Summary
Adherens juctions and Tight junctions comprise two modes of cell-cell adhesion that provide different functions. Both junctional complexes are proposed to associate with the actin cytoskeleton, and formation and maturation of cell-cell contacts involves reorganization of the actin cytoskeleton. Adherens junctions initiate cell-cell contacts, and mediate the maturation and maintenance of the contact. Adherens junctions consist of the transmembrane protein E-cadherin, and intracellular components, p120-catenin, β-catenin and α-catenin. Tight junctions regulate the paracellular pathway for the movement of ions and solutes in-between cells. Tight junctions consist of the transmembrane proteins occludin and claudin, and the cytoplasmic scaffolding proteins ZO-1,-2, and -3. This review discusses the binding interactions of the most studied proteins that occur within each of these two junctional complexes and possible modes of regulation of these interactions, and the different mechanisms that connect and regulate interactions with the actin cytoskeleton.
Introduction
The Adherens junction (AJ) and Tight junction (TJ) provide important adhesive contacts between neighboring epithelial cells. Although these junctions comprise different proteins, there are similarities in the roles of specialized transmembrane proteins in forming extracellular adhesive contacts between cells, and intracellular links to the actin cytoskeleton and signaling pathways including the regulation of gene transcription.
Classical cadherins, such as E-cadherin, are the major transmembrane protein of the Adherens junction and initiate intercellular contacts through trans-pairing between cadherins on opposing cells [1]. Classical cadherins also bind directly and indirectly to many cytoplasmic proteins, particularly members of the catenin family, which locally regulate the organization of the actin cytoskeleton, cadherin stability and intracellular signaling pathways that control gene transcription [2]. Formation of the Adherens junction leads to assembly of the Tight junction, but E-cadherin is not required to maintain Tight junction organization [3]. Surprisingly, HepG2-AJ- cells unable to form Adherens junctions slowly form functional Tight junctions [4]. The occludin and claudin family of transmembrane proteins form the core of the Tight junction and control ion selectivity and permeability of the paracellular pathway between adhering cells. Occludin and claudins bind to members of the MAGUK family of cytoplasmic proteins that interact with the actin cytoskeleton and other signaling proteins that also localize to the nucleus [5].
Recent studies of these adhesion complexes have provided new insights into molecular mechanisms involved in the formation, maintenance and function of protein components. This review will focus on the formation and interactions of these two junctional complexes, mechanisms of regulation and dynamics of protein interactions, and how they interact with and regulate the actin cytoskeleton.
1. Adherens Junction
The Adherens junction performs multiple functions including initiation and stabilization of cell-cell adhesion, regulation of the actin cytoskeleton, intracellular signaling and transcriptional regulation. The core of the Adherens junction includes interactions among transmembrane glycoproteins of the classical cadherin superfamily, such as E-cadherin, and the catenin family members including p120-catenin, β-catenin, and α-catenin. Together, these proteins control the formation, maintenance and function of adherens junctions.
1.1 E-cadherin
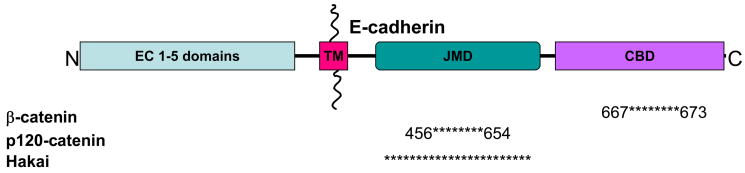
E-cadherin is a single-pass, transmembrane glycoprotein (Fig. 1) that belongs to the classical cadherin family of Ca2+-dependent adhesion proteins; other members of this family include N-, P, and R-cadherin [6]. Classical cadherins have five characteristic extracellular cadherin (EC) repeat domains. These domains form trans-cadherin interactions between neighboring cells and initiate weak cell-cell adhesion and formation of the Adherens junction [7]. Binding of Ca2+ to each EC domain is required for the correct conformational organization of the cadherin extracellular domain [8]. Recent structural studies indicate that trans-pairing of EC1 domains is critical for cadherin adhesion [9]. Indeed, expression of two cadherin mutant proteins in which the EC1 domains were swapped revealed that the correct EC1 domain determined aggregation and sorting of specific motor neuron pools in the spinal cord [9, 10]. It remains possible, however, that other EC domains are important since a monoclonal antibody DECMA-1, against an epitope mapped to the EC4/EC5 region [11], blocks cell-cell adhesion [12].
Figure 1.
Adherens junctions are comprised of the single pass transmembrane protein, E-cadherin. The extracellular domain is proposed to form trans-interactions with E-cadherin on neighboring cells. The intracellular domain has two binding regions; juxtamembrane domain (JMD) and catenin binding domain (CBD). (TM; Transmembrane) The protein-protein interactions presented are limited to those involved in connections with the actin cytoskeleton. The asterisks represent the region proteins have been shown to bind. Not drawn to scale. Information was gathered based on mutational analysis or co-immunoprecipitation studies; all references are cited within the text.
The cytoplasmic domain of E-cadherin binds proteins that regulate E-cadherin endocytosis, recycling and degradation, intracellular signaling and gene transcription, and local control of the actin cytoskeleton [2, 7]. Upon formation of intercellular contacts, cadherins cluster and spread laterally thereby strengthening the contact [13–15]. Clustering of cadherins in a maturing contact requires the cytoplasmic juxtamembrane domain [16]. Within the cytoplasmic domain there are two relatively well-defined catenin binding domains (CBD) encompassing a 94 amino acid juxtamembrane domain (JMD) that binds p120-catenin [16], and an extended region to the C-terminal that binds β-catenin [17] (Fig. 1).
Cadherin-mediated cell-cell adhesion is highly dynamic enabling the reorganization and dispersal of cells, for example, during epithelial-to-mesenchymal transition in normal development and carcinogenesis [18]. In epithelial derived tumors, loss of cell-cell adhesion is correlated with down-regulation of E-cadherin as well as increased proliferation and tumor invasiveness [19–23]. For example, E-cadherin regulates normal cell-cell adhesion in the mammary gland and expression of E-cadherin in breast cancer has been studied in relation to prognosis, diagnosis, and potential therapy [21]. Lobular carcinoma, accounting for ~15% of breast cancer, is characterized by a reduction or elimination of E-cadherin expression [24–26]. Approximately 85% of cases are associated with a loss of heterozygocity (LOH) of the E-cadherin gene on chromosome 16q [27, 28]. Ductal carcinoma, accounting for ~80% of breast cancer, is associated with a reduction in both E-cadherin and α-catenin [29], moreover loss of α-catenin was associated with advanced stages and poor patient survival [30]. Together these observations provide strong evidence that regulation of E-cadherin and associated protein expression and localization are factors involved in carcinogenesis.
