Abstract
Loss-of-function mutations of DAP12 and TREM2 cause a recessively inherited disease PLOSL, manifesting in brain white matter. The genes of the DAP12-TREM2 signaling receptor are located on 19q13.12 and 6p21.1, to which linkage has been observed also in families affected by another immune-mediated demyelinating disease, MS. We have tested if allelic variation in DAP12 or TREM2 predisposes also to MS by monitoring carrier frequency of the Finnish PLOSL mutation in Finnish MS cases and by studying DAP12 and TREM2 in MS by linkage and association. To conclude, the DAP12-TREM2 complex unlikely has a role in genetic susceptibility of MS.
Keywords: MS, PLOSL, DAP12, TREM2, Association
1. Introduction
The quite recently identified signalling receptor complex, formed by DAP12 (the gene also known as TYROBP, TYRO protein tyrosine kinase binding protein) and TREM2 (triggering receptor expressed on myeloid cells 2), is an important regulator of the innate immune system (Klesney-Tait et al., 2006) The DAP12 and TREM2 genes are expressed in various cell types of the myeloid lineage. The exact cellular processes as well as the ligands for TREM2 are still largely unrecognized.
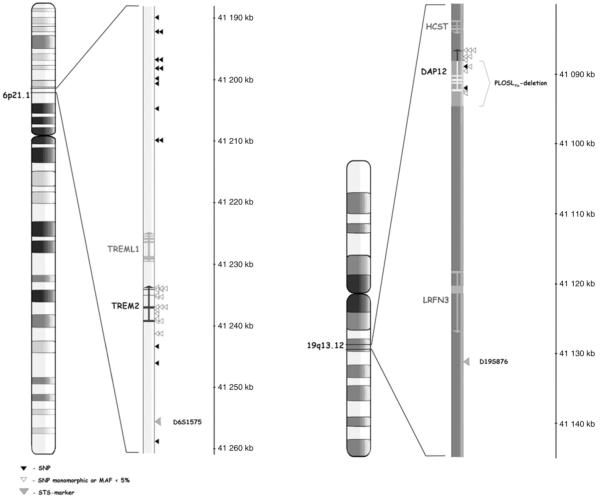
Mutations of the DAP12 and TREM2 genes cause a recessively inherited rare disease of the bone and brain white matter called polycystic lipomembranous osteodysplasia with sclerosing leucoencephalopathy (PLOSL) (Paloneva et al., 2000, 2002). All Finnish PLOSL patients carry a 5.3 kb deletion of DAP12 (PLOSLFin-deletion, Fig. 1), and inactivating point mutations of TREM2 have been found in PLOSL patients of other populations (Paloneva et al., 2002). Thefirst symptoms of the disease are typically pain and fractures in wrists and ankles at early adulthood, followed by neuropsychiatric symptoms, dementia and premature death. The most prominent feature of PLOSL, as well as Dap12 deficient mice, is myelin loss in the central nervous system (CNS) (Paloneva et al., 2001; Kaifu et al., 2003; Nataf et al., 2005). Both microglia, the resident immune cells of the CNS, and oligodendrocytes, myelin forming cells of the CNS, express DAP12 and TREM2 proteins (Kaifu et al., 2003; Roumier et al., 2004; Kiialainen et al., 2005; Takahashi et al., 2005). Interestingly, TREM2 overexpression has been observed to protect mice from autoimmune demyelination; when experimental autoimmune encephalomyelitis (EAE) mice were injected with TREM2-transduced myeloid precursor cells, the clinical symptoms of the mice alleviated, and there was less axonal damage in the mice CNS (Takahashi et al., 2007).
Fig. 1.
Polymorphisms in TREM2 and DAP12. Alignment of the studied polymorphisms in the genes of interest. SNPs with MAF≥0.05 according to HapMap (CEU) or our validation sample set of 260 Finnish individuals are marked as black triangles, other SNPs as white triangles, and STS markers as grey triangles.
