Abstract
The molecular mechanisms involved in human aging are complicated. Two progeria syndromes, Werner’s syndrome (WS) and Hutchinson-Gilford progeria syndrome (HGPS), characterized by clinical features mimicking physiological aging at an early age, provide insights into the mechanisms of natural aging. Based on recent findings on WS and HGPS, we suggest a model of human aging. Human aging can be triggered by two main mechanisms, telomere shortening and DNA damage. In telomere-dependent aging, telomere shortening and dysfunction may lead to DNA damage responses which induce cellular senescence. In DNA damage-initiated aging, DNA damage accumulates, along with DNA repair deficiencies, resulting in genomic instability and accelerated cellular senescence. In addition, aging due to both mechanisms (DNA damage and telomere shortening) is strongly dependent on p53 status. These two mechanisms can also act cooperatively to increase the overall level of genomic instability, triggering the onset of human aging phenotypes.
Keywords: human aging, Hutchinson-Gilford Progeria syndrome, Werner syndrome
Introduction
Aging is an extremely complicated process and is known to be driven by a variety of different mutually interacting mechanisms. Conventionally, it is seen as a process of progressive failure of homeostasis involving genes for maintenance and repair, environmental factors leading to molecular damage and molecular heterogeneity, and chance events with potentially significant consequences for death (Alberts et al 2002). Since several human progeroid synqdromes (PSs) (human premature aging-like syndromes) are characterized by features resembling precocious aging, the identification of the genes involved in PSs has provided important clues to understanding the molecular mechanisms underlying normal human aging (Hutchinson 1886, Gilford 1904; Martin 1978, 1985).
The classic example of PS is Werner’s syndrome (WS), a progeria of the adult, which is caused by a mutation in a gene coding for a member of the RecQ helicase family, WRN (Yu et al 1996) and is characterized by features resembling precocious aging, appearing as a variety of visible features associated with aging, such as graying of the hair and skeletal changes, which occur much earlier than normal (Goto 1997). WRN acts as a “caretaker of the genome” and functions in both DNA repair and transcription, suggesting that breakdown of these processes is critical in promoting aging (Epstein et al 1966; Goto 1997; Bachrati et al 2003). Hutchinson-Gilford progeria syndrome (HGPS), a progeria of the child, is the other classic example, and is caused by mutations in the gene LMNA (1q21.2) (Eriksson et al 2003), encoding a nuclear envelope protein, lamin A, which has been shown to affect RNA polymerase II transcription, probably by alterations in chromatin organization (Martin and Oshima 2000; Spann et al 2002; Hickson 2003; Andressoo and Hoeijmakers 2005). In addition, skin fibroblast biopsies from patients with HGPS fail to show evidence of normal DNA strand rejoining in vitro after exposure to irradiation, suggesting that DNA repair may be absent or greatly reduced in the cells (Epstein et al 1973). Thus, these two PSs directly mimic the clinical and molecular features of natural aging, providing unique insights into the molecular mechanisms underlying human aging (Martin 1978, 1985). In this review, we will focus on these two best characterized PSs, with particular emphasis on the functions of the mutated syndrome-causing genes, and their potential implications for resemble aging. Based on the current information on these two syndromes, we also propose a model to explain human aging.
Werner’s syndrome
Werner’s syndrome is a rare autosomal recessive disorder characterized by premature aging (Epstein et al 1966; Martin et al 1970; Goto et al 1996; Yu et al 1996). The WRN gene responsible for WS encodes the 1432 amino acid WRN protein, which is a member of the RecQ DNA helicase family (Yu et al 1996). The C-terminal region of WRN contains the conserved RQC domain, which includes a nucleolar targeting sequence (NTS) (Marciniak et al 1998; von Kobbe and Bohr 2002), and the helicase and RNaseD C-terminal (HRDC) domain (Liu et al 1999). WRN has DNA-dependent ATPase, DNA strand annealing, DNA helicase, and exonuclease activities (Gray et al 1997; Suzuki et al 1997, 1999; Huang et al 1998). WRN, as a “caretaker of the genome”, is needed to prevent telomere dysfunction and consequent genomic instability (Opresko et al 2003; Crabbe et al 2004; Du et al 2004). Since most of the mutations identified in WRN cause premature termination of translation (Goto 1997; Yu et al 1997), resulting in impaired nuclear import of the protein (Matsumoto et al 1997), the clinical features and cellular phenotypes of WS patients are major due to an absolute lack of normal WRN in the nucleus. WS cells have defects in DNA replication, resulting in dysfunction in multiple cellular DNA metabolic processes, such as DNA replication initiation, replication foci establishment, and the resolution of stalled replication forks during replication and DNA repair (Martin et al 1970; Hasty et al 2003). In addition, a large number of reports have shown that many cellular events, including transcription and apoptosis, are affected in WS cells (Ogburn et al 1997; Balajee et al 1999; Spillare et al 1999, 2006; Sommers et al 2005). The loss of WRN function in WS appears as chromosome instability, a shorter life span in culture (Martin 1977), and accelerated telomere shortening (Schulz et al 1996), suggesting that breakdown of WRN function is important in promoting aging (Martin et al 1970; Hasty et al 2003).
Multiple functions of the WRN protein
Biochemistry and catalytic activities
WRN, a multifunctional nuclear protein, interacts with a number of proteins to catalyze four major DNA-dependent enzymatic activities, acting (1) as a DNA-dependent ATPase, (2) as an intrinsic 3′→5′ helicase (Gray et al 1997), (3) as a 3′→5′ exonuclease (Huang et al 1998, 2000), and (4) in DNA strand annealing (Machwe et al 2005), and interacts with factors with established roles in DNA metabolic pathways (Brosh and Bohr 2002). WRN contains three distinct structure-specific DNA-binding domains, one N-terminal domain and two different C-terminal fragments (the RecQ and HRDC domains), each of which plays roles in distinct DNA metabolic pathways (von Kobbe et al 2003a).
ATP-stimulated activities are required for the successive steps in the hydrolytic reaction, such as structure recognition, DNA binding, and 3′-terminal hydrolysis. In this context, WRN plays a DNA-dependent ATPase, using the energy from ATP hydrolysis to unwind double-stranded DNA, and its exonuclease and helicase activities act in concert to catalyze structure-dependent DNA degradation in resolving aberrant DNA structures (Shen and Loeb 2000). As a helicase, WRN is active on the forked end of a DNA duplex, while, as an exonuclease, it acts on the blunt end of the same duplex (Gray et al 1997; Huang et al 1998; Opresko et al 2001). WRN exonuclease can efficiently remove a mismatched nucleotide at a 3′ recessed terminus and can initiate DNA degradation from a 12-nucleotide gap or a nick (Huang et al 2000). Furthermore, WRN forms a trimer, which interacts with the proliferating cell nuclear antigen (PCNA) to participate in the replication restart process (Huang et al 2000; Rodríguez-López et al 2003; Jeziorny et al 2006). WRN exonuclease activity is suppressed by interaction with p53 (Brosh et al 2001a) or BLM (von Kobbe et al 2002) and stimulated by interaction with Ku70/80 (Li and Comai 2001) or phosphorylation (Karmakar et al 2002). WRN helicase activity is stimulated by interaction with replication protein A (RPA) (Shen et al 1998) or telomere repeat binding factor 2 (TRF2) (Opresko et al 2002) and phosphorylation (Karmakar et al 2002). Moreover, WRN colocalizes and interacts with RAD52 and has strand annealing activity in addition to its DNA unwinding activity (Baynton et al 2003; Machwe et al 2005, 2006). WRN-dependent unwinding activity is significantly stronger than previously believed (Machwe et al 2006).
Post-translational modifications
In response to DNA damage, WRN can be modulated by post-translational modifications, including phosphorylation, sumoylation, and acetylation. It is phosphorylated at serine/threonine and tyrosine residues in vivo after bleomycin treatment or after replication stress (Karmakar et al 2002; Cheng et al 2003; Pichierri et al 2003). WRN phosphorylation at serine/threonine residues is primarily dependent on DNA-PKcs and c-Abl kinases (Karmakar et al 2002; Cheng et al 2003; Pichierri et al 2003). Importantly, the serine/threonine phosphorylation of WRN by the DNA-PK complex results in inhibition of both WRN helicase and exonuclease activities, whereas dephosphorylation of WRN enhances both these activities. Thus, the serine/threonine phosphorylation status of WRN plays a role in the regulation of its catalytic activities (Karmakar et al 2002; Cheng et al 2003). c-Abl was also found to phosphorylate WRN (Cheng et al 2003). c-Abl kinase, a regulator of the DNA damage response, mediates WRN nuclear localization and catalytic activities in response to DNA damage. WRN directly binds to c-Abl and this interaction is disrupted in the early cellular response to bleomycin (Cheng et al 2003). As with phosphorylation by the DNA-PK complex, c-Abl phosphorylation inhibits WRN helicase and exonuclease activities (Cheng et al 2003).
