Abstract
The Lc petunia system, which displays enhanced, light-induced vegetative pigmentation, was used to investigate how high light affects anthocyanin biosynthesis, and to assess the effects of anthocyanin pigmentation upon photosynthesis. Lc petunia plants displayed intense purple anthocyanin pigmentation throughout the leaves and stems when grown under high-light conditions, yet remain acyanic when grown under shade conditions. The coloured phenotypes matched with an accumulation of anthocyanins and flavonols, as well as the activation of the early and late flavonoid biosynthetic genes required for flavonol and anthocyanin production. Pigmentation in Lc petunia only occurred under conditions which normally induce a modest amount of anthocyanin to accumulate in wild-type Mitchell petunia [Petunia axillaris×(Petunia axillaris×Petunia hybrida cv. ‘Rose of Heaven’)]. Anthocyanin pigmentation in Lc petunia leaves appears to screen underlying photosynthetic tissues, increasing light saturation and light compensation points, without reducing the maximal photosynthetic assimilation rate (Amax). In the Lc petunia system, where the bHLH factor Leaf colour is constitutively expressed, expression of the bHLH (Lc) and WD40 (An11) components of the anthocyanin regulatory system were not limited, suggesting that the high-light-induced anthocyanin pigmentation is regulated by endogenous MYB transcription factors.
Keywords: Anthocyanin, bHLH, flavonol, Lc, Leaf colour, light, MYB, photosynthesis, vegetative pigmentation
Introduction
Anthocyanins are plant pigments produced by the flavonoid biosynthetic pathway. They are often present in flowers, fruit, leaves, and stems and range in colour from orange/red to purple/blue. Anthocyanins function in flowers and fruits primarily to attract pollinators and seed distributors (Gould and Lister, 2006). The roles for vegetative pigmentation, however, are less clearly understood. Anthocyanins are often produced in vegetative tissues under stress conditions, such as high-light, cold temperature, nutrient deficiency or pathogen attack (Dixon and Paiva, 1995; Chalker-Scott, 1999). This induction suggests anthocyanins act as some form of protectant, but whether this is due to their antioxidant activities or their optical properties (or both) has yet to be resolved. There is evidence that anthocyanins can protect photosynthetic tissues from photoinhibition by absorbing blue-green light and, thereby, reducing the amount of light reaching the chloroplasts (Feild et al., 2001; Neill and Gould, 2003; Hughes et al., 2005; Merzlyak et al., 2008). Flavonoids, including anthocyanins, are potent antioxidants, raising the possibility they may scavenge reactive oxygen species (ROS) generated during photosynthesis, particularly under conditions of photoinhibition (typically high light and low temperature). Although anthocyanins are usually sequestered in vacuoles, colourless cytosolic anthocyanins may act to scavenge ROS generated by chloroplasts and mitochondria (Neill and Gould, 2003). In addition, ROS such as H2O2 diffuse rapidly through membranes, which may allow vacuolar anthocyanins to scavenge ROS (Hatier and Gould, 2009). However, a major limitation for the investigation of anthocyanin functions in vegetative tissues has been the lack of an appropriate system to make physiological comparisons between anthocyanic and acyanic leaves. Most studies have relied on natural colour variants within species, or compared leaves of different developmental stages.
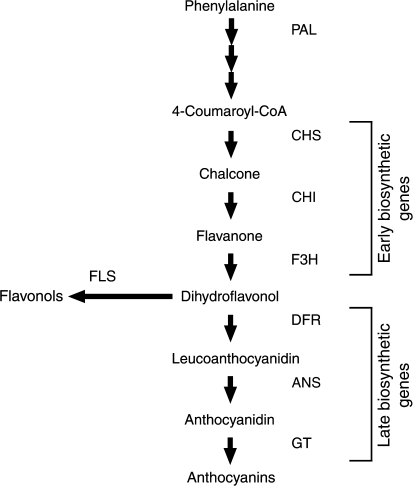
Flavonoid and anthocyanin biosynthesis is well characterized, and most of the biosynthetic genes have been identified and cloned (Fig. 1) (Grotewold, 2006). The three major anthocyanidins, pelargonidin (orange/red), cyanidin (pink), and delphinidin (violet/blue) differ in their hydroxylation pattern, which contributes to their optical and chemical properties. Other flavonoids such as flavonols are also produced by this pathway, and have specific roles distinct from anthocyanins. Flavonols often accumulate to high levels in leaves and flowers, providing protection from UV light (Li et al., 1993; Middleton and Teramura, 1993). In addition, they are often associated with anthocyanins as co-pigments, forming complex inter-molecular interactions (Mol et al., 1998; Aida et al., 2000).
Fig. 1.
