Abstract
Methionine synthase is a key enzyme poised at the intersection of folate and sulfur metabolism and functions to reclaim homocysteine to the methionine cycle. The 5′ leader sequence in human MS is 394 nucleotides long and harbors two open reading frames (uORFs). In this study, regulation of the main open reading frame by the uORFs has been elucidated. Both uORFs downregulate translation as demonstrated by mutation of the upstream AUG codons (uAUG) either singly or simultaneously. The uAUGs are capable of recruiting the 40S ribosomal complex as revealed by their ability to drive reporter expression in constructs in which the luciferase is fused to the uORFs. uORF2, which is predicted to encode a 30 amino acid long polypeptide, has a clustering of rare codons encoding arginine and proline. Mutation of a tandemly repeated rare codon for arginine at positions 3 and 4 in uORF2 to either common codons for the same amino acid or common codons for alanine, result in complete alleviation of translation inhibition. This suggests a mechanism for ribosome stalling and demonstrates that the cis-effects on translation by uORF2 is dependent on the nucleotide sequence but is apparently independent of the sequence of the encoded peptide. This study reveals complex regulation of the essential housekeeping gene, methionine synthase, by the uORFs in its leader sequence.
Keywords: methionine, upstream open reading frame, methionine synthase, gene regulation
Homocysteine is an intermediate in methionine metabolism and is cleared via the concerted actions of three enzymes, methionine synthase (MS)1, cystathionine β-synthase and betaine homocysteine methyltransferase (1). Of these, only MS is present ubiquitiously and reclaims homocysteine to the methionine cycle via a transmethylation reaction (2). In contrast, betaine homocysteine methyltransferase, which also catalyzes the remethylation of homocysteine, has a very restricted tissue distribution being present only in liver and kidney. Cystathionine β-synthase commits homocysteine to the transsulfuration pathway that leads to synthesis of cysteine, and is present in some but not all tissues (2).
MS is an essential gene as indicated by the embryonic lethality of the null genotype in transgenic mice (3). It performs two significant cellular functions. First, by salvaging homocysteine to the methionine cycle, it helps maintain low intracellular levels of this nonprotein amino acid. Elevated levels of homocysteine are correlated with numerous complex diseases including cardiovascular diseases (4), neural tube defects (5) and some neurodegenerative diseases (6). Second, by demethylating 5-CH3-tetrahydrofolate, the methyl group donor, MS liberates tetrahydrofolate that is then used widely as a template for one-carbon transfers to support purine and amino acid syntheses. In mammals, MS utilizes methylcobalamin, a derivative of vitamin B12 as a cofactor.
In humans, this housekeeping gene appears to be subjected to complex regulation and the open reading frame (ORF) is framed by a 394 base-long leader sequence that precedes it and an ~3000-base long 3′ UTR that follows it. Eukaryotic leader sequences are on average 50–70 bases in length and longer leaders are invariably involved in regulation (7). Indeed recent studies have identified regulatory elements in the 5′-leader sequence of the MS mRNA (8). In contrast, the role, if any, of the long 3′ UTR in mediating message stability and/or translational regulation is unknown.
B12 supplementation to cells in culture increases the activity of MS (9, 10) and this regulation is exerted at a translational level (8, 11). Addition of B12 to normal medium results in a reequilbration of the MS message from the inactive ribonucleoprotein pool to the active polysomal pool (8). The long 5′ leader sequence is predicted to be highly structured posing obvious problems for cap-dependent commencement of translation and suggests the presence of an alternative initiation strategy, i.e. via an IRES element. Indeed, studies with a bicistronic reporter construct confirmed the presence of an IRES element and localized the boundaries to 70 bases immediately upstream of the AUG (12). Curiously, the boundaries of the B12 responsive element coincide with those of the IRES element (Figure 1) and reveal that IRES-dependent translation of MS is modulated by a nutrient, i.e. the cofactor of the encoded enzyme.
Figure 1.
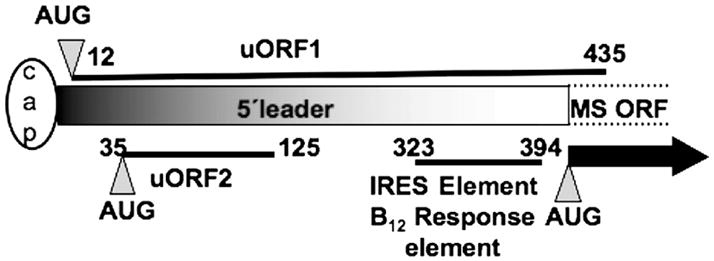
A schematic representation of the 5′ leader of the human MS mRNA. The relative positions of the uORFs, the IRES element and the B12-response element are indicated by solid lines. The numbers refer to the positions of nucleotides with respect to the 5′ end of the leader sequence (full length is 394 nucleotides). The MS-ORF refers to the main open reading frame shown by the thick arrow, and the cap depicts the me7G cap at the 5′ end of the message.
Sequence analysis of the 5′-leader sequence reveals the presence of two ORFs that are upstream of the MS ORF (Figure 1). The first, uORF1, commences at uAUG1 at position 12 and overlaps with the MS ORF, albeit in a different frame. It is predicted to encode a polypeptide that is 141 residues in length. The second, uORF2, commences at uAUG2 at position 35 and is predicted to encode a shorter polypeptide of 30 amino acids in length. It is estimated that <10% of eukaryotic mRNAs contain uORFs in their leader sequences (13) but they are prominent in oncogenes, two-thirds of which have this regulatory element (14). In addition, leader sequences containing uORFs are prevalent in other genes involved in control of cellular growth and differentiation (15). In this study, we have evaluated the influence of the uORFs in the leader sequence of human MS and demonstrate that they are negative modulators of translation.