Phosphorylation of WRN may influence other forms of WRN post-translational modification, such as sumoylation and/or acetylation. Modification of proteins by the small ubiquitin-related modifier 1 (SUMO-1) conjugating system requires a set of enzymes, including SUMO-activating (E1), conjugating (Ubc9), and ligating enzymes (Muller et al 2004). WRN interacts with Ubc9, which is required for conjugation of SUMO-1 to the N-terminal fragment (amino acids 272–514) of WRN (Kawabe et al 2000). In addition, binding of p14 Arf to WRN is multivalent and resembles the binding of p14 Arf to Mdm2, promoting sumoylation of WRN in a synergistic manner with the SUMO-conjugating enzyme Ubc9 (Woods et al 2004). p14 Arf causes redistribution of WRN within the nucleus, and this effect is reversed by expression of a SUMO-specific protease, thus implicating the SUMO conjugation pathway in controlling WRN relocalization (Woods et al 2004). Besides phosphorylation and sumoylation, WRN can be acetylated after mitomycin C or methyl methane-sulfonate treatment (Sharma et al 2005). On exposure to UV or ionizing radiation, WRN is acetylated by acetyltransferase p300, a transcriptional coactivator, which has acetylation activity (Blander et al 2002).
DNA replication
WRN helicase unwinds replication fork structures very efficiently (Mohaghegh et al 2001), acting to resolve the block and/or in the replication restart process (Rodríguez-López et al 2003). Compared to normal cells, cells from WS patients undergo premature replicative senescence (Martin et al 1970; Salk et al 1985), display an extended S-phase (Poot et al 1992), and show a reduced frequency of replication initiation sites (Takeuchi et al 1982; Hanaoka et al 1985), thus exhibiting defects in DNA replication consistent with the inability to properly recover from DNA replication fork demise (Hickson 2003).
WRN is able to interact with PCNA, DNA topoisomerase I (topo I), polδ, and RPA (Wold 1997; Lebel and Leder 1998; Brosh et al 1999; Lebel et al 1999; Constantinou et al 2000; Huang et al 2000; Laine et al 2003) and catalyze DNA unwinding in vitro for DNA replication, recombination, and repair (Brosh and Bohr 2002).
WRN and PCNA colocalize at replication foci, suggesting a physiological interaction between them in cycling primary cells (Rodríguez-López et al 2003). In addition, WRN is as part of the 17S multiprotein DNA replication complex, and establishes PCNA and topoisomerase I as the two WRN-interacting components (Warbrick 1998, 2000; Lebel et al 1999; Rodríguez-López et al 2003).
Polδ participates in DNA replication and repair of DNA damage. WRN interacts specifically with the p50 subunit of polδ, and WRN directly modifies DNA replication via its interaction with p50 and is involved in the dynamic relocalization of polδ complexes within the nucleus (Kamath-Loeb et al 2000; Szekely et al 2000). Moreover, WRN enhances the rate of nucleotide incorporation in polδ-mediated replication in the absence of PCNA and its helicase activity enables polδ to overcome hairpin and G-quadruplex DNA structures (Fry and Loeb 1999; Szekely et al 2000; Kamath-Loeb et al 2000, 2001; Brosh et al 2001b; Mohaghegh et al 2001). Thus, WRN may facilitate polδ-mediated DNA replication, and disruption of the WRN-polδ interaction in WS cells may contribute to the S-phase defects.
Human RPA is a heterotrimeric, single stranded DNA-binding protein required for DNA replication, recombination, and repair (Wold 1997; Brosh et al 1999, 2002). RPA directly interacts with to WRN, markedly increasing the DNA helicase activity of WRN (Brosh et al 1999; Shen et al 2003) and its ability to unwind forked telomeric DNA structures (Ohsugi et al 2000; Opresko et al 2001). After DNA damage, WRN can colocalize with BLM and RPA (Constantinou et al 2000; Bischof et al 2001; Sakamoto et al 2001). RPA binds to WRN and BLM to stimulate their unwinding of long DNA duplexes (Bachrati and Hickson 2003).
Another interesting replication protein, FEN-1, interacts with the 144-amino acid RQC domain on the C-terminal region of WRN (Brosh et al 2001c). FEN-1, a DNA structure-specific nuclease, participates in pathways of DNA metabolism that are important for genomic stability (Brosh et al 2002) and is involved in the maturation of Okazaki fragments during lagging strand DNA replication (Bambara et al 1997; Merrill and Holm 1998). WRN stimulates FEN-1-mediated cleavage activity of displaced flaps that occur during lagging strand DNA synthesis at Okazaki fragments (Brosh et al 2001c; Sharma et al 2004). WRN-FEN-1 complex colocalizes in foci associated with arrested replication forks, and this complex plays a role in the unwinding and degradation of Holliday junction structures associated with regressed replication forks (Sharma et al 2004). Defective Okazaki fragment processing causes DSBs, which may lead to the genomic instability in WS (Brosh et al 2002).
WRN colocalizes with, and directly interacts with, human topo I (Laine et al 2003). WRN stimulates the ability of topo I to relax negatively supercoiled DNA and specifically stimulates the religation step of the relaxation reaction, and cell extracts from WS fibroblasts exhibit a decreased ability to unwind negatively supercoiled DNA (Laine et al 2003). These findings provide the interrelationship between WRN helicases and topoisomerases in the maintenance of genomic integrity.
DNA repair
WS cells display sensitivity to 4-nitroquinoline 1-oxide (Ogburn et al 1997; Poot et al 2002), a carcinogen which causes the formation of DNA strand breaks and bulky DNA adducts (Nagao and Sugimura 1976), and are also hypersensitive to O6-methylguanine, a site-specific alkylating agent that can block DNA replication (Blank et al 2004). WS cells are sensitive to DNA crosslinking drugs (Poot et al 2001, 2002) and, since DNA crosslinks are normally repaired by homologous repair (HR), this suggests a defect in this repair pathway. WS cells display extensive deletions at nonhomologous joined ends (Oshima et al 2002; Chen et al 2003), while expression of wild-type WRN prevents excessive DNA deletions (Oshima et al 2002; Chen et al 2003). These findings suggest that lack of WRN may increase DNA damage and disrupt the regulatory processes controlling DNA repair, for example, nonhomologous end joining (NHEJ), HR, and base excision repair (BER).
Double-strand break repair
The first evidence for a link between WRN and double-strand break repair was the discovery that WRN interacts with both Ku and DNA-dependent protein kinase catalytic subunits (DNA-PKcs) (Cooper et al 2000; Li and Comai 2000; Cheng et al 2003; Opresko et al 2003) to participate in NHEJ (Li and Comai 2002). Assembly of DNA-PK and WRN at DNA ends allows DNA-PKcs to phosphorylate WRN, thus stimulating WRN enzymatic activity and facilitating efficient processing of double-strand breaks (DSBs) prior to ligation (Yannone et al 2001). Deficiencies in either Ku or DNA-PKcs result in sensitivity to ionizing radiation due to defects in DSB repair. Complementation with the exonuclease/helicase double mutant or wild-type WRN restores NHEJ activity, suggesting that WRN is necessary for normal repair of DSBs (Chen et al 2003).
Chromatographic studies showed that WRN is bound to PARP-1 in a complex that contains Ku70/80 (Li et al 2004). PARP-1 can induce apoptosis or necrosis in cells with extensive DNA damage. Absence of functional WRN prevents activation of PARP-1 in response to DNA damage caused by oxidative stress and alkylating agents (von Kobbe et al 2003b).
Some proteins that participate in the recombinational repair pathway have been found to functionally interact with WRN. WRN is involved in resolving recombination intermediates in RAD51-dependent HR (Prince et al 2001; Saintigny et al 2002) and forms distinct nuclear foci that partially overlap with the RAD51 nuclear foci formed in response to DNA damage (including DSBs) (Sakamoto et al 2001). In vivo data show that WRN interacts functionally with NBS1 (Cheng et al 2004), which is thought to act downstream of RAD51 (Saintigny et al 2002; Tauchi et al 2002; Monnat and Saintigny 2004). WRN colocalizes with, and interacts with, RAD52 (Baynton et al 2003), NBS1 in the MRN complex (Cheng et al 2004), and flap endonuclease 1 (FEN-1) (Brosh et al 2001c; Sharma et al 2004). In addition, it interacts physically with the Mre11-RAD50-NBS1 complex, which also functions in HR for DSB processing (Cheng et al 2004).
