Abstract
SCWRL and MolIDE are software applications for prediction of protein structures. SCWRL is designed specifically for the task of prediction of side-chain conformations given a fixed backbone usually obtained from an experimental structure determined by X-ray crystallography or NMR. SCWRL is a command-line program that typically runs in a few seconds. MolIDE provides a graphical interface for basic comparative (homology) modeling using SCWRL and other programs. MolIDE takes an input target sequence, and uses PSI-BLAST to identify and align templates for comparative modeling of the target. The sequence alignment to any template can be manually modified within a graphical window of the target-template alignment and visualization of the alignment on the template structure. MolIDE builds the model of the target structure based on the template backbone, predicted side-chain conformations with SCWRL, and a loop-modeling program for insertion-deletion regions with user-selected sequence segments. SCWRL and MolIDE can be obtained at http://dunbrack.fccc.edu/Software.php.
Keywords: Computational methods, Protein structure prediction, Comparative (homology) modeling
INTRODUCTION
To understand basic biological processes such as cell division, cell signaling, development, metabolism, and cell death, detailed knowledge of the three-dimensional structures of the active participants is necessary. Structures of a large number of proteins have been determined experimentally using primarily X-ray crystallography and NMR spectroscopy. There are currently more than 50,000 entries in the Protein Data Bank (PDB)1, which archives experimentally determined structures of proteins and protein complexes, as well as nucleic acids and other biological macromolecules. However, a list of sequences of proteins in the PDB such that no two sequences are more than 20% identical to each other, contains only about 4000 sequences2. Many of these proteins can be grouped further into about 1000 folds in 2000 superfamilies3.
Since it was first recognized that proteins can share similar structures4, computational methods have been developed to build models of proteins of unknown structure based on related proteins of known structure5. Most such modeling efforts, referred to as homology modeling or comparative modeling, follow a basic protocol: 1) for a target sequence of unknown structure, identify a template structure with sequence related to the target and align the target sequence to the template sequence and structure; 2) for core secondary structures and all well-conserved parts of the alignment, borrow the backbone coordinates of the template according to the sequence alignment of the target and template; 3) build side chains onto the backbone model according to the target sequence; 4) for segments of the target sequence for which coordinates cannot be borrowed from the template because of insertions and deletions in the alignment (usually in loop regions of the protein) or because of missing coordinates in the template, rebuild these regions using loop modeling methods or other ab initio structure prediction methods; 5) refine the structure, modeling likely differences in the relative positions of α helices, β sheet strands, and other elements of structure.
The identification step necessarily involves sequence alignment, but even once a template has been identified and aligned to the target, a number of different methods may be used to improve the alignment, including fold recognition methods6 and profile-profile alignment7. Manual editing based on visualization of the template structure is frequently used to improve the alignment. Steps 3 and 4, side-chain and backbone modeling, may be coupled, since certain backbone conformations may be unable to accommodate the required side chains in any low-energy conformation. The refinement step involves moving all parts of the structure, including the backbone model produced in Step 2, allowing them to adjust to the new sequence. For instance, two helices packed against each other may move apart to accommodate larger side chains in the target than in the template. Many methods have been proposed to perform each of the steps in the homology modeling process. Other procedures based on reconstructing structures (rather than perturbing a starting structure) by satisfying spatial restraints using distance geometry8 or molecular dynamics and energy minimization9-12 have also been developed. The popular program Modeller is one of these9.
Homology modeling of proteins has been of great value in interpreting the relationships of sequence, structure, and function. In particular, orthologous proteins usually show a pattern of conserved residues that can be interpreted in terms of three-dimensional models of the proteins. Orthologues are genes and proteins in two different organisms that have descended from a single common ancestor without duplication. Conserved residues often form a contiguous active site or interaction surface of the protein, even if they are distant from each other in the sequence. With a structural model, a multiple alignment of orthologous proteins can be interpreted in terms of the constraints of natural selection in terms of protein folding, stability, dynamics, and function. Paralogues, on the other hand, arise from gene duplication and subsequent divergence of sequence and function. For paralogous proteins, three-dimensional models can be used to interpret the similarities and differences in the sequences in terms of the related structure but usually different functions of the proteins concerned.13 In many cases, there are significant insertions and deletions and amino acid changes in the active or binding site between paralogues. Indeed, homology models can serve to help us identify which protein belongs to which functional group by the conservation of important residues in the active or binding site14. A number of groups have used comparative modeling to predict protein function.15-19
Another important use of homology modeling is to understand functional changes due to point mutations in protein sequences that arise either by natural processes or by experimental manipulation. The human genome project has produced significant amounts of data concerning polymorphisms and other mutations potentially related to differences in susceptibility, prognosis, and treatment of human disease. There are now many such examples, including the Factor V/Leiden R506Q mutation20 that causes increased occurrence of thrombosis, mutations in the serine protease HTRA2 associated with Parkinson's disease21, and BRCA1 for which many sequence differences are known, some of which may lead to breast cancer22. At the same time, there are many polymorphisms in important genes that have no discernible effect on those who carry them. At least for some of these, there may be some effect that has yet to be measured in a large enough population of patients, and therefore the risk of cancer, heart disease, or other illness to these patients is unknown. This is yet another important application of homology modeling, since a good model may indicate readily which mutations pose a likely risk and which do not23.
Homology models may also be used in computer-aided drug design, especially when a closely related template structure is available for the target sequence. In such cases, the active site may be sufficiently conserved such that a model of the protein provides a reasonable target for computer programs that can suggest the most likely compounds that will bind to the active site. This has been used successfully in the early development of HIV protease inhibitors24,25 and in the development of anti-malarial compounds that target the cysteine protease of P. falciparum26. It has been used recently for high-throughput computational screening of alpha glucosidase inhibitors for treatment of diabetes27.
In this paper, we provide protocols for using two tools for protein structure prediction. The first is SCWRL, which is a computer program for side-chain prediction28,29. SCWRL is perhaps the most popular program for modeling of side chains (2,636 licenses in 72 countries as of 12 August 2008), and offers a good tradeoff between accuracy and speed as described in a recent independent benchmark30. SCWRL may be used for homology modeling by inputting a sequence different from the input backbone coordinates. It may also be used to complete a structure that is missing some or all of its side chains. This may happen if some side chains are not present in a PDB structure because of lack of electron density, and yet it would be useful to have at least a predicted position for these residues. SCWRL can be used to build mutations into proteins by converting one side-chain type into another, although it does not model any changes in backbone structure that might result.
The second program we describe is MolIDE31. MolIDE provides a graphical interface to the basic protocol of homology modeling, except for the final refinement step. That is, it provides for identification and alignment of the target to a template, modeling of the backbone according to this alignment, building of side chains of the target sequence, and loop modeling of insertion-deletion regions. For many purposes, such modeling is sufficient, since it provides rough locations of all amino acids within the protein: whether they are on the surface or buried, in active sites or not, and proximity in space versus proximity in sequence, etc. The refinement step is quite difficult and time consuming in computational resources and may provide little added benefit for many users of homology models. This is an area of active research32.
Experimental Design
SCWRL itself has been designed to be an easy-to-use command-line program that builds side chains onto an input backbone structure. The current version, SCWRL3.029, is based on a graph-theory algorithm that represents the interactions of side chains within a protein as a graph. It then uses the graph and an energy function to determine the lowest energy conformations of all of the side chains. Most side-chain types in proteins have a limited number of discrete conformations referred to as rotamers. These arise from steric repulsions of atoms separated by three or four covalent bonds. The shortest side chains, serine, threonine, valine, and cysteine, have only three available conformations. Leucine and isoleucine have nine conformations, although some of these are high in energy and are rare in proteins. The longer side chains have many possible conformations, although in practice many of these are also high in energy and many such residues are exposed to the solvent and sample many conformations in a dynamically active structure. SCWRL uses a backbone-dependent rotamer library33,34, which expresses the frequency of rotamers as a function of the backbone dihedral angles ϕ and Ψ for each of the amino acid types. It also contains information on the average side-chain dihedral angles in a backbone-dependent manner. SCWRL3.0 uses the 2002 version of the backbone-dependent rotamer library35. This library was based on 850 chains in the PDB with resolution better than or equal to 1.7 Å. Residues with high B-factors or atomic clashes were removed from the data set, as suggested by Lovell et al.36, and the terminal dihedrals of Asn, Gln, and His residues were flipped if there was a clear hydrogen bond formed by doing so37.
