Abstract
CIRL (the calcium-independent receptor of α-latrotoxin), a neuronal cell surface receptor implicated in the regulation of exocytosis, is a member of the GPS family of chimeric cell adhesion/G protein-coupled receptors. The predominant form of CIRL is a membrane-bound complex of two subunits, p120 and p85. Extracellularly oriented p120 contains hydrophilic cell adhesion domains, whereas p85 is a heptahelical membrane protein. Both subunits are encoded by the same gene and represent products of intracellular proteolytic processing of the CIRL precursor. In this study, we demonstrate that a soluble form of CIRL also exists in vitro and in vivo. It results from the further cleavage of CIRL by a second protease. The site of the second cleavage is located in the short N-terminal extracellular tail of p85, between the GPS domain and the first transmembrane segment of CIRL. Thus, the soluble form of CIRL represents a complex of p120 non-covalently bound to a 15 amino acid residue N-terminal peptide fragment of p85. We have previously shown that mutations of CIRL in the GPS domain inhibit intracellular proteolytic processing and also result in the absence of the receptors from the cell surface. Our current data suggest that although CIRL trafficking to the cell membrane is impaired by mutations in the GPS region, it is not blocked completely. However, at the cell surface, the non-cleaved mutants are preferentially targeted by the second protease that sheds the extracellular subunit. Therefore, the two-step proteolytic processing may represent a regulatory mechanism that controls cell surface expression of membrane-bound and soluble forms of CIRL.
The adhesion G protein-coupled receptors (GPCRs) also known as LNB-TM7 receptors are related to the family B(II) of the GPCR superfamily. The hallmark of this recently defined family of heptahelical cell surface receptors is their unusually large extracellular N-terminal domains with mosaics of cell adhesion modules. Thus they are thought to couple cell-to-cell contacts with intracellular G protein signaling (1-4), and the physiological significance of some of them was determined by in vivo analysis of their mutations (5-7). Despite a decade effort, no natural agonist has been found yet for any of about 30 adhesion GPCRs present in the human and other mammalian genomes raising a possibility that they may be activated by unusual mechanisms substantially different from other heptahelical receptors (8). However, several proteins were identified that bind their long N-terminal extracellular regions suggesting that the adhesion-like ectodomains of these receptors may function separately and independently of the heptahelical receptor core (9-11).
Ample experimental data indicate that most if not all adhesion GPCRs are proteolytically cleaved in two parts that structurally correspond to their two putative functions, adhesion-like and receptor-like. This posttranslational modification is constitutive and yields two associated fragments, the hydrophilic N-terminal ectodomain and heptahelical polypeptide resembling canonical GPCRs. The cleavage site has been localized precisely for some receptors and appears to be about 20 amino acid residues away from the first transmembrane segment (5, 12-19). Interestingly, this site lies within the conserved motif comprised of a four-cysteine box with two internal conserved tryptophans, and a short hydrophobic segment C-terminally from the cysteine residues. This domain, named GPS (GPCR Proteolysis Site) is found at the same juxtamembrane position in all homologous adhesion GPCRs with the only exception of GPR123 (20). Less conserved GPS-like domains were also found in just a few non-heptahelical receptors, such as polycystic kidney disease protein and sea urchin egg jelly sperm receptor (21, 22). These receptors are also cleaved, similarly to the GPS-containing GPCRs.
The GPS-targeted processing is different from the typical posttranslational proteolytic processing of receptors by furin-like proteases, not only by the characteristics of the cleavage site, but also because it proceeds early in the biosynthetic pathway, either in the endoplasmic reticulum or in the early compartment of Golgi, prior to the addition of complex carbohydrate chains (14, 15, 23). Moreover, because of the presence of a cis-proteolytic motif in the GPS domain, the hypothesis was formulated that the GPS proteolysis has an autocatalytic mechanism (24).
Most of the accumulated data suggest that the two cleavage fragments of adhesion GPCR remain tightly but non-covalently bound and thus can be viewed as receptor subunits. The primary evidence is that the extracellular adhesion-like hydrophilic subunit can be detected bound to the membrane, although it behaves as a soluble secreted protein when exogenously expressed (13). Also, in detergent extracts, both subunits can be efficiently co-precipitated by fragment-specific antibodies (12). However, an examination of the expression and trafficking of CIRL/latrophilin (CL), a neuronal adhesion GPCR implicated in the regulation of neurosecretion (25), with subunit-specific antibodies indicated that its cleavage products do not co-localize completely at the cell surface; they also internalize in a separate manner. It was therefore proposed that, both hydrophilic and heptahelical fragments remain membrane bound, but they dissociate and function independently at the cell membrane; still they can re-associate by an unknown mechanism upon the detergent extraction or ligand binding (23).
To reconcile these contradictory observations, we tested the possibility that the two-subunit complex of adhesion GPCR can dissociate under physiological conditions and the resulting subunits would exist and function independently. Our data demonstrate that a minor portion of the receptor complexes does dissociate producing the soluble receptor ectodomain, and this dissociation is due to the second cleavage at the site between the site of primary proteolysis and the first transmembrane domain.
Materials and Methods
Miscellaneous Procedures
CIRL-encoding plasmids, anti-CIRL-1 and anti-CIRL-2 antibodies were described previously (12, 26). α-Latrotoxin was purified from lyophilized Black Widow Spider glands and radioactively labeled with 125I by chloramine T procedure. The toxin was immobilized on BrCN-agarose as described (27). COS or HEK 293 cells were transfected using lipofectamine reagent according to standard manufacturer’s protocol (Life Technology). The Western blot analysis of cells and conditioned media was performed essentially as described (14, 28).
Detection of CIRL Soluble Form in vivo
10 g of rat brains were homogenized in 100 ml ice cold extraction buffer, containing 50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA and 0.5 mM PMSF, pH 8.0 in a Blender homogenizer for 2 min. The insoluble material was removed by centrifugation for 30 min at 50,000g and the obtained supernatant was batch-adsorbed onto 1 ml of α-latrotoxin-agarose overnight at 4°C. The matrix was further washed with 100 ml of extraction buffer in a chromatography column and the adsorbed proteins were eluted with 5 ml of 1M NaCl, 50 mM Tris-HCl and 2 mM EDTA pH 8.0 for 1h. In a parallel experiment, the extract was similarly processed with 1 ml of BSA-agarose as a negative control.
