Abstract
AIM: To verify that CD markers are available for detecting cancer stem cell populations and to evaluate their clinical significance in colon cancer.
METHODS: Immunohistochemistry for CD133, CD24 and CD44 was performed on the tissue microarray of 523 colorectal adenocarcinomas. Medical records were reviewed and clinicopathological analysis was performed.
RESULTS: In colorectal adenocarcinoma, 128 of 523 cases (24.5%) were positive and 395 cases (75.5%) were negative for CD133 expression. Two hundred and sixty-four of 523 cases (50.5%) were positive and 259 cases (49.5%) were negative for CD24 expression. Five hundred and two of 523 cases (96%) were negative and 21 cases (4%) were positive for CD44 expression. Upon clinicopathological analysis, CD133 expression was present more in male patients (P = 0.002) and in advanced T stage cancer (P = 0.024). Correlation between CD24 expression and clinicopathological factors was seen in the degree of differentiation (P = 0.006). Correlation between CD44 expression and clinicopathological factors was seen in the tumor size (P = 0.001). Survival was not significantly related to CD133, CD24 and CD44 expression.
CONCLUSION: CD markers were related to invasiveness and differentiation of colorectal adenocarcinoma. However, CD expression was not closely related to survival.
Keywords: CD133, CD24, CD44, Colon cancer stem cells, Colorectal adenocarcinoma
INTRODUCTION
Colorectal adenocarcinoma is the second most common type of cancer and a major cause of cancer-related morbidity and mortality in the Western world[1]. The incidence of colorectal cancer has increased from 5.8% in 1980 to 10.3% in 2000 in South Korea, in part because of Westernization of the diet[2].
Countless treatment protocols, including chemotherapy and radiation, have been applied to colorectal cancer and a number of studies have identified conventional prognostic factors[3]. However, a complete cure of colorectal cancer has not been accomplished despite numerous efforts. Recently, the prospective identification of colon cancer stem cells has received major attention because of their potential for colon cancer treatment[4,5]. Current colon cancer treatment modalities target proliferating cells, but colon cancer stem cells are thought to be slowly cycling cells; therefore, they may escape present targeted interventions because they are not actively proliferating. This may be one of the most important reasons behind colon cancer treatment failure and recurrence. It is important to validate in vitro/in vivo colon cancer stem cell findings in clinical samples. This will be a critical step toward the development of effective targeted colon cancer treatment, but thus far, no data are available on the clinical implications of the suggested colon cancer stem cells in clinical samples.
Recently, several CD markers have been identified as solid cancer stem cell markers. CD133, also known as PROML1 or prominin, is a stem cell surface antigen that has been recently identified as a potential cancer stem cell marker in brain, colon and prostate cancer[4–7]. CD44, also known as homing cell adhesion molecule, is a cell surface glycoprotein expressed on lymphocytes, monocytes and granulocytes, which has been identified as a stem cell marker in breast and head and neck cancer[8,9]. CD24, a cell surface marker, is a single chain sialoglycoprotein with a molecular mass of 42 kDa. CD24- and CD44-expressing pancreatic cancer cells show cancer stem cell characteristics[10]. Here, we report the identification of CD133-, CD24- and CD44-positive tumor cells in colon tumor sections by an immunohistochemistry-based technique, and discuss the findings in conjunction with clinicopathological data.
MATERIALS AND METHODS
Patients and specimens
This retrospective study consisted of a consecutive series of 523 colorectal adenocarcinomas with complete histopathological data available. Patients were diagnosed and treated at the Hanyang University Hospital, Seoul, Korea, from January 1991 to August 2001. There were 295 male and 228 female patients, with ages ranging from 17 to 87 years (mean, 59.0 years). The adenocarcinomas were located in the cecum (n = 18), ascending colon (n = 77), hepatic flexure (n = 12), transverse colon (n = 26), splenic flexure (n = 4), descending colon (n = 24), sigmoid colon (n =112), and rectum (n = 250). Their sizes ranged from 0.3 to 15 cm (mean, 5.7 cm).
All tissue samples were formalin-fixed and paraffin-embedded. Hematoxylin and eosin (HE)-stained slides, pathological reports, and other medical records were reviewed to confirm the diagnosis and clinicopathological parameters, including age, gender, tumor location, tumor size, depth of invasion, lymph node metastasis, distant metastasis, American Joint Committee on Cancer (AJCC) stage, Dukes’ stage, degree of differentiation, lymphovascular invasion and patient survival.
