Abstract
Context: In mice, scavenger receptor class B, type I (SR-BI) receptor protein deficiency is associated with elevated high-density lipoprotein (HDL)-cholesterol (HDL-C) levels.
Objective: Our objective was to determine the relationship between SR-BI protein and HDL-C levels in humans.
Design: This was a prospective study of adults with hyperalphalipoproteinemia. Fasting blood was obtained for lipid and lipoprotein measurement, genomic DNA, and monocyte-derived macrophages. SR-BI protein levels were measured by Western blots, and SR-BI activity was measured by cholesteryl ester (CE) uptake of each donor’s radiolabeled HDL with their monocyte-derived macrophages, or by degradation and specific cell association of dual-labeled HDL in vitro.
Setting: The study was performed in a tertiary university teaching hospital.
Results: The mean age was 57.2 ± 10.9 yr (n = 65). SR-BI protein levels were inversely associated with HDL-C levels (P < 0.002), HDL particle size (P < 0.05), and positively associated with CE uptake (P < 0.004); there was no association with plasma apolipoprotein levels. SR-BI protein levels (P = 0.01) were independent predictors of HDL-C levels. Subjects who were carriers of the A allele for the rs4238001 (glycine to serine at position 2) polymorphism [single nucleotide polymorphism (SNP)] had lower SR-BI protein levels (P = 0.01), whereas carriers of the C allele for the rs2278986 SNP also had lower SR-BI protein levels (P = 0.02). Body mass index (P = 0.05), rs4238001 (P = 0.01), and rs2278986 (P = 0.01) SNPs were independent predictors of SR-BI protein levels. In vitro studies of murine macrophages stably expressing the glycine to serine at position 2 SNP showed less degradation (P < 0.0004) and specific cell association (P < 0.0004) of [125I, 3H]-CE-labeled HDL.
Conclusions: SR-BI protein has an independent effect on HDL-C levels in women with hyperalphalipoproteinemia. Two SNPs were significantly associated with lower SR-BI protein levels.
Scavenger receptor, class B, type I protein is a predictor of HDL levels in hyperalphalipoproteinemia subjects.
A number of epidemiological studies have shown an inverse relationship between high-density lipoprotein (HDL)-cholesterol (HDL-C) levels and cardiovascular disease (1,2,3). Among the number of proteins and receptors shown to influence HDL-C levels, especially in rodent models, is the scavenger receptor, class B, type I (SR-BI) receptor. SR-BI was isolated and characterized as a physiologically relevant lipoprotein receptor in murine tissues by Acton et al. (4). Although SR-BI has been shown to mediate triglyceride (TG) and phospholipid uptake from HDL, its major role is to mediate the selective uptake of cholesteryl esters (CEs) from HDL.
Targeted deletions of the murine SR-BI gene have shown its importance in lipoprotein metabolism (5). Total cholesterol levels were significantly increased in homozygous SR-BI−/− mice, and analysis of the lipoprotein profiles by fast protein liquid chromatography showed a significant increase in HDL-C levels and a shift of the HDL particles to a larger size. This finding was due to enrichment of the core of the HDL particle with CE and was thought to occur because of the inability of hepatic SR-BI to remove selectively this core CE. The fraction of intermediate-density lipoproteins and low-density lipoprotein-cholesterol (LDL-C) was higher in the SR-BI−/− mice but with a greater variability in the absolute values (5).
In humans, polymorphisms within the SR-BI gene (SCARB1) were first identified by Acton et al. (6) in a white European population, and were associated with plasma lipid levels and body mass index (BMI). These investigators identified five variants within SCARB1, with three variants at exons 1 and 8 and intron 5 having minor allele frequencies greater than 10%. The exon 1 variant [rs4238001, which is a nonsynonymous single nucleotide polymorphism (SNP) that encodes an amino acid change from glycine to serine at position 2 (G2S)] was significantly associated with higher HDL-C and lower LDL-C levels in men, but not in women. The exon 8 variant (rs5888, a synonymous SNP) was associated with lower LDL-C levels in women, but not in men, and the intron 5 variant showed an association with BMI in women.
Thus far the relationship between SR-BI and HDL-C levels has been linked by epidemiological associations. For instance, the association between the G2S SNP and higher HDL-C levels suggested deficiency of SR-BI protein. However, this relationship was not directly tested in the study by Acton et al. (6). Therefore, the purpose of our study was to examine the association of SR-BI protein with HDL-C levels in subjects with hyperalphalipoproteinemia (HALP). The selection of macrophage SR-BI protein for study was based on the accessibility of peripheral monocytes (which when differentiated into macrophages, abundantly express SR-BI protein), whereas SR-BI protein is highly expressed in liver and steroidogenic tissues, but these sources are impossible to obtain in otherwise healthy subjects. We hypothesized that a HALP population would be ideal in identifying subjects with low-tissue SR-BI protein levels, given the fact that mice deficient in SR-BI protein have markedly elevated HDL-C levels (5). Moreover, to date, there have not been any published studies demonstrating a significant association between SR-BI protein levels and lipids in humans.
