Abstract
Plants have two isoprenoid biosynthetic pathways: the cytosolic mevalonate (MVA) pathway and the plastidic 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway. Since the discovery of the MEP pathway, possible metabolic cross-talk between these pathways has prompted intense research. Although many studies have shown the existence of such cross-talk using feeding experiments, it remains to be determined if native cross-talk, rather than exogenously applied metabolites, can compensate for complete blockage of the MVA pathway. Previously, Arabidopsis mutants for HMG1 and HMG2 encoding HMG-CoA reductase (HMGR) were isolated. Although it was shown that HMGR1 is a functional HMGR, the enzyme activity of HMGR2 has not been confirmed. It is demonstrated here that HMG2 encodes a functional reductase with similar activity to HMGR1, using enzyme assays and complementation experiments. To estimate the contribution of native cross-talk, an attempt was made to block the MVA pathway by making double mutants lacking both HMG1 and HMG2, but no double homozygotes were detected in the progeny of self-pollinated HMG1/hmg1 hmg2/hmg2 plants. hmg1 hmg2 male gametophytes appeared to be lethal based on crossing experiments, and microscopy indicated that ∼50% of the microspores from the HMG1/hmg1 hmg2/hmg2 plant appeared shrunken and exhibited poorly defined endoplasmic reticulum membranes. In situ hybridization showed that HMG1 transcripts were expressed in both the tapetum and microspores, while HMG2 mRNA appeared only in microspores. It is concluded that native cross-talk from the plastid cannot compensate for complete blockage of the MVA pathway, at least during male gametophyte development, because either HMG1 or HMG2 is required for male gametophyte development.
Keywords: Anther, cross-talk, HMG-CoA reductase, isoprenoid, male gametophyte, MEP pathway, MVA pathway, pollen, sterol, tapetum
Introduction
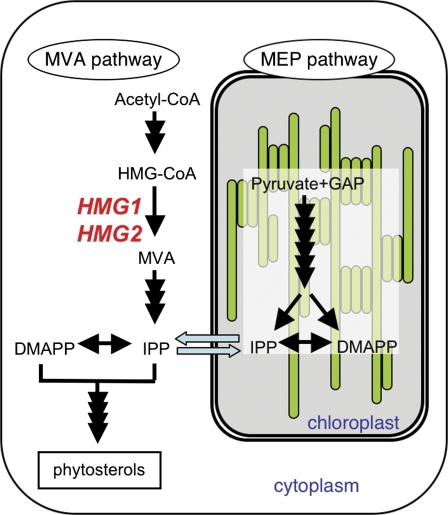
Plants produce a wide variety of isoprenoids by the condensation of common five-carbon intermediates, isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP). Over the course of evolution, plants have maintained the well-known eukaryotic mevalonate (MVA) pathway for cytosolic synthesis of IPP and DMAPP, and this pathway was considered to be the sole synthetic source of these compounds for several decades. However, plants have recently been discovered also to make use of the prokaryotic 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway from the endosymbiotic ancestral cyanobacteria in plastids (Rohdich et al., 2001; Fig. 1). Under normal physiological conditions, the two pathways are compartmentalized. Cytoplasmic isoprenoids (e.g. sterols and brassinosteroids) and mitochondrial isoprenoids (e.g. side chains of ubiquinone) are synthesized via the MVA pathway, while the MEP pathway is used to synthesize plastidic isoprenoids (e.g. carotenoids and the side chains of chlorophylls and plastoquinones) and some isoprenoid-type phytohormones (e.g. gibberellins, abscisic acid, and cytokinins).
Fig. 1.
Outline of the two isoprenoid biosynthetic pathways in an Arabidopsis cell: the MVA and MEP pathways. Blue arrows indicate metabolic flow between the cytosol and plastid. Two genes, HMG1 and HMG2, encoding HMGR are shown in red letters. MVA, mevalonate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GAP, glyceraldehyde 3-phosphate.
Cytosolic IPP and DMAPP are synthesized in seven enzymatic steps from acetyl-CoA via the MVA pathway, and the HMG-CoA reductase (HMGR) reaction is considered to be rate-limiting in this pathway (Schaller et al., 1995; Manzano et al., 2004; Suzuki et al., 2004). The biological importance of HMGR is further emphasized by the existence of highly specific natural inhibitors of the enzyme including mevinolin (Alberts et al., 1980), also known as lovastatin, and its analogues from various rhizopheric ascomycetes (Bach et al., 1990), and their use as growth inhibitors and antibiotics. Earlier work revealed that mevinolin was an efficient plant growth regulator primarily through its inhibition of de novo phytosterol biosynthesis (Bach and Lichtenthaler, 1987; Bach et al., 1990). Mevinolin and its analogues blocked cell proliferation in tobacco (Bach and Lichtenthaler, 1987; Hata et al., 1987; Crowell and Salaz, 1992; Hemmerlin and Bach, 1998; Randall et al., 1993; Morehead et al., 1995), Helianthus tuberosus (Ceccarelli and Lorenzi, 1984), Medicago sativa (Hata et al., 1987), and Acer pseudoplantanus (Ryder and Goad, 1980). Plant HMGRs are encoded by a small family of genes that are differentially expressed upon various internal and external stimuli, and the transcriptional and post-transcriptional regulation of HMGRs have been investigated (Dale et al., 1995; Learned, 1996; Newman and Chappell, 1997). Genetic approaches have demonstrated some of the novel HMGR regulatory mechanisms in plants relative to animals (Rodríguez-Concepción et al., 2004; Kobayashi et al., 2007).
Feeding experiments in plants have indicated the occurence of cross-talk between the MVA and MEP pathways. Labelled mevalonolactone (MVL) and 1-deoxy-D-xylulose (DX) are incorporated into plastidic and cytosolic isoprenoids, respectively, in Lemna gibba (Schwender et al., 2001), Arabidopsis (Kasahara et al., 2002), and tobacco Bright Yellow-2 (BY-2) cells (Hemmerlin et al., 2003). Exogenously applied MVL partially rescues developmental arrest in cla1-1, an Arabidopsis mutant with a disrupted MEP pathway (Nagata et al., 2002), and partially rescues growth inhibition of BY-2 cells treated with fosmidomysin, a DX reductoisomerase inhibitor (Hemmerlin et al., 2003). Exogenously applied DX partially rescues the growth inhibition in Arabidopsis plants (Kasahara et al., 2002) and BY-2 cells (Hemmerlin et al., 2003) treated with mevinolin. However, these feeding studies using exogenously applied metabolites were insufficient to determine whether mutual and native cross-talk can compensate for complete blockage of one of the isoprenoid biosynthetic pathways.
