Abstract
The human concentrative nucleoside transporter 2 (CNT2) plays an important role in the absorption, disposition, and biological effects of endogenous nucleosides and nucleoside analog drugs. We identified genetic variation in the basal promoter region of CNT2 and characterized the function of the variants. We screened DNA from an ethnically diverse population and identified five basal promoter variants in CNT2. Three major haplotypes in the CNT2 basal promoter region were identified and were found at different allele frequencies in various ethnic groups. The common promoter variants and haplotypes were constructed and characterized for their promoter activity using luciferase reporter assays. One polymorphic variant, rs2413775 (-146T>A), with an allele frequency >20% in all populations, showed a gain of function in luciferase activity. Furthermore, in vivo mouse promoter assays of these nucleotide variants using the hydrodynamic tail vein injection, leading to their expression in the liver, demonstrated similar results. Transcription factor binding site (TFBS) analysis indicated this variant alters a hepatic nuclear factor (HNF) 1 TFBS. Electrophoretic mobility shift assay demonstrated stronger binding of HNF1α and weaker binding of HNF1β to the -146T and -146A regions, whereas the single nucleotide polymorphism (SNP), -146A, exhibited enhanced binding to both HNF1α and HNF1β, consistent with its greater activity in reporter assays. The data collectively suggest that the common variant, -146T>A, in the proximal promoter of CNT2 may result in an enhanced transcription rate of the gene and, thus, expression levels of CNT2. This SNP may play a role in variation in the pharmacokinetics and pharmacological effects of nucleoside analogs.
Naturally occurring nucleosides and synthetic nucleoside analogs such as anticancer and antiviral drugs are transported across cell membranes by multiple nucleoside transporters. The concentrative nucleoside transporter family (SLC28A) has three members, which interact with various nucleoside analog drugs. In particular, CNT1 prefers pyrimidine-based analogs (e.g., zidovudine, gemcitabine, cytarabine), CNT2 prefers purine-based analogs (e.g., ribavirin and didanosine), and CNT3 accepts both purine and pyrimidine-based analogs (e.g., cladribine, gemcitabine, and fludarabine) (Gray et al., 2004). Human CNT2 is localized to the apical membranes of absorptive epithelial cells of the intestine, kidney, and liver and is also expressed in lymphocytes (Gray et al., 2004; Fernández-Veledo et al., 2006; Meier et al., 2007; Minuesa et al., 2008). The role of CNT2 in the disposition and response to anticancer and antiviral nucleoside drugs has been characterized in intestinal tissues (Shin et al., 2006), renal epithelium (Damaraju et al., 2007; Elwi et al., 2009), and lymphocytes (Molina-Arcas et al., 2003; Minuesa et al., 2008). In addition, the expression of the CNT2 in hepatocytes and macrophages has led to an understanding of the endogenous physiological role of CNT2 in the salvage of nucleosides during cell proliferation (Aymerich et al., 2006). The other roles of CNT2 in cell physiology, for example in modulating purinergic responses during cell activation and/or apoptosis, have also been investigated (Soler et al., 2003; Fernández-Veledo et al., 2006).
Previously, our group has functionally characterized coding region variants of CNT2 that were identified in an ethnically diverse sample population (Owen et al., 2005). Another group recently reported the frequencies and activities of CNT2 coding variants in an ethnically diverse population sample consisting of Chinese, Malays, and Indians residing in Singapore (Li et al., 2007). Many novel nonsynonymous variants were discovered and functionally investigated in both studies. However, none of the nonsynonymous variants studied revealed significant differences in functional uptake and kinetics of model nucleoside substrates with the exception of one singleton (a singleton SNP that occurs only once in the sample), E385K, which showed significant reduced uptake of nucleoside substrates (Li et al., 2007).
In this study, we set forth to analyze whether nucleotide variation in the basal promoter region of CNT2 (250 bp upstream of the gene transcription start site and 50 bp downstream from the transcription start site) may lead to differences in functional uptake and kinetics of model nucleoside substrates. In particular, we identified novel variants in the CNT2 basal promoter region and studied their functional effects in cells and in vivo using a luciferase reporter assay. In addition to identifying promoter region variants with altered activity, our study revealed that HNF1α and HNF1β are involved in the transcriptional activation of CNT2.
Materials and Methods
Identification of CNT2 Promoter Variants. The CNT2 promoter variants were identified in the study of Leabman et al. (2003) through a combination of direct sequencing and denaturing high-performance liquid chromatography from an ethnically diverse population of 247 individuals (100 African Americans, 100 European Americans, 30 Asian Americans, 10 Mexican Americans, and seven Pacific Islanders). A different set of the genomic DNA samples was collected from unrelated healthy individuals in the San Francisco Bay area as part of the Studies of Pharmacogenetics in Ethnically Diverse Populations (SOPHIE) project. The DNA samples from 60 unrelated Asian-Americans from the SOPHIE project were used for genotyping the common haplotypes in the CNT2 promoter region.
Cloning of the Human CNT2 Basal Promoter Region. The transcriptional start site of the human CNT2 gene at position chr15: 43331726 was identified and reported in the genome browser [http://genome.ucsc.edu; Human Mar. 2006 (hg18)] (Fig. 1). The CNT2 promoter fragment (-265/+100 bp from the transcriptional start site; Chr15:43331461-43331825) was PCR-amplified from the genomic DNA of the HepG2 cell line using the primers listed in Table 1. Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA) was used for the PCR amplification of the CNT2 promoter fragment. The resulting PCR product (-265/+100) was digested with MluI and BglII and ligated into pGL4.11b[luc2] that had been predigested with MluI and BglII, which resulted in the CNT2-luciferase promoter constructs. The pGL4.11b[luc2] vector was a gift from Dr. Richard M. Myers (Stanford University, Palo Alto, CA). Restriction enzymes and T4 ligase were purchased from New England Biolabs (Ipswich, MA). PCR products were cleaned up using the QIAGEN PCR clean-up kit (QIAGEN, Valencia, CA), and the plasmid DNA was prepared using the QIAprep spin miniprep kit (QIAGEN). The sequences of the promoter constructs were verified by DNA sequencing.
Fig. 1.
The genomic DNA sequence of the CNT2 promoter region (-265/+100; Chr15:43331461-43331825) that was PCR-amplified and cloned into pGL4.11b[luc2] vector. The nucleotide positions are numbered relative to the transcription start sites (arrow). The nucleotide position and nucleotide change of the five SNPs discovered in this resequencing are presented in this figure (boldface). The underlined cis-acting elements in this CNT2 promoter region contain an HNF1 consensus element.
