Abstract
Background
Human anxiety disorders are complex diseases with largely unknown etiology. We have taken a cross-species approach to identify genes that regulate anxiety-like behavior with inbred mouse strains that differ in their innate anxiety levels as a model. We previously identified 17 genes with expression levels that correlate with anxiety behavior across the studied strains. In the present study, we tested their 13 known human homologues as candidate genes for human anxiety disorders with a genetic association study.
Methods
We describe an anxiety disorder study sample derived from a Finnish population-based cohort and consisting of 321 patients and 653 carefully matched control subjects, all interviewed to obtain DSM-IV diagnoses. We genotyped altogether 208 single nucleotide polymorphisms (SNPs) (all non-synonymous SNPs, SNPs that alter potential microRNA binding sites, and gap-filling SNPs selected on the basis of HapMap information) from the investigated anxiety candidate genes.
Results
Specific alleles and haplotypes of six of the examined genes revealed some evidence for association (p ≤ .01). The most significant evidence for association with different anxiety disorder subtypes were: p = .0009 with ALAD (δ-aminolevulinate dehydratase) in social phobia, p = .009 with DYNLL2 (dynein light chain 2) in generalized anxiety disorder, and p = .004 with PSAP (prosaposin) in panic disorder.
Conclusions
Our findings suggest that variants in these genes might predispose to specific human anxiety disorders. These results illustrate the potential utility of cross-species approaches in identification of candidate genes for psychiatric disorders.
Key Words: Anxiety disorders, association study, genetic isolate, SNP
Fear and anxiety are normal emotional responses to threatening situations. However, when exaggerated and causing distress, disability, and loss of quality of life, they manifest as anxiety disorders as classified in the DSM-IV (1) and ICD-10 (2). In DSM-IV, anxiety disorders include panic disorder (with or without agoraphobia), obsessive-compulsive disorder, generalized anxiety disorder, post-traumatic stress disorder, social phobia, agoraphobia, and specific phobias. The combined lifetime prevalence of these disorders in the general population has been estimated to be 16.6% on average (3). Anxiety disorders are also highly comorbid with each other and other psychiatric disorders such as depression and substance abuse (4).
Anxiety disorders are complex diseases, influenced by a combination of both genetic and environmental predisposing factors. For example, the heritability of panic disorder and generalized anxiety disorder has been estimated to be 48% and 32%, respectively (5). Models suggest that the genetic architecture of anxiety disorders can be best explained by one shared set of genetic factors mainly influencing predisposition to panic disorder, generalized anxiety disorder, and agoraphobia and another set primarily underlying specific phobias (6). Furthermore, there is evidence for considerable overlap between the genetic factors influencing anxiety disorders and those influencing depression and the personality traits neuroticism and introversion (7,8).
Because the pursuit of human anxiety disorder susceptibility genes by conventional linkage and association mapping has proved to be demanding (reviewed in 9), there is a need to consider the aid of alternative approaches, such as ones relying on animal models for identification of genes and loci with relevance for complex traits (reviewed in 9,10). Several paradigms for the measurement of anxiety in mice have been developed and pharmacologically validated, and they are considered to model aspects of human anxiety (e.g., 11–13). We have previously combined behavioral testing with gene expression profiling of specific brain regions to identify genes whose expression levels might regulate anxiety-related behavior in mice (14). This approach takes advantage of the different innate levels of anxiety-like behavior of well-characterized inbred mouse strains. Briefly, by combining behavioral analysis of six inbred strains with gene expression profiling of seven brain regions involved in the regulation of anxiety, we identified 17 genes with expression levels that correlated significantly with the anxiety phenotype across all strains in at least one brain region. We further showed functionally that 2 of the identified genes, glyoxalase 1 (Glo1) and glutathione reductase 1 (Gsr), regulate anxiety behavior in mice in vivo. Although the 15 remaining genes were not tested functionally, they remain strong candidate genes for anxiety. In the present study, we tested these genes as candidate genes for human anxiety disorders with a genetic association analysis in a Finnish population-based study sample.
Methods and Materials
Study Sample
The individuals in this study derive from the Finnish population-based Health 2000 Study carried out in 2000–2001. This work was approved by the institutional ethics committee of the National Public Health Institute, and written consent was obtained. Identification of individuals with anxiety disorders has been described in detail by Pirkola et al. (15,16). Briefly, the 12-month prevalence of DSM-IV mental disorders was estimated from a representative sample (n = 6005) of the Finnish general adult (≥ 30 years of age) population with the Composite International Diagnostic Interview (CIDI). Finland represents a well-characterized genetic isolate (17), with approximately 2% of individuals with foreign descent. No ethnic groups were excluded during recruitment to obtain a representative epidemiological cohort, but the interview was conducted in Finnish, excluding all non-fluent foreigners. A computer-aided version of the mental health interview (M-CIDI) was administered by trained non-psychiatric healthcare professionals and designed to determine the 12-month prevalence of a set of clinically and epidemiologically relevant mental disorders. These included major depressive disorder, dysthymia, generalized anxiety disorder, panic disorder with or without agoraphobia, agoraphobia, social phobia, and alcohol abuse and dependence. The total number of reliably performed mental health interviews was 6005, amounting to 75% of the original sample. Compared with CIDI participants, the dropouts (n = 981) had somewhat higher scores in the Beck Depression Inventory (BDI), indicating depressive symptoms, and General Health Questionnaire-12 (GHQ-12), indicating psychic distress. They also had slightly older age, lower education, and were more frequently single. Although this might bias the final results toward underestimation of prevalence of mental disorders in the general population, we believe that the most common anxiety disorders are well-represented in our cohort. Due to the structure of the interview, we were not able to reliably diagnose obsessive-compulsive disorder and post-traumatic stress disorder and chose to focus on a subset of the most clinically relevant anxiety disorders in the Finnish population.