Mutants lacking one functional isoprenoid biosynthetic pathway are very useful to examine native cross-talk. The identification in Arabidopsis of the isoprenoid biosynthesis mutant cla1-1, a null mutant for a DX synthase (DXS) gene, is one important example (Mandel et al., 1996; Araki et al., 2000; Estévez et al., 2000). Despite the albino phenotype of cla1-1, two DXS homologous genes exist in the Arabidopsis genome in addition to CLA1 (Lange and Ghassemian, 2003). It remains unclear whether a cla1 mutation is sufficient for complete shutdown of the MEP pathway. Some MEP pathway mutants have been reported, including ispF-1 for 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MCS; Hsieh and Goodman, 2006), clb4 for hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase (HDS; Gutiérrez-Nava et al., 2004), and clb6 for 1-hydroxy-2-methyl-butenyl 4-diphosphate reductase (HDR; Guevara-García et al., 2005; Hsieh and Goodman, 2005). These three enzymes are encoded by a single gene in Arabidopsis. When grown on sucrose-supplemented medium, even homozygous mutants completely lacking the MEP pathway are viable and capable of germinating.
The seven enzymatic reactions from acetyl-CoA to IPP and DMAPP are encoded by 12 genes in Arabidopsis (Suzuki and Muranaka, 2007). Among these 12 genes, HMG1 and HMG2, encoding HMGR (Suzuki et al., 2004), as well as IPI1 and IPI2 (also refered to as IDI1 and IDI2), encoding IPP isomerase (Okada et al., 2008; Phillips et al., 2008; Sapir-Mir et al., 2008) have been analysed genetically. Whereas IPI1 has been found in multiple subcellular locations: the cytosol (Okada et al., 2008), the plastid (Phillips et al., 2008) or the peroxisome (Sapir-Mir et al., 2008), IPI2 appears in the three cases to be mainly localized in the mitochondria. Thus, it is still not well understood which IPI isoform is devoted to the MVA pathway. On the contrary, HMGR is a key enzyme of the MVA pathway. The isolation and characterization of hmg1 and hmg2 mutants for the two HMGR genes in Arabidopsis have previously been reported (Suzuki et al., 2004). Although hmg1 mutants show pleiotropic phenotypes (dwarfism, early senescence, and male sterility), hmg2 mutants exhibit no morphological phenotypes (Suzuki et al., 2004). HMGR1 enzyme activity has been identified using yeast complementation (Learned and Fink, 1989) and in vitro analysis using the catalytic domain of HMGR1 expressed in Escherichia coli (Dale et al., 1995). The function of HMGR2 is thought to be almost equal to that of HMGR1 based on the findings that hmg2 has slightly higher sensitivity to lovastatin than do wild-type (WT) plants (Suzuki et al., 2004) and that the sterol and triterpenoid levels of hmg2 are slightly lower than those of WT plants (Ohyama et al., 2007); however, direct evidence that HMG2 encodes a functional HMGR has not yet been presented. To examine the influence of complete blockage of the MVA pathway on the plant phenotype, the generation and characterization of an hmg1 hmg2 double mutant is important. Prior to attempting to generate the double mutant, the biochemical and physiological functions of HMGR2 were examined by an enzyme assay and hmg1 complementation test using HMG2. It was found that the hmg1 hmg2 double mutant is lethal during male gametophyte development.
Materials and methods
Plant growth conditions
Arabidopsis thaliana plants (ecotype Wassilewskija (WS), ecotype Columbia (Col), hmg1-1 (WS background; Suzuki et al., 2004), hmg1-2 (WS background; Suzuki et al., 2004), hmg2-1 (Col background; Suzuki et al., 2004), and qrt1-1 (Landsberg erecta background; Preuss et al., 1994) were germinated on 1× Murashige-Skoog (MS) plates containing 3% sucrose and 0.8% agar. For the HMGR enzyme assay and sterol analysis, 1-week-old seedlings were transplanted into 1× MS liquid medium containing 3% sucrose and cultured for one additional week. For RNA extraction, seedlings were grown on MS plates for 2 weeks.
HMGR enzyme assay and sterol analysis
The preparation of the microsomal fraction was performed as described by Kobayashi et al. (2007). HMGR activity was measured as described by Chappell et al. (1995). Sterols were extracted from freeze-dried plant tissues (2-week-old seedlings; 10 mg) and quantified as described by Suzuki et al. (2004).
RT-PCR analysis
RT-PCR analysis of XTR9 and extensin-like protein was performed as described by Suzuki et al. (2004). The relative quantification of HMG1 and HMG2 was performed using an Applied Biosystems 7500 Real Time PCR System, using the following primers and probes: HMG1-fwd, 5′-GGCGTGACAAGATCCGTTACA-3′; HMG1-rev, 5′-GCAATAATGGCGCCGAGTT-3′; HMG1-probe, 5′-CACGTCGTCACTATCACA-3′; HMG2-fwd, 5′-TACTCGGTGTGAAAGGATCAAACA-3′; HMG2-rev, 5′-CCGAACCAGCCACTATTCTTG-3′; HMG2-probe, 5′-AGAAACCTGGCTCGAACGCACAGC-3′; 18SrRNA-fwd, 5′-CGGCTACCACATCCAAGGAA-3′; 18SrNA-rev, 5′-GCTGGAATTACCGCGGCT-3′ and 18SrRNA probe, 5′-TGCTGGCACCAGACTTGCCCTC-3′.
Construction of chimeric genes with HMG1 promoter::HMG1S and HMG1 promoter::HMG2 and transformation of Arabidopsis
The HMG1 promoter (Suzuki et al., 2004) was connected to HMG1S or HMG2 cDNA using PCR. These gene cassettes were digested with EcoRI and XhoI and cloned into these sites in pENTR™1A (Invitrogen, Carlsbad, CA, USA). The resultant entry clones were used for the attL×attR (LR) recombination reaction between GATEWAY™ -converted binary vectors pBCR112 (H Seki et al. unpublished data). The transformation of Arabidopsis was performed as described previously (Suzuki et al., 2004).