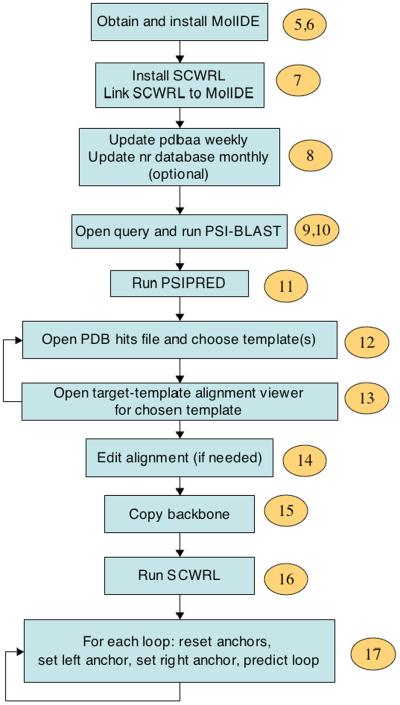
MolIDE performs homology modeling of single proteins in a visual environment. The basic protocol, described step-by-step below, is shown in Figure 1 (Please note that the description for MolIDE starts at step 5 in the PROCEDURES section). Its first step is to perform a database search with the program PSI-BLAST38. PSI-BLAST is an iterative program that searches a database for sequences similar to the target or query sequence. Usually the sequence database for this search is the non-redundant database (“nr”) of protein sequences provided by the NCBI39; currently this database contains over 6 million sequences. The first iteration of PSI-BLAST is just like the BLAST program, finding the most closely related sequences in the database related to the query. PSI-BLAST creates a multiple sequence alignment of the target sequence with these sequences. This multiple sequence alignment is then transformed into a sequence profile, which expresses the frequency of each of the 20 amino acid types in each column of the multiple sequence alignment. That is, for each residue of the query, the profile contains a numerical value for each of the 20 amino acids, depending on how often that residue type is found aligned to the query residue amongst homologues.
Figure 1.
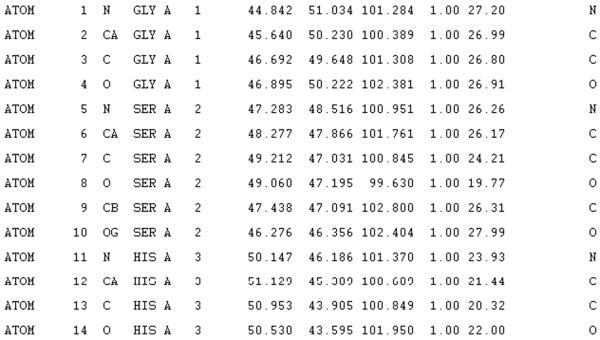
An excerpt from a PDB file.
The second iteration of PSI-BLAST searches the database with the profile instead of just the query sequence, and scores each sequence in the database by how well it aligns to the profile. For instance, if the profile shows that a particular position in the multiple sequence is 100% glycines, then the profile will score glycine in a database sequence very highly, and all other residue types either neutrally or negatively. If another column is a mixture of hydrophobic residues, then all hydrophobic residues will be scored positively but charged residues will be scored quite negatively. Some positions in the multiple sequence alignment contain most or all of the 20 amino acids, and such positions will score all of the amino acids neutrally. The second iteration will produce a new list of hits. Many of these will be the same as in the first round, but the alignments may be different. True hits will usually have longer alignments, and the expectation values or E-values will be much better. An E-value for a hit is the number of hits expected with a raw score the same as or better than the hit; thus a very small E-value (<0.001) indicates a high statistical significance. In the second round, many new hits with good E-values will be found that were not found in the first round. PSI-BLAST then builds a new multiple sequence alignment and profile from the hits in the second iteration, and then performs a search with this profile. The number of iterations is controlled by the user from within MolIDE. We have added to the most recent version of MolIDE (version 1.6) the ability to open the PSI-BLAST output file against the nr database. This produces a table that is sortable by E-value, sequence identity, starting and stopping residues of the alignment, protein name, and species. As such, MolIDE provides a graphical interface for PSI-BLAST searches of large databases such as nr, in addition to tools for structure prediction.
Once PSI-BLAST has created these profiles, each profile is used to search a database of sequences of proteins of known structure, called pdbaa. We provide access to this database from our website each week as new structures are added to the PDB2. This PSI-BLAST of the PDB runs automatically after the search of nr to create the profiles. The resulting files, one for each round of PSI-BLAST search of nr, contain lists of possible template structures for modeling the target sequence as well as alignments between the target and the template sequences. The version of PSI-BLAST provided in MolIDE is modified from that provided by NCBI. In particular, it outputs a profile matrix for each round of PSI-BLAST with a different filename (e.g. file1, file2, etc.) rather than overwriting a single filename. It also outputs a profile after the last search round of PSI-BLAST, while NCBI's version does not.
It is helpful at this point to perform a prediction of secondary structure within MolIDE using PSIPRED40. The PSIPRED predictions can be used to identify folded domains of the proteins in regions where there is significant amounts of predicted secondary structure, and disordered regions where little secondary structure is predicted. Also, the PSIPRED predictions will later be displayed in the alignments of the target sequence to a template. In this situation, they can be used to help determine whether the template is correct (at least some agreement of predicted and experimental secondary structure is expected) and to identify regions that may be misaligned (poor agreement of major secondary structure elements). PSIPRED uses the sequence profiles produced by PSI-BLAST to predict the positions of α helices and β sheets in the target sequence.
The next step in modeling is to choose a template by opening the list of hits from the PDB. MolIDE parses each PSI-BLAST output file and creates a table of hits, including their experimental source (XRAY or NMR), the resolution of X-ray structures, the E-value, sequence identity, the beginning and ending points in the target sequence, and the alignment length. This table is sortable by any of these elements, which provides a rapid way of finding the highest resolution structure or the one with the highest sequence identity. Quite frequently, a multi-domain protein may have homologues of known structure that cover non-overlapping regions of the target sequence. The table can be sorted by beginning or ending residue numbers of the target sequence in order to locate templates that cover the domain of interest among a list of possibly hundreds of hits. The template for modeling is chosen based on the best combination of a number of factors. First, it must cover the region or regions of interest in the target protein. From among those hits, usually the best E-value or highest sequence identity template is chosen. When there are a number of templates with about the same evolutionary distance from the target sequence, the one with highest resolution is usually the best choice of template. Another consideration may be the number and positions of gaps in the sequence alignment. Often one template or another may contain a ligand or binding partner of interest while others will not. This ligand may be nucleic acid, small organic molecules, ions, or other proteins. Our database program ProtBuD41 can be used to search within a protein family for particular ligands. Structures of such complexes may be used to identify important binding residues in the model of the target.
With a simple click within the list of templates, MolIDE downloads the template structure from the PDB and then displays the structure of the template and the sequence alignment of the target and template. The predicted secondary structure of the target is shown above the target sequence and the experimental secondary structure of the template is shown below the template sequence. At this point, the alignment can be manually edited. PSI-BLAST alignments are reasonably accurate at sequence identities above 30%42, but even then the positions of gaps of insertions or deletions in the alignment may be placed within regular secondary structures of the template. The visualization tool within MolIDE allows the user to see the placement of deletions from the structure (deleted residues marked by red balls) and insertions into the structure (marked by yellow balls at the point of insertion). The gaps can be moved within the alignment as described in the protocol and the changes are marked in real time on the structure. For deletions from the structure, it is best if the end points of the deletion are relatively close to one another in space on the template structure.
Once the alignment has been edited, the modeling process consists of three steps performed within the MolIDE graphical interface. The first is simply to copy the backbone coordinates to a new file and renumber the sequence and the residue names to the target sequence according to the alignment. Only aligned residues are copied. If the residue types are the same at a given position in the target and template sequences, the side-chain coordinates are also copied to the new file. If the template PDB contains modified residues (selenomethionine or phosphorylated residues), only the backbone is copied. The second step is to run SCWRL to build the side chains of the target sequence onto the model backbone. SCWRL is able to preserve the Cartesian coordinates of side chains that are conserved, and this generally results in more accurate side-chain prediction in homology modeling. The third step is to model each loop in turn. MolIDE uses the program Loopy43 which is a relatively fast program for loop structure prediction. It is one of the few stand-alone loop-modeling programs. To model the first loop, the user selects positions for the left and right anchors of the region to be remodeled. These are positions in the structure that will be kept fixed while the residues in between will be modeled by Loopy. The left anchor can be chosen as the last residue of the secondary structure preceding the gap, while the right anchor is the first residue of the secondary structure following the gap. Alternatively, longer or shorter regions may be chosen in order to choose the anchor positions as the closest conserved residues to the gap. Once the anchors are set, the loop can be modeled with a click. The anchors are then set around the next gap in the same manner and so on, until all the insertion and deletion regions have been remodeled with Loopy.
