Abstract
Otubain 1 belongs to the ovarian tumor (OTU) domain class of cysteine protease deubiquitinating enzymes. We show here that human otubain 1 (hOtu1) is highly linkage-specific, cleaving Lys48 (K48)-linked polyubiquitin but not K63-, K29-, K6-, or K11-linked polyubiquitin, or linear α-linked polyubiquitin. Cleavage is not limited to either end of a polyubiquitin chain, and both free and substrate-linked polyubiquitin are disassembled. Intriguingly, cleavage of K48-diubiquitin by hOtu1 can be inhibited by diubiquitins of various linkage types, as well as by monoubiquitin. NMR studies and activity assays suggest that both the proximal and distal units of K48-diubiquitin bind to hOtu1. Reaction of Cys23 with ubiquitin-vinylsulfone identified a ubiquitin binding site that is distinct from the active site, which includes Cys91. Occupancy of the active site is needed to enable tight binding to the second site. We propose that distinct binding sites for the ubiquitins on either side of the scissile bond allow hOtu1 to discriminate among different isopeptide linkages in polyubiquitin substrates. Bidentate binding may be a general strategy used to achieve linkage-specific deubiquitination.
Keywords: deubiquitination, isopeptide, linkage specificity, otubain, polyubiquitin
Introduction
Covalent modification of proteins by ubiquitin (Ub) is used to signal protein degradation by the 26S proteasome and is also essential for many proteasome-independent aspects of cellular regulation 1; 2; 3. Ub is conjugated to a protein substrate in a series of enzymatic steps, usually resulting in an isopeptide bond between the C-terminal α-carboxylate of Ub and the ε-amine of a lysine within the protein substrate. Ub itself can be modified in this way at one of its seven lysine residues, giving rise to a polyubiquitin (polyUb) chain. All seven types of polyUb linkages, which are distinguished by isopeptide linkages with Ub residues K48, K63, K33, K29, K27, K6, or K11, have been identified in vivo 4; 5. The fate of ubiquitinated substrates depends, in part, on the length and linkage-type of the polyUb chain. Substrates modified with K48-linked polyUb of n ≥ 4 usually are targeted for degradation by the 26S proteasome 6, whereas K63-linked polyUb plays a non-degradative role in DNA damage tolerance, NF-κB activation, and other pathways 2; 7.
Ubiquitination is reversible. Deubiquitinating enzymes (DUBs) can disassemble polyUb chains or cleave Ub (or polyUb) from conjugated proteins, as well as non-protein adducts. Deubiquitination plays important roles in regulating Ub-dependent pathways including processing of Ub precursors, editing of polyUb chains, release of proteins from (poly)Ub conjugates, and recycling of Ub 8; 9; 10; 11. Individually, many, and possibly most, DUBs regulate a limited number of proteins and pathways, which suggests that they target specific substrates 11. However, few specific substrates have been identified 12; 13. Current evidence also suggests that the specificities of some DUBs are modulated in vivo by their interactions with scaffolds or adaptor proteins 14; 15; 16; 17; 18. Six classes of deubiquitinating enzymes have been identified11; 19: the Ub C-terminal hydrolases (UCHs), Ub specific proteases (UBPs), ovarian tumor proteases (OTUs) 20, Machado-Joseph disease protein domain proteases (MJDs) 21, JAMM/MPN+ motif proteases 22; 23, and viral DUBs 24. With the exception of the JAMM/MPN+ DUBs, which are Zn2+-metalloproteases, all known DUBs are cysteine proteases.
Of the more than 100 OTU-domain proteins identified to date, the otubains 18; 25; 26 are among only a few shown to have DUB activity in vitro or in vivo 17; 26; 27; 28; 29; 30; 31; 32; 33. Otubain proteins from different species share a high degree of sequence homology (Supplementary Figure 1A). Human otubain 1 (hOtu1) has been reported to cleave isopeptide-linked tetraUb 26 but not diUb 33. Human otubain 2, on the other hand, is inactive in vitro against peptide and isopeptide-linked substrates 26, but cleaves Ub-AMC 29.
Two isoforms of hOtu1 were found to regulate T cell anergy by forming a complex with GRAIL1, an E3 ligase 25. hOtu1 affects both GRAIL1 expression and GRAIL-mediated ubiquitination. The yeast homolog of otubain 1, Otu1, recently was reported to bind to Cdc48, a chaperone-like AAA ATPase. Otu1 binding to Cdc48 counteracts the association of Cdc48 with Ufd2, an “E4” Ub ligase that extends polyUb chains on conjugates 18. In addition, the transcription factor Spt23 was identified as an Otu1 substrate in vivo 18. In vitro, yeast Otu1 can cleave Ub from Ub-AMC and disassemble K48-linked, but not K29- or K63-linked, polyUb 18; 34.
Other OTU proteins known to have DUB activity are Cezanne (cellular zinc finger anti-NF-κB), TRABID, A20, VCIP135, DUBA, and viral OTU-containing proteases (Supplementary Figure 1B). Overall, the substrate specificities of these DUBs appear to be very diverse, although in no case has polyUb linkage specificity been explored in detail. A20 removes K63-linked polyUb from proteins in vivo, but A20 or its N-terminal OTU domain can cleave both K48 and K63 linked polyUb chains in vitro 28; 31; 35. The Cezanne catalytic domain will cleave Ub-AMC and linear or branched (K48 or K63-linked) polyUb 27; 31. VCIP135 cleaves K48-linked polyUb in vitro 30. DUBA selectively cleaves K63-linked polyUb on TRAF3 17. A viral OTU-containing protease cleaves Ub and ISG15 from cellular target proteins 32.
We report here a detailed study of the specificity of hOtu1. We find that hOtu1 cleaves only K48 linkages, can disassemble K48-linked chains that are conjugated to substrate proteins, and can remove both proximal and distal ubiquitins from an unattached polyUb chain. In contrast to its highly specific cleavage of K48 isopeptide linkages, hOtu1 binds equally well to a variety of polyUb chains of different linkage types. NMR chemical shift perturbation studies and activity assays with mutant diUbs indicate that the proximal and distal Ubs bind to separate sites on hOtu1. Additional evidence for a proximal Ub binding site was obtained by affinity-labeling of the hOtu1-ubiquitin-aldehyde (Ubal) complex with ubiquitin-vinylsulfone (UbVS). Our results suggest how otubain 1 is able to recognize K48-linked polyUb selectively; moreover, our model offers a general strategy whereby different types of polyUb chains might be distinguished in the cell for disassembly by specific DUBs.