TABLE 1.
Sequences of oligonucleotides for functional assays and cloning
Underlined nucleotide sequences in the primer pairs for cloning represent the specified restriction enzyme sites.
| Oligonucleotide Sequences | |
|---|---|
| Site-Directed Mutagenesis Experiment | |
| rs60273882 (− 241C>T) | 5′-GTCTCAAGCGATCTGCCCGTCTCGGCC-3′ |
| rs2413775 (− 146A>T) | 5′-CCTGCAAAACAGTCTTAAAAACATATAATATTTAACTTGAGAGGTGCA-3′ |
| rs2899376 (− 115G>T) | 5′-GGTGCAGTCCTCCTCTACATTGAGGGCAGG-3′ |
| rs60354957 (− 14A>T) | 5′-CCTCCCTCGCAGCTGTGCTTTTCTTTCAGTC-3′ |
| rs61553676 (− 1G>A) | 5′-CTCGCAGCTGAGCTTTTCTTTCAATCCTTCACTGAG-3′ |
| EMSA probes and competitors | |
| −146-A | 5′-GGGCTTAAAAACATATAA TAATTAACTTGAGAGGTGCAGTC −3′ |
| −146-T | 5′-GGGCTTAAAAACATATAATATTTAACTTGAGAGGTGCAGTC −3′ |
| HNF1α per | 5′-GGGCGCAAAAGAAAGTTAATCATTAACCCGGGAAACAGC-3′ (Kikuchi et al., 2006) |
| Cloning | |
| CNT2 basal promoter | 5′-ATAACGCGTTATCCTGGCTGGTCTCAAGCG-3′ (MluI) |
| (−265/+ 100) | 5′-ATAAGATCTATCTCTTTAAAAATTTGTAAAT-3′ (BglII) |
| HNF1α | 5′-AAGCTTGCCATGGTTTCTAAACTGAGCC −3′ (HindIII) |
| 5′-GAATTCTGGTTACTGGGAGGAAGAGGCC −3′ (EcoRI) | |
| HNF1β | 5′-AAGCTTGAAAATGGTGTCCAAGCTCACG −3′ (HindIII) |
| 5′-GAATTCGGCATCACCAGGCTTGTAGAGG −3′ (EcoRI) |
Cloning of Human HNF1α and HNF1β cDNA to Plasmid Vectors. The coding regions of HNF1α and HNF1β were PCR-amplified from human liver cDNA, which was reverse transcribed from total liver RNA (Clontech, Mountain View, CA) using the primers described by Kikuchi et al. (2006). The oligonucleotide sequences used to clone HNF1α and HNF1β are shown in Table 1. HNF1α and HNF1β were created by cloning the coding sequence region of the genes, NM_000545.4 and NM_000458.2, respectively. The amplified products were cloned in TA vector pCRII-TOPO (Invitrogen). Positive clones were subjected to direct sequencing to verify the sequence. BamHI/XhoI and BamHI/NotI restriction enzymes were used to double-digest the TOPO constructs containing HNF1α and HNF1β, respectively, and the fragments were then subcloned into pCITE-2B and pCITE-2C vectors (Novagen, Madison, WI), respectively, and into the pcDNA3.1(+) vector (Invitrogen).
Site-Directed Mutagenesis. To obtain the identified variants, site-directed mutagenesis was performed using PfuUltra high-fidelity DNA polymerase following the manufacturer's instructions for site-directed mutagenesis (Stratagene, La Jolla, CA). The oligonucleotide sequences used for site-directed mutagenesis are listed in Table 1. The sequences of the resulting CNT2 promoter variants were verified by direct sequencing. The CNT2 variant construct was excised from the pGL4.11b[luc2] vector using the restriction enzymes MluI and BglII and religated into these same sites within the pGL4.11b[luc2] vector again to ensure that the vector backbone used for all the promoter constructs remained the same.
Cell Culture and Transfections. ACHN, human renal cell adenocarcinoma cell line, was purchased from the American Type Culture Collection (Manassas, VA). The human hepatoblastoma cell line, HepG2, and the human colorectal carcinoma cell line, HCT-116, were supplied by the University of California (San Francisco, CA) Cell Culture Facility and were maintained in a culture medium consisting of Dulbecco's modified Eagle's medium with 4500 mg/l glucose supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. In the in vitro functional assay, the three cell lines, ACHN, HCT-116, or HepG2, were seeded into 48-well culture plates (5 × 104 ACHN cells/well, 5 × 104 HCT-116 cells/well, and 3 × 104 HepG2 cells/well) and 16 to 24 h later were transfected with 1.0 μl/well Lipofectamine LTX (Invitrogen). pGL4.11b[luc2] vector (192 ng) with or without the promoter region constructs and 8 ng of pGL4.74[hRluc/TK] vector in 40 μl of Opti-MEM I reduced-serum medium (Invitrogen) as recommended by Invitrogen protocol for Lipofectamine LTX transfection were used. The pGL4.74[hRluc/TK] vector was cotransfected with the pGL4.11b[luc2] vector to normalize the transfection efficiency. For the cotransfection assay, 200 ng of pcDNA3.1(+) vector or pcDNA3.1(+) containing HNF1α or HNF1β was added to the transfection reaction with 100 ng of pGL4.11b[luc2] vector alone or pGL4.11b[luc2] containing the CNT2 basal promoter sequence. Cells were lysed with passive lysis buffer 24 or 48 h after transfection and were assayed for luciferase and Renilla activity in a 96-well plate using the GloMax 96 Microplate Luminometer with Dual Injectors (Promega, Madison, WI), according to the protocol in the Dual-Luciferase Reporter Assay system kit (Promega). The luciferase activity was measured as the relative light units of firefly (Photinus pyralis) luciferase activity per unit of Renilla reniformis luciferase. In each experiment, three wells were transfected with the same constructs, and the experiments were repeated three independent times.
In Vivo Functional Assay of CNT2 Basal Promoter in Mice. For the in vivo functional assay, each separate tail vein injection experiment was performed on at least seven mice using the TransIT In Vivo Gene Delivery System (Mirus Bio, LLC, Madison, WI) following the manufacturer's protocol. In brief, 10 μg of each pGL4.11b[luc2] vector encompassing either no promoter or the CNT2 promoter variants constructs alongside 2 μg of pGL4.74[hRluc/TK] vector, to correct for injection efficiency, was injected into the tail vein of each mouse. After 24 h, animals were euthanized, and livers were dissected and homogenized in Passive Lysis Buffer (Promega) followed by centrifugation for 10 min at 4000 rpm. Firefly luciferase and Renilla luciferase activity of the 1:20 diluted supernatant were measured in triplicate for each liver using the Dual-Luciferase Reporter Assay system (Promega) and quantified using the Lumat LB 9507 (Berthold Technologies, Bad Wildbad, Germany). The firefly luciferase to Renilla luciferase ratios were determined and expressed as relative luciferase activity. The experiments for each construct were repeated at least twice as independent experiments.