A stylized diagram of the flavonoid biosynthetic pathway. CHS is the first committed step towards flavonoid production, leading to the production of the major flavonoids, flavonols, and anthocyanins. Abbreviations: PAL, phenylalanine ammonia lyase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; GT, glycosyltransferases.
The regulation of flavonoid metabolism is achieved primarily through transcriptional regulation of the biosynthetic genes (Martin et al., 2001; Davies and Schwinn, 2003), and several regulatory genes required for anthocyanin regulation have been identified, cloned, and characterized in several species. These transcription factors principally belong to two classes, MYB and basic-Helix-Loop-Helix (bHLH), and together with a WD40 protein, are thought to regulate the anthocyanin biosynthetic genes co-operatively (Koes et al., 2005). Flavonol regulation in Arabidopsis occurs by a separate system from anthocyanin regulation, which is independent of bHLH factors, and involves a subset of MYB factors distinct from those known to regulate anthocyanin synthesis. AtMYB12 and two closely related transcription factors, AtMYB11 and AtMYB111, have been shown co-ordinately to regulate the early flavonoid biosynthetic genes CHS, CHI, and F3H, as well as the flavonol-specific gene FLS, in response to light for flavonol production in different tissues throughout the plant (Mehrtens et al., 2005; Stracke et al., 2007).
The ability of transcription factors to regulate an entire pathway has led to the use of anthocyanin-regulating transcription factors to alter anthocyanin production and pigmentation patterns in several plant systems (Boase et al., 1998; Bradley et al., 1998, 1999; Bovy et al., 2002). Leaf colour (Lc) is a bHLH anthocyanin regulator from maize (Ludwig et al., 1989) and, when expressed in petunia, resulted in enhanced anthocyanin production throughout the vegetative tissues (Bradley et al., 1998). However, this enhanced pigmentation was dependent upon the environmental conditions (particularly light) to which the plants were exposed.
Ectopic expression of Lc in petunia (Bradley et al., 1998), alfalfa (Ray et al., 2003), tomato (Bovy et al., 2002), and Arabidopsis (Lloyd et al., 1992) enhanced anthocyanin production throughout the vegetative tissues, with each of these systems displaying some degree of light-induction. The mechanism responsible for determining these light-induced anthocyanin accumulation responses is unknown, especially with respect to the interaction between endogenous and transgenic transcription factors. Light-induced vegetative anthocyanin production is also a common feature of many non-transgenic plants under a range of environmental conditions, and developmental stages; particularly in seedlings, juvenile shoots at bud-burst, and stressed-plants. Our aims for this study were to characterize the light-induced anthocyanin pigmentation response in Lc and wild-type Mitchell petunia further, to understand better the regulation of the flavonoid pathway and possible interactions between transgenic and endogenous transcription factors, and to investigate the effect of intense anthocyanin accumulation upon photosynthesis in a near-isogenic system where anthocyanins could be induced to enhanced levels.
Materials and methods
Plant material and growth conditions
Mitchell petunia [Petunia axillaris×(Petunia axillaris×Petunia hybrida cv. ‘Rose of Heaven’)] (Ausubel et al., 1980) was originally obtained from Professor Richard Gardner at the School of Biological Sciences, University of Auckland, New Zealand. Mitchell petunia (also known as W115) is an2– an4–. The Lc petunia seed line 118C was derived from the primary transformant 118 described in Bradley et al. (1998), and is hemizygous for the transgene (Mitchell background). Seeds for both Mitchell and Lc petunia were sown and germinated under ambient summer greenhouse conditions (February 2004). Plants were grown under 50% shade cloth, to prevent the induction of pigmentation, until they had approximately 10 nodes. Axillary buds were removed to prevent a branched architecture which would result in self-shading. Five replicates of both Mitchell and Lc plants were transferred to either high-light or shade conditions. Plants were maintained under each lighting condition for 12 d. Plants were grown in bark:pumice (60:40 v/v) potting mix.
In a second experiment investigating the effects of anthocyanin pigmentation upon photosynthesis (January 2007), 10 homozygous (118C×118C) Lc petunia and 10 Mitchell plants were grown until they had reached seven nodes, at which time they were transferred to a growth cabinet set up as the low-light treatment.
Light treatments
In the first experiment, plants in the ‘shade’ condition were grown in the greenhouse under a tent of 50% shade cloth. The existing photoperiod was approximately 14 h, and the greenhouse was heated at 15 °C and vented at 25 °C. The ‘high light’ growth treatment was provided within a Contherm Cat 640 controlled environment growth cabinet and plants were arranged randomly within the cabinet and spaced to prevent shading. Plants were grown under a 14 h photoperiod, at a constant 22 °C and 65% humidity. White light in the growth cabinet was provided by 12 HPI-T metal halide bulbs (Philips). The light levels varied between 50– 350 μmol m−2 s−1 in the shade treatment, depending on the time of day and weather, and were constant at 750 μmol m−2 s−1 in the high-light treatment.