Results
Otubain 1 specifically cleaves K48-linked polyUb
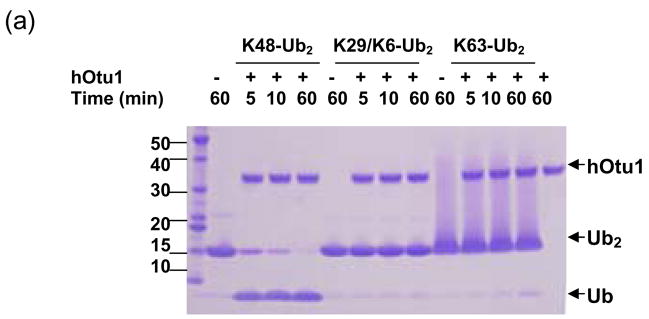
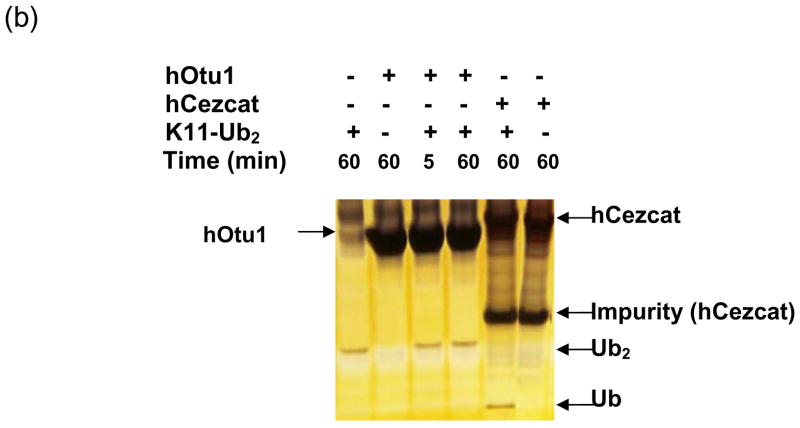
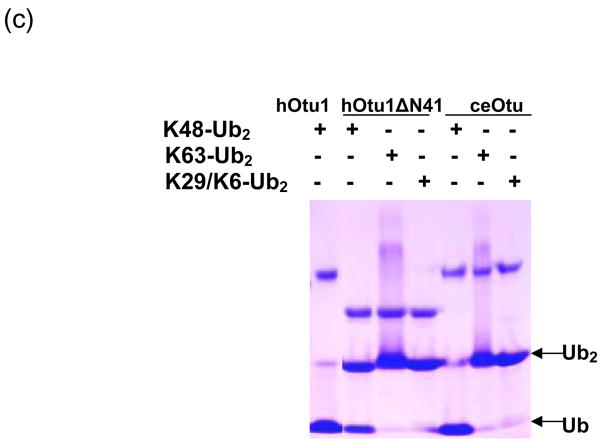
hOtu1 has been reported to cleave tetraUb but not LRGG-AMC or Ub fused to GFP 26. A yeast otubain protein, Otu1, cleaves Ub-AMC and K48-linked polyUb chains but not K63-linked chains 18. A recent paper also showed preference of hOtu1 for cleavage of K48-linked over K63-linked tetraUb 33. We suspected that hOtu1 could be a highly selective isopeptidase and therefore further investigated its specificity. hOtu1 was cloned from a cDNA library, and the recombinant protein was expressed in E. coli and purified for biochemical studies. We first examined the ability of hOtu1 to cleave forms of diUb (Ub2) that differ in their isopeptide linkages. Among the five Ub2 substrates that we tested, only K48-Ub2 was processed significantly. hOtu1 readily cleaved K48-Ub2, but negligible K63-Ub2 and no K29 or K6-linked Ub2 were hydrolyzed (Figure 1a). The same results were obtained when 10-fold more hOtu1 was used, or when the incubations were extended to 20 h (data not shown). Similarly, hOtu1 could not cleave K11-Ub2 (Figure 1b), further indicating that the isopeptidase activity is highly specific for K48 linkages. We also tested the specificity of hOtu1 lacking the N-terminal 41 residues (hOtu1ΔN41), as well as a putative full-length otubain from C. elegans (ceOtu). Our assays show that ceOtu and hOtu1ΔN41 share the same specificity for K48-linked polyUb as hOtu1, although the full-length hOtu1 and ceOtu appear to cleave more efficiently than the truncated hOtu1ΔN41 (Figure 3). These results indicate that the determinants of K48 cleavage specificity lie within the conserved enzymatic domain (Figure 1c).
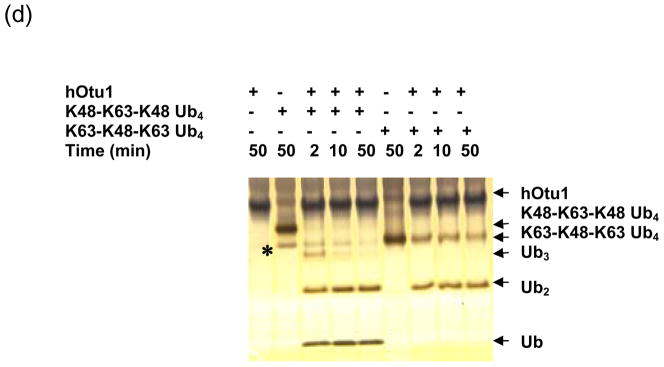
Figure 1. Otubain 1 cleaves K48-linked isopeptide bonds in polyubiquitin chains specifically.
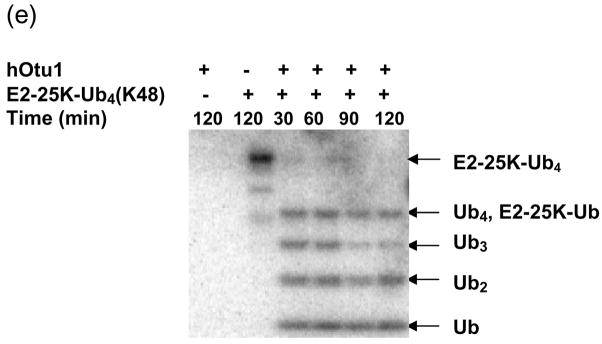
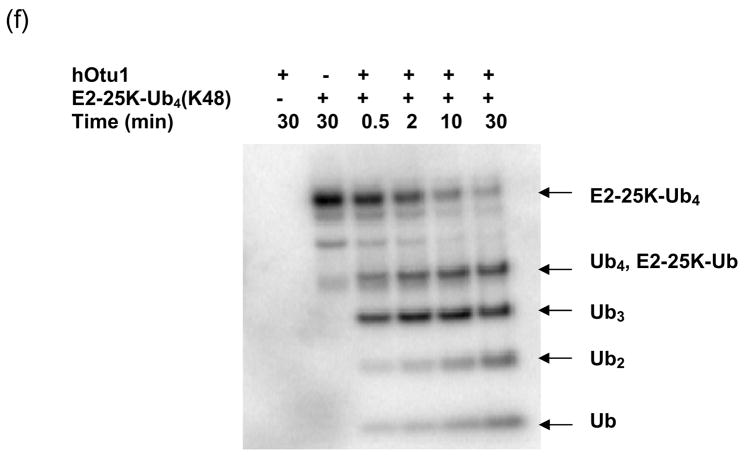
(a) Recombinant full-length hOtu1 cleaves K48-Ub2 but not K63-, K29-, or K6-Ub2. hOtu1 (1 μg) was incubated with 5 μg of K48-Ub2, K63-Ub2, or mixed K29/K6-Ub2 (see Materials and Methods). Proteins were detected after SDS-PAGE with Coomassie Blue. (b) hOtu1 does not cleave K11-Ub2. The reaction mixtures included 5 μg of hOtu1 or 5 μg of recombinant Cezanne catalytic domain (hCezcat, 125–455 a. a.), and 0.5 μg of K11-Ub2. hCezcat has a major impurity band at ~20 kDa. Proteins were detected after SDS-PAGE by silver-staining. (c) Linkage specificity of polyubiquitin chain cleavage by hOtu1ΔN41 and ceOtu. Samples were resolved by SDS-PAGE followed by staining with Coomassie Blue. Recombinant human otubain 1 fragment hOtu1ΔN41 (i.e., hOtu1 lacking the N-terminal 41 residues) and putative C. elegans otubain (ceOtu) cleave K48-Ub2 but not K63-Ub2 or K29/K6-Ub2. In each 10 μl reaction, 1 μg enzyme and 5 μg of substrate were used. (d) hOtu1 selectively cleaves K48-linked isopeptide bonds in mixed-linkage Ub4 chains Ub-K48-Ub-K63-Ub-K48-Ub and Ub-K63-Ub-K48-Ub-K63-Ub. hOtu1 (0.2 μg) and 0.05 μg of Ub4 were used. An asterisk marks an impurity in Ub-K48-Ub-K63-Ub-K48-Ub, which is a cyclic form of the tetraubiquitin. (e) hOtu1 cleaves isopeptide bonds in E2-25 kDa-(K48-linked)Ub4 but not isopeptide bonds in Ubc13-(K63-linked)Ub4. 125I labeled E2-25K-(K48-linked)Ub4 and Ubc13-(K63-linked)Ub4 were used (see Materials and Methods). There are contaminant bands in E2-25K-(K48-linked)Ub4 that migrated between E2-25K-(K48-linked)Ub4 and Ub4 or monoUb-E2-25K. (f) The proximal Ub in E2-25K-(K48-linked)Ub4 is slowly removed by hOtu1. 125I labeled E2-25K-(K48-linked)Ub4 was incubated with hOtu1 as in 1e for the times indicated.