Electrophoretic Mobility Shift Assay. Electrophoretic mobility shift assay (EMSA) was performed as described by De Val et al. (2004). Double-stranded oligonucleotides were labeled with [32P]dCTP, using Klenow to fill in overhanging 5′ ends, and purified on a nondenaturing polyacrylamide-Tris borate-EDTA gel. The binding reactions were preincubated at room temperature in 1× binding buffer (40 mM KCl, 15 mM HEPES, pH 7.9, 1 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol) containing equal amounts of recombinant HNF1α or HNF1β protein, 1 μg of poly(dI-dC) (polydeoxyinosinic-deoxycytidylic acid; to reduce nonspecific binding), and competitor DNA (50-fold excess unless indicated otherwise) for 10 min before the addition of the radiolabeled DNA probe. Reactions were incubated for an additional 20 min at room temperature and then electrophoresed on an 8% nondenaturing polyacrylamide gel. The HNF1α and HNF1β recombinant proteins were generated from plasmid pCITE-2B and pCITE-2C (Novagen), respectively, using the TnT Quick Coupled Transcription/Translation System as described in the manufacturer's directions (Promega).
TaqMan SNP Genotyping. Genotypes from 60 unrelated Asian Americans recruited for the SOPHIE project and from 90 unrelated Ashkenazi Jews were determined for the SNP rs2413775 and rs2899376. The TaqMan SNP genotyping assays were used (assay identification C__282297_10 for rs2413775 or assay identification C__15846818_10 for rs2899376) for genotyping on the ABI 7900 Fast HT Sequence Detection Systems (Applied Biosystems, Foster City, CA). Ten nanograms of genomic DNA was amplified by Taq-Man Genotyping Master Mix (Applied Biosystems). The PCR conditions were as follows: one cycle of 95°C for 10 min followed by 60 cycles of two-step PCR with denaturation at 95°C for 15 s and annealing and extension at 60°C for 1.5 min.
Statistical Analysis. Data are expressed as mean ± S.D. For statistical analysis, multiple comparisons were analyzed using one-way analysis of variance followed by Dunnett's two-tailed test. CNT2 basal promoter reference was used as the basis for comparison unless stated otherwise.
The data were analyzed using GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA). A p value less than 0.05 was considered statistically significant.
Results
Genetic Polymorphism and Haplotype Analysis of the CNT2 Basal Promoter Region. A total of five polymorphisms in the CNT2 basal promoter were identified in samples from 240 unrelated individuals from four major ethnic groups. The five polymorphisms are: -1G>A, -14A>T, -115T>G, -146T>A, and -241C>T (the numbers are the nucleotide position relative to the transcription start site). The allele frequencies of each SNP in each ethnic group in this cohort are presented in Table 2. Two common SNPs, -146T>A (rs2413775) and -115T>G (rs2899376), were identified (Table 2). Three less common variants were discovered in this resequencing study of the CNT2 basal promoter: -1G>A (rs61553676), -14A>T (rs60354957), and -241C>T (rs60273882) (Table 2). One of the SNPs, -14C>T, was exclusively found in African Americans, with a 2% allele frequency. These discovered SNPs have been submitted to the dbSNP build 129. Haplotypes were constructed from the CNT2 promoter variant positions using PHASE, a Bayesian statistical method (Stephens et al., 2001). Four unambiguous basal promoter haplotypes were identified and are shown in Table 3. The most common haplotype in European Americans and African Americans is the A-G-A (-146A/-115G/-14A) haplotype, whereas the T-G-A (-146T/-115G/-14A) is more common in the Asian and Mexican ethnic groups. We also genotyped the two common SNPs, rs2413775 and rs2899376, in a large sample of Asian Americans and Ashkenazi Jews. Table 4 shows the allele frequencies of all the combined genotyped data in four different ethnic groups. The Asian Americans (which includes Japanese, Chinese, and South East Asians) have higher allele frequency for the haplotype T-G (64.0% in Asian Americans versus 26-36% in the other three ethnic groups), although the haplotype A-G is more common in the other three ethnic groups, namely the African Americans (61.2%), European Americans (72.5%), and Ashkenazi Jews (61.9%). In addition, the least common haplotype, T-T, which only presents at low frequency (1-3.1%) in African Americans, European Americans, and Ashkenazi Jews, is present at a higher frequency in the Asian Americans (16.9% in Asian Americans).
TABLE 2.
Identity and frequency of SNPs in the proximal promoter region of CNT2
0* implies that the SNP was not detected in a particular ethnic group. n = Number of chromosomes examined.
| Nucleotide Position from the TSSa | Nucleotide Changeb | Frequency in AA (n = 200) | Frequency in EA (n = 200) | Frequency in AS (n = 60) | Frequency in ME (n = 20) | Frequency in PA (n = 14) | Ref SNP Identification (dbSNP Build 129) |
|---|---|---|---|---|---|---|---|
| −241 | C→T | 0.005 | 0.005 | 0* | 0* | 0* | rs60273882 |
| −146 | T→A | 0.600 | 0.725 | 0.267 | 0.400 | 0.500 | rs2413775 |
| −115 | T→G | 0.970 | 0.990 | 0.817 | 1.000 | 0.929 | rs2899376 |
| −14 | A→T | 0.020 | 0* | 0* | 0* | 0* | rs60354957 |
| −1 | G→A | 0.005 | 0.005 | 0* | 0* | 0* | rs61553676 |
AA, African American; ME, Mexican American; AS, Asian American; EA, European American; PA, Pacific Islander.
The number refers to the nucleotide position upstream from the TSS, and the information is obtained from the University of California Santa Cruz genome browser, hg18 (http://genome.ucsc.edu).
The nucleotide change of the reference allele to the variant allele is obtained from the University of California Santa Cruz genome browser.
TABLE 3.