Subjects and Methods
Study subjects
Adult men and women between the ages of 18 and 80 yr were recruited from the greater Baltimore, MD, area. Subjects presented to The Johns Hopkins Bayview Medical Center General Clinical Research Center in the fasting state on two separate visits held 2 wk apart, and the mean fasting plasma HDL-C had to be 1.55 mmol/liter or more (60 mg/dl) for inclusion in this study. Subjects were not taking cholesterol medications (statins, fibrates, or niacin), and had normal liver, renal, and thyroid function tests. Study subjects donated 150 ml blood after an overnight fast for the purposes of isolating plasma, serum, lipoproteins, circulating monocytes for in vitro macrophage differentiation and genomic DNA for sequencing of the SCARB1 gene. Study subjects provided written consent, and the protocol was approved by The Johns Hopkins Institutional Review Board.
Lipid and lipoprotein analysis
See supplemental materials, which are published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org, for details.
Cell culture
See supplemental materials for details.
Western blotting
See supplemental materials for details.
Transfection of cells
See supplemental materials for details.
CE uptake and cell association assays
See supplemental materials for details.
SCARB1 sequencing
See supplemental materials for details.
Statistics
The distribution profile was determined for each variable ascertained, and values that did not exhibit a normal distribution were log transformed. To evaluate the association of SR-BI protein levels with quantitative traits, stepwise multiple regression analyses were performed with adjustment for a number of covariates. JMP Start Statistics, Third Edition software, SAS Institute Inc. (Cary, NC), was used to perform the statistical analyses, and P values of 0.05 or less were considered significant. Threshold significance values for selection and retention of covariates in the stepwise selection procedure were 0.25 and 0.10, respectively.
Results
The characteristics of the study population are shown in Table 1, stratified by sex. The majority of the participants were Caucasian women, with a mean age of 55.7 ± 10.3 yr for this subgroup. Nine women indicated a history of hormone therapy (HT) use, but only three women were current users. Women had significantly higher total cholesterol (P = 0.04) and HDL-C levels (P = 0.004) compared with men. The mean SR-BI protein levels were not different between men and women. The range of SR-BI protein levels for the entire group was 0.03–0.67 OD, and the protein distribution was normal (supplemental Fig. I, which is published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org). We performed all of the analyses with and without the inclusion of men (n = 14), and found that the inclusion or exclusion of men did not affect the final results (supplemental Figs. II–V, and supplemental Tables I and II, which are published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org). Because women were the majority of our population, we have confined the primary analysis to the results generated in the group of women.
Table 1.
Characteristics of the study population (n = 65)
| Men | Women | |
|---|---|---|
| n | 14 | 51 |
| Age (yr) | 62.2 ± 11.7 | 55.7 ± 10.3a |
| Race | ||
| White (%) | 5 (36) | 40 (78)b |
| Black (%) | 7 (50) | 10 (20) |
| Asian (%) | 2 (14) | 0 (0) |
| Hispanic (%) | 0 (0) | 1 (2) |
| BMI (kg/m2) | 25.2 ± 3.5 | 25.7 ± 6.0 |
| Total cholesterol (mmol/liter) | 5.2 ± 0.98 | 5.8 ± 0.85c |
| TGs (mmol/liter) | 0.85 ± 0.34 | 0.88 ± 0.31 |
| HDL-C (mmol/liter) | 1.92 ± 0.28 | 2.36 ± 0.54b |
| LDL-C (mmol/liter) | 2.9 ± 0.96 | 3.0 ± 0.75 |
| Apolipoprotein A-I (mmol/liter) | 1.53 ± 0.18 | 1.61 ± 0.17 |
| SR-BI protein (OD) | 0.27 ± 0.12 | 0.30 ± 0.04 |
All P values are compared with men.
P = 0.03.
P = 0.004.
P = 0.04.
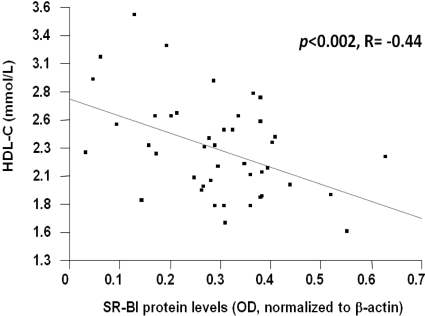
Our primary goal was to determine the relationship between SR-BI protein and HDL-C levels. The rationale for isolating monocyte-derived macrophages (MDMs) from each subject was that this tissue is readily accessible (in contrast to other tissues highly expressing SR-BI such as liver, adrenals, and gonads) and has SR-BI protein (7,8). As shown in Fig. 1, SR-BI protein levels were inversely associated with HDL-C levels [P = 0.002; r = −0.44; Pearson’s correlation with outliers excluded based on values above and below the 95th and 5th percentile (n = 4)].
Figure 1.
HDL-C is significantly inversely associated with SR-BI protein levels. SR-BI protein levels were measured by Western blotting of cell lysates isolated from MDMs from each subject (n = 43). P = 0.002.
In our HALP population, we did not observe an association between HDL-C levels and cholesteryl ester transfer protein (CETP), hepatic lipase/endothelial lipase, lipoprotein lipase, or lecithin-cholesterol acyltransferase (LCAT) activities (data not shown). When SR-BI protein levels were stratified by percentile, with low expression defined as below the 25th percentile and high expression defined as above the 75th percentile, HDL-C levels were significantly higher in subjects with low SR-BI protein expression (2.68 ± 0.13 mmol/liter) (103.5 ± 5.0 mg/dl) compared with subjects with high SR-BI expression (2.21 ± 0.10 mmol/liter) (85.3 ± 3.7 mg/dl) (P = 0.01, Tukey-Kramer; n = 43) (data not shown). We did not observe an association between SR-BI protein and LDL-C or intermediate-density lipoprotein levels, nor was there an association between SR-BI protein and any of the apolipoprotein measurements (data not shown).
