR gene showed similar distributions of the allelic variations and protein isoforms in PCOS and control subjects in Chinese Singapore [11] and Caucasian women [48]. 50 Italian women with PCOS harbored the A307T polymorphic variant, 56% harbored N680S, 30% S680S and 14% N680N polymorphisms [10]. In our study, the percentages of 680Ser carriers (Ser/Asn + Ser/Ser) are nearly the same in cases (70%) and controls (72%). In addition, the percentages of 307Ala carriers (Ala/Thr + Ala/Ala) are nearly the same in cases (64%) and controls (68%). We found that the frequency of “Ala” allele was 0.58 in cases and 0.57 in controls. The distribution patterns of the two alleles (“Ala” and “Thr”) were similar in both groups. Moreover, in our study, distribution of the cases with N680S (45%), S680S (25%) and N680N (13%) was found to be similar to findings of Orio et al. Mutations of the FSHR were found to be rare in Italian women and the only mutation found did not appear to have any pathophysiological significance in PCOS [10]. We found that these common polymorphisms did not seem to play a role in development of PCOS in adolescent girls (P > 0.05). Furthermore, clinical and biochemical profiles of the cases did not change the results. Our results were consistent with those of the three above-mentioned studies. However, Sudo et al reported that the “S” allele was more prevalent in PCOS cases than in normal subjects in Japanese women and that there were associations between the genotype and some aspects of patient status [9].
We then examined the SNPs of CYP17 −34 C/T and CYP1A1 T6235C. We found that the genotype and allele frequencies were not different between the cases and controls in both polymorphisms (P > 0.05). Park et al found seven SNPs of the gene CYP17, which is active in estrogen biosynthesis and located at 10q24.3, and found no significant association between these SNPs and PCOS [16]. For CYP17 −34 C/T, we found that CC genotype was the least common genotype both in cases and controls. TC genotype for CYP17 was the most common both in cases and controls. Genotype distribution of controls and cases were very close in our study. We further found that CC genotype was the least common both in cases and controls. These findings were consistent with those of some previous studies [15, 17]. Diamanti-Kandarakis et al showed that homozygosity of the polymorphic A2 allele (or CC genotype) was not observed in controls, but lower percentage (8%) was observed in PCOS women. Therefore, this difference was found statistically significant [14]. On the other hand, in our study, we found the frequency of “C” allele as 0.44 in cases and 0.36 in controls. The distribution pattern of the two alleles (“C” and “T”) was similar in both groups. This result is also in agreement with previous studies [14, 15, 17, 49]. Echiburú et al found that PCOS carriers of A2 (or C) allele presented a greater BMI and waist circumference than PCOS non-carriers [17]. However, we found no evidence of a relationship between clinical and biochemical characteristics and CYP17 −34 C/T polymorphism in PCOS patients (P > 0.05). Contrary to previous observations of Babu et al [18], who suggested that CYP1A1 (T6235C) polymorphism may represent a risk factor for PCOS, we did not find any evidence to support an association between CYP1A1 polymorphism and PCOS. Furthermore, Esinler et al examined the correlation between CYP1A1 genotypes and PCOS women [19]. They found that the patients with PCOS had a 7.8-fold higher frequency of CYP1A1 Ile/Val genotype and a 7.4-fold higher frequency of CYP1A1 of any Val genotype (Ile/Val or Val/Val) in Turkish population. This discrepancy between the two studies was due to the fact that different regions of the CYP1A1 gene were studied in the two studies. We examined the polymorphic variants in the 3′-flanking region (T6235C) of the gene, whereas Esinler et al examined the Ile462Val (A/G) variants in the exon 7 of the gene.