1.2 Catenins
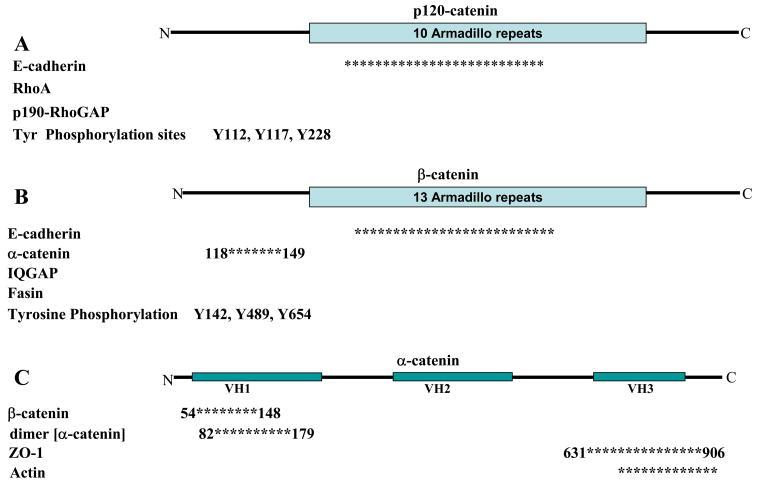
E-cadherin is the core transmembrane protein of the Adherens junction and is required for binding and localization of a number of important cytoplasmic proteins termed catenins that connect the cadherin complex to the actin cytoskeleton and several signaling pathways (Fig. 2). The catenin family comprises p120-catenin, β-catenin and α-catenin. An experimentally induced decrease in E-cadherin expression by siRNA [3] causes a delay in the correct localization of adherens junction core proteins, α- and β-catenin, and the tight junction-associated protein ZO-1.
Figure 2.
Adherens junctions are comprised of A) p120-catenin, B) β-catenin, and C) α-catenin. All three proteins interact with additional proteins known to regulate actin cytoskeleton. Asterisks represent regions known to bind the respective protein. Numbers associated with the asterisks correspond to amino acids flanking the binding site. Not drawn to scale. Information was collected either through mutational analysis or co-immunoprecipitation. Proteins without asterisks represent interactions demonstrated by co-immunoprecipitation. References are cited within the text.
1.2.1 p120-catenin
p120-catenin was first identified as a substrate for Src- tyrosine receptor kinase [31], and later defined as a member of the catenin family based on sequence homology to an armadillo domain of β-catenin [32] (Fig. 2A). There are four isoforms of p120 resulting from either post-translational modifications or different internal translation start sites [33]. p120-catenin binds E-cadherin [34] at a highly conserved octapeptide sequence (YDEEGGGE) [35] within the juxtamembrane domain [16, 36, 37]. Mutations of the E-cadherin JMD have shown that this domain is both necessary and sufficient for recruitment of p120-catenin to Adherens junctions [38] (Fig. 1).
Association of p120-catenin with the JMD of E-cadherin has been proposed to stabilize E-cadherin at the plasma membrane during the formation of cell-cell contacts. Expression of different cadherin cytoplasmic domains [16] and mutation analysis of the JMD [35, 38] demonstrate the p120-catenin-E-cadherin interaction is required for increased adhesiveness of cells. Furthermore, siRNA-mediated knock-down [39] and competitive expression of other cadherins [40–42] suggest that p120-catenin increases the retention of the cadherin complex at the plasma membrane. Loss of p120-catenin-induced stabilization of E-cadherin is linked to tumor progression and invasion [19, 20]. Phosphorylation of p120-catenin increases binding affinity to E-cadherin [43]. Binding of p120-catenin to the JMD may prevent cadherins from being internalized and degraded [39, 41, 42] or lead to the recycling of internalized cadherin back to the plasma membrane [44]. One possible mechanism of targeting cadherin for degradation involves Hakai, an E3-ubiquitin ligase, which binds E-cadherin in a Src-dependent manner [45] (Fig. 1). Expression of Hakai increased both the ubiquitination and rate of E-cadherin endocytosis [45], but it is not known if p120-catenin binding is involved in this degradation pathway. Note, however, that loss of p120-catenin in an E-cadherin null background has also been shown to increase cell-cell adhesion, raising the possibility that p120-catenin plays additional roles in modulating cell-cell adhesion [46].
p120-catenin also functions as a regulator of cell motility through the actin cytoskeleton by interacting with Rho family GTPases [47] (Fig. 2A). Over-expression of p120-catenin in fibroblasts [37] or MDCK cells [47] increased membrane extensions and cell migration. These effects correlated with an increase in activated Rac and Cdc42 [47]. In p120-catenin knock-down experiments, invasiveness of A431 cells in a three-dimensional matrix was reduced, while re-expression of p120-catenin that could not be phosphorylated restored cell motility. While migration of cells was restored, an increase in Rac activity was not observed even though Rac activation is generally associated with increased cell migration [48]. Differences in p120-catenin effects on cell motility could be due to differences in cell lines, and further studies are needed to characterize the downstream effects of p120-catenin on Rac activity. Moreover, p120-catenin may also regulate cell motility and invasiveness by inhibiting RhoA activity independent of p120-catenin-E-cadherin binding [46, 47]. In vitro, p120-catenin binds RhoA-GDP which would have the effect of sequestering RhoA from activation by a guanine exchange factor (GEF) [49]. The affinity of the p120-catenin-RhoA interaction is reduced by Fyn-mediated phosphorylation of p120-catenin (Y112), and increased by Src-dependent phosphorylation of p120-catenin (Y117, Y228) [50]. In addition, p120-catenin interacts with p190RhoGAP, as a downstream effect of Rac activation, and may recruit p190RhoGAP to the plasma membrane resulting in local inhibition of Rho mediated contractility and antagonizing Rac and Rho signaling [51].
1.2.2 β-catenin
Beta-catenin, which was originally identified in Drosophila as the segment polarity protein armadillo [52, 53], contains 13 repeats of a characteristic armadillo domain of ~42 amino acids that form triple α-helix [54] (Fig. 2B). Beta-catenin binds the C-terminal cytoplasmic domain of E-cadherin (Fig. 1) in a phospho-regulated manner [2]. Three serine residues in the cadherin cytoplasmic domain (S684, S686, S692) are phosphorylated by CKII and GSK-3β kinases which create additional interactions between β-catenin and E-cadherin resulting in a large increase in the affinity of the interaction (~9pM affinity; [55, 56]). In contrast, tyrosine phosphorylation of β-catenin at Y489 or Y654 disrupts binding to cadherin, and at Y142 binding to α-catenin is weakened [57]. The structural basis for these effects is due to β-catenin Y654 forming a hydrogen bond with E-cadherin Asp665, which stabilizes the interaction of the cadherin region 2 helix with the last two armadillo repeats of β-catenin [55]; phosphorylation of Y654 would prevent this interaction thereby eliminating binding of this region of cadherin and sharply reducing the cadherin/β-catenin affinity. The kinases involved in β-catenin phosphorylation have been identified and include: Src phosphorylation at Y654 [58, 59], Abl kinase phosphorylation at Y489 [60], EGF receptor phosphoylation of Y654 [61], and Fer kinase phosphorylation at Y142 [43]. Recently, a detailed analysis of the thermodynamics of the β-catenin/E-cadherin interaction proposed that the C-terminal tail of β-catenin (post-armadillo domain) regulates the binding affinity for β-catenin and its ligands [62].