Base excision repair
Oxidative DNA lesions are repaired primarily by base excision repair (BER), and the accumulation of oxidative products and the resulting phenotypic changes have been implicated in the aging process (Beckman et al 1998). In general, BER can be divided into short patch repair and long patch repair. WRN has been shown to physically interacts with polβ and participate in short patch BER (Harrigan et al 2003). The active WRN helicase domain stimulates polβ strand displacement DNA synthesis at a nick on a BER substrate (Harrigan et al 2003). In addition, it has been shown to interact physically and/or functionally with several replication proteins which participate in long patch BER, including PCNA, RPA, polδ, and FEN-1 (Shen et al 1998; Brosh et al 1999, 2002; Kamath-Loeb et al 2001). PCNA is part of a sliding clamp which forms a ring which maintains the connection between polymerase and its DNA template, allowing uninterrupted synthesis. WRN has been shown to directly interact with PCNA in vitro, suggesting a unique role for WRN in DNA synthesis (Huang et al 2000). WRN stimulates FEN-1 flap cleavage (Brosh et al 2001c, 2002) and nucleotide incorporation by polδ (Kamath-Loeb et al 2000, 2001). RPA stimulates WRN helicase unwinding of long substrates (Shen et al 1998; Brosh et al 1999) and the poly(ADP-ribosyl)ation state of PARP-1 regulates WRN helicase and exonuclease activities (von Kobbe et al 2004). PARP-1 binds strongly to strand breaks and acts in the DNA damage surveillance network, partly by ribosylating a variety of nuclear proteins in response to DNA damage.
DNA recombination
Cellular DNA recombination can occur physiologically during meiotic DNA replication and V(D)J recombination, or can be induced by DNA damaging agents. Some reports define a physiological role for WRN RecQ helicase in recombination via RAD51-dependent HR (Prince et al 2001; Saintigny et al 2002). WRN and BLM colocalize to DNA damage-induced RAD51 foci, implicated in HR (Bischof et al 2001; Sakamoto et al 2001; Wu et al 2001; Saintigny et al 2002; Von Kobbe et al 2002; Wu and Hickson 2003; Spillare et al 2006). The WRN and BLM helicase activities are possible in a synergistic manner to intermediate DNA recombination, since a coimmunoprecipitation and colocalization study showed that the exonuclease domain of WRN interacts with BLM (von Kobbe and Bohr 2002). In addition, a biochemical study showed that WRN interacts with the homologous recombination mediator protein RAD52, and that WRN and RAD52 form a complex, should be a general response to replication forks arrested by DNA damage (Baynton et al 2003).
WRN interacts physically and functionally with the MRN complex via NBS1 and they colocalize in response to ionizing radiation or mitomycin C treatment (Cheng et al 2004). WS cells display a deficiency in resolving DNA recombination intermediates which contributes to DNA damage hypersensitivity, limited cell growth, and genomic instability (Prince et al 2001). The generation of viable mitotic recombinant progeny was rescued by the expression of WRN, which also improved WS cell survival after DNA damage (Saintigny et al 2002). These results define a physiological role for the WRN RecQ helicase protein in RAD51-dependent HR and identify a mechanistic link between defective recombination resolution and limited cell division potential, DNA damage hypersensitivity, and genetic instability in human somatic cells (Saintigny et al 2002). WRN also colocalizes with RAD51 and RPA in response to DNA damaging agents (Constantinou et al 2000; Sakamoto et al 2001), and both WRN and BLM interact with RPA (Brosh et al 1999, 2000).
Telomere maintenance
Telomeres are specialized nucleoprotein structures consisting of G-rich repetitive sequences that cap the ends of eukaryotic chromosomes and are crucial for the maintenance of chromosomal integrity and cell viability (McClintock et al 1941). They are maintained by the enzyme telomerase, which consists of an essential telomerase RNA component (TERC), which serves as a template for the addition of telomere repeats, and a protein component, the telomerase reverse transcriptase catalytic subunit (TERT). Telomere shortening eventually results in diverse pathophysiological consequences, primarily through accelerated telomere erosion, and triggers entry into premature senescence (Wright and Shay 1992; Blasco 2002; Chang et al 2004; Du et al 2004). Defects in telomere structure can initiate a DNA damage response and may lead to telomeric end fusions and chromosome breakage if not properly repaired (De Lange 2002).
Biochemical and cellular evidence suggest that WRN may dissociate secondary structures at the telomere to allow replication, repair, and telomerase activity at the telomere end (Opresko et al 2003). Under normal conditions, WRN associates with telomeres in S phase to prevent loss of individual telomeres (Crabbe et al 2004). Accelerated loss of telomere reserves and activation of cellular checkpoints appear integral to the decreased replicative potential seen in WS, as evidenced by the capacity of enforced TERT expression to impart unlimited replicative potential (Wyllie et al 2000).
WRN functionally interacts with a number of proteins involved in telomere length maintenance, including Ku 70/86 (Orren et al 2001), RPA (Brosh et al 1999, 2000; Sanz et al 2000), TRF1, and TRF2 (Opresko et al 2002, 2004; Machwe et al 2004). RPA is a single strand DNA-binding protein that is required for all aspects of DNA metabolism (Brosh et al 2000), while TRF1 and TRF2 are homodimeric proteins that have been shown to bind exclusively to double stranded telomeric DNA throughout the cell cycle and are thought to be involved in the regulation of telomeric length (Fairall et al 2001). TRF2 as a promoter for the helicase activity of WRN, the interaction between WRN and TRF2 may serve to stabilize TRF2 in its active form or to improve TRF2’s interaction with DNA (Opresko et al 2002). Due to the absence of WRN in WS patients, TRF2 is unable to perform its duty and telomeric-specific structures that need to be degraded are not (Griffith 1999; Opresko 2002; Jeziorny 2006). These structures then act as barriers against various transcription factors and telomeres are left without being fully transcribed.
Taken together, these results suggest that WRN is necessary for the efficient replication of G-rich telomeric DNA and that WRN deficiency and dysfunctional telomeres appear to act cooperatively to increase the overall level of genomic instability, triggering the onset of premature aging phenotypes (Orren et al 2001; Opresko et al 2002, 2004).
DNA apoptosis
A link between WRN and apoptosis was first proposed by studies demonstrating that WS fibroblasts exhibit a decreased p53- mediated apoptotic response, and this deficiency can be rescued by expression of wild-type WRN (Spillare et al 1999). p53 is a key cellular component in maintaining genomic stability either by regulating the cell cycle to allow DNA repair or by inducing apoptosis (Haupt et al 2003; Hofseth et al 2004; Lane 2004). The C-terminus of WRN binds to the C-terminal domain of p53 to induce p53-mediated apoptosis (Clarke et al 1993; Symonds et al 1994; Wang et al 1996, 2001; Spillare et al 1999; Brosh et al 2001a).
Fibroblasts from WS patients have a decreased ability to undergo p53-mediated apoptosis (Spillare et al 1999), so the absence of WRN-p53 direct interaction could serve as a signal for programmed cell death. Moreover, the expression of wild-type WRN is sufficient to rescue WS−/− cells from the attenuation of p53-mediated apoptosis (Spillare et al 1999; Blander et al 2001). p53 may exert its effect on WRN by its interaction with RPA. Overexpression of p53 results in a decrease in Sp-1-mediated transcription of the WRN gene, suggesting that p53 regulates WRN expression (Yamabe et al 1998). Thus, the interaction of WRN with p53 and/or the WRN-RPA complex may be critical in preventing entry into S phase or in directing S phase cells towards apoptosis. The absence of a p53-WRN helicase interaction may disrupt the signal to direct S-phase cells into apoptosis for programmed cell death and contribute to the pronounced genomic instability and cancer predisposition seen in WS cells (Sommers et al 2005). In addition, epigenetic inactivation (promoter hypermethylation) of WRN can lead to the loss of WRN-associated exonuclease activity and increased chromosomal instability and apoptosis induced by topoisomerase inhibitors (Agrelo et al 2006).