If a protein is very long (>500 amino acids) and contains multiple domains and long disordered regions, it is sometimes helpful to use target sequences of the single domains of interest. Many proteins contain long regions that are intrinsically disordered. Several web servers are available to predict these regions, including DISOPRED44. Such regions can be removed from the target sequence before the PSI-BLAST search step.
The homology modeling procedure provided by MolIDE is simple and straightforward. It produces models assuming that aligned regions of the target to the template do not change backbone conformation. While this is not in general true, it is a reasonable approximation when the sequence identity is above 30%. Even below this value, the model is still useful in understanding the relative positions of residues in the protein. In any sequence alignment, some regions are more conserved than others, and these regions usually have functional or structural significance. Thus, even such a simple modeling procedure will predict whether residues are on the surface or buried: mutation of buried residues may lead to unfolding, while mutations on the surface may abolish binding to other molecules. For protein-protein interactions, a patch of conserved surface residues may be close together on the surface but far apart in the sequence. Such a conserved patch may be used to locate likely binding surfaces, which can be tested using site-directed mutagenesis.
It should be noted that there are web servers that will also produce homology models, including SwissModel45 as well as databases of models, including ModBase46. These are certainly valid alternatives to performing homology modeling with SCWRL and MolIDE. However, there are many choices in homology modeling that a user may wish to make with consideration of particular biological questions in mind, and MolIDE is designed to allow the user to make these choices while handling the nitty-gritty computational steps with a few clicks. This is especially true in the choice of template. Some templates may be preferred because they contain ligands of interest, including other proteins, small molecules, or nucleic acids. Some templates may have better conservation near residues that the user is interested in, for instance given existing mutation data. Other templates may have fewer insertions or deletions in regions of interest, for instance near an interface. Further, MolIDE provides user interaction during the model-building process that may be highly beneficial. This is especially true of manual editing of the target-template alignment using additional information that the user may possess, including multiple sources for target-template alignment and visualization of the template structure. Choosing the positions where loop modeling begins and ends (the loop anchors) by visualizing the structure may also lead to better structure predictions.
MATERIALS
Equipment Setup
Computer
Any computer running Windows XP or Vista or Linux may be used. PSI-BLAST will run more quickly with 512 MBytes of RAM or more.
Software
All of the software and various components described here can be downloaded from http://dunbrack.fccc.edu as described in the procedures below.
Databases
The sequence databases required can be downloaded from our web site and publicly available websites as described in the procedures below.
PROCEDURES
SCWRL: Obtaining and installing SCWRL
1 | From the SCWRL webpage, http://dunbrack.fccc.edu/SCWRL3.php, follow the link labeled “Download” and fill out the license form. SCWRL is free to non-profit institutions. Commercial institutions should contact Roland.Dunbrack@fccc.edu. Fill out the form and click the “I agree” button at the bottom of the page. This leads to a verification page for the input information. Click “Send request.” The request is sent to the Fox Chase Cancer Center for approval. On approval, the user will receive an e-mail message with the subject heading “SCWRL3.0 Download.” Click the link in this e-mail message to obtain SCWRL3.0 for various platforms, including Windows (both XP and Vista), Linux, Mac OS X, SGI Irix, and SunOS. Click “download” to begin downloading of an archive that contains the SCWRL program and the binary rotamer library file used by SCWRL.
The installation kits for each operating system have the following names respectively:
scwrl3_win.msi
scwrl3_lin.tar.gz
scwrl3_mac.tar.gz
scwrl3_sgi.tar.gz
scwrl3_sun.tar.gz
2 | The procedure for installing SCWRL is slightly different on Windows (Option A) and the Unix-related platforms (Option B).
(A) For Windows XP and Vista
Double-click on scwrl3_win.msi and follow the instructions in the installer. By default, the installer will place SCWRL3 in the folder C:/FCCC/scwrl3_win/. This is where MolIDE expects to find SCWRL, and so placing it in this location makes installation of MolIDE simpler.
(B) Unix-based operating systems (Linux, MacOS X, SGI Irix, and SunOS)
(i) Move the archive to a location on your hard drive where you want to keep the SCWRL program. From a terminal window, give the following commands (for example, for the Linux distribution):
cd scwrl_path/
gzip -d scwrl3_lin.tar.gz
tar -xvf scwrl3_lin.tar
where “scwrl_path/” is the name of the directory that contains the file scwrl3_lin.tar.gz. Ordinarily on Linux systems, a typical directory for SCWRL might be /usr/local/bin.
This previous step will create a new directory, scwrl3_lin/, and unpacks four files and a folder into that directory:
BBDep.bin
setup
scwrl3_
README.scwrl3
examples/
On the command line of the terminal window, now type:
cd scwrl3_lin
./setup
This command will modify the executable file scwrl3_ and move it to the filename scwrl3. This executable now contains within it the location of the rotamer library file, BBDep.bin. The executable can be moved elsewhere on the computer and can be executed in any directory, as long as the rotamer library remains in its location where ./setup was run. If you decide to move the BBDep.bin to a different location, repeat the installation procedure for that directory, beginning with uncompressing the kit.
SCWRL: Using SCWRL from the command line
3 | SCWRL is a command-line program. That is, a command must be issued from a console window on Windows (Option A) or a terminal window on Unix systems (Option B) or a
(A) Windows
Open a console window by selecting “Command Prompt” from the “Start” menu. On some systems, the Command Prompt may be found by selecting “All Programs” from the “Start” menu, and then selecting “Accessories” and then “Command Prompt.”
(B) Unix systems
Unix systems vary on how to open a terminal window. On Mac OS X, for example, the Terminal program is located in the Utilities folder within the Applications folder. Consult your system administrator for assistance if necessary.
4 | SCWRL may be used in various ways using some optional flags on the command line. SCWRL can predict side-chain conformations for an input backbone structure without modification of the sequence. In this case, SCWRL removes all side-chain atoms from the input file (if any), and rebuilds all of the side chains according to the residue names of the backbone atom coordinates (Option A). SCWRL can predict side chain conformation in the presence of non-protein atoms, such as ions, ligands, and nucleic acids. The ligand atoms are treated only with a simple steric repulsive energy function, so that the predicted side chains will not overlap the ligand atoms. SCWRL determines the element (N, C, Zn, Mg, etc.) from the atom name, and assigns a radius to each atom based on its element type. The procedure for modeling in the presence of ligands is as described in Option B. SCWRL can change the sequence of the input file by reading an additional file containing the new sequence. This sequence file should contain one-letter codes for the new sequence, and must contain exactly the same number of residues that the input PDB file contains. If it does not, SCWRL will report an error. The new sequence is placed on the backbone, retaining the input chain identifiers (A, B, C, etc.) and residue numbering. The input file may contain multiple chains, as long as the input sequence file contains the new sequence for each chain in the same order as the input coordinate file. The input sequence file also may contain information to indicate whether the Cartesian coordinates of the side chain in the input file should be kept. This is useful in homology modeling, since better predictions will usually be produced when conserved side chains (same residue type in the target and template sequences) are kept fixed during side-chain prediction (Option C).
(A) Running SCWRL without modifying the sequence and without ligands
Prepare the file containing the backbone coordinates. This is a PDB format file, which typically looks something like the example shown in Figure 1. The element types on the end of the line are ignored and are not necessary in the input. The chain ID (“A”) is also not necessary, as long as this character is replaced by a space. The spacing is critical and should adhere to the standard PDB format (see http://www.wwpdb.org/docs.html). For instance, the atom names for the backbone atoms (N, CA, C, O) should begin in column 14 and are left-justified. The three-letter residue type of the 20 standard amino acids begins in column 18. All other residue types are ignored. The residue number is right-justified with the right-most digit in column 16. The last two numbers on the line are the occupancy (usually 1.0) and the atomic B-factor or temperature factor. SCWRL ignores these, replacing the occupancy with 1.0 and the B-factor with 0.0. The file can contain REMARK records or other record types besides the ATOM records of the x,y,z coordinates of the backbone. These other record types will also be ignored. This file can have any name or extension, although typically PDB format files have the extension .pdb or .ent.