While there is a great deal of information on the interaction of β-catenin and E-cadherin, there is little demonstrating whether β-catenin dissociates from E-cadherin (for example during E-cadherin internalization), in part because the affinity of this interaction is very high [55]. The regulation of cytosolic β-catenin is critical as β-catenin can bind to the transcription factor Tcf/Lef and mediate the transcription of a genes involved in cell proliferation, a signaling pathway activated by Wnt [63]. It is proposed that the E-cadherin/β-catenin interaction occurs in the endoplasmic reticulum (ER) and is required for cadherin exit from the ER [64]. Normally cytosolic levels of β-catenin are low due to rapid targeting of excess β-catenin to the proteosome [65, 66]. However, recent studies have identified a role for BCL9-2, a transcription factor involved in epithelial-mesenchymal transition, in mediating a switch between the adhesive and transcriptional functions of β-catenin. This switch is caused by phosphorylation of Y142 on β-catenin, which favors BCL9-2 binding and precludes other protein-protein interactions, and results in translocation of β-catenin to the nucleus and induction of specific gene transcription [67]. Significantly, BCL9-2 RNAi induces an epithelial phenotype in the colon cancer cell line SW480 and causes β-catenin to translocate from the nucleus to the plasma membrane [67].
β-catenin binds IQGAP, fascin, and α-catenin (see also α-catenin sub-section; Fig. 2B). The α-/β-catenin interaction dissociates upon binding to IQGAP, an actin binding protein activated by the small GTPases Rac1 and Cdc42 [68]. Activation of IQGAP by Rac1 or Cdc42 disrupts IQGAP binding to β-catenin resulting in rebinding of α-catenin to β-catenin and, hence, functional assembly of the cadherin core complex and initiation of cell-cell adhesion. β-catenin has also been identified by yeast 2-hybrid as a directly interaction partner of the actin bundling protein fascin, whose binding site within β-catenin overlaps that of E-cadherin, and therefore competes with E-cadherin for binding β-catenin [69].
1.2.3 α-catenin
The textbook model of the adherens junction states that α-catenin is the link between the cadherin/beta-catenin complex and the actin cytoskeleton. Indeed, α-catenin binds and bundles actin filaments in vitro [29] and binds to β-catenin [17] (Fig. 2C), but a ternary complex of E-cadherin/β-catenin/α-catenin and actin had not been tested [70]. However, recent evidence demonstrated that this simultaneous interaction could not be reconstituted in vitro [70].
Alpha-catenin exists in either a monomeric or homo-dimeric state. The β-catenin/α-catenin binding domain and the α-catenin homodimerization domains overlap within amino acids 57–143 on α-catenin [71], but the actin and β-catenin binding domains on α-catenin do not (Fig. 2C). In vitro binding assays demonstrated that monomeric α-catenin binds β-catenin, but not actin. Conversely, homo-dimeric α-catenin binds actin filaments but not β-catenin [70]; a-catenin homo-dimer binding to actin also appears to compete binding of the Arp2/3 complex to actin filaments thereby suppresses actin polymerization [72]. This allosteric switch between monomeric and dimeric states appears to be the molecular explanation for the lack of simultaneous binding of α-catenin to both β-catenin and actin filaments. In vitro studies showed that dimerization of α-catenin occurs at a 10-fold higher concentration than that of the monomeric pool of α-catenin in the cytoplasm of epithelial cells, indicating that α-catenin must be locally concentrated prior to dimerization perhaps by clustering the cadherin-catenin complex during cell-cell adhesion. A dynamic crosstalk between the α-catenin plasma membrane pool (monomeric, β-catenin bound) the cytoplasmic pool (monomeric) and cytoskeleton pool (dimeric, actin-bound) has been proposed where α-catenin switches between the adherens complex and binding the actin cytoskeleton [72].
A new model for Adherens junction connection to the cytoskeleton has been proposed [70, 72]. It is suggested that the increase in local concentration of α-catenin at the membrane during clustering of the cadherin-catenin complex at cell-cell contacts provides a local increase in α-catenin concentration sufficient to drive α-catenin dimerization in the cytoplasm. α-catenin dimers would locally inhibit Arp2/3 and thereby the formation of branching networks of actin filaments characteristic of lamellipodia of migrating cells. At the same time, α-catenin dimers bind to and bundle existing actin filaments, resulting in actin reorganization from branched to bundled arrays. This model predicts that the interaction of α-catenin with the cadherin/β-catenin complex is labile such that α-catenin can dissociate from the cadherin complex and dimerize in the cytoplasm. Indeed, the interaction between α- and β-catenin may be concentration-dependent (see above) or phospho-regulated. Two large-scale proteomic analyses identified S641 and S652/S655 as phosphorylation sites on α-catenin [73, 74]. Phosphorylation of tyrosine 148 on α-catenin has been shown to increase binding to β-catenin [75]. Further investigation into the dynamics of different α-catenin pools is needed to verify the physiological relevance of these different α-catenin binding states. In addition, further studies are required to test whether an increased local concentration of α-catenin is sufficient to drive the switch from branched to bundled actin cables, and whether additional factors such as kinases and phosphatases are necessary for this switch to occur.
2. Tight Junctions
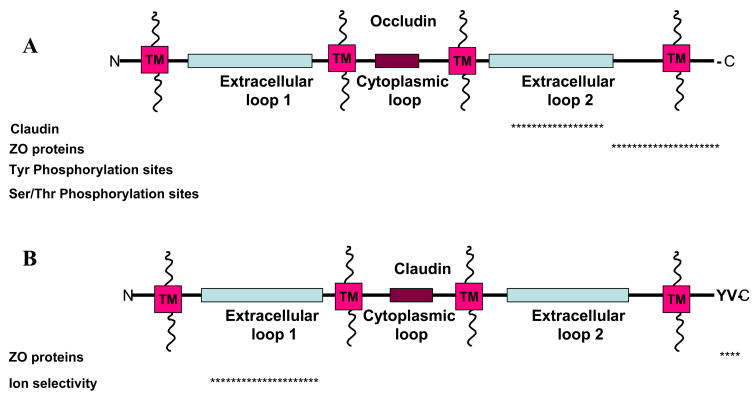
Tight junctions have been proposed to have two mutually exclusive functions: a fence function which prevents the mixing of membrane proteins between the apical and basaolateral membranes; and a gate function which controls the paracellular passage of ions and solutes in-between cells. Tight junctions contain two types of transmembrane proteins, occludins and claudins, which confer these functions (Fig. 3), and associated cytoplasmic proteins (Fig. 4) that may link tight junctions to the actin-cytoskeleton and the adherens junction.
Figure 3.
Tight Junction transmembrane proteins A) occludin and B) claudin, and proposed binding partners with corresponding binding regions. The asterisks represent the region in which the proteins have been shown to bind with the occludin and claudin. Not drawn to scale. Information was gathered based on mutational analysis or co-immunoprecipitation. All references are cited within the text.