Hutchinson-Gilford progeria syndrome
Hutchinson-Gilford progeria syndrome, a childhood progeroid disorder, is a rare, fatal genetic disorder characterized by segmental accelerated aging. Affected children appear normal at birth, but within a year develop characteristic features of failure to thrive, delayed dentition, alopecia, atherosclerosis, prominent scalp veins, a high pitched voice, and sclerodermatous skin changes, with death at approximately 13 years from atherosclerosis of the coronary and cerebrovascular arteries. Eriksson and colleagues (2003) identified the disease causing mutations in the LMNA gene (encoding lamin A/C). The vast majority of HGPS cases are caused by a single-base substitution (GGC > GGT), which does not cause an amino acid change (G608G), but results in deletion of 150 nucleotides in exon 11, causing an alternatively spliced truncated variant of lamin A mRNA and an in-frame deletion of 50 amino acids near the carboxy terminus, leading to changes in the nuclear architecture (De Sandre-Giovannoli et al 2003; Eriksson et al 2003). Other HGPS mutations that have been described in LMNA include E145K, R471C, R527C, G608S, T623S, and 1824C > T (Cao and Hegele 2003; Csoka et al 2004a; Fukuchi et al 2004). Lamins form microfilaments in the nucleus and are important in maintaining the proper structure of the nuclei, but they also influence on chromatin structure, regulation of gene expression, localization and probably protein degradation (Ly et al 2000; Goldman et al 2002; Scaffidi and Misteli 2005). Immunofluorescence of HGPS fibroblasts with antibodies directed against lamin A revealed that many cells show visible abnormalities of the nuclear membrane (Eriksson et al 2003). In addition, HGPS cells have also altered histone modification patterns, including reduced heterochromatin-specific trimethylation of Lys9 on histone H3 (Tri-Me-K9H3) (Scaffidi and Misteli 2005, 2006). A recent study implicated lamin A in physiological aging, showing that the molecular mechanism responsible for the premature aging (Hasty and Vijg 2004; Scaffidi and Misteli 2006). Thus, accelerated aging in HGPS might thus reflect an exaggerated lamin A-dependent mechanism, which contributes to physiological aging.
Defects in prelamin A processing
Lamin A has a conserved C-terminal CAAX motif, which is a potential target for subsequent processing steps. To generate mature lamin A, prelamin A undergoes substantial post-translational modification of its CAAX motif via four processing steps (Zhang and Casey 1996; Young et al 2006). First, a 15-carbon farnesyl lipid is added to the thiol group of the cysteine by a cytosolic enzyme, protein farnesyltransferase. Second, the last three amino acids of the protein (ie, the -AAX) are clipped off by the metalloprotease ZMP-STE24 and/or RCE1 (Bergo et al 2002; Corrigan et al 2005). Third, the newly exposed farnesylcysteine is carboxylmethylated by ICMT, a prenylprotein-specific methyltransferase in the endoplasmic reticulum (Clarke et al 1988; Dai et al 1998). Fourth, the last 15 amino acids of the protein, including the farnesylcysteine methyl ester, are clipped off by ZMPSTE24 and degraded, releasing mature lamin A (Weber et al 1989; Beck et al 1990; Corrigan et al 2005; Young et al 2006). In HGPS, a 50-amino acid deletion in the C-terminus of the protein (amino acids 607–656) lacks an important endoprotease cleavage site recognized by ZMPSTE24 during prelamin A post-translational processing, so no mature lamin A is formed and a farnesylated mutant prelamin A (progerin) accumulates in cells (De Sandre-Giovannoli et al 2003; Eriksson et al 2003; Navarro et al 2006; Sun and Schatten 2006; Tsai et al 2006; Young et al 2006).
Progerin accumulation and nuclear morphology abnormalities
The mutant prelamin A is targeted to the nuclear rim, where it disrupts the integrity of the nuclear lamina, leading to premature cell death (Eriksson et al 2003; Goldman et al 2004; Varela et al 2005; Yang et al 2005; Fong et al 2006). The GGC > GGT (G608G) mutation of LMNA causes accumulation of farnesylprelamin A in the nucleus in a cellular age-dependent manner, and the cells display irregular nuclear shapes, including lobulation of the nuclear envelope, thickening of the nuclear lamina, loss of peripheral hetero-chromatin, and clustering of nuclear pores (Eriksson et al 2003; Goldman et al 2004). Aberrant nuclear morphology is also reported with other LMNA mutations, which have been linked to other “laminopathies”, such as Emery–Dreifuss muscular dystrophy, dilated cardiomyopathy-1A, Dunnigan-type familial partial lipodystrophy, mandibuloacral dysplasia, and atypical WS (Goldman et al 2002; Chen et al 2003). In HGPS cells, the nuclei and lamina appeared normal at early passages, but, at later passages, the nuclei are severely misshapen and contain an abnormally thick lamina (de Sandre-Giovannoli et al 2003; Eriksson et al 2003). These structural defects worsen as HGPS cells age in culture, and their severity correlates with an obvious increase in mutant lamin A (Goldman et al 2004). An abnormal distribution of nuclear pore complexes is seen in late passage HGPS cells. The changes in nuclear pore complexes gradually affect various aspects of the normal trafficking of protein and RNA across the nuclear envelope, having severe effects on the physiological state of HGPS cells (Yoshida and Blobel 2001). In addition, mutant lamin A induces decreased cellular proliferation, premature senescence, and altered motility (Goldman et al 2004). The highly lobulated late passage HGPS cells primarily exhibit PCNA patterns resembling early S phase, suggesting that there is a block in the transition from the early chain-elongation phase of DNA replication to the mid- and later phases of replication. Mutant lamin A (progerin) progressively accumulates in the nucleus with cellular age, resulting in premature cessation of growth in the later passages of HGPS cells (Goldman et al 2004).
Universal transcriptional alterations
Lamin A is a major constituent of the nuclear membrane, and an immunofluorescence study of HGPS fibroblast nuclei demonstrated abnormalities (Goldman et al 2004). Given the prominent structural role of lamin A in the nuclear membrane, it is suggested that this protein has diverse roles in DNA metabolism, including DNA replication and transcription. Gene expression in HGPS was investigated by measuring mRNA levels in fibroblasts isolated from young, middle aged, and old humans with or without progeria (Ly et al 2000). Of the 152 genes studied, 47 (31%) were differentially transcribed in both old and HGPS compared to young. The direction of the change was the same in old and HGPS for all coregulated genes. Genes involved in mitosis were downregulated and the observed changes might result in increased rates of somatic mutation, leading to chromosome aberrations and mutations manifesting as an aging phenotype. Csoka and colleagues (2004b) found that the genes differentially expressed in HGPS fibroblasts compared to age-matched control cell lines are involved in a variety of biological processes. Of the approximately 33,000 genes analyzed, 361 (1.1%) showed at least a 2 fold change in HGPS compared to the aged controls. The most prominent categories encoded transcription factors and extracellular matrix proteins, many of which are known to function in the tissues severely affected in HGPS. The most affected gene was MEOX/GAX, a homeobox gene that functions as a negative regulator of proliferation. Several genes involved in DNA replication and chromatin remodeling were downregulated. These changes were interpreted as contributing to depression of cellular proliferation. Some of the transcription changes suggested excess extracellular matrix deposition through the increased expression of extracellular matrix components and decreased expression of extracellular matrix remodeling enzymes. Of the 58 genes examined by both of these groups, the expression of 17 (29%) changed in the same direction, demonstrating a reasonable agreement between the two studies considering their different designs. The studies by Kyng and Bohr (2005) and Csoka and colleagues (2004b) point to a shared mechanism of aging acceleration in PSs ie, misregulated transcription. The idea that aging is due to the loss of the proper transcriptional state of the cell followed by “dysdifferentiation” had been previously proposed by other investigators (Kator et al 1985; Zs-Nagy et al 1988; Fossel 2003; Prolla 2005). These findings that HGPS patients show inappropriate transcriptional patterns provides new evidence that transcriptional deregulation can contribute to the aging process in humans.
Genomic instability
DNA damage accumulation and the effects of repair defects can lead to genomic instability associated with premature aging and have causal roles in normal aging (Lombard et al 2005; von Zglinicki et al 2005). Indeed, defective recruitment of 53BP1 and RAD51 to sites of DNA lesion is seen in HGPS fibroblasts, resulting in a delayed checkpoint response and defective DNA repair (Liu et al 2005). Wild-type mouse embryonic fibroblasts ectopically expressing unprocessible prelamin A show similar defects in checkpoint response and DNA repair (Liu et al 2005). These results indicate that unprocessed prelamin A and truncated lamin A act dominant negatively to perturb the DNA damage response and repair, resulting in genomic instability, which might contribute to laminopathy-based premature aging (Liu et al 2005). In HGPS cells, DNA damage checkpoints are persistently activated, and inactivation of checkpoint kinases ATM and ATR can partially restore cell cycle progression into S-phase (Cortez et al 2001; Liu et al 2006), suggesting that senescence can be suppressed by inactivating DNA damage response pathways in HGPS cells. Organismal aging has been linked to activation of p53-dependent signaling pathways and initiation of the senescence program in a premature aging mouse model (Varela et al 2005). Inhibition of aberrant splicing of lamin A results in significant downregulation of p21, IGFBP3, and GADD45B compared with mock-treated cells (Scaffidi and Misteli 2006). Consistent with the reduction in p53 activation, upon elimination of Δ50 lamin A from old cells, the fraction of 5-bromo-2′-deoxyuridine-positive proliferating cells increased by 30% and was similar to that in mock-treated young cells. Over the past few years, there have been reports that progerin leads to defective DNA repair and genome instability (Liu et al 2005), overexpression of p53 target genes (Varela et al 2005), and changes in histone methylation that affect heterochromatin organization (Shumaker et al 2006).