- To predict the side chains for the input backbone conformation, issue this command in the terminal window or console window:(for Windows)
(for Unix systems)scwrl_path\scwrl3.exe -i inputpdbfile -o outputpdbfile > logfile
The filenames can be any desired names for the input file (“inputpdbfile”), the output PDB-format file (“outputpdbfile”), and the log file (“logfile”). So for example, the actual commands on Windows or Unix systems, respectively, might be:scwrl_path/scwrl3 -i inputpdbfile -o outputpdbfile > logfile
The log file will contain some output from SCWRL about how the prediction problem was solved, including details about the graph theory algorithm process as described in the paper29. For most users, the information in this file is not relevant.C:\FCCC\scwrl3_win\scwrl3.exe -i myfile.pdb -o mymodel.pdb > mylog.log/usr/local/bin/scwrl3_lin/scwrl3 -i myfile.pdb -o mymodel.pdb > mylog.log
(B) Running SCWRL in with input ligand coordinates
Prepare the input files. The backbone file is prepared in the same way as in part (A)(i). Place the ligand coordinates into a separate PDB-format file. These might come from an experimental structure that contains non-protein atoms, and can simply be cut and pasted into a new file, which we call the frame file. These ligand coordinates must be in the same Cartesian frame as the inputpdbfile. That is, they must be in the right location in space. This would already be true, for instance, if the coordinates in inputpdbfile and framefile come from the same PDB entry. If they do not, then the desired template (inputpdbfile) can be superimposed onto a structure of a protein containing the ligand of interest (or vice versa). The FATCAT server47, for instance, will take two PDB files and calculate the structure alignment and provide the coordinates of protein A (and its ligands) rotated and translated in order to superimpose onto protein B, or vice versa.
- Type the appropriate command into the console or terminal window:(for Windows)
(for Unix-based systems)scwrl_path\scwrl3.exe -i inputpdbfile -o outputpdbfile -f framefile > logfile
where framefile contains the ligand coordinates.scwrl_path/scwrl3 -i inputpdbfile -o outputpdbfile -f framefile > logfile
(C) Running SCWRL with an input sequence different from the input coordinates
- Prepare the input files. The backbone file is prepared in the same way as in part (A)(i). The sequence file is a text file containing the new sequence. To indicate that the input side-chain coordinates should be kept, the residue type is put in lower-case. If SCWRL is to predict the side-chain coordinates, the residue is in upper-case. SCWRL will ignore spaces, carriage-returns, and numbers in the sequence file. For instance, to remodel the side chains in the protein crambin, PDB entry 1CRN, but keeping the cysteines fixed in their input coordinates, the sequence file would appear as:
If the lower-case residue type does not agree with the input PDB file residue type in the same position within the structure, then the input PDB file residue type will be used and the coordinates will be predicted by SCWRL.TTccPSIVARSNFNVcRLPGTPEAIcATYTGcIIIPGATcPGDYAN - Type the appropriate command into the console or terminal window:(for Windows)
(for Unix-based systems)scwrl_path\scwrl3.exe -i inputpdbfile -o outputpdbfile -s sequencefile > logfile
where sequencefile contains the new sequence.SCWRL can combine the optional sequencefile and framefile by using both flags on the command line. It has two further flags, -u and -d. The flag -u tells SCWRL not to predict disulfides. This is useful for proteins in reducing environments, especially if they contain cysteines in proximity of one another around a metal ion such as zinc. The other option is -d, which tells SCWRL to print out a file with the dihedral angles of the predicted structure in a file called outputpdbfile.dihed. The order of flags is not important.scwrl_path/scwrl3 -i inputpdbfile -o outputpdbfile -s sequencefile > logfile
Homology modeling with MolIDE
Obtaining and installing MolIDE (version 1.6)
5 | From the MolIDE webpage, http://dunbrack.fccc.edu/molide, follow the link labeled “Download” and fill out the license form. MolIDE is free to both non-profit and commercial institutions. Fill out the form and click the “I agree” button at the bottom of the page. This leads to a verification page for the input information. Click “Send request.” The request is sent to the Fox Chase Cancer Center for approval. On approval, the user will receive an e-mail message with the subject heading “MolIDE Download.” Click the link in this e-mail message to obtain MolIDE for either Windows or Linux. Click “download” to begin downloading of an archive that contains MolIDE and its associated files. Because of incompatibilities in the wxWindows framework used in MolIDE to build a Windows/Linux cross-platform graphical user interface, MolIDE is not available for other systems, such as Mac OS X, although it can be installed on Intel-based Macs that are running Windows, and run from the Windows operating system.
The installation kits for each operating system have the following names
molide1.6_win.msi
molide1.6_lin.tar.gz
6 | Installing and setting up MolIDE on Windows (Option A) and Linux systems (Option B) follow different procedures. The procedure on Windows is significantly simpler.
(A) Installing MolIDE on Windows XP or Vista
MolIDE on Windows now comes with an automatic installer. Just double-click on the icon for molide1.6_win.msi, and follow the instructions of the installer. By default, the installer will place MolIDE and its associated files in the directory C:\FCCC\MolIDE.!Caution. MolIDE cannot be installed in folders with names that contain space characters. This is due to the Unix-based programs that are part of the MolIDE distribution; these programs cannot handle spaces in file names or file paths. On Windows, the default folder for applications is usually “C:\Program Files.” However, because of the space, MolIDE will not work if located in this folder.
To start MolIDE, double-click on the MolIDE icon in C:\FCCC\MolIDE\molide.exe.
(B) Installing and setting up MolIDE on Linux (i)
-
Move the archive file, molide1.6_lin.tar.gz, to a location on your hard drive where you want to keep the MolIDE archive. From a terminal window, give the following commands:
where “molide_path/” is the name of the directory that contains the file molide1.6_lin.tar.gz. Ordinarily on Linux systems, a typical directory for MolIDE might be /usr/local/bin.cd molide_path/gzip -d molide1.6_lin.tar.gztar -xvf molide1.6_lin.tarThe previous step will create a new directory, molide1.6_lin/, and unpacks directories and files into that directory.
- MolIDE uses the wxWindows cross-platform library, which must be installed. For convenience we have included in the Linux distribution the rpm files for version 2.4.2-1, located in the subdirectory “molide_path/molide1.6_lin/wx”. To install the wxWindows library on Linux, the user must be logged into the computer as “root.” If you do not have root privileges on your machine, ask a system administrator to perform this step. Type the following in a terminal window:
!Caution. If a later version of wxWindows is already installed, the system may return error messages with the previous command. To override this, the flag “-f” can also be given in the rpm command. However, this may compromise other programs on your system that may use wxWindows. It is unlikely that most users will face this problem, because wxWindows is not that common on Linux machines.cd molide_path/molide1.6_lin/wxrpm -U *.rpm - To set up MolIDE, type the following in a terminal window:
! Caution If you already have the NCBI package for BLAST installed on your machine, make back-up copies of the file .ncbirc in your home directory, if it exists. It will be overwritten by setup.cd molide_path/molide1.6_lin/./setup To start MolIDE, on the command line of a terminal window, type:
cd molide_path/molide1.6_lin/
./molide
7 | MolIDE uses PSI-BLAST38, PSIPRED40, SCWRL29, and Loopy43 to perform the basic steps in homology modeling. MolIDE comes with PSI-BLAST, PSIPRED, and Loopy in default locations. However, SCWRL must be installed as a separate step since it requires a separate license. It does not matter which program is installed first. After installing MolIDE on each system, SCWRL must be installed if it is not already installed and the location of SCWRL must be set within MolIDE. To obtain and install SCWRL, follow the instructions above. On Windows, the SCWRL installer will place SCWRL within C:\FCCC\scwrl3_win. This default location is already set in MolIDE on Windows. In both Windows and Linux, from within the Tools menu, select “Options” and then “Scwrl.” The location of the executable for SCWRL can be entered into the box using the “browse” button. If it is already correct, then this step can be skipped.
8 | MolIDE depends on two sequence databases for producing homology models. The first is the non-redundant protein sequence database, “nr,” from NCBI, currently about 6 million sequences. This sequence database is used to produce sequence profiles for the target sequence based on multiple sequence alignment of many homologues. The second is the PDB protein sequence database, “pdbaa,” which must be obtained from our website. This version of pdbaa is different from NCBI's version in a number of ways. It is more up-to-date than NCBI's, and contains additional information on the header line for each sequence, including experiment type, resolution, sequence length, and R-factors. It also has distinct names for different sequences in a single PDB file, based on the gene name for that protein.