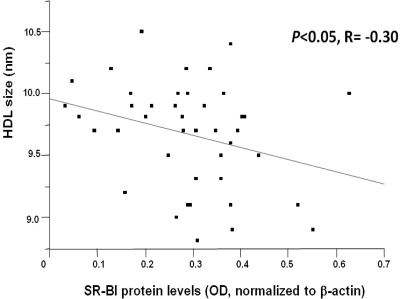
We next determined the relationship between SR-BI protein levels and HDL size as measured by nuclear magnetic resonance spectroscopy. As shown in Fig. 2, HDL particle size was inversely correlated with SR-BI protein level (P = 0.05; r = −0.30; n = 42). We did not observe an association between SR-BI protein levels and very low-density lipoprotein particle number or size, or LDL particle number or size (data not shown).
Figure 2.
HDL particle size is inversely associated with SR-BI protein levels. HDL particle size was determined by nuclear magnetic resonance spectroscopy (n = 42). P < 0.05.
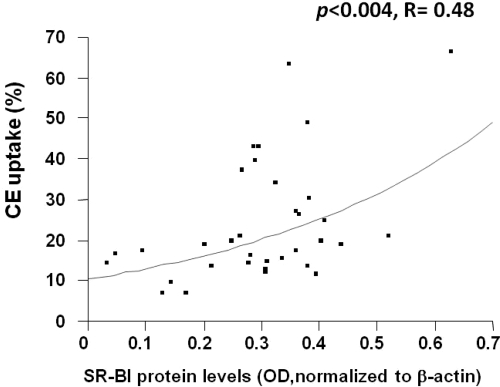
We then measured the uptake of [3H]cholesteryl-oleyl ether mediated by HDL in MDMs isolated from the study subjects. The results in Fig. 3 showed that the percent CE uptake mediated by HDL was significantly associated with SR-BI protein levels (P = 0.004; r = 0.48; n = 34).
Figure 3.
CE uptake from HDL is positively associated with SR-BI protein levels. HDL isolated from each study subject was radiolabeled with [1,2-3H]cholesteryl oleyl ether, and then incubated with each donor’s respective MDMs (n = 34). P = 0.004.
We next determined whether SR-BI protein was an independent predictor of HDL-C. We considered the additional covariates of age, BMI, smoking, alcohol use, exercise, HT, TGs, hemoglobin A1c, fish oil use, and SR-BI protein levels, which together explained 55% of the variance in HDL-C levels (model P = 0.001). In a stepwise multiple regression analysis, SR-BI remained a significant predictor of HDL-C levels (P = 0.01) (Table 2). TG levels (P = 0.01), exercise (P = 0.03), and fish oil use (P = 0.01) were also significant independent predictors of HDL-C levels in this HALP population.
Table 2.
Multiple regression of independent predictors for HDL-C levels
| A. Initial full model (P < 0.001; r = 0.74) |
| Covariates: age (P = 0.48), BMI (P = 0.40), exercise (P = 0.09), alcohol use (P = 0.34), HT use (P = 0.55), TG (P = 0.05), hemoglobin A1c (P = 0.13), fish oil use (P = 0.008), smoking (P = 0.39), and SR-BI protein (P = 0.06) |
| B. Final model (P < 0.0002; r = 0.71) |
| 1. TG (P = 0.01) |
| 2. Exercise (P = 0.03) |
| 3. Fish oil use (P = 0.01) |
| 5. SR-BI (P = 0.01) |
We next examined the prevalence of SCARB1 gene variants by directly sequencing the coding regions, intron-exon junctions, the 5′- and 3′-untranslated regions, and 1-kB upstream of the initiation start site. The results in supplemental Table III showed synonymous and nonsynonymous coding SNPs, as well as noncoding SNPs found in SCARB1 with a frequency greater than 10%. We have identified five HALP subjects with the previously reported G2S SNP. We identified two subjects with a nonsynonymous SNP in exon 3. This SNP encodes a change from valine to isoleucine at amino acid 135. We also identified a novel nonsynonymous SNP in exon 10 (Gly→Arg, G404R) in one subject with SR-BI protein levels below the 25th percentile. In none of the HALP subjects have we identified the 11-bp deletion within the SCARB1 promoter described by Hsu et al. (9). In addition, none of the subjects with SR-BI protein levels above the 75th percentile was a carrier for any of the nonsynonymous SNPs identified in subjects with the lowest SR-BI protein levels.