We then examined the SNPs of CAPN10 (UCSNP44, UCSNP43, UCSNP19 and UCSNP63), INSR exon 17 C/T, SERPINE1 4G/5G. Although Wiltgen et al [24] did not find a direct association between UCSNP43, UCSNP19 and UCSNP63 and susceptibility to PCOS in Brazilian patients, they found an association between higher prevalence of the UCSNP43 polymorphic allele of the CAPN10 gene and metabolic syndrome in PCOS women. Márquez et al [25] suggested that the presence of uncommon (A) allele for UCSNP43 was significantly associated to PCOS. Similarly, we found that the percentages of “A” allele (19% for cases vs. 18% for controls) were quite less than those of “G” allele (81% for cases vs. 82% for controls). However, we did not find an association between UCSNP43 and PCOS as well as between UCSNP44, UCSNP19 and UCSNP63 and PCOS (P > 0.05). Clinical and biochemical profiles of the cases did not change the results. Genotype frequencies of UCSNP44, UCSNP43, UCSNP19 and UCSNP63 polymorphisms are similar both in cases and controls to findings of Gonzales et al. Gonzalez et al [50] found that UCSNP43, UCSNP19 and UCSNP63 polymorphisms were not associated with PCOS in Spanish population, but that there was an association between UCSNP44 and PCOS. Our genotype frequencies of the UCSNP43 and UCSNP63 were in agreement with previous studies [25, 27, 50]. Vollmert et al investigated sample of eight variants (UCSNP-44, -43, -56, ins/del-19, -110, -58, -63, and -22) and none of the variants alone showed any significant association with PCOS [51]. Our frequency of “Del” allele for UCSNP19 was 45% in PCOS and 44% in controls. This was consistent with findings of some previous studies [25, 51]. On the other hand, the polymorphic allele 2R (or Del) of UCSNP19 polymorphism showed a trend toward higher frequency in PCOS patients presenting metabolic syndrome, without, however, reaching statistical significance [24]. Our finding regarding the frequency of UCSNP19 “Del” allele was not in agreement with this study. We also did not find any significant differences in the genotype distribution between cases and controls of exon 17 C/T, exon 12 G/A and exon 12 G/C variants of the INSR gene (P > 0.05). The INSR exon 17 C/T genotype distribution indicated a narrow departure from Hardy-Weinberg equilibrium due to observation of excess heterozygotes and non-observation of the CC genotype in controls and cases. We found the frequency of the TT genotype to be similar in cases (50%) and controls (42%). This was consistent with findings of Lee et al [52]. Although Lee et al found that the CC genotype was presented even more frequently than TT genotype in both cases and controls, they did not observe an association between the gene and susceptibility to PCOS in Korean population [52]. On the other hand, the polymorphism of INSR gene is one of the susceptibility factors in patients with PCOS, especially in non-obese PCOS patients [30]. The frequency of the uncommon “T” allele of the INSR was significantly increased in lean patients with PCOS compared with lean controls [29]. Recently, nine candidate SNPs were found in the INSR gene, but the minor allele of the novel SNP 176477 C>T has an association with the pathogenesis of PCOS in the Korean population [32]. We found no evidence of a relationship between clinical and biochemical characteristics and exon 17 C/T in the INSR gene polymorphism in PCOS patients (P > 0.05). When we sequenced INSR exon 12 region, we identified 2 SNPs (for both polymorphism one in cases and one in controls) in exon 12 G/A and exon 12 G/C. This is the first study to sequence this region; therefore, we could not compare our results to those of studies on other populations. We detected no statistically significant differences between the genotype and allele frequencies of SERPINE1 (or PAI-1) promotor 4G/5G gene polymorphism (P > 0.05). We found that the 4G allele frequency was 43.2% in adolescent girls with PCOS and 50% in controls. We found 4G/5G heterozygote to be the most common genotype both in cases and controls. This was also consistent with the findings of a previous study, which evaluated the 4G/5G polymorphism in the promoter region of PAI-1 gene in Turkish population [36]. Similarly, Karadeniz et al [36] did not find an association between 4G/5G gene polymorphism and PCOS in Turkish population. These results are also in agreement with the previous observations of San Millán et al [34] and Walch et al [35], who also did not find any differences in the 4G/5G polymorphism between PCOS patients and controls. In our study, presence of the 4G/5G polymorphism of SERPINE1 was not associated with clinical and biochemical characteristics (P > 0.05). Our findings are similar to those of Walch [35]. In contrast to the above mentioned studies, Diamanti-Kandarakis et al [33] found evidence that the 4G/5G polymorphism is associated to PCOS in the Greek population.