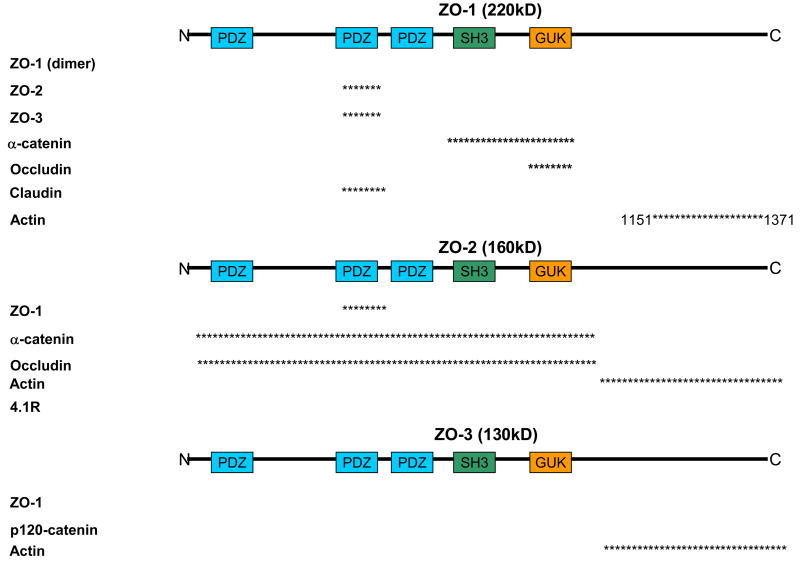
Figure 4.
ZO proteins are proposed to be a scaffolding protein that could link Tight junctions to the actin cytoskeleton through a direct interaction with actin or through additional protein interactions. In addition, ZO proteins may link Adherens junctions to Tight junctions through protein linkages. Proteins represented are proposed for these functions. Asterisks represent the region in which the proteins have been shown to bind with ZO protein. Numbers correspond with the amino acids on the parent ZO protein; not drawn to scale. Information was gathered based on mutational analysis or co-immunoprecipitation. Proteins without asterisks represent interactions demonstrated by co-immunoprecipitation but regions were not determined. All references are cited within the text.
2.1 Transmembrane components of tight junctions
2.1.1 Occludin
Occludin is a ~65 kDa tetraspan protein with two extracellular loops [5, 76] (Fig. 3A). There are two isoforms of occludin that result from alternative mRNA splicing, but have similar tissue distributions [77]. Localization of occludin to tight junctions is regulated by phosphorylation in both epithelial and endothelial cells [78]. Western blot analysis of occludin reveals a range of molecular weights (62–82 kDa) that are sensitive to alkaline phosphatase treatment. Multiple phosphorylation sites have been identified on tyrosine [79], serine, and threonine residues [78]. Non-phosphorylated occludin is localized to both the basolateral membrane and in cytoplasmic vesicles, whereas phosphorylated occludin is localized to tight junctions [78]. Multiple kinase and phosphatases are proposed to regulate occludin phosphorylation states and its localization and function within the tight junction. The non-receptor tyrosine kinase c-Yes co-localizes and co-immunoprecipitates with occludin in Ras-transformed MDCK cells [80]. In addition, when c-Yes is inhibited by CGP77675 occludin phosphorylation and tight junction localization are decreased, and trans-epithelial resistance (TER), a measure of paracellular permeability, is increased [80]. Protein Kinase C (PKC) stimulated by phorbol 12-myristate 13-acetate and 1,2 dioctanoylglycerol in low calcium increases occludin phosphorylation and localization to tight junctions [81]. Incubation of C-terminal occludin with purified PKC identified Ser 338 of occludin as a phosphorylation site [81]. However, stimulation of PKC with 12-0-tertradecanoylphorbol-13-acetate (TPA, another phorbol ester) led to a decrease in threonine phosphorylation which correlated with an increase in TER although the subcellular localization of occludin was not affected [82]. TPA has further been shown to increase gene transcription of occludin [83]. The difference in stimulation of PKC by phorbol esters may depend on additional proteins that are being stimulated including chimaerins [84], protein kinase D, and/or diaclglycerol kinases [85]. The catalytic subunit of protein phosphatase 2A (PP2A), a serine/threonine phosphatase, decreases occludin phosphorylation and increases TER while okadaic acid, a PP2A inhibitor, increases occludin phosphorylation and decreases TER [86].. Thus, multiple regulation pathways may provide redundancy to ensure that occludin localizes correctly to the Tight junction.
The extracellular domains of occludin are shown to function in localization of occludin to tight junctions and in regulating the paracellular permeability barrier between cells (Fig. 3). Synthetic peptides corresponding to a 20 amino acid sequence within the second extracellular loop of occludin, but not other adherens or tight junction proteins, increased TER and decreased occludin levels, as a result of increased protein turnover, and localization to the tight junction [87]. Photoactivatable crosslinking indicates that second loop extracellular loop of occludin interacts with claudin and junction adhesion molecule (JAM), and disruption of these interactions inhibited reformation of tight junctions after calcium repletion [88]. It should be noted however that the occludin null mouse does not exhibit deficiencies in barrier function, but does have an abnormal gastric morphology [89]. Tricellulin a tight junction protein localized at tricellular junctions, may provide functional redundancy that allows for intact barrier function in the occludin null mouse [90].
2.1.2 Claudin
The claudin family consists of at least 24 members ranging from 20–27 kDa [5, 91] (Fig. 3B). Although claudins do not share sequence similarity with the occludin family, they also comprise a tetra-span transmembrane protein [92] with two extracellular loops [5, 91]. Claudins recruit occludin to tight junctions [92]. With the exception of claudin 12, the intracellular C-terminal of all other claudin family members ends in the dipeptide sequence YV [93]. This sequence binds in the groove of PDZ domain proteins. Inhibition of this domain of does not affect localization of claudin to tight junctions but inhibits the association of ZO-1,-2, and -3 proteins [94]; this interaction will be discussed in the next sub-section.
Claudins form the protein strands of the tight junction that were observed previously by freeze-fracture electron microscopy [95]. When claudins were overexpressed in L fibroblasts lacking endogenous claudin they formed “paired” strands within areas of over-lapping cell-cell interactions [92]. These ‘paired strands’ were dynamic, breaking and re-annealling, and forming end-to-end and side-to side interactions. It is thought that these strands provide a physical barrier between apical and basolateral membrane; the fence function.
The gate function of the tight junction controls the paracellular pathway for ion movement in-between cells in an epithelial layer. Claudins directly regulate the gate function as paracellular tight junction channels (PTJC) that have biophysical properties similar to those of traditional ion channel including ion charge selectivity, permeability dependence on ion concentration, and competition for movement of permeative molecules [96]. While the majority of channels established by claudin interactions allow the passage of cations, the passage of anions has also been observed [5]. Remarkably, changes in the type of claudin expressed, or single amino acid substitutions in claudins effects claudin ion selectivity. For example, expression of claudin 8 in MDCK II cells, which lack endogenous claudin 8, reduced the paracellular movement of mono- and divalent cations while not affecting the movement of anions or uncharged solutes [97]. A single substitution of amino acid 65 from a negative to positive charge within the first extracellular loop of claudin 15 caused an increase in Na2+ permeability. Mutating three positive charges to negative within the same region switched the ion selectivity of the claudin channel from Na2+ to Cl− [98]. Swapping the extracellular loops of claudin 4 and claudin 2 revealed that the first extracellular loop is sufficient to determine charge selectivity of the ion channel [99]. Claudin-16, which is expressed solely in the kidney [100], forms a non-selective cation channel, and mutant claudin-16 results in renal wasting of magnesium and calcium [101].