MATERIALS AND METHODS
Materials
Eagle’s minimum essential medium (EMEM) and Anti-rabbit IgG antibody were purchased from Sigma. FBS (fetal bovine serum) was from HyClone. Cos-1 cells (monkey kidney fibroblast) were obtained from Dr. Charles Wood at the University of Nebraska, Lincoln. Radiolabeled [α-32P] ATP (5000 mCi/mmol) was purchased from Amersham Pharmacia. Restriction enzymes were from Invitrogen or New England Biolabs. PCR reagents and enzymes were from Invitrogen or Stratagene. Primers were purchased from Fisher Scientific.
Cell Culture Conditions
Cells were grown in Eagle’s MEM supplemented with 10% FBS and incubated at 37°C, 5% CO2. For luciferase reporter studies and Western and Northern analyses of MS, cells were grown in 100 mm plates to 60–80% confluency and transfection was performed as described previously (8).
Plasmid Construction
The recombinant plasmids used in this study are shown in Table I. Primer sequences used to generate individual constructs are shown in Table II. All PCR reactions were performed using conditions described previously (8) and the high fidelity Pfu polymerase (Stratagene). The DNA sequences of all constructs were verified by nucleotide sequence determination at the Center for Biotechnology’s Genomics Core Facility (University of Nebraska-Lincoln). Mutants were generated using the Quickchange Site-Directed Mutagenesis kit (Stratagene).
Table I.
Plasmids used or constructed in this study
| Plasmid name | Template | Primer pair | Description | Reference |
|---|---|---|---|---|
| pBC110 | pSO102 | BCOL3-BCOL4 | Luc-AUG mutated to AAG | This study |
| pBC112 | pSO104 | BCOL3-BCOL4 | Luc-AUG mutated to AAG | This study |
| pBC111 a | pSO103 | BCOL3-BCOL4 | uAUG1 intact; uAUG2 mutated; Luc-AUG mutated to AAG | This study |
| pBC116 a | pSO104 | BCOL3-BCOL4 | Negative control of pBC111 | This study |
| pBC121a b | pBC110 | BCOL5-BCOL6 | uAUG2 intact; Luc-AUG mutated to AAG | This study |
| pBC123 b | pBC112 | BCOL5-BCOL6 | Negative control of pBC121a | This study |
| pGL3 promoter | NA | NA | the plasmid contaning the firefly Luciferase | Promega |
| pSO101 c | i | 5′ UTR sense-5′UTR antisense | 5′UTR of MS gene cloned into pGL3p | This study |
| pSO102 d | pSO101 | Mut Start 1 sense-Mut Start 1 antisense | uORF2 is intact, uAUG1 mutated | This study |
| pSO103 e | pSO101 | Mut Start 2 sense-Mut Start 2 antisense | uORF1 is intact/truncated, uAUG2 mutated | This study |
| pSO104f | pSO102 | Mut Start 2 sense-Mut Start 2 antisense | uAUG1 and uAUG2 are mutated | This study |
| pBC220g | pSO101 | BCOL20-BCOL21 | In vitro transcription/translation construct | This study |
| pBC221g | pSO104 | BCOL20-BCOL21 | In vitro transcription/translation construct | This study |
| pBC222g | pSO101 | BCOL20-BCOL22 | In vitro transcription/translation construct | This study |
| pBC224a | pSO101 | BCOL23-BCOL24 | Codons 3 and 4 in uORF2 are converted from Arg (rare) to Arg (common) | This study |
| pBC226d | pSO101 | BCOL25-BCOL26 | Codons 3 and 4 in uORF2 are converted from Arg to Ala | This study |
| phRGTK | NA | NA | Renilla luciferase vector | Promega |
| pGL3lucR or pBC777h | NA | NA | Renilla luciferase gene was cloned into pGL3p lacking Firefly luciferase gene | This study |
| pBC207bh | NA | NA | No uORFs present. The (pSO104 insert) cloned into pGL3lucR vector (cis/trans experiment) | This study |
NA: Not-applicable.
pBC111 and pBC116: Only the uAUG1 is present in pBC111 and the uORF1 sequence is in the same frame as the luciferase gene (the uORF1 fusion). pBC116 contains no AUGs (negative control).
pBC121a and pBC123: Only uAUG2 is present in pBC121a. Also, the stop codon of uORF2 (TAG) was eliminated by addition of 2 nucleotides so that uORF2 is in frame with the luciferase gene. pBC123 contains no AUGs (negative control).
pSO101: The wild type 5′ leader was cloned into pGL3p (using the HindIII-NcoI sites) and resulted in an extra C due to NcoI cloning. Both uAUGs are intact however the uORF1 sequence was truncated as a result of cloning into the pGL3p vector. In the MS sequence, the uORF1 overlaps with the main ORF.
pSO102: uORF1 is mutated, uORF2 intact. uAUG1 was mutated to UUG while keeping uAUG2 unchanged.
pSO103: uAUG2 mutated to UUG, uORF1 intact/truncated (note that uORF1 is in frame with luciferase).
pSO104: Both AUGs for the uORFs are mutated (serves as a negative control).
pBC220-pBC223: These constructs were used in the in vitro transcription/translation experiments to determine if there is any peptide production by the uORFs. The 5′ Primer (BCOL220) has a T7 promoter sequence. The sequence is the same as the sequence in the region 960–990 nt from the pGEMEX-2 vector (Promega). The PCR products were cloned into the TOPO vector (Invitrogen) to create the desired plasmids. In pBC220–221, inserts only had the 5′ leader region and the 3′ primer had a stop codon for the uORF1. uORF2 already has its own stop codon (UAG). In pBC222–223, inserts had the 5′ leader sequence in addition to the luciferase gene.
The plasmid containing the Renilla luciferase (phRGTK) was cut with NcoI and XbaI to excise out the Renilla Luc gene. Then, pGL3p was cut using NcoI and XbaI. The Renilla luciferase gene was cloned into the pGL3 promoter lacking Firefly luciferase to create pGL3lucR (pBC777). pBC202a and pBC207b were created by cloning the SO102 and SO104 insert (HindIII-NcoI) into pGL3lucR, respectively.