Since the main limitation of this study is the small sample size due to the inclusion of only adolescent girls in the study population, further studies in larger populations are needed to come up with supporting data. Furthermore, differences in ethnicity, sample size, criteria in selecting cases and controls, effects of the SNPs in the genes could account for some of the conflicting results.
To conclude, many genes have been investigated as possible susceptibility loci but if PCOS is indeed a complex genetic disorder, the effect of any one gene may be small. To our knowledge, this is the first report to explore the possible association between adolescent girls with PCOS and 17 different polymorphic loci on the FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 genes in Turkish population. By investigating the functional aspects of these SNPs in adolescence and building up more information about the underlying genetic basis for PCOS, it may be possible to manage the impact on PCOS through controlling environmental factors.
Acknowledgements
This study has been supported by the Gazi University Research Fund, with the project code number 01/2006-05.
Appendix
Supporting document
Gene structure and linkage disequilibrium plot for CAPN10. The locations of the genotyped SNPs (UCSNP44, 43, 19 and 63) relative to the introns are indicated. The linkage disequilibrium plot at the bottom displays D' values (percent) for each pair of SNPs in the box at the intersection of the diagonals from each SNP.
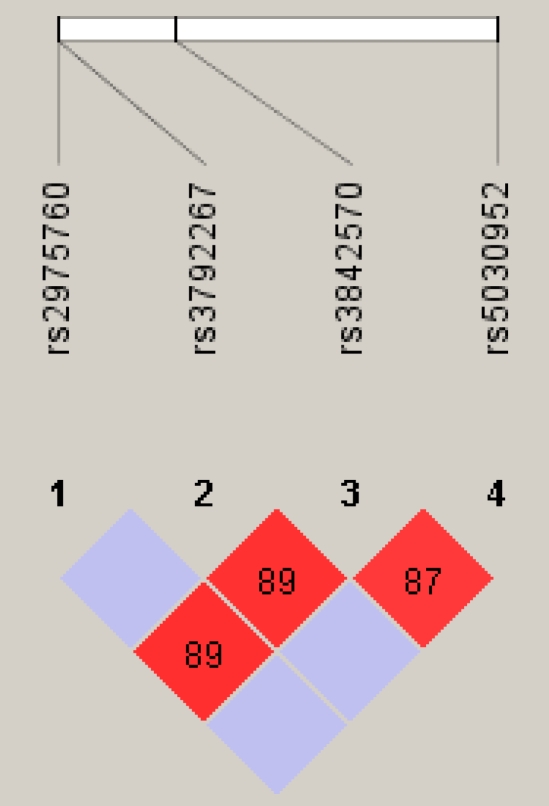
Pair wise linkage disequilibrium between the SNPs were carried out using Haploview version 4.0 (http://www.broad.mit.edu/mpg/haploview). However, CAPN10 UCSNP44 was not in linkage disequilibrium with UCSNP 43 and UCSNP63. In addition, CAPN10 UCSNP43 was not in linkage disequilibrium with UCSNP 44 and UCSNP63. Therefore, neither conducting a haplotype analyses nor calculating potential associations among these four different polymorphisms of CAPN10 gene is necessary.