Tissue Distribution of Soluble CIRL Ectodomain
1 g of fresh rat tissue (heart, muscle, brain, liver, kidney, lung, spleen) were cut in small pieces and homogenized in 3 ml ice cold 50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA and 0.5 mM PMSF buffer, pH 8.0, with a Polytron homogenizer. Insoluble matter was removed by centrifugation for 30 min at 50,000g and 1 ml portions of the supernatants were incubated with 25 μl aliquots of α-latrotoxin-agarose overnight at 4°C with gentle rotation. One ml of rat serum was supplemented with protease inhibitors and incubated with the matrix directly. The affinity matrices were then washed four times with 1.25 ml of the extraction buffer and the adsorbed proteins were eluted with the SDS sample buffer followed by electrophoresis and Western blotting with anti-p120 antibodies.
CIRL Mutants
The plasmids encoding full-length GPS mutants of CIRL, pCDR7-C834/W, pCDR7-T838/P and pCDR7-W815/S (14), and the soluble ectodomain of CIRL, pSTR7-2 (13), have been previously described (12, 26). To generate soluble, single residue GPS mutants, pCDR7-C834/W, pCDR7-T838/P and pCDR7-W815/S were digested with Bgl II, the 2337 bp fragments were isolated from the agarose gel and ligated with the dephosphorylated 5225 bp product of partial digestion of pSTR7-2 with Bgl II. Correct insert orientation of isolated plasmids pSTR7-2-C834/W, pSTR7-2-T838/P and pSTR7-2-W815/S was verified by restriction mapping. The soluble ectodomain, single residue mutants at the second cleavage site, were generated with the QuikChange Site-Directed Mutagenesis Kit (Stratagene), according to the manufacturer’s protocol. The oligonucleotides CACCGAGAGATCTACCAACCCCGTATTAATGAGCTGTTGCTG and CGAGAGATCTACCAAGGCCCTATTAATGAGCTGTTGCTG were used to introduce G852/P and R853/P mutations, respectively. Short fragments with introduced mutations were excised with AgeI and XhoI and ligated back into the original plasmid to avoid spontaneous mutations. The final plasmids were named pSTR7-2-G852/P and pSTR7-2-R853/P. The correct sequence of all constructs obtained by PCR was additionally verified by DNA sequencing.
Mass Spectrometry
COS cells were transfected with plasmids coding CIRL-1 or CIRL-2 and grown in serum-free medium. On day 3 the medium was harvested and clarified by centrifugation at 40,000g for 30 min. 100 ml of that medium was concentrated on ice by ultrafiltration using Amicon P-10 filter up to final volume of 5 ml. The supernatant fluid was centrifuged at 40,000g for 30 min one more time and then mixed with 100 μl α-latrotoxin-agarose. After overnight incubation at +4°C α-latrotoxin-agarose was collected by brief centrifugation and washed 3 times with 15 ml ice cold buffer containing 50 mM Tris-HCl, 150 mM NaCl and 2 mM EDTA, pH 8.0. The final wash buffer was removed entirely and 200 μl of elution buffer (3 M MgCl2, 50 mM Tris-HCl, pH 8.0) was added. After incubation with agitation for 15 min the mixture was centrifuged and supernatant was collected. For isolation of the cleavage peptide produced in vivo, the supernatant fraction from brain lysates was subjected to the same purification protocol. Peptides were concentrated and contaminants removed by micro reverse phase chromatography on C18 silica resin (600 nl bed volume) pipette tip columns (Millipore C18 ZipTips™). Samples in 0.1% trifluoroacetic acid (TFA) were bound to the columns, washed in 0.1% TFA, and eluted in 1 to 2 μl 90% acetonitrile and 0.1% TFA. Crystals were formed using the dried droplet method by allowing mixtures of 0.5 μl sample and 0.5 μl matrix solution consisting of 10 mg/ml α-cyano-4-hydroxycinnamic acid (Aldrich Chemical Company) in 50% ethanol and 50% acetonitrile to dry at room temperature. For COS-cell derived peptides, positive ion mass spectra were acquired in linear mode using a TofSpec-2E MALDI-TOF mass spectrometer (Micromass, Milford, MA) with time lag focusing using standard instrument settings. For peptides isolated from brain lysates, 150 μl of the LTX column eluate was concentrated by micro reverse phase solid phase extraction (ZipTip™) and analyzed using LTQ-Orbitrap nanoflow LC/MS/MS. Samples were loaded onto a Symmetry C18 precolumn (Milford, MA), then washed 5 min with 1% acetonitrile in 0.1% formic acid at a flow rate of 20 μl/min. After washing, peptides were eluted and passed through a 75 μm × 150 mm 1.7μm particle size BEH300 C18 analytical column (Waters, Milford, MA) with a gradient of 1-45 % acetonitrile in 0.1% formic acid. The gradient was delivered over 30 min by a nanoACQUITY UPLC (Waters) at a flow rate of 275 nl/min, to a fused silica distal end-coated tip nano-electrospray needle (New Objective, Woburn, MA). Mass spectra were acquired with an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) with data dependent acquisition using Xcalibur software Version 2.0 as follows: 6 scan events per second were used, 1 Orbitrap accurate mass scan of resolution of 60,000 with simultaneous MS/MS acquisition on the 5 highest peaks with charge state 2 to 4 as determined by a 15,000 resolution preview scan, and each MS/MS scan took up to 150ms.