Light microscopy
For anther observations, flower buds were selected just before flowering. The anthers were fixed in 2% glutaraldehyde, and a coverslip was placed over the sample and pressed onto the slide. For pollen grain observations, flowers were selected just after flowering. The pollen grains were spread on a glass slide and fixed in 2% glutaraldehyde, and then stained with 2 μg ml−1 DAPI. The samples were observed using an I× 70 fluorescence microscope (Olympus, Tokyo, Japan).
Electron microscopy
Several days before flowering, the anthers of flower buds were fixed for 20 h in 4% glutaraldehyde and 4% paraformaldehyde and then post-fixed with 2% osmium tetroxide for 20 h. The fixed samples were dehydrated through an ethanol series and embedded in Spurr's resin. Ultra-thin sections (70 nm) were double-stained with uranyl acetate and lead citrate, and then observed using a JEM-1200 EX transmission electron microscope (Jeol, Tokyo, Japan).
In situ hybridization
Arabidopsis (ecotype WS) flower buds were fixed with paraformaldehyde, dehydrated through a conventional ethanol series, and embedded in paraffin. Sections (5 μm) were made with a microtome (RM2135; Leica, Vienna, Austria) and deparaffinized with xylene. The sections were probed with sense or antisense RNA probes. For the HMG1 probe, a 356 bp HMG1 cDNA 3′ untranslated region (UTR) fragment was amplified using the primers 5′-CATGATCTGAATCTGAATCATCATCCT-3′ and 5′-CCAAACGCAACTGCACCATACA-3′. For the HMG2 probe, a 420 bp HMG2 cDNA 3’ UTR fragment was amplified using the primers 5′-AGACATTGGCCCTTCGTCTCAAGTC-3′ and 5′-ACCATATCACTCCTCCATCGTCTGC-3′. The amplified fragments were cloned into pCR-II TOPO vectors (Invitrogen; Carlsbad, CA, USA). Digoxigenin (DIG)-labelled HMG1 and HMG2 sense and antisense RNA probes were synthesized using a DIG RNA labelling kit (Roche, Basel, Switzerland). After hybridization and washing, the sections were incubated in a 1:2000 dilution of anti-DIG antibody coupled with alkaline phosphatase. The colour was developed using a detection buffer (100 mM TRIS-HCl, pH 9.5, 100 mM NaCl, 50 mM MgCl2, and 0.1% Tween-20) containing 18 μg ml−1 of 4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate (NBT/BCIP) solution (Roche).
Results
HMG2 encodes a functional HMGR
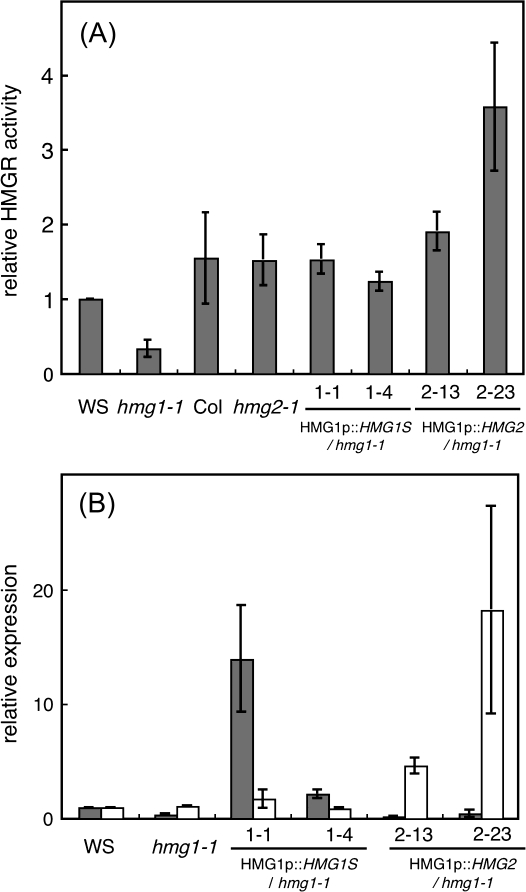
To examine the activity of HMGR1 and HMGR2 in plants, HMGR assays were performed using microsomal fractions prepared from 2-week-old seedlings of WS, Col, hmg1-1, and hmg2-1. The HMGR activity of hmg1-1 was approximately 20% that of the WT (151±6.9 pmol min−1 mg−1 protein), whereas the HMGR activity of hmg2-1 was comparable to that of the WT (Fig. 2A). The remaining 20% activity in hmg1-1 should be derived from HMGR2. This result biochemically indicates that HMGR2 has HMGR activity, as does HMGR1.
Fig. 2.
HMGR enzyme activity and expression of hmgr mutants and complementation lines of hmg1-1. (A) Relative HMGR activity in microsome fractions prepared from 2-week-old seedlings of WS, hmg1-1, Col, hmg2-1, lines 1.1 and 1.4, and lines 2.13 and 2.23. The activity in WS plants, 151±6.9 pmol min−1 mg−1 protein, was given a value of 1. Values are the average of three experiments. (B) Relative expression of HMG1 (gray box) and HMG2 (white box) in 2-week-old seedlings of WS, hmg1-1, lines 1.1 and 1.4 (both have HMG1 promoter::HMG1S in an hmg1-1 background), and lines 2.13 and 2.23 (both have HMG1 promoter::HMG2 in an hmg1-1 background). The expression in WS plants was given a value of 1.