References
- 1.Franks S. Polycystic ovary syndrome in adolescents. Int J Obes. 2008;32(7):1035–41. doi:10.1038/ijo.2008.61. [DOI] [PubMed]
- 2.Ehrmann DA. Polycystic ovary syndrome. N Engl J Med. 2005;352(12):1223–36. doi:10.1056/NEJMra041536. [DOI] [PubMed]
- 3.Luque-Ramírez M, San Millán JL, Escobar-Morreale HF. Genomic variants in polycystic ovary syndrome. Clin Chim Acta. 2006;366(1–2):14–26. doi:10.1016/j.cca.2005.10.017. [DOI] [PubMed]
- 4.Valdés P, Cerda A, Barrenechea C, Kehr M, Soto C, Salazar LA. No association between common Gly972Arg variant of the insulin receptor substrate-1 and polycystic ovary syndrome in Southern Chilean women. Clin Chim Acta. 2008;390(1–2):63–6. doi:10.1016/j.cca.2007.12.018. [DOI] [PubMed]
- 5.Simoni M, Tempfer CB, Destenaves B, Fauser BC. Functional genetic polymorphisms and female reproductive disorders: Part I: Polycystic ovary syndrome and ovarian response. Hum Reprod Update. 2008;14(5):459–84. doi:10.1093/humupd/dmn024. [DOI] [PMC free article] [PubMed]
- 6.Davis D, Liu X, Segaloff DL. Identification of the sites of N-linked glycosylation on the follicle-stimulating hormone (FSH) receptor and assessment of their role in FSH receptor function. Mol Endocrinol. 1995;9(2):159–70. doi:10.1210/me.9.2.159. [DOI] [PubMed]
- 7.Hipkin RW, Liu X, Ascoli M. Truncation of the C-terminal tail of the follitropin receptor does not impair the agonist- or phorbol ester-induced receptor phosphorylation and uncoupling. J Biol Chem. 1995;270(44):26683–9. doi:10.1074/jbc.270.44.26683. [DOI] [PubMed]
- 8.de Castro F, Ruiz R, Montoro L, et al. Role of follicle-stimulating hormone receptor Ser680Asn polymorphism in the efficacy of follicle-stimulating hormone. Fertil Steril. 2003;80(3):571–6. doi:10.1016/S0015-0282(03)00795-7. [DOI] [PubMed]
- 9.Sudo S, Kudo M, Wada S, Sato O, Hsueh AJ, Fujimoto S. Genetic and functional analyses of polymorphisms in the human FSH receptor gene. Mol Hum Reprod. 2002;8(10):893–9. doi:10.1093/molehr/8.10.893. [DOI] [PubMed]
- 10.Orio F Jr, Ferrarini E, Cascella T, et al. Genetic analysis of the follicle stimulating hormone receptor gene in women with polycystic ovary syndrome. J Endocrinol Invest. 2006;29(11):975–82. [DOI] [PubMed]
- 11.Tong Y, Liao WX, Roy AC, Ng SC. Absence of mutations in the coding regions of follicle-stimulating hormone receptor gene in Singapore Chinese women with premature ovarian failure and polycystic ovary syndrome. Horm Metab Res. 2001;33(4):221–6. doi:10.1055/s-2001-14941. [DOI] [PubMed]
- 12.Lazar L, Kauli R, Bruchis C, Nordenberg J, Galatzer A, Pertzelan A. Early polycystic ovary-like syndrome in girls with central precocious puberty and exaggerated adrenal response. Eur J Endocrinol. 1995;133(4):403–6. doi:10.1530/eje.0.1330403. [DOI] [PubMed]
- 13.Carey AH, Waterworth D, Patel K, et al. Polycystic ovaries and premature male pattern baldness are associated with one allele of the steroid metabolism gene CYP17. Hum Mol Genet. 1994;3(10):1873–6. doi:10.1093/hmg/3.10.1873. [DOI] [PubMed]