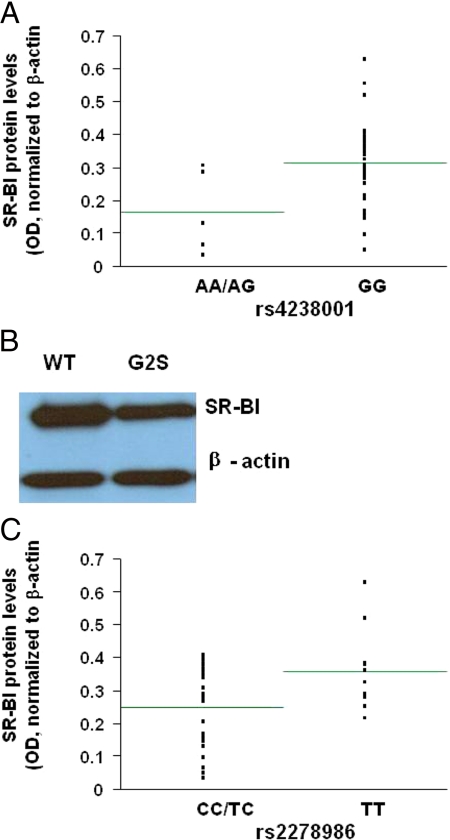
We then examined the association of SR-BI protein levels with each individual SNP shown in supplemental Table III, and found that the subjects who were carriers of the minor A allele (AA/GA) for the G2S SNP had significantly lower SR-BI protein levels (0.16 ± 0.05, 48% lower) compared with noncarriers (0.31 ± 0.02) [P = 0.01; r = 0.40; n = 40 (Fig. 4A)]. Results of a sliding window haplotype analysis (data not shown) were also consistent with an association with low SR-BI levels driven by the G2S variant. In addition, the single minor allele carrier for synonymous rs2070242 in exon 1 had a higher SR-BI level. We next transiently transfected COS-7 cells with wild-type or G2S-SR-BI plasmids. The results shown in Fig. 4B are representative of two independent experiments and showed that SR-BI protein levels were approximately 37% lower in cells transfected with the G2S construct compared with wild-type cells. The results in Fig. 4C also showed that subjects who were carriers of the minor C allele (CC/TC) for the intron 3 rs2278986 SNP had significantly lower SR-BI protein levels (0.25 ± 0.02, 31% lower) compared with subjects who were noncarriers (0.36 ± 0.03) (P = 0.02; r = 0.40; n = 36).
Figure 4.
The association of macrophage SR-BI protein levels with the rs4238001 and rs2278986 SNPs. A, SR-BI protein levels were significantly lower in carriers of the A allele (AA/AG) for the rs4238001 SNP compared with noncarriers (GG) (P = 0.01; r = 0.40; n = 40). B, COS-7 cells transiently transfected with plasmid expressing the rs4238001 or G2S SNP had approximately 37% lower SR-BI protein expression compared with cells transfected with the wild-type (WT) SR-BI plasmid. The Western blot is representative of two independent experiments. C, SR-BI protein levels were significantly lower in carriers of the C allele (CC/TC) for the rs2278986 SNP compared with noncarriers (TT) (P = 0.02; r = 0.40; n = 36).
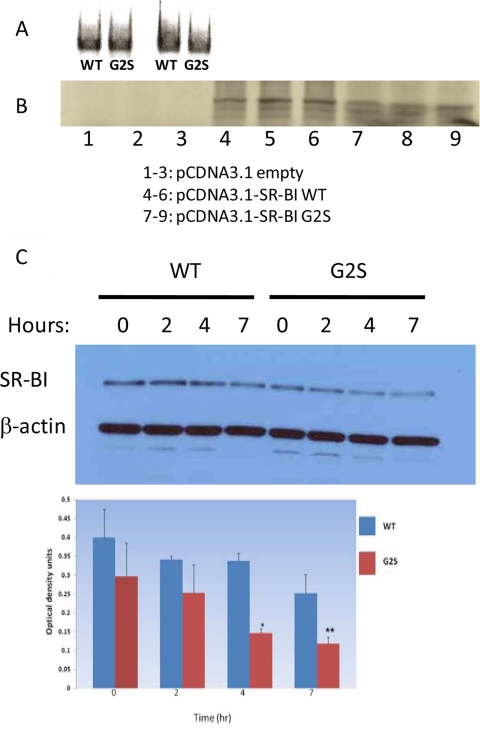
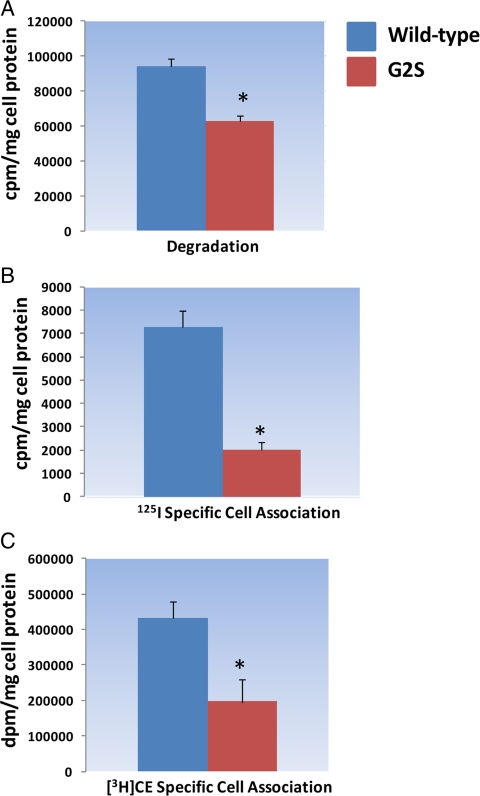
To gain insight into the mechanism of how the G2S variant affected SR-BI protein expression, we performed experiments that measured SR-BI mRNA transcription, translation, protein degradation, and specific cell association of dual-radiolabeled HDL. As shown in Fig. 5A, there was no difference in the amount of mRNA transcribed in rabbit reticulocytes transfected with SR-BI wild-type or G2S variant. However, translation of SR-BI protein in the rabbit reticulocytes was lower in cells transfected with the G2S variant (Fig. 5B). We next examined SR-BI protein degradation in murine RAW macrophages that stably expressed wild-type or the G2S variant by incubating cells in the presence of cycloheximide for varying periods of time. As shown in Fig. 5C, by 4 and 7 h, the turnover of SR-BI protein was greater in cells expressing the G2S variant (P < 0.0007 and P < 0.04, respectively). Finally, we examined degradation and specific cell association of [125I, 3H]HDL in murine RAW macrophages. As shown in Fig. 6, degradation and specific [125I] and [3H] cell association were significantly lower in cells expressing the G2S variant (P < 0.0004).