In the second experiment for photosynthetic measurements, low-light and high-light conditions were provided in Contherm growth cabinets set at a 14 h photoperiod, 22 °C, 65% humidity, with either 80 μmol m−2 s−1 or 600 μmol m−2 s−1 lighting intensity, respectively. All light measurements were determined using a Li-Cor Li-250 light meter, using the LI-190SA quantum sensor.
Pigment extraction and analysis
Leaf tissue for pigment and RNA analysis was collected from nodes 7–12 from each plant and frozen immediately in liquid nitrogen. Sampling was performed at midday to standardize any circadian or diurnal effects on flavonoid biosynthetic gene transcript abundance. Flavonoids were extracted from 50 mg DW of ground leaf tissue in 2 ml methanol:acetic acid:water (70:3:27 by vol.) for 72 h at 4 °C. The supernatant was removed and the pellet re-extracted in 2 ml methanol:acetic acid:water (90:1:9 by vol.) overnight at 4 °C. The combined supernatants were dried under vacuum and made up to 0.5 ml with methanol:acetic acid:water (80:2:18 by vol.). HPLC analysis of flavonoids was performed as described in Bradley et al. (1998). Anthocyanin levels were determined as cyanidin 3-O-glucoside (Extrasynthese, Genay, France) equivalents and other flavonoids as quercetin-3-O-rutinoside (Apin Chemicals, Abingdon, Oxon, UK) equivalents by integrating peak areas.
Assessment of photosynthetic parameters
In experiment two, the plants were allowed to acclimatize for 24 h after transfer to the low-light treatment before photosynthetic measurements were taken. Five Lc and five Mitchell plants were measured, and then transferred to the high-light treatment. The same measurements were taken on the remaining Lc and Mitchell plants the following day, before these plants were returned to the low-light treatment. The plants were maintained under the different light regimes for 7 d, at which time their photosynthetic parameters were measured. Photosynthetic measurements were performed on the first fully expanded leaf. This leaf was marked at the beginning of the experiment and was measured again after the 7 d light treatment. The marked leaf and the next four initiated leaves were collected for pigment extractions.
Photosynthetic measurements
In experiment two, light response curves, were performed using a CIRAS-2 Infra-red gas analyser (PP systems, Hitchin, UK), coupled with a PLC6 leaf cuvette that maintained constant temperature (22±0.2 °C) and CO2 (1200 μl l−1), and excluded external light from the enclosed area of the leaf. A 15-point CO2 saturated light response curve was then conducted, using a range of light levels appropriate to the physiological properties of the plants, low-light-grown plants at 0–800 μmol m−2 s−1; high-light-grown plants at 0–1200 μmol m−2 s−1. The maximal photosynthetic assimilation rate, Amax, quantum efficiency, QE, the light compensation point, and the light saturation levels were calculated for each plant using the ‘Photosyn Assistant’ software (Dundee Scientific, Dundee, UK).
Chlorophyll and carotenoids were extracted from 20 mg DW tissue with 2 ml acetone:methanol (7:3 v/v) containing 200 mg ml−1 CaCO3. The extract was centrifuged at 10 000 g for 2 min, the supernatant removed, and the pellet re-extracted in 2 ml acetone:methanol (7:3 v/v). This procedure was repeated until the tissue was colourless. The combined supernatants were partitioned with equal volumes of diethyl ether and water, and the ether phase removed. The diethyl ether fractions were dried under O2-free N2 and the carotenoids and chlorophylls were dissolved in 1 ml ethyl acetate. Chlorophylls and carotenoids were quantified by measuring their absorbance at 480 nm, 648 nm, and 666 nm in chloroform, using a Jasco V-530 UV/Vis spectrophotometer (Jasco, Tokyo, Japan) and the pigment concentrations calculated using the equation described by Wellburn (1994). Flavonoids were extracted from 30 mg DW ground leaf tissue as described above. Total flavonoids were measured spectrophotometrically at 350 nm (in methanol), and anthocyanins at 530 nm (in 0.1 N HCl methanol). Total flavonoids were determined as quercetin-3-O-rutinoside equivalents (ε=14300, Mr=610) and anthocyanins as cyanidin 3-O-glucoside equivalents (ε=22750, Mr=484.4).