- 14.Diamanti-Kandarakis E, Bartzis MI, Zapanti ED, et al. Polymorphism T-->C (-34 bp) of gene CYP17 promoter in Greek patients with polycystic ovary syndrome. Fertil Steril. 1999;71(3):431–5. doi:10.1016/S0015-0282(98)00512-3. [DOI] [PubMed]
- 15.Marszalek B, Laciński M, Babych N, et al. Investigations on the genetic polymorphism in the region of CYP17 gene encoding 5'-UTR in patients with polycystic ovarian syndrome. Gynecol Endocrinol. 2001;15(2):123–8. doi:10.1080/713602803. [DOI] [PubMed]
- 16.Park JM, Lee EJ, Ramakrishna S, Cha DH, Baek KH. Association study for single nucleotide polymorphisms in the CYP17A1 gene and polycystic ovary syndrome. Int J Mol Med. 2008;22(2):249–54. [PubMed]
- 17.Echiburú B, Pérez-Bravo F, Maliqueo M, Sánchez F, Crisosto N, Sir-Petermann T. Polymorphism T-->C (-34 base pairs) of gene CYP17 promoter in women with polycystic ovary syndrome is associated with increased body weight and insulin resistance: a preliminary study. Metabolism. 2008;57(12):1765–71. doi:10.1016/j.metabol.2008.08.002. [DOI] [PubMed]
- 18.Babu KA, Rao KL, Kanakavalli MK, Suryanarayana VV, Deenadayal M, Singh L. CYP1A1, GSTM1 and GSTT1 genetic polymorphism is associated with susceptibility to polycystic ovaries in South Indian women. Reprod Biomed Online. 2004;9(2):194–200. [DOI] [PubMed]
- 19.Esinler I, Aktas D, Otegen U, Alikasifoglu M, Yaralı H, Tuncbilek E. CYP1A1 gene polymorphism and polycystic ovary syndrome. Reprod Biomed Online. 2008;16(3):356–60. [DOI] [PubMed]
- 20.Gaasenbeek M, Powell BL, Sovio U, et al. Large-scale analysis of the relationship between CYP11A promoter variation, polycystic ovarian syndrome, and serum testosterone. J Clin Endocrinol Metab. 2004;89(5):2408–13. doi:10.1210/jc.2003-031640. [DOI] [PubMed]
- 21.Wang Y, Wu X, Cao Y, Yi L, Chen J. A microsatellite polymorphism (tttta)n in the promoter of the CYP11a gene in Chinese women with polycystic ovary syndrome. Fertil Steril. 2006;86(1):223–6. doi:10.1016/j.fertnstert.2005.12.037. [DOI] [PubMed]
- 22.Horikawa Y, Oda N, Cox NJ, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26(2):163–75. doi:10.1038/79876. [DOI] [PubMed]
- 23.Baier LJ, Permana PA, Yang X, et al. A calpain-10 gene polymorphism is associated with reduced muscle mRNA levels and insulin resistance. J Clin Invest. 2000;106:R69–73. doi:10.1172/JCI10665. [DOI] [PMC free article] [PubMed]
- 24.Wiltgen D, Furtado L, Kohek MB, Spritzer PM. CAPN10 UCSNP-43, UCSNP-19 and UCSNP-63 polymorphisms and metabolic syndrome in polycystic ovary syndrome. Gynecol Endocrinol. 2007;23(3):173–8. doi:10.1080/09513590701233661. [DOI] [PubMed]
- 25.Márquez JL, Pacheco A, Valdés P, Salazar LA. Association between CAPN10 UCSNP-43 gene polymorphism and polycystic ovary syndrome in Chilean women. Clin Chim Acta. 2008;398(1–2):5–9. doi:10.1016/j.cca.2008.07.028. [DOI] [PubMed]
- 26.Ehrmann DA, Schwarz PE, Hara M, et al. Relationship of calpain-10 genotype to phenotypic features of polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87(4):1669–73. doi:10.1210/jc.87.4.1669. [DOI] [PubMed]