Figure 5.
The rs4238001 (G2S) variant is associated with alterations in SR-BI protein translation and degradation, but not SR-BI mRNA transcription. A, Transcription of SR-BI RNA was similar in rabbit reticulocytes transfected with plasmids expressing either SR-BI wild-type (WT) or GS2 variant. B, Translation of SR-BI protein was reduced in rabbit reticulocytes transfected with plasmids expressing the G2S variant. C, Stably expressing wild-type and G2S variant murine RAW macrophages were incubated with cycloheximide (140 μg/ml) for varying periods of time (0–7 h). SR-BI turnover was significantly greater in G2S cells by 4 and 7 h compared with wild-type cells (P < 0.0007 and P < 0.04, respectively). *, P < 0.0007 and **, P < 0.04 compared to murine RAW macrophages transfected with wild-type SR-BI.
Figure 6.
Degradation and cell association of HDL are reduced in murine RAW macrophages stably expressing the SR-BI G2S variant. A, Degradation of [125I, 3H]-labeled HDL was significantly lower in G2S expressing cells (P < 0.0004). B, Specific 125I and [3H]CE cell association (panel C) were significantly lower in G2S expressing cells (*, P < 0.0004 compared to wild-type).
The next question was to determine the variables that were independent predictors of SR-BI protein levels. We considered the covariates of BMI, HT use, CETP activity, LCAT activity, G2S SNP, rs2278986 (intron 3), and rs5888 (exon 8) polymorphisms, which together explained 42% of the variance in SR-BI protein levels (model P = 0.04) (Table 3). In a stepwise multiple regression analysis, BMI (P = 0.05), G2S SNP (P = 0.01), and rs2278986 (intron 3) (P = 0.01) remained as significant independent predictors of SR-BI protein levels. We also did not observe linkage disequilibrium between the G2S SNP and rs2278986 SNPs.
Table 3.
Multiple regression of independent predictors for SR-BI protein levels
| A. Initial full model (P = 0.04; r = 0.65) |
| Covariates: BMI (P = 0.06), HT use (P = 0.36), CETP activity (P = 0.78), LCAT activity (P = 0.37), rs4238001 (G2S) (P = 0.05), rs2278986 (intron 3) (P = 0.04), and rs5888 (exon 8) (P = 0.50) |
| B. Final model (P = 0.004; r = 0.62) |
| 1. BMI (P = 0.05) |
| 2. rs4238001 (P = 0.01) |
| 3. rs2278986 (P = 0.01) |
Discussion
The purpose of our study was to examine the relationship between SR-BI protein and HDL-C levels in subjects with HALP. We found that SR-BI protein levels were significantly inversely associated with HDL-C levels and HDL size. In a multiple linear regression analysis, SR-BI protein was an independent predictor of HDL-C levels after adjustment for other covariates. Subjects who were carriers of the minor A allele of a previously identified nonsynonymous SNP, G2S, had significantly lower SR-BI protein levels compared with subjects who were carriers of the major G allele, whereas subjects who were carriers of the C allele of the intron 3 SNP also had significantly lower SR-BI protein levels. In addition, BMI, the G2S SNP, and rs2278986 polymorphisms were independent predictors of SR-BI protein levels. In vitro studies of murine macrophages stably expressing the G2S variant showed significantly less degradation and specific cell association of dual-radiolabeled HDL compared with cells stably expressing wild-type SR-BI.
Mice completely deficient in SR-BI have significantly higher HDL-C levels, with the HDL particles also being much larger in size (5). To the best of our knowledge, no humans have been identified that are completely deficient in SR-BI protein. We hypothesized that the probability of identifying subjects deficient in SR-BI would be greatest in those subjects with HALP. As was shown in Fig. 1, there was a significant inverse relationship between macrophage expression of SR-BI and HDL-C levels. Although we could not assess intraindividual variation in SR-BI protein levels, the results from our HALP population nonetheless demonstrated a significant relationship between SR-BI protein level in macrophages and plasma HDL-C. We selected MDMs because this tissue is readily accessible and has been shown to express SR-BI protein (7,8).
We have also shown a significant inverse relationship between SR-BI protein levels and HDL size. Our findings are consistent with data derived from others using in vitro and animal models that showed the inverse relationship between SR-BI protein level and HDL size (10,11,12,13). The results in Fig. 3 showed a positive association between SR-BI protein levels and CE uptake from HDL. We did not perform selective CE uptake assays due to logistical problems in radiolabeling each donor’s HDL with 125I before incubation with each donor’s MDMs. However, because we also observed no association between SR-BI protein levels and any of the plasma apolipoproteins, this suggested that our findings were consistent with previous observations that the major function of SR-BI is to mediate the uptake of CE, and not the apolipoprotein moieties of HDL (4). It should be noted that the results presented herein are not from subjects completely deficient in SR-BI protein, and that the primary analysis was confined to women with HALP. Moreover, whereas the relationship between SR-BI and CE uptake was significant, the results did not suggest that SR-BI was the only factor influencing CE uptake in human macrophages.