RNA isolation and analysis
Total RNA was extracted from frozen ground leaf material, using a modified hot borate protocol (Hunter et al., 2002). Northern blots were performed with 15 μg of total RNA separated on a 1.2% (w/v) denaturing agarose gel (1× MOPS, 0.66 M formaldehyde), and transferred to Hybond N+ membrane (GE Healthcare) with 10× SSC by capillary action. RNA was cross-linked to the membrane by exposure to UV-C at 70 000 μl cm−2 (Hoefer, San Francisco, CA) for 1 min. Radiolabelled probes ([α-32P]dCTP) were generated using the HighPrime® (Roche Applied Science) labelling kit, using inserts isolated from the following plasmids: pLc349 (Lc), pCGP701 (CHSa), pCGP62 (CHIa), pCGP481 (FLS), pCGP1402 (DFRa), pCGP1407 (ANS), and pTIP6 (25/26S rRNA). Hybridization and washing conditions were performed as described in Bradley et al. (1998).
Semi-quantitative RT-PCR
First strand cDNA was prepared from 5 μg total RNA from shade and high-light-grown Mitchell leaves using SuperscriptIII Reverse Transcriptase and oligo dT12-18 (Invitrogen). Semi-quantitative reverse-transcription-PCR was performed for An11 (primers K135 5′-AGCTGGTACCATGGAAAATTCAAGTCAAG-3′; K136 5′-CTGATCTAGATTCAATC TTTCAATCACCT-3′), and Actin (NA22 5′-TTCAGCCACTTGTCTGTGAC-3′; NA23 5′-CGACATCACATTTCATGATGG-3′) from 1 μl of first strand cDNA with Taq polymerase. Cycling conditions were 94 °C for 2 min, then (94 °C for 30 s, 60 °C for 30 s, 72 °C for 45 s) with 23 or 30 cycles for Actin and An11, respectively. PCR reactions were run on 1.5% (w/v) agarose gels with ethidium bromide, and fluorescent images were captured using the FLA-5100 imaging system (Fujifilm).
Leaf disc transformation of petunia
Leaf discs of shade-grown (acyanic) Lc and Mitchell leaves were transformed by inoculation with Agrobacterium tumefaciens (LBA4404, Invitrogen) carrying the binary vector pLN83 (CaMV35S:Rosea1), expressing either an anthocyanin-regulating MYB gene from Antirrhinum (Schwinn et al., 2006) or an ER-tagged GFP reporter construct pBIN-m-gfp5-ER (CaMV35S:GFP-ER) (Haselhoff et al., 1997). Leaf discs were dipped into the bacterial culture, blotted, and transferred on to half-strength solid MS media (Murashige and Skoog, 1962). Leaf discs were cultured at 25 °C with a photoperiod of 16 h supplied by cool fluorescent tubes (25 μmol m−2 s−1). Anthocyanin pigments were readily detectable by light microscopy after 3 d. Images were collected with an Olympus SZX12 light microscope and a Leica Microsystems DC500 digital camera. GFP detection was performed with an Olympus SZX-RFL coaxial fluorescence attachment, consisting of a mercury lamp, blue wavelength excitation filter (BP460-490), a dichroic mirror (DM505), and a long-pass barrier filter that blocks wavelengths below 510 nm (BA510IF).
Statistical calculations
Statistical significance for pigment and photosynthetic measurements was determined by two-way analysis of variance (ANOVA), using Genstat version 10 (2007) for Windows (VSN International Ltd, Hemel Hempstead, UK). For pigment measurements, light saturation and light compensation point, ANOVA was performed upon log10 transformed means, as the variance was higher for treatments with higher means. Least significant difference (LSD) values are reported.
Results
High light induces vegetative anthocyanin pigmentation in petunia
The vegetative anthocyanin pigmentation phenotypes for Mitchell and Lc petunia plants grown under shade (50–350 μmol m−2 s−1) and high-light (750 μmol m−2 s−1) are shown in Fig. 2A and C, B and D, respectively. When Mitchell plants were grown under high-light conditions, weak anthocyanin pigmentation was observed, with purple anthocyanin pigmentation visible in the leaves, especially the veins (Fig. 2B). The stems of the plant were also purple with anthocyanin pigments. The modest amount of anthocyanin developed slowly over the light-treatment period. By contrast, the Lc plants exposed to high-light were dark purple with anthocyanin pigments throughout the leaves and stems (Fig. 2D), with the pigmentation visible within 12 h of exposure to high-light. Pigmentation intensity increased throughout the light treatment and, therefore, the older leaves appeared more intensely purple than newly exposed leaves at the apex.
Fig. 2.
Wild-type Mitchell and Lc petunia grown under shade and high-light treatments, showing vegetative pigmentation phenotypes. (A, C) Mitchell and Lc petunia grown under shade conditions (50–350 μmol m−2 s−1). (B, D) Mitchell and Lc petunia grown under high-light conditions (750 μmol m−2 s−1). The individual plants shown are representative of the five plants grown per treatment.
Mitchell petunia plants grown under shade conditions (Fig. 2A), did not produce visible levels of anthocyanins, and Lc petunia appeared largely non-pigmented (Fig. 2C). Upon close inspection, shade-grown Lc petunia had a border, 1–2 cells wide, at the leaf margins that were pigmented with anthocyanins. Both Mitchell and Lc petunia grown under the shade conditions were taller than plants grown under high light.