- 27.Haddad L, Evans JC, Gharani N, et al. Variation within the type 2 diabetes susceptibility gene calpain-10 and polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87(6):2606–10. doi:10.1210/jc.87.6.2606. [DOI] [PubMed]
- 28.Seino S, Seino M, Bell GI. Human insulin-receptor gene. Diabetes. 1990;39(2):129–33. doi:10.2337/diabetes.39.2.129. [DOI] [PubMed]
- 29.Siegel S, Futterweit W, Davies TF, et al. A C/T single nucleotide polymorphism at the tyrosine kinase domain of the insulin receptor gene is associated with polycystic ovary syndrome. Fertil Steril. 2002;78(6):1240–3. doi:10.1016/S0015-0282(02)04241-3. [DOI] [PubMed]
- 30.Chen ZJ, Shi YH, Zhao YR, et al. Correlation between single nucleotide polymorphism of insulin receptor gene with polycystic ovary syndrome. Zhonghua Fu Chan Ke Za Zhi. 2004;39(9):582–5. [PubMed]
- 31.Jin L, Huang HF, Jin F, Qian YL. Polymorphism in insulin receptor gene exon 17 in women with polycystic ovary syndrome. Zhonghua Fu Chan Ke Za Zhi. 2005;40(5):323–6. [PubMed]
- 32.Lee EJ, Oh B, Lee JY, Kimm K, Lee SH, Baek KH. A novel single nucleotide polymorphism of INSR gene for polycystic ovary syndrome. Fertil Steril. 2008;89(5):1213–20. doi:10.1016/j.fertnstert.2007.05.026. [DOI] [PubMed]
- 33.Diamanti-Kandarakis E, Palioniko G, Alexandraki K, Bergiele A, Koutsouba T, Bartzis M. The prevalence of 4G5G polymorphism of plasminogen activator inhibitor-1 (PAI-1) gene in polycystic ovarian syndrome and its association with plasma PAI-1 levels. Eur J Endocrinol. 2004;150(6):793–8. doi:10.1530/eje.0.1500793. [DOI] [PubMed]
- 34.San Millán JL, Cortón M, Villuendas G, Sancho J, Peral B, Escobar-Morreale HF. Association of the polycystic ovary syndrome with genomic variants related to insulin resistance, type 2 diabetes mellitus, and obesity. J Clin Endocrinol Metab. 2004;89(6):2640–6. doi:10.1210/jc.2003-031252. [DOI] [PubMed]
- 35.Walch K, Grimm C, Huber JC, Nagele F, Kolbus A, Hefler LA. A polymorphism of the plasminogen activator inhibitor-1 gene promoter and the polycystic ovary syndrome. Eur J Obstet Gynecol Reprod Biol. 2005;123(1):77–81. doi:10.1016/j.ejogrb.2005.07.002. [DOI] [PubMed]
- 36.Karadeniz M, Erdogan M, Berdeli A, Saygili F, Yilmaz C. 4G/5G polymorphism of PAI-1 gene and Alu-repeat I/D polymorphism of TPA gene in Turkish patients with polycystic ovary syndrome. J Assist Reprod Genet. 2007;24(9):412–8. doi:10.1007/s10815-007-9160-7. [DOI] [PMC free article] [PubMed]
- 37.Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81(1):19–25. doi:10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed]
- 38.Sundblad V, Chiauzzi VA, Escobar ME, Dain L, Charreau EH. Screening of FSH receptor gene in Argentine women with premature ovarian failure (POF). Mol Cell Endocrinol. 2004;222(1–2):53–9. doi:10.1016/j.mce.2004.05.002. [DOI] [PubMed]
- 39.Yang CQ, Chan KY, Ngan HY, et al. Single nucleotide polymorphisms of follicle-stimulating hormone receptor are associated with ovarian cancer susceptibility. Carcinogenesis. 2006;27(7):1502–6. doi:10.1093/carcin/bgl014. [DOI] [PubMed]