The DAP12 and TREM2 genes are located on chromosomes 19q13.12 and 6p21.1, respectively. Interestingly, linkage to these loci has been observed also in families affected by another, more common demyelinating disease, multiple sclerosis (MS) (Ebers et al., 1996; Haines et al., 1996; Sawcer et al., 1996; Kuokkanen et al., 1997; Haines et al., 2002; Reunanen et al., 2002; Haghighi et al., 2006). MS is a chronic inflammatory disease of the CNS with complex inheritance and a putative autoimmune pathogenesis (Sadovnick et al., 1993; Ebers et al., 1995). The etiology of MS is still poorly understood, but both twin and population studies demonstrate a significant genetic component (Sadovnick et al., 1993; Oksenberg and Barcellos, 2005).
We wanted to test if allelic variation of the genes coding for the DAP12-TREM2 receptor complex predisposes also to MS, which shares some common features with PLOSL. Since homozygous mutation of DAP12 results in severe white matter changes and premature death in Finnish PLOSL, we hypothesized that heterozygous PLOSLFin-mutation could lead to a milder, relapsing phenotype like MS. We also wanted to study the role of allelic variation of DAP12 and TREM2 in MS by linkage and association analyses.
2. Materials and methods
2.1. Study sample
Of the 905 unrelated Finnish MS cases (Table 1), 172 originate from the Southern Ostrobothnian MS high-risk region (Wikstrom and Palo, 1975; Sumelahti et al., 2000, 2001) and the rest are from other parts of Finland. 744 unrelated MS cases representing equally our full study set were selected randomly from these cases and genotyped for the PLOSLFin-deletion. Of the 1348 Finnish population controls (Table 1), 389 were from Southern Ostrobothnia MS high-risk region and the rest were from other parts of Finland. Genotypes of the SNP rs3817624 in 1376 population controls were obtained from Finnish genome wide association studies (unpublished data). Of the 24 Finnish multiplex MS families (Table 1), nine did not carry the HLA-DR15 risk allele. The diagnosis of MS has strictly followed Poser’s diagnostic criteria. All individuals have given their informed consent and the study has been approved by the Ethics Committee for Ophthalmology, Otorhinolaryngology, Neurology and Neurosurgery in the Hospital District of Helsinki and Uusimaa (Decision 46/2002, Dnro 192/E9/02).
Table 1.
Study sample
| PLOSLFin-deletion | SNPs | STS-markers | |
|---|---|---|---|
| Number of affected | 744 | 905 | 55 |
| Number of families | - | - | 24 |
| Number of unrelated controls | 1348 | 1348 | - |
2.2. Testing carrier frequency of the PLOSLFin-deletion
We developed a high throughput method to monitor for the carrier frequency of the PLOSLFin-deletion using the ABI 3730xl DNA Analyzer and GeneMapper 4.0 software (Applied Biosystems, Foster City, CA, USA) (Fig. 2). All the pipeting procedures were automatized to Tecan pipeting robots (Tecan Group Ltd., Männerdorf, Switzerland). A single PCR was performed as a multiplex reaction using a FAM-6 labelled Reverse-primer (5′-GATGAGACTAGGAGGCTGATGCT-3′) annealing upstream from the telomeric deletion brake point. The Forward-primers were different for the two alleles; we used a primer (5′-TAGTATGTCCAG TCTCGAGTTCTCA-3′) that annealed to the region deleted in the PLOSLFin-mutation for the control allele, and another primer (5′-CAGAGAGGGGGATAGATGACTA-3′) for the mutant allele that annealed to the sequence downstream from the centromeric deletion breakpoint. 5.0 ng of genomic DNA and 0.28 U of Amplitaq Gold enzyme (Applied Biosystems) was used in the PCR with concentrations of 200 μM of dNTP, 1.50 mM of MgCl2, 1× Amplitaq PCR Buffer, 300 nM of each Forward-primer and 200 nM of the FAM-6 labelled Reverse-primer. The reaction was carried out in conditions as follows: initial denaturation in 94 °C for 12 min; 10 cycles of 94 °C for 30 s, 60 °C for 15 s reduced by 0.5 °C per cycle, and 72 °C for 30 s; 25 cycles of 94 °C for 30 s, 55 °C for 15 s, and 72 °C for 30 s; 72 °C for 10 min, and cooling to 4 °C until further use. Only one PCR-product was produced from the control allele, since the putative product containing the deleted segment would have been over 5 kb and thus did not amplify in the multiplex reaction. PCR-products were diluted in ddH2O and run on ABI 3730xl DNA Analyzer in Hi-Di containing the GeneScan-500 LIZ size standard. The ABI Analyzer separated the PCR-products in capillary gel electrophoresis according to size, and measured the FAM-6 signal peaks. The size for the control allele PCR-product was 409 bp and for the mutant allele 540 bp. Allele calling was done using the GeneMapper 4.0-software (Applied Biosystems). Either a Finnish PLOSL patient or a known mutation carrier was used as homozygous or heterozygous positive control, respectively, on every 96-well plate analyzed. The success rate for the assay was of 94%. Genotypes for all positive and negative (water) controls were called correctly. Genotypes of two identified deletion carriers among the MS cases were further verified by agarose gel electrophoresis.