Farnesyltransferase inhibitor
Farnesyl-prelamin A is targeted to the nuclear envelope, where it interferes with the integrity of the nuclear envelope and causes misshapen cell nuclei. It has also been shown to affect the mechanical stability of the nucleus (Dahl et al 2006). Farnesyltransferase inhibitors (FTIs) can block prelamin A processing and reduce the percentage of cells with misshapen nuclei (Capell et al 2005; Glynn and Glover 2005; Mallampalli et al 2005; Toth et al 2005). Thus, the favorable effects of FTIs raise the question whether an FTI might improve disease phenotypes in HGPS (Young et al 2006). It is also important to define the extent to which these abnormalities are affected by an FTI and to determine whether FTIs will be a useful therapy in children with HGPS. A recent study found that treatment of patient’s cells with an FTI did not result in a reduction in DNA DSBs and damage checkpoint signaling, although it significantly reversed the aberrant shape of their nuclei (Liu et al 2006), suggesting that DNA damage accumulation and aberrant nuclear morphology are independent phenotypes arising from prelamin A accumulation in these PSs. Yang and colleagues (2006) created gene-targeted mice with an HGPS mutation (LmnaHG/+) and examined the effect of an FTI on the disease phenotypes. The LmnaHG/+ mice exhibited phenotypes similar to those in human HGPS patients, including retarded growth, reduced amounts of adipose tissue, micrognathia, osteoporosis, and osteolytic lesions in bone. In addition, osteolytic lesions in the ribs led to spontaneous bone fractures. Treatment with an FTI ameliorated the disease phenotypes, resulting in an increased adipose tissue mass, improved body weight curves, a reduction in the number of rib fractures, and improved bone mineralization and bone cortical thickness, suggesting that FTIs could be useful for treating HGPS patients (Yang et al 2006). Though FTIs fall short of curing the disease (Yang et al 2006), these findings have established a paradigm for ameliorating the most obvious cellular pathology in HGPS and suggest a potential strategy for treating this disease.
A hypothetical model of aging
The molecular mechanisms involved in human senescence are complicated. Two canonical PSs, WS and HGPS, characterized by clinical features mimicking physiological aging at an early age, have provided insights into the mechanisms of natural aging. In these PSs, several cellular pathways are affected, resulting in the formation of endogenous and exogenous sources of oxidative stress, telomere attrition, and a decline in DNA repair, which can jointly contribute to genomic instability, and subsequently result in growth arrest and apoptosis, leading to the human aging phenotypes (Karanjawala and Lieber 2004; Prolla 2005; Collado 2007). The gene defective in WS, WRN, encodes a helicase of the RecQ family and possesses an exonuclease domain. WRN is involves in multiple DNA repair pathways and plays a significant role in the maintenance of overall genomic stability (Bachrati and Hickson 2003) (as seen in WS). The clinical features of HGPS show similarities to WS, but progress more rapidly (Hennekam 2006). LMNA, the gene defective in HGPS, affects the structure or post-translational maturation of lamin A, a major nuclear component (Eriksson et al 2003). Recently, several studies have established a functional link between DNA repair and A-type lamin-associated syndromes, which are associated with transcriptional alterations, abnormal DNA replication, changed organization of higher order chromatin structure, and genomic instability (Serrano and Blasco 2007), suggesting a link between these syndromes and physiological aging (as seen in HGPS). DNA damage is generated throughout life and causes continuous damage to the macromolecular components of cells. Importantly, the rate of DNA damage production increases with ageing (Hasty and Vijg 2004; Serrano and Blasco 2007).
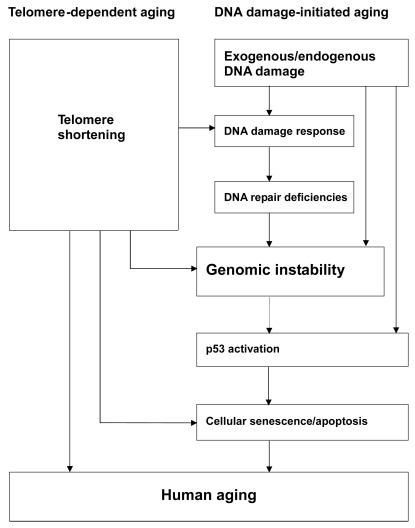
Thus, the causal relationship between the DNA damage response and cellular senescence suggests that DNA damage-initiated genomic instability can induce human aging phenotypes (Harman 1956). p53-mediated senescence and apoptosis in response to DNA damage also probably contribute to aging. Indeed, p53, as a master integrator of cellular stress, is able to respond to a wide range of DNA damage (Horn and Vousden 2007), then, if the stress persists, prevent propagation of the damaged cells (by apoptosis or senescence) (Vousden and Lane 2007). Following DNA damage, activation of p53 leads to transcription and upregulation of p53 target genes, among which P21 acts as the major effector of p53-induced cellular senescence. P21 levels increase gradually as cells pass into senescence (Alcorta et al 1996). Collectively, these results suggest that p53 activation is at least partially responsible for the induction of cellular senescence in response to DNA damage. On the other hand, various lines of evidence have shown that telomere shortening and dysfunction can also trigger DNA damage responses and are sufficient to induce cellular senescence (d’Adda di Fagagna et al 2003; Takai et al 2003; Chang et al 2004; Du et al 2004). Some HGP fibroblasts also appear refractory to telomerase-mediated immortalization (Wallis et al 2004), and most cultures show elevated apoptosis and senescence (Bridger and Kill 2004). Numerous studies indicate that, in the setting of WRN deficiency, dysfunctional telomeres trigger the onset of premature aging phenotypes, suggesting a link between increased telomere dysfunction and the genomic instability associated with the aging process (d’Adda di Fagagna et al 2003; Takai et al 2003; Smogorzewska and de Lange 2004; Chang et al 2004; Du et al 2004). On the basis of the information available for WS and HGPS, we suggest a model of human aging (Figure 1). Human aging can be triggered by two main mechanisms, telomere shortening and DNA damage. In telomere-dependent aging, telomere shortening and dysfunction can lead to DNA damage responses, inducing cellular senescence. In DNA damage-initiated aging, DNA damage accumulates, along with DNA repair deficiencies, resulting in genomic instability and accelerated cellular senescence. Both aging mechanisms depend strongly on p53 status. These two mechanisms can act cooperatively to increase the overall level of genomic instability and trigger the onset of human aging phenotypes.
Figure 1.
A hypothetical model: Genomic instability plays a central role during the aging process, triggered by two main stimuli, telomere shortening and DNA damage. (1) Telomere-dependent aging: Telomeres are essential for chromosomal stability. Telomere shortening and dysfunction can trigger DNA damage responses and are sufficient to induce cellular senescence. (2) DNA damage-initiated aging: DNA damage accumulates, along with DNA repair deficiencies, resulting in genomic instability and accelerated cellular senescence. Both mechanisms depend strongly on p53. These two mechanisms can act cooperatively to increase the overall level of genomic instability and trigger the onset of human aging phenotypes.
Footnotes
Conflict of interest
The authors report no conflicts of interest in this work.