The PDB is updated weekly on Wednesdays, and the pdbaa database is updated on our website within a couple of days. The pdbaa database within MolIDE therefore can be updated as often as weekly when MolIDE is in use. Over 100 structures are added to the PDB every week. To update pdbaa, select from the Tools menu, “Update DB”. A window appears with the option to update either “PDBAA” or “NR.” Select “PDBAA,” click “OK”, and then “Download.” The PSI-BLAST formatted files will be installed in the appropriate location.
After MolIDE is installed for the first time, download the nr database via the “Update DB” option in the Tools menu. The nr sequence database will be downloaded from the NCBI ftp site, and automatically formatted for PSI-BLAST using the NCBI program formatdb, included with MolIDE. Depending on download speed, it may take 30 minutes or more to download nr, and 10 minutes or more to uncompress it and format it for PSI-BLAST. All of this will be done automatically by MolIDE. The nr database can be updated periodically. A monthly update is more than sufficient.
Other databases may be used instead of the nr database from NCBI. For instance the UniRef databases from UniProt are suitable. To use the uniref100 database from uniprot:
under Tools->Servers, change “NR ftp Server” to ftp.uniprot.org;
under Tools->Servers, change “NR ftp server Path (.gz file)” to /pub/databases/uniprot/current_release/uniref/uniref100/uniref100.fasta.gz”
under Tools->Psiblast, change “NR Seq DB File and Path” to C:\FCCC\MolIDE\db\nr\uniref100 or wherever the uniref100 database is located
!Caution. The PDB sequence database pdbaa must be obtained from our website for use in MolIDE rather than NCBI's file of the same name.
? TROUBLESHOOTING
Modeling with MolIDE : Running PSI-BLAST and PSIPRED
9 | Prepare a target sequence for modeling. This should be placed in a single file in FASTA format with the extension “.seq”. Such a sequence can be obtained from NCBI by keyword search. NCBI's site can format a sequence in FASTA format. The target sequence file should look something like this:
>P53 [Homo sapiens]
MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDIEQWFTEDPGPDEAPRMPEAA PRVAPAPAAPTPAAPAPAPSWPLSSSVPSQKTYQGSYGFRLGFLHSGTAKSVTCTYSPALNKMFCQLAKT CPVQLWVDSTPPPGTRVRAMAIYKQSQHMTEVVRRCPHHERCSDSDGLAPPQHLIRVEGNLRVEYLDDRN TFRHSVVVPYEPPEVGSDCTTIHYNYMCNSSCMGGMNRRPILTIITLEDSSGNLLGRNSFEVHVCACPGR DRRTEEENLRKKGEPHHELPPGSTKRALSNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALEL KDAQAGKEPGGSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD
A FASTA-formatted sequence file includes “>Name” as the first line in the file with the sequence following, starting on the next line. The sequence can be spread over as many lines as necessary and the lines can be of any length. Other text can follow the name on the first line, and there should only be one sequence in the file. Spaces and numbers within the sequence will be ignored.
Within MolIDE, open a sequence file via the File menu, selecting “Open” and then “Sequence.” In what follows, we describe choosing items under one of the menus with the following notation, as in this case: “File->Open->Sequence”.
10 | While a sequence file is open, run a multiple-round PSI-BLAST by selecting PSIBLAST from the Tools menu. PSI-BLAST is run first against the non-redundant protein sequence database (or a database of the user's choosing that can be set under Tools->Options->PSIBLAST) with a customized version of PSI-BLAST that comes with MolIDE. This version of PSI-BLAST outputs profiles after every round (including the last round), each with a unique name. Once the non-redundant sequence database search is completed, the PDB is automatically searched with each of the profiles and a separate PDB alignment file is created.
You can change other parameters used during the PSI-BLAST runs using Tools->Options->PSIBLAST. For instance, for common protein domains such as kinases or immunoglobulins, it may be desirable to use a stricter E-value cutoff for creating the profiles in PSI-BLAST. If you know that you have close homologues to your target sequence in the PDB, then rather than searching nr to create profiles, you can use pdbaa for this step as well.
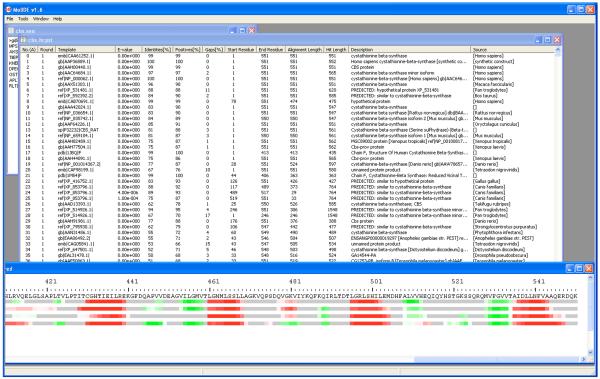
Once PSI-BLAST is finished, close the PSI-BLAST run window by clicking “Done.” The PSI-BLAST output of the search against the non-redundant protein database can be opened by choosing “Open->Seq HITS Alignment”. A window opens with a table of all the hits found in the non-redundant database. This is shown in Figure 3.
Figure 3.
Viewing PSI-BLAST output from the non-redundant sequence database search and secondary structure predictions with PSIPRED within MolIDE. The target sequence file remains open (upper left) and the table of PSI-BLAST results from the non-redundant database is shown (upper right). This table can be sorted by clicking on any of the headers at the top of the table, once for ascending order (A) and once again for descending order (D). The secondary structure predictions are shown in the lower window. Predictions are shown in red (helix), green (sheet strand), and coil (gray). The intensity of the color is in proportion to the confidence values (from 0 to 9) given by PSIPRED. The region shown is for the C-terminal Bateman domain49 of the protein cystathionine beta synthase (CBS). Each line contains a secondary structure prediction from a round of PSI-BLAST. While the longer helices are well predicted, the beta sheet strands change as more remote homologues are added to the multiple sequence alignment used by PSI-BLAST (top to bottom). The beta sheet strand around 510 is predicted in the first 3 rounds but fades out at round 4. In fact, the early predictions are more likely to be accurate in this case than the earlier ones50.
? TROUBLESHOOTING
11 | Run PSIPRED to predict the secondary structure of the target by selecting Tools->PSIPRED. PSIPRED uses the output PSI-BLAST sequence profiles from the nr database search. PSIPRED should therefore be run only after the PSI-BLAST run in the previous step is completed. A secondary structure prediction for each round of PSI-BLAST is created.
To view the secondary structure predictions, select File->Open->Sec Struct Pred. After choosing a “.psipred” file, you are given the option to display all of the predictions based on the matrices generated by PSI-BLAST after each round. These are displayed in a single window with each prediction on a separate line. A predicted sheet is colored in green and a predicted helix is in red. Predicted coil regions (loops and unstructured regions) are depicted in gray. A screenshot of predicted secondary structure from several rounds of PSI-BLAST is also shown in Figure 3.
The intensity of the color is proportional to the prediction confidence; the darker the color, the higher the prediction confidence. This view allows you to see if the secondary structure prediction changes as more remotely related sequences are added to the profiles. For proteins with few close relatives, the predictions may be more accurate in later rounds as distantly related sequences provide information on likely secondary structure patterns. However, for proteins with a good number of close relatives (>50), the addition of distantly related sequences with potentially large structural changes (additional secondary structure or missing secondary structure) may degrade the secondary structure prediction.
Modeling with MolIDE: Choosing a template and editing the alignment
12 | Open the PSI-BLAST file containing the alignments of your query sequence with sequences of proteins from PDB. There will be a separate hits file for each round of PSI-BLAST. Open a file of hits from the PDB by selecting “Open->PDB Hits Alignment”. Only files with the correct extension, .pdbout, will be shown.
The results are displayed as a table, as shown in Figure 4. To sort the table by some feature, click on the column header of that feature, such as E-value, sequence identity, or starting and ending positions of the alignment. Clicking again on the same column header will reverse the sorting order for that column. In a multi-domain protein, for instance, it is common to have many available templates for one domain, but only a few templates for a different domain. These are hard to locate in the raw PSI-BLAST output text file, but the sorting feature in MolIDE makes them easy to find.
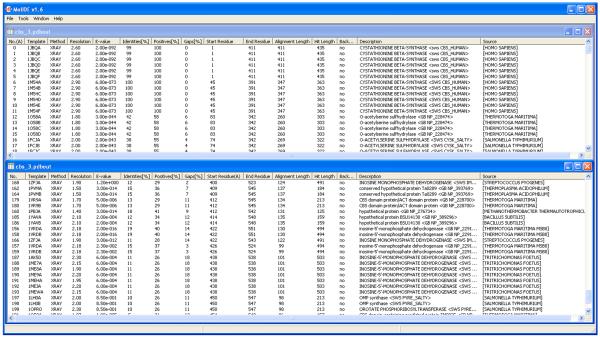
Figure 4.