A clone containing the 5′ leader sequence of MS was generously provided by Dr. Barry Shane (University of California, Berkeley). This clone was used as the template to amplify the 5′ leader sequence and clone it into the pGL3p to create pSO101.
Table II.
Primer sequences employed in this study
| Primer name | Sequence (5′→3′)a |
|---|---|
| BCOL3 | GGAGACTCGACAACCAAGGAAGACGCCAAA |
| BCOL4 | TTTGGCGTCTTCCTTGGTTGTCGAGTCTCC |
| BCOL5 | CGTGTCTGGCTGCTTTAGGCCGACACCAAGG |
| BCOL6 | CCTTGGTGTCGGCCTAAAGCAGCCAGACACG |
| BCOL20 | GTGGTCTCCCTATAGTGAGTCGTATTAATTAAAGGTTCTAAATGTCTGCG |
| BCOL21 | TTTGGCGTCTTCTTAGGTTGTC |
| BCOL22 | CTCTAGAATTACACGGCGAT |
| BCOL23 | AGCCGGATGTCACGCCGCCCTCCTCTGCCG |
| BCOL24 | CGGCAGAGGAGGGCGGCGTGACATCCGGCT |
| BCOL25 | AGCCGGATGTCAGCAGCACCTCCTCTGCCG |
| BCOL26 | CGGCAGAGGAGGTGCTGCTGACATCCGGCT |
| 5′UTR sense | CCCAAGCTTAAAGGTTCTAAAT (HindIII) |
| 5′UTR antisense | CATGCCATGGTTGTCGAGTCTC (NcoI) |
| Mut Start 1 sense | AAAGGTTCTAATTGTCTGCGGGGCTCAGAG |
| Mut Start 1 antisense | CTCTGAGCCCCGCAGACAATTAGAACCTTT |
| Mut Start 2 sense | CTCAGAGCCGGTTGTCACGTCGTC |
| Mut Start antisense | GACGACGTGACAACCGGCTCTGAG |
Mutated nucleotides and restriction enzyme sites are underlined. Restriction enzymes are shown in parentheses where applicable.
Transient transfection and luciferase Assays
Transfections were performed using the GeneJammer reagent (Stratagene) for Cos-1 cells. Briefly, 2–8 μg of plasmid DNA was mixed with 10–50 μl of transfection reagent according to the manufacturer’s specifications, and the mixture was added to 100 mm plates containing cells at 60% confluency. Reporter gene activities (firefly and Renilla luciferase) were determined according to the vendor’s protocol (Promega).
Northern Analysis
Analyses were performed as previously described (8). A 32P-labeled probe consisting of a DNA fragment encoding the luciferase gene was used (1.1 and 0.9 kb fragment for Firefly or Renilla luciferase, respectively). The bands were quantified by densitometry using Quantity One software (BioRad). The luciferase band intensities were normalized versus 18S rRNA in the same samples. At least three independent experiments were performed for each sample.
In vitro transcription-translation
The plasmids or PCR fragments containing a T7 promoter upstream of the 5′ UTR was created using the primers and templates shown in Tables I and II (plasmids pBC220-pBC223). Primer BCOL20 contains the T7 promoter sequence. In vitro transcription-translation was performed according to the manufacturer’s protocol (Promega).
RESULTS
Mutation of the uAUGs stimulates translation of the main ORF
There are two initiation codons in the 5′ leader sequence of the MS mRNA preceding the initiation codon for the main ORF. Both uAUGs are followed by ORFs. To evaluate the role of these uORFs on the expression of MS, reporter constructs were prepared by cloning the MS 5′ leader sequence upstream of the luciferase gene (construct pSO101) and mutating uAUG1 and uAUG2 to UUG either singly or together (constructs pSO102, pSO103 and pSO104 respectively) (Figure 2). In order to study translational effects, the reporter luciferase activities were normalized to the luciferase mRNA levels determined by Northern analysis. Elimination of either uAUG resulted in higher reporter activity (Figure 2). Specifically, loss of uAUG1 or uAUG2 resulted in 2- or 2.5 fold increase in luciferase activity respectively, while concomitant elimination of both uAUGs resulted in a 3.5-fold increase in reporter activity. These results indicate that the uORFs inhibit translation of the downstream ORF.
Figure 2.