Results
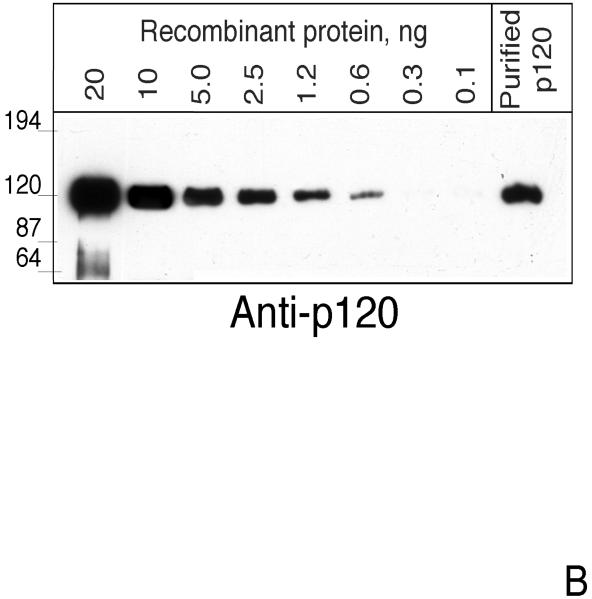
To test whether a soluble form of CIRL may exist in vivo, we subjected aqueous extracts of brain tissue to α-latrotoxin-agarose chromatography. To minimize artifactual protein degradation, freshly frozen tissue was processed with ice-cold buffers containing protease inhibitors. The adsorbed proteins were eluted and analyzed by Western blotting. As a negative control, chromatography on BSA-agarose was performed. Staining with anti-p120 antibody revealed significant amount of p120 in the toxin column eluate but not in the control preparation (Fig. 1A). To estimate the size of the soluble fraction of CIRL in vivo, we performed semi-quantitative blotting for p120 precipitated with α-latrotoxin-agarose from non-detergent extracts of freshly-frozen brain tissue (Fig. 1B). As a standard, known concentrations of secreted recombinant p120 protein (pCDR7N) (13) produced in COS cells were used. By comparing the signal strength, we estimated that the concentration of p120 was about 8 fmol/mg of membrane protein. This is a lower estimate based on the assumption that recovery from the affinity column was 100%. From α-latrotoxin binding experiments, we know that the concentration of CIRL in brain membranes is about 200 fmol/mg (29). Thus, at least 4 % of CIRL in the brain exists in the form of soluble p120.
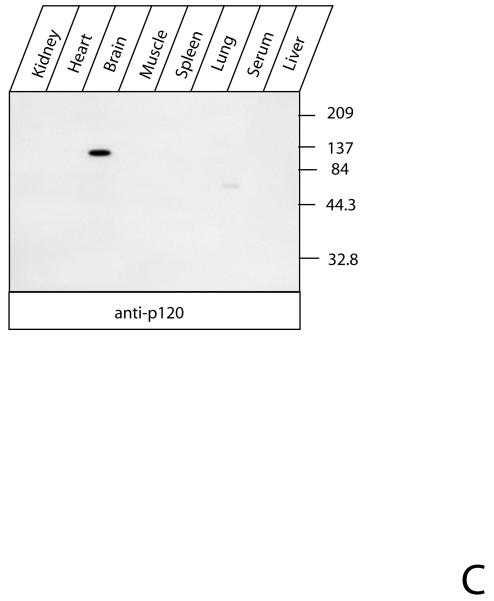
Fig. 1. The soluble form of CIRL in brain.
A. p120 in the soluble fraction of brain protein extract. Rat brains were extracted, precipitated with either α-latrotoxin-agarose or BSA-agarose and analyzed by Western blotting with anti-p120 antibody as described in Experimental Procedures. B. Semi-quantitative assay of soluble p120. One ml of rat brain extract equivalent to 8 mg of membrane protein was precipitated with 25 μl of α-latrotoxin-agarose for 15 hr at 4°C and analyzed by Western blotting with anti-p120 antibody. The indicated amounts of purified recombinant p120 were used as standards. C. Tissue distribution of soluble CIRL. Rat tissues were extracted and analyzed for the presence of p120 as described in the Experimental Procedures. The position of molecular mass protein standards (Gibco/Invitrogen) and their molecular mass in kDa are shown.
Northern blotting and α-latrotoxin-binding experiments indicated that CIRL is expressed almost exclusively in neural tissues (12, 30). To analyze the tissue distribution of soluble CIRL, we chromatographed aqueous non-detergent extracts of different tissues on α-latrotoxin-agarose. The analysis of the adsorbed protein by Western blotting with anti-p120 antibody revealed that the soluble form of CIRL was present only in brain extracts (Fig. 1C), which correlated with its mRNA distribution.
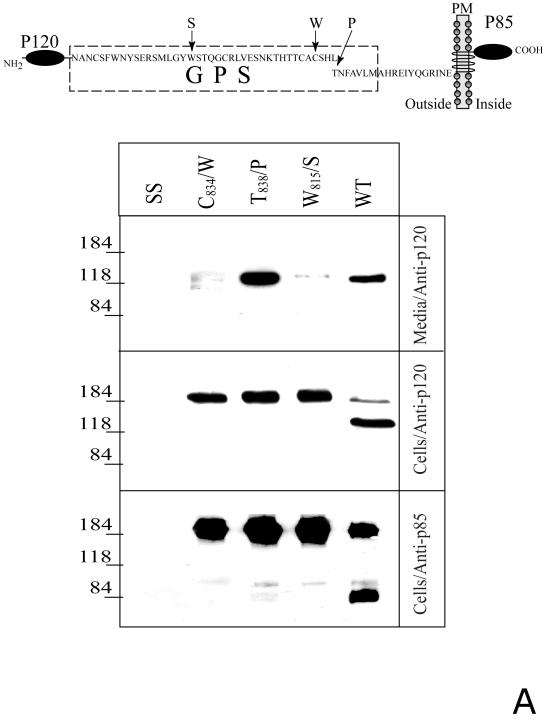
We further analyzed the expression of soluble and membrane forms of CIRL and its proteolysis-resistant mutants (Fig. 2A, upper scheme) in transfected COS cells. A significant amount of soluble p120 was detected in the medium of the cells expressing the wild-type CIRL. Unexpectedly, an even larger amount of p120 was present in the medium of T838/P mutant-expressing cells (Fig. 2A, lower panels). However, in agreement with our previously published data (14), no CIRL presence at the cell surface could be detected by α-latrotoxin binding assay with intact cells. Thus, a large amount of secreted T838/P p120 contrasted with the absence of the full-length membrane mutant at the cell surface.