2.2 Cytoskelatal connectors
2.2.1 ZO proteins
ZO-1, ZO-2, and ZO-3 are members of the MAGUK (membrane-associated guanylate kinase homologs) family with binding domains to Adherens and Tight junction proteins in addition to the actin cytoskeleton (Fig. 4). The MAGUK family is characterized by their PDZ domain, SH3 domain and guanylate kinase homologous domain [102]. ZO-1 [103–105] and ZO-2 [106] have both been shown to bind α-catenin, while the C-terminus of ZO-3 is sufficient to bind to p120-catenin in vitro [107]. In addition, in vitro binding assays demonstrated ZO-1 [108, 109] and ZO-2 [106] bind directly to occludin. ZO-1 has been isolated as a homodimer and is proposed to dimerize through the second PDZ domain [110]. Upon tyrosine phosphorylation of occludin, interactions between all ZO proteins and occludin are reduced [111]. The proline rich C-terminal of ZO-1 [108] and ZO-3 [112] bind F-actin in co-sedimentation assays, while ZO-2 does not bind actin [108, 113] (Fig. 4). ZO-2 does, however, bind the actin-associated protein 4.1R [114]. ZO-1 can bind ZO-2 or ZO-3 independently, but ZO-2 and ZO-3 cannot form a binary complex [113]. In addition, in a ZO-1 knock-out/ZO-2 knock-down cell line, exogenous expression of either ZO-1 or ZO-2 alone could restore claudin localization to tight junctions as seen by immunofluorescence [115]. The multiple interactions between the ZO proteins may provide different scaffolds and/or connections to the actin-cytoskeleton (Fig. 4).
ZO-1 has been proposed to be a scaffolding protein between transmembrane and cytoplasmic proteins, and possibly form a link between the Adherens and Tight juctions. While ZO-1 can bind α-catenin, evidence is lacking that ZO-1 can bind actin and α-catenin simultaneously. In addition, homo-dimerization of ZO-1 has been proposed to provide a link between the ZO-1/ZO-2 and ZO-1/ZO-3 complexes, which would link occludin (TJ) to p120-catenin (AJ) although the interaction between these two ZO-1 complexes has yet to be investigated [110] (Fig. 4). It should be noted, however, the C-terminal of ZO-3, which binds to p120-catenin, overlaps with the binding region to N-terminal ZO-3 (Fig. 4). It has been proposed that this switch in binding could sequester p120-catenin from regulation of RhoA activity thus affecting actin polymerization [107].
Formation of the Adherens junction through E-cadherin is associated with the formation and localization of the Tight junction proteins, particularly ZO-1 [105, 116]. Conversely, expression of mutated ZO-1 in a ZO null cell line significantly delayed the maturation of the Adherens junction from “fibroblastic” AJs to “polarized epithelial” AJs [117]. The region necessary for proper localization of tight junctions has been mapped to the SH3-U5-GUK-U6 on ZO-1 [118]. Exogenous expression of the N-terminal half of ZO-3 can delay the localization of E-cadherin, β-catenin, and ZO-1 [112]. Further investigation is necessary to elucidate the connections amongst ZO proteins during contact formation and maturation.
3. Conclusions
The structure of Adherens and Tight junctions is well established. While a number of proteins that comprise these junctions have been presented in this review, additional proteins are present within these junctions. For example formin, an actin nucleator binds α-catenin [119] and cortactin, an actin assembly regulator, binds p120-catenin [120]. In addition, Ankrin-G binds to the juxtamembrane domain of E-cadherin and recruits beta-2 spectrin to E-cadherin/β-catenin complexes providing another potential link to the actin cytoskeleton [121]. While this review focused on the role of conformational states of α-catenin in regulating actin cytoskeleton reorganization, both formin [119] and cortactin [122] are necessary for actin reorganization in mature cell-cell contacts. In addition, Ankrin-G and beta-2 spectrin are required for E-cadherin localization at the plasma membrane in both cultured cells and mouse embryos [121]. The proteins presented in this review provide evidence for the formation and maintenance of these junctional complexes. Evidence is beginning to elucidate the physical connections that are made between the junctions and the actin-cytoskeleton. Questions remain, however, concerning these physical connections, and the dynamics and regulation of these proposed connections in vivo.
Acknowledgments
Work from the Nelson laboratory is supported by a grant from the NIH (GM 35527); Andrea Hartsock is additionally supported under the NIH Cell and Molecular Biology Training Program (5 T32 GM007276).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–34. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Moreno M, Fuchs E. Catenins: keeping cells from getting their signals crossed. Dev Cell. 2006;11:601–12. doi: 10.1016/j.devcel.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capaldo CT, Macara IG. Depletion of E-cadherin disrupts establishment but not maintenance of cell junctions in Madin-Darby canine kidney epithelial cells. Mol Biol Cell. 2007;18:189–200. doi: 10.1091/mbc.E06-05-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theard D, Steiner M, Kalicharan D, Hoekstra D, van Ijzendoorn SC. Cell polarity development and protein trafficking in hepatocytes lacking E-cadherin/beta-catenin-based adherens junctions. Mol Biol Cell. 2007;18:2313–21. doi: 10.1091/mbc.E06-11-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol. 2004;286:C1213–28. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 6.Gooding JM, Yap KL, Ikura M. The cadherin-catenin complex as a focal point of cell adhesion and signalling: new insights from three-dimensional structures. Bioessays. 2004;26:497–511. doi: 10.1002/bies.20033. [DOI] [PubMed] [Google Scholar]
- 7.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 8.Pokutta S, Herrenknecht K, Kemler R, Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem. 1994;223:1019–26. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- 9.Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–68. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 10.Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell. 2002;109:205–16. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- 11.Ozawa M, Hoschutzky H, Herrenknecht K, Kemler R. A possible new adhesive site in the cell-adhesion molecule uvomorulin. Mech Dev. 1990;33:49–56. doi: 10.1016/0925-4773(90)90134-8. [DOI] [PubMed] [Google Scholar]
- 12.Vestweber D, Kemler R. Identification of a putative cell adhesion domain of uvomorulin. Embo J. 1985;4:3393–8. doi: 10.1002/j.1460-2075.1985.tb04095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams CL, Chen YT, Smith SJ, Nelson WJ. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J Cell Biol. 1998;142:1105–19. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrlich JS, Hansen MD, Nelson WJ. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev Cell. 2002;3:259–70. doi: 10.1016/s1534-5807(02)00216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev Cell. 2002;3:367–81. doi: 10.1016/s1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 16.Yap AS, Niessen CM, Gumbiner BM. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J Cell Biol. 1998;141:779–89. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci. 1994;107(Pt 12):3655–63. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- 18.D’Souza-Schorey C. Disassembling adherens junctions: breaking up is hard to do. Trends Cell Biol. 2005;15:19–26. doi: 10.1016/j.tcb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 2001;3:289–93. doi: 10.1186/bcr309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conacci-Sorrell M, Zhurinsky J, Ben-Ze’ev A. The cadherin-catenin adhesion system in signaling and cancer. J Clin Invest. 2002;109:987–91. doi: 10.1172/JCI15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowin P, Rowlands TM, Hatsell SJ. Cadherins and catenins in breast cancer. Curr Opin Cell Biol. 2005;17:499–508. doi: 10.1016/j.ceb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–5. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 23.Van Aken E, De Wever O, Correia da Rocha AS, Mareel M. Defective E-cadherin/catenin complexes in human cancer. Virchows Arch. 2001;439:725–51. doi: 10.1007/s004280100516. [DOI] [PubMed] [Google Scholar]