Haplotypes of the CNT2 proximal promoter in ethnically diverse population samples
Only haplotypes from 238 assignable samples (unambiguous or occur >70% PHASE runs) from a total of 247 samples are used. 0* implies that the haplotype was not detected in a particular ethnic group.
| Haplotypes | Percentage of Chromosomes with the Specific Haplotypes Described | |||||||
|---|---|---|---|---|---|---|---|---|
| Nucleotide positiona | −146 | −115 | −14 | |||||
| Nucleotide changeb | T>A | T>G | A>T | AA | EA | AS | ME | PA |
| Haplotypes in CNT2 basal promoter | T | T | A | 3.0 | 1.0 | 18.3 | 0* | 0* |
| T | G | A | 34.0 | 25.5 | 55.0 | 60.0 | 50.0 | |
| A | G | A | 60.0 | 72.5 | 26.7 | 40.0 | 50.0 | |
| T | G | T | 2.0 | 0* | 0* | 0* | 0* | |
AA, African American; EA, European American; AS, Asian American; ME, Mexican American; PA, Pacific Islander.
The number refers to the nucleotide position upstream from the TSS, and the information is obtained from University of California Santa Cruz genome browser (http://genome.ucsc.edu).
The nucleotide change of the reference allele to the variant allele is obtained from the University of California Santa Cruz genome browser.
TABLE 4.
The allele frequencies of the common haplotypes in the CNT2 basal promoter region (−146T>A and −115T>G) in different ethnic groups n = Number of chromosomes analyzed.
| Nucleotide Position from TSS | −146T>A | −115T>G | The Allele Frequency of the Common Haplotypes in Each Ethnic Group | |||
|---|---|---|---|---|---|---|
| % | ||||||
| dbSNP/Hapmap SNP | rs2413775 | rs2899376 | AA (n = 200) | EA (n = 200) | AS (n = 172) | AJ (n = 176) |
| Common haplotypes in CNT2 basal promoter | T | T | 3.1 | 1.0 | 16.9 | 2.8 |
| T | G | 35.7 | 26.5 | 64.0 | 35.2 | |
| A | G | 61.2 | 72.5 | 19.2 | 61.9 | |
AA, African American; EA, European American; AS, Asian American; AJ, Ashkenazi Jew.
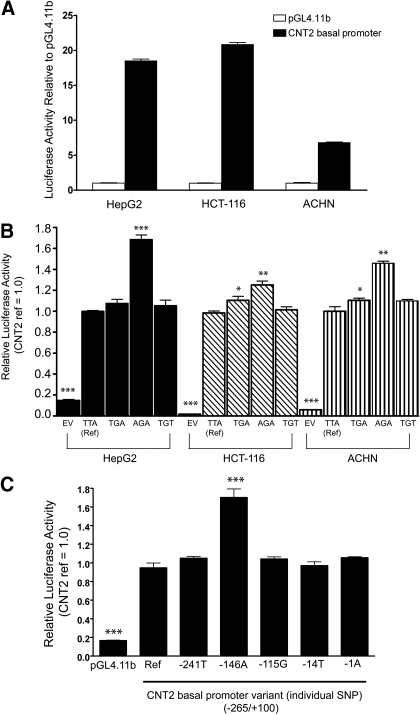
Functional Analysis of Human CNT2 Basal Promoter Region Variants. The presence of the CNT2 transporter on the apical membrane of the kidney, intestine, and liver influenced our decision to perform the functional analysis in three different cell lines, namely the human renal cell adenocarcinoma cell line, ACHN, the human colorectal carcinoma cell line, HCT-116, and the human hepatoblastoma cell line, HepG2. The activity of the CNT2 promoter was determined as the ratio between the activity of firefly luciferase and Renilla luciferase. Luciferase activity of CNT2 basal promoter (-265/+100) was highest in HCT-116 followed by ACHN and HepG2 (Fig. 2A). The luciferase activities of the common CNT2 promoter variant constructs were compared within the same cell lines (Fig. 2B). In all three cell lines, the CNT2 promoter variant construct containing the variant -146A in the haplotype-AGA showed enhanced luciferase activity compared with the haplotypes-TTA (reference), -TGA, and -TGT (Fig. 2B). This observation was most apparent in the HepG2 cell line, in which the haplotype-AGA exhibited 1.8-fold greater activity compared with the reference haplotype-TTA (Fig. 2B). To identify the SNP in the various promoter haplotypes that was responsible for the altered luciferase activity, the experiments were reproduced examining the individual SNPs comprising the haplotypes. The analysis demonstrated that -146A was responsible for the altered luciferase activities in the various promoter haplotypes in HepG2 cell lines. Similar results were obtained in the other two cell lines (data not shown). In contrast, the three less common SNPs (-1A, -14T, and -241T) did not show significant differences (p > 0.05) in the luciferase activity compared with the reference (Fig. 2C).
Fig. 2.
A, luciferase activity in three cell lines transfected with reporter constructs of the proximal promoter of CNT2. The CNT2 promoter constructs (-265/+100) (192 ng) with Renilla constructs (8 ng) were transiently transfected into HepG2, HCT116, and ACNH cell lines for analysis of luciferase activity. Firefly luciferase activity was normalized to Renilla luciferase activity. B, effect of the CNT2 haplotypes on CNT2 transporter promoter activity. The CNT2 haplotype promoter constructs (192 ng) with Renilla construct (8 ng) were transiently transfected into HepG2 (24 h), HCT116 (48 h), and ACHN (48 h) cell lines and assayed for luciferase activity. C, CNT2 individual SNP promoter constructs (192 ng) with Renilla construct (8 ng) were transiently transfected into HepG2 cells and assayed for luciferase activity (24 h). Firefly luciferase activity was normalized to Renilla luciferase activity. Data are reported as the relative -fold increase compared with the pGL4.11b vector containing the CNT2 promoter (reference). The results here are shown as the mean ± S.D. of three separate experiments performed in triplicate. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Multiple comparisons were analyzed using one-way analysis of variance followed by Dunnett's two-tailed test. CNT2 basal promoter reference was used as control.