Fig. 2. Proteolytic processing of the full-length and soluble deletion constructs of CIRL mutated in the GPS domain.
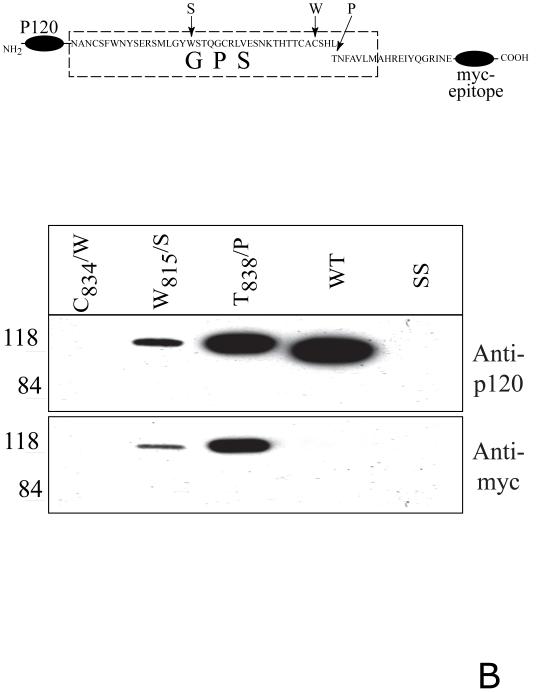
A. Soluble forms of CIRL GPS mutants. COS cells were transfected with either wild-type CIRL or its GPS mutants with single residue substitution within the GPS domain (C834/W, W815/S, and T838/P), schematically described at the top panel, PM - plasma membrane. The cells were harvested and analyzed by Western blotting with anti-p120 or anti-p85 antibody. The conditioned media were precipitated with α-latrotoxin-agarose followed by Western blotting with anti-p120 antibody. Salmon sperm DNA transfected cells (SS) were used as control. B. Secretion of soluble GPS mutants of the CIRL ectodomain. COS cells were transfected with the plasmids encoding either the wild type CIRL ectodomain or three single-residue mutant constructs (C834/W, W815/S, and T838/P), schematically described at the top panel. The conditioned media were precipitated with α-latrotoxin-agarose followed by Western blotting with either anti-p120 or anti-myc antibody. Note an increase in the apparent size of p120 in the non-cleaved mutants W815/S, and T838/P due to the 3.8 kDa myc-tag addition. The pictures shown are representative of five independent transfection, precipitation and blotting experiments that produced essentially similar results.
In a complementary approach, we analyzed the trafficking of soluble deletion mutants of CIRL. We expressed fusion proteins of the entire CIRL extracellular N-terminal domain (p120 plus 19 residues of p85 up to the first transmembrane segment) with a myc-epitope at the C-terminus. In parallel, similar constructs were designed that had the GPS point mutations (Fig. 2B, upper scheme). All proteins, except the C834/W mutant, were secreted into the medium. The wild-type tagged p120 was completely cleaved whereas T838/P and W815/S p120 mutants were not cleaved at all, similarly to the full-length membrane mutants (Fig. 2B, lower panels). Therefore, the intracellular cleavage is not required for the secretion of truncated, soluble CIRL.
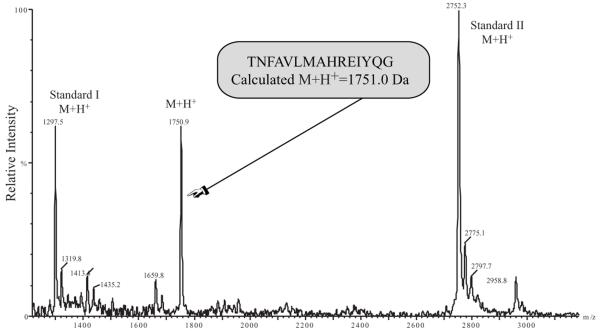
The simplest explanation of the presence of soluble CIRL in media would be dissociation of p120 and p85. However, this mechanism fails to explain soluble forms of non-cleaved GPS mutants. Therefore, we postulated that CIRL can be additionally cleaved and that this second proteolysis results in the dissociation of the p120/p85 complex. Apparently, the first and second proteases have different specificity towards CIRL and its GPS mutants. Thus it is unlikely that they share the same cleavage and the double cleavage of the wild-type CIRL should result in a peptide fragment in addition to processed p120 and p85. We succeeded in identifying this fragment bound to soluble p120. When p120 was purified from the CIRL-transfected COS cells and analyzed by mass spectrometry, a peptide of m/z 1750.9 was observed (Fig. 3). This mass corresponds to the predicted average mass (1751.0) of a protonated 15-residue peptide that would result from two cleavages, one N-terminal to T838, and the other C-terminal to G852.
Fig. 3. Secondary proteolysis of CIRL-1.
MALDI-TOF spectrum of the peptide product of the dual cleavage of CIRL by intracellular and extracellular proteases. The medium of CIRL-transfected COS cells (100 ml) was concentrated and precipitated with α-latrotoxin-agarose. The adsorbed material was eluted and analyzed by MALDI-TOF mass spectrometry. Internal mass standards were angiotensin I (average M+H+ 1297.5) and a synthetic peptide (average M+H+ 2752.3).
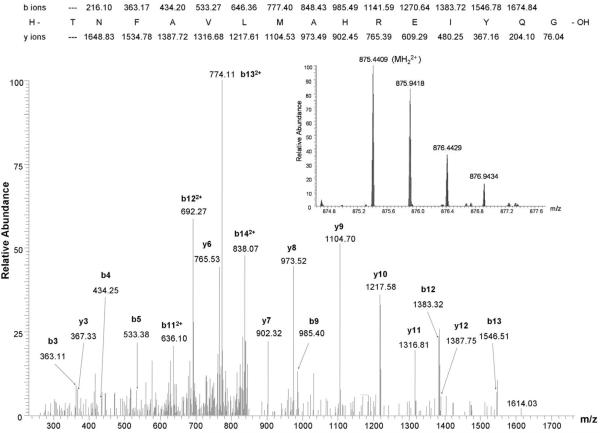
To confirm the physiological relevance of the observed proteolysis, we purified the soluble CIRL fragment from aqueous brain extracts and analyzed in the same manner. The peptide with the same predicted mass was observed and its sequence was further confirmed by the MS/MS analysis (Fig. 4).