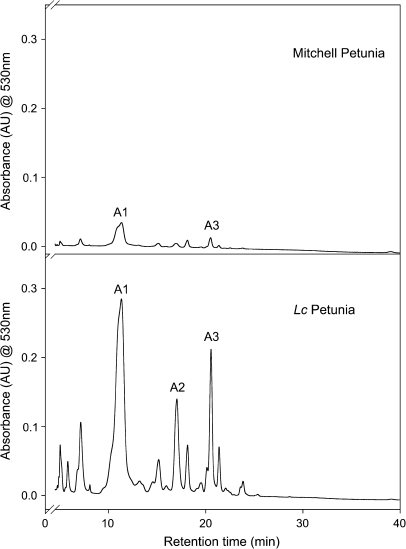
Lc petunia grown under high-light conditions showed a large increase in anthocyanin concentration in leaf tissue up to approximately 3 mg g−1 DW (Table 1), while Mitchell plants grown under the same conditions exhibited much lower anthocyanin concentrations, (<0.5 mg g−1 DW). Three major anthocyanin peaks were detected by HPLC analysis of high-light Mitchell and Lc petunias (Fig. 3). These were identified as malvidin and petunidin glycosides, consistent with previous studies (Bloor et al., 1998). The major anthocyanins with known structure are labelled. The same anthocyanin peaks detected in Lc petunia were present in wild-type Mitchell petunia, but at lower levels. Anthocyanins were not detected in Mitchell or Lc plants grown under shade conditions.
Table 1.
Leaf anthocyanin and flavonoid content (mg g−1 DW) in Mitchell (MP) and Lc petunia grown under shade or high light
| Shade |
High light |
LSD | |||
| MP | Lc | MP | Lc | ||
| Anthocyanins | |||||
| Petunidin-3-rutinoside-5-glucoside, p-coumaric acid (A1) | NDa | ND | 0.17 | 1.61 | |
| Petunidin-3-rutinoside-5-glucoside, caffeic acid (A2) | ND | ND | 0.03 | 0.39 | |
| Petunidin-3-rutinoside-5-glucoside, 4-O-glucosyl-p-coumaric acid (A3) | ND | ND | 0.15 | 0.40 | |
| Other anthocyanins | ND | ND | 0 | 0.54 | |
| Total (mean) | ND | ND | 0.35 | 2.94 | |
| Log10 total | – | – | (–0.46) | (0.47) | (0.32)b |
| Other flavonoids (flavonols) | |||||
| Quercetin-3-O-(caffeoyl diglucoside) (F1) | 4.70 | 4.61 | 6.11 | 6.34 | |
| Quercetin-3-O-(2-O-caffeoyl 6-O- malonyl diglucoside) (F2) | ND | ND | 3.90 | 3.62 | |
| Kaempferol 3-O-(feruloyl diglucoside) (F3) | 0.50 | 0.51 | 3.18 | 4.37 | |
| Other flavonols | 1.89 | 1.99 | 9.26 | 10.82 | |
| Total (mean) | 7.09 | 6.79 | 22.45 | 25.15 | |
| Log10 total | (0.85) | (0.83) | (1.35) | (1.40) | (0.12)b |
Data presented are the mean values for individual anthocyanins or flavonols as well as a total concentration (n=3). Individual compounds (A1-3 or F1-3) correspond to the labelled peaks for the chromatograms shown in Figs 3 and 4, respectively.
ND, not detected.
LSD reported is for the Log10 total mean value only. LSD is at 5% significance level; residual df=8 for other flavonoids, df=4 for anthocyanins.
Fig. 3.
HPLC chromatograms for leaf extracts from Mitchell and Lc petunia grown under high light. The absorbance was monitored at 530 nm to detect anthocyanins. The major anthocyanin peaks are indicated. The major anthocyanin peaks are petunidin-3-rutinoside-5-glucoside acylated with p-coumaric acid (A1), caffeic acid (A2), or 4-O-glucosyl-p-coumaric acid (A3).