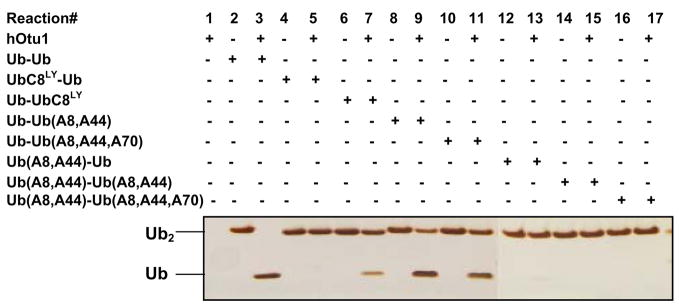
Figure 3. Both proximal and distal ubiquitins of K48-Ub2 bind to otubain 1.
Mutations in K48-Ub2 affect its cleavage by hOtu1. In a 10 μl reaction, 1.0 μg of wild-type or mutant K48-Ub2 was used in the deubiquitination assay containing 1.0 μg hOtu1 (see Materials and Methods for details). The reactions were for 30 min. Protein bands after SDS-PAGE were detected by silver staining.
We then asked if hOtu1 could selectively cut K48-linked Ub2 in the context of polyUb chains with mixed linkages. TetraUb chains having either K48-K63-K48 or K63-K48-K63 isopeptide linkages were incubated with hOtu1 (Figure 1d). In each case, the products obtained were consistent with cleavage at the K48 linkage(s) only; processing of K63-K48-K63 tetraUb yielded only diUb, whereas K48-K63-K48 tetraUb was cleaved into triUb, diUb, and Ub monomers. The K48-K63-K48 tetraUb substrate contained an additional and unexpected faster-migrating band (asterisk in Figure 1d). This species is a cyclized side-product of the K48-K63-K48 tetraUb synthesis (EMC and REC, unpublished) that, in analogy with cyclic K48-linked chains 36, can form in vitro. hOtu1 could cleave both the linear and cyclized forms of K48-K63-K48 tetraUb.
The high K48-linkage selectivity of hOtu1 on free polyUb chains led us to investigate deubiquitination of polyUb-protein conjugates. We tested the activity of hOtu1 against a polyUb-protein conjugate, E2-25K-(K48-linked)Ub4. Using a strategy similar to that of Liu et al.37, we synthesized E2-25K-(K48-linked)Ub4 in which the Ub-conjugating (E2) enzyme E2-25K was self-ubiquitinated with a preformed K48-linked Ub4 chain. As shown in Figure 1e, hOtu1 cleaved E2-25K-(K48-linked)Ub4 at each Ub-Ub bond, with Ub3, Ub2 and Ub produced. In addition, a protein band was generated that corresponds to ~34 kDa, the size of either free Ub4 or monoUb-E2-25K. Even with prolonged incubation, this species persisted longer than Ub3, which suggested that at least some and perhaps all of it was monoUb-E2-25K. We identified this product as monoUb-E2-25K by its retention by Q-Sepharose resin at high salt, at conditions where free Ub or polyUb does not bind (data not shown). At short incubations (i.e., 0.5–10 min; Figure 1f), the Ub4/monoUb-E2-25K band was much weaker than the Ub3 band. Therefore, cleavage at the proximal Ub to release Ub4 from the E2-25K, if it occurred at all, must have been much slower than cleavage at Ub-Ub linkages.
Otubain 1 can cleave at either end of a K48-linked polyUb chain
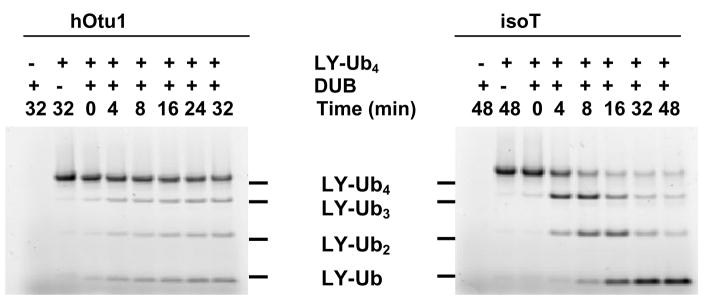
Some DUBs release Ub preferentially from distal ends of free or protein-conjugated polyUb chains 38; 39. Other DUBs reverse Ub conjugation by cleaving the isopeptide bond that links the proximal Ub of polyUb to a protein 22 or disassemble free chains by releasing monomers from the proximal end 40. To see whether hOtu1 cleavage of K48-linked polyUb proceeds from the distal or proximal end, we used as a substrate K48-linked Ub4 labeled with a Lucifer Yellow fluorophore on the distal Ub (UbC66LY-Ub3) 39. When hOtu1 was incubated with UbC66LY-Ub3, fluorescently-labeled products UbC66LY-Ub2, UbC66LY-Ub, and UbC66LY appeared simultaneously (Figure 2). In contrast, when the proximal end-specific DUB isopeptidase T 40 was used with the same substrate, mostly UbC66LY-Ub2 was generated first; UbC66LY-Ub and UbC66LY appeared only as the reaction progressed further. Thus, unlike isopeptidase T, cleavage by hOtu1 is not specific for either end of a polyUb chain. Instead, hOtu1 recognizes and cleaves wherever there is a Ub-Ub K48 isopeptide linkage.
Figure 2. Otubain 1 cleaves K48-linked polyubiquitin chains at both ends.
UbC48LY-Ub3, with the fluorophore Lucifer Yellow attached to the distal end of the tetraubiquitin, was incubated with the hOtu1 or isopeptidase T (isoT) deubiquitinating enzymes; see Materials and Methods for details.
The OTU domain proteins or fragments do not cleave α-linked peptide bonds
Information as to whether OTU domain proteins can act on non-isopeptide bonds is limited 18; 26; 27; 29. hOtu1 has been reported to cleave Ub-AMC 18 but not LRGG-AMC 26. We examined the activity of OTU domain proteins against pentaUb connected by α-amide linkages between the Ub C-termini and the N-termini of each succeeding Ub (i.e., linear Ub5). When incubated with hOtu1, ceOtu, or hOtu1ΔN41, the linear Ub5 was not cleaved (Supplementary Figure S2). In contrast, linear Ub5 was processed, albeit slowly 40, by isopeptidase T, which was included as a positive control in the assay. These data contradict a report 27 that human Cezanne catalytic domain can cleave linear polyUb.
PolyUb chains of different linkage types bind to hOtu1
We asked whether the strong preference by hOtu1 for cleaving within K48-linked polyUb is due to selective binding to a K48-Ub2 unit. Inhibition of the hOtu1 cleavage reaction by K48-Ub2, K63-Ub2, K29/K6-Ub2, or monoUb was examined in assays using radioiolabeled K48-linked Ub2 as the substrate (Supplementary Figure S3). Surprisingly, monoUb and each of the diUb species inhibited the reaction. Moreover, the extent of inhibition by these different forms of Ub was similar in all cases. Thus, in contrast to the cleavage reaction, binding of hOtu1 to substrate is not specific for a particular type of Ub-Ub isopeptide linkage. The cleavage specificity of hOtu1 for K48 linkages therefore is not determined simply by selective binding of the enzyme to K48-linked Ub-Ub.
hOtu1 binds to the proximal and distal ubiquitins of K48-Ub2 simultaneously
At least two possible mechanisms can account for both the high cleavage specificity of otubain 1 and the ability of various types of diUb, as well as monoUb, to inhibit the reaction. One is that otubain 1 specificity could be at the level of catalysis rather than initial selective substrate binding. Another is that otubain 1 contains a pair of monoUb binding sites, and that the proximal and distal ubiquitins flanking a K48 linkage each binds to a distinct site in the vicinity of the enzyme active site. According to this bidentate binding model, monoUb or different forms of diUb could occupy one or the other site and thereby inhibit the reaction, but only K48-linked Ub2 would bind in an orientation that positions the isopeptide linkage in the active site for cleavage.