Circulating HDL-C levels are influenced by a number of factors (14). Although there have been a number of epidemiological studies (15,16,17,18,19,20) showing significant associations between certain SCARB1 genotypes and HDL-C levels, the influence of SR-BI protein on plasma HDL-C levels has not been directly assessed. Therefore, we selected a model that included a number of known independent variables that affect HDL-C levels, and found that SR-BI protein levels remained as independent predictors of HDL-C levels in this HALP population.
We have now shown stronger associations between known SCARB1 SNPs, SR-BI protein expression, and HDL-C levels. Acton et al. (6) was the first to show that a common SNP within exon 1, G2S, was significantly associated with higher HDL-C levels in men, suggesting that this SNP might be associated with lower SR-BI protein levels. The frequency of this SNP in our HALP cohort was 12%, and it contributed 13% of the variation in SR-BI protein levels, suggesting that it is a relatively common SNP that exerts a major influence on SR-BI protein levels. In contrast to Acton et al. (6), in our HALP cohort, the vast majority of the carriers with the minor A allele were women (five of seven). We are now the first to show that the G2S variant decreased SR-BI protein levels by increasing the rate of SR-BI protein turnover, and as a consequence of lower protein expression, there was significantly lower specific cell association of HDL.
When we next evaluated significant influences on SR-BI protein expression, we found that BMI, the G2S variant, and rs2278986 polymorphisms were independent predictors of SR-BI protein levels. With regard to the relationship of BMI and SR-BI, Acton et al. (6) had earlier observed that certain SCARB1 SNPs were significantly associated with BMI. Moreover, Perez-Martinez et al. (19) examined the effect of dietary fat content on lipid levels in subjects with the G2S SNP and reported that subjects expressing the minor allele had significantly higher LDL-C levels. This suggests the possibility that dietary fat might also have an important affect on SR-BI expression. In vitro studies have also shown a significant influence of fatty acids on SR-BI expression (21,22,23). Alternatively, adipokines such as leptin have increased hepatic SR-BI expression (24). In animals, estrogen has regulated the expression of SR-BI and its isoform SR-BII (25,26). In humans, we were the first to report a significant association between low SR-BI RNA levels in granulosa cells and low plasma estradiol levels in infertile women undergoing in vitro fertilization (27).
The limitations in our study merit further discussion. The rationale for recruiting HALP subjects was to enrich the pool for possible SR-BI deficient subjects, given that SR-BI deficient mice have significantly increased HDL-C levels and that HDL-C levels above 1.55 mmol/liter (60 mg/dl) in humans are considered a negative cardiovascular risk factor. It is not known whether a similar relationship would be found in subjects with HDL-C levels less than 1.55 mmol/liter. However, we are embarking on similar studies in subjects with low to normal HDL-C levels. The choice of measuring SR-BI protein in MDMs was based on the accessibility of these cells. Although it might be possible that human macrophage SR-BI directly affects HDL-C levels, this has not been observed in rodent macrophages (28,29), likely due to the fact that rodent macrophages express low levels of SR-BI protein (11). However, it has been shown that murine hepatic SR-BI expression affects HDL-C levels and reverse cholesterol transport (30). Thus, until such time that we are able to obtain liver samples from healthy donors, we believe that MDMs are likely surrogates for hepatic SR-BI protein because it is more likely than not that hepatic SR-BI protein levels are exerting an influence on circulating HDL-C levels. Nonetheless, we feel that it would have been extremely difficult to obtain genomic DNA, liver tissue, and plasma HDL-C levels from healthy donors.
The studies examining genetic associations with lipid levels and the influence of sex on these associations are inconsistent. For instance, Acton et al. (6) did not observe an association between the rs5888 (exon 8) polymorphism and HDL-C levels in women, whereas we (20) and Richard et al. (31) did report a significant association between this SNP and HDL-C levels in women. A definitive answer of whether there exists sex specificity in SR-BI protein expression and associations with lipid levels or BMI remains to be determined. Our study was underpowered to determine whether or not there was a sex difference in the association of SR-BI protein and HDL-C levels.
Another important aspect of SR-BI receptor expression/function has been its role in coronary artery disease, particularly in light of the accelerated atherosclerosis observed in SR-BI knockout mice with a background of apolipoprotein E deficiency (32). In our HALP population, we did not observe a significant association between SR-BI protein expression and carotid intimal-medial thickening, or a family or personal history of coronary artery disease (data not shown). Our lack of findings might be due to the small sample size or alternatively that SR-BI does not directly affect the vascular tree. Svensson et al. (33) examined the regulation and splicing of SR-BI in human macrophages and atherosclerotic plaques, and reported that SR-BI RNA expression was similar in MDMs isolated from subjects with atherosclerotic disease as compared with healthy controls. In contrast, Ritsch et al. (34) recently reported that three common SCARB1 variants were positively associated with increased risk for peripheral arterial disease.
In summary, in a cross-sectional study of women with HALP, we have shown that SR-BI protein is inversely associated with HDL-C levels and HDL size. SR-BI protein levels were also found to be independent predictors of HDL-C levels, and two SNPs, rs4238001 (G2S) and rs2278986, were independent predictors of SR-BI protein levels. Further study is needed to determine whether there is a causal relationship between SR-BI protein levels and cardiovascular outcomes.