From this cohort, we initially selected all individuals with a DSM-IV anxiety disorder diagnosis for analysis (core diagnostic group). In addition, we broadened our definition of anxiety disorder subjects to include individuals with sub-threshold diagnoses as defined by the CIDI (extended diagnostic group). For each case, we selected two control subjects who lacked diagnosed anxiety or major mental disorders and were matched according to gender, age (± 1 year), and hospital catchment area. The DNA was not available from 34 subjects (15 cases and 19 control subjects), and therefore, the final analyzed study sample consisted of 321 cases and 653 control subjects (Table 1). Forty-one core subjects were diagnosed with more than one anxiety disorder (5 with three disorders, and 36 with two disorders). The most frequent combination of diagnoses was panic disorder comorbidity with social phobia (18 subjects).
Table 1.
Demographic Characteristics of the Health 2000 Anxiety Disorder Study Sample
| Diagnostic Group | DSM-IV Casesa | DSM-IV Sub-Threshold Cases | Cases Totalb | Controls | Total | Men | Women | Mean Age ± SD |
|---|---|---|---|---|---|---|---|---|
| Panic Disorderc | 108 | 0 | 108 | 218 | 326 | 106 (32.5%) | 220 (67.5%) | 46.7 ± 11.3 |
| Generalized Anxiety Disorder | 73 | 30 | 103 | 206 | 309 | 122 (39.5%) | 187 (60.5%) | 50.6 ± 12.6 |
| Social Phobia | 58 | 7 | 65 | 133 | 198 | 99 (50.0%) | 99 (50.0%) | 45.4 ± 10.2 |
| Agoraphobia | 31 | 15 | 46 | 94 | 140 | 60 (42.9%) | 80 (57.1%) | 52.8 ± 13.0 |
| Phobia, not otherwise specified | 58 | 0 | 58 | 121 | 179 | 54 (30.2%) | 125 (69.8%) | 54.6 ± 13.3 |
| All Studied Anxiety Disorders | 282 | 39 | 321 | 653 | 974 | 357 (36.7%) | 617 (63.3%) | 49.8 ± 12.7 |
Core diagnostic group.
Extended diagnostic group.
Includes both panic disorder with and without agoraphobia.
Single Nucleotide Polymorphism Selection
The association study was carried out in two stages. In stage I, we examined the known human homologues of 13 anxiety candidate genes identified in the mouse (14) with a set of 139 carefully selected single nucleotide polymorphisms (SNPs; Table 2). In stage II, we followed up on positive findings by genotyping 69 additional markers from genes showing evidence for association after stage I. The 208 markers selected for genotyping represented three different variation groups: non-synonymous SNPs, haplotype-tagging SNPs, and putative polymorphic microRNA (miRNA) binding sites.
Table 2.
Investigated Candidate Genes and Number of Analyzed SNPs/Gene
| Symbol | Gene Information |
Stage I |
Stage II |
Total | ||||
|---|---|---|---|---|---|---|---|---|
| Name | Position | Size (kb) | Non-Synonymous | tagSNPs | miRNA Sitesa | tagSNPs | ||
| ALAD | Aminolevulinate, δ-, dehydratase | 9q33.1 | 15.0 | 1 (1) | 7 | 2 (3) | 4 | 14 (4) |
| CDH2 | Cadherin-2, type 1, N-cadherin (neuronal) | 18q11.2 | 226.3 | 1 (2) | 10 | — | 10 | 21 (2) |
| CPSF4 | Cleavage and polyadenylation specificity factor 4, 30kDa | 7q22.1 | 18.3 | — | 4 | — | — | 4 |
| DYNLL2 | Dynein, light chain, LC8-type 2 | 17q22 | 6.0 | — | 4 | — | 2 (1) | 6 (1) |
| EPB41L4A | Erythrocyte membrane protein band 4.1 like 4A | 5q22.2 | 256.7 | 3 | 10 (1) | — | 14 | 27 (1) |
| EPHX1 | Epoxide hydrolase 1, microsomal (xenobiotic) | 1q42.1 | 20.3 | 2 (5) | 6 | — | — | 8 (5) |
| GLO1 | Glyoxalase I | 6p21.3-p21.1 | 27.2 | 1 (2) | 6 | - (1) | 5 | 12 (3) |
| GSR | Glutathione reductase | 8p21.1 | 49.0 | 2 (6) | 7 | — | 7 | 16 (6) |
| PSAP | Prosaposin (variant Gaucher disease and metachromatic leukodystrophy) | 10q21-q22 | 35.0 | - (3) | 9 | 1 (1) | 5 | 15 (4) |
| PTGDS | Prostaglandin D2 synthase 21kDa (brain) | 9q34.2-q34.3 | 4.2 | - (3) | 4 | — | 2 (1) | 6 (4) |
| S100A10 | S100 calcium binding protein A10 | 1q21 | 21.0 | - (2) | 3 | — | — | 3 (2) |
| SCN1B | Sodium channel, voltage-gated, type I, β | 19q13.1 | 9.8 | - (2) | 3 | — | — | 3 (2) |
| SLC15A2 | Solute carrier family 15 (H+/peptide transporter), member 2 | 3q13.33 | 47.2 | 2 (3) | 7 | — | — | 9 (3) |
| Total | 12 (29) | 80 (1) | 3 (5) | 49 (2) | 144 (37) | |||
Number of genotyped single nucleotide polymorphisms (SNPs) that were monomorphic in our study sample is shown in parenthesis.
Putative polymorphic microRNA (miRNA) binding sites selected with the Patrocles database.
First, we selected all known validated and unvalidated non-synonymous SNPs with SNPper (18; http://snpper.chip.org), with the aim to identify SNPs that potentially alter the function of the gene products. Second, we retrieved HapMap (19; http://www.hapmap.org) phase I genotype information for the European-derived Caucasian/European ancestry (CEU) population and used the Tagger algorithm as implemented in Haploview (20; http://www.broad.mit.edu/mpg/haploview) to identify complementary tagSNPs for genotyping. TagSNPs were required to have minor allele frequencies ≥ .05, when possible and to capture the variation of other SNPs with minor allele frequencies ≥ .05 by a minimum r2 = .8. For 11 genes, we selected tagging SNPs every 5 kb, on average. For the two largest genes, CDH2 and EPB41L4A, we selected one tagging SNP/exon. In stage II of the study, we selected supplementary tagSNPs with HapMap phase II data. Third, to select SNPs that potentially alter gene expression, we used the Patrocles database (http://www.patrocles.org) to identify validated and unvalidated SNPs resulting in putative polymorphic miRNA binding sites within the 3'-untranslated regions (UTRs) of our candidate genes and genotyped them in stage II.