Tissue microarray (TMA) construction
The most representative area was carefully selected and marked on an H&E-stained slide. The TMA was assembled using a tissue-array instrument (AccuMac arrayer; ISU ABXIS Co. Ltd., Seoul, Korea) that consisted of thin-walled stainless steel punches and stylets used to empty and transfer the needle content. The assembly was held in an X-Y position guide equipped with semiautomatic micrometers, with a 1-mm increment between individual samples and a 3-mm punch depth stop device. Briefly, the instrument was used to create holes in a recipient block with defined array cores. A solid stylet, which closely fits the needle, was used to transfer the tissue cores into the recipient block. Taking into account the limitations of the representative areas of the tumor, we used triplicate 1-mm diameter tissue cores from each donor block.
HE and immunohistochemical staining
Multiple 4-μm sections were cut with a Leica microtome. Sections were transferred to adhesive-coated slides. One section was routinely deparaffinized with standard xylene and hydrated through graded ethanol in water, stained with HE, and covered with a coverslip. For immunohistochemical staining, the TMA slides were dewaxed by heating at 55°C for 30 min and by three 5-min washes with xylene. Tissues were rehydrated by a series of 5-min washes in 100%, 90% and 70% ethanol and phosphate buffered saline (PBS). Antigen retrieval was performed by microwaving the samples for 4 min 20 s at full power in 250 mL 10 mmol/L sodium citrate (pH 6.0). Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxidase for 20 min. The primary polyclonal rabbit anti-CD133 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was diluted 1:200 using goat serum and incubated at room temperature for 1 h. The primary monoclonal mouse anti-CD24 antibody (Santa Cruz Biotechnology) was diluted 1:50 and the primary monoclonal mouse anti-CD44s antibody (Neomarkers, CA, USA) was diluted 1:100. After three 2-min washes with PBS, the sections were incubated with a biotinylated goat secondary antibody for 30 min (DAKO, Carpinteria, CA, USA). After three 2-min washes with PBS, horseradish peroxidase-streptoavidin (DAKO) was added to the section for 30 min, followed by another three 2-min washes with PBS. The samples were developed with 3,3’-diaminobenzidine substrate (Vector Laboratories, Burlington, Ontario, Canada) for 1 min and counterstained with Mayer’s hematoxylin. The slides were dehydrated following a standard procedure and sealed with coverslips. We used the glioblastoma tissue as a positive control of CD133, the tonsillar lymphoid tissue as a positive control of CD24, and the tonsillar mucosal epithelial tissue as a positive control of CD44. Negative controls were performed by omitting CD133, CD24 and CD44 antibodies during the primary antibody incubation. The representative sections of CD133, CD24 and CD44 immunostaining are shown in Figure 1.
Figure 1.
Representative photograph of the TMA slides with immunohistochemical staining. A: CD133; B: CD24; C: CD44.
Interpretation of CD133, CD24 and CD44 expression
CD133, CD24 and CD44 expression was evaluated semi-quantitatively by two independent pathologists (Paik SS and Song YS), in a blinded fashion without knowledge of clinical and pathological information. The sections were scanned at high magnification to assess the positivity of staining in tumor cells. We regarded the staining as positive in cases with cytoplasmic positivity. In cases of discrepant assessments, slides were reinvestigated by both pathologists under a multi-head microscope and an agreement was obtained.
Statistical analysis
Statistical analysis was performed using SPSS version 12.0 software (SPSS, Chicago, IL, USA). The χ2 test was used to examine the association between CD133, CD24 and CD44 expression and various clinicopathological characteristics, including age, gender, tumor location, tumor size, TNM category, AJCC stage, Dukes’ stage, degree of differentiation, and lymphovascular invasion. The Kaplan-Meier method was used to calculate overall survival curves. Univariate survival analysis with the log-rank test was used to compare the difference between the survival rates of the patients’ subgroups. Multivariate survival analysis with Cox’s proportional hazard regression model was used to evaluate the independent prognostic factors. A difference of P < 0.05 between groups was considered significant.
RESULTS
Pattern of CD marker expression in colorectal adenocarcinoma
CD marker expression was evaluated in colorectal adenocarcinoma. One hundred and twenty-eight of 523 cases (24.5%) were positive and 395 cases (75.5%) were negative for CD133 expression (Figure 2A and B). Two hundred and sixty-four of 523 cases (50.5%) were positive and 259 cases (49.5%) were negative for CD24 expression (Figure 2C and D). However, 21 of 523 cases (4%) were positive and 502 cases (96%) were negative for CD44 expression (Figure 2E and F).