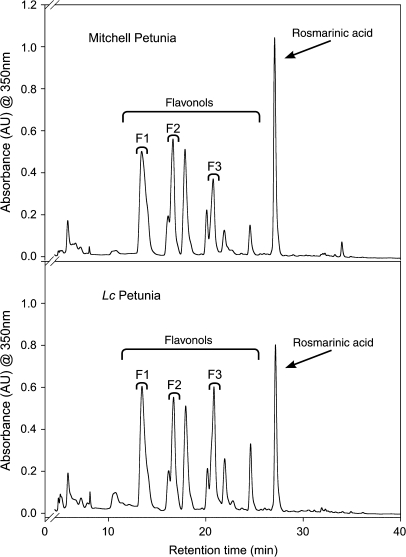
Flavonols unaffected by anthocyanin induction
Mitchell and Lc petunia plants had equivalent levels of other flavonoids (flavonols) in all light treatments (Table 1). Flavonol concentration was higher at increased light intensity, increasing from approximately 7 mg g−1 DW in shade-grown plants, to 25 mg g−1 DW in high-light plants. Mitchell and Lc petunia accumulated similar flavonoid compounds (Fig. 4), with the major peaks matching the various flavonols. The major flavonols with a known structure are labelled. A non-flavonoid compound was also present, and was identified as rosmarinic acid, after comparison with published data (Bloor et al., 1998; Troncoso et al., 2005). Rosmarinic acid was excluded from the calculations of other flavonoid total concentration (Table 1) as it is not derived from the flavonoid biosynthetic pathway.
Fig. 4.
HPLC chromatograms for leaf extracts from Mitchell and Lc petunia grown under high light. Absorbance was monitored at 350 nm. The major flavonoids, the flavonols, and a non-flavonoid compound, rosmarinic acid, are indicated. F1, quercetin-3-O-(caffeoyl diglucoside); F2, quercetin-3-O-(2-O-caffeoyl 6-O-malonyl diglucoside); F3, kaempferol 3-O-(feruloyl diglucoside).
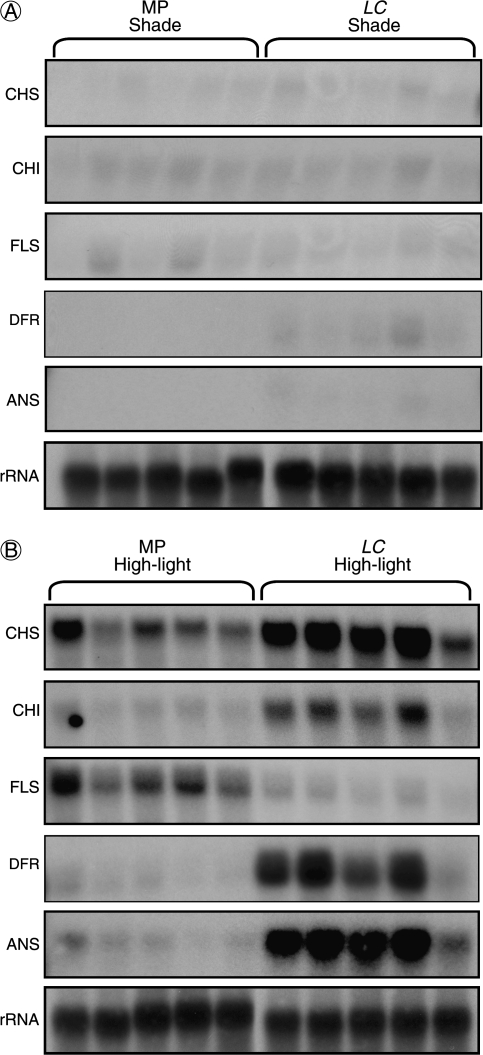
Changes in flavonoid gene expression underpin induction of anthocyanin synthesis
Flavonoid gene expression in Mitchell and Lc plants grown under high-light and shade conditions is shown in Fig. 5. The flavonoid biosynthetic genes CHS, CHI, and FLS showed high transcript abundance in high-light Mitchell plants, while weak signals for DFR and ANS transcripts were also detected. Lc plants show enhanced transcript levels for CHS and CHI levels compared with Mitchell, while FLS transcripts were reduced, and strong signals for transcripts of the late biosynthetic genes DFR and ANS were clearly observed. Weak signals for CHS, CHI, and FLS were detected in Mitchell and Lc petunia plants grown under shade. Faint signals for DFR and ANS were detected in Lc petunia, but not in Mitchell.
Fig. 5.
Northern blot analysis of flavonoid structural gene expression in Mitchell (MP) and Lc petunia plants under (A) shade or (B) high-light treatments. Each lane represents a different individual plant within each treatment. 25/26S rRNA is shown as a loading control.
Expression of a MYB anthocyanin regulator overcomes the high-light requirement for anthocyanin accumulation in Lc petunia
Light-induced anthocyanin pigmentation in Lc petunia raised questions about the regulation of the anthocyanin biosynthetic genes in petunia leaves. The current model for anthocyanin regulation links three components: bHLH, WD40, and MYB factors.