Supplementary Material
Footnotes
This work was supported by a National Institutes of Health grant to A.R. (HL075646) and from The Johns Hopkins Bayview Medical Center General Clinical Research Center (M01-RR-02179). None of the authors report a financial conflict of interest.
Disclosure Summary: The authors have nothing to declare.
First Published Online January 21, 2009
Abbreviations: BMI, Body mass index; CE, cholesteryl ester; CETP, cholesteryl ester transfer protein; G2S, glycine to serine at position 2; HALP, hyperalphalipoproteinemia; HDL, high-density lipoprotein; HDL-C, high-density lipoprotein-cholesterol; HT, hormone therapy; LCAT, lecithin-cholesterol acyltransferase; LDL-C, low-density lipoprotein-cholesterol; MDM, monocyte-derived macrophage; SNP, single nucleotide polymorphism; SR-BI, scavenger receptor class B, type I; TG, triglyceride.
References
- Castelli WP, Garrison RJ, Wilson PW, Abbott RD, Kalousdian S, Kannel WB 1986 Incidence of coronary heart disease and lipoprotein-cholesterol levels: the Framingham study. JAMA 256:2835–2838 [PubMed] [Google Scholar]
- Gordon D, Rifkind BM 1989 Current concepts: high density lipoproteins-the clinical implications of recent studies. N Engl J Med 321:1311–1315 [DOI] [PubMed] [Google Scholar]
- Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR 1977 High density lipoprotein as a protective factor against coronary heart disease. Am J Med 62:707–714 [DOI] [PubMed] [Google Scholar]
- Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M 1996 Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271:518–520 [DOI] [PubMed] [Google Scholar]
- Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, Krieger M 1997 A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc Natl Acad Sci USA 94:12610–12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acton S, Osgood D, Donoghue M, Corella D, Pocovi M, Cenarro A, Mozas P, Keilty J, Squazzo S, Woolf EA, Ordovas JM 1999 Association of polymorphisms at the SR-BI gene locus with plasma lipid levels and body mass index in a white population. Arterioscler Thromb Vasc Biol 19:1734–1743 [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Wee SB 1999 The HDL receptor, SR-BI/CLA-1 does not mediate cholesterol efflux from human macrophage foam cells. Circulation Suppl 100:I-538 [Google Scholar]
- Hirano K, Yamashita S, Nakagawa Y, Ohya T, Matsuura F, Tsukamoto K, Okamoto Y, Matsuyama A, Matsumoto K, Miyagawa J, Matsuzawa Y 1999 Expression of human scavenger receptor class B type I in cultured human monocyte-derived macrophages and atherosclerotic lesions. Circ Res 85:108–116 [DOI] [PubMed] [Google Scholar]
- Hsu LA, Ko YL, Wu S, Teng MS, Peng TY, Chen CF, Chen CF, Lee YS 2003 Association between a novel 11-base pair deletion mutation in the promoter region of the scavenger receptor class B type gene and plasma HDL cholesterol levels in Taiwanese Chinese. Arterioscler Thromb Vasc Biol 23:1869–1874 [DOI] [PubMed] [Google Scholar]
- Jian B, de la Llera-Moya M, Ji Y, Wang N, Phillips MC, Swaney JB, Tall AR, Rothblat GH 1998 Scavenger receptor class B type I as a mediator of cellular cholesterol efflux to lipoproteins and phospholipid acceptors. J Biol Chem 273:5599–5606 [DOI] [PubMed] [Google Scholar]
- Ji Y, Jian B, Wang N, Sun Y, de la Llera-Moya M, Phillips MC, Rothblat GH, Swaney JB, Tall AR 1997 Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J Biol Chem 272:20982–20985 [DOI] [PubMed] [Google Scholar]
- Ueda Y, Royer L, Gong E, Zhang J, Cooper PN, Francone O, Rubin EM 1999 Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B type I transgenic mice. J Biol Chem 274:7165–7171 [DOI] [PubMed] [Google Scholar]
- Kozarsky KF, Donahee MH, Glick JM, Krieger M, Rader DJ 2000 Gene transfer and hepatic overexpression of the HDL receptor SR-BI reduces atherosclerosis in the cholesterol-fed LDL receptor-deficient mouse. Arterioscler Thromb Vasc Biol 20:721–727 [DOI] [PubMed] [Google Scholar]
- de Backer G, de Bacquer D, Kornitzer M 1998 Epidemiological aspects of high density lipoprotein cholesterol. Atherosclerosis 137(Suppl):S1–S6 [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Lewitzky S, Reeves C, Permutt A, Glaser B, Groop LC, Lehner T, Meyer JM 2003 Polymorphisms of the HDL receptor gene associated with HDL cholesterol levels in diabetic kindred from three populations. Hum Hered 55:163–170 [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Lehner T, Reeves C, Moliterno DJ, Newby LK, Rogers WJ, Topol EJ, Genequest investigators 2003 Association of genetic variants in the HDL receptor, SR-B1, with abnormal lipids in women with coronary artery disease. J Med Genet 40:453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Esparragon F, Rodriguez-Perez JC, Hernandez-Trujillo Y, Macias-Reyes A, Medina A, Caballero A, Ferrario CM 2005 Allelic variants of the human scavenger receptor class B type 1 and paraoxonase 1 on coronary heart disease: genotype-phenotype correlations. Arterioscler Thromb Vasc Biol 25:854–860 [DOI] [PubMed] [Google Scholar]