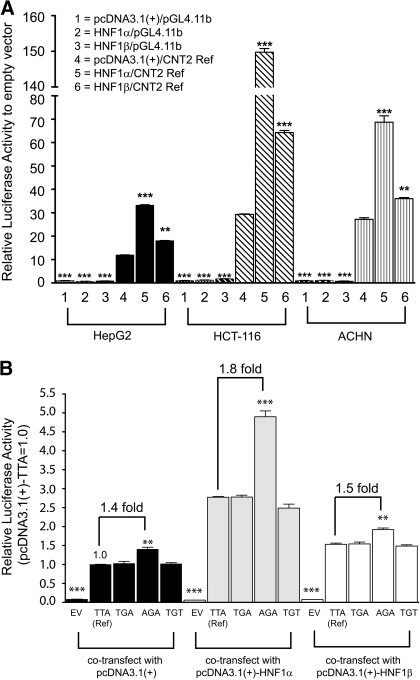
Next, we examined the effect of exogenously expressed HNF1α and HNF1β on the CNT2 basal promoter activity by cotransfecting each in the HepG2, HCT116, and ACHN cells. Cotransfection of HNF1α or HNF1β enhanced the luciferase activity of the CNT2 promoter construct (-265/+100 reference) compared with the pcDNA3.1(+)-transfected control (Fig. 3A). The luciferase activity of the reference CNT2 promoter construct was greater in cells cotransfected with HNF1α compared with those cotransfected with HNF1β, suggesting that the transactivation effect of HNF1β is lower than that of HNF1α. The stimulatory effect of both transcription factors was greater in cells expressing promoter constructs containing the variant -146A (in haplotype-AGA) compared with cells expressing the -146T (in haplotypes -TTA, -TGA, and -TGT) (Fig. 3B). A 1.4-, 1.8-, and 1.5-fold greater luciferase activity was obtained in HepG2 cells expressing constructs with haplotype-AGA (with -146A allele) compared with haplotype-TTA (with -146T allele) and simultaneously cotransfected with the pcDNA3.1(+) empty vector, pcDNA3.1(+)-HNF1α, and pcDNA3.1(+)-HNF1β, respectively. This stimulatory effect by HNF1α and HNF1β were reproducible in ACHN cells, the results showed 1.2-, 1.6-, and 1.3-fold greater activity in the haplotype-AGA with respect to the haplotype-TTA in ACHN cells cotransfected with the pcDNA3.1(+) empty vector, pcDNA3.1(+)-HNF1α, and pcDNA3.1(+)-HNF1β, respectively (data not shown). However, the effects were less apparent in HCT116 cells where there was 1.1-, 1.2-, and 1.2-fold greater activity, respectively (data not shown). The transactivation effect by HNF1β was lower than HNF1α for all the CNT2 basal promoter haplotype constructs in all the cells (data in HepG2 cells are shown in Fig. 3B).
Fig. 3.
A, effect of cotransfection of HNF1α and HNF1β on CNT2 basal promoter function in HepG2, HCT116, and ACHN cells. B, stimulatory effects of HNF1α and HNF1β on CNT2 haplotype promoter constructs in HepG2 cells. In both experiments, the cells were transfected with 200 ng of CNT2 haplotype promoter constructs (-265/+100 reference) or the promoterless pGL4.11b vector with 100 ng of pCDNA3.1(+) vector alone or with 100 ng of pcDNA3.1(+) vector containing HNF1α or HNF1β and with Renilla constructs (8 ng). The cells were assayed for luciferase activity after 24 h. The results here are shown as the mean ± S.D. of two separate experiments performed in triplicate. **, p < 0.01; ***, p < 0.001. Multiple comparisons were analyzed using one-way analysis of variance followed by Dunnett's two-tailed test. CNT2 basal promoter reference (TTA-haplotype) was used as control.
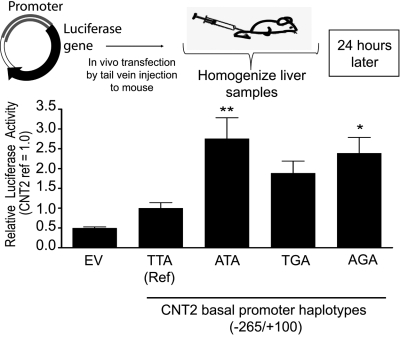
To corroborate our findings using the luciferase assays, we assessed the functional activity of the CNT2 promoter variants in vivo using tail vein injection assays in mice. This technique utilizes intravascular injection of DNA into the mouse tail vein in a large volume and rapid manner such that it ends up predominantly in the liver, leading to high expression levels of the foreign DNA in the mouse liver (Zhang et al., 1999). The results from these in vivo studies are shown in Fig. 4 and are consistent with the in vitro luciferase assay, in which SNP -146A and haplotype-AGA showed significantly higher luciferase activities compared with the reference. Similar to cellular assays, the in vivo studies demonstrated that the activity of haplotype-TGA was not significantly different compared with the reference haplotype.
Fig. 4.
The in vivo tail vein injection assay showing CNT2 proximal promoter activity. The CNT2 proximal promoter reference or variant constructs upstream of luciferase gene in pGL4.11b vector were transfected in vivo into mouse hepatocytes using the hydrodynamic tail vein injection method. The means and S.D. of the relative luciferase activity are shown as -fold difference compared with the CNT2 reference region. Multiple comparisons were analyzed using one-way analysis of variance followed by Dunnett's two-tailed test. CNT2 basal promoter reference was used as control. *, p < 0.05; **, p < 0.01.
Effect of -146T>A on the Binding of HNF1 Transcription Factors to the CNT2 Promoter Region. To assess the effect of the -146T>A promoter variant on putative transcription-factor binding sites, we performed an analysis using the Delta-MATCH Query Tool developed by Dr. David W. Williamson (personal communication). This tool retrieves human polymorphisms that are predicted to create allele-specific transcription factor binding sites. It was observed that the sequence at position -146 upstream from the transcription start site of CNT2 contains an HNF1 consensus element and is located within a region of high conservation (chr15:43331576-43331587) in the human genome (University of California Santa Cruz browser hg18). The analysis also revealed that the matrix match score of the -146A allele is higher than that of the reference allele T (minor allele), suggesting that the HNF1 transcription factors have higher binding affinity to the site containing the A allele in comparison with the T allele.
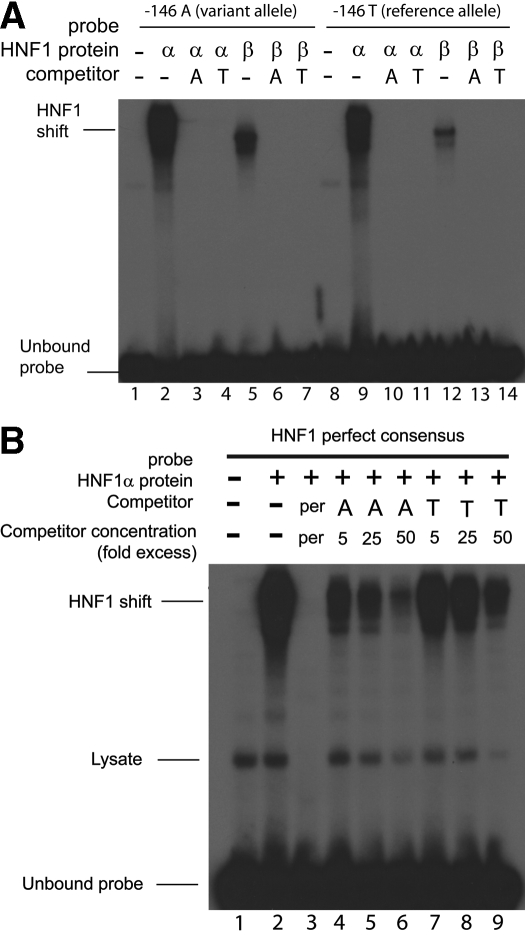
Validation of the in silico transcription factor analysis was performed using EMSAs to determine whether the nucleotide substitutions altered DNA-transcription factor interactions. HNF1α and HNF1β each bound to a synthetic double-stranded oligonucleotide encoding the region between -163 and -125, which spans the putative HNF1 binding site (Fig. 5A).