Viewing and sorting the list of templates. The hits shown are from a PSI-BLAST search of pdbaa from profiles built from searching NCBI's nr database. The query is the same as in Figure 2, human CBS. The top window is sorted by hit number (the order in the PSI-BLAST output), and shows that there is an experimental structure of human CBS that covers residues 1 to 413 of the query (length 551). PDB entry 1JBQ is a longer structure than PDB entry 1M54. The bottom window is sorted by starting residue in the query (A=ascending order; D=descending order). The window shows that there are a number of templates for the C-terminal regulatory Bateman domain. From this window, target-template alignments can be viewed by double-clicking on the hit number in the first column (“No.”). Some Bateman domains in the PDB are insertions into inosine monophosphate dehydrogenase (IMPDH). The Bateman domains in these structures are largely disordered. This is evident once the target-template alignment is opened up. After looking through this list, and examining them visually, the PDB entry 1PVM51 appears to be the best template for the Bateman domain of CBS. It has only 2 gaps in the alignment, and is a high-resolution structure (1.9 Å) with no missing residues in the aligned region.
Because there is a file of hits for each round of PSI-BLAST, one may ask which file should be used. There are no strict rules for determining this. A rough rule of thumb would be the earliest iteration that produces a complete alignment of the target sequence or domain of the target to a known structure. For example, if the target is a single-domain protein, then the earliest round that aligns the entire target domain to a complete domain of known structure should be used. In many cases, the first or second round may only align a central well-conserved region, but not the whole domain. In that case, later rounds should be examined. This can be determined by observing the alignment and the template structure simultaneously, as detailed in the next step.
13 | While sorting the table enables the user to locate the best templates by resolution, sequence identity, and other features, viewing the alignment of the target to a possible template is a key step in this choice. From the hits table, double-click on the hit number in the first column for a template of interest. The alignment of the target to that structure in the PDB will be extracted from the PSI-BLAST output file, and saved in a separate file in the same directory where the sequence file resides. The extension of this file is .alnonet, which stands for “alignment with onetemplate.”
At the same time, a window will appear for downloading the coordinates of the template structure from the PDB. Click “Download.” The default ftp server, ftp.wwpdb.org, should work well (at times it may be slow), but the user can change the ftp server by selecting Tools->Options->Servers from the Tools menu. MolIDE uses the XML-format files from the PDB and converts these to PDB format. It also extracts information from the XML file on how the template sequence (numbered from 1 to N, its length) corresponds to residues in the coordinates. PDB files are sometimes missing coordinates due to disorder, and the coordinates may be numbered starting on any number. This information is critical in converting a template into a target model given an alignment, and MolIDE handles it automatically from information contained in the XML format. This information is not present in the PDB's PDB-format files. For a PDB entry with accession code 1abc, for example, the coordinates will appear in file 1abc.pdb and the sequence-coordinate residue correspondence will appear in file 1abc.sc.
Once the XML file is downloaded, read, and converted, a window appears with a view of the target-template sequence alignment, the secondary structure prediction of the target, and the experimental secondary structure of the template. Above the alignment, the backbone of the template structure appears. The whole template protein is displayed in gray, while the part of the structure used in the alignment is displayed in green. Insertions in the target (target longer than template) are marked by 2 adjacent yellow spheres on the template structure Cα atoms surrounding the insertion point. Deletions from the template (target shorter than template) are represented by red spheres on the Cα atom of residues to be deleted from the template structure.
Viewing the alignment and the structure simultaneously allows the user to determine visually whether the alignment covers an entire template domain and to identify where insertions and deletions are located on the structure. MolIDE's viewer is quite simple and is not suitable for making images for publication. Its purpose is to examine and edit the target-template sequence alignment. The PDB files produced by MolIDE can be read into any molecular viewer, such as PyMol (W. L. Delano, http://pymol.org).
To manipulate the structure view in MolIDE:
| Left_button_drag | Rotates the structure |
| Right_button_drag | Zoom in and out (Z-direction) |
| Middle_button_drag | Move in plane of screen (X-Y plane) |
| Double_left_click | On an atom in the structure displays the residue numbers at the bottom of the window |
| Left_click | Show spacefill in template viewer |
| Middle_click | Spacefill additional residues in viewer |
In the View Menu, the options are:
| Backbone | Displays connected Cα atoms |
| Spacefill | Displays spheres on each atom |
| Aligned fragment | Displays only that part of the template structure that aligns with the template |
| Whole template | Displays the entire template with the part that is aligned in green and the rest in gray |
? TROUBLESHOOTING
14 | Generally it is a good idea to edit the target-template sequence alignment manually. An example of alignment editing is shown in Figure 5. Deletions from the structure are least disruptive if the N- and C-terminal endpoints of the deletion are nearby each other in space. Insertions are best placed in the middle of loop regions, not immediately next to regular secondary structure. The correspondence of predicted secondary structure of the target and the experimental secondary structure of the template can be used to guide the alignment. Often PSI-BLAST may fail to align some regions correctly, so if there is other information available, on conserved residues for instance, then the alignment can be edited accordingly. For sequence identities below 30%, it is advisable to seek alternative alignments from servers that provide profile-profile alignments, which are generally more accurate than PSI-BLAST. One of the best and most usable of these is FFAS7. The MolIDE alignment can be edited according to the alignment provided by FFAS.
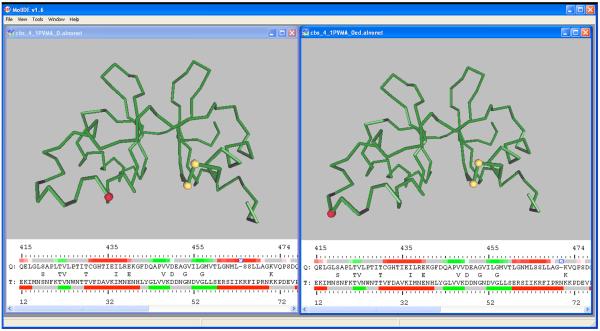
Figure 5.
Manual editing of a target-template sequence alignment. The unedited alignment is shown at left, and the partially edited alignment is shown at right. In each image, the alignment shows the target sequence and its predicted secondary structure above the template sequence and its experimental structure, with the same color coding as in Figure 2. The original alignment (left) shows a gap in the middle of an alpha helix. To edit the alignment, a ctrl-click on the gap in the left figure removes the gap. Shift-click on a position six residues to the right inserts a gap, and produces the alignment at right. The editing does not reduce the similarity of the aligned residues, and is more consistent with structural changes in homologous proteins.
Moving the mouse over the alignment will display in the status bar at the bottom of the window the sequence numbers for query and template sequences, as well as the corresponding PDB coordinate residue number in the template PDB. The color-coding scheme for the secondary structure of the template is the same one used for the secondary structure prediction (helix=red; sheet=green). The third column of the status bar displays the number of identities in the alignment.
To move a gap over several residues, delete it first, then move to the place of insertion and insert the appropriate number of gap characters as follows:
| Shift+Left_click | Insert gap |
| Ctrl+Left_click | Delete gap |
These operations can be performed on either the target sequence or the template sequence. Only gap characters can be inserted or deleted.
Modeling with MolIDE : Building the model
15 | Once the alignment editing is completed, choose “Copy backbone” from the Tools menu. This step produces a file with extension .model that contains a model of the target sequence based on the aligned residues in the current target-template alignment window. Side chains for conserved residues (identical and aligned in the target and template alignment) are also copied to the model.
16 | Select “Build Side Chains (SCWRL)” from the Tools menu. Click “Run SCWRL” in the window that appears. The conserved side chains are left in the original conformation from the template crystal structure. This option can be changed with Tools->Options->Scwrl. SCWRL should run very quickly (seconds). If it takes a long time (>5 minutes), the run should be canceled in the window, and another template selected. This occurs when the backbone of the template will not accommodate the target side chains very easily.
17 | Loop building is done by first selecting residues for the left and right anchors. These are residues that will be kept fixed while the intervening sequence is modeled using the Loopy program43. It is usually a good idea to allow at least 2-3 residues on either side of the insertion or deletion to move during the loop-building process. One option is to make the left and right anchors the last and first residues of the flanking secondary structures respectively. However, if part of a long loop is well conserved, it may be better to select a smaller region that contains less conserved segments. Loopy will sometimes be unable to build a loop if the loop length is too short and the distance to be spanned by the predicted loop is too large. In this case the anchors should be reset (cleared) and then selected again further apart and Loopy should be run again.