Figure 2.
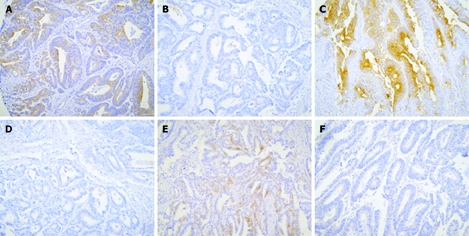
Representative photographs of CD marker expression in colorectal adenocarcinoma. Positive staining (A) and negative staining (B) for CD133. Positive staining (C) and negative staining (D) for CD24. Positive staining (E) and negative staining (F) for CD44.
Correlation of CD marker expression and clinicopathological parameters in colorectal adenocarcinoma
Upon clinicopathological analysis, CD133 expression was present more in male patients (P = 0.002) and in advanced T stage cancer (P = 0.024) (Table 1). Correlation between CD24 expression and clinicopathological factors was seen in the degree of differentiation (P = 0.006) (Table 1). Correlation between CD44 expression and clinicopathological factors was seen for the tumor size (P = 0.001) (data not shown).
Table 1.
Correlation between CD133 and CD24 expression and clinicopathological factors in colorectal cancer (n = 523)
| n |
Expression of CD133 |
Pvalue |
Expression of CD24 |
Pvalue |
|||
| Negative (n = 395) | Positive (n = 128) | (χ2-test) | Negative (n = 259) | Positive (n = 264) | (χ2-test) | ||
| Age (yr) | 0.431 | 0.999 | |||||
| < 59 | 261 | 201 | 60 | 129 | 132 | ||
| ≥ 59 | 262 | 194 | 68 | 130 | 132 | ||
| Gender | 0.002 | 0.261 | |||||
| Male | 295 | 208 | 87 | 140 | 155 | ||
| Female | 228 | 187 | 41 | 120 | 108 | ||
| Tumor location | 0.735 | 0.315 | |||||
| Right colon | 133 | 99 | 34 | 71 | 62 | ||
| Left colon | 390 | 296 | 94 | 189 | 201 | ||
| Tumor size | 0.436 | 0.658 | |||||
| < 5.5 cm | 254 | 188 | 66 | 124 | 130 | ||
| ≥ 5.5 cm | 269 | 207 | 62 | 136 | 133 | ||
| T category | 0.0241 | 0.219 | |||||
| Tis | 12 | 12 | 0 | 8 | 4 | ||
| T1 | 9 | 8 | 1 | 4 | 5 | ||
| T2 | 37 | 29 | 8 | 20 | 17 | ||
| T3 | 452 | 337 | 115 | 223 | 229 | ||
| T4 | 13 | 9 | 4 | 5 | 8 | ||
| N category | 0.890 | 0.525 | |||||
| N0 | 234 | 178 | 56 | 113 | 121 | ||
| N1 | 132 | 96 | 36 | 65 | 67 | ||
| N2 | 157 | 121 | 36 | 81 | 76 | ||
| M category | 0.555 | 0.482 | |||||
| M0 | 502 | 378 | 124 | 248 | 254 | ||
| M1 | 21 | 17 | 4 | 12 | 9 | ||
| AJCC stage | 0.259 | 0.998 | |||||
| 0 | 12 | 12 | 0 | 8 | 4 | ||
| I | 34 | 29 | 5 | 18 | 16 | ||
| IIA, IIB | 185 | 134 | 51 | 85 | 100 | ||
| IIIA, IIIB, IIIC | 271 | 203 | 68 | 137 | 134 | ||
| IV | 21 | 17 | 4 | 12 | 9 | ||
| Dukes’ stage | 0.560 | 0.515 | |||||
| A | 17 | 16 | 1 | 10 | 7 | ||
| B1, B2 | 210 | 157 | 53 | 98 | 112 | ||
| C1, C2 | 275 | 205 | 70 | 139 | 136 | ||
| D | 21 | 17 | 4 | 12 | 9 | ||
| Degree of differentiation | 0.084 | 0.0061 | |||||
| Well | 23 | 16 | 7 | 10 | 13 | ||
| Moderate | 393 | 300 | 93 | 183 | 210 | ||
| Poorly | 100 | 74 | 26 | 61 | 39 | ||
| Undifferentiated | 7 | 5 | 2 | 5 | 2 | ||
| Lymphatic invasion | 0.848 | 0.772 | |||||
| Absent | 225 | 169 | 56 | 110 | 115 | ||
| Present | 298 | 226 | 72 | 150 | 148 | ||
| Vascular invasion | 0.740 | 0.508 | |||||
| Absent | 513 | 387 | 126 | 254 | 259 | ||
| Present | 10 | 8 | 2 | 6 | 4 | ||
χ2 test for linear trend. AJCC: American Joint Committee on Cancer; CD: Cluster of differentiation.