Genotyping and Quality Control
The DNA was extracted from whole blood at the DNA extraction core facility of the National Public Health Institute with the Puregene manual DNA extraction kit (Gentra Systems, Minneapolis, Minnesota). The DNA quality control (QC) measures consisted of gender-specific polymerase chain reaction to verify that observed gender matched database information and genotyping of five polymorphic microsatellites to verify that samples were not contaminated (21).
Genotyping was done with Sequenom MassARRAY genotyping technology (Sequenom, San Diego, California) with iPLEX chemistry in stage I and iPLEX Gold chemistry in stage II. Assay conditions were as recommended by the manufacturer, and assay information including primer sequences is available from the authors upon request. Genotyping QC measures consisted of checking no-template control samples for allele peaks; reviewing all marker data and verifying that genotype clusters were well distinguished and outliers removed; verifying consistencies in genotype calls of randomly selected duplicate samples (approximately 2% of samples across, and 2% of samples within sample plates); disregarding marker assays with low success rates (< 85%); and discarding all genotype information for individual samples with genotype calls for < 75% of the analyzed markers. Furthermore, Hardy-Weinberg equilibrium was evaluated for each marker in the control sample with a χ2 test in addition to confirming Mendelian inheritance of marker alleles by genotyping an additional sample of 60 anonymous parent-offspring trios.
Statistical Analysis
We estimated the magnitude of genotypic relative risk possible to detect with the studied sample size (321 cases and 653 control subjects) with the Genetic Power Calculator tool (22; http://pngu.mgh.harvard.edu/∼purcell/gpc). Disease prevalence was set at 5%, corresponding to the Finnish annual anxiety disorder prevalence (15), inheritance to follow a dominant model, and the frequencies of disease and marker alleles were defined as equal with D' = 1. The simulation was run with a range of marker and disease allele frequencies (.01–.6) to illustrate how they influence power to detect association.
Statistical analyses were carried out for core and extended diagnostic groups for all anxiety disorders combined as well as separately for the subgroups panic disorder, generalized anxiety disorder, and social phobia. Two subgroups, agoraphobia and phobia not otherwise specified, were included in the analysis of all anxiety disorders combined but not analyzed separately due to small sample size and phenotypic heterogeneity, respectively.
A conventional 2 × 2 contingency table likelihood-ratio test (LRT) of independence of the SNP allele counts in cases and control subjects (23), with p values determined from permutation testing, was performed to test individual SNP alleles for association with anxiety disorders. We analyzed 10,000 permutations of the dataset to obtain empirical p values. In addition, we performed haplotype analyses on 2- and 3-marker “sliding windows” with the UNPHASED software (24; http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased) that estimates haplotype frequencies from unphased genotype data with an expectation-maximization algorithm. Haplotypes were analyzed with the default full model implemented by the software that tests the null hypothesis that there is no difference in odds ratios between haplotypes using a likelihood-ratio test. A zero frequency threshold of .01 was used to exclude haplotypes with estimated frequencies below 1% in both cases and control subjects. Empirical p values were calculated with 10 000 permutations.
To order the genes according to their significance with regard to anxiety disorders, a supplementary analysis was performed to examine whether significant trends in associations could be detected across individual genes. The obtained pointwise p values for all SNPs were ranked from most to least significant (1–1008), after which they were divided into four classes representing the top and bottom findings (top decile, second decile, second quintile, and bottom 60%). For each gene, we then determined the distribution of findings across these classes. A similar analysis was done in which the p value for each of the 144 markers was minimized over all tests performed to eliminate redundancy.
Results
Power Calculations
With marker and disease allele frequencies of 1%–60%, the power calculations implied that our entire study sample would provide > 80% power to reject the null hypothesis of no association at p < .05 when a genotypic relative risk was higher than 1.48–2.62, with the exact value depending on the allele frequency (Supplement 1).
Genotyping QC
Out of the total 208 SNPs selected for genotyping, 181 marker assays (87%) met our QC criteria. Thirty-seven markers, typically unvalidated or rare non-synonymous polymorphisms or unvalidated putative polymorphic miRNA binding sites, were monomorphic, making the total number of statistically analyzed SNPs 144 (92 in stage I, and 52 in stage II; Table 2). The average genotyping success rate for the analyzed markers was 98.7% (median 99.1%), and it varied from 86.1% (rs16933168) to 99.9% (13 markers). The average genotyping success rate in individual samples for the analyzed markers was 98.7% (median 100%), and it varied from 78.1% to 100%. Eight markers included in the analysis exhibited deviation from Hardy-Weinberg equilibrium in the control sample at p < .05, with the most significant p value being p = .015 for marker rs16933168. Detailed results and allele frequencies for all analyzed markers are available in Supplement 2.
Association Analysis, Stage I
Statistical analysis of single-marker associations for 92 SNPs in stage I identified markers from several genes exhibiting empirical pointwise p values of ≤ .05 (Supplement 2). The following criteria were used to select genes for further investigation in stage II: 1) at least two markers in the gene showing p ≤ .05 in allele-based association tests in any diagnostic group, or 2) a haplotype window associated at p ≤ .05 in any diagnostic group. On the basis of these criteria, eight genes were studied further: ALAD (δ-aminolevulinate dehydratase), CDH2 (cadherin 2; N-cadherin), DYNLL2 (dynein light chain 2), EPB41L4A (erythrocyte membrane protein band 4.1 like 4a), GLO1 (glyoxalase I), GSR (glutathione reductase), PSAP (prosaposin), and PTGDS (prostaglandin D2 synthase). We did not find any association with SNPs in CPSF4 (cleavage and polyadenylation specificity factor 4), EPHX1 (epoxide hydrolase 1), S100A10 (s100 calcium binding protein A10), SCN1B (voltage gated sodium channel, type I, β), or SLC15A2 (solute carrier family 15 [H+/peptide transporter], member 2) genes and did not examine them further after stage I.