To see if there are two binding sites in hOtu1, we first tested whether mutations in either the proximal or distal Ub of K48-Ub2 affect the cleavage by hOtu1. K48-Ub2 was synthesized as UbR48-UbD77 (i.e., distal UbR48 and proximal UbD77). Leu8, Ile44, or Val70 in either the proximal Ub (UbD77), the distal Ub (UbR48), or both were mutated. These residues comprise a hydrophobic patch on the Ub surface that is critical for most non-covalent Ub-protein interactions 41; 42. When Leu8 alone was mutated, it was changed to cysteine followed by alkylation with Lucifer Yellow-iodoacetamide (i.e., UbC8LY) to disrupt interactions at the hydrophobic patch. When two or three residues were mutated simultaneously, each was changed to Ala. As shown in Figure 3, the seven K48-Ub2 mutants were cleaved by hOtu1 far less efficiently than the wild-type K48-Ub2. In addition, the mutations in the distal Ub of K48-Ub2 affected the cleavage more significantly. Whereas UbC8LY-Ub remained uncut, hOtu1 was still able to cleave a small amount of Ub-UbC8LY. These results suggest that hOtu1 binds to both the proximal and the distal ubiquitins of K48-Ub2, most likely at different binding sites.
To examine further the binding between hOtu1 and K48-Ub2, we measured the NMR chemical shift perturbations that resulted from titration of catalytically-inactive hOtu1C91S into 15N-labeled K48-Ub2. The Ub2 was labeled uniformly on either the proximal or the distal Ub; titration into 15N-labeled monoUb was done for comparison. As shown in Figure 4, hOtu1C91S interacts with both the proximal and the distal Ub units in K48-Ub2. Like the interaction between hOtu1C91S and monoUb, the hydrophobic patches in both the proximal and distal ubiquitins in K48-Ub2 are involved.
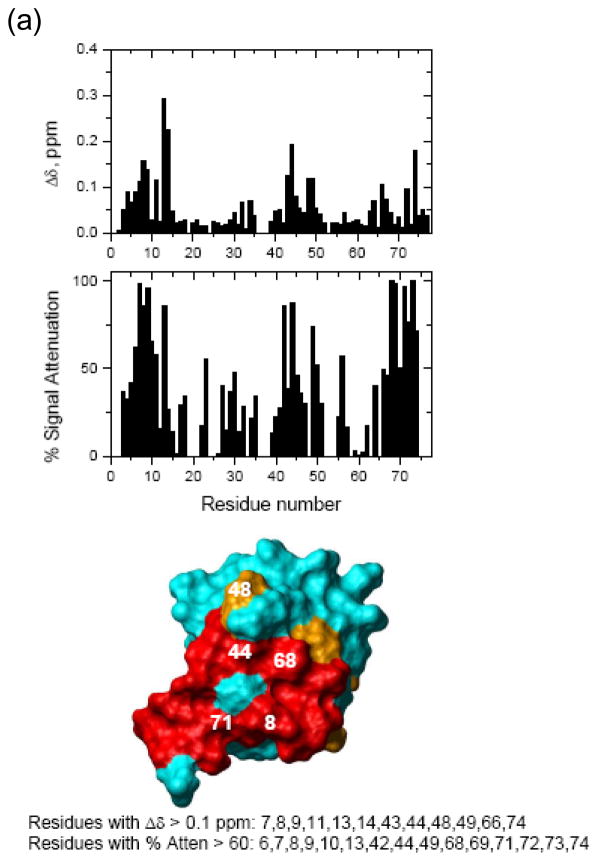
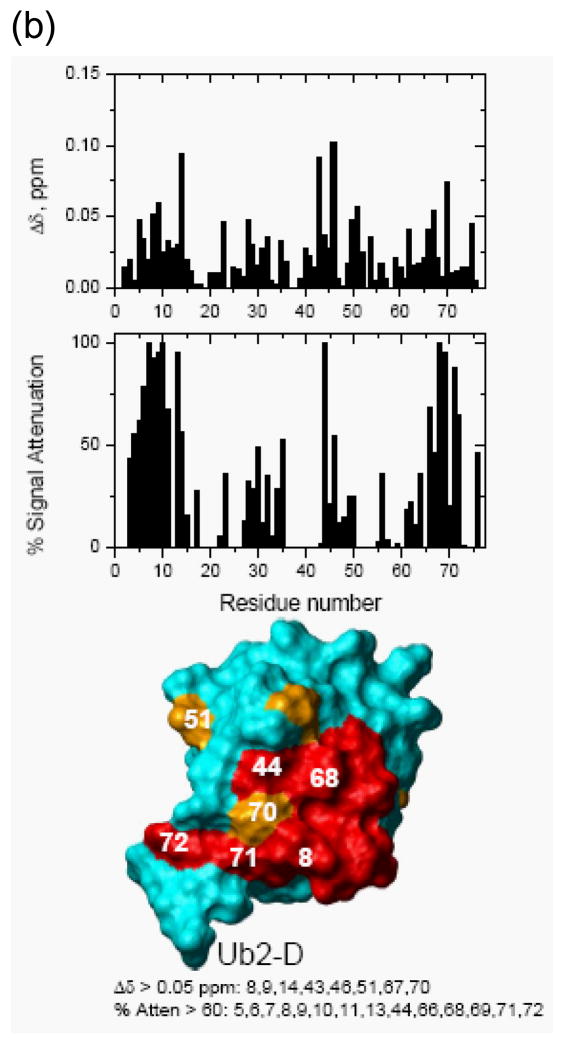
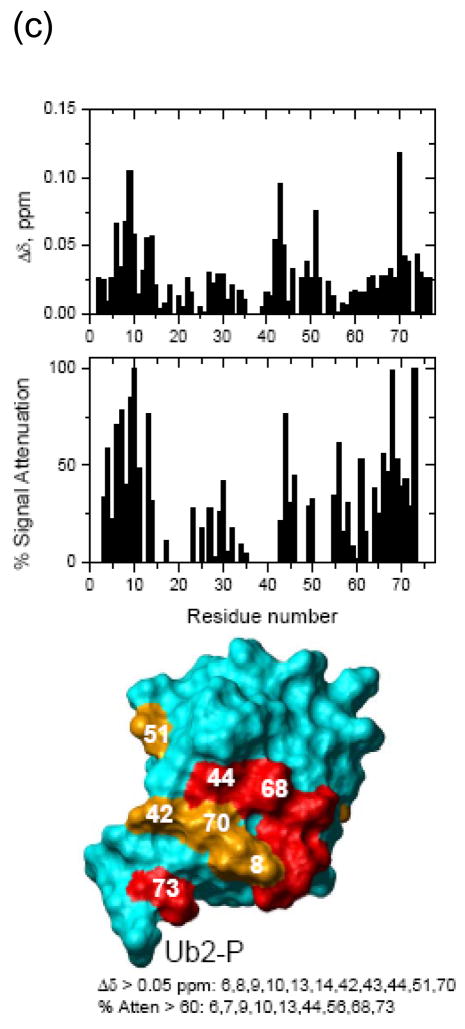
Figure 4. NMR chemical shift perturbation maps of the hOtu1-binding interface on the two Ub units in K48-Ub2.
NMR mapping revealed the hOtu1-binding interface on (a) monoUb, (b) distal Ub of K48-Ub2, and (c) proximal Ub of K48-Ub2. The upper panels show chemical shift perturbations as a function of residue number, middle panels show signal attenuations at the endpoint of titration as a function of residue number, and bottom panels show cartoon representations of the surfaces of monoUb, and the distal (Ub2-D) and proximal (Ub2-P) ubiquitins of K48-Ub2. Perturbed residues (listed beneath the drawings) are colored orange (Δδ> 0.05 ppm) and red (% attenuation > 60%); red color is also used for those residues that showed both perturbations. Note that the signal corresponding to the amide group in the isopeptide bond was strongly attenuated and disappeared upon titration with hOtu1.