Fig. 5.
A, binding affinity of HNF1α and HNF1β protein to the CNT2 proximal promoter region, -163/-125 bp upstream of the TSS, containing either the -146A allele or -146T allele. Recombinant HNF1α (lanes 2-4 and 9-11) and HNF1β (lanes 5-7 and 12-14) were transcribed and translated in vitro, and equal amounts of each protein were used in EMSA with [γ-32P]ATP-labeled double-stranded oligonucleotides (-163/-125 bp) containing the -146A allele (lanes 2-7) or the -146T allele (lanes 9-14). Lanes 1 and 8 contain reticulocyte lysate without recombinant HNF1α or HNF1β protein. HNF1α and HNF1β efficiently bound to the CNT2 basal promoter oligonucleotide (-163/-125 bp) (lanes 2, 5, 9, and 12), and these interactions were efficiently competed by an excess of unlabeled oligonucleotide (-163/-125 bp) containing the -146A allele (lanes 3, 6, 10, and 13) and the -146T allele (lanes 4, 7, 11, and 14). The oligonucleotide (-163/-125 bp) containing the -146A allele has greater binding than -146T allele for HNF1α (comparing lanes 2 and 9) and HNF1β (comparing lanes 5 with 12). B, competition assay showing the difference in the binding affinity of -146A allele and -146T allele in the -163-/-125-bp oligonucleotide with the HNF1α perfect consensus oligonucleotide (HNF1α per). Recombinant HNF1α (lanes 2-9) was transcribed and translated in vitro and used in EMSA with [γ-32P]ATP-labeled double-stranded oligonucleotide (-163/-125 bp) containing the cold -146A allele (lanes 4-6) or the -146T allele (lanes 7-9). Lane 1 contains reticulocyte lysate without recombinant HNF1α protein. HNF1α protein bound efficiently to the HNF1α oligonucleotide probe containing a perfect binding sequence element for HNF1α (HNF1α per), and this binding was more efficiently competed by the cold oligonucleotide-containing -146A allele (lanes 4-6) than the -146T allele (lanes 7-9).
The EMSA also suggested that the -146A allele has a higher binding affinity than the T allele for HNF1α (Fig. 5A, comparing lanes 2 and 9) and that this region has a higher binding affinity to HNF1α than HNF1β (Fig. 5A, comparing lanes 2 and 5 and 9 and 12). Competition experiments were performed with an oligonucleotide corresponding to the perfect HNF1α consensus (Fig. 5B). In this competition assay, 50-fold excess of the competitor containing the HNF1α perfect consensus (Kikuchi et al., 2006) (Fig. 5B, lane 3) was able to compete with the labeled probe containing the HNF1α perfect (per) consensus. In lanes 4 to 9 (Fig. 5B), increasing amounts (5-, 25-, or 50-fold excess) of the unlabeled oligonucleotide sequence containing the -146A allele (lanes 4-6) or -146T allele (lanes 7-9) competed with the labeled probe containing the HNF1α perfect consensus sequence for the HNF1α recombinant protein. The binding of HNF1α perfect consensus element was more efficiently competed off by the oligonucleotide containing the -146A allele (lanes 4-6) in comparison with the oligonucleotide containing the -146T allele (lanes 7-9). Even at high concentrations of -146T oligonucleotide (50-fold excess, lane 9), the effect of competition was less effective compared with the -146A oligonucleotide (50-fold excess, lane 6).
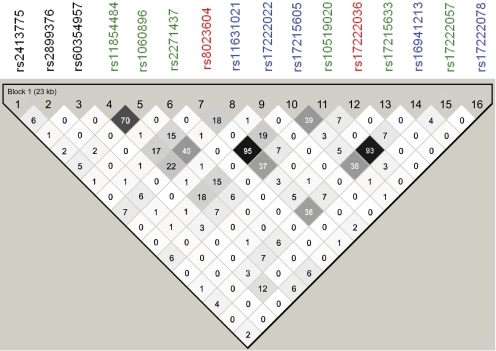
Linkage of CNT2 Basal Promoter Region Variants with the Coding and Intronic Regions Variant. Haplo-view 4.1 was used to evaluate the linkage of the CNT2 basal promoter variants with the CNT2 coding and intronic region variants identified previously by our group (Owen et al., 2005). The LD plot in Fig. 6 demonstrated that there is a lack of linkage between the CNT2 basal promoter region variants and the coding and intronic region variants.
Fig. 6.
An LD plot showing lack of linkage of CNT2 basal promoter region variant and coding and noncoding region variants. Top lane, rs number of the identified polymorphisms in the CNT2 proximal promoter, coding, and intronic regions. Black, rs number in promoter region; green, rs number in coding region (nonsynonymous); red, rs number in coding region (synonymous); blue, rs number in intronic region. The number displayed in the LD plot is the r2 number, e.g., 95 = r2 = 0.95; 93 = r2 = 0.93; 70 = r2 = 0.70.
Discussion
The purpose of our study was to identify variants in the CNT2 basal promoter (from 250 bp upstream to 50 bp downstream of the transcription start site) and to characterize their functional effects. The three main haplotypes found in the basal promoter of CNT2 accounted for more than 97% of the haplotypes in the chromosomes from each ethnic group examined. In all cell lines tested, the haplotype containing the -146A allele demonstrated higher relative luciferase activity compared with the reference haplotype, which contained the -146T allele. An in vivo assay involving tail vein injection of the CNT2 proximal promoter reporter construct (Fig. 4) produced similar results as the reporter assays carried out in cell lines (Fig. 2, A-C). That is, the -146A allele and the haplotype containing the -146A allele (haplotype-AGA) had higher luciferase activities compared with the reference construct (haplotype-TTA). In general, SNPs of potentially high functional importance, such as nonsynonymous SNPs, SNPs in splicing sites, and SNPs in 5′- and 3′-untranslated region are thought to have lower minor allele frequencies (Urban et al., 2006; Ke et al., 2008). This is suggested to be due to selective pressure that is keeping these functional regions of the genome at low allele frequencies. Our results here with variants in a noncoding region of CNT2 are not consistent with this general observation. That is, the -146A and the -146T alleles are both present at high allele frequencies in all populations samples and demonstrate distinct functional properties. In contrast, the two rare SNPs that were discovered in this sequencing analysis (rs60273882 and rs61553676) did not result in any changes in luciferase reporter assays in comparison with the reference alleles (Fig. 2C).