Association Analysis, Stage II
Genotype data for 52 additional stage II SNPs was added to the stage I information. Upon final analysis, we chose to use pointwise and global haplotypic p values of p ≤ .01 (empirically evaluated) as criteria for evidence for association. Five genes (ALAD, CDH2, EPB41L4A, PSAP, and PTGDS) showed evidence for association at this level in single-point analyses (Table 3), whereas associations to GLO1 and GSR remained suggestive (.01 < p < .05; Supplement 2).
Table 3.
SNPs with p Value ≤ .01 in the LRT for Allelic Association
| Gene | SNP rs # | Alleles [A1/A2] | Group | Allele Frequencies |
Allelic LRT p | Diagnostic Group | |
|---|---|---|---|---|---|---|---|
| A1 | A2 | ||||||
| ALAD | rs818702 | A/G | Cases | .884 | .116 | .008 | SOCPHa |
| Controls | .761 | .239 | |||||
| CDH2 | rs7240351b | A/G | Cases | .333 | .667 | .006 | SOCPHc |
| Controls | .488 | .513 | |||||
| EPB41L4A | rs7719346 | A/G | Cases | .140 | .860 | .008 | GADa |
| Controls | .248 | .752 | |||||
| rs1464766 | A/G | Cases | .255 | .745 | .010 | PD | |
| Controls | .350 | .651 | |||||
| rs12657885b | C/T | Cases | .918 | .083 | .010 | GADc | |
| Controls | .849 | .151 | |||||
| PSAP | rs4746097 | C/T | Cases | .729 | .271 | .004 | PD |
| Controls | .616 | .384 | |||||
| rs11597008b | C/T | Cases | .615 | .385 | .008 | PD | |
| Controls | .502 | .498 | |||||
| PTGDS | rs4880179 | A/G | Cases | .053 | .947 | .010 | GADc |
| Controls | .015 | .985 | |||||
Empirical p values from 10,000 permutations are shown. SNP, single nucleotide polymorphism; LRT, likelihood-ratio test; SOCPH, social phobia; GAD, generalized anxiety disorder; PD, panic disorder; refer to Table 2 for gene names.
DSM-IV core diagnosis.
Rs-numbers of markers genotyped in stage II.
DSM-IV extended diagnosis.
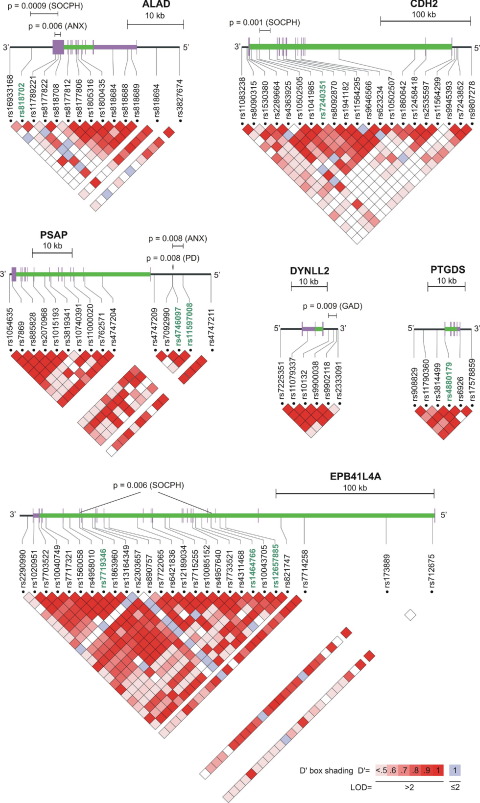
Examination of two- and three-marker haplotypes supported the single-point findings in ALAD, CDH2, and PSAP (Table 4). In addition, DYNLL2 was implicated by the haplotype-based analysis, even though SNPs within this gene did not show evidence for association at p ≤ .01 in the single marker analysis. Haplotypes in ALAD and PSAP were associated not only with social phobia and panic disorder, respectively, but also with all anxiety disorders combined. We did not find haplotype associations with empirical p values of < .01 for any combination of adjacent markers in the genes PTGDS and EPB41L4A in analysis of sliding windows. However, when performing a combined analysis of two non-adjacent EPB41L4A markers showing single-point associations (rs7719346 and rs1464766, 83 kb apart), we identified haplotypes associated with either increased or decreased risk for social phobia. All genes with SNPs and/or haplotypes associated to anxiety disorders with p ≤ .01 are depicted in Figure 1.
Table 4.
Two- and Three-Marker Haplotypes with Global p Value ≤ .01 in Sliding Window Analysis
| Gene | Markers | Haplotype | Haplotype Frequencies |
Global p | Diagnostic Group | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| ALAD | rs11789221 - rs8177822 - rs818708 | C-A-C | .014 | .002 | .006 | ANXa |
| C-C-C | .578 | .607 | ||||
| C-C-T | .408 | .391 | ||||
| rs818702 - rs11789221 - rs8177822 | A-C-A | .036 | .000 | .0009 | SOCPHa | |
| A-C-C | .848 | .766 | ||||
| G-C-C | .116 | .234 | ||||
| CDH2 | rs1041985 - rs7240351 | C-A | .333 | .487 | .001 | SOCPHb |
| C-G | .317 | .160 | ||||
| T-G | .350 | .353 | ||||
| DYNLL2 | rs10132 - rs9900038 - rs9902118 | A-G-C | .131 | .061 | .009 | GADb |
| A-G-T | .621 | .717 | ||||
| G-A-C | .248 | .222 | ||||
| EPB41L4A | rs7719346 - rs1464766 | A-A | .053 | .197 | .006 | SOCPHa |
| A-G | .174 | .124 | ||||
| G-A | .201 | .159 | ||||
| G-G | .572 | .521 | ||||
| PSAP | rs7092990 - rs4746097 | C-C | .729 | .616 | .008 | PD |
| C-T | .196 | .310 | ||||
| T-T | .075 | .074 | ||||
| rs7092990 - rs4746097 - rs11597008 | C-C-C | .612 | .552 | .008 | ANXb | |
| C-C-T | .100 | .116 | ||||
| C-T-T | .197 | .259 | ||||
| T-T-T | .091 | .073 | ||||
Empirical p values from 10 000 permutations are shown. ANX, all studied anxiety disorders; SOCPH, social phobia; GAD, generalized anxiety disorder; PD, panic disorder; refer to Table 2 for gene names.