Fig. 2.
Testing Carrier Frequency of the PLOSLFin-deletion in Finnish MS. An in-house developed high throughput method was developed to monitor for the carrier frequency of the PLOSLFin-deletion. The star refers to the FAM-6 labelled Reverse-primer. Two different Forward-primers were used, producing only one PCR-product from the control allele (the other product would have been over 5 kb) and two PCR-products from the mutant allele (409 bp and 540 bp).
2.3. Marker selection and genotyping
All putative SNPs mapping to DAP12 and TREM2 were initially selected from UCSC Genome Browser, hg18 assembly, Mar2006 (Fig. 1, triangles; Supplementary table 1). None of the SNPs mapping to TREM2 had MAF≥0.05 according to HapMap (CEU) or our validation sample set of 260 Finnish individuals. To capture possible variation in this highly conserved locus, additional 15 polymorphic SNPs flanking TREM2 were selected for genotyping. To identify more polymorphisms we resequenced most of the DAP12 gene and identified four novel non-coding SNPs: DAP12_1, DAP12_2, DAP12_3, and DAP12_4 (positions on Chr19 according to UCSC Genome Browser, hg18 assembly, Mar2006 are 41 087 962, 41 089 190, 41 091 004 and 41 091 073, respectively) (Supplementary table 1). Only SNP DAP12_2 had MAF≥0.05 in our validation sample set and was selected for genotyping.
The final set of 17 SNPs (Fig. 1, black triangles) was genotyped using the Sequenom’s MassARRAY iPLEX system (Sequenom, San Diego, California, United States). PCR and extension-reaction primers were designed using Assay Designer 3.1 (Sequenom). 15 ng of genomic DNA was used in PCR, and PCR-product purification and extension reactions were carried out according to manufacturer’s protocol. Automatic allele calling was done with SpectroCALLER-software (Sequenom), and the genotypes were further manually checked as described elsewhere (Silander et al., 2003). The SNPs were genotyped in two multiplex reactions in the set of MS cases, and in one multiplex reaction in the controls. SNPs DAP12_2 and rs3817624 did not work in the genotyping multiplex for controls, thus DAP12_2 was re-genotyped in 1 104 controls (from Southern Ostrobothnia, Helsinki and Kuopio) in a singleplex reaction, and genotype data from Finnish genome wide association studies was utilized for rs3817624.
Two STS markers nearby DAP12 and TREM2 (D19S876 and D6S1575; Fig. 1, grey triangles) were genotyped in fragment analysis on ABI 3730xl DNA Analyzer (Applied Biosystems). Primers stated at the UCSC Genome Browser were used in genotyping, and allele calling was done with GeneMapper 4.0 software (Applied Biosystems).