Correlation of CD marker expression and patient overall survival
We examined the effect of CD marker expression on clinicopathological prognostic factors in colorectal adenocarcinoma. A significant prognostic influence of age, histological grade, AJCC stage, lymphatic invasion and vascular invasion on overall survival was found by univariate and/or multivariate analyses. However, no impact of CD133, CD24 and CD44 expression on overall survival was observed in univariate and multivariate survival analyses. Kaplan-Meier survival curves and log-rank tests showed no significant correlation between patient survival and CD133, CD24 and CD44 expression (P = 0.774, P = 0.775 and P = 0.636, respectively) (Figure 3).
Figure 3.
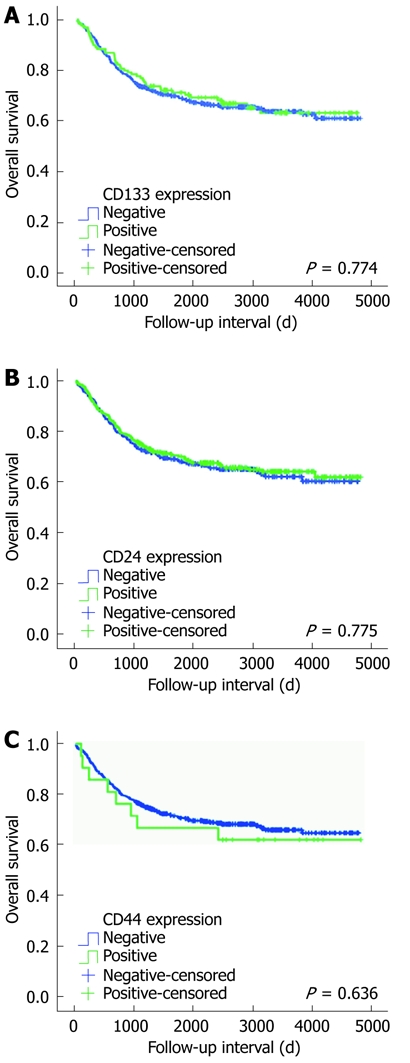
Cumulative survivals according to CD133 (P = 0.774) (A), CD24 (P = 0.775) (B) and CD44 (P = 0.636) (C) expression in colorectal cancer patients (Kaplan-Meier method).
DISCUSSION
Cancer stem cells have recently been proposed to be the cancer initiating cells that are responsible for tumorigenesis and for contributing to cancer resistance in leukemia[11]. Compared to leukemia, evidence for the existence of cancer stem cells in solid tumors has been more difficult to obtain for several reasons. Cells within solid tumors are less accessible, and functional assays suitable for detecting and quantifying normal stem cells from many organs have not yet been developed, and the cell surface markers required to isolate such cells have not been identified fully. However, there have been some impressive studies in this area recently. Advances have been made in identifying and enriching cancer stem cells in several solid tumors, including breast, brain and colon cancers[4–10].
Lapidot et al[12] have shown that leukemia-initiating stem cells present in the peripheral blood of acute myelogenous leukemia (AML) patients can induce AML when transplanted into severe combined immunodeficient mice. In 2003, Al-Hajj et al[8] isolated human breast cancer stem cells that can cause breast cancer in non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice through serial transplantations, which suggests a capacity for self-renewal. The following year, Singh et al[6] have found evidence of stem cell involvement in brain cancer. Recently, O’Brien et al[4] have demonstrated that CD133-positive colon-cancer-initiating cells in the human colon cancer specimen generate tumors in the renal capsule of pre-irradiated NOD/SCID mice. More recently, cells have been isolated from human prostate cancer patients, which can produce serially transplantable prostate tumors in NOD/SCID mice[10]. Even though definitive cancer stem cell markers have not been found in all the previously mentioned studies, these studies have revealed that only a small subset of cells in different tumor types is capable of tumor formation and several candidate stem cell markers have been evaluated. While CD133, CD24 and CD44 have been tested as cancer stem cell markers for serial transplantation studies in various cancers, their prognostic value has not been elucidated clearly[4–10].