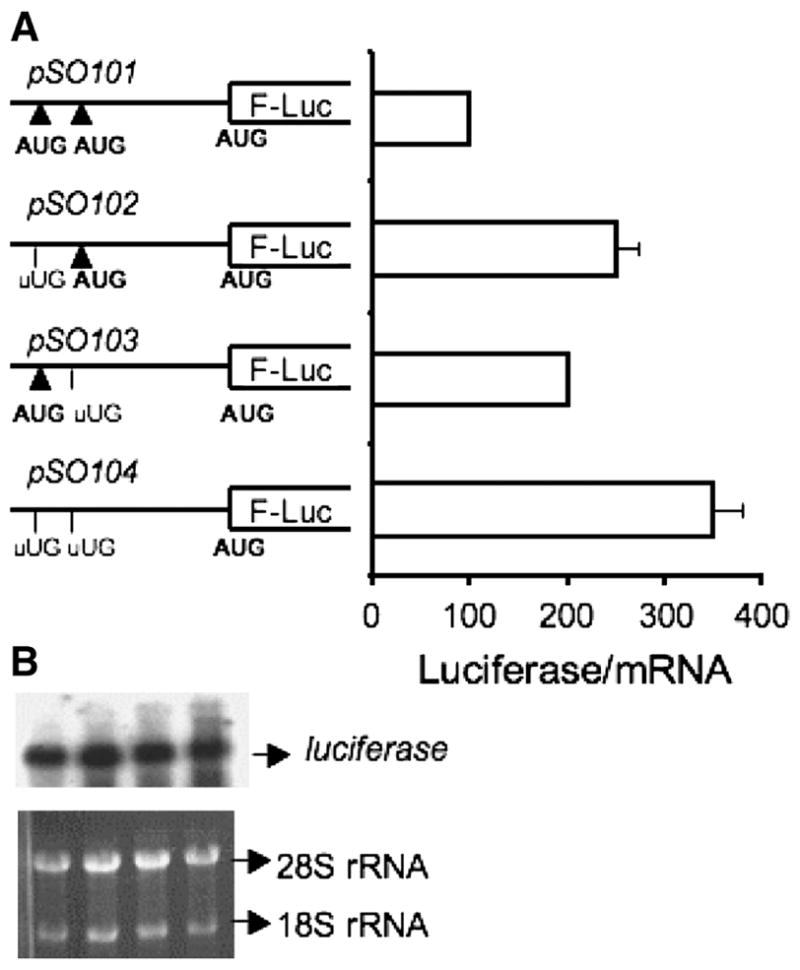
uORFs inhibit translation of the main ORF. A. Reporter constructs in which the uAUGs were mutated singly or together to UUG codons were used to test the effect of the uORFs on firefly luciferase (F-Luc) expression. The 5′ leader sequences were cloned into the pGL3-promoter vector as described under Materials and Methods. The luciferase activities were normalized to luciferase mRNA levels in each experiment. B. Northern analysis of each reporter construct used in A demonstrates that the mutations do not affect luciferase mRNA levels. Total RNA was isolated from the transfected Cos-1 cells and probed with a 32P- labeled fragment of the luciferase gene as described under Methods. Equal loading was ensured by monitoring the 18S and 28S ribosomal RNAs in each lane as detected by ethidium bromide staining.
Translation can be initiated at the AUGs of uORF1 and uORF2
For optimal cap-dependent translation, an AUG codon in a Kozak sequence context is used (13). Although the uAUGs in the MS 5′ leader do not reside in a conserved Kozak sequence, it is still possible that they recruit ribosomes and thus compete with the initiation codon of the main ORF. To evaluate whether translation can be initiated at the uAUGs, an indirect approach was employed. Fusion constructs were prepared in which the AUG of the reporter ORF and uAUG2 were each mutated to AAG. In the resulting construct, pBC111 (Figure 3), uAUG1 is in frame with the luciferase gene and reporter expression is dependent on translation initiation at uAUG1. As a consequence of using the HindIII and NcoI restriction enzyme sites to insert the 5′ leader sequence, an additional nucleotide was inserted, which also had the desired consequence of bringing uORF1 in frame with the reading frame for the downstream luciferase reporter. As a negative control, the pBC116 construct lacking all three AUGs was employed. A similar strategy was used to test whether uAUG2 can serve as an initiation codon. However, two additional bases (tt at positions 123 and 124 immediately preceding the stop codon for uORF2) were added to bring uAUG2 in frame with the luciferase ORF (construct pBC121a). The corresponding negative control, pBC123 also had the additional two-base insertion and lacked all three initiator AUGs (Figure 3).
Figure 3.
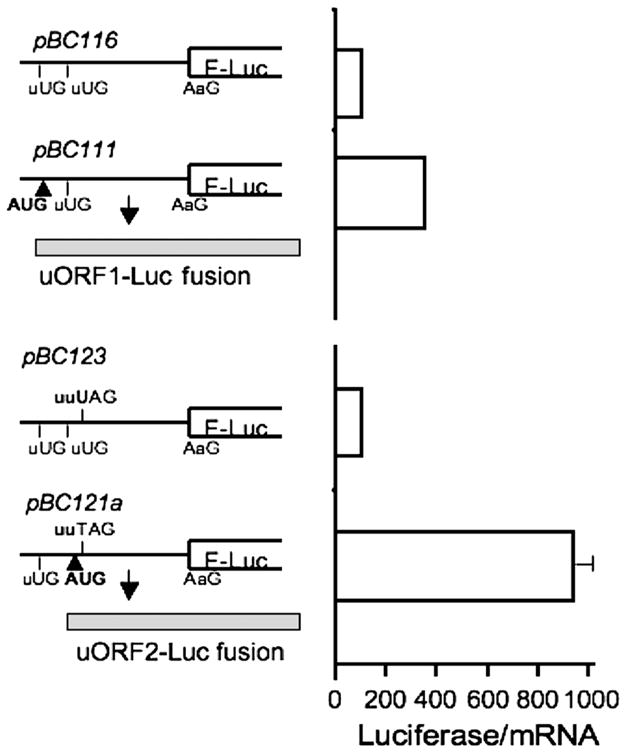
Translation can be initiated at the uORFs. The upper panel shows the construct (pBC111) in which the reporter is fused to uORF1. Compared to the negative control, i.e. a construct lacking all AUGs (pBC116). luciferase activity was 3.5 fold higher. The lower panel shows the construct pBC121a in which the reporter is fused in frame to uORF2. To bring uORF2 in frame with the luciferase gene, two nucleotides (tt) were added at positions 123–124 just preceding the stop codon (UAG) of uORF2. The corresponding negative control lacking all AUGs (pBC123) but containing the additional tt nucleotides was employed. Luciferase activity in the uORF2 fusion was ~10 fold higher than in the negative control. Luciferase activities are reported relative to the negative control (set at 100) and normalized to mRNA levels.
The uORF1-luciferase fusion (pBC111) exhibited a 3.5-fold higher reporter activity than the corresponding negative control (pBC116), whereas the uORF2 fusion (pBC121a) resulted in an ~10 fold higher reporter activity than the corresponding negative control (pBC123) (Figure 3). The luciferase activity associated with the uORF1 and uORF2 fusions were 35- and 13–fold lower respectively than the activity of the native luciferase enzyme. This could result from less efficient translation initiation at the uAUGs and/or inhibitory effects of the resulting N-terminal extensions in the uORF-luciferase fusions. These results are nonetheless consistent with the potential of both uAUGs in the MS 5′ leader sequence to support translation initiation. Furthermore, they suggest that translation initiation is more efficient from uAUG2 than from uAUG1, which could result from the very close proximity of uAUG1 to the 5′ end of the message.