2.4. Sequencing
We resequenced 78% of the DAP12 gene (2 000 bases of the DAP12 promoter, exons 1, 2 and 5, introns 1 and 2, 1500 bases of intron 4 and 600 bases downstream of DAP12) in 12 Finnish MS cases, three healthy family members and four unrelated controls. Due to technical difficulties caused by the intense short interspersed nuclear elements (SINEs) and ALU-elements in intronic and promoter regions of DAP12, we were not able to sequence the whole gene. The primer design was done using Primer3, 10ul PCR reactions were cleaned up using ExoSAP-IT (USB Corporation, Cleveland Ohio, United States), and detection was done using ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
2.5. Statistical analyses
Deviation from the Hardy-Weinberg equilibrium was tested using Pearson’s chi-square test. Only SNPs with genotyping success rate >90% and validity of Hardy-Weinberg equilibrium in control samples were chosen into analyses. SNP allele frequencies and haplotype frequencies were estimated from the data, and the case-control p-values were obtained from the Haploview custom association test (Barrett et al., 2005) (Table 2 and Supplementary Table 2, represented as uncorrected p-values). Linkage disequilibrium (r2) between the SNPs in the Finnish case-control sample was illustrated using Haploview (Supplementary Figure 1). Twopoint LOD scores for the STS markers were calculated using the MLINK program (Lathrop and Lalouel, 1984; Cottingham et al., 1993) (recessive model of inheritance, f=0.76).
Table 2.
Association analysis of the polymorphic variants of DAP12 and TREM2 and the flanking regions
| Chr | dbSNP | Positiona | Location | Case/Control MAF | P-valueb |
|---|---|---|---|---|---|
| 6 | rs878998 | 41 187 979 | 46 kb upstream from TREM2 | 0.10/0.09 | 0.348 |
| 6 | rs10456499 | 41 191 330 | 43 kb” | 0.40/0.40 | 0.643 |
| 6 | rs6926018 | 41 191 485 | 43 kb” | 0.07/0.07 | 0.881 |
| 6 | rs11968153 | 41 197 288 | 37 kb” | 0.10/0.09 | 0.400 |
| 6 | rs722442 | 41 197 659 | 37 kb” | 0.35/0.33 | 0.238 |
| 6 | rs6924867 | 41 198 148 | 36 kb ” | 0.11/0.09 | 0.150 |
| 6 | rs6905117 | 41 198 227 | 36 kb ” | 0.11/0.10 | 0.158 |
| 6 | rs6926079 | 41 198 660 | 36 kb ” | 0.10/0.09 | 0.425 |
| 6 | rs11969526 | 41 199 285 | 35 kb ” | 0.10/0.10 | 0.406 |
| 6 | rs11964090 | 41 207 143 | 27 kb ” | 0.11/0.09 | 0.154 |
| 6 | rs6918289 | 41 209 806 | 24 kb ” | 0.10/0.09 | 0.310 |
| 6 | rs6923053 | 41 210 061 | 24 kb ” | 0.11/0.10 | 0.179 |
| 6 | rs774877 | 41 241 784 | 3 kb downstream from TREM2 | 0.46/0.43 | 0.067 |
| 6 | rs7759295 | 41 243 828 | 5 kb ” | 0.09/0.10 | 0.097 |
| 6 | rs9357347 | 41 258 569 | 20 kb ” | 0.35/0.32 | 0.054 |
| 19 | DAP12_2 | 41 089 190 | DAP12 | 0.31/0.30 | 0.564 |
| 19 | rs3817624 | 41 090 739 | DAP12 | 0.11/0.09 | 0.064 |
Position according to UCSC Genome Browser, Mar 2006 freeze.
Haploview custom association test.