References
- Agrelo R, Cheng WH, Setien F, et al. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci U S A. 2006;103:8822–7. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, et al. Development of multicellular organisms. In: Alberts B, editor. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. [Google Scholar]
- Alcorta DA, Xiong Y, Phelps D, et al. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A. 1996;93:13742–7. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andressoo JO, Hoeijmakers JH. Transcription-coupled repair and premature ageing. Mutat Res. 2005;577:179–94. doi: 10.1016/j.mrfmmm.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee AS, Machwe A, May A, et al. The Werner syndrome protein is involved in RNA polymerase II transcription. Mol Biol Cell. 1999;10:2655–68. doi: 10.1091/mbc.10.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambara RA, Murante RS, Hendrickson LA. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–50. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- Baynton K, Otterlei M, Bjøras M, et al. WRN interacts physically and functionally with the recombination mediator protein RAD52. J Biol Chem. 2003;278:36476–86. doi: 10.1074/jbc.M303885200. [DOI] [PubMed] [Google Scholar]
- Beck LA, Hosick TJ, Sinensky M. Isoprenylation is required for the processing of the lamin A precursor. J Cell Biol. 1990;110:1489–99. doi: 10.1083/jcb.110.5.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–81. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bergo MO, Gavino B, Ross J, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc Natl Acad Sci U S A. 2002;99:13049–54. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof O, Kim SH, Irving J, et al. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153:367–80. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Zalle N, Daniely Y, et al. DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J Biol Chem. 2002;277:50934–40. doi: 10.1074/jbc.M210479200. [DOI] [PubMed] [Google Scholar]
- Blank A, Bobola MS, Gold B, et al. The Werner syndrome protein confers resistance to the DNA lesions N3-methyladenine and O(6)-methyl-guanine: implications for WRN function. DNA Repair. 2004;3:629–38. doi: 10.1016/j.dnarep.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Blasco MA. Telomerase beyond telomeres. Nat Rev Cancer. 2002;2:627–33. doi: 10.1038/nrc862. [DOI] [PubMed] [Google Scholar]
- Bridger JM, Kill IR. Aging of Hutchinson-Gilford progeria syndrome fibroblasts is characterised by hyperproliferation and increased apoptosis. Exp Gerontol. 2004;39:717–24. doi: 10.1016/j.exger.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Orren DK, Nehlin JO, et al. Functional and physical interaction between WRN helicase and human replication protein. A J Biol Chem. 1999;274:18341–50. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Li JL, Kenny MK, et al. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–8. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Karmakar P, Sommers JA, et al. p53 modulates the exonuclease activity of Werner syndrome protein. J Biol Chem. 2001a;276:35093–102. doi: 10.1074/jbc.M103332200. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Majumdar A, Desai S, et al. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J Biol Chem. 2001b;276:3024–30. doi: 10.1074/jbc.M006784200. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, von Kobbe C, Sommers JA, et al. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001c;20:5791–801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr, Bohr VA. Roles of the Werner syndrome protein in pathways required for maintenance of genome stability. Exp Gerontol. 2002;37:491–506. doi: 10.1016/s0531-5565(01)00227-3. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Driscoll HC, Dianov GL, et al. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry. 2002;41:12204–16. doi: 10.1021/bi026031j. [DOI] [PubMed] [Google Scholar]
- Cao H, Hegele RA. LMNA is mutated in Hutchinson-Gilford progeria (MIM 176670) but not in Wiedemann- Rautenstrauch progeroid syndrome (MIM 264090) J Hum Genet. 2003;48:271–4. doi: 10.1007/s10038-003-0025-3. [DOI] [PubMed] [Google Scholar]
- Capell BC, Erdos MR, Madigan JP, et al. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2005;102:12879–84. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Multani AS, Cabrera NG, et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nature Genetics. 2004;36:877–82. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- Chen L, Huang S, Lee L, et al. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell. 2003;2:191–9. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- Cheng WH, von Kobbe C, Opresko PL, et al. Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol Cel Biol. 2003;23:6385–95. doi: 10.1128/MCB.23.18.6385-6395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, von Kobbe C, Opresko PL, et al. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J Bio Chem. 2004;279:21169–76. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- Clarke S, Vogel JP, Deschenes RJ, et al. Posttranslational modification of the Haras oncogene protein: evidence for a third class of protein carboxyl methyltransferases. Proc Natl Acad Sci, USA. 1988;85:4643–7. doi: 10.1073/pnas.85.13.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Purdie CA, Harrison DJ, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–52. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–33. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, et al. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–4. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MP, Machwe A, Orren DK, et al. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–12. [PMC free article] [PubMed] [Google Scholar]
- Corrigan DP, Kuszczak D, Rusinol AE, et al. Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem J. 2005;387:129–38. doi: 10.1042/BJ20041359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, et al. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–16. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, et al. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–3. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- Csoka AB, Cao H, Sammak PJ, et al. Novel lamin A/C gene (LMNA) mutations in atypical progeroid syndromes. J Med Genet. 2004a;41:304–8. doi: 10.1136/jmg.2003.015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csoka AB, English SB, Simkevich CP, et al. Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 2004b;3:235–43. doi: 10.1111/j.1474-9728.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–8. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- Dahl KN, Scaffidi P, Islam MF, et al. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2006;103:10271–6. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Choy E, Chiu V, et al. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem. 1998;273:15030–4. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- De Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–40. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, Bernard R, Cau P, et al. Lamin A truncation in Hutchinson–Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- Du X, Shen J, Kugan N, et al. Telomere shortening exposes functions for the Werner and Bloom syndrome genes in mice. Mol Cell Biol. 2004;24:8437–46. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ, Martin GM, Schultz AL, et al. Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine. 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- Epstein J, Williams JR, Little JB. Deficient DNA repair in human progeroid cells. Proc Natl Acad Sci U S A. 1973;70:977–81. doi: 10.1073/pnas.70.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Brown WT, Gordon LB, et al. Recurrent de novo point mutations in lamin a cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–8. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L, Chapman L, Moss H, et al. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol Cell. 2001;8:351–61. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- Fong LG, Ng JK, Lammerding J, et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J Clin Invest. 2006;116:743–52. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossel M. The progerias. J Antiaging Med. 2003;6:123–38. doi: 10.1089/109454503769684793. [DOI] [PubMed] [Google Scholar]
- Fry M, Loeb LA. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- Fukuchi K, Katsuya T, Sugimoto K, et al. LMNA mutation in a 45 year old Japanese subject with Hutchinson-Gilford progeria syndrome. J Med Genet. 2004;41:e67. doi: 10.1136/jmg.2003.014688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilford H. Ateleiosis and progeria: continous youth and premature old age. Br Med J. 1904:914–8. [Google Scholar]
- Glynn MW, Glover TW. Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum Mol Genet. 2005;14:2959–69. doi: 10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Gruenbaum Y, Moir RD, et al. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–47. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci, USA. 2004;101:8963–8. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Miller RW, Ishikawa Y, et al. Excess of rare cancers in Werner’s syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–46. [PubMed] [Google Scholar]
- Goto M. Hierarchical deterioration of body systems in Werner’s syndrome: Implications for normal ageing. Mech Ageing Dev. 1997;98:239–54. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, et al. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–3. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–14. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Halaschek-Wiener J, Brooks-Wilson A. Progeria of stem cells: stem cell exhaustion in hutchinson-gilford progeria syndrome. J Gerontol A Biol Sci Med Sci. 2007;62:3–8. doi: 10.1093/gerona/62.1.3. [DOI] [PubMed] [Google Scholar]
- Hanaoka F, Yamada M, Takeuchi F, et al. Autoradiographic studies of DNA replication in Werner’s syndrome cells. Adv Exp Med Biol. 1985;190:439–57. doi: 10.1007/978-1-4684-7853-2_22. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harrigan JA, Opresko PL, von Kobbe C, et al. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J Biol Chem. 2003;278:22686–95. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- Hasty P, Campisi J, Hoeijmakers J, et al. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–9. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- Hasty P, Vijg J. Accelerating aging by mouse reverse genetics: a rational approach to understand longevity. Aging Cell. 2004;3:55–65. doi: 10.1111/j.1474-9728.2004.00082.x. [DOI] [PubMed] [Google Scholar]