Fig. 4. LTQ-Orbitrap LC-MS/MS spectrum of 7% of the eluate from the LTX affinity column using the soluble fraction of brain homogenate as starting material.
Observed b and y ions from the peptide of sequence TNFAVLMAHREIYQG are labeled in the MS/MS spectrum. Inset shows a portion of the Orbitrap MS survey scan containing the doubly charged precursor ion of the sequenced peptide, which has a calculated m/z of 875.4407 (mass error 0.2 ppm).
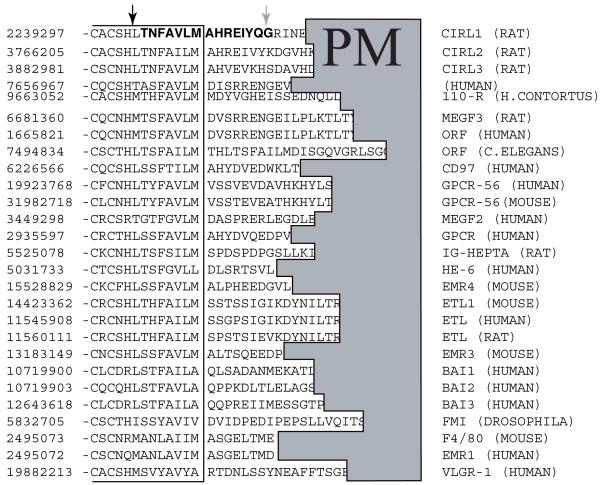
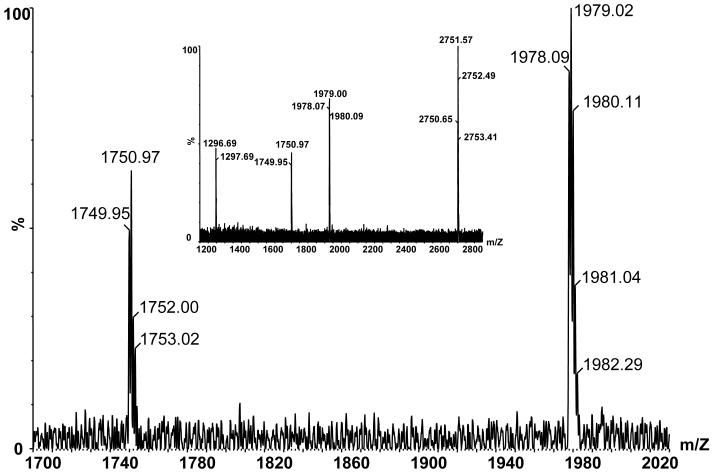
The site of the second cleavage was not characteristic of any particular protease. The multiple alignment of GPS receptor sequences did not reveal any significant conservation in this region (Fig. 5). We tested if another GPS receptor, CIRL-2, which is a close homolog of CIRL (also referred to as CIRL-1) can be cleaved in the same manner. COS cells were co-transfected with CIRL-1 and CIRL-2, and the soluble forms of the receptors p120 were precipitated with α-latrotoxin-agarose. The precipitate was further analyzed by MALDI-TOF mass spectrometry, and a monoisotopic peak of m/z 1978.1, corresponding to the17-residue peptide resulting from cleavages N-terminally to T822 and C-terminally to G838 was observed (Fig. 6). Therefore, the cell surface processing of CIRL-2 occurred essentially similarly to CIRL-1 in a close vicinity to the first transmembrane segment.
Fig. 5. Multiple alignment of GPS receptors in the region of the sites of intracellular and extracellular proteolysis.
GenBank™ protein accession numbers are shown in the left column. The intracellular cleavage site identified in CIRL-1 and several other GPS receptors is shown by a black arrow. The site of the extracellular cleavage of CIRL-1 only is marked by a gray arrow. PM, plasma membrane, denotes the region of hydrophobic residues of the first transmembrane segments of the aligned receptors.
Fig. 6. Secondary proteolysis of CIRL-2.
Reflectron mode MALDI-TOF MS spectra of CIRL-2 cleavage peptide (calculated m/z of [M+H]+ ion = 1978.02). Large figure shows expanded view of an externally calibrated spectrum, inset shows a spectrum acquired with angiotensin I (calculated m/z of [M+H]+ ion = 1296.69) and a synthetic peptide (calculated m/z of [M+H]+ ion = 2750.65) as internal calibrants.
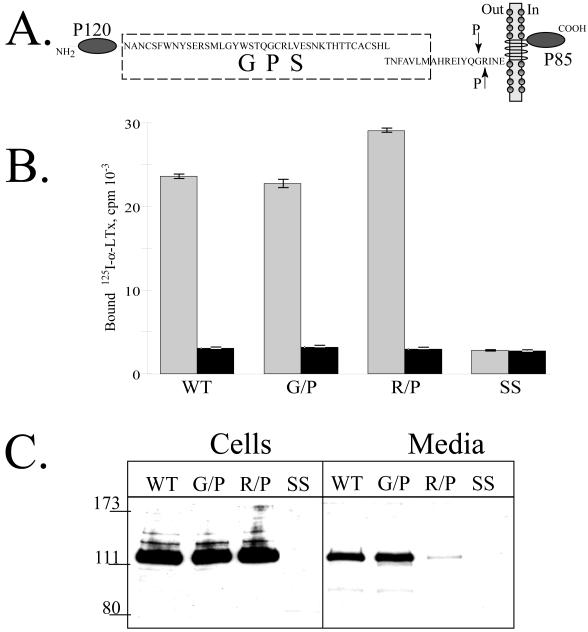
To analyze the specificity of the extracellular cleavage, we mutated the residues surrounding this site in the full-length CIRL. Either N-terminal G852 or C-terminal R853 was replaced with a proline residue (Fig. 7A). The analysis of α-latrotoxin binding to intact cells indicated that the mutants expressed at the cell surface in amounts comparable to the wild-type CIRL (Fig. 7B). However, the soluble form of the R/P receptor mutant was present in the medium in a very low concentration compared to the wild type and G/P mutant of CIRL (Fig. 7C). In agreement with these data, the cell surface density of the R/P receptors was slightly higher that may reflect the absence of the cleavage (Fig. 7B).