3. Results
3.1. Carrier frequency of the Finnish PLOSL mutation in Finnish MS cases
The 5.3 kb PLOSLFin-deletion of DAP12 (chr19: 41 088 604-41 093 869, UCSC Genome Browser, hg18 assembly, Mar2006) (Fig. 1) was genotyped in 744 unrelated Finnish MS cases and 1348 population controls (Table 1) using an in-house developed high throughput method (Fig. 2). Two carriers of the deletion were identified among the MS cases, corresponding to carrier frequency of 2.7/1000. The clinical picture of these MS patients did not differ from that of the non-carriers. The other carrier had a relapsing, remitting MS with only mild symptoms, as the other had a more severe disease course of primary progressive MS. A common feature between the two patients was relatively wide changes in the brain white matter in the magnetic resonance imaging. No cognitive or psychiatric symptoms were observed in their disease history. DNA was available for the mother and a sibling of one of the two carriers. Thesefirst-degree relatives were also genotyped for the mutation, and both were observed to be carriers. Six carriers of the deletion were identified among the controls, corresponding to carrier frequency of 4.9/1000. According to the prevalence of PLOSL, carrier frequency of the deletion has previously been estimated to be 2.4/1000 in Finland (Hakola, 1990), which was now showed to be slightly underestimated. Frequency of the deletion was further compared between the Southern Ostrobothnian MS high-risk region, where the prevalence and familial occurrence of MS are exceptionally high due to a founder effect and isolation (Wikstrom and Palo, 1975; Sumelahti et al., 2000, 2001), and other parts of Finland. Of the studied MS cases 138 and of the controls 389 originated from Southern Ostrobothnia. The carrier frequency was observed to be comparable in the geographical MS high-risk region: the deletion was carried by three population controls (corresponding to regional carrier frequency of 5.7/1000). Importantly, the DAP12 deletion was not observed to be over-represented among the MS cases.
3.2. Allelic variation of DAP12 and TREM2 in MS
We further studied the role of allelic variation in DAP12 and TREM2 in MS by linkage and association analyses. All putative SNPs (n=24) mapping to DAP12 and TREM2 were initially selected from public databases (UCSC Genome Browser, hg18 assembly, Mar2006) (Supplementary Table 1). Of these SNPs only one (rs3817624 in DAP12) had a minor allele frequency (MAF)≥0.05 according to HapMap (CEPH individuals of European origin) or our validation sample of 260 Finnish individuals and was further selected for genotyping. Due to the low number of polymorphic SNPs in these genes, parts of DAP12 were re-sequenced in 19 Finnish individuals. Four novel non-coding SNPs were observed, but only one of these had a MAF≥0.05 in the validation sample, and this SNP (Supplementary Table 1, DAP12_2) was included in the genotyping panel. Further, 15 polymorphic SNPs (MAF≥0.05 in our validation sample set) flanking the TREM2 gene were selected for genotyping to capture possible variation in this highly conserved locus (Fig. 1).
The final set of 17 polymorphic SNPs (Fig. 1) was genotyped in 905 Finnish MS cases and 1348 population controls (Table 1). No evidence for association was observed with any of the SNPs or the haplotypes: the allele frequencies were almost identical between the MS cases and the controls (Table 2, Supplementary Table 2). Two of the genotyped SNPs mapped to the PLOSLFin-deletion region (Fig. 1). As expected, the genotypes of these SNPs were homozygous in individuals known to carry the deletion. Linkage disequilibrium between the SNPs is shown in Supplementary Fig. 1.
Due to a low number of polymorphic SNPs in TREM2 and DAP12, two STS markers nearby these genes (D6S1575 and D19S876, respectively) (Fig. 1) were genotyped in 24 multiplex MS families (Table 1). No evidence for linkage was observed (maximum pairwise LOD score 0.18 for D6S1575 and 0.06 for D19S876). On the basis of previous reports (Reunanen et al., 2002; Dyment et al., 2004) we performed linkage analysis also after HLA-DR15 stratification, but again, no evidence for linkage was observed: a maximum pairwise LOD score of 0.16 was observed with the marker D19S876 in families not carrying the HLA-DR15 risk allele (9 multiplex MS families).
4. Discussion
The lack of polymorphisms makes DAP12 and TREM2 difficult to study. None of the SNPs listed in public databases within exonic or intronic regions of TREM2 were observed to have a MAF≥0.05, and only two polymorphic intronic SNPs were observed in DAP12. The four novel SNPs identified by re-sequencing were also located on non-coding regions of DAP12, and only one of those had a MAF≥0.05. As has been demonstrated previously, the low number of variants in DAP12 and TREM2 most probably indicates the crucial role of this receptor complex in immune response modulation (Fenoglio et al., 2007).