DSM-IV core diagnosis.
DSM-IV extended diagnosis.
Figure 1.
Genomic structure, relative single nucleotide polymorphism (SNP) positions, and linkage disequilibrium (LD) plots for the six genes showing evidence for association. The SNP haplotype windows with global p values ≤ .01 are indicated above the genomic structure of the gene. The shown haplotype-based p value for EPB41L4A is from an analysis of two non-adjacent SNPs. The rs-numbers of SNPs showing p ≤ .01 in the single-point analysis are highlighted in green. The triangular plots represent pairwise comparisons of intermarker LD, measured in D'. The span of the coding sequence of each gene is depicted with a green line. Figure modified from Locusview2.0 output (T. Petryshen, A. Kirby, and M. Ainscow, unpublished software).
Trend Analysis
Examination of trends in pointwise association across individual genes with a 3-df LRT revealed most noticeably that EPB41L4A was overrepresented among the top findings, with a clear trend toward lower representation as one descends in ranked p values (all ranked p values = .0003; p minimized p values = .008; Supplement 3). We further explored the significance of this finding, focusing on the degree of overrepresentation of this gene in the top quintile with a one-sided LRT (pall = 9.46 × 10−6; pminimized = .002). We focused the analysis in this way for the following reasons: 1) if a gene is influencing the trait, one would expect overrepresentation in the top ranked set of markers; 2) significance testing can be performed without extra df when the primary focus of interest is on the top decile; and 3) in this way, the test can be made one-sided, because a paucity of results in this tail would not be of inferential importance. The observed deviation from the expected distribution of ranks supports the evidence for etiological involvement of EPB41L4A, with the rank-test p value remaining significant after Bonferroni-correction (pall = .0001; pminimized = .028).
Linkage disequilibrium among markers might create correlation in their results. However, the average pairwise marker linkage disequilibrium in each gene is similar, and only EPB41L4A shows a clear trend in ranks. Furthermore, the average r2 between markers is quite low, implying that correlation among marker signals would be minimal under the null hypothesis.
Discussion
Adaptive responses related to anxiety, fear, and stress seem evolutionarily conserved in mammals, and genes influencing anxiety-like behavior in mice can therefore be considered excellent candidate genes also for human anxiety disorders (see e.g., 9). Several quantitative trait loci for anxiety-like behavior have been identified in mouse crosses (e.g., 25,26). Syntenic regions in humans have been tested as candidate regions for panic disorder (27) and personality trait behavioral inhibition (28). In this study, we tested whether variation in 13 genes previously shown to be differentially expressed between non-anxious and anxious inbred mouse strains predisposes humans to anxiety disorders. Six of the genes (ALAD, CDH2, DYNLL2, EPB41L4A, PSAP, and PTGDS) showed empirical p values of ≤ .01 in either single marker, haplotype, or both analyses. To our knowledge, this is the first study showing evidence for association between anxiety disorders and variants in these genes. Neither have any conclusive functional connections between them and anxiety been established so far, although they all seem important for proper functioning of the central nervous system.
Delta-aminolevulinate dehydratase catalyzes conversion of δ-aminolevulinate into porphobilinogen within the heme biosynthetic pathway (29). It is markedly inhibited by lead (30), and a functional polymorphism in ALAD might modulate the adverse effects of lead burden on phobic anxiety symptoms (31). Our most significant association for ALAD also is for a phobic phenotype but not for the same polymorphism (rs1800435). This difference might be explained by differences in study population, cohort demography, and analyzed phenotype. We found a rare risk haplotype (rs818702, rs11789221, and rs8177822), enriched among social phobia subjects. Intriguingly, the rare A allele of rs817782 creates a putative miRNA binding site for miR-204 and miR-211 (www.patrocles.org), within the ALAD 3'-UTR. We previously found Alad expressed at a higher level in the hippocampus and periaqueductal grey of anxious mice (14).
The CDH2 gene encodes a cell-cell adhesion molecule (32) with several functions, including establishment of embryonic left-right asymmetry (33), synaptic adhesion (34), and dendritic spine (35) and cortical morphology (36). We have shown that increased expression of Cdh2 in the pituitary gland correlates with increased anxiety-related behavior in mice (14). This study implicates the CDH2 intron 14 as a region influencing risk for social phobia.
The DYNLL (dynein light chain LC8) proteins are components of the motor protein complexes of dynein and myosin Va (37,38). In neurons, the dynein complex is involved in axonal retrograde transport (39). The DYNLL2 binds multiple proteins in the brain, including synaptic scaffolding proteins and enzymes of glutamate metabolism (40). Interestingly, DISC1, a susceptibility factor for schizophrenia, schizoaffective disorder, bipolar disorder, depression, and autism spectrum disorders (41), is also part of the dynein complex (42). Our results suggest that haplotypes of DYNLL2, which is expressed at a higher level in the periaqueductal grey of anxious mice (14), modulate susceptibility to generalized anxiety disorder.
The functions of EPB41L4A are largely unknown, but it might regulate interactions between the plasma membrane and the cytoskeleton (43). The EPB41L4A is upregulated in vitro by activation of β-catenin (44). Beta-catenin and the Wnt signaling pathway have been implicated in the pathophysiology and treatment of mood disorders, and recent characterization of forebrain-specific conditional β-catenin knock-out mice revealed some evidence for a depression-like phenotype (45). The Epb41l4a is expressed at a higher level in the pituitary gland of anxious mice (14). In this study, we found that genetic variants within EPB41L4A might influence susceptibility to several anxiety disorders. Our most significant association was to a common His→Tyr polymorphism (rs7719346) in generalized anxiety disorder.