Fig. 7. Cell surface expression of CIRL mutated at the second cleavage site.
A. Schematic description of the CIRL constructs with single residue mutations (G852/P and R853/P, indicated by arrows) at the second cleavage site. B. Cell surface expression of the second cleavage site CIRL mutants. Intact COS cells transfected with either wild-type CIRL or its mutants (G852/P and R853/P) were assayed for binding of 125I-α-latrotoxin either in the presence (black bars) or absence of excess non-labeled α-latrotoxin. Measurements were performed in triplicates. C. Secretion of the soluble forms of CIRL mutants. One ml of conditioned media of the same cells as in B was precipitated with 10 μl of α-latrotoxin-agarose and the eluates were analyzed by Western blotting with anti-p120 antibody. SS, salmon sperm DNA transfected cells. The picture shown is representative of three independent transfection, precipitation and blotting experiments that produced essentially similar results.
Discussion
CIRL is an orphan cell surface receptor with the structural features of a cell adhesion protein and GPCR. A key step of the CIRL biosynthesis is proteolytic processing in the lumen of the endoplasmic reticulum by an unidentified protease. The site of this cleavage is located in the C-terminal region of the GPS domain of CIRL, in about 20 residues from the first transmembrane segment. The resulting two fragments, the cell adhesion-like p120 and heptahelical p85, remain tightly bound in a non-covalent manner as indicated by localization of hydrophilic p120 at the cell surface and co-precipitation of both subunits in detergent extracts. Our findings together with published observations (23) suggest that under certain conditions, the subunits can dissociate; as a result, p120 secretes to the medium. The existence of soluble forms has also been demonstrated for other GPS-containing receptors (15, 31, 32).
In this study we identified the mechanism of such dissociation. It involves a second, membrane-associated protease that cleaves the N-terminus of p85 close to the membrane core. This cleavage does not occur frequently and may represent a regulated event. As a result of this cleavage, p120 dissociates from the membrane in a complex with a small peptide fragment of p85. These soluble complexes are secreted extracellularly where they can potentially bind other cell adhesion proteins or membrane receptors.
This mechanism of CIRL dissociation is supported by our experiments on expression of full-length and truncated CIRL mutated at the sites of the cleavage. We showed earlier that the T838/P mutation of the first cleavage site results in the complete resistance to the intracellular proteolysis (14). While no p120 fragment of this mutant could be detected in the transfected cells, a significant amount of p120 was found in the conditioned medium. In fact, there was more p120 in the medium of the mutant-expressing cells than of the wild-type ones. This finding suggested the mechanism of p120 secretion that is not based on its dissociation from p85 but rather on the secondary cleavage by another protease.
The direct evidence of the second proteolysis was the observation by mass spectrometry of a peak corresponding in mass to the peptide product derived from the two cleavages (Fig. 3). It was isolated by precipitation with α-latrotoxin-agarose and therefore should be bound to p120. The N-terminus of this peptide corresponds to the previously determined site of the cleavage by the intracellular protease. Subsequently, its C-terminus defined the second cleavage site between G852 and R853.
Importantly, the same peptides were found to be complexed to p120 in brain extracts and the conditioned media of CIRL-1-transfected cells. A highly similar peptide was also found the media of cells transfected with CIRL-2, a ubiquitously expressed close homolog of CIRL-1. Thus, the in vivo and in vitro mechanisms of CIRL processing are essentially similar and the proteases involved are likely to be ubiquitously expressed. This is not surprising in the view that, although individual GPS adhesion GPCRs are typically confined to specific tissues, the overall family is present in the majority if not all tissues (3).
The specificity of the extracellular proteolysis was further confirmed by mutating the second cleavage site. When R853 was replaced with P, the surface expression of the membrane form of the mutant did not change significantly. However, virtually no p120 could be detected in the cell medium suggesting the importance of this residue for the second proteolysis.
The analysis of cell surface expression of the wild-type CIRL and its intracellular proteolysis-resistant T838/P mutant led to an unexpected finding. While the soluble form of mutant was found at a higher concentration in the medium compared to the wild-type receptor, the full-length membrane form of the mutant could not be detected at the cell surface at all, indicating its complete cleavage by the second protease (Fig. 2A). Thus, the wild-type CIRL is processed completely by the intracellular protease and only to a minor extent by the second, extracellular protease. The T838/P mutant is not cleaved by the first protease at all but seems to be quite efficiently processed by the second extracellular protease because no p120 can be detected on the cell surface while plenty of soluble p120 is present in the medium. This observation can be explained by either higher susceptibility of the non-cleaved mutant to the extracellular proteolysis or by the activation of the extracellular protease as a result of mutant expression. Either possibility suggests the second proteolysis is a regulated event and there may be a functional link between the primary and secondary proteolysis.
The experiments with the soluble CIRL constructs encoding its N-terminal ectodomain further confirm our model of CIRL processing and trafficking (Fig. 2B). They also provide additional information about the localization of the two CIRL-processing proteases. Similarly to the full-length membrane CIRL, its ectodomain fused with myc-epitope is processed by the first intracellular protease while the corresponding T838/P mutant is not cleaved. The secreted wild-type soluble form is cleaved completely. The detected complexes of p120 and a small myc-tagged N-terminal fragment of p85 indicate the intracellular processing at the first cleavage site. The T838/P soluble mutant (as well as the W815/S mutant) is not cleaved by either protease. We may conclude that the first intracellular protease cleaves efficiently both membrane and soluble forms of CIRL whereas the second protease cleaves only membrane proteins. The presence of soluble CIRL form in the medium and absence of p120 in the cells expressing the T838/P mutant suggest the extracellular location of the second protease. On the basis of the described characteristics of the second cleavage we may speculate that the second protease may be one of the “sheddases” that work to remodel cell surface and to release hormones that derive from membrane-anchored proteins (33).