We also compared shifts in the peak positions in Ub2 observed upon addition of hOtu1C91S with the shifts in the same residues that result from opening of the Ub/Ub interface in free K48-Ub2 43; the latter can be represented by the difference in peak positions between Ub2 and monoUb. The interface opening and ligand binding are distinct events and are expected to result in different directions of the shifts in amide 15N resonances. Our data indicate that the perturbations in both Ub units in Ub2 are a result of direct hOtu1C91S binding. Thus, we can exclude the possibility that only one specific Ub unit of K48-Ub2 binds hOtu1C91S and that the perturbations observed in the other Ub are due to the opening of the Ub/Ub interface that accompanies (and is required for) hOtu1C91S binding. The chemical shift and attenuation data for each residue are included in Figure 4.
To probe further the binding between hOtu1 and K48-Ub2, we measured the substrate concentration dependence of the cleavage rate. Velocities of K48-Ub2 cleavage by hOtu1 were determined under steady-state conditions and fit to Michaelis-Menten kinetics (Supplementary Figure S4). The Km was determined to be 78 ± 15 μM, which suggests that binding between hOtu1 and K48-Ub2 is relatively weak and is consistent with the results of the NMR titrations.
Evidence for the hOtu1 proximal Ub-binding site from affinity-labeling with ubiquitin-vinylsulfone
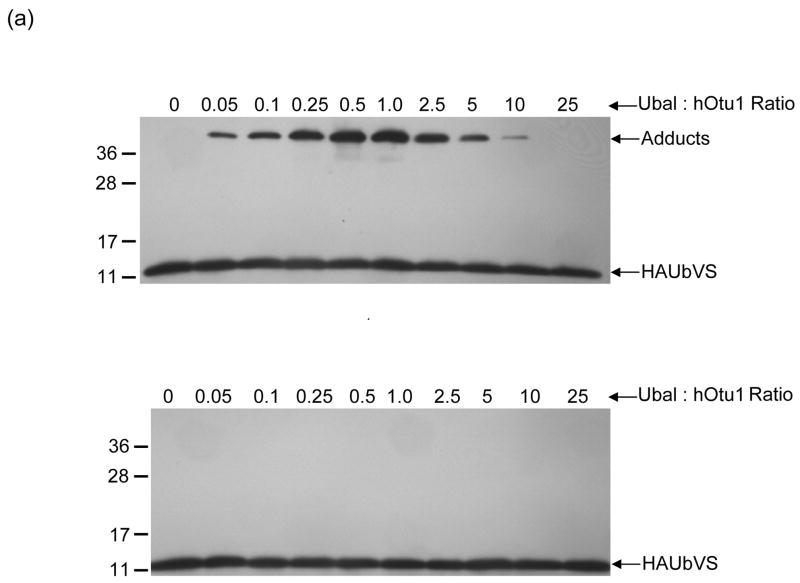
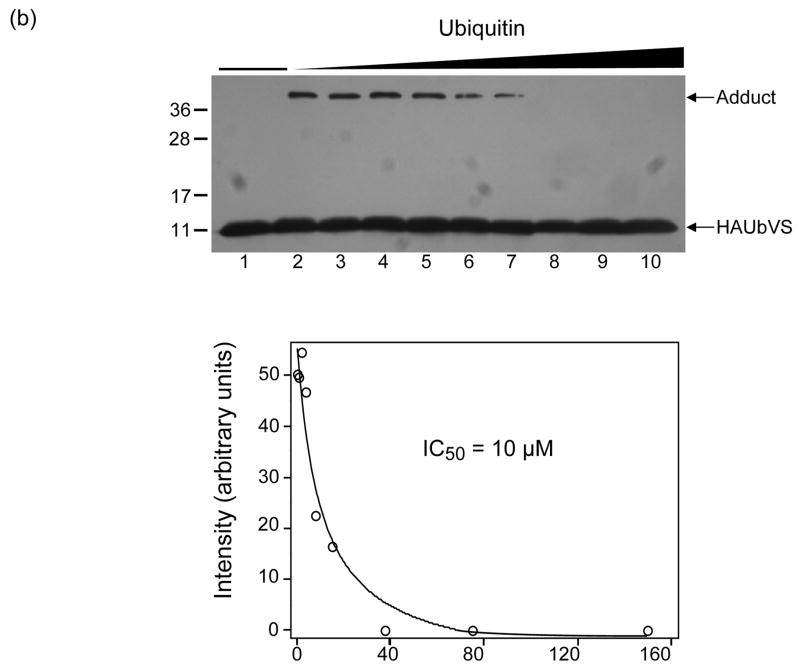
Ubiquitin-aldehyde (Ubal) and ubiquitin-vinylsulfone (UbVS) are generally regarded as mechanism-based DUB inhibitors that react with the active site cysteine of a susceptible DUB to form, respectively, a reversible thiohemiacetal or an irreversible thioether adduct. hOtu1 has been reported to be inhibited strongly by Ubal 26 but insensitive to UbVS 44; we have confirmed these results (data not shown). Therefore, we were surprised to discover that hOtu1 could rapidly form an adduct with UbVS if Ubal was included in the reaction (Figure 5a, upper panel). Notably, the yield of this adduct increased with increasing Ubal until the molar ratio of Ubal:hOtu1 reached ~1, whereupon it decreased progressively.
Figure 5. Affinity-labeling with UbVS reveals a second Ub-binding site on hOtu1.
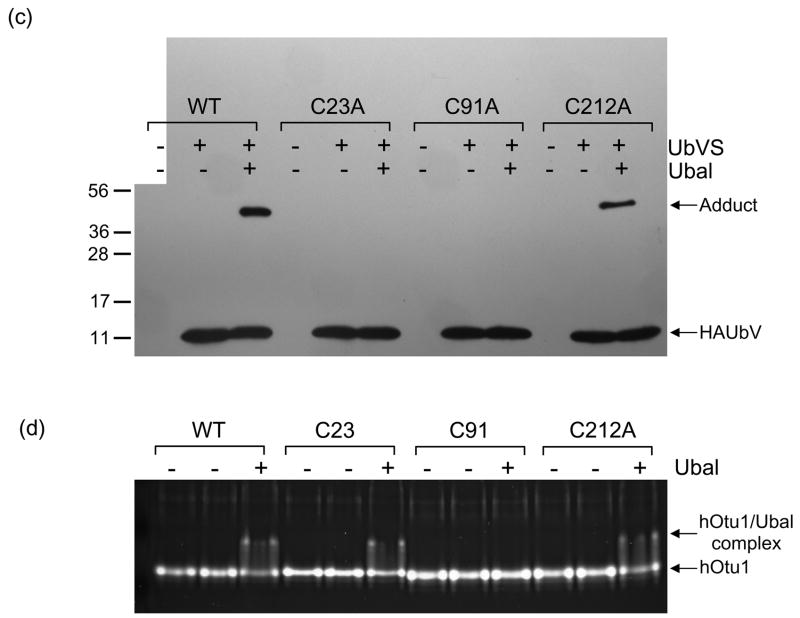
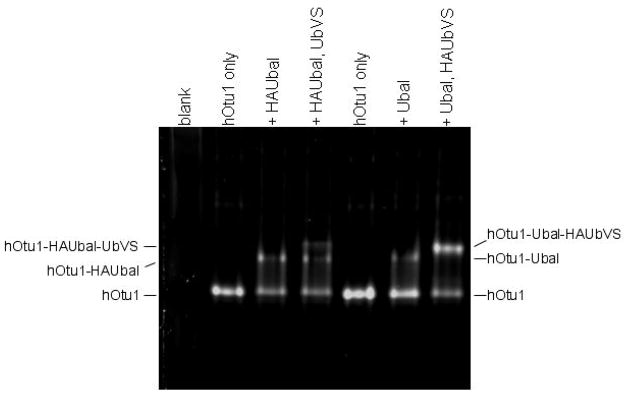
(a) Ubal, but not free Ub, activates hOtu1 for reaction with UbVS. hOtu1 (1.7 μM) was preincubated 10 min with Ubal (top panel) or Ub (bottom panel) at the indicated molar ratios; HA-tagged UbVS (HAUbVS; 0.6 μM) was added and incubation continued for 7 min before analysis by SDS-PAGE and immunoblotting with anti-HA antibody. (b) Free Ub inhibits HAUbVS reaction with hOtu1. Mixtures of hOtu1 (0.84 μM) and Ubal (1 μM) were preincubated with Ub (0 – 150 μM; lanes 2 – 10) for 30 min; a sample without hOtu1 was a negative control (lane 1). HAUbVS (0.12 μM) was then added and, after 5 min, the mixtures analyzed as in A. hOtu1-HAUbVS adduct formation was quantified by densitometry of the immunoblot (upper panel) and the results, when fit to a model for Ub binding, yielded an IC50 of 10 μM (lower panel). (c) Ubal-activated hOtu1-HAUbVS adduct formation requires both C23 and C91 residues. Reactions as in 5b, but without added Ub, were done to compare HAUbVS affinity-labeling of wild-type (WT), C23A, C91A, and C212A forms of hOtu1. (d) Tight binding of Ubal to hOtu1 requires active-site cysteine C91, but not C23 or C212. hOtu1 incubated with excess Ubal was analyzed by native gel electrophoresis to resolve the hOtu1-Ubal complex from the free proteins; see Supplementary Data for details. (e) Simultaneous binding of Ubal and UbVS to hOtu1. Following preincubation of hOtu1 with HA-tagged (or untagged) Ubal and subsequent incubation with UbVS (or HAUbVS), protein complexes were resolved by native electrophoresis and stained with SYPRO Ruby. hOtu1 complexes containing both (HA)Ubal and (HA)UbVS are evident as the slowest migrating, supershifted bands.