The cellular function of HMGR2 was examined. Because hmg1 shows pleiotropic phenotypes such as dwarfism, early senescence, and male sterility (Suzuki et al., 2004), it was determined whether HMG2 could complement hmg1. Transgenic hmg1-1 plants expressing HMG2 cDNA driven by an HMG1 promoter were generated and characterized. Transgenic hmg1-1 plants expressing HMG1S cDNA driven by an HMG1 promoter were used as positive controls. Two kinds of transcript encoded by HMG1, i.e. HMG1S and HMG1L, occur in Arabidopsis (Lumbreras et al., 1995). Because HMG1S is the major transcript of HMG1, HMG1S cDNA was used as a positive control for the complementation test with hmg1-1. Several independent lines of these transgenic plants were generated in an hmg1-1 homozygous background. Segregation analysis of the kanamycin resistance trait indicated that these lines contained a single T-DNA insertion and were homozygous with respect to the transgene. Among these lines, lines 1.1 and 1.4 for HMG1 promoter::HMG1S and lines 2.13 and 2.23 for HMG1 promoter::HMG2 were selected for further characterization. The expression of HMG1 and HMG2 in each line was analysed using quantitative RT-PCR. The expression of HMG1 in line 1.1 was much higher than that of WT plants, and HMG1 expression in line 1.4 was comparable to that of WT plants (Fig. 2B). Low levels of HMG1 expression were detected in hmg1-1 (Fig. 2B). The primer set of HMG1 for this quantitative RT-PCR was designed prior to the T-DNA insertion. Northern hybridization showed no transcript for HMG1 in hmg1-1 (Suzuki et al., 2004). T-DNA was inserted before the catalytic domain of HMGR1. Although small amounts of transcript derived from HMG1 were detected in hmg1-1, it is considered that no functional HMGR1 exists in hmg1-1. The expression of HMG2 in lines 2.13 and 2.23 was much higher than that of WT plants (Fig. 2B). The HMGR assay demonstrated that the HMGR activity of transgenic lines was comparable to or higher than that of WT plants (Fig. 2A).
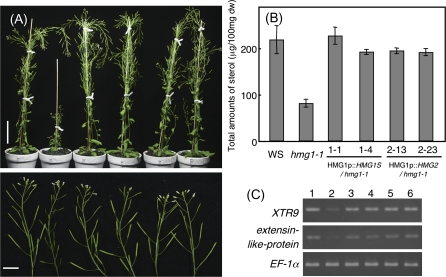
The expression of HMG2 driven by the HMG1 promoter complemented the characteristic hmg1 phenotype in seedlings, such as the inhibition of root growth, small cotyledons, and delayed development of rosette leaves, similar to the expression of HMG1S (data not shown). In mature plants, the height and fertility of hmg1-1 plants expressing HMG2 were comparable to those of WT plants and of hmg1-1 plants expressing HMG1S (Fig. 3A). Because sterols are the major products of the MVA pathway, the total amount of sterols in hmg1 plants is approximately 50% that of WT plants (Suzuki et al., 2004; Ohyama et al., 2007). To test whether HMG2 expression in hmg1-1 rescues the sterol level, the sterols were quantified. The total amount of sterols in hmg1-1 was restored to that of WT plants and of hmg1-1 plants expressing HMG1S by the expression of HMG2 under the control of the HMG1 promoter (Fig. 3B). The sterol profile was not altered in these transgenic plants (data not shown).
Fig. 3.
Phenotypic analyses of hmg1-1 complementation lines. (A) Left to right, upper figure: mature WS, hmg1-1, 1.1, 1.4, 2.13, and 2.23 plants. Scale bar indicates 5 cm. Left to right, lower figure: close-up photos of inflorescences of WS, hmg1-1, 1.1, 1.4, 2.13, and 2.23 plants. Scale bar indicates 1 cm. (B) Total sterols (μg 100mg−1 dry weight) in WS, hmg1-1, 1.1, 1.4, 2.13, and 2.23 plants. (C) The effect of HMG1S and HMG2 expressed in hmg1-1 plants on XTR9 and extensin-like-protein expression. Total RNA was extracted from 2-week-old seedlings of WS (1), hmg1-1 (2), 1.1 (3), 1.4 (4), 2.13 (5), and 2.23 (6) plants. EF-1 transcripts were amplified as a control.
The expression of XTR9 and extensin-like protein, which are thought to be related to cell elongation, is lower in hmg1 plants than in WT plants (Suzuki et al., 2004). The expression of these genes in transgenic hmg1-1 plants was examined. The expression of both genes down-regulated in hmg1 plants was restored, with expression comparable to that in WT plants, in transgenic hmg1-1 plants expressing HMG2 or HMG1S (Fig. 3C). The complementation of the hmg1 mutation by HMG2 indicates that the biochemical and cellular functions of HMGR2 are equal to those of HMGR1.
An hmg1 hmg2 double mutant was not obtained
As mentioned above, two genes encoding functional HMGR isoforms occur in the Arabidopsis genome. To examine the contribution of the MEP pathway to the biosynthesis of cytosolic isoprenoids, an attempt was made to generate a hmg1-1 hmg2-1 double mutant that completely lacked MVA pathway activity. However, a double homozygous mutant was not produced, even in the progeny of self-pollinated HMG1/hmg1-1 hmg2-1/hmg2-1 plants.
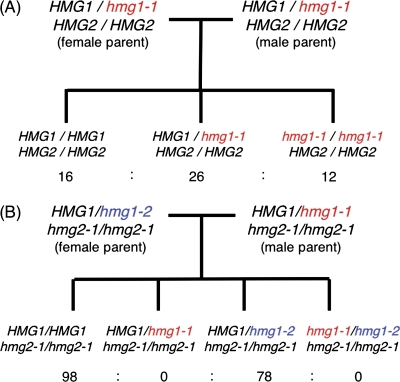
To determine why double mutants were not obtained, crossing experiments were performed. An hmg1-1 homozygous mutant was obtained from the progeny of self-pollinated hmg1-1 heterozygous parents in the expected 1:2:1 Mendelian segregation ratio (Fig. 4A). This indicates that hmg1-1 HMG2 male gametophytes were functional in hmg1-1 heterozygous parents. If HMG1 deficiency is a gametophytic mutation that affects the haploid life cycle, segregation distortion should be evident. It has already been shown that hmg1 homozygous plants exhibit male sterility (Suzuki et al., 2004). Because the segregation was not distorted, HMG1 deficiency of male sterility in hmg1 homozygous plants is considered to be a sporophytic mutation that affects diploid cells such as tapetum cells or microsporocytes.
Fig. 4.
Outline of the crossing experiment. The results of self-pollination of HMG1/hmg1-1 HMG2/HMG2 plants (A) and crossing of HMG1/hmg1-2 hmg2-1/hmg2-1 as the female parent pollinated with pollen from HMG1/hmg1-1 hmg2-1/hmg2-1 plants (B) are shown. The resultant segregation of the F1 generation is shown.