Also note that if residues are missing from the structure due to poor electron density, they will be marked with blue squares below the template sequence. These regions should also be rebuilt with Loopy.
To build loops: Right_Click on a Query residue in the sequence alignment will display a pop-up menu:
Set Loop Left Anchor
Set Loop Right Anchor
Reset Anchors
Build Loop
!Caution. Click on the query sequence not the template sequence to select anchors, to reset the anchors, and to build the loop.
After choosing the loop's anchor residues, proceed with “Build Loop”. Click “Run Loopy” in the window that appears. Loopy should take less than a minute or so to build the loop for loops up to lengths 15 residues.
Proceed with loop building of each insertion-deletion region in turn until all the insertions/deletions/missing residues are modeled. The model is contained in a PDB-format file. The file name follows this convention: ProteinName_x_TemplatePDBChain_y.pdb where x is the round number of PSI-BLAST run and y is the fragment number of the query sequence that is aligned with that particular template PDB. This file is first generated after the side chains are built with SCWRL3. It is subsequently overwritten by Loopy output after each loop is built. When all loops are built, this file will contain the final homology model.
Adding Ligands to the Model (Optional)
18 | It may be desirable to remodel the side chains in the presence of a ligand using SCWRL on the command line. MolIDE creates a second sequence file when the “Copy backbone” command is given. This sequence file contains the complete target sequence over the region of the target-template alignment. So once loops are built, this sequence file can be used as input to SCWRL. To perform this step, first copy and paste the ligand coordinates of interest from the template PDB file produced by MolIDE into a new file, called a frame file (also see SCWRL instructions above). The sequence file has extension .s3seqall. To remodel the side chains with SCWRL, type this command in the console window (on Windows) or the terminal window (on Linux):
(Windows)
cd modeling_directory\
scwrl_path\scwrl3.exe -i inputpdbfile -o outputfile -f framefile -s file.s3seqall > logfile
(Linux)
cd modeling_directory/
scwrl_path/scwrl3 -i inputpdbfile -o outputfile -f framefile -s file.s3seqall > logfile
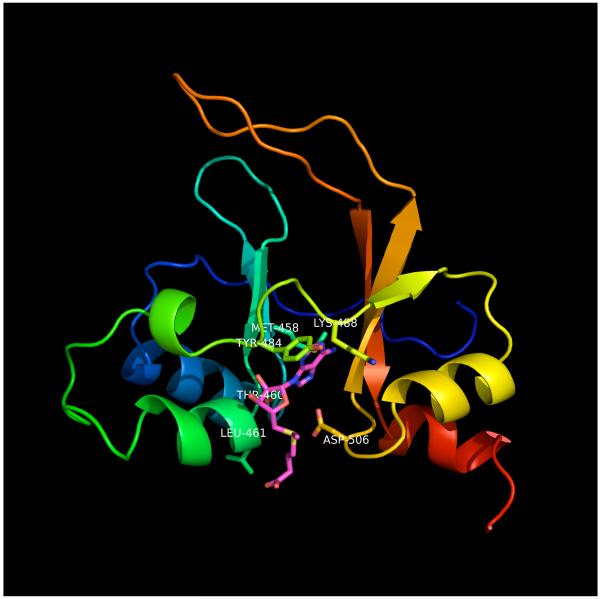
where framefile contains the ligand coordinates. An example of this is shown in Figure 6.
Figure 6.
Image of model of Bateman domain of human cystathionine beta synthase with S-adenosyl methionine (SAM). Because the template, 1PVM, does not contain SAM or any other nucleotide, the model of CBS was superimposed on PDB entry 2YZQ (Kanagawa et al., unpublished), which does contain SAM. The side chains of the model were then remodeled as described in Step 14. Side chains near SAM are shown in stick representation. SAM is colored magenta. The backbone ribbon is colored from blue to red from N to C terminus. The image was produced with PyMOL (W. L. Delano, www.pymol.org).
Timing
To give a brief idea about the length of the homology modeling procedure with SCWRL and MolIDE, we list below the number of minutes required in each step. The estimated time is based on our modeling experience on a machine with an AMD Dual Core Processor and 2GB RAM, given a query sequence of 200 ~ 500 residues.
Step 1: 2 ~ 5 minutes.
Step 2: 1 ~ 2 minutes.
Step 3: < 1 minute.
Step 4: 1 ~ 2 minutes.
Step 5: 2 ~ 5 minutes.
Step 6: 1 ~ 2 minutes.
Step 7: 1 ~ 5 minutes.
Step 8: 20 ~ 40 minutes
Step 9: < 1 minute.
Step 10: 10 ~ 20 minutes.
Step 11: < 1 minute.
Step 12: 1 ~ 2 minutes.
Step 13: 1 ~ 2 minutes.
Step 14: 5 ~ 10 minutes.
Step 15: < 1 minute.
Step 16: 1 ~ 2 minutes.
Step 17: 1 ~ 10 minutes.
Step 18: 1 ~ 2 minutes.
ANTICIPATED RESULTS and LIMITATIONS
The accuracy of protein structure prediction depends critically on sequence similarity between the target and the template. When the sequence identity is higher than 30%, usually most or all of the alignment is correct, and the relative positions of structural elements are therefore reliable. Below 30%, this is no longer true, and there may be significant changes in structure between the template structure and the target structure (if it were known). On native backbones SCWRL3 is able to predict about 83% of side chains with the first dihedral angle of the side chain (χ1) within 40° of the experimental structure29. At 50% sequence identity between target and template and keeping conserved side chains fixed according to the template structure, SCWRL3 predicts about 72% of side chains correctly (unpublished data).
MolIDE provides a simple and fast modeling procedure based on sequence alignments with PSI-BLAST. At sequence identities above 30%, PSI-BLAST alignments are reasonably accurate42. Below this value, there may be poorly conserved regions that are not accurately aligned, or the alignment may not be complete. In this case, it may be advisable to use profile-profile alignment methods to obtain a more accurate alignment. For instance, the FFAS server7 produces more accurate alignments than PSI-BLAST. The alignment that MolIDE produces can then be edited to conform to that provided by FFAS or any other server or program.
Also, MolIDE does not take account of any structural changes, other than side chains and loops, between the target and template. Therefore parts of the modeled structure produced by alignments with large and/or frequent gaps and/or low sequence conservation are therefore quite suspect. At lower sequence identities, the backbone model will not be very accurate. In these cases, SCWRL may predict side-chain conformations with significant steric overlaps with other side chains or the backbone. An energy minimization with CHARMM48 or other programs will remove these steric overlaps, although the resulting model will not likely be any closer to the target structure, if it were known. However, sequence conservation may vary substantially in different parts of the alignment. Often a few well-conserved motifs are noticeable in the alignment, and these are likely to be modeled reasonably well based on the template structure.
Troubleshooting
1) Trouble downloading database files (Step 8)
It is possible that some users may have trouble downloading the database and PDB XML files because of local IT security policies. In the case of the nr database, the FASTA formatted file can be manually downloaded from NCBI by putting this address into a web browser:
The file must first be uncompressed (ungzipped). On Linux, this is accomplished by typing:
gzip -d nr.gz
On Windows, the user must obtain software for uncompressing files such as FreeZip (http://www.versiontracker.com/dyn/moreinfo/win/10360) and follow the instructions in the program.
Once downloaded and uncompressed, the file should be put in the MolIDE database directory, C:\FCCC\MolIDE\db on Windows or molide_path/db on Linux. Then the database can be formatted using the program formatdb distributed with MolIDE from the console window in Windows and a command-line terminal window on Linux:
(on Windows)
cd C:\FCCC\MolIDE\db
C:\FCCC\MolIDE\bin_aux\NCBI\formatdb.exe -i nr -t nr
(on Linux)
cd molide_path/db
molide/path/bin_aux/NCBI/formatdb -i nr -t nr -o
The PDBAA file can be downloaded using a browser from this address:
and the same procedure followed to format the database for PSI-BLAST.
2) Running PSI-BLAST (Step 10)
If running PSI-BLAST fails for some reason, copy the PSI-BLAST command given in the PSI-BLAST runtime window, and paste it into a console window (Windows) or terminal window (Linux), and hit “return.” PSI-BLAST will now run, and any error messages it sends to the window will now be visible. These messages may help to diagnose the problem.