- Haupt S, Berger M, Goldberg Z, et al. Apoptosis - the p53 network. J Cell Sci. 2003;116:4077–85. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- Hennekam RC. Hutchinson-Gilford progeria syndrome: Review of the phenotype. Am J Med Genet A. 2006;140:2603–24. doi: 10.1002/ajmg.a.31346. [DOI] [PubMed] [Google Scholar]
- Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–78. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- Hofseth LJ, Hussain SP, Harris CC. p53: 25 years after its discovery. Trends Pharmacol Sci. 2004;25:177–81. doi: 10.1016/j.tips.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–16. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, et al. The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nature Genet. 1998;20:114–6. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Beresten S, Li B, et al. Characterization of the human and mouse WRN 3′→5′ exonuclease. Nucleic Acids Res. 2000;28:2396–405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. Case of congenital absence of hair, with atrophic condition of the skin and its apendages. Lancet. 1886:923. doi: 10.1177/095952878606900127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeziorny L, McCurdy L, Michael K, et al. The Heli-CASE of the missing WRN gene. Eukaryon. 2006;2:34–9. [Google Scholar]
- Kamath-Loeb AS, Johansson E, Burgers PM, et al. Functional interaction between the Werner syndrome protein and DNA polymerase delta. Proc Natl Acad Sci USA. 2000;97:4603–8. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Loeb LA, Johansson E, et al. Interactions between the Werner syndrome helicase and DNA polymerase d specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J Biol Chem. 2001;276:16439–46. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- Karanjawala ZE, Lieber MR. DNA damage and aging. Mech Ageing Dev. 2004;125:405–16. doi: 10.1016/j.mad.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Karmakar P, Snowden CM, Ramsden DA, et al. Ku heterodimer binds to both ends of the Werner protein and functional interaction occurs at the Werner N-terminus. Nucleic Acids Res. 2002;30:3583–91. doi: 10.1093/nar/gkf482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kator K, Cristofalo V, Charpentier R, et al. Dysdifferentiative nature of aging: passage number dependency of globin gene expression in normal human diploid cells grown in tissue culture. Gerontology. 1985;31:355–61. doi: 10.1159/000212724. [DOI] [PubMed] [Google Scholar]
- Kawabe Y, Seki M, Seki T, et al. Covalent modification of the Werner’s syndrome gene product with the ubiquitin-related protein, SUMO-1. J Biol Chem. 2000;275:20963–6. doi: 10.1074/jbc.C000273200. [DOI] [PubMed] [Google Scholar]
- Kyng KJ, Bohr VA. Gene expression and DNA repair in progeroid syndromes and human aging. Ageing Res Rev. 2005;4:579–602. doi: 10.1016/j.arr.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Laine JP, Opresko PL, Indig FE, et al. Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer Res. 2003;63:7136–46. [PubMed] [Google Scholar]
- Lane D. Anthony Dipple Carcinogenesis Award. p53 from pathway to therapy. Carcinogenesis. 2004;25:1077–81. doi: 10.1093/carcin/bgh186. [DOI] [PubMed] [Google Scholar]
- Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc Natl Acad Sci USA. 1998;95:13097–102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M, Spillare EA, Harris CC, et al. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–9. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- Li B, Comai L. Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J Biol Chem. 2000;275:28349–52. doi: 10.1074/jbc.C000289200. [DOI] [PubMed] [Google Scholar]
- Li B, Comai L. Requirements for the nucleoytic processing of DNA ends by the Werner syndrome protein: Ku70/80 complex. J Biol Chem. 2001;276:9896–902. doi: 10.1074/jbc.M008575200. [DOI] [PubMed] [Google Scholar]
- Li B, Comai L. Displacement of DNA-PKcs from DNA ends by the Werner syndrome protein. Nucleic Acids Res. 2002;30:3653–61. doi: 10.1093/nar/gkf488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Navarro S, Kasahara N, et al. Identification and biochemical characterization of a Werner’s syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J Biol Chem. 2004;279:13659–67. doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- Liu B, Wang J, Chan KM, et al. Genomic instability in laminopathy-based premature aging. Nat Med. 2005;11:780–5. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- Liu Y, Rusinol A, Sinensky M, et al. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J Cell Sci. 2006;119:4644–9. doi: 10.1242/jcs.03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Macias MJ, Bottomley MJ, et al. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Struct Fold Des. 1999;7:1557–66. doi: 10.1016/s0969-2126(00)88346-x. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, et al. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Ly DH, Lockhart DJ, Lerner RA, et al. Mitotic misregulation and human aging. Science. 2000;287:2486–92. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Orren DK. TRF2 recruits the Werner syndrome (WRN) exonuclease for processing of telomeric DNA. Oncogene. 2004;23:149–56. doi: 10.1038/sj.onc.1206906. [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, et al. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem. 2005;280:23397–407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- Machwe A, Lozada EM, Xiao L, et al. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol Biol. 2006;7:1. doi: 10.1186/1471-2199-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–19. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallampalli MP, Huyer G, Bendale P, et al. Inhibiting farnesylation reverses the nuclear morphology defect in a HeLa cell model for Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci, USA. 2005;102:14416–21. doi: 10.1073/pnas.0503712102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Lombard DB, Johnson FB, et al. Nucleolar localization of the Werner syndrome protein in human cells. Proc Natl Acad Sci USA. 1998;95:6887–92. doi: 10.1073/pnas.95.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM, Sprague CA, Epstein CJ. Replicative life-span of cultivated human cells. Effects of donor’s age, tissue, and genotype. Lab Inves. 1970;23:86–92. [PubMed] [Google Scholar]
- Martin GM. Cellular aging-clonal senescence. A review (Part I) Am J Pathol. 1977;89:484–511. [PMC free article] [PubMed] [Google Scholar]
- Martin GM. Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects Orig Artic Ser. 1978;14:5–39. [PubMed] [Google Scholar]
- Martin GM. Genetics and aging: the Werner syndrome as a segmental progeroid syndrome. Adv Exp Med Biol. 1985;190:161–70. doi: 10.1007/978-1-4684-7853-2_5. [DOI] [PubMed] [Google Scholar]
- Martin GM, Oshima J. Lessons from human progeroid syndromes. Nature. 2000;408:263–6. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Shimamoto A, Goto M, et al. Impaired nuclear localization of defective DNA helicases in Werner’s syndrome. Nat Genet. 1997;16:335–6. doi: 10.1038/ng0897-335. [DOI] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–82. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BJ, Holm C. The RAD52 recombinational repair pathway is essential in pol30 (PCNA) mutants that accumulate small single-stranded DNA fragments during DNA synthesis. Genetics. 1998;148:611–24. doi: 10.1093/genetics/148.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh JR, Jr, et al. The Bloom’s and Werner’s syndrome proteins are DNA structurespecific helicases. Nucleic Acids Res. 2001;29:2843–49. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnat RJ, Jr, Saintigny Y. Werner syndrome protein-unwinding function to explain disease. Sci Aging Knowledge Environ. 2004;2004:3. doi: 10.1126/sageke.2004.13.re3. [DOI] [PubMed] [Google Scholar]
- Müller S, Ledl A, Schmidt D. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- Nagao M, Sugimura T. Molecular biology of the carcinogen, 4- nitroquinoline 1-oxide. Adv Cancer Res. 1976;23:131–69. doi: 10.1016/s0065-230x(08)60545-x. [DOI] [PubMed] [Google Scholar]
- Navarro CL, Cau P, Levy N. Molecular bases of progeroid syndromes. Human Molecular Genetics. 2006;15:R151–61. doi: 10.1093/hmg/ddl214. [DOI] [PubMed] [Google Scholar]
- Ogburn CE, Oshima J, Poot M, et al. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum Genet. 1997;101:121–5. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- Ohsugi I, Tokutake Y, Suzuki N, et al. Telomere repeat DNA forms a large non-covalent complex with unique cohesive properties which is dissociated by Werner syndrome DNA helicase in the presence of replication protein A. Nucleic Acids Res. 2000;28:3642–8. doi: 10.1093/nar/28.18.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL, Laine J-P, Brosh RM, Jr, et al. Coordinate action of the helicase and 3′ to 5′ exonuclease of Werner syndrome protein. J Biol Chem. 2001;276:44677–87. doi: 10.1074/jbc.M107548200. [DOI] [PubMed] [Google Scholar]
- Opresko PL, von Kobbe C, Laine JP, et al. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicase. J Biol Chem. 2002;277:41110–19. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Cheng WH, von Kobbe C, et al. Werner syndrome and the function of the Werner protein: what they can teach us about the molecular aging process. Carcinogenesis. 2003;24:791–802. doi: 10.1093/carcin/bgg034. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Otterlei M, Graakjaer J, et al. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–74. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Orren DK, Machwe A, Karmakar P, et al. A functional interaction of Ku with Werner exonuclease facilitates digestion of damaged DNA. Nucleic Acids Res. 2001;29:1926–34. doi: 10.1093/nar/29.9.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima J, Huang S, Pae C, et al. Lack of WRN results in extensive deletion at nonhomologous joining ends. Cancer Res. 2002;62:547–51. [PubMed] [Google Scholar]
- Pichierri P, Rosselli F, Franchitto A. Werner’s syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene. 2003;22:1491–500. doi: 10.1038/sj.onc.1206169. [DOI] [PubMed] [Google Scholar]
- Poot M, Hoehn H, Runger TM, et al. Impaired S phase transit of Werner syndrome cells expressed in lymphoblastoid cells. Exp Cell Res. 1992;202:267–73. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- Poot M, Yom JS, Whang SH, et al. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–6. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- Poot M, Gollahon KA, Emond MJ, et al. Werner syndrome diploid fibroblasts are sensitive to 4-nitroquinoline-N-oxide and 8-methoxypsoralen: implications for the disease phenotype. FASEB J. 2002;16:757–8. doi: 10.1096/fj.01-0906fje. [DOI] [PubMed] [Google Scholar]