There are two hmg1 alleles: hmg1-1 and hmg1-2 (Suzuki et al., 2004). When HMG1/hmg1-2 hmg2-1/hmg2-1 plants were pollinated with pollen from HMG1/hmg1-1 hmg2-1/hmg2-1 plants, 98 plants with hmg2-1 homozygous single mutants and 78 plants with HMG1/hmg1-2 hmg2-1/hmg2-1 were obtained in the F1 generation (Fig. 4B). No HMG1/hmg1-1 hmg2-1/hmg2-1 plants or hmg1 hmg2 double homozygotes were obtained. When HMG1/hmg1-1 hmg2-1/hmg2-1 plants were pollinated with pollen from HMG1/hmg1-2 hmg2-1/hmg2-1 plants, four plants with hmg2-1 homozygous single mutants and four plants with HMG1/hmg1-1 hmg2-1/hmg2-1 were obtained in the F1 generation. No HMG1/hmg1-2 hmg2-1/hmg2-1 plants or hmg1 hmg2 double homozygotes were obtained. These crossing experiments demonstrated that the hmg1 mutation in the F1 generation was inherited only from the female parent. These results suggest that the hmg1 hmg2 male gametophytes were lethal and that the genotype could not be inherited from the male parent. While HMG1 deficiency in hmg1 homozygotes sporophytically cause male sterility, double deficiency of HMG1 and HMG2 may gametophytically cause defects of the male gametophyte.
hmg1 hmg2 male gametophytes are shrunken
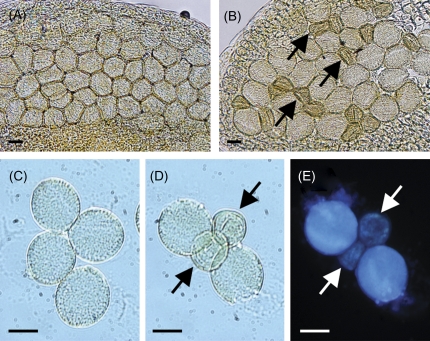
To examine the influence of the hmg1 hmg2 double mutation on the male gametophyte, the anthers of HMG1/HMG1 hmg2-1/hmg2-1 and HMG1/hmg1-1 hmg2-1/hmg2-1 plants were compared microscopically just before flowering. Although all pollen grains were globular in HMG1/HMG1 hmg2-1/hmg2-1 anthers, approximately half of the pollen grains were shrunken in HMG1/hmg1-1 hmg2-1/hmg2-1 anthers (Fig. 5A, B). The hypothesis that the shrunken pollen grains correspond to those of the hmg1 hmg2 genotype is consistent with the crossing data. To confirm this, tetrad analysis was attempted using quartet1 (qrt1) (Preuss et al., 1994). In microsporogenesis, a microsporocyte becomes a tetrad of four haploid cells after meiosis. The tetrad is then separated into four microspores, which develop into mature male gametophytes. In the qrt1 mutant, the tetrad does not separate during male gametophyte development (Preuss et al., 1994). If HMGR deficiency is sporophytically expressed, the numbers of normal and shrunken pollen might vary in a given quartet. By contrast, if the mutation is gametophytic, the ratio of normal to shrunken pollen in each quartet should be 2:2. To determine the genotype of these shrunken pollen grains, an HMG1/hmg1-1 hmg2-1/hmg2-1 qrt1-1/qrt1-1 triple mutant was generated, and tetrad analysis using this triple mutant was performed. Tetrads of qrt1-1 single mutants produced four normal pollen grains (Fig. 5C). By contrast, tetrads of HMG1/hmg1-1 hmg2-1/hmg2-1 qrt1-1/qrt1-1 triple mutants produced two normal and two shrunken pollen grains (Fig. 5D). No nuclei were observed in DAPI-stained shrunken pollen grains (Fig. 5E). These results strongly suggest that the two shrunken pollen grains were hmg1-1 hmg2-1 qrt1-1 and that the male gametophyte of the hmg1 hmg2 genotype exhibits lethality, whereas that of single hmg2 does not.
Fig. 5.
Photographs of the anthers and pollen grains. (A) Anther of HMG1/HMG1 hmg2-1/hmg2-1 plant. (B) Anther of a HMG1/hmg1-1 hmg2-1/hmg2-1 plant. (C) Tetrad in HMG1/HMG1 HMG2/HMG2 qrt1-1/qrt1-1 plant. (D) Tetrad in HMG1/hmg1-1 hmg2-1/hmg2-1 qrt1-1/qrt1-1 plant. (E) DAPI-stained tetrad shown in (D). Arrows indicate abnormal, shrunken pollen grains. Scale bars indicate 10 μm.
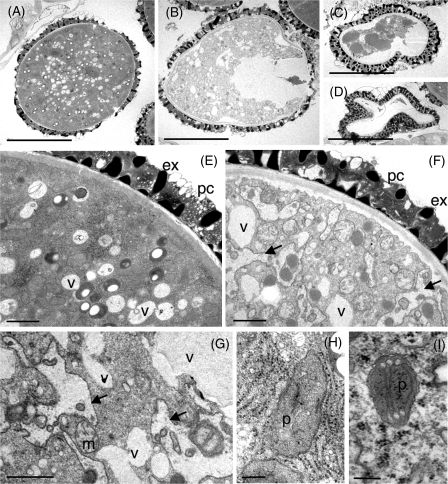
To determine why the hmg1 hmg2 pollen grains were shrunken, the ultrastructure of the male gametophyte was next observed at the tricellular stage in HMG1/hmg1-1 hmg2-1/hmg2-1 plants using a transmission electron microscope. The inside of the globular male gametophyte was densely packed, and its cytoplasm, organelles, exine, and pollen coat were perfectly normal (Fig. 6A), whereas the inside of the shrunken male gametophyte was hollow and less packed (Fig. 6B, C, D). However, the exine and pollen coat were normal in the shrunken male gametophyte. Magnified views of the shrunken male gametophyte showed that the vacuole was larger than normal, and the membraneous structures that seemed to originate from the endoplasmic reticulum (ER) were hypertrophic (Fig. 6E, F, G), whereas symbiontic organelles (plastids and mitochondria) appeared nearly normal (Fig. 6G, H, I). The most remarkable aberration in shrunken male gametophytes is the poor condition of the membraneous element in the membrane traffic system. Thus, HMGR is essential for the development of male gametophytes, especially the formation of membranes originating from the ER. This indicates that the contribution of metabolic cross-talk from the MEP pathway is not very effective in male gametophyte development.