We were unable to detect the fusion proteins generated by constructs pBC111 and pBC121a by Western analysis using commercial anti-luciferase antibody (Promega). This could have resulted from a lack of sensitivity of our analysis due to the apparently lower expression level of the fusion proteins versus native luciferase. As an alternative, we attempted to detect the peptides encoded by the uORFs in an vitro transcription/translation system using constructs pBC220-pBC223 described in Table I. Despite repeated attempts, we were unable to detect peptides under these conditions. This could be due to the highly structured leader sequence, which is known to inhibit translation efficiency in the in vitro transcription-translation system. Due to the more efficient translation from uAUG2 observed for the fusion protein (Figure 3), further analysis of regulation by uORFs focused on the effects of uORF2 in this study.
Cis effects of uORF2
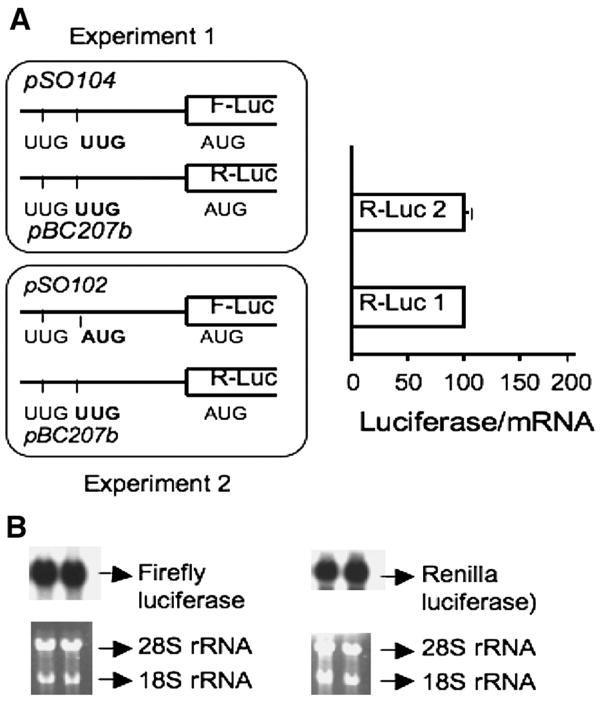
In principle, uORFs can regulate translation of the main ORF in trans, i.e. via a translated peptide product or in cis (in a sequence-dependent or independent fashion). To distinguish between these two possibilities, the following strategy was employed. Cos-1 cells were co-transfected with plasmids harboring two different reporter genes (firefly and Renilla luciferase, respectively) containing the MS 5′ leader sequence in which a functional uORF2 was either present in cis or trans (Figure 4).
Figure 4.
Cis effect of uORF2 on translation of the main ORF. Two reporter genes (firefly and Renilla luciferase respectively) were cotransfected using the strategy outlined in the figure. In experiment 1, expression of uORF2 is suppressed by mutation of uAUG2 to uUUG, whereas in experiment 2, a functional uORF2 is present upstream of the firefly luciferase reporter. F-luc and R-luc denote firefly luciferase and Renilla luciferase, respectively. R-Luc 1 and R-Luc 2 represent the relative Renilla luciferase activities of the extracts observed in experiments 1 and 2, respectively. R-Luc activity in the two experiments was comparable, indicating that the effect of uORF2 is in cis. B. Northern analysis demonstrates that mRNA levels were unaffected by the mutations introduced into the different constructs. Two different probes were used to detect the firefly and Renilla luciferase messages respectively as described under Methods and normalized against the density of 18S rRNA in each lane.
In the first cotransfection experiment, neither plasmid (pSO101 or pBC207b) had a functional uORF since both uAUG codons were mutated to UUG. In the second experiment, a functional uORF2 is present upstream of the firefly but not the Renilla luciferase. If the peptide encoded by uORF2 acts in trans, it is expected to inhibit translation of the Renilla luciferase resulting in lower activity of this reporter in experiment 2 versus 1. However, the presence of uORF2 in trans had no effect on Renilla luciferase activity. Consistent with cis regulation by uORF2, firefly luciferase activity was lower in experiment 2 containing a functional uORF2 versus in experiment 1 in which the leader sequence lacked uAUG2 (not shown).
Rare tandemly repeated arginine codons modulate uORF2-mediated translation inhibition
Analysis of codon usage in uORF2 revealed the presence of six rare codons. Of these, two represent the rarest codons for arginine (CGU) based on the frequency of their usage in the human transcriptome (16), and occur at codons 3 and 4, respectively. The remaining rare codons are found at positions 8, 19 and 25 (CCG encoding proline) and 16 (CGU encoding ariginine). The presence of rare Arg codons early in the encoded sequence is striking and suggested the possibility that they may cause ribosomes to stall at these positions during translation of uORF2. To test this hypothesis, the CGU-CGU sequence was substituted by synonymous mutations representing the most commonly used Arg codon in humans, CGC (construct pBC224a, Figure 5). In a second construct, the rare Arg codons (CGU-CGU) were substituted by common alanine codons (GCA-GCA) (construct pBC226d). Both the synonymous and missense mutations increased translation efficiency of the reporter gene ~3- to 4-fold (Figure 5). Alleviation of translation inhibition by both sets of mutations is consistent with a mechanism for “ribosome stalling” at the rare Arg codons in uORF2.
Figure 5.