- Osgood D, Corella D, Demissie S, Cupples LA, Wilson PW, Meigs JB, Schaefer EJ, Coltell O, Ordovas JM 2003 Genetic variation at the scavenger receptor class B type I gene locus determines plasma lipoprotein concentrations and particle size and interacts with type 2 diabetes: the Framingham Study. J Clin Endocrinol Metab 88:2869–2879 [DOI] [PubMed] [Google Scholar]
- Perez-Martinez P, Perez-Jimenez F, Bellido C, Ordovas JM, Moreno JA, Marin C, Gomez P, Delgado-Lista J, Fuentes F, Lopez-Miranda J 2005 A polymorphism exon 1 variant at the locus of the scavenger receptor class B type I (SCARB1) gene is associated with differences in insulin sensitivity in healthy people during the consumption of an olive oil-rich diet. J Clin Endocrinol Metab 90:2297–2300 [DOI] [PubMed] [Google Scholar]
- Roberts CGP, Shen H, Mitchell BD, Damcott CM, Shuldiner AR, Rodriguez A 2007 Variants in scavenger receptor class B type I gene are associated with HDL cholesterol levels in younger women. Hum Hered 64:107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spady DK, Kearney DM, Hobbs HH 1999 Polyunsaturated fatty acids up-regulate hepatic scavenger receptor BI (SR-BI) expression and HDL cholesteryl ester uptake in the hamster. J Lipid Res 40:1384–1394 [PubMed] [Google Scholar]
- Loison C, Mendy F, Serougne C, Lutton C 2002 Dietary myristic acid modifies the HDL- cholesterol concentration and liver scavenger receptor BI expression in the hamster. Br J Nutr 87:199–210 [DOI] [PubMed] [Google Scholar]
- Loison C, Mendy F, Serougne C, Lutton C 2002 Increasing amounts of dietary myristic acid modify the plasma cholesterol level and hepatic mass of scavenger receptor BI without affecting bile acid biosynthesis in hamsters. Reprod Nutr Dev 42:101–114 [DOI] [PubMed] [Google Scholar]
- Lundasen T, Liao W, Angelin B, Rudlilng M 2003 Leptin induces the hepatic high density lipoprotein receptor scavenger receptor B type I (SR-BI) but not cholesterol 7α-hydroxylase (Cyp7a1) in leptin-deficient (ob/ob) mice. J Biol Chem 278:43224–43228 [DOI] [PubMed] [Google Scholar]
- Graf GA, Roswell KL, Smart EJ 2001 17β-estradiol promotes the up-regulation of SR-BII in HepG2 cells and in rat livers. J Lipid Res 42:1444–1449 [PubMed] [Google Scholar]
- Stangl H, Graf GA, Yu L. Cao G, Wyne K 2002 Effect of estrogen on scavenger receptor BI expression in the rat. J Endocrinol 175:663–672 [DOI] [PubMed] [Google Scholar]
- Velasco M, Alexander C, King J, Zhao Y, Garcia J, Rodriguez A 2006 Association of lower plasma estradiol levels and low expression of scavenger receptor class B type I in infertile women. Fertil Steril 85:1391–1397 [DOI] [PubMed] [Google Scholar]
- Adorni MP, Zimetti F, Billheimer JT, Wang N, Rader DJ, Phillips MC, Rothblat GH 2007 The roles of different pathways in the release of cholesterol from macrophages. J Lipid Res 48:2453–2462 [DOI] [PubMed] [Google Scholar]
- Wang X, Collins HL, Ranalletta M, Fuki IV, Billheimer JT, Rothblat GH, Tall AR, Rader DJ 2007 Macrophage ABCA1 and ABCG1, but not SR-BI, promote macrophage reverse cholesterol transport in vivo. J Clin Invest 117:2216–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Da Silva R, Reilly M, Billheimer JT, Rothblat GH, Rader DJ 2005 Hepatic expression of scavenger receptor class B type I (SR-BI) is a positive regulator of macrophage reverse cholesterol transport in vivo. J Clin Invest 115:2870–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard E, von Muhlen D, Barrett-Connor E, Alcaraz J, Davis R, McCarthy JJ 2005 Modification of the effects of estrogen therapy on HDL cholesterol levels by polymorphisms of the HDL-C receptor, SR-BI: the Rancho Bernardo Study. Atherosclerosis 180:255–262 [DOI] [PubMed] [Google Scholar]
- Trigatti B, Rayburn H, Vinals M, Braun A, Miettinen H, Penman M, Hertz M, Schrenzel M, Amigo L, Rigotti A, Krieger M 1999 Influence of the high density lipoprotein receptor SR-BI on reproductive and cardiovascular pathophysiology. Proc Natl Acad Sci USA 96:9322–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson PA, Englund MC, Snackestrand MS, Hagg DA, Ohlsson BG, Stemme V, Mattsson- Hulten L, Thelle DS, Fagerberg B, Wiklund O, Carlsson LM, Carlsson B 2005 Regulation and splicing of scavenger receptor class B type I in human macrophages and atherosclerotic plaques. BMC Cardiovasc Disord 5:25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsch A, Sonderegger G, Sandhofer A, Stanzl U, Tancevski I, Eller P, Schgoer W, Wehinger A, Mueller T 2007 Scavenger receptor class B type I polymorphisms and peripheral arterial disease. Metabolism [Erratum (2007) 56:1599] 56:1135–1141 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.