The first MS genome-wide association (GWA) study was published recently (International Multiple Sclerosis Genetics Consortium et al., 2007), but the Affymetrix 550K SNP panel used in the study includes only one SNP in TREM2 (rs7748513, MAF 0.02 in our validation study set, Supplementary Table 1) and no SNPs in DAP12. Neither do the latest GWA panels, SNP Array 6.0 from Affymetrix and Human1M BeadChip from Illumina, cover these genes properly: The TREM2 SNPs of both panels have been proven to have a MAF<0.01 (at least in populations of European origin) by either HapMap (CEU population, rs2234250, rs2234253, rs2234255, rs2234256) or us (rs2234258, rs7748513). There are no SNPs on SNP 6.0 array to DAP12, and the SNPs on Illumina BeadChip have a MAF<0.01 (rs1802029, rs7260613, rs8110231, rs8113524, validated by HapMap or us) or have been studied here (rs3817624). Thus, the future genome wide association studies might bring some enlightenment for potential involvement of copy number variations in these genes in MS pathogenesis, but the role of genetic variation will remain unknown. Further, deep sequencing of DAP12 and TREM2 in multiple MS cases could potentially reveal rare sporadic mutations, which are not detectable with conventional linkage and association studies.
Evidence for linkage to chromosomes 6p21 and 19q13 has been observed in several studies and in multiple populations, including Finns. Linkage to 6p21 is generally thought to be, at least mainly, due to HLA-DRB1, and this was supported by our data; STS markers near DAP12 and TREM2 provided no evidence for linkage in Finnish multiplex MS families. However we cannot exclude the possibility that rare, even family-specific variants of these loci could potentially contribute to the linkage previously detected to these chromosomes.
Notably, only homozygous deletion of DAP12 causes PLOSL in Finland, and no similar neurological symptoms have been observed in heterozygous carriers of this mutation (P. Hakola, personal communication). Moreover, as we stated here, no differences were observed in the clinical picture of the MS cases carrying the Finnish PLOSL mutation when compared to that of the non-carriers. Further, no MS cases are known to exist in the Finnish PLOSL pedigrees (P. Hakola, personal communication). However, number of Finnish PLOSL families is too small to make any final conclusions.
To summarize, we have studied the known polymorphisms of DAP12 and TREM2. No evidence for association with MS was observed, and the PLOSLFin-mutation was not enriched in MS patients. The highly conserved molecules DAP12 and TREM2, loss of function mutations of which cause a lethal disease PLOSL, unlikely have a role in genetic susceptibility of multiple sclerosis.
Supplementary Material
Acknowledgements
We wish to thank all participating MS patients, the Finnish Genome Center, Professor Panu Hakola for his expert advice concerning PLOSL families, and Dr. Anna Kiialainen for the PLOSL samples used as positive controls in PLOSLFin-deletion genotyping. This work was supported by NIH grant RO1 NS 43559, grants from the Center of Excellence for Disease Genetics of the Academy of Finland, research grant of the Finnish Academy, the Sigrid Juselius Foundation, Helsinki University Central Hospital, and a grant LSHM-CT-2005-018637 from Neuropromise EU project.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jneuroim.2008.10.005
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Cottingham RW, Jr, Idury RM, Schäffer AA. Faster sequential genetic linkage computations. Am. J. Hum. Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- Dyment DA, Ebers GC, Sadovnick AD. Genetics of multiple sclerosis. Lancet Neurol. 2004;3:104–110. doi: 10.1016/s1474-4422(03)00663-x. [DOI] [PubMed] [Google Scholar]