CD133, which is one of the most important cancer stem cell markers[4–7], was stained fairly well in 24.5% of our colon cancer patients. In terms of clinicopathological parameters, CD133 expression was related to gender and T stage. The present study revealed that male gender was positively related to CD133 expression. T0 and T1 colon cancers showed lower incidence of CD133 protein expression compared to advanced colon cancer.
CD24, another important cancer stem cell marker, was expressed in 50.5% of the colon cancer patients. Correlation between CD24 expression and clinicopathological factors was seen in degree of differentiation. CD24 consists of a small protein core comprising 27 amino acids, which is extensively glycosylated and is bound to the membrane via a phosphatidylinositol anchor[13–15]. Several reports have shown that CD24 can be expressed on several solid tumors such as small cell lung cancer, neuroblastoma, rhabdomyosarcoma and renal cell cancer[16,17]. Lim et al[18] have reported that CD24 expression is related to lymph node metastasis in colon cancer. Weichert et al[19] have shown that cytoplasmic CD24 expression in colorectal cancer is independently correlated with shortened patient survival. We have not seen any positive correlation between CD24 expression and nodal status and patient survival.
CD44 is an unique adhesion molecule and several studies have revealed that CD44 is overexpressed at the mRNA and protein levels in colon cancer[20,21]. In our study, large tumors bigger than 5.5 cm showed more frequent CD44 expression, which indicated that CD44 expression was related to clinical tumor burden.
Our results in human colon cancer specimens showed various expression patterns of CD markers. This is believed to be the first trial to verify the relationship between well-known prognostic factors of colon cancer and conventional cancer stem cell markers. Tumor invasiveness and differentiation were identified as clinicopathological factors related to cancer stem cell markers, especially CD133 and CD24.
For further study, other colon stem cell markers which are related to patient survival should be found for clinical application of colon cancer stem cells. Several studies have indicated that pleuripotency-related factors, including Oct3/4, are related to cancer development[22–24]. Beside CD markers, other cancer stem cell markers may help distinguish cancer stem cells from cancer cells.
In summary, we have presented some evidence that several conventional clinical factors were related to cancer stem cell markers in colorectal adenocarcinoma. However, CD133, CD24 and CD44 expression did not show a close relationship with the survival outcome of colorectal adenocarcinoma. These results warrant further careful and well-designed studies of colon cancer stem cells as markers for clinical application.
COMMENTS
Background
Colorectal adenocarcinoma is the second most common type of cancer and a major cause of cancer-related morbidity and mortality in the Western world. Countless treatment protocols including chemotherapy and radiation have been applied to colorectal cancer treatment and a number of studies have identified conventional prognostic factors. Recently, the prospective identification of colon cancer stem cells has received major attention because of their potential for colon cancer treatment.
Research frontiers
This study focused on the identification of CD133-, CD24- and CD44-positive tumor cells in colon tumor sections by an immunohistochemistry-based technique, and the findings are discussed in conjunction with clinicopathological data.
Innovations and breakthroughs
Results in human colon cancer specimens showed various expression patterns of CD markers. This is believed to be the first trial for verifying the relationship between well-known prognostic factors of colon cancer and cancer stem cell markers. Tumor invasiveness and differentiation were identified as clinicopathological factors related to cancer stem cell markers.
Applications
The authors have presented some evidence that several conventional clinical factors were related to cancer stem cell markers in colorectal adenocarcinoma. However, CD133, CD24 and CD44 expression did not show a close relationship with survival. These results warrant further careful and well-designed studies of colon cancer stem cells as markers for clinical application.
Terminology
CD133, also known as PROML1 or prominin, is a stem cell surface antigen that has been identified recently as a potential cancer stem cell marker in brain, colon and prostate cancer. CD44, also known as homing cell adhesion molecule, is a cell surface glycoprotein expressed on lymphocytes, monocytes and granulocytes, which has been identified as a stem cell marker in breast and head and neck cancer. CD24, a cell surface marker, is a single chain sialoglycoprotein with a molecular mass of 42 kDa.