CNT2 is known to have a high affinity for adenosine and has been shown to modulate extracellular levels of adenosine (Km values for adenosine = 8 μM) (Mangravite et al., 2003; Molina-Arcas et al., 2008). The transcription factors, CCAAT/enhancer-binding protein α and HNF3γ, were demonstrated to affect CNT2 expression in human liver parenchymal cells (Fernández-Veledo et al., 2007). In silico methods (Delta-Match Query tool, developed by Dr. David W. Williamson; TFSearch version 3.1, http://www.cbrc.jp/research/db/TFSEARCH.html) revealed that the -146 position contains an HNF1 binding site. In addition, the in silico methods did not predict the CNT2 promoter region cloned to have CCAAT/enhancer-binding protein α or HNF3γ binding sites, although this does not preclude their existence in the remainder of the promoter. HNF1α is predominantly expressed in the liver and kidney and is required for liver-specific expression of a variety of genes (Courtois et al., 1987). HNF1β is highly homologous to HNF1α, and in addition to the liver and kidney, it is expressed in the gut and lung. HNF1α and HNF1β act either as homodimers or as heterodimers (Bach et al., 1991). Because HNF1α is predominantly expressed in the liver, this may explain the greater -fold difference in the reporter assay of the CNT2 promoter construct. This hypothesis is also strengthened by the observation that the -146A allele that is speculated to have higher affinity to HNF1α compared with the construct containing the -146T allele demonstrated increased luciferase activity in the HepG2 cell line (1.8-fold greater luciferase activity, p < 0.001; Fig. 2, B and C) and in the in vivo tail vein injection assay, in which the reporter construct is expressed in the liver (Fig. 4). HNF-1 regulates transcription of genes by binding as a dimer to the cis-acting elements that match the inverted palindrome GTTAATNATTAAC (Tomei et al., 1992). Examination of the nucleotide sequence within the -160/-125 segment of the CNT2 basal promoter region revealed the presence of an HNF1-like sequence. The cis-acting elements ATAATAATTAA (-152- to -142-bp regions) of the CNT2 basal promoter contain the elements that match the described inverted palindrome GTTAATNATTAAC. On the other hand, the cis-acting elements with the variant allele -146T results in the sequence, ATAATATTTAA (-152- to -142-bp regions), which is not as good of a match as the -146A allele-containing sequence. The predictions are consistent with our experimental observations demonstrating a reduced binding affinity of the oligonucleotide containing the -146T allele to both HNF1α and HNF1β in comparison with the oligonucleotide containing the -146A allele (Fig. 5A). This finding is also supported by results in the experiments involving overexpression of HNF1α and HNF1β (Fig. 3, A and B). The overexpression assays showed HNF1α has a greater effect than HNF1β in enhancing the CNT2 basal promoter activity in the three cell lines (Fig. 3A), and this effect remained the same for the different haplotypes of the CNT2 basal promoter (Fig. 3B). In addition, the stimulatory effect of HNF1α and HNF1β was greater in the haplotype containing the -146A allele (AGA-haplotype) compared with the -146T allele (TTA, TGA, and TGT haplotypes) (Fig. 3B).
Interindividual variation affecting clearance and bioavailability of ribavirin, an antiviral agent used in the treatment of hepatitis C, is a very common phenomenon (Wade et al., 2006). In previous functional studies of nonsynonymous variants of CNT2, only a single nonsynonymous nucleotide variant appearing in one individual of the 288 samples sequenced, E385K, resulted in reduced uptake of endogenous nucleoside substrates and the antiviral drug, ribavirin (Owen et al., 2005; Li et al., 2007). The lack of linkage between the CNT2 basal promoter region variants with variants in the coding and intronic regions of the CNT2 gene that we observed here suggests that recombination in the CNT2 gene regions has occurred. Our studies here revealed that within the CNT2 promoter region, one of the two common SNPs (-146T>A, rs2413775), which is present in all ethnic groups, resulted in significant functional differences in both in vitro and in vivo functional assays. This SNP potentially may result in altered transcription rates of the gene in vivo and correspondingly altered expression levels of CNT2 in various tissues, such as in the liver. With this consideration, the 146T>A SNP has the potential to influence the pharmacologic effect of ribavirin. This SNP may also be associated with variation in the pharmacokinetics of ribavirin, including the intestinal absorption and the renal elimination of the drug, both of which may be mediated by CNT2. It is important that variation in the frequency of the -146T>A among ethnic populations (as shown in Tables 2 and 3) may produce interethnic differences in the expression of the transporter and ultimately in drug disposition and response. It is clear that the clinical consequences of this functional SNP -146T>A (rs2413775) require further investigation.
Acknowledgments
We thank Dr. Richard M. Myers for the pGL4.11b[luc2] and pGL4.74[hRluc/TK] vectors, Dr. Richard M. Myers and Yuya Kobayashi for guidance with experimental procedures, and Dr. David Williamson for guidance and suggestions in using the Delta-Match Query tool.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM61390]; by the National Institutes of Health National Cancer Institute [Grant P50-CA58207]; and by the Sandler Family Supporting Foundation.
A grant from the New Energy and Industrial Technology Development Organization supported some of the methods of development in this study.
Data showing allele frequencies of CNT2 promoter variants are available at http://www.pharmgkb.org.
doi:10.1124/jpet.108.147207.
ABBREVIATIONS: CNT, concentrative nucleoside transporter; SNP, single nucleotide polymorphism; bp, base pair; HNF, hepatic nuclear factor; SOPHIE, Studies of Pharmacogenetics in Ethnically Diverse Populations; PCR, polymerase chain reaction; EMSA, electrophoretic mobility shift assay; TSS, transcription start site; LD, linkage disequilibrium.