- 24.Dillon DA, D’Aquila T, Reynolds AB, Fearon ER, Rimm DL. The expression of p120ctn protein in breast cancer is independent of alpha- and beta-catenin and E-cadherin. Am J Pathol. 1998;152:75–82. [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez MA, Pinder SE, Wencyk PM, Bell JA, Elston CW, Nicholson RI, Robertson JF, Blamey RW, Ellis IO. An immunohistochemical examination of the expression of E-cadherin, alpha- and beta/gamma-catenins, and alpha2- and beta1-integrins in invasive breast cancer. J Pathol. 1999;187:523–9. doi: 10.1002/(SICI)1096-9896(199904)187:5<523::AID-PATH296>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Zschiesche W, Schonborn I, Behrens J, Herrenknecht K, Hartveit F, Lilleng P, Birchmeier W. Expression of E-cadherin and catenins in invasive mammary carcinomas. Anticancer Res. 1997;17:561–7. [PubMed] [Google Scholar]
- 27.Berx G, Cleton-Jansen AM, Nollet F, de Leeuw WJ, van de Vijver M, Cornelisse C, van Roy F. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. Embo J. 1995;14:6107–15. doi: 10.1002/j.1460-2075.1995.tb00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berx G, Cleton-Jansen AM, Strumane K, de Leeuw WJ, Nollet F, van Roy F, Cornelisse C. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 1996;13:1919–25. [PubMed] [Google Scholar]
- 29.Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci U S A. 1995;92:8813–7. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakopoulou L, Gakiopoulou-Givalou H, Karayiannakis AJ, Giannopoulou I, Keramopoulos A, Davaris P, Pignatelli M. Abnormal alpha-catenin expression in invasive breast cancer correlates with poor patient survival. Histopathology. 2002;40:536–46. doi: 10.1046/j.1365-2559.2002.01392.x. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds AB, Roesel DJ, Kanner SB, Parsons JT. Transformation-specific tyrosine phosphorylation of a novel cellular protein in chicken cells expressing oncogenic variants of the avian cellular src gene. Mol Cell Biol. 1989;9:629–38. doi: 10.1128/mcb.9.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds AB, Herbert L, Cleveland JL, Berg ST, Gaut JR. p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors beta-catenin, plakoglobin and armadillo. Oncogene. 1992;7:2439–45. [PubMed] [Google Scholar]
- 33.Reynolds AB, Roczniak-Ferguson A. Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene. 2004;23:7947–56. doi: 10.1038/sj.onc.1208161. [DOI] [PubMed] [Google Scholar]
- 34.Jou TS, Stewart DB, Stappert J, Nelson WJ, Marrs JA. Genetic and biochemical dissection of protein linkages in the cadherin-catenin complex. Proc Natl Acad Sci U S A. 1995;92:5067–71. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferber A, Yaen C, Sarmiento E, Martinez J. An octapeptide in the juxtamembrane domain of VE-cadherin is important for p120ctn binding and cell proliferation. Exp Cell Res. 2002;274:35–44. doi: 10.1006/excr.2001.5436. [DOI] [PubMed] [Google Scholar]
- 36.Ohkubo T, Ozawa M. p120(ctn) binds to the membrane-proximal region of the E-cadherin cytoplasmic domain and is involved in modulation of adhesion activity. J Biol Chem. 1999;274:21409–15. doi: 10.1074/jbc.274.30.21409. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds AB, Daniel JM, Mo YY, Wu J, Zhang Z. The novel catenin p120cas binds classical cadherins and induces an unusual morphological phenotype in NIH3T3 fibroblasts. Exp Cell Res. 1996;225:328–37. doi: 10.1006/excr.1996.0183. [DOI] [PubMed] [Google Scholar]
- 38.Thoreson MA, Anastasiadis PZ, Daniel JM, Ireton RC, Wheelock MJ, Johnson KR, Hummingbird DK, Reynolds AB. Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J Cell Biol. 2000;148:189–202. doi: 10.1083/jcb.148.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–34. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyer S, Ferreri DM, DeCocco NC, Minnear FL, Vincent PA. VE-cadherin-p120 interaction is required for maintenance of endothelial barrier function. Am J Physiol Lung Cell Mol Physiol. 2004;286:L1143–53. doi: 10.1152/ajplung.00305.2003. [DOI] [PubMed] [Google Scholar]
- 41.Maeda M, Johnson E, Mandal SH, Lawson KR, Keim SA, Svoboda RA, Caplan S, Wahl JK, 3rd, Wheelock MJ, Johnson KR. Expression of inappropriate cadherins by epithelial tumor cells promotes endocytosis and degradation of E-cadherin via competition for p120(ctn) Oncogene. 2006;25:4595–604. doi: 10.1038/sj.onc.1209396. [DOI] [PubMed] [Google Scholar]
- 42.Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, Kowalczyk AP. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J Cell Biol. 2003;163:535–45. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piedra J, Miravet S, Castano J, Palmer HG, Heisterkamp N, Garcia de Herreros A, Dunach M. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin Interaction. Mol Cell Biol. 2003;23:2287–97. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao K, Garner J, Buckley KM, Vincent PA, Chiasson CM, Dejana E, Faundez V, Kowalczyk AP. p120-Catenin regulates clathrin-dependent endocytosis of VE-cadherin. Mol Biol Cell. 2005;16:5141–51. doi: 10.1091/mbc.E05-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T, Birchmeier W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol. 2002;4:222–31. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- 46.Yanagisawa M, Anastasiadis PZ. p120 catenin is essential for mesenchymal cadherin-mediated regulation of cell motility and invasiveness. J Cell Biol. 2006;174:1087–96. doi: 10.1083/jcb.200605022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noren NK, Liu BP, Burridge K, Kreft B. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J Cell Biol. 2000;150:567–80. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macpherson IR, Hooper S, Serrels A, McGarry L, Ozanne BW, Harrington K, Frame MC, Sahai E, Brunton VG. p120-catenin is required for the collective invasion of squamous cell carcinoma cells via a phosphorylation-independent mechanism. Oncogene. 2007 doi: 10.1038/sj.onc.1210334. [DOI] [PubMed] [Google Scholar]
- 49.Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford HC, Zheng Y, Reynolds AB. Inhibition of RhoA by p120 catenin. Nat Cell Biol. 2000;2:637–44. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- 50.Castano J, Solanas G, Casagolda D, Raurell I, Villagrasa P, Bustelo XR, Garcia de Herreros A, Dunach M. Specific phosphorylation of p120-catenin regulatory domain differently modulates its binding to RhoA. Mol Cell Biol. 2007;27:1745–57. doi: 10.1128/MCB.01974-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, Reynolds AB. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–39. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