Unlike Ubal, free Ub did not stimulate hOtu1-UbVS adduct formation (Figure 5a, lower panel); instead, Ub actually inhibited the Ubal-promoted reaction with an IC50 of ~10 μM (Figure 5b). We tested the possibility that the inhibition by Ub was due to competition with Ubal for binding to the hOtu1 active site as assayed by native gel electrophoresis. Even when added in a 100-fold excess over Ubal, Ub had no discernable effect on the amount of hOtu1-Ubal complex (Supplementary Figure S5). This suggested that the inhibition by Ub was due to competition with UbVS at a second site and not with Ubal at the active site; inhibition by binding at a second site also would explain why activation by Ubal was reversed when Ubal:hOtu1 exceeded 1.
These results presented a puzzle. Tight binding of Ubal to a DUB is expected to depend upon a reaction with the active-site cysteine, but that is true as well for the formation of a DUB-UbVS adduct. When mass spectrometry of the hOtu1-UbVS adduct was done to determine the site of UbVS attachment, we found that UbVS had reacted with C23 and not C91, the active-site cysteine (Supplementary Figure S6). We then compared wild-type hOtu1 with the C23A, C91A, and (as a control) C212A mutant proteins for reaction with UbVS and Ubal binding (Figure 5c, d). As expected, C91 was essential both for Ubal binding and adduct formation with UbVS, but the C23A mutation prevented only hOtu1-UbVS adduct formation. These data support our identification of C23 as the UbVS attachment site and confirm that Ubal-binding was a prerequisite to the UbVS reaction. The results prove that hOtu1 has two distinct Ub-binding sites.
Discussion
Although the OTU-class DUBs share a conserved enzymatic core, their substrate specificities appear to be quite diverse. As with many other DUBs, little is known about OTU deubiquitination substrates in vivo, and no clear picture of their specificity has emerged 17; 18; 26; 29. Our results show that cleavage by hOtu1 is highly specific for K48 isopeptide linkages. Remarkably, this specificity held even with polyUb substrates containing mixtures of K48 and K63 linkages within the same chain. To our knowledge, this is the clearest example of a DUB that cleaves only K48 linkages. Previous studies of UbpY (USP8), USP14, and yeast Otu1 had shown that K63 or K29-linked polyUb is a very poor substrate as compared to K48-linked polyUb 18; 34; 45; 46. However, UbpY also can disassemble linear polyUb and Ub-protein fusions 47. While USP14 does prefer K48 over K63 linkages, the activity was low in both cases and reflected the enzyme in an inactivated state 45. Similarly, yeast Otu1 prefers K48 over K63 linkages, but even overnight incubations that resulted in disassembly of longer K48-linked polyUb chains did not eliminate K48-Ub2 18.
The overall fold of OTU enzyme family members is known from crystal structures of OTU domain proteins including human otubain 2 29, yeast otu1 34, human A20 48; 49, and human otubain 1 33. The structures show conserved catalytic sites among all OTU domain proteins but give little information on how the OTU domain DUBs achieve their enzyme activity and substrate specificity. Despite the differences among the OTU domain DUBs, the substrate specificity we have described for hOtu1 may well be shared among related proteins in evolutionarily distant species. We have shown here that a putative otubain from C. elegans that we call ceOtu has the same polyUb cleavage specificity as hOtu1 (Figure 1c). Previously, S. cerevisiae Otu1 was shown to cleave K48 but not K63 or K29-linked polyUb 18; 34. We suspect that otubain 1 homologs from other species will be similarly selective. The exquisite specificity of hOtu1 for K48-linked chains is likely to be important for its function in regulating T cell anergy 25. The specificity of this enzyme, and the ease by which the recombinant enzyme can be purified, makes hOtu1 an attractive biochemical tool for selective cleavage of K48 isopeptide linkages within polyUb conjugates.
A surprising conclusion from our studies is that the discrimination by hOtu1 among polyUb substrates cannot be attributed simply to selective binding to K48-linked polyUb. Diubiquitins of different linkage types inhibited K48-Ub2 cleavage by hOtu1 equally well, and even monoUb could inhibit. Insight into the basis for linkage selectivity was obtained by substrate mutation studies and NMR chemical shift perturbation assays that indicated that both the proximal and distal ubiquitins of K48-Ub2 contact hOtu1. The simplest interpretation is that there are two sites on hOtu1 for Ub-binding. That this is indeed the case is evident from our observation that UbVS binds to hOtu1 at a site distinct from the catalytic site occupied by Ubal (Figure 5).
Our unexpected finding that UbVS formed a covalent adduct with C23 of hOtu1, and especially that the reaction depended upon addition of Ubal, provides strong support for a bidentate model for K48-Ub2 binding. UbVS has been regarded as a highly selective, mechanism-based DUB inhibitor that reacts with the active-site cysteine 44, which is C91 in hOtu1. However we discovered that UbVS does not target the DUB active site, but instead reacts with a different cysteine residue, C23. In addition, we found that the substitution, C23A, affects binding of UbVS only, while a mutation of the active site residue, C91A, decreases Ubal as well as UbVS binding. Taken together, these results point strongly to the presence of two distinct ubiquitin binding sites in hOtu1.
The targeting and rapid reaction of UbVS with a residue outside of a DUB active site signals the need for caution in using this reagent. The combination of moderately tight binding and proximity of a cysteine sidechain, independent of the associated base (i.e., histidine) normally found at a DUB active site, are likely to suffice for reaction with UbVS. Thus, in principle, UbVS adducts could form with non-DUB proteins that have Ub binding sites and appropriately positioned cysteines. Caution is needed as well with the use of other Ub and Ub-like protein derivatives with C-terminal electrophilic groups, such as Ub-vinylmethyl ester and SUMO-vinylsulfone.
In hOtu1, Ubal binding presumably promotes a conformational rearrangement that either exposes a new Ub binding site, repositions hOtu1 residue C23 to enable reaction with UbVS, or both. Because tight-binding of Ubal to hOtu1 requires the active-site C91 (Figure 5d), and free Ub cannot replace Ubal to activate UbVS-adduct formation (Figure 5a), we conclude that Ubal occupies the hOtu1 catalytic site preferentially. This site would function as the “S1” site in hOtu1 and would accommodate the distal Ub of a diUb substrate; presumably, UbVS binds at the corresponding proximal Ub or S1′ site.