References
- Aymerich I, Foufelle F, Ferré P, Casado FJ, and Pastor-Anglada M (2006) Extracellular adenosine activates AMP-dependent protein kinase (AMPK). J Cell Sci 119 1612-1621. [DOI] [PubMed] [Google Scholar]
- Bach I, Mattei MG, Cereghini S, and Yaniv M (1991) Two members of an HNF1 homeoprotein family are expressed in human liver. Nucleic Acids Res 19 3553-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois G, Morgan JG, Campbell LA, Fourel G, and Crabtree GR (1987) Interaction of a liver-specific nuclear factor with the fibrinogen and alpha 1-antitrypsin promoters. Science 238 688-692. [DOI] [PubMed] [Google Scholar]
- Damaraju VL, Elwi AN, Hunter C, Carpenter P, Santos C, Barron GM, Sun X, Baldwin SA, Young JD, Mackey JR, et al. (2007) Localization of broadly selective equilibrative and concentrative nucleoside transporters, hENT1 and hCNT3, in human kidney. Am J Physiol Renal Physiol 293 F200-F211. [DOI] [PubMed] [Google Scholar]
- De Val S, Anderson JP, Heidt AB, Khiem D, Xu SM, and Black BL (2004) Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev Biol 275 424-434. [DOI] [PubMed] [Google Scholar]
- Elwi AN, Damaraju VL, Kuzma ML, Baldwin SA, Young JD, Sawyer MB, and Cass CE (2009) Human concentrative nucleoside transporter 3 is a determinant of fludarabine transportability and cytotoxicity in human renal proximal tubule cell cultures. Cancer Chemother Pharmacol 63 289-301. [DOI] [PubMed] [Google Scholar]
- Fernández-Veledo S, Huber-Ruano I, Aymerich I, Duflot S, Casado FJ, and Pastor-Anglada M (2006) Bile acids alter the subcellular localization of CNT2 (concentrative nucleoside cotransporter) and increase CNT2-related transport activity in liver parenchymal cells. Biochem J 395 337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Veledo S, Jover R, Casado FJ, Gómez-Lechón MJ, and Pastor-Anglada M (2007) Transcription factors involved in the expression of SLC28 genes in human liver parenchymal cells. Biochem Biophys Res Commun 353 381-388. [DOI] [PubMed] [Google Scholar]
- Gray JH, Owen RP, and Giacomini KM (2004) The concentrative nucleoside transporter family, SLC28. Pflugers Arch 447 728-734. [DOI] [PubMed] [Google Scholar]
- Ke X, Taylor MS, and Cardon LR (2008) Singleton SNPs in the human genome and implications for genome-wide association studies. Eur J Hum Genet 16 506-515. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kusuhara H, Hattori N, Shiota K, Kim I, Gonzalez FJ, and Sugiyama Y (2006) Regulation of the expression of human organic anion transporter 3 by hepatocyte nuclear factor 1alpha/beta and DNA methylation. Mol Pharmacol 70 887-896. [DOI] [PubMed] [Google Scholar]
- Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, et al. (2003) Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci U S A 100 5896-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tan CM, Koo SH, Chong KT, and Lee EJ (2007) Identification and functional analysis of variants in the human concentrative nucleoside transporter 2, hCNT2 (SLC28A2) in Chinese, Malays and Indians. Pharmacogenet Genomics 17 783-786. [DOI] [PubMed] [Google Scholar]
- Mangravite LM, Badagnani I, and Giacomini KM (2003) Nucleoside transporters in the disposition and targeting of nucleoside analogs in the kidney. Eur J Pharmacol 479 269-281. [DOI] [PubMed] [Google Scholar]
- Meier Y, Eloranta JJ, Darimont J, Ismair MG, Hiller C, Fried M, Kullak-Ublick GA, and Vavricka SR (2007) Regional distribution of solute carrier mRNA expression along the human intestinal tract. Drug Metab Dispos 35 590-594. [DOI] [PubMed] [Google Scholar]
- Minuesa G, Purcet S, Erkizia I, Molina-Arcas M, Bofill M, Izquierdo-Useros N, Casado FJ, Clotet B, Pastor-Anglada M, and Martinez-Picado J (2008) Expression and functionality of anti-human immunodeficiency virus and anticancer drug uptake transporters in immune cells. J Pharmacol Exp Ther 324 558-567. [DOI] [PubMed] [Google Scholar]
- Molina-Arcas M, Bellosillo B, Casado FJ, Montserrat E, Gil J, Colomer D, and Pastor-Anglada M (2003) Fludarabine uptake mechanisms in B-cell chronic lymphocytic leukemia. Blood 101 2328-2334. [DOI] [PubMed] [Google Scholar]
- Molina-Arcas M, Trigueros-Motos L, Casado FJ, and Pastor-Anglada M (2008) Physiological and pharmacological roles of nucleoside transporter proteins. Nucleosides Nucleotides Nucleic Acids 27 769-778. [DOI] [PubMed] [Google Scholar]
- Owen RP, Gray JH, Taylor TR, Carlson EJ, Huang CC, Kawamoto M, Johns SJ, Stryke D, Ferrin TE, and Giacomini KM (2005) Genetic analysis and functional characterization of polymorphisms in the human concentrative nucleoside transporter, CNT2. Pharmacogenet Genomics 15 83-90. [DOI] [PubMed] [Google Scholar]
- Shin HC, Kim JS, Vig BS, Song X, Drach JC, and Amidon GL (2006) Interaction of intestinal nucleoside transporter hCNT2 with amino acid ester prodrugs of floxuridine and 2-bromo-5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole. Biol Pharm Bull 29 247-252. [DOI] [PubMed] [Google Scholar]
- Soler C, Felipe A, García-Manteiga J, Serra M, Guillén-Gómez E, Casado FJ, MacLeod C, Modolell M, Pastor-Anglada M, and Celada A (2003) Interferongamma regulates nucleoside transport systems in macrophages through signal transduction and activator of transduction factor 1 (STAT1)-dependent and -independent signalling pathways. Biochem J 375 777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, and Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68 978-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomei L, Cortese R, and De Francesco R (1992) A POU-A related region dictates DNA binding specificity of LFB1/HNF1 by orienting the two XL-homeodomains in the dimer. EMBO J 11 4119-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban TJ, Sebro R, Hurowitz EH, Leabman MK, Badagnani I, Lagpacan LL, Risch N, and Giacomini KM (2006) Functional genomics of membrane transporters in human populations. Genome Res 16 223-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JR, Snoeck E, Duff F, Lamb M, and Jorga K (2006) Pharmacokinetics of ribavirin in patients with hepatitis C virus. Br J Clin Pharmacol 62 710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Budker V, and Wolff JA (1999) High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther 10 1735-1737. [DOI] [PubMed] [Google Scholar]