The PSAP gene encodes a glycoprotein precursor that is cleaved into four soluble saposins A–D, which stimulate the hydrolysis of sphingolipids (46). Unprocessed PSAP exists also as an integral surface membrane protein in neuronal cells (47). Prosaposin has been suggested to function as a neurotrophic factor (48,49) and to be a neural injury-repair protein (50). Consistently, Psap knockout mice show neurodegeneration (51). Suggestive evidence for association between PSAP and schizophrenia was recently reported (52). In this study, we found two SNPs upstream of PSAP associating with panic disorder, in a region containing putative regulatory elements (53). We previously observed increased expression of Psap in periaqueductal grey of anxious mice (14).
Prostaglandin D2 synthase is the enzyme responsible for the biosynthesis of brain prostaglandin D2, which functions as a neuromodulator and/or trophic factor (54) and participates in the regulation of sleep (55). Changes in cerebrospinal fluid PTGDS quantity have been observed in multiple sclerosis, schizophrenia, and Parkinson's disease (56). Anxious mice show decreased expression of Ptgds in the bed nucleus of the stria terminalis and periaqueductal grey (14). We found here association of rs4880179 with generalized anxiety disorder.
The results of this study should be interpreted in the context of certain limitations including: 1) limited diagnostic subgroup sample size, 2) problems related to multiple testing correction when a large number of SNPs have been investigated, and 3) differences between mouse and human studies. Each of these is briefly discussed in the following text.
There is a disadvantage in studying a complex phenotype with a limited sample size. Nevertheless, we feel that both combined and separate analyses of specific anxiety disorders are warranted, because these diseases seem to be influenced by both shared and disorder-specific genes (6). Our restricted sample size is exemplified by some fluctuation in p values in the analyses of core and extended diagnostic groups (e.g., rs7719346 and rs4880179). However, our cohort is comparatively well-characterized. It is derived from a population-based study and represents the general Finnish population, which has reduced genetic and environmental heterogeneity (17,57). Both cases and control subjects were interviewed, and because we carefully matched control subjects we do not consider population stratification to be a source of false positives.
Due to the large number of SNPs studied (n = 144) in several diagnostic subgroups (n = 7), we have carried out 1008 statistical tests. Therefore, none of our results would survive correction for multiple testing across all tests with a conventional Bonferroni-correction. For this reason, replication in other cohorts is needed to assess the significance of these genes to anxiety disorders in general. Nevertheless, a potential role for particularly EPB41L4A in susceptibility to anxiety disorders was suggested by an analysis of trends in pointwise association between genes.
Although mice can be used to model some aspects of human anxiety fairly well (e.g., 9), it is possible that cognitive differences account for discrepancies between our mouse and human study, besides the type of variation (gene expression vs. DNA polymorphisms) investigated. Although Glo1 and Gsr were functionally shown to regulate anxiety in mice (14), it seems unlikely on the basis of this study that common genetic variants within these genes play a major role in the etiology of human anxiety disorders. Others examining a functional Ala/Glu polymorphism (rs2736654) within GLO1 similarly failed to establish a convincing association with panic disorder (58). However, the genes for which no associations were discovered might nevertheless be important modulators of anxiety, because the role of genetic variation in their hypothetical trans-acting regulatory elements remains to be assessed. In fact, considering that the investigated genes were identified on the basis of differential expression, it is appealing that two of our strongest associations, in ALAD and PSAP, are within potential regulatory regions that might regulate gene expression. Because functional characterization of candidate genes in animal models is laborious and time-consuming, we have not shown causality for all genes studied in this work. However, we have now potentially identified the subset of our candidate genes that is most relevant for human anxiety disorders. This information will enable us to prioritize genes for future functional testing in mice.
In conclusion, we report here that six candidate genes first identified as being differentially expressed between anxious and non-anxious mouse strains are also implicated as possible susceptibility genes for human anxiety disorders. Future replication of our results in independent study samples as well as functional studies are needed to shed light on the role of these genes in the regulation of normal and pathological anxiety. Nevertheless, our results illustrate the potential utility of cross-species approaches in the identification of susceptibility genes for human psychiatric disorders.
Acknowledgments
This study was supported by the Center of Excellence of the Academy of Finland (LP), the Biocentrum Helsinki Foundation (IH and LP), the NEURO research program of the Academy of Finland (IH), Sigrid Jusélius Foundation (IH), Yrjö Jahnsson Foundation (IH), Finnish L'Oréal-UNESCO Women in Science fellowship (IH), Helsinki Biomedical Graduate School (JD), the Academy of Finland (JDT), and the US National Institute of Mental Health (JDT). We thank Minna Suvela, Siv Knaappila, and Olli Kiviruusu for their valuable assistance and all participants in the Health 2000 Study for their kind co-operation.
The authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online.