- Prince PR, Emond MJ, Monnat RJ., Jr Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 2001;15:933–8. doi: 10.1101/gad.877001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla TA. Multiple roads to the aging phenotype: insights from the molecular dissection of progerias through DNA microarray analysis. Mech Ageing Dev. 2005;126:461–5. doi: 10.1016/j.mad.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Rodríguez-López AM, Jackson DA, Nehlin JO, et al. Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner’s syndrome, and an essential replication factor, PCNA. Mech Ageing Dev. 2003;124:167–74. doi: 10.1016/s0047-6374(02)00131-8. [DOI] [PubMed] [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, et al. Homologous recombination resolution defect in Werner syndrome. Mol Cell Biol. 2002;22:6971–8. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S, Nishikawa K, Heo SJ, et al. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells. 2001;6:421–30. doi: 10.1046/j.1365-2443.2001.00433.x. [DOI] [PubMed] [Google Scholar]
- Salk D, Bryant E, Hoehn H, et al. Growth characteristics of Werner syndrome cells in vitro. Adv Exp Med Biol. 1985;190:305–11. doi: 10.1007/978-1-4684-7853-2_14. [DOI] [PubMed] [Google Scholar]
- Sanz MM, Proytcheva M, Ellis NA, et al. BLM, the Bloom’s syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet Cell Genet. 2000;91:217–23. doi: 10.1159/000056848. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–5. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–63. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz VP, Zakian V A, Ogburn CE, et al. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum Genet. 1996;97:750–4. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- Serrano M, Blasco MA. Cancer and ageing: convergent and divergent mechanisms. Nat Rev Mol Cell Biol. 2007;8:715–22. doi: 10.1038/nrm2242. [DOI] [PubMed] [Google Scholar]
- Sharma S, Otterlei M, Sommers JA, et al. WRN helicase and FEN-1 form a complex upon replication arrest and together process branch-migrating DNA structures associated with the replication fork. Mol Biol Cell. 2004;15:734–50. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Sommers JA, Gary RK, et al. The interaction site of Flap Endonuclease-1 with WRN helicase suggests a coordination of WRN and PCNA. Nucleic Acids Res. 2005;33:6769–81. doi: 10.1093/nar/gki1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Gray MD, Oshima J, et al. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998;26:2879–85. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Loeb LA. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000;28:3260–8. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Lao Y, Kamath-Loeb A, et al. The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech Ageing Dev. 2003;124:921–30. doi: 10.1016/s0047-6374(03)00164-7. [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Sommers JA, Sharma S, Doherty KM, et al. p53 modulates RPA-dependent and RPA-independent WRN helicase activity. Cancer Res. 2005;65:1223–33. doi: 10.1158/0008-5472.CAN-03-0231. [DOI] [PubMed] [Google Scholar]
- Spann TP, Goldman AE, Wang C, et al. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J Cell Biol. 2002;156:603–8. doi: 10.1083/jcb.200112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillare EA, Robles AI, Wang XW, et al. p53-mediated apoptosis is attenuated in Werner syndrome cells. Genes Dev. 1999;13:1355–60. doi: 10.1101/gad.13.11.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillare EA, Wang XW, von Kobbe C, et al. Redundancy of DNA helicases in p53-mediated apoptosis. Oncogene. 2006;25:2119–23. doi: 10.1038/sj.onc.1209242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun QY, Schatten H. Role of NuMA in vertebrate cells: review of an intriguing multifunctional protein. Front Biosci. 2006;11:1137–46. doi: 10.2741/1868. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Shimamoto A, Imamura O, et al. DNA helicase activity in Werner’s syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25:2973–8. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Shiratori M, Goto M, et al. Werner syndrome helicase contains a 5′→3′ exonuclease activity that digests DNA and RNA strands in DNA/DNA and RNA/DNA duplexes dependent on unwinding. Nucleic Acids Res. 1999;27:2361–8. doi: 10.1093/nar/27.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds H, Krall L, Remington L, et al. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell. 1994;78:703–11. doi: 10.1016/0092-8674(94)90534-7. [DOI] [PubMed] [Google Scholar]
- Szekely AM, Chen YH, Zhang C, et al. Werner protein recruits DNA polymerase δ to the nucleolus. Proc Natl Acad Sci, USA. 2000;97:11365–70. doi: 10.1073/pnas.97.21.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–56. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, Hanaoka F, Goto M, et al. Altered frequency of initiation sites of DNA replication in Werner’s syndrome cells. Hum Genet. 1982;60:365–8. doi: 10.1007/BF00569220. [DOI] [PubMed] [Google Scholar]
- Tauchi H, Kobayashi J, Morishima K, et al. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–8. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- Toth JI, Yang SH, Qiao X, et al. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc Natl Acad Sci U S A. 2005;102:12873–8. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–93. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Varela I, Cadinanos J, Pendas AM, et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–8. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Bohr VA. A nucleolar targeting sequence in the Werner syndrome protein resides within residues 949–1092. J Cell Sci. 2002;115:3901–7. doi: 10.1242/jcs.00076. [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Karmakar P, Dawut L, et al. Colocalization, physical and functional interaction between Werner and Bloom syndrome proteins. J Biol Chem. 2002;277:22035–44. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Thoma NH, Czyzewski BK, et al. Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem. 2003a;278:52997–3006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Harrigan JA, May A, et al. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol. 2003b;23:8601–13. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C, Harrigan JA, Schreiber V, et al. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic Acids Res. 2004;32:4003–14. doi: 10.1093/nar/gkh721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T, Saretzki G, Ladhoff J, et al. Human cell senescence as a DNA damage response. Mech Ageing Dev. 2005;126:111–17. doi: 10.1016/j.mad.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nature Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Wallis CV, Sheerin AN, Green MH, et al. Fibroblast clones from patients with Hutchinson-Gilford progeria can senesce despite the presence of telomerase. Exp Gerontol. 2004;39:461–7. doi: 10.1016/j.exger.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Wang XW, Vermeulen W, Coursen JD, et al. The XPB and XPD DNA helicases are components of the p53-mediated apoptosis pathway. Genes Dev. 1996;10:1219–32. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- Wang XW, Tseng A, Ellis NA, et al. Functional interaction of p53 and BLM DNA helicase in apoptosis. J Biol Chem. 2001;276:32948–55. doi: 10.1074/jbc.M103298200. [DOI] [PubMed] [Google Scholar]
- Warbrick E. PCNA binding through a conserved motif. BioEssays. 1998;20:195–9. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA’s many partners. BioEssays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Weber K, Plessmann U, Traub P. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor: implications for the structure of the nuclear lamina. FEBS Lett. 1989;257:411–4. doi: 10.1016/0014-5793(89)81584-4. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single stranded DNA binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Woods YL, Xirodimas DP, Prescott AR, et al. p14 Arf promotes small ubiquitin-like modifier conjugation of Werners helicase. J Biol Chem. 2004;279:50157–66. doi: 10.1074/jbc.M405414200. [DOI] [PubMed] [Google Scholar]
- Wright WE, Shay JW. The two-stage mechanism controlling cellular senescence and immortalization. Exp Gerontol. 1992;27:383–9. doi: 10.1016/0531-5565(92)90069-c. [DOI] [PubMed] [Google Scholar]
- Wu L, Davies SL, Levitt NC, et al. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276:19375–81. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–4. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wyllie FS, Jones CJ, Skinner JW, et al. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat Genet. 2000;24:16–7. doi: 10.1038/71630. [DOI] [PubMed] [Google Scholar]
- Yamabe Y, Shimamoto A, Goto M, et al. Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Mol Cell Biol. 1998;18:6191–200. doi: 10.1128/mcb.18.11.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Bergo MO, Toth JI, et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc Natl Acad Sci U S A. 2005;102:10291–6. doi: 10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Meta M, Qiao X, et al. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J Clin Invest. 2006;116:2115–21. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannone SM, Roy S, Chan DW, et al. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem. 2001;276:38242–8. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Blobel G. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol. 2001;152:729–40. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SG, Meta M, Yang SH, et al. Prelamin A farnesylation and progeroid syndromes. J Biol Chem. 2006;281:39741–5. doi: 10.1074/jbc.R600033200. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, et al. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–62. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Wijsman EM, et al. Mutations in the consensus helicase domains of the Werner syndrome gene. Werner’s Syndrome Collaborative Group. Am J Hum Genet. 1997;60:330–41. [PMC free article] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–69. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- Zs-Nagy I, Cutler RG, Semsei I. Dysdifferentiation hypothesis of aging and cancer: a comparison with the membrane hypothesis of aging. Ann NY Acad Sci. 1988;521:215–25. doi: 10.1111/j.1749-6632.1988.tb35280.x. [DOI] [PubMed] [Google Scholar]