Fig. 6.
Ultrastructural analysis of the male gametophyte. Normal male gametophytes (A, E, H) and abnormal male gametophytes (B, C, D, F, G, I) in HMG1/hmg1-1 hmg2-1/hmg2-1 plants. Abbreviations: ex, exine; pc, pollen coat; v, vacuole; m, mitochondria; p, plastid. Arrows indicate abnormal membrane structures of apparent endoplasmic reticulum origin. Scale bars indicate 10 μm (A, B, C, D), 1 μm (E, F, G), or 200 nm (H, I).
HMG1 and HMG2 are expressed in microspores
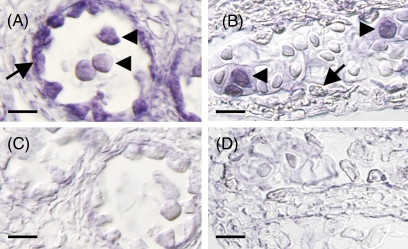
The spatial distribution of HMG1 and HMG2 in WT anthers was examined using in situ hybridization. HMG1 expression was observed in both the tapetum and microspores. HMG2 expression was detected only in the microspores (Fig. 7). Thus, both HMGR genes are expressed in microspores. In contrast to the double mutant (microspores with a hmg1 hmg2 genotype), the microspores in hmg1 and hmg2 plants develop normally. In the single mutant, the remaining HMGR may compensate for one HMGR mutation during microspore and male gametophyte development.
Fig. 7.
Comparison of HMG1 and HMG2 expression in anthers using in situ hybridization. Sections of an anther hybridized with antisense HMG1 (A) or HMG2 (B) probes. Sections of an anther hybridized with sense HMG1 (C) or HMG2 (D) probes as a control. Arrows and arrowheads indicate the tapetum and microspores, respectively. Scale bars indicates 10 μm.
Discussion
Since the discovery of the MEP pathway, metabolic cross-talk between the cytosol and plastid has been a central topic in isoprenoid biosynthesis research. Despite numerous endeavours, it has remained unclear whether native cross-talk, rather than exogenously applied metabolites, can compensate for the complete blockage of one of the isoprenoid biosynthetic pathways.
The biochemical and cellular functions of HMGR2 were analysed and the hmg1-1 hmg2-1 double mutant was characterized to assess the contribution of the native metabolic flow from the MEP pathway. The hmg1-1 plants have approximately 20% of the activity of WT plants, which is thought to correspond to HMGR2 activity (Fig. 2A). HMG2 expression under an HMG1 promoter complemented the hmg1 phenotype in terms of HMGR activity, sterol accumulation, gene expression, and morphology (Fig. 3). Although the physiological function(s) of HMG1 and HMG2 differ from each other (because of the difference in the phenotypes of hmg1 and hmg2), it was demonstrated that HMG2 encodes a functional HMGR. Although HMG1 expression is not affected in hmg2 plants (Suzuki et al., 2004), the HMGR activity of hmg2-1 plants was comparable to that of WT plants. HMGR activity of transgenic hmg1-1 lines with HMG1 or HMG2 genes was not necessarily parallel to the expression level of HMG1 or HMG2 genes in these lines. These results suggest that HMGR activity is post-transcriptionally and post-translationally regulated. Post-transcriptional and post-translational feedback regulation of HMGR was suggested by the experiments using Arabidopsis treated with lovastatin (Kobayashi et al., 2007) and with squalestatin and terbinafine, inhibitors of squalene synthase and of squalene epoxidase, respectively (Nieto et al., 2009). The feedback regulation was also suggested by the experiment using tobacco BY-2 cells treated with squalestatin and terbinafine (Wentzinger et al., 2002). Genetic studies have also demonstrated the regulatory mechanisms of Arabidopsis HMGR (Rodríguez-Concepción et al., 2004; Kobayashi et al., 2007). Since the occurrence of multiple genes encoding HMGR is a general feature of higher plants, elucidation of the regulatory mechanisms is a complicated but challenging research area.
Because HMGR2 activity was confirmed, an attempt was made to generate an hmg1-1 hmg2-1 double mutant. However, double homozygotes were not obtained, due to a defect in the male gametophyte of the hmg1 hmg2 genotype (Fig. 4B). A tetrad analysis of HMG1/hmg1-1 hmg2-1/hmg2-1 in a qrt1-1 background demonstrated that the male gametophyte of the hmg1 hmg2 genotype was shrunken and lethal (Fig. 5). Ultrastructural observations revealed that these disrupted male gametophytes are hollow and that ER-derived membranes are in a poor condition (Fig. 6). The deficiency in male gametophytes might be caused by a defect in the build-up of membrane systems that depend on essential components such as sterols, or on the presence of MVA-derived storage lipids like sterol esters. In contrast to the ER-derived membrane, plastids in shrunken gametophytes appeared normal, which may suggest that blockage of the MVA pathway does not affect plastid morphology. Furthermore, metabolic flow from the MEP pathway in the plastids cannot rescue deficiency of the male gametophyte in the blockage of the MVA pathway, although the plastid appeared normal.
Interestingly, the crossing experiment suggested that complete blockage of the MVA pathway does not affect female gametophyte development (Fig. 4B). It is possible that a sufficient amount of MVA or MVA-derived metabolites is accumulated in the embryo-sac cell after meiosis during female gametophyte development. It is also possible that native cross-talk of isoprenoid compounds from the plastid might be able to compensate for the blockage of the MVA pathway in the embryo-sac cell. Recently, it was shown that complete deficiency of cycloartenol synthase 1 (CAS1) leads to male gametophyte lethality (Babiychuk et al., 2008). Since the embryo-sac cell of the cas1-2 mutant is viable, the viability of the hmg1 hmg2 embryo-sac cell may not be derived from the native cross-talk from the plastid. Although important genes (e.g. housekeeping genes) are generally required for both male and female gametophyte development, it is interesting that HMGR, a key enzyme for cytosolic isoprenoid biosynthesis, is required only for male gametophyte development.