3) Downloading PDB XML files (Step 13)
For users at some institutions, the IT security setup may not allow MolIDE to access the PDB's ftp site for the XML files. In this case, the user can go to http://www.rcsb.org, search for the PDB code, and download the “PDBML/XML gz” or “PDBML/XML text” files and place them in the working directory. In this case, clicking on the row number in the PDB Hits Alignment (.pdbout) table will use the manually downloaded XML file instead of going to the ftp server to get it. The rest of the processing is the same.
Figure 2.
Flowchart for homology modeling with MolIDE. Step numbers to the right of each step correspond to the protocol described in the text, beginning with Step 5.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01-HG02302 and R01-GM84453 (to R.L.D) and P30-CA06927 to Fox Chase Cancer Center. We thank Mark Andrake and Radka Stoyanova for testing MolIDE 1.6.
REFERENCES
- 1.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–3. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang G, Dunbrack RL., Jr. PISCES: recent improvements to a PDB sequence culling server. Nucleic Acids Res. 2005;33:W94–8. doi: 10.1093/nar/gki402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Conte L, et al. SCOP: a structural classification of proteins database. Nucleic Acids Res. 2000;28:257–9. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perutz MF, Kendrew JC, Watson HC. Structure and function of haemoglobin. Journal of Molecular Biology. 1965;13:669–678. [Google Scholar]
- 5.Browne WJ, North AC, Phillips DC. A possible three-dimensional structure of bovine alpha-lactalbumin based on that of hen's egg-white lysozyme. Journal of Molecular Biology. 1969;42:65–86. doi: 10.1016/0022-2836(69)90487-2. [DOI] [PubMed] [Google Scholar]
- 6.Jones DT, Taylor WR, Thornton JM. A new approach to protein fold recognition. Nature. 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 7.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile--profile sequence alignments. Nucleic Acids Res. 2005;33:W284–8. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Havel TF, Snow ME. A new method for building protein conformations from sequence alignments with homologues of known structure. Journal of Molecular Biology. 1991;217:1–7. doi: 10.1016/0022-2836(91)90603-4. [DOI] [PubMed] [Google Scholar]
- 9.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez R, Sali A. Evaluation of comparative protein structure modeling by MODELLER-3. Proteins. 1997;Suppl:50–8. doi: 10.1002/(sici)1097-0134(1997)1+<50::aid-prot8>3.3.co;2-w. [DOI] [PubMed] [Google Scholar]
- 11.Li H, et al. Homology modeling using simulated annealing of restrained molecular dynamics and conformational search calculations with CONGEN: application in predicting the three-dimensional structure of murine homeodomain Msx-1. Protein Sci. 1997;6:956–70. doi: 10.1002/pro.5560060502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahasrabudhe PV, Tejero R, Kitao S, Furuichi Y, Montelione GT. Homology modeling of an RNP domain from a human RNA-binding protein: Homology-constrained energy optimization provides a criterion for distinguishing potential sequence alignments. Proteins. 1998;33:558–66. [PubMed] [Google Scholar]
- 13.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. Journal of Molecular Biology. 1996;257:342–58. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 14.Fetrow JS, Godzik A, Skolnick J. Functional Analysis of the Escherichia coli Genome Using the Sequence- to-Structure-to-Function Paradigm: Identification of Proteins Exhibiting the Glutaredoxin/Thioredoxin Disulfide Oxidoreductase Activity. Journal of Molecular Biology. 1998;282:703–711. doi: 10.1006/jmbi.1998.2061. [DOI] [PubMed] [Google Scholar]
- 15.Whisstock JC, Lesk AM. Prediction of protein function from protein sequence and structure. Q Rev Biophys. 2003;36:307–40. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 16.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Najmanovich RJ, Torrance JW, Thornton JM. Prediction of protein function from structure: insights from methods for the detection of local structural similarities. Biotechniques. 2005;38:847, 849, 851. doi: 10.2144/05386TE01. [DOI] [PubMed] [Google Scholar]
- 18.Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr Opin Struct Biol. 2005;15:275–84. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Kim SH, et al. Structure-based functional inference in structural genomics. J Struct Funct Genomics. 2003;4:129–35. doi: 10.1023/a:1026200610644. [DOI] [PubMed] [Google Scholar]
- 20.Zoller B, Dahlback B. Linkage between inherited resistance to activated protein C and factor V gene mutation in venous thrombosis. Lancet. 1994;343:1536–8. doi: 10.1016/s0140-6736(94)92940-8. [DOI] [PubMed] [Google Scholar]
- 21.Bogaerts V, et al. Genetic variability in the mitochondrial serine protease HTRA2 contributes to risk for Parkinson disease. Hum Mutat. 2008 doi: 10.1002/humu.20713. [DOI] [PubMed] [Google Scholar]
- 22.Couch FJ, Weber BL. Mutations and polymorphisms in the familial early-onset breast cancer (BRCA1) gene. Breast Cancer Information Core. Hum Mutat. 1996;8:8–18. doi: 10.1002/humu.1380080102. [DOI] [PubMed] [Google Scholar]
- 23.Karchin R, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–20. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 24.Weber IT, et al. Molecular modeling of the HIV-1 protease and its substrate binding site. Science. 1989;243:928–31. doi: 10.1126/science.2537531. [DOI] [PubMed] [Google Scholar]
- 25.Weber IT. Evaluation of homology modeling of HIV protease. Proteins. 1990;7:172–84. doi: 10.1002/prot.340070206. [DOI] [PubMed] [Google Scholar]
- 26.Ring CS, et al. Structure-based inhibitor design by using protein models for the development of antiparasitic agents. Proc. Natl. Acad. Sci. USA. 1993;90:3583–7. doi: 10.1073/pnas.90.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park H, et al. Discovery of novel alpha-glucosidase inhibitors based on the virtual screening with the homology-modeled protein structure. Bioorg Med Chem. 2008;16:284–92. doi: 10.1016/j.bmc.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 28.Bower M, Cohen FE, Dunbrack RL. SCWRL: A program for building sidechains onto protein backbones. University of California; San Francisco: 1997. J. http://www.cmpharm.ucsf.edu/~bower/scwrl.html. [Google Scholar]
- 29.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–14. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallner B, Elofsson A. All are not equal: a benchmark of different homology modeling programs. Protein Sci. 2005;14:1315–27. doi: 10.1110/ps.041253405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canutescu AA, Dunbrack RL., Jr. MolIDE: a homology modeling framework you can click with. Bioinformatics. 2005;21:2914–6. doi: 10.1093/bioinformatics/bti438. [DOI] [PubMed] [Google Scholar]
- 32.Qian B, et al. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–64. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunbrack RL., Jr. Ph. D. dissertation. Harvard University; 1993. [Google Scholar]
- 34.Dunbrack RL, Jr., Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–81. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunbrack RL., Jr. Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12:431–40. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 36.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 37.Lovell SC, Word JM, Richardson JS, Richardson DC. Asparagine and glutamine rotamers: B-factor cutoff and correction of amide flips yield distinct clustering. Proc Natl Acad Sci U S A. 1999;96:400–5. doi: 10.1073/pnas.96.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of database programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 41.Xu Q, Canutescu A, Obradovic Z, Dunbrack RL., Jr. ProtBuD: a database of biological unit structures of protein families and superfamilies. Bioinformatics. 2006;22:2876–82. doi: 10.1093/bioinformatics/btl490. [DOI] [PubMed] [Google Scholar]
- 42.Sauder JM, Arthur JW, Dunbrack RL., Jr. Large-scale comparison of protein sequence alignment algorithms with structure alignments. Proteins. 2000;40:6–22. doi: 10.1002/(sici)1097-0134(20000701)40:1<6::aid-prot30>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 43.Xiang Z, Soto CS, Honig B. Evaluating conformational free energies: the colony enegy and its application to the problem of protein loop prediction. Proc. Natl. Acad. Sci. USA. 2002;99:7432–7437. doi: 10.1073/pnas.102179699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–9. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 45.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pieper U, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–5. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32:W582–5. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacKerell AD, Jr., Bashford D, Bellott M, Dunbrack RL, Jr., Evanseck, M.J.F. J, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. 1998;B102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 49.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–3. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 50.Shan X, Dunbrack RL, Jr., Christopher SA, Kruger WD. Mutations in the regulatory domain of cystathionine beta synthase can functionally suppress patient-derived mutations in cis. Hum Mol Genet. 2001;10:635–43. doi: 10.1093/hmg/10.6.635. [DOI] [PubMed] [Google Scholar]
- 51.Proudfoot M, et al. Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain. J Mol Biol. 2008;375:301–15. doi: 10.1016/j.jmb.2007.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]