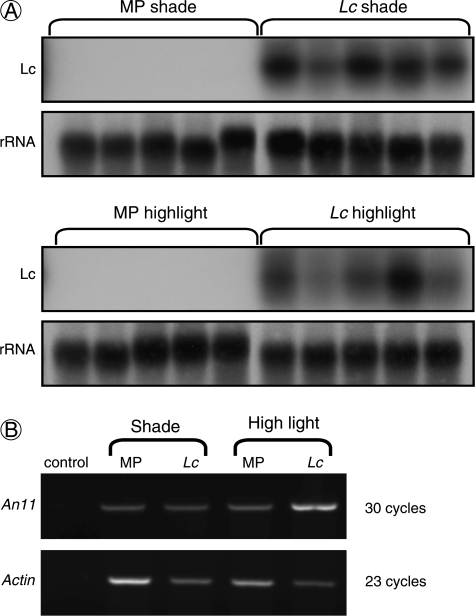
Expression of the bHLH Lc transgene in Lc petunia was shown to be insensitive to the light treatment (Fig. 6A), and therefore not responsible for the light-induced phenotype. A WD40 factor (An11) is required for floral anthocyanin regulation in petunia in addition to MYB and bHLH factors. Semi-quantitative RT-PCR showed similar transcript levels for An11 in both Mitchell and Lc petunia leaves, in either shade or high-light conditions (Fig. 6B).
Fig. 6.
Transcript abundance for anthocyanin regulation components. (A) Northern blot showing Leaf colour transcript abundance in Lc and Mitchell petunia (MP) grown under shade or high-light treatments. 25/26S rRNA is shown as a loading control. (B) Semi-quantitative RT-PCR of An11 transcripts in shade and high-light-grown Mitchell and Lc petunia leaves. Actin was amplified as a cDNA loading control. PCR cycles are indicated.
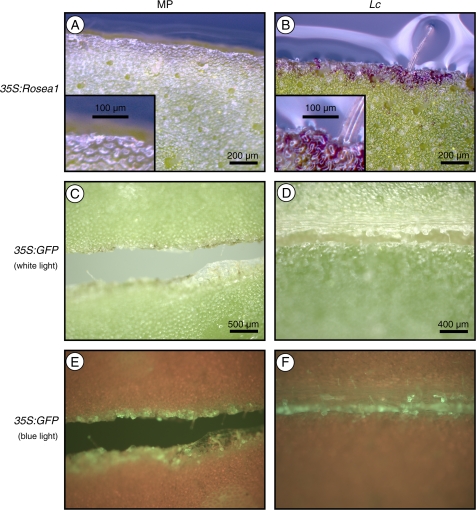
The anthocyanin-regulating MYB transcription factor Rosea1, introduced by Agrobacterium-mediated transformation into acyanic leaf discs of Mitchell and Lc petunia, was able to complement the requirement for high-light to induce pigmentation in Lc petunia. Transformation of Mitchell petunia with CaMV35S:Rosea1 did not induce anthocyanin production (Fig. 7A), but scarlet anthocyanin-producing cells were clearly visible along the cut surface of the leaf disc in Lc petunia (Fig. 7B). Transformation with CaMV35S:GFP was performed as a negative control. Anthocyanins were not induced in Mitchell or Lc petunia (Fig. 7C, D) but GFP expression was clearly visible (Fig. 7E, F).
Fig. 7.
Light complementation experiments, utilizing Agrobacterium-mediated transformation of Mitchell (MP) and Lc petunia leaves. (A, B) MP and Lc leaf explants, respectively, transformed with CaMV35S:Rosea1. Inset images in (A) and (B) show higher magnification of cells at the cut surface of the leaves. MP and Lc leaf explants transformed with CaMV35S:GFP, viewed under white light (C, D) and blue light (E, F), respectively.
Vegetative anthocyanins screen photosynthetic tissues
Lc and Mitchell plants were grown under different light conditions and leaf photosynthetic measurements were made to determine whether the accumulation of anthocyanin pigments in Lc petunia affected photosynthetic capacity.
Light response curves and photosynthetic CO2 assimilation rates were determined for Lc and Mitchell plants before and after being subjected to either low- or high-light treatments. No differences in the CO2 assimilation rates, light compensation points or light saturation points were observed in low-light plants (Table 2). However, once anthocyanins accumulation was induced by high-light in Lc petunia, the light saturation point was increased by 46% compared with Mitchell plants, and the light compensation point was raised by 68%. Despite the changes in light compensation and saturation points, the maximum photosynthetic rate, Amax, was not significantly different between Lc and Mitchell plants, although it was higher in the high-light than low-light plants.
Table 2.