It is also interesting the HMG1 deficiency in hmg1 homozygous plants is a sporophytic mutation, while complete blockage of HMGR causes a gametophytic mutation. This may be caused by a difference in the expression pattern of HMG1 and HMG2 (Fig. 7). In male gametophyte development, both HMG1 and HMG2 are expressed, such that at least one HMGR isoenzyme is required. By contrast, diploid tapetum cells, which are essential for normal pollen formation, express only HMG1, so that hmg1 homozygous plants may exhibit male sterility.
Since prenyl diphosphates are thought to flow between the cytosol and plastids (Bick and Lange, 2003), HMGR is located upstream of the cross-talk in the MVA pathway (Fig. 1). Our data demonstrated that plants completely lacking the MVA pathway did not survive, even if they could biosynthesize small amounts of cytosolic isoprenoids using IPP and DMAPP from plastids. This indicates that the metabolic flow from the MEP pathway is insufficient, at least during male gametophyte development, to compensate for a non-functional MVA pathway. This may reflect the fact that the cytosol and plastid originated from different organisms, a primitive eukaryote and a cyanobacterium, respectively. Even if the contribution of native cross-talk is low, the regulatory mechanisms for isoprenoid biosynthesis in different organelles are interesting and challenging research areas.
Acknowledgments
We thank Dr. Hikaru Seki (RIKEN, PSC) for helpful discussions, and Mizuki Magoshiro, Misaki Yano, Chiho Yamazaki, and Naomi Suzuki for technical assistance with in situ hybridization and TEM. This work was supported in part by a Grant-in-Aid for Young Scientists (B) (grant 18770048), a Scientific Research on Priority Areas grant (grant 17051028) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and the Research and Development Program for New Bio-industry Initiatives, Japan.
References
- Alberts AW, Chen J, Kuron G, et al. Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proceedings of the National Academy Sciences, USA. 1980;77:3957–3961. doi: 10.1073/pnas.77.7.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki N, Kusumi K, Masamoto K, Niwa Y, Iba K. Temperature-sensitive Arabidopsis mutant defective in 1-deoxy-D-xylulose 5-phosphate synthase within the plastid non-mevalonate pathway of isoprenoid biosynthesis. Physiologia Plantarum. 2000;108:19–24. [Google Scholar]
- Babiychuk E, Bouvier-Navé P, Compagnon V, Suzuki M, Muranaka T, Montagu MV, Kushnir S, Schaller H. Alleric mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proceedings of the National Academy Sciences, USA. 2008;105:3163–3168. doi: 10.1073/pnas.0712190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach TJ, Lichtenthaler HK. Plant growth regulation by mevinolin and other sterol biosynthesis inhibitors. In: Fuller G, Nes WD, editors. Ecology and metabolism of plant lipids. American Chemical Society Symposium Ser. 325. Washington: American Chemical Society; 1987. pp. 109–139. [Google Scholar]
- Bach TJ, Weber T, Motel A. Some properties of enzymes involved in the biosynthesis and metabolism of 3-hydroxy-3-methylglutaryl-CoA in plants. Recent Advances in Phytochemistry. 1990;24:1–82. [Google Scholar]
- Bick JA, Lange BM. Metabolic cross-talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelop membrane. Archives of Biochemistry and Biophysics. 2003;415:146–154. doi: 10.1016/s0003-9861(03)00233-9. [DOI] [PubMed] [Google Scholar]
- Ceccarelli N, Lorenzi R. Growth inhibition by competitive inhibitors of 3-hydroxy-3-methylglutaryl-CoA reductase in Helianthus tuberosus tissue explants. Plant Science Letters. 1984;34:269–276. [Google Scholar]
- Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C. Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme A reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiology. 1995;109:1337–1343. doi: 10.1104/pp.109.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell DN, Salaz MS. Inhibition of growth of cultured tobacco cells at low concentrations of lovastatin is reversed by cytokinin. Plant Physiology. 1992;100:2090–2095. doi: 10.1104/pp.100.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S, Arró M, Becerra B, Morrice NG, Boronat A, Hardie DG, Ferrer A. Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. European Journal of Biochemistry. 1995;233:506–513. doi: 10.1111/j.1432-1033.1995.506_2.x. [DOI] [PubMed] [Google Scholar]
- Estévez JM, Cantero A, Romero C, Kawaide H, Jiménez LF, Kuzuyama T, Seto H, Kamiya Y, León P. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiology. 2000;124:95–103. doi: 10.1104/pp.124.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-García A, Roman CS, Arroyo A, Cortés ME, Gutiérrez-Nava MDL, León P. Characterization of Arabidopsis clb6 mutant illustrates the importance of post-transcriptional regulation of the methyl-D-erythritol 4-phosphate pathway. The Plant Cell. 2005;17:628–643. doi: 10.1105/tpc.104.028860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Nava MDL, Gillmor CS, Jiménez LF, Guevara-García A, León P. CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiology. 2004;135:471–482. doi: 10.1104/pp.103.036996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata S, Shirata K, Takagishi H, Kouchi H. Accumulation of rare phytosterols in plant cells on treatment with metabolic inhibitors and mevalonic acid. Plant and Cell Physiology. 1987;28:715–722. [Google Scholar]
- Hemmerlin A, Bach TJ. Effects of mevinolin on cell cycle progression and viability of tobacco BY-2 cells. The Plant Journal. 1998;14:65–74. doi: 10.1046/j.1365-313X.1998.00095.x. [DOI] [PubMed] [Google Scholar]
- Hemmerlin A, Hoeffler J-F, Meyer O, Tritsch D, Kagan IA, Grosdemange-Billiard C, Rohmer M, Bach TJ. Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. Journal of Biological Chemistry. 2003;278:26666–26676. doi: 10.1074/jbc.M302526200. [DOI] [PubMed] [Google Scholar]