Peer review
The authors have presented some evidence that several conventional clinical factors were related to cancer stem cell markers in colorectal adenocarcinoma. This study is modestly reported with respect to a careful and large study. The approach needs to be encouraged, despite the relatively negative nature of the study with respect to the utility of the markers as a prognostic markers.
Footnotes
Supported by The Research fund of Hanyang University (HY-2007-C) to Paik SS
Peer reviewer: Finlay A Macrae, MD, Professor, Royal Melbourne Hospital, Po Box 2010, Victoria 3050, Australia
S- Editor Cheng JX L- Editor Kerr C E- Editor Yin DH
References
- 1.Compton CC. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod Pathol. 2003;16:376–388. doi: 10.1097/01.MP.0000062859.46942.93. [DOI] [PubMed] [Google Scholar]
- 2.Bae JM, Won YJ, Jung KW, Park JG. Annual report of the Korea central cancer registry program 2000: based on registered data from 131 hospitals. Cancer Res Treat. 2001;34:77–83. doi: 10.4143/crt.2002.34.2.77. [DOI] [PubMed] [Google Scholar]
- 3.Chung KY, Saltz LB. Adjuvant therapy of colon cancer: current status and future directions. Cancer J. 2007;13:192–197. doi: 10.1097/PPO.0b013e318074d26e. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 5.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 6.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 7.Miki J, Furusato B, Li H, Gu Y, Takahashi H, Egawa S, Sesterhenn IA, McLeod DG, Srivastava S, Rhim JS. Identification of putative stem cell markers, CD133 and CXCR4, in hTERT-immortalized primary nonmalignant and malignant tumor-derived human prostate epithelial cell lines and in prostate cancer specimens. Cancer Res. 2007;67:3153–3161. doi: 10.1158/0008-5472.CAN-06-4429. [DOI] [PubMed] [Google Scholar]
- 8.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 11.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 12.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 13.Pirruccello SJ, LeBien TW. The human B cell-associated antigen CD24 is a single chain sialoglycoprotein. J Immunol. 1986;136:3779–3784. [PubMed] [Google Scholar]
- 14.Fischer GF, Majdic O, Gadd S, Knapp W. Signal transduction in lymphocytic and myeloid cells via CD24, a new member of phosphoinositol-anchored membrane molecules. J Immunol. 1990;144:638–641. [PubMed] [Google Scholar]
- 15.Kay R, Rosten PM, Humphries RK. CD24, a signal transducer modulating B cell activation responses, is a very short peptide with a glycosyl phosphatidylinositol membrane anchor. J Immunol. 1991;147:1412–1416. [PubMed] [Google Scholar]
- 16.Jackson D, Waibel R, Weber E, Bell J, Stahel RA. CD24, a signal-transducing molecule expressed on human B cells, is a major surface antigen on small cell lung carcinomas. Cancer Res. 1992;52:5264–5270. [PubMed] [Google Scholar]
- 17.Akashi T, Shirasawa T, Hirokawa K. Gene expression of CD24 core polypeptide molecule in normal rat tissues and human tumor cell lines. Virchows Arch. 1994;425:399–406. doi: 10.1007/BF00189578. [DOI] [PubMed] [Google Scholar]
- 18.Lim SC, Oh SH. The role of CD24 in various human epithelial neoplasias. Pathol Res Pract. 2005;201:479–486. doi: 10.1016/j.prp.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Weichert W, Denkert C, Burkhardt M, Gansukh T, Bellach J, Altevogt P, Dietel M, Kristiansen G. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin Cancer Res. 2005;11:6574–6581. doi: 10.1158/1078-0432.CCR-05-0606. [DOI] [PubMed] [Google Scholar]
- 20.Wielenga VJ, Heider KH, Offerhaus GJ, Adolf GR, van den Berg FM, Ponta H, Herrlich P, Pals ST. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993;53:4754–4756. [PubMed] [Google Scholar]
- 21.Woodman AC, Sugiyama M, Yoshida K, Sugino T, Borgya A, Goodison S, Matsumura Y, Tarin D. Analysis of anomalous CD44 gene expression in human breast, bladder, and colon cancer and correlation of observed mRNA and protein isoforms. Am J Pathol. 1996;149:1519–1530. [PMC free article] [PubMed] [Google Scholar]
- 22.Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 23.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]