- Ebers GC, Sadovnick AD, Risch NJ. A genetic basis for familial aggregation in multiple sclerosis. Nature. 1995;377:150–151. doi: 10.1038/377150a0. [DOI] [PubMed] [Google Scholar]
- Ebers GC, Kukay K, Bulman DE, Sadovnick AD, Rice G, Anderson C, Armstrong H, Cousin K, Bell RB, Hader W, Paty DW, Hashimoto S, Oger J, Duquette P, Warren S, Gray T, O’Connor P, Nath A, Auty A, Metz L, Francis G, Paulseth JE, Murray TJ, Pryse-Phillips W, Nelson R, Freedman M, Brunet D, Bouchard JP, Hinds D, Risch N. A full genome search in multiple sclerosis. Nat. Genet. 1996;13:472–476. doi: 10.1038/ng0896-472. [DOI] [PubMed] [Google Scholar]
- Fenoglio C, Galimberti D, Piccio L, Scalabrini D, Panina P, Buonsanti C, Venturelli E, Lovati C, Forloni G, Mariani C, Bresolin N, Scarpini E. Absence of TREM2 polymorphisms in patients with Alzheimer’s disease and frontotemporal lobar degeneration. Neurosci. Lett. 2007;411:133–137. doi: 10.1016/j.neulet.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Haghighi S, Andersen O, Nilsson S, Rydberg L, Wahlström J. A linkage study in two families with multiple sclerosis and healthy members with oligoclonal CSF immunopathy. Mult. Scler. 2006;12:723–730. doi: 10.1177/1352458506070972. [DOI] [PubMed] [Google Scholar]
- Haines JL, Ter-Minassian M, Bazyk A, Gusella JF, Kim DJ, Terwedow H, Pericak-Vance MA, Rimmler JB, Haynes CS, Roses AD, Lee A, Shaner B, Menold M, Seboun E, Fitoussi R-P, Gartioux C, Reyes C, Ribierre F, Gyapay G, Weissenbach J, Hauser SL, Goodkin DE, Lincoln R, Usuku K, Garcia-Merino A, Gatto N, Young S, Oksenberg JR. A complete genomic screen for multiple sclerosis underscores a role for the major histocompatability complex. Nat. Genet. 1996;13:469–471. doi: 10.1038/ng0896-469. [DOI] [PubMed] [Google Scholar]
- Haines JL, Bradford Y, Garcia ME, Reed AD, Neumeister E, Pericak-Vance MA, Rimmler JB, Menold MM, Martin ER, Oksenberg JR, Barcellos LF, Lincoln R, Hauser SL, Multiple Sclerosis Genetics Group Multiple susceptibility loci for multiple sclerosis. Hum. Mol. Genet. 2002;11:2251–2256. doi: 10.1093/hmg/11.19.2251. [DOI] [PubMed] [Google Scholar]
- Hakola P. Polycystic lipomembranous osteodysplasia with sclerosing leukoence-phalopathy (membranous lipodystrophy): a neuropsychiatric follow-up study. Monogr. Psych. Fenn. 1990;17:1–114. [Google Scholar]
- International Multiple Sclerosis Genetics Consortium. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- Kaifu T, Nakahara J, Inui M, Mishima K, Momiyama T, Kaji M, et al. Osteopetrosis and thalamic hypomyelinosis with synaptic degeneration in DAP12-deficient mice. J. Clin. Invest. 2003;111:323–332. doi: 10.1172/JCI16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiialainen A, Hovanes K, Paloneva J, Kopra O, Peltonen L. Dap12 and Trem2, molecules involved in innate immunity and neurodegeneration, are co-expressed in the CNS. Neurobiol. Dis. 2005;18:314–322. doi: 10.1016/j.nbd.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Klesney-Tait J, Turnbull IR, Colonna M. The TREM receptor family and signal integration. Nat. Immunol. 2006;7:1266–1273. doi: 10.1038/ni1411. [DOI] [PubMed] [Google Scholar]
- Kuokkanen S, Gschwend M, Rioux JD, Daly MJ, Terwilliger JD, Tienari PJ, Wikström J, Palo J, Stein LD, Hudson TJ, Lander ES, Peltonen L. Genomewide scan of multiple sclerosis in Finnish multiplex families. Am. J. Hum. Genet. 1997;61:1379–1387. doi: 10.1086/301637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM. Easy calculations of LOD scores and genetic risks on small computers. Am. J. Hum. Genet. 1984;36:460–465. [PMC free article] [PubMed] [Google Scholar]