Supplementary data
References
- 1.American Psychiatric Association . 4th ed. American Psychiatric Association; Washington DC: 2000. Diagnostic and Statistical Manual of Mental Disorders: DSM-IV-TR. [Google Scholar]
- 2.World Health Organization . World Health Organization; Geneva: 1993. The ICD-10 Classification of Mental and Behavioral Disorders: Diagnostic Criteria for Research, 10th ed. [Google Scholar]
- 3.Somers J.M., Goldner E.M., Waraich P., Hsu L. Prevalence and incidence studies of anxiety disorders: A systematic review of the literature. Can J Psychiatry. 2006;51:100–113. doi: 10.1177/070674370605100206. [DOI] [PubMed] [Google Scholar]
- 4.Kessler R.C., Chiu W.T., Demler O., Walters E.E. Prevalence, severity and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey replication. Arch Gen Psychiatry. 2005;62:617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hettema J.M., Neale M.C., Kendler K.S. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 2001;158:1568–1578. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- 6.Hettema J.M., Prescott C.A., Myers J.M., Neale M.C., Kendler K.S. The structure of genetic and environmental risk factors for anxiety disorders in men and women. Arch Gen Psychiatry. 2005;62:182–189. doi: 10.1001/archpsyc.62.2.182. [DOI] [PubMed] [Google Scholar]
- 7.Hettema J.M., Neale M.C., Myers J.M., Prescott C.A., Kendler K.S. A population based twin study of the relationship between neuroticism and internalizing disorders. Am J Psychiatry. 2006;163:857–864. doi: 10.1176/ajp.2006.163.5.857. [DOI] [PubMed] [Google Scholar]
- 8.Bienvenu O.J., Hettema J.M., Neale M.C., Prescott C.A., Kendler K.S. Low extraversion and high neuroticism as indices of genetic and environmental risk for social phobia, agoraphobia, and animal phobia. Am J Psychiatry. 2007;164:1714–1721. doi: 10.1176/appi.ajp.2007.06101667. [DOI] [PubMed] [Google Scholar]
- 9.Hovatta I., Barlow C. Molecular genetics of anxiety in mice and men. Ann Med. 2008;40:92–109. doi: 10.1080/07853890701747096. [DOI] [PubMed] [Google Scholar]
- 10.Flint J., Valdar W., Shifman S., Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 11.Bourin M., Hascoet M. The mouse light/dark box test. Eur J Pharmacol. 2003;463:55–65. doi: 10.1016/s0014-2999(03)01274-3. [DOI] [PubMed] [Google Scholar]
- 12.Dulawa S.C., Hen R. Recent advances in animal models of chronic antidepressant effects: The novelty-induced hypophagia test. Neurosci Biobehav Rev. 2005;29:771–783. doi: 10.1016/j.neubiorev.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Kalueff A.V., Wheaton M., Murphy D.L. What's wrong with my mouse model?: Advances and strategies in animal modeling of anxiety and depression. Behav Brain Res. 2007;179:1–18. doi: 10.1016/j.bbr.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Hovatta I., Tennant R.S., Helton R., Marr R.A., Singer O., Redwine J.M. Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature. 2005;438:662–666. doi: 10.1038/nature04250. [DOI] [PubMed] [Google Scholar]
- 15.Pirkola S.P., Isometsä E., Suvisaari J., Aro H., Joukamaa M., Poikolainen K. DSM-IV mood-, anxiety- and alcohol use disorders and their comorbidity in the Finnish general population. Soc Psychiatry Psychiatr Epidemiol. 2005;40:1–10. doi: 10.1007/s00127-005-0848-7. [DOI] [PubMed] [Google Scholar]
- 16.Pirkola S., Isometsa E., Aro H., Kestila L., Hamalainen J., Veijola J. Childhood adversities as risk factors for adult mental disorders: Results from the Health 2000 study. Soc Psychiatry Psychiatr Epidemiol. 2005;40:769–777. doi: 10.1007/s00127-005-0950-x. [DOI] [PubMed] [Google Scholar]
- 17.Varilo T., Peltonen L. Isolates and their potential use in complex gene mapping efforts. Curr Opin Genet Dev. 2004;14:316–323. doi: 10.1016/j.gde.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Riva A., Kohane I.S. SNPper: Retrieval and analysis of human SNPs. Bioinformatics. 2002;18:1681–1685. doi: 10.1093/bioinformatics/18.12.1681. [DOI] [PubMed] [Google Scholar]
- 19.The International HapMap Consortium The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 20.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 21.Silander K., Komulainen K., Ellonen P., Jussila M., Alanne M., Levander M. Evaluating whole genome amplification via multiply-primed rolling circle amplification for SNP genotyping of samples with low DNA yield. Twin Res Hum Genet. 2005;8:368–375. doi: 10.1375/1832427054936664. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S., Cherny S.S., Sham P.C. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 23.Edwards A.W.F. Johns Hopkins University Press; Baltimore, Maryland: 1992. Likelihood: Expanded Edition. [Google Scholar]
- 24.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 25.Turri M.G., Datta S.R., DeFries J., Henderson N.D., Flint J. QTL analysis identifies multiple behavioral dimensions in ethological tests of anxiety in laboratory mice. Curr Biol. 2001;11:725–734. doi: 10.1016/s0960-9822(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 26.Henderson N.D., Turri M.G., DeFries J.C., Flint J. QTL analysis of multiple behavioral measures of anxiety in mice. Behav Genet. 2004;34:267–293. doi: 10.1023/B:BEGE.0000017872.25069.44. [DOI] [PubMed] [Google Scholar]
- 27.Smoller J.W., Acierno J.S., Rosenbaum J.F., Biederman J., Pollack M.H., Meminger S. Targeted genome screen of panic disorder and anxiety disorder proneness using homology to murine QTL regions. Am J Med Genet. 2001;105:195–206. doi: 10.1002/ajmg.1209. [DOI] [PubMed] [Google Scholar]
- 28.Smoller J.W., Rosenbaum J.F., Biederman J., Susswein L.S., Kennedy J., Kagan J. Genetic association analysis of behavioral inhibition using candidate loci from mouse models. Am J Med Genet. 2001;105:226–235. doi: 10.1002/ajmg.1328. [DOI] [PubMed] [Google Scholar]
- 29.Wetmur J.G., Bishop D.F., Cantelmo C., Desnick R.J. Human delta-aminolevulinate dehydratase: Nucleotide sequence of a full-length cDNA clone. Proc Natl Acad Sci U S A. 1986;83:7703–7707. doi: 10.1073/pnas.83.20.7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haeger-Aronsen B., Abdulla M., Fristedt B.I. Effect of lead on -aminolevulinic acid dehydrase activity in red blood cells. Arch Environ Health. 1971;23:440–445. doi: 10.1080/00039896.1971.10666033. [DOI] [PubMed] [Google Scholar]