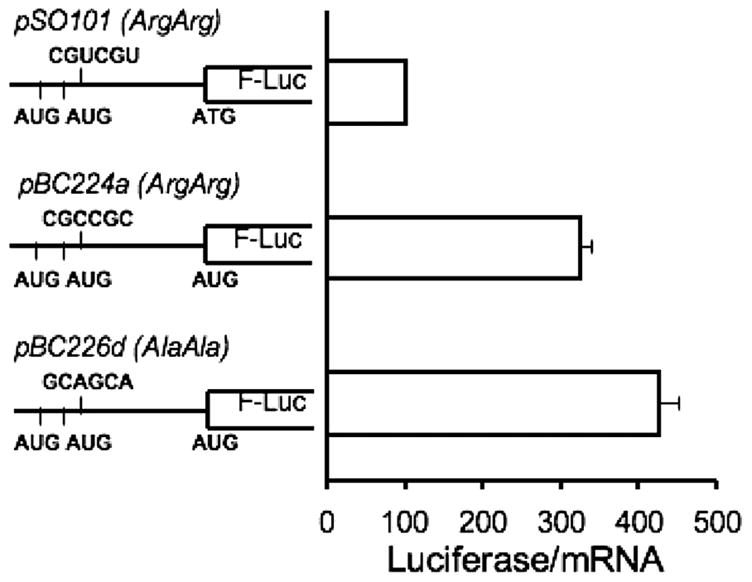
The effect of rare arginine codons on regulation by uORF2. The rare arginine codons (at positions 3 and 4) in uORF2 were either changed to common arginine codons (CGC-CGC in construct pBC224a) or to common alanine codons (GCA-GCA in construct pBC226d). The relative luciferase activities indicate that both the synonymous and the missense mutations of the rare arginine codons result in a 3- to 4-fold increase in translation of the downstream reporter.
DISCUSSION
Although uORFs are present in a minority of eukaryotic messages, they impact translation of the downstream ORFs in a wide range of organisms extending from viruses to fungi and vertebrates. It is estimated that uAUG codons are present in at least 3% of Saccharomyces cerevisiae mRNAs (17) and in <10% of vertebrate mRNAs (13). uORFs can exert their regulatory effects in a sequence-dependent or independent fashion. In the MS leader, both uAUGs reside in a suboptimal Kozak sequence context, an established determinant of translation efficiency in vertebrates (18). However, even relatively inefficient initiation at an uAUG codon can result in substantial inhibition of the main ORF translation since multiple factors viz. efficiency of translation elongation, termination, and the properties of the uORF-encoded peptide itself can influence downstream translation (14). The termination codon for the MS uORF2 is followed by a GC-rich sequence while that for uORF1 is A-rich. The sequence immediately following the termination codon of uORFs can have a bearing on the efficiency of ribosomal reinitiation at the downstream ORF as exemplified by the case of the yeast transcription factor, GCN4 (19).
The generally accepted experimental criterion for ascribing a regulatory role to uORFs is demonstrated modulation of expression of the downstream cistron following mutation of the uAUG codon. Mutation of the uAUGs in the MS 5′ leader sequence resulted in a 2- to 2.5-fold higher expression of the reporter luciferase activity without altering mRNA levels (Figure 2). Ablation of both uORFs by simultaneous mutation of the uAUGs resulted in a 3.5-fold higher activity of the downstream reporter. Together, these results support a regulatory role for the uORFs in MS.
In order to function as regulatory elements, uAUGs must be recognized by a scanning 40S ribosomal subunit and the associated factors in the initiation complex. To investigate the potential of the uAUGs in the MS mRNA to support translation initiation, the sequence of the MS 5′ leader was engineered to force translation of the downstream luciferase reporter starting at either uAUG1 or uAUG2 (Figure 3). In both instances, luciferase activity, albeit lower than that in the control nonfusion construct, was observed. These results are consistent with the ability of both uAUGs in the MS 5′ leader to support translation initiation. The efficacy of initiation at uAUG1 is apparently lower based on the lower activity of the uORF1-luciferase fusion versus the uORF2-luciferase reporter. This could be due to the closer proximity of uAUG1, which starts at position 12, to the 5′ end of the message. Translation of the uORF in S-adenosylmethionine decarboxylase is initiated 14 bases downstream from the 5-me7G cap (20). Extending the distance between the cap and uAUG to 47 nucleotides enhanced translation repression of the downstream cistron in nonlymphoid cells (21). Alternatively, the lower activity of the fusion protein generated by initiation at uAUG1 could be due to a greater inhibitory effect of the longer N-terminal extension on the luciferase.
In principle, the peptide product of a uORF, if produced, could exert its effect in cis or in trans. There are several examples of uORFs in which translation regulation is exerted in a sequence-dependent fashion and the resulting peptide interacts with the ribosome. To distinguish between a cis versus trans effect, we cotransfected cells with dual reporter constructs encoding the firefly and Renilla luciferases respectively downstream of the MS 5′ leader. We interrogated the ability of the putative product of the uORF furnished by the firefly luciferase construct to modulate translation of the Renilla luciferase one (Figure 4). However, no trans effects were seen in contrast to the observed cis effect of the uORF on the downstream firefly reporter. In instances where the mechanism by which the encoded peptide transduces the regulatory effects of uORFs have been studied, they appears to function in cis. Both elongation and termination phases are affected by uORF-encoded peptides and the peptidyltransferase is believed to be the likely target in some instances (14, 22).
The density of rare codons in uORF2 is striking and the role of a pair of tandemly repeated arginine codons in the mechanism of translation inhibition was investigated. Surprisingly both missense as well as synonymous mutations of the rare codons to common codons encoding alanine and arginine respectively resulted in complete ablation of uORF2 inhibition of the reporter luciferase (Figure 5). These results are consistent with cis effects and suggest that stalling of ribosomes at the rare codons impedes ribosomal scanning and diminishes loading at the downstream initiation codon. They also reveal that the sequence per se of the encoded peptide is not important. A caveat inherent to such experiments bears note, i.e. mutations can lead to changes in secondary and tertiary interactions in the RNA, which in turn could be responsible for the observed effects on translation. However, the correspondence in the magnitude of the observed effects between mutation of uAUG2 (Figure 2) and of the rare arginine codons (Figure 5) suggests that codon-specific changes are being registered on the efficiency of translation of the main ORF.