- Nataf S, Anginot A, Vuaillat C, Malaval L, Fodil N, Chereul E, Langlois JB, Dumontel C, Cavillon G, Confavreux C, Mazzorana M, Vico L, Belin MF, Vivier E, Tomasello E, Jurdic P. Brain and bone damage in KARAP/DAP12 loss-of-function mice correlate with alterations in microglia and osteoclast lineages. Am. J. Pathol. 2005;166:275–286. doi: 10.1016/S0002-9440(10)62251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg JR, Barcellos LF. Multiple sclerosis genetics: leaving no stone unturned. Genes Immun. 2005;6:375–387. doi: 10.1038/sj.gene.6364237. [DOI] [PubMed] [Google Scholar]
- Paloneva J, Kestilä M, Wu J, Salminen A, Böhling T, Ruotsalainen V, Hakola P, Bakker AB, Phillips JH, Pekkarinen P, Lanier LL, Timonen T, Peltonen L. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat. Genet. 2000;25:357–361. doi: 10.1038/77153. [DOI] [PubMed] [Google Scholar]
- Paloneva J, Autti T, Raininko R, Partanen J, Salonen O, Puranen M, Hakola P, Haltia M. CNS manifestations of Nasu-Hakola disease: a frontal dementia with bone cysts. Neurology. 2001;56:1552–1558. doi: 10.1212/wnl.56.11.1552. [DOI] [PubMed] [Google Scholar]
- Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, Tranebjaerg L, Konttinen Y, Peltonen L. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reunanen K, Finnilä S, Laaksonen M, Sumelahti ML, Wikström J, Pastinen T, Kuokkanen S, Saarela J, Uimari P, Ruutiainen J, Ilonen J, Peltonen L, Tienari PJ. Chromosome 19q13 and multiple sclerosis susceptibility in Finland: a linkage and two-stage association study. J. Neuroimmunol. 2002;126:134–142. doi: 10.1016/s0165-5728(02)00051-6. [DOI] [PubMed] [Google Scholar]
- Roumier A, Béchade C, Poncer JC, Smalla KH, Tomasello E, Vivier E, Gundelfinger ED, Triller A, Bessis A. Impaired synaptic function in the microglial KARAP/DAP12-deficient mouse. J. Neurosci. 2004;24:11421–11428. doi: 10.1523/JNEUROSCI.2251-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovnick AD, Armstrong H, Rice GP, Bulman D, Hashimoto L, Paty DW, Hashimoto SA, Warren S, Hader W, Murray TJ. A population-based study of multiple sclerosis in twins: update. Ann. Neurol. 1993;33:281–285. doi: 10.1002/ana.410330309. [DOI] [PubMed] [Google Scholar]
- Sawcer S, Jones HB, Feakes R, Gray J, Smaldon N, Chataway J, Robertson N, Clayton D, Goodfellow PN, Compston A. A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat. Genet. 1996;13:464–468. doi: 10.1038/ng0896-464. [DOI] [PubMed] [Google Scholar]
- Silander K, Axelsson T, Widen E, Dahlgren A, Palotie A, Syvänen AC. Analysis of genetic variation in the GenomEUtwin project. Twin Res. 2003;6:391–398. doi: 10.1375/136905203770326394. [DOI] [PubMed] [Google Scholar]
- Sumelahti ML, Tienari PJ, Wikstrom J, Palo J, Hakama M. Regional and temporal variation in the incidence of multiple sclerosis in Finland 1979-1993. Neuroepidemiology. 2000;19:67–75. doi: 10.1159/000026241. [DOI] [PubMed] [Google Scholar]
- Sumelahti ML, Tienari PJ, Wikstrom J, Palo J, Hakama M. Increasing prevalence of multiple sclerosis in Finland. Acta Neurol. Scand. 2001;103:153–158. doi: 10.1034/j.1600-0404.2001.103003153.x. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Rochford CD, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 2005;201:647–657. doi: 10.1084/jem.20041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Prinz M, Stagi M, Chechneva O, Neumann H. TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 2007;4:e124. doi: 10.1371/journal.pmed.0040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom J, Palo J. Studies on the clustering of multiple sclerosis in Finland I: Comparison between the domiciles and places of birth in selected subpopulations. Acta Neurol. Scand. 1975;51:85–98. doi: 10.1111/j.1600-0404.1975.tb01362.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.