C23 in hOtu1 clearly is required for UbVS-adduct formation, but we think that it is unlikely to have a role in the polyUb linkage-specificity of hOtu1. One reason is that C23 is not conserved evolutionarily (Supplementary Figure 1a), and a second is that deletion of the N-terminal 41 residues of hOtu1 had no effect on diUb cleavage specificity in vitro (Figure 1c).
Because K48-Ub2 is structurally distinct from Ub2 assembled through other linkages 50, non-K48-linked forms of diUb may bind to only one of the two Ub binding sites in hOtu1 whereas K48-Ub2 binds to both. Alternatively, each Ub moiety in a non-K48-Ub2 may be able to associate with the enzyme, but their simultaneous binding would constrain the position of the intervening isopeptide bond which, as a result, may not be oriented to permit attack by the OTU active-site cysteine (see Figure 5). It also is possible that interactions with residues adjacent to the isopeptide-linked K48 of the proximal Ub contribute to catalysis.
We consider it likely that our bidentate-binding model for linkage selectivity will apply to other DUBs as well. Significantly, the model can account for specificity that is not manifested at the level of initial substrate binding. Although contacts with multiple ubiquitins within polyUb might enhance affinity, avid binding would not necessarily promote linkage selectivity. Indeed, a hallmark of our model is that it can explain linkage-selective polyUb cleavage despite linkage-independent binding. Based on an analysis of the conserved surface regions of the DUB A20, the OTU domain in A20 was recently proposed to have binding sites for both the proximal and the distal units of Ub2 49. Whether or not the linkage-specificity of A20 is manifested at the level of substrate binding remains to be determined. Results with a non-OTU DUB also might be explained by the bidentate-binding model. A recent crystal structure of human AMSH-LP DUB complexed with K63-linked Ub2 indicates binding sites for both the proximal and distal Ubs 51. This DUB, a JAMM/MPN+ domain enzyme, preferentially disassembles K63-linked polyUb. Other JAMM/MPN+ DUBs also are K63-linkage specific and, like hOtu1, are inhibited by monoUb or diubiquitins having other linkages (EMC and REC, unpublished data). Structural studies of the aforementioned DUBs bound to polyUb, as well as of polyUb with different isopeptide linkages, will be needed to establish the detailed mechanisms that underlie their extraordinary specificity.
Materials and methods
Plasmid construction
DNAs encoding full-length human otubain 1 (hOtu1) and Cezanne were cloned by the polymerase chain reaction (PCR) from a human fetus cDNA library (Clontech). DNA encoding full-length putative otubain from Caenorhabditis elegans (ceOtu) was cloned from a cDNA library (Invitrogen). Dynazyme (MJ Research) was used as the DNA polymerase for PCR. DNAs encoding hOtu1, hOtu1 with an N-terminal 41-residue truncation (hOtu1ΔN41), and ceOtu were subcloned into a pProEx-c vector (Life Technologies) through NcoI (5′) and BamHI (3′) sites.
Protein expression and purification
For expression of hOtu1, plasmid pProEx-hOtu1 was transformed into Rosetta E. coli cells (Novagen). The transformed cells were grown overnight at 37 °C in 10 ml LB medium containing 100 μg/ml ampicillin and 33 μg/ml chloramphenicol, diluted 100-fold into the same medium, and then grown at 37 °C to an OD600 of 1.0. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was then added to 1.0 mM to induce protein expression and the temperature was reduced to 15 °C; after overnight growth, the cells were harvested by centrifugation. The cells were resuspended in 20 mM Tris pH 7.8, 4 mM MgCl2, 60 μg/ml DNAse I, 0.5 M NaCl, 5 mM imidazole, 5% glycerol, 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and protease inhibitor cocktail (Complete™ without EDTA; Roche), and lysed using a microfluidizer. The lysate was clarified by centrifugation and applied to a Talon column (Clontech), which retained the His6-tagged hOtu1. The column was washed with 20 mM Tris pH 7.8, 0.5 M NaCl, 10 mM imidazole. His6-tagged hOtu1 was eluted with 150 mM imidazole. After cleavage of the His6 tag with His6-TEV protease, the protein solutions were dialyzed against 20 mM Tris, pH 7.8, 0.1 M NaCl, and 1 mM TCEP. The His6-tag and His6-TEV were removed by applying the protein solutions to a Talon column. The flow-through was applied to a Hiload 26/60 Superdex 75 gel filtration column eluted with 50 mM Tris-HCl (pH 7.8), 0.2 M NaCl, 1 mM TCEP, and 1 mM EDTA. Peak fractions were combined, concentrated to 10 mg/ml, and stored in small aliquots at −80 °C. As estimated by SDS-PAGE and Coomassie blue staining, purity of the hOtu1 was 99%. hOtu1ΔN41 and ceOtu were expressed and purified to ~99% homogeneity using the protocol described above.
Preparation of polyubiquitin conjugates
DiUb substrates K48-Ub2 (proximal UbD77) 52, K63-Ub2 (proximal UbD77) 53, linear Ub5 54, and distal-end Lucifer Yellow-labeled (LY) K48-Ub4 (UbC66LY-Ub3) 39 were prepared as described. Two mixed-linkage tetraUb chains, K48-K63-K48 Ub4 (catalog #UCM310) and K63-K48-K63 Ub4 (catalog #UCM210), were from Boston Biochem. Other polyUb conjugates were made as described in Supplementary data.
Deubiquitination assays
Specified amounts of substrate were incubated in a 10 μl volume with hOtu1, hOtu1ΔN41, ceOtu, or hCezcat (see Supplement) in 20 mM Tris pH 7.5 (at 37 °C), 50 mM NaCl, and 5 mM DTT; 0.05% BSA was included as a carrier protein in some reactions. Reactions were quenched with SDS sample buffer and applied to a 4–12% acrylamide gel for SDS-PAGE. Fluorescent LY-tagged substrates and 125I-labeled substrates were detected with a phosphorimager (Typhoon 4001, Molecular Dynamics). Otherwise, protein bands were detected by silver staining or Coomassie Blue staining.
For competition assays, 125I-labeled K48-Ub2 (7 μM; 2.3×105 cpm/nmole) was incubated with 0.16 μM hOtu1 in 50 mM Tris, pH 7.5, 5 mM DTT, and 0.05% BSA at 25 °C. Unlabeled K48-Ub2, K63-Ub2, (K29/K6)-Ub2, or Ub was added at molar concentrations 1, 5, and 10-fold above the amount of 125I-labeled substrate. Reactions were quenched with SDS sample buffer and applied to 4–12% gels for SDS-PAGE. The gels were dried and the 125I-labeled Ub and Ub2 were quantified using a phosphorimager. The assays were done in duplicate. Competition assays with 0.27 μM hCezcat were performed similarly, using 8.4 μM 125I-labeled K48-Ub2 as the substrate and K48-Ub2, K63-Ub2, (K29/K6)-Ub2, or monoUb as potential inhibitors.
Reactions with ubiquitin-vinylsulfone (UbVS)
Ub-aldehyde (Ubal) and HA-tagged UbVS were prepared using intein-mediated ligation as described previously 55. All incubations were at 0 °C in 50 mM Tris-Cl, pH 7.5, with 6 – 10 mM DTT; other details are described in the figure legends. Adduct formation was evaluated by SDS-PAGE (4–20% gel) and either staining with SYPRO Ruby (Invitrogen) or detection on immunoblots with anti-HA antibody. Mass spectrometry of hOtu1 and the hOtu1-UbVS adduct was done as described in Supplementary data.