A uORF-encoded arginine attenuator peptide regulates translation of the carbamoyl phosphate synthetase small subunit gene in fungi albeit by a different mechanism (23). When arginine is in surplus, translation of this uORF reduces synthesis of the polypeptide from the main ORF by causing ribosomal stalling and reducing scanning-based initiation downstream. Ribosomal stalling at a tandem repeat of rare arginine codons and release of peptidyl tRNAArg has been reported for the lambda int gene as a mechanism for its translational regulation (24).
We have previously reported that translation initiation of the MS mRNA is mediated by a B12-responsive IRES element, which has been localized to a 70 bp region immediately upstream of the initiation codon (8, 12). Reporter constructs in which the MS 5′-UTR sequence was inserted upstream of a reporter gene, induced B12 responsiveness. Deletion of the first 172 bases of the MS 5′-UTR sequence resulted in higher reporter gene activity compared to the construct containing the full-length 394 bp 5′-UTR sequence (8). This is consistent with the inhibitory effect of the MS uORFs on translation initiation from a downstream AUG codon.
The unusually high prevalence of uORFs in transcripts of genes involved in controlling cellular growth suggests that they may play a general role in sensing translational capacity and thereby modulating cellular growth and division. Under conditions of amino acid deprivation or stress, global protein synthesis is downregulated by phosphorylation of eIF2, which inhibits GDP⇔GTP exchange. Under these conditions, a select population of mRNAs with built-in strategies for circumventing global arrest of protein synthesis, is translated and confers a survival advantage. A well-studied example of this is the yeast transcription activator, GCN4, which controls a large number of genes involved in amino acid biosynthesis. In glucose or amino acid starved cells, the reduced initiation efficiency at uORF4 alleviates reinitiation-dependent inhibition of translation at the main ORF (25). A second example is the cationic amino acid transporter, Cat-1, in which elegant studies have revealed the complex and dynamic interplay between uORF and IRES elements (26). Under conditions of amino acid starvation, translation of the uORF leads to mRNA remodeling and unmasking of the IRES-element, which in turn, promotes translation of the main ORF. MS contributes to homeostasis of methionine, an essential amino acid that initiates translation of all mRNAs and the potential role of methionine-dependent modulation of uORF function warrants investigation.
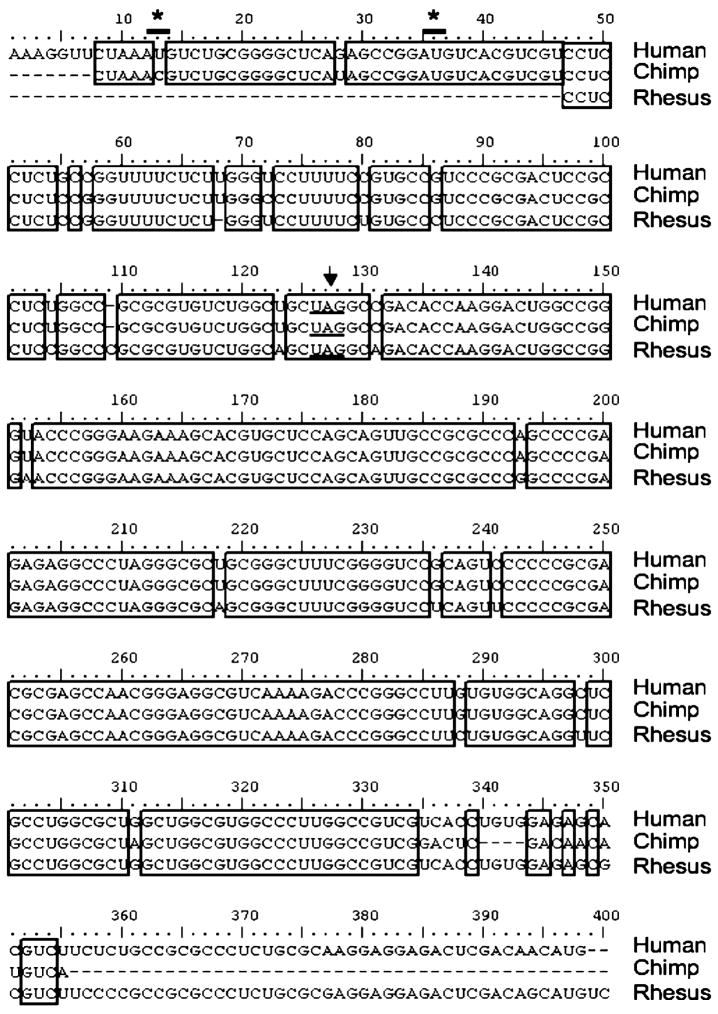
The evolution of uORF-based regulation of MS appears to be relatively recent and restricted to primates (Figure 6). Comparison of the human MS leader sequence with all available DNA sequences identified homology with only the chimp and Rhesus monkey leader sequences. Unfortunately, the first 46 residues where the uAUG codons if present would be located, are missing in the reported monkey sequence, which limits inferences that can be drawn. uAUG2 is present in the chimp sequence as is the termination codon for ORF2 in both chimp and monkey sequences, and strongly suggests the existence of ORF2 in all three organisms. In contrast, the human uAUG1 codon corresponds to ACG in chimp, encoding threonine. However, given the high level of sequence identity between the chimp and human sequences immediately following human uAUG1, the possibility of a sequencing error at this position needs to be considered. Based on the very high degree of conservation in the MS 5′ leader sequences of human, chimp and monkey, which is unexpected for a noncoding leader sequence, we speculate that all three organisms exhibit a similar mode of uORF-dependent translational regulation.
Figure 6.