- 52.McCrea PD, Gumbiner BM. Purification of a 92-kDa cytoplasmic protein tightly associated with the cell-cell adhesion molecule E-cadherin (uvomorulin). Characterization and extractability of the protein complex from the cell cytostructure. J Biol Chem. 1991;266:4514–20. [PubMed] [Google Scholar]
- 53.McCrea PD, Turck CW, Gumbiner B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science. 1991;254:1359–61. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- 54.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–82. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 55.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 56.Lickert H, Bauer A, Kemler R, Stappert J. Casein kinase II phosphorylation of E-cadherin increases E-cadherin/beta-catenin interaction and strengthens cell-cell adhesion. J Biol Chem. 2000;275:5090–5. doi: 10.1074/jbc.275.7.5090. [DOI] [PubMed] [Google Scholar]
- 57.Lilien J, Balsamo J, Arregui C, Xu G. Turn-off, drop-out: functional state switching of cadherins. Dev Dyn. 2002;224:18–29. doi: 10.1002/dvdy.10087. [DOI] [PubMed] [Google Scholar]
- 58.Piedra J, Martinez D, Castano J, Miravet S, Dunach M, de Herreros AG. Regulation of beta-catenin structure and activity by tyrosine phosphorylation. J Biol Chem. 2001;276:20436–43. doi: 10.1074/jbc.M100194200. [DOI] [PubMed] [Google Scholar]
- 59.Roura S, Miravet S, Piedra J, Garcia de Herreros A, Dunach M. Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J Biol Chem. 1999;274:36734–40. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- 60.Rhee J, Mahfooz NS, Arregui C, Lilien J, Balsamo J, VanBerkum MF. Activation of the repulsive receptor Roundabout inhibits N-cadherin-mediated cell adhesion. Nat Cell Biol. 2002;4:798–805. doi: 10.1038/ncb858. [DOI] [PubMed] [Google Scholar]
- 61.Hoschuetzky H, Aberle H, Kemler R. Beta-catenin mediates the interaction of the cadherin-catenin complex with epidermal growth factor receptor. J Cell Biol. 1994;127:1375–80. doi: 10.1083/jcb.127.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi HJ, Huber AH, Weis WI. Thermodynamics of beta-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J Biol Chem. 2006;281:1027–38. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- 63.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen YT, Stewart DB, Nelson WJ. Coupling assembly of the E-cadherin/beta-catenin complex to efficient endoplasmic reticulum exit and basal-lateral membrane targeting of E-cadherin in polarized MDCK cells. J Cell Biol. 1999;144:687–99. doi: 10.1083/jcb.144.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. Embo J. 1997;16:3797–804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272:24735–8. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 67.Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. Essential role of BCL9-2 in the switch between beta-catenin’s adhesive and transcriptional functions. Genes Dev. 2004;18:2225–30. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukata M, Kuroda S, Nakagawa M, Kawajiri A, Itoh N, Shoji I, Matsuura Y, Yonehara S, Fujisawa H, Kikuchi A, Kaibuchi K. Cdc42 and Rac1 regulate the interaction of IQGAP1 with beta-catenin. J Biol Chem. 1999;274:26044–50. doi: 10.1074/jbc.274.37.26044. [DOI] [PubMed] [Google Scholar]
- 69.Tao YS, Edwards RA, Tubb B, Wang S, Bryan J, McCrea PD. beta-Catenin associates with the actin-bundling protein fascin in a noncadherin complex. J Cell Biol. 1996;134:1271–81. doi: 10.1083/jcb.134.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pokutta S, Weis WI. Structure of the dimerization and beta-catenin-binding region of alpha-catenin. Mol Cell. 2000;5:533–43. doi: 10.1016/s1097-2765(00)80447-5. [DOI] [PubMed] [Google Scholar]
- 72.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–15. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Collins MO, Yu L, Coba MP, Husi H, Campuzano I, Blackstock WP, Choudhary JS, Grant SG. Proteomic analysis of in vivo phosphorylated synaptic proteins. J Biol Chem. 2005;280:5972–82. doi: 10.1074/jbc.M411220200. [DOI] [PubMed] [Google Scholar]
- 75.Burks J, Agazie YM. Modulation of alpha-catenin Tyr phosphorylation by SHP2 positively effects cell transformation induced by the constitutively active FGFR3. Oncogene. 2006;25:7166–79. doi: 10.1038/sj.onc.1209728. [DOI] [PubMed] [Google Scholar]
- 76.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–88. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muresan Z, Paul DL, Goodenough DA. Occludin 1B, a variant of the tight junction protein occludin. Mol Biol Cell. 2000;11:627–34. doi: 10.1091/mbc.11.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sakakibara A, Furuse M, Saitou M, Ando-Akatsuka Y, Tsukita S. Possible involvement of phosphorylation of occludin in tight junction formation. J Cell Biol. 1997;137:1393–401. doi: 10.1083/jcb.137.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y, Lu Q, Schneeberger EE, Goodenough DA. Restoration of tight junction structure and barrier function by down-regulation of the mitogen-activated protein kinase pathway in ras-transformed Madin-Darby canine kidney cells. Mol Biol Cell. 2000;11:849–62. doi: 10.1091/mbc.11.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen YH, Lu Q, Goodenough DA, Jeansonne B. Nonreceptor tyrosine kinase c-Yes interacts with occludin during tight junction formation in canine kidney epithelial cells. Mol Biol Cell. 2002;13:1227–37. doi: 10.1091/mbc.01-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Andreeva AY, Krause E, Muller EC, Blasig IE, Utepbergenov DI. Protein kinase C regulates the phosphorylation and cellular localization of occludin. J Biol Chem. 2001;276:38480–6. doi: 10.1074/jbc.M104923200. [DOI] [PubMed] [Google Scholar]
- 82.Clarke H, Soler AP, Mullin JM. Protein kinase C activation leads to dephosphorylation of occludin and tight junction permeability increase in LLC-PK1 epithelial cell sheets. J Cell Sci. 2000;113(Pt 18):3187–96. doi: 10.1242/jcs.113.18.3187. [DOI] [PubMed] [Google Scholar]
- 83.Weiler F, Marbe T, Scheppach W, Schauber J. Influence of protein kinase C on transcription of the tight junction elements ZO-1 and occludin. J Cell Physiol. 2005;204:83–6. doi: 10.1002/jcp.20268. [DOI] [PubMed] [Google Scholar]