- 31.Rajan P., Kelsey K.T., Schwartz J.D., Bellinger D.C., Weuve J., Sparrow D. Lead burden and psychiatric symptoms and the modifying influence of the delta-aminolevulinic acid dehydratase (ALAD) polymorphism: The VA Normative Aging Study. Am J Epidemiol. 2007;166:1400–1408. doi: 10.1093/aje/kwm220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeichi M. The cadherins: Cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Castro M.I., Vielmetter E., Bronner-Fraser M. N-cadherin, a cell adhesion molecule involved in establishment of embryonic left-right asymmetry. Science. 2000;288:1047–1051. doi: 10.1126/science.288.5468.1047. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka H., Shan W., Phillips G.R., Arndt K., Bozdagi O., Shapiro L. Molecular modification of N-cadherin in response to synaptic activity. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 35.Togashi H., Abe K., Mizoguchi A., Takaoka K., Chisaka O., Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 36.Kadowaki M., Nakamura S., Machon O., Krauss S., Radice G.L., Takeichi M. N-cadherin mediates cortical organization in the mouse brain. Dev Biol. 2007;304:22–33. doi: 10.1016/j.ydbio.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 37.King S.M., Barbarese E., Dillman J.F., III, Patel-King R.S., Carson J.H., Pfister K.K. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem. 1996;271:19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- 38.Espindola F.S., Suter D.M., Partata L.B., Cao T., Wolenski J.S., Cheney R.E. The light chain composition of chicken brain myosin-Va: Calmodulin, myosin-II essential light chains, and 8-kDa dynein light chain/PIN. Cell Motil Cytoskeleton. 2000;47:269–281. doi: 10.1002/1097-0169(200012)47:4<269::AID-CM2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 39.Goldstein L.S., Yang Z. Microtubule-based transport systems in neurons: The roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- 40.Navarro-Lerida I., Martinez Moreno M., Roncal F., Gavilanes F., Albar J.P., Rodriguez-Crespo I. Proteomic identification of brain proteins that interact with dynein light chain LC8. Proteomics. 2004;4:339–346. doi: 10.1002/pmic.200300528. [DOI] [PubMed] [Google Scholar]
- 41.Chubb J.E., Bradshaw N.J., Soares D.C., Porteous D.J., Millar J.K. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 42.Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi K., Kawashima A., Nagafuchi A., Tsukita S. Structural diversity of band 4.1 superfamily members. J Cell Sci. 1994;107:1921–1928. doi: 10.1242/jcs.107.7.1921. [DOI] [PubMed] [Google Scholar]
- 44.Ishiguro H., Furukawa Y., Daigo Y., Miyoshi Y., Nagasawa Y., Nishiwaki T. Isolation and characterization of human NBL4, a gene involved in the beta-catenin/tcf signaling pathway. Jpn J Cancer Res. 2000;91:597–603. doi: 10.1111/j.1349-7006.2000.tb00987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gould T.D., O'Donnell K.C., Picchini A.M., Dow E.R., Chen G., Manji H.K. Generation and behavioral characterization of beta-catenin forebrain-specific conditional knock-out mice. Behav Brain Res. 2008;189:117–125. doi: 10.1016/j.bbr.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morimoto S., Martin B.M., Yamamoto Y., Kretz K.A., O'Brien J.S., Kishimoto Y. Saposin A: Second cerebrosidase activator protein. Proc Natl Acad Sci U S A. 1989;86:3389–3393. doi: 10.1073/pnas.86.9.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Q., Carson G.S., Hiraiwa M., Grafe M., Kishimoto Y., O'Brien J.S. Occurrence of prosaposin as a neuronal surface membrane component. J Mol Neurosci. 1994;5:59–67. doi: 10.1007/BF02736694. [DOI] [PubMed] [Google Scholar]
- 48.O'Brien J.S., Carson G.S., Seo H.C., Hiraiwa M., Kishimoto Y. Identification of prosaposin as a neurotrophic factor. Proc Natl Acad Sci U S A. 1994;91:9593–9596. doi: 10.1073/pnas.91.20.9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sikora J., Harzer K., Elleder M. Neurolysosomal pathology in human prosaposin deficiency suggests essential neurotrophic function of prosaposin. Acta Neuropathol. 2007;113:163–175. doi: 10.1007/s00401-006-0148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiraiwa M., Liu J., Lu A.G., Wang C.Y., Misasi R., Yamauchi T. Regulation of gene expression in response to brain injury: Enhanced expression and alternative splicing of rat prosaposin (SGP-1) mRNA in injured brain. J Neurotrauma. 2003;20:755–765. doi: 10.1089/089771503767869980. [DOI] [PubMed] [Google Scholar]
- 51.Fujita N., Suzuki K., Vanier M.T., Popko B., Maeda N., Klein A. Targeted disruption of the mouse sphingolipid activator protein gene: A complex phenotype, including severe leukodystrophy and wide-spread storage of multiple sphingolipids. Hum Mol Genet. 1996;5:711–725. doi: 10.1093/hmg/5.6.711. [DOI] [PubMed] [Google Scholar]
- 52.Jungerius B.J., Hoogendoorn M.L., Bakker S.C., Van't Slot R., Bardoel A.F., Ophoff R.A. An association screen of myelin-related genes implicates the chromosome 22q11 PIK4CA gene in schizophrenia [published online ahead of print September 25] Mol Psychiatry. 2007 doi: 10.1038/sj.mp.4002080. [DOI] [PubMed] [Google Scholar]
- 53.Sun Y., Jin P., Witte D.P., Grabowski G.A. Isolation and characterization of the human prosaposin promoter. Gene. 1998;218:37–47. doi: 10.1016/s0378-1119(98)00391-6. [DOI] [PubMed] [Google Scholar]
- 54.Nagata A., Suzuki Y., Igarashi M., Eguchi N., Toh H., Urade Y. Human brain prostaglandin D synthase has been evolutionarily differentiated from lipophilic-ligand carrier proteins. Proc Natl Acad Sci U S A. 1991;88:4020–4024. doi: 10.1073/pnas.88.9.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qu W.M., Huang Z.L., Xu X.H., Aritake K., Eguchi N., Nambu F. Lipocalin-type prostaglandin D synthase produces prostaglandin D2 involved in regulation of physiological sleep. Proc Natl Acad Sci U S A. 2006;103:17949–17954. doi: 10.1073/pnas.0608581103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrington M.G., Fonteh A.N., Biringer R.G., R, Huhmer A.F., Cowan R.P. Prostaglandin D synthase isoforms from cerebrospinal fluid vary with brain pathology. Dis Markers. 2006;22:73–81. doi: 10.1155/2006/241817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Service S., DeYoung J., Karayiorgou M., Roos J.L., Pretorious H., Bedoya G. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- 58.Politi P., Minoretti P., Falcone C., Martinelli V., Emanuele E. Association analysis of the functional Ala111Glu polymorphism of the glyoxalase I gene in panic disorder. Neurosci Lett. 2006;396:163–166. doi: 10.1016/j.neulet.2005.11.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.