- Hsieh M-H, Goodman HM. The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiology. 2005;138:641–653. doi: 10.1104/pp.104.058735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M-H, Goodman HM. Functional evidence for the involvement of Arabidopsis IspF homolog in the nonmevalonate pathway of plastid isoprenoid biosynthesis. Planta. 2006;223:779–784. doi: 10.1007/s00425-005-0140-9. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Hanada A, Kuzuyama T, Takagi M, Kamiya Y, Yamaguchi S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. Journal of Biological Chemistry. 2002;277:45188–45194. doi: 10.1074/jbc.M208659200. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Suzuki M, Tang J, et al. LOVASTATIN INSENSITIVE 1, a novel pentatricopeptide repeat protein, is a potential regulatory factor of isoprenoid biosynthesis in Arabidopsis. Plant and Cell Physiology. 2007;48:322–331. doi: 10.1093/pcp/pcm005. [DOI] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M. Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Molecular Biology. 2003;51:925–948. doi: 10.1023/a:1023005504702. [DOI] [PubMed] [Google Scholar]
- Learned RM. Light suppresses 3-hydroxy-3-methylglutaryl-coenzyme A reductase gene expression in Arabidopsis thaliana. Plant Physiology. 1996;110:645–655. doi: 10.1104/pp.110.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned RM, Fink GR. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Arabidopsis thaliana is structurally distinct from the yeast and animal enzymes. Proceedings of the National Academy of Sciences, USA. 1989;86:2779–2783. doi: 10.1073/pnas.86.8.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Campos N, Boronat A. The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. The Plant Journal. 1995;8:541–549. doi: 10.1046/j.1365-313x.1995.8040541.x. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, León P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. The Plant Journal. 1996;9:649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- Manzano D, Fernández-Busquets X, Schaller H, González V, Boronat A, Arró M, Ferrer A. The metabolic imbalance underlying lesion formation in Arabidopsis thaliana overexpressing farnesyl diphosphate synthase (isoform 1S) leads to oxidative stress and is triggered by the developmental decline of endogenous HMGR activity. Planta. 2004;219:982–992. doi: 10.1007/s00425-004-1301-y. [DOI] [PubMed] [Google Scholar]
- Morehead TA, Biermann BJ, Crowell DN, Randall SK. Changes in protein isoprenylation during growth of suspension-cultured tobacco cells. Plant Physiology. 1995;109:277–284. doi: 10.1104/pp.109.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Suzuki M, Yoshida S, Muranaka T. Mevalonic acid partially restores chloroplast and etioplast development in Arabidopsis lacking the non-mevalonate pathway. Planta. 2002;216:345–350. doi: 10.1007/s00425-002-0871-9. [DOI] [PubMed] [Google Scholar]
- Newman JD, Chappell J. Isoprenoid biosynthesis in plants: carbon partitioning within the cytoplasmic pathway. In: Parish EJ, Nes WD, editors. Biochemistry and function of sterols. Boca Raton, FL: CRC Press; 1997. pp. 123–134. [DOI] [PubMed] [Google Scholar]
- Nieto B, Forés O, Arró M, Ferrer A. Arabidopsis 3-hydroxy-3-methylglutaryl-CoA reductase is regulated at the post-translational level in response to alterations of the sphingolipid and the sterol biosynthetic pathways. Phytochemistry. 2009;70:58–64. doi: 10.1016/j.phytochem.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Suzuki M, Masuda K, Yoshida S, Muranaka T. Chemical phenotypes of the hmg1 and hmg2 mutants of Arabidopsis demonstrate the in planta role of HMG-CoA reductase in triterpene biosynthesis. Chemical and Pharmaceutical Bulletin. 2007;55:1518–1521. doi: 10.1248/cpb.55.1518. [DOI] [PubMed] [Google Scholar]
- Okada K, Kasahara H, Yamaguchi S, Kawaide H, Kamiya Y, Nojiri H, Yamane H. Genetic evidence for the role of isopentenyl diphosphate isomerases in the mevalonate pathway and plant development in Arabidopsis. Plant and Cell Physiology. 2008;49:604–616. doi: 10.1093/pcp/pcn032. [DOI] [PubMed] [Google Scholar]
- Phillips MA, D'Auria JC, Gershenzon J, Pichersky E. The Arabidopsis thaliana type 1 isopentenyl diphosphate isomerase are targeted to multiple subcellular compartments and gave overlapping functions in isoprenoid biosynthesis. The Plant Cell. 2008;20:677–696. doi: 10.1105/tpc.107.053926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science. 1994;264:1458–1460. doi: 10.1126/science.8197459. [DOI] [PubMed] [Google Scholar]
- Randall SK, Marshall MS, Crowell DN. Protein isoprenylation in suspension-cultured tobacco cells. The Plant Cell. 1993;5:433–442. doi: 10.1105/tpc.5.4.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F, Kis K, Bacher A, Eisenreich W. The non-mevalonate pathway of isoprenoids: genes, enzymes and intermediates. Current Opinion in Chemical Biology. 2001;5:535–540. doi: 10.1016/s1367-5931(00)00240-4. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Forés O, Martínez-García JF, González V, Phillips MA, Ferrer A, Boronat A. Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. The Plant Cell. 2004;16:144–156. doi: 10.1105/tpc.016204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder NS, Goad LJ. The effect of the 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor ML-236 B on phytosyterol synthesis in Acer pseudoplantanus tissue culture. Biochimica et Biophysica Acta. 1980;619:424–427. doi: 10.1016/0005-2760(80)90092-2. [DOI] [PubMed] [Google Scholar]
- Sapir-Mir M, Mett A, Belausov E, Tal-Meshulam T, Frydman A, Gidoni D, Eyal Y. Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiology. 2008;148:1219–1228. doi: 10.1104/pp.108.127951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller H, Grausem B, Benveniste P, Chye ML, Tan CT, Song YH, Chua NH. Expression of the Hevea brasiliensis (H.B.K.) Müll. Arg. 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant Physiology. 1995;109:761–770. doi: 10.1104/pp.109.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Gemünden C, Lichtenthaler HK. Chlorophyta exclusively use the 1-deoxyxylulose 5-phosphate/2-C-methylerythritol 4-phosphate pathway for the biosynthesis of isoprenoids. Planta. 2001;212:416–423. doi: 10.1007/s004250000409. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kamide Y, Nagata N, et al. Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. The Plant Journal. 2004;37:750–761. doi: 10.1111/j.1365-313x.2004.02003.x. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Muranaka T. Molecular genetics of plant sterol backbone synthesis. Lipids. 2007;42:47–54. doi: 10.1007/s11745-006-1000-5. [DOI] [PubMed] [Google Scholar]
- Wentzinger LF, Bach TJ, Hartmann MA. Inhibition of squalene synthase and squalene epoxidase in tobacco cells triggers an up-regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Physiology. 2002;130:334–346. doi: 10.1104/pp.004655. [DOI] [PMC free article] [PubMed] [Google Scholar]