The functional importance of the CIRL proteolytic processing remains a puzzle. The primary intracellular GPS cleavage of CIRL is constitutive and is most likely required for the correct folding and/or trafficking of this as well as other GPS-containing receptors. The experiments with the GPS-containing receptor polycystin 1 revealed ablation of its function resulting from proteolysis-arresting mutations both in vivo and in vitro (31). Although most published data indicate that the adhesion GPCRs undergo complete primary cleavage in vivo, in vitro cell expression results only in partial processing. We observed regulation of the primary CIRL cleavage in transfected cells by protein kinase C activators (unpublished experiments). The proposed autocatalytic mechanism of the GPS processing (24) may be more complex and involve accessory proteins that facilitate or block the cleavage (34).
The secondary cleavage results in the shedding of the receptor ectodomain of only some cell surface expressed CIRL that opens several possibilities. One is that the shedded fragment functions independently as a ligand of other receptors, as was proposed for BAI receptor (32). Another possibility is that the sheddase, as with other membrane receptors, serves to remove all membrane proteins and thus to renew the cell surface. A possible mechanism of this event is that the receptor is activated by the second cleavage and further endocytosed. Finally, our data on the CIRL GPS mutants suggest that the shedding may work as a checkpoint to remove uncleaved, and therefore misfolded and unfunctional receptor precursors from the cell surface.
Acknowledgments
Supported by Public Health Service grants R01NS35098 from the NINDS and R03 TW007210 from the FIC, Molecular and Cellular Biology Program and Basic Sciences to Medicine Program of the Russian Academy of Sciences, and RFBR grant 06-04-49706-a (to A.G.P.) and by NIH Shared Instrumentation Grant 1 S10 RR14662-01 (to T.A.N.).
Abbreviations used
- CIRL
calcium-independent receptor of α-latrotoxin
- GPCR
G protein-coupled receptor
- GPS
GPCR Proteolysis Site
- EDTA
ethylenediaminetetraacetic acid
- PMSF
phenylmethylsulphonyl fluoride
- TFA
trifluoroacetic acid
- BSA
bovine serum albumin
- LTX
latrotoxin
References
- 1.Hayflick JS. A family of heptahelical receptors with adhesion-like domains: a marriage between two super families. J Recept Signal Transduct Res. 2000;20:119–131. doi: 10.3109/10799890009150640. [DOI] [PubMed] [Google Scholar]
- 2.Stacey M, Lin HH, Gordon S, McKnight AJ. LNB-TM7, a group of seven-transmembrane proteins related to family-B G-protein-coupled receptors. Trends in biochemical sciences. 2000;25:284–289. doi: 10.1016/s0968-0004(00)01583-8. [DOI] [PubMed] [Google Scholar]
- 3.Yona S, Lin HH, Siu WO, Gordon S, Stacey M. Adhesion-GPCRs: emerging roles for novel receptors. Trends in biochemical sciences. 2008;33:491–500. doi: 10.1016/j.tibs.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Kwakkenbos MJ, Kop EN, Stacey M, Matmati M, Gordon S, Lin HH, Hamann J. The EGF-TM7 family: a postgenomic view. Immunogenetics. 2004;55:655–666. doi: 10.1007/s00251-003-0625-2. [DOI] [PubMed] [Google Scholar]
- 5.Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 6.Wang T, Tian L, Haino M, Gao JL, Lake R, Ward Y, Wang H, Siebenlist U, Murphy PM, Kelly K. Improved antibacterial host defense and altered peripheral granulocyte homeostasis in mice lacking the adhesion class G protein receptor CD97. Infection and immunity. 2007;75:1144–1153. doi: 10.1128/IAI.00869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piao X, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, Voit T, Ross ME, Michaud JL, Descarie JC, Barkovich AJ, Walsh CA. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 8.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Molecular pharmacology. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 9.Lea S. Interactions of CD55 with non-complement ligands. Biochemical Society transactions. 2002;30:1014–1019. doi: 10.1042/bst0301014. [DOI] [PubMed] [Google Scholar]
- 10.Stacey M, Chang GW, Davies JQ, Kwakkenbos MJ, Sanderson RD, Hamann J, Gordon S, Lin HH. The epidermal growth factor-like domains of the human EMR2 receptor mediate cell attachment through chondroitin sulfate glycosaminoglycans. Blood. 2003;102:2916–2924. doi: 10.1182/blood-2002-11-3540. [DOI] [PubMed] [Google Scholar]
- 11.Wang T, Ward Y, Tian L, Lake R, Guedez L, Stetler-Stevenson WG, Kelly K. CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood. 2005;105:2836–2844. doi: 10.1182/blood-2004-07-2878. [DOI] [PubMed] [Google Scholar]
- 12.Krasnoperov VG, Bittner MA, Beavis R, Kuang Y, Salnikow KV, Chepurny OG, Little AR, Plotnikov AN, Wu D, Holz RW, Petrenko AG. alpha-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron. 1997;18:925–937. doi: 10.1016/s0896-6273(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 13.Krasnoperov V, Bittner MA, Holz RW, Chepurny O, Petrenko AG. Structural requirements for alpha-latrotoxin binding and alpha-latrotoxin-stimulated secretion. A study with calcium-independent receptor of alpha-latrotoxin (CIRL) deletion mutants. The Journal of biological chemistry. 1999;274:3590–3596. doi: 10.1074/jbc.274.6.3590. [DOI] [PubMed] [Google Scholar]