Alignment of the 5′ MS leader sequence from human, chimp and Rhesus monkey. The DNA sequences for the alignment were obtained from the UCSC Genome Browser (27) using the BLAT alignment tool (28) (http://genome.ucsc.edu/cgi-bin/hgBlat) using the 5′ leader of human MS as the query sequence. Only two significant matches were obtained, both in primates. It should be noted that the BLAST search at the NCBI site (http://www.ncbi.nlm.nih.gov/BLAST/) did not pick up any significant matches in any genomes except the human genome. The three RNA sequences were aligned using Clustal W Multiple Alignment. Identical bases are boxed and the locations of uAUG1 and uAUG2 in the human sequence are shown by asterices while the termination codon for uORF2 is underlined.
Footnotes
This work was supported by grants from the National Institutes of Health (DK64959) and by a predoctoral fellowship from the American Heart Association from the Heartland Affiliate (to S.O).
Abrreviations Used: MS, methionine synthase; ORF, open reading frame; UTR, untranslated region; uORF, upstream open reading frame; uAUG1 and 2, initiation codons for uORFs 1 and 2; IRES, internal ribosome entry site.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zou CG, Banerjee R. Homocysteine and Redox Signaling. Antiox Redox Signal. 2005;7:547–559. doi: 10.1089/ars.2005.7.547. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein JD. Methionine metabolism in mammals. J Nutr Biochem. 1990;1:228–237. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- 3.Swanson DA, Liu ML, Baker PJ, Garrett L, Stitzel M, Wu J, Harris M, Banerjee R, Shane B, Brody LC. Targeted Disruption of the Methionine Synthase Gene in Mice. Mol Cell Biol. 2001;21:1058–1065. doi: 10.1128/MCB.21.4.1058-1065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boushey CJ, Beresford SA, Omenn GS, Motulsky AG. A quantitative assesment of plasma homocysteine as a risk factor for vascular disease. Probable benefits of increasing folic acid intakes. JAMA. 1995;274:1049–1057. doi: 10.1001/jama.1995.03530130055028. [DOI] [PubMed] [Google Scholar]
- 5.Mills JL, McPartlin JM, Kirke PN, Lee YJ, Conle MR, Weir DG. Homocysteine metabolism in pregnancies complicated by neural tube defects. Lancet. 1995;345:149–151. doi: 10.1016/s0140-6736(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 6.Clarke R, Smith AD, Jobst KA, Refsum H, Sutton L, Ueland PM. Folate, vitmain B12, and serum total homocysteine levels in confirmed Alzheimers disease. Arch Neruol. 1998;55:1449–1455. doi: 10.1001/archneur.55.11.1449. [DOI] [PubMed] [Google Scholar]
- 7.Hershey JWB, Merrick WC. The pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. New York: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- 8.Oltean S, Banerjee R. Translational regulation of human methionine synthase by vitamin B12. J Biol Chem. 2003;278:20778–20784. doi: 10.1074/jbc.M300845200. [DOI] [PubMed] [Google Scholar]
- 9.Mangum JH, North JA. Vitamin B12-dependent methionine biosynthesis in HEp-2 cells. Biochem Biophys Res Commun. 1968;32:105–110. doi: 10.1016/0006-291x(68)90433-6. [DOI] [PubMed] [Google Scholar]
- 10.Mangum JH, Murray BK, North JA. Vitamin B12 dependent methionine biosynthesis in cultured mammalian cells. Biochemistry. 1969;8:3496–3499. doi: 10.1021/bi00837a002. [DOI] [PubMed] [Google Scholar]
- 11.Gulati S, Brody LC, Banerjee R. Post transcriptional regulation of mammalian methionine synthase by B12. BBRC. 1999;259:436–442. doi: 10.1006/bbrc.1999.0696. [DOI] [PubMed] [Google Scholar]
- 12.Oltean S, Banerjee R. A B12-responsive IRES element in human methionine synthase. J Biol Chem. 2005;280:32662–8. doi: 10.1074/jbc.M501964200. [DOI] [PubMed] [Google Scholar]
- 13.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–48. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–42. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris DR. Growth control of translation in mammalian cells. Prog Nucleic Acid Res Mol Biol. 1995;51:339–63. doi: 10.1016/s0079-6603(08)60883-1. [DOI] [PubMed] [Google Scholar]
- 16.Lavner Y, Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene. 2005;345:127–38. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 17.Vilela C, Linz B, Rodrigues-Pousada C, McCarthy JE. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 1998;26:1150–9. doi: 10.1093/nar/26.5.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 19.Hinnesbusch AG. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Tanslational Control. New York: Cold Spring Harbor Press; 1996. pp. 199–244. [Google Scholar]
- 20.Hill JR, Morris DR. Cell-specific translation of S-adenosylmethionine decarboxylase mRNA. Regulation by the 5′ transcript leader. J Biol Chem. 1992;267:21886–93. [PubMed] [Google Scholar]
- 21.Ruan H, Hill JR, Fatemie-Nainie S, Morris DR. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Influence of the structure of the 5′ transcript leader on regulation by the upstream open reading frame. J Biol Chem. 1994;269:17905–10. [PubMed] [Google Scholar]
- 22.Geballe AP, Sachs MS. Translational control by upstream open reading frames. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 2000. pp. 595–614. [Google Scholar]
- 23.Wang Z, Sachs MS. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–13. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olivares-Trejo JJ, Bueno-Martinez JG, Guarneros G, Hernandez-Sanchez J. The pair of arginine codons AGA AGG close to the initiation codon of the lambda int gene inhibits cell growth and protein synthesis by accumulating peptidyl-tRNAArg4. Mol Microbiol. 2003;49:1043–9. doi: 10.1046/j.1365-2958.2003.03611.x. [DOI] [PubMed] [Google Scholar]
- 25.Gaba A, Wang Z, Krishnamoorthy T, Hinnebusch AG, Sachs MS. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. Embo J. 2001;20:6453–63. doi: 10.1093/emboj/20.22.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE, Zhou L, Snider MD, Scheuner D, Kaufman RJ, Hatzoglou M. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–31. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- 27.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]