- 40.Onen IH, Ekmekci A, Eroglu M, Polat F, Biri H. The association of 5alpha-reductase II (SRD5A2) and 17 hydroxylase (CYP17) gene polymorphisms with prostate cancer patients in the Turkish population. DNA Cell Biol. 2007;26(2):100–7. doi:10.1089/dna.2006.0534. [DOI] [PubMed]
- 41.Hirata H, Hinoda Y, Okayama N, et al. CYP1A1, SULT1A1, and SULT1E1 polymorphisms are risk factors for endometrial cancer susceptibility. Cancer. 2008;112(9):1964–73. doi:10.1002/cncr.23392. [DOI] [PubMed]
- 42.Daimon M, Oizumi T, Saitoh T, et al. Calpain 10 gene polymorphisms are related, not to type 2 diabetes, but to increased serum cholesterol in Japanese. Diabetes Res Clin Pract. 2002;56(2):147–52. doi:10.1016/S0168-8227(01)00372-2. [DOI] [PubMed]
- 43.Shima Y, Nakanishi K, Odawara M, Kobayashi T, Ohta H. Association of the SNP-19 genotype 22 in the calpain-10 gene with elevated body mass index and hemoglobin A1c levels in Japanese. Clin Chim Acta. 2003;336(1–2):89–96. doi:10.1016/S0009-8981(03)00320-6. [DOI] [PubMed]
- 44.Orho-Melander M, Klannemark M, Svensson MK, Ridderstråle M, Lindgren CM, Groop L. Variants in the calpain-10 gene predispose to insulin resistance and elevated free fatty acid levels. Diabetes. 2002;51(8):2658–64. doi:10.2337/diabetes.51.8.2658. [DOI] [PubMed]
- 45.Brown NJ, Murphey LJ, Srikuma N, Koschachuhanan N, Williams GH, Vaughan DE. Interactive effect of PAI-1 4G/5G genotype and salt intake on PAI-1 antigen. Arterioscler Thromb Vasc Biol. 2001;21(6):1071–7. [DOI] [PubMed]
- 46.Seino S, Seino M, Bell GI. Human insulin-receptor gene. Partial sequence and amplification of exons by polymerase chain reaction. Diabetes. 1990;39(1):123–8. doi:10.2337/diabetes.39.1.123. [DOI] [PubMed]
- 47.Amato P, Simpson JL. The genetics of polycystic ovary syndrome. Best Pract Res Clin Obstet Gynaecol. 2004;18(5):707–18. doi:10.1016/j.bpobgyn.2004.05.002. [DOI] [PubMed]
- 48.Conway GS, Conway E, Walker C, Hoppner W, Gromoll J, Simoni M. Mutation screening and isoform prevalence of the follicle stimulating hormone receptor gene in women with premature ovarian failure, resistant ovary syndrome and polycystic ovary syndrome. Clin Endocrinol (Oxf). 1999;51(1):97–9. doi:10.1046/j.1365-2265.1999.00745.x. [DOI] [PubMed]
- 49.Liović M, Prezelj J, Kocijancic A, Majdic G, Komel R. CYP17 gene analysis in hyperandrogenised women with and without exaggerated 17-hydroxyprogesterone response to ovarian stimulation. J Endocrinol Invest. 1997;20(4):189–93. [DOI] [PubMed]
- 50.Gonzalez A, Abril E, Roca A, et al. Comment: CAPN10 alleles are associated with polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87(8):3971–6. doi:10.1210/jc.87.8.3971. [DOI] [PubMed]
- 51.Vollmert C, Hahn S, Lamina C, et al. Calpain-10 variants and haplotypes are associated with polycystic ovary syndrome in Caucasians. Am J Physiol Endocrinol Metab. 2007;292(3):E836–44. doi:10.1152/ajpendo.00584.2005. [DOI] [PubMed]
- 52.Lee EJ, Yoo KJ, Kim SJ, Lee SH, Cha KY, Baek KH. Single nucleotide polymorphism in exon 17 of the insulin receptor gene is not associated with polycystic ovary syndrome in a Korean population. Fertil Steril. 2006;86(2):380–4. doi:10.1016/j.fertnstert.2005.12.073. [DOI] [PubMed]