- 14.Krasnoperov V, Lu Y, Buryanovsky L, Neubert TA, Ichtchenko K, Petrenko AG. Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. The Journal of biological chemistry. 2002;277:46518–46526. doi: 10.1074/jbc.M206415200. [DOI] [PubMed] [Google Scholar]
- 15.Gray JX, Haino M, Roth MJ, Maguire JE, Jensen PN, Yarme A, Stetler-Stevenson MA, Siebenlist U, Kelly K. CD97 is a processed, seven-transmembrane, heterodimeric receptor associated with inflammation. J Immunol. 1996;157:5438–5447. [PubMed] [Google Scholar]
- 16.Nechiporuk T, Urness LD, Keating MT. ETL, a novel seven-transmembrane receptor that is developmentally regulated in the heart. ETL is a member of the secretin family and belongs to the epidermal growth factor-seven-transmembrane subfamily. The Journal of biological chemistry. 2001;276:4150–4157. doi: 10.1074/jbc.M004814200. [DOI] [PubMed] [Google Scholar]
- 17.Abe J, Fukuzawa T, Hirose S. Cleavage of Ig-Hepta at a “SEA” module and at a conserved G protein-coupled receptor proteolytic site. The Journal of biological chemistry. 2002;277:23391–23398. doi: 10.1074/jbc.M110877200. [DOI] [PubMed] [Google Scholar]
- 18.Stacey M, Chang GW, Sanos SL, Chittenden LR, Stubbs L, Gordon S, Lin HH. EMR4, a novel epidermal growth factor (EGF)-TM7 molecule up-regulated in activated mouse macrophages, binds to a putative cellular ligand on B lymphoma cell line A20. The Journal of biological chemistry. 2002;277:29283–29293. doi: 10.1074/jbc.M204306200. [DOI] [PubMed] [Google Scholar]
- 19.Obermann H, Samalecos A, Osterhoff C, Schroder B, Heller R, Kirchhoff C. HE6, a two-subunit heptahelical receptor associated with apical membranes of efferent and epididymal duct epithelia. Mol Reprod Dev. 2003;64:13–26. doi: 10.1002/mrd.10220. [DOI] [PubMed] [Google Scholar]
- 20.Fredriksson R, Gloriam DE, Hoglund PJ, Lagerstrom MC, Schioth HB. There exist at least 30 human G-protein-coupled receptors with long Ser/Thr-rich N-termini. Biochem Biophys Res Commun. 2003;301:725–734. doi: 10.1016/s0006-291x(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 21.Ponting CP, Hofmann K, Bork P. A latrophilin/CL-1-like GPS domain in polycystin-1. Curr Biol. 1999;9:R585–588. doi: 10.1016/s0960-9822(99)80379-0. [DOI] [PubMed] [Google Scholar]
- 22.Mengerink KJ, Moy GW, Vacquier VD. suREJ3, a polycystin-1 protein, is cleaved at the GPS domain and localizes to the acrosomal region of sea urchin sperm. The Journal of biological chemistry. 2002;277:943–948. doi: 10.1074/jbc.M109673200. [DOI] [PubMed] [Google Scholar]
- 23.Volynski KE, Silva JP, Lelianova VG, Atiqur Rahman M, Hopkins C, Ushkaryov YA. Latrophilin fragments behave as independent proteins that associate and signal on binding of LTX(N4C) Embo J. 2004;23:4423–4433. doi: 10.1038/sj.emboj.7600443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin HH, Chang GW, Davies JQ, Stacey M, Harris J, Gordon S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. The Journal of biological chemistry. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 25.Sudhof TC. alpha-Latrotoxin and its receptors: neurexins and CIRL/latrophilins. Annu Rev Neurosci. 2001;24:933–962. doi: 10.1146/annurev.neuro.24.1.933. [DOI] [PubMed] [Google Scholar]
- 26.Ichtchenko K, Bittner MA, Krasnoperov V, Little AR, Chepurny O, Holz RW, Petrenko AG. A novel ubiquitously expressed alpha-latrotoxin receptor is a member of the CIRL family of G-protein-coupled receptors. The Journal of biological chemistry. 1999;274:5491–5498. doi: 10.1074/jbc.274.9.5491. [DOI] [PubMed] [Google Scholar]
- 27.Petrenko AG, Kovalenko VA, Shamotienko OG, Surkova IN, Tarasyuk TA, Ushkaryov Yu A, Grishin EV. Isolation and properties of the alpha-latrotoxin receptor. Embo J. 1990;9:2023–2027. doi: 10.1002/j.1460-2075.1990.tb08331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krasnoperov V, Bittner MA, Mo W, Buryanovsky L, Neubert TA, Holz RW, Ichtchenko K, Petrenko AG. Protein-tyrosine phosphatase-sigma is a novel member of the functional family of alpha-latrotoxin receptors. The Journal of biological chemistry. 2002;277:35887–35895. doi: 10.1074/jbc.M205478200. [DOI] [PubMed] [Google Scholar]
- 29.Krasnoperov VG, Beavis R, Chepurny OG, Little AR, Plotnikov AN, Petrenko AG. The calcium-independent receptor of alpha-latrotoxin is not a neurexin. Biochem Biophys Res Commun. 1996;227:868–875. doi: 10.1006/bbrc.1996.1598. [DOI] [PubMed] [Google Scholar]
- 30.Lelianova VG, Davletov BA, Sterling A, Rahman MA, Grishin EV, Totty NF, Ushkaryov YA. Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. The Journal of biological chemistry. 1997;272:21504–21508. doi: 10.1074/jbc.272.34.21504. [DOI] [PubMed] [Google Scholar]
- 31.Qian F, Boletta A, Bhunia AK, Xu H, Liu L, Ahrabi AK, Watnick TJ, Zhou F, Germino GG. Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16981–16986. doi: 10.1073/pnas.252484899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur B, Brat DJ, Devi NS, Van Meir EG. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene. 2005;24:3632–3642. doi: 10.1038/sj.onc.1208317. [DOI] [PubMed] [Google Scholar]
- 33.Werb Z, Yan Y. A cellular striptease act. Science. 1998;282:1279–1280. doi: 10.1126/science.282.5392.1279. [DOI] [PubMed] [Google Scholar]
- 34.Wei W, Hackmann K, Xu H, Germino G, Qian F. Characterization of cis-autoproteolysis of polycystin-1, the product of human polycystic kidney disease 1 gene. The Journal of biological chemistry. 2007;282:21729–21737. doi: 10.1074/jbc.M703218200. [DOI] [PubMed] [Google Scholar]