- 84.Kazanietz MG. Targeting protein kinase C and “non-kinase” phorbol ester receptors: emerging concepts and therapeutic implications. Biochim Biophys Acta. 2005;1754:296–304. doi: 10.1016/j.bbapap.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 85.Brose N, Betz A, Wegmeyer H. Divergent and convergent signaling by the diacylglycerol second messenger pathway in mammals. Curr Opin Neurobiol. 2004;14:328–40. doi: 10.1016/j.conb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 86.Nunbhakdi-Craig V, Machleidt T, Ogris E, Bellotto D, White CL, 3rd, Sontag E. Protein phosphatase 2A associates with and regulates atypical PKC and the epithelial tight junction complex. J Cell Biol. 2002;158:967–78. doi: 10.1083/jcb.200206114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wong V, Gumbiner BM. A synthetic peptide corresponding to the extracellular domain of occludin perturbs the tight junction permeability barrier. J Cell Biol. 1997;136:399–409. doi: 10.1083/jcb.136.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nusrat A, Brown GT, Tom J, Drake A, Bui TT, Quan C, Mrsny RJ. Multiple protein interactions involving proposed extracellular loop domains of the tight junction protein occludin. Mol Biol Cell. 2005;16:1725–34. doi: 10.1091/mbc.E04-06-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schulzke JD, Gitter AH, Mankertz J, Spiegel S, Seidler U, Amasheh S, Saitou M, Tsukita S, Fromm M. Epithelial transport and barrier function in occludin-deficient mice. Biochim Biophys Acta. 2005;1669:34–42. doi: 10.1016/j.bbamem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 90.Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol. 2005;171:939–45. doi: 10.1083/jcb.200510043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–29. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 92.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–50. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147:1351–63. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCarthy KM, Francis SA, McCormack JM, Lai J, Rogers RA, Skare IB, Lynch RD, Schneeberger EE. Inducible expression of claudin-1-myc but not occludin-VSV-G results in aberrant tight junction strand formation in MDCK cells. J Cell Sci. 2000;113(Pt 19):3387–98. doi: 10.1242/jcs.113.19.3387. [DOI] [PubMed] [Google Scholar]
- 95.Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang VW, Goodenough DA. Paracellular ion channel at the tight junction. Biophys J. 2003;84:1660–73. doi: 10.1016/S0006-3495(03)74975-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu AS, Enck AH, Lencer WI, Schneeberger EE. Claudin-8 expression in Madin-Darby canine kidney cells augments the paracellular barrier to cation permeation. J Biol Chem. 2003;278:17350–9. doi: 10.1074/jbc.M213286200. [DOI] [PubMed] [Google Scholar]
- 98.Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol. 2002;283:C142–7. doi: 10.1152/ajpcell.00038.2002. [DOI] [PubMed] [Google Scholar]
- 99.Colegio OR, Van Itallie C, Rahner C, Anderson JM. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am J Physiol Cell Physiol. 2003;284:C1346–54. doi: 10.1152/ajpcell.00547.2002. [DOI] [PubMed] [Google Scholar]
- 100.Kiuchi-Saishin Y, Gotoh S, Furuse M, Takasuga A, Tano Y, Tsukita S. Differential expression patterns of claudins, tight junction membrane proteins, in mouse nephron segments. J Am Soc Nephrol. 2002;13:875–86. doi: 10.1681/ASN.V134875. [DOI] [PubMed] [Google Scholar]
- 101.Hou J, Shan Q, Wang T, Gomes AS, Yan Q, Paul DL, Bleich M, Goodenough DA. Transgenic RNAi Depletion of Claudin-16 and the Renal Handling of Magnesium. J Biol Chem. 2007;282:17114–22. doi: 10.1074/jbc.M700632200. [DOI] [PubMed] [Google Scholar]
- 102.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–45. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 103.Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. J Cell Biol. 1997;138:181–92. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Muller SL, Portwich M, Schmidt A, Utepbergenov DI, Huber O, Blasig IE, Krause G. The tight junction protein occludin and the adherens junction protein alpha-catenin share a common interaction mechanism with ZO-1. J Biol Chem. 2005;280:3747–56. doi: 10.1074/jbc.M411365200. [DOI] [PubMed] [Google Scholar]
- 105.Rajasekaran AK, Hojo M, Huima T, Rodriguez-Boulan E. Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J Cell Biol. 1996;132:451–63. doi: 10.1083/jcb.132.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Itoh M, Morita K, Tsukita S. Characterization of ZO-2 as a MAGUK family member associated with tight as well as adherens junctions with a binding affinity to occludin and alpha catenin. J Biol Chem. 1999;274:5981–6. doi: 10.1074/jbc.274.9.5981. [DOI] [PubMed] [Google Scholar]
- 107.Wittchen ES, Haskins J, Stevenson BR. NZO-3 expression causes global changes to actin cytoskeleton in Madin-Darby canine kidney cells: linking a tight junction protein to Rho GTPases. Mol Biol Cell. 2003;14:1757–68. doi: 10.1091/mbc.E02-08-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fanning AS, Jameson BJ, Jesaitis LA, Anderson JM. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem. 1998;273:29745–53. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- 109.Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–26. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Utepbergenov DI, Fanning AS, Anderson JM. Dimerization of the scaffolding protein ZO-1 through the second PDZ domain. J Biol Chem. 2006;281:24671–7. doi: 10.1074/jbc.M512820200. [DOI] [PubMed] [Google Scholar]
- 111.Kale G, Naren AP, Sheth P, Rao RK. Tyrosine phosphorylation of occludin attenuates its interactions with ZO-1, ZO-2, and ZO-3. Biochem Biophys Res Commun. 2003;302:324–9. doi: 10.1016/s0006-291x(03)00167-0. [DOI] [PubMed] [Google Scholar]
- 112.Wittchen ES, Haskins J, Stevenson BR. Exogenous expression of the amino-terminal half of the tight junction protein ZO-3 perturbs junctional complex assembly. J Cell Biol. 2000;151:825–36. doi: 10.1083/jcb.151.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wittchen ES, Haskins J, Stevenson BR. Protein interactions at the tight junction. Actin has multiple binding partners, and ZO-1 forms independent complexes with ZO-2 and ZO-3. J Biol Chem. 1999;274:35179–85. doi: 10.1074/jbc.274.49.35179. [DOI] [PubMed] [Google Scholar]
- 114.Mattagajasingh SN, Huang SC, Hartenstein JS, Benz EJ., Jr Characterization of the interaction between protein 4.1R and ZO-2. A possible link between the tight junction and the actin cytoskeleton. J Biol Chem. 2000;275:30573–85. doi: 10.1074/jbc.M004578200. [DOI] [PubMed] [Google Scholar]
- 115.Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M, Tsukita S. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741–54. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 116.Siliciano JD, Goodenough DA. Localization of the tight junction protein, ZO-1, is modulated by extracellular calcium and cell-cell contact in Madin-Darby canine kidney epithelial cells. J Cell Biol. 1988;107:2389–99. doi: 10.1083/jcb.107.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ikenouchi J, Umeda K, Tsukita S, Furuse M, Tsukita S. Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J Cell Biol. 2007;176:779–86. doi: 10.1083/jcb.200612080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fanning AS, Little BP, Rahner C, Utepbergenov D, Walther Z, Anderson JM. The unique-5 and -6 motifs of ZO-1 regulate tight junction strand localization and scaffolding properties. Mol Biol Cell. 2007;18:721–31. doi: 10.1091/mbc.E06-08-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kobielak A, Pasolli HA, Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boguslavsky S, Grosheva I, Landau E, Shtutman M, Cohen M, Arnold K, Feinstein E, Geiger B, Bershadsky A. p120 catenin regulates lamellipodial dynamics and cell adhesion in cooperation with cortactin. Proc Natl Acad Sci U S A. 2007;104:10882–7. doi: 10.1073/pnas.0702731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kizhatil K, Davis JQ, Davis L, Hoffman J, Hogan BL, Bennett V. Ankyrin-G is a molecular partner of E-cadherin in epithelial cells and early embryos. J Biol Chem. 2007 doi: 10.1074/jbc.M703158200. [DOI] [PubMed] [Google Scholar]
- 122.Helwani FM, Kovacs EM, Paterson AD, Verma S, Ali RG, Fanning AS, Weed SA, Yap AS. Cortactin is necessary for E-cadherin-mediated contact formation and actin reorganization. J Cell Biol. 2004;164:899–910. doi: 10.1083/jcb.200309034. [DOI] [PMC free article] [PubMed] [Google Scholar]