Supplementary Material
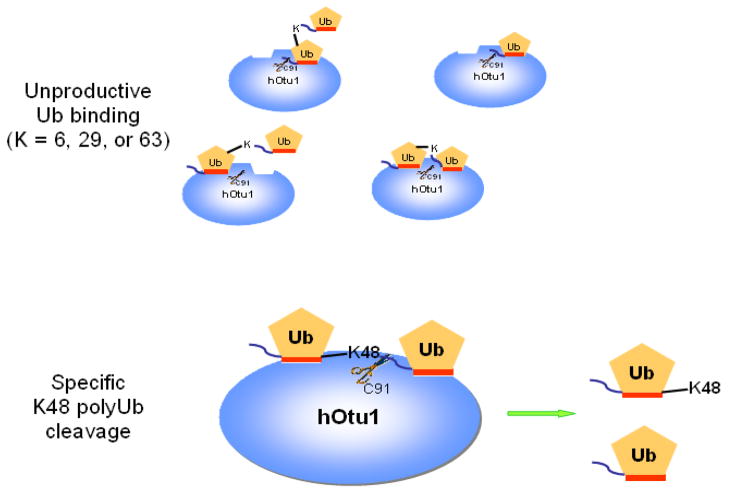
Figure 6. A bidentate-binding model representing interactions between hOtu1 and diubiquitin can account for specific cleavage of K48-Ub2 by hOtu1.
Two Ub binding sites in hOtu1 are postulated to bind both the proximal and distal Ubs of K48-Ub2 simultaneously via their hydrophobic surfaces (orange stripes). This bidentate binding of substrate positions the K48 isopeptide linkage for attack by the C91 thiolate within the catalytic site of hOtu1. Other Ub2 forms, i.e., K63-, K29-, or K6-linked Ub2, either can bind to only one of the sites on hOtu1 or, if both Ub moieties occupy the two sites simultaneously, the conformation is constrained so as to preclude proper positioning of the isopeptide linkage for cleavage.
Acknowledgments
We thank Michael Eddins for providing K48-Ub2, Matthew Steele for providing 125I-labeled K48-Ub2 and isopeptidase T, and Junmin Peng for LC-MS analyses. This work was funded in part by NIH Roadmap grant RR020839 (CW and REC), NIH postdoctoral fellowship 5F32GM075712 (EMC), and NIH grants R01GM065334 (DF) and R01GM066355 (KDW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data are available at Journal of Molecular Biology Online. Also included in the supplement are additional methods for polyUb conjugate syntheses and mass spectrometry, and assays for steady-state deubiquitination kinetics, Ubal binding, and NMR chemical shift perturbation.
References
- 1.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haglund K, Dikic I. Ubiquitylation and cell signaling. Embo J. 2005;24:3353–9. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer JA. Deubiquitinating enzymes: their roles in development, differentiation, and disease. Int Rev Cytol. 2003;229:43–72. doi: 10.1016/s0074-7696(03)29002-1. [DOI] [PubMed] [Google Scholar]
- 9.Wing SS. Deubiquitinating enzymes--the importance of driving in reverse along the ubiquitin-proteasome pathway. Int J Biochem Cell Biol. 2003;35:590–605. doi: 10.1016/s1357-2725(02)00392-8. [DOI] [PubMed] [Google Scholar]
- 10.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–53. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 13.Mizuno E, Kitamura N, Komada M. 14-3-3-dependent inhibition of the deubiquitinating activity of UBPY and its cancellation in the M phase. Exp Cell Res. 2007;313:3624–34. doi: 10.1016/j.yexcr.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 14.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–22. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 15.Li K, Zhao K, Ossareh-Nazari B, Da G, Dargemont C, Marmorstein R. Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor. J Biol Chem. 2005;280:29176–85. doi: 10.1074/jbc.M502975200. [DOI] [PubMed] [Google Scholar]
- 16.Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 17.Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O’Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, Zhang Z, Arnott D, Dixit VM. DUBA: A Deubiquitinase That Regulates Type I Interferon Production. Science. 2007 doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 18.Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell. 2006;21:261–9. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Schlieker C, Weihofen WA, Frijns E, Kattenhorn LM, Gaudet R, Ploegh HL. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol Cell. 2007;25:677–87. doi: 10.1016/j.molcel.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova KS, Aravind L, Koonin EV. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci. 2000;25:50–2. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- 21.Scheel H, Tomiuk S, Hofmann K. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum Mol Genet. 2003;12:2845–52. doi: 10.1093/hmg/ddg297. [DOI] [PubMed] [Google Scholar]
- 22.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 23.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–5. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 24.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–51. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 25.Soares L, Seroogy C, Skrenta H, Anandasabapathy N, Lovelace P, Chung CD, Engleman E, Fathman CG. Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat Immunol. 2004;5:45–54. doi: 10.1038/ni1017. [DOI] [PubMed] [Google Scholar]
- 26.Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517–22. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans PC, Smith TS, Lai MJ, Williams MG, Burke DF, Heyninck K, Kreike MM, Beyaert R, Blundell TL, Kilshaw PJ. A novel type of deubiquitinating enzyme. J Biol Chem. 2003;278:23180–6. doi: 10.1074/jbc.M301863200. [DOI] [PubMed] [Google Scholar]
- 28.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 29.Nanao MH, Tcherniuk SO, Chroboczek J, Dideberg O, Dessen A, Balakirev MY. Crystal structure of human otubain 2. EMBO Rep. 2004;5:783–8. doi: 10.1038/sj.embor.7400201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164:973–8. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans PC, Ovaa H, Hamon M, Kilshaw PJ, Hamm S, Bauer S, Ploegh HL, Smith TS. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J. 2004;378:727–34. doi: 10.1042/BJ20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, Garcia-Sastre A, Virgin HWt. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–16. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edelmann MJ, Iphoefer A, Akutsu M, Altun M, di Gleria K, Kramer HB, Fiebiger E, Dhe-Paganon S, Kessler BM. Structural basis and specificity of human otubain 1 mediated deubiquitylation. Biochem J. 2008 doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 34.Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, Shanks JR, Reyes-Turcu FE, Wilkinson KD, Marmorstein R. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–49. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 36.Yao T, Cohen RE. Cyclization of polyubiquitin by the E2-25K ubiquitin conjugating enzyme. J Biol Chem. 2000;275:36862–8. doi: 10.1074/jbc.M006050200. [DOI] [PubMed] [Google Scholar]
- 37.Liu CW, Li X, Thompson D, Wooding K, Chang TL, Tang Z, Yu H, Thomas PJ, DeMartino GN. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam YA, DeMartino GN, Pickart CM, Cohen RE. Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26 S proteasomes. J Biol Chem. 1997;272:28438–46. doi: 10.1074/jbc.272.45.28438. [DOI] [PubMed] [Google Scholar]
- 39.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–40. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson KD, Tashayev VL, O’Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–46. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 41.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci U S A. 1996;93:861–6. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–72. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–47. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 44.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–59. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 45.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. Embo J. 2005;24:3747–56. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–92. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naviglio S, Mattecucci C, Matoskova B, Nagase T, Nomura N, Di Fiore PP, Draetta GF. UBPY: a growth-regulated human ubiquitin isopeptidase. Embo J. 1998;17:3241–50. doi: 10.1093/emboj/17.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin SC, Chung JY, Lamothe B, Rajashankar K, Lu M, Lo YC, Lam AY, Darnay BG, Wu H. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J Mol Biol. 2008;376:526–40. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komander D, Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- 50.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–63. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 51.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–62. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J Biol Chem. 1990;265:21835–42. [PubMed] [Google Scholar]
- 53.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–53. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 54.Jonnalagadda S, Butt TR, Marsh J, Sternberg EJ, Mirabelli CK, Ecker DJ, Crooke ST. Expression and accurate processing of yeast penta-ubiquitin in Escherichia coli. J Biol Chem. 1987;262:17750–6. [PubMed] [Google Scholar]
- 55.Wilkinson KD, Gan-Erdene T, Kolli N. Derivitization of the C-terminus of ubiquitin and ubiquitin-like proteins using intein chemistry: methods and uses. Methods Enzymol. 2005;399:37–51. doi: 10.1016/S0076-6879(05)99003-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.