Photosynthetic characteristics of Mitchell (MP) and Lc petunia after 7 d of low-light or high-light treatment, n=5
| Amax (μmol CO2 m−2 s−1) | QE (×10−2) | Light saturation point (μmol m−2 s−1) |
Light compensation point (μmol m−2 s−1) |
|||
| Mean | Mean | Mean | (Log10) | Mean | (Log10) | |
| MP low light | 12.2 | 6.11 | 216 | (2.334) | 15 | (1.176) |
| Lc low light | 14.2 | 6.89 | 225 | (2.352) | 18 | (1.255) |
| MP high light | 33.1 | 5.93 | 590 | (2.771) | 28 | (1.447) |
| Lc high light | 30.1 | 3.71 | 863 | (2.936) | 47 | (1.672) |
| LSDa | 4.3 | 0.77 | (0.061) | (0.187) | ||
LSD is at the 5% significance level; residual df=16.
Photosynthetic pigment content
Vegetative pigment levels were also measured in the plants used to examine photosynthetic parameters. This included chlorophylls and carotenoids, as well as anthocyanins and flavonoids. No significant differences in pigment levels between Lc or Mitchell plants grown under low-light were noted (Table 3). Under high-light conditions, however, anthocyanins accumulated to 10-fold higher levels in Lc petunia and total flavonoid levels were slightly higher than in Mitchell (Table 3), consistent with the results from the first experiment (Table 1). Chlorophyll and carotenoid levels in Mitchell plants grown under high-light were lower than in the low-light treatment, and there was an increase in the chlorophyll a/b ratio. Lc petunia plants under high-light did not show the same reduction in chlorophyll and carotenoid levels or change in chlorophyll a/b ratio observed for the Mitchell plants.
Table 3.
Leaf pigment analysis of Mitchell (MP) and Lc petunia plants grown under low or high light
| Total flavonoids |
Anthocyanins |
Total chlorophylls |
Chla/Chlb |
Total carotenoids |
||||||
| Mean | (Log10) | Mean | (Log10) | Mean | (Log10) | Mean | (Log10) | Mean | (Log10) | |
| MP low light | 5.61 | (0.749) | NDa | – | 6.85 | (0.836) | 4.26 | (0.629) | 1.91 | (0.281) |
| Lc low light | 5.94 | (0.774) | ND | – | 6.04 | (0.781) | 4.08 | (0.611) | 1.85 | (0.267) |
| MP high light | 16.88 | (1.227) | 0.04 | (–1.398) | 1.93 | (0.286) | 8.16 | (0.912) | 0.74 | (–0.131) |
| Lc high light | 20.43 | (1.310) | 0.91 | (–0.041) | 3.05 | (0.484) | 7.02 | (0.846) | 1.02 | (0.009) |
| LSDb | (0.077) | (0.130) | (0.072) | (0.086) | (0.055) | |||||
Pigment concentrations are expressed as mg g−1 DW, n=5.
ND, not detected.
LSD is at the 5% significance level; residual df=16, except for anthocyanins; df=8.
Discussion
Light-induced anthocyanin pigmentation
Light-induced vegetative anthocyanin pigmentation was confirmed in transgenic Lc petunia plants. Despite constitutive expression of the bHLH transgene Lc, increased pigmentation was only observed in the high-light treatment. This pigmentation was limited to those tissues which normally accumulate a modest amount of anthocyanin in Mitchell petunia (Fig. 2), in sub-epidermal cell layers overlying the photosynthetic cells in the leaves of both Mitchell and Lc petunia (Bradley et al., 1998). Transgenic alfalfa (Medicago sativa) (Ray et al., 2003), tomato (Solanum lycopersicum) (Goldsbrough et al., 1996), and Arabidopsis plants (Lloyd et al., 1992) ectopically expressing Lc also showed light-induced anthocyanin pigmentation, and light has been shown to enhance proanthocyanidin (PA) production in lotus plants (Lotus corniculatus) ectopically expressing Sn, a bHLH transcription factor homologous to Lc (Paolocci et al., 2005). These findings indicate that, in these systems, the bHLH transgene alone was insufficient to induce anthocyanin (or PA) biosynthesis, and it is acting with other endogenous regulatory factors in each of these systems to activate the biosynthetic genes, and that these endogenous factors are expressed in vegetative tissues during high-light or stress conditions.
Lc petunia under high-light conditions show enhanced expression of flavonoid biosynthetic genes required for anthocyanin production. The early genes CHS and CHI, as well as the late genes DFR and ANS, were induced in Mitchell petunia leaves, matching the modest accumulation of anthocyanin. Transcript levels for the early and late biosynthetic genes were greatly enhanced in Lc plants, matching the accumulation of anthocyanin, suggesting that LC was co-operatively activating the biosynthetic genes normally targeted by endogenous anthocyanin regulators. The specific anthocyanins which accumulated in Lc petunia were the same as those in Mitchell (Bloor et al., 1998) (Fig. 3), indicating that the same genes, including those for anthocyanin modification (acylation, methylation, glycosylation) were being targeted. This suggests that the specificity of target gene recognition was provided by the endogenous regulators that act with LC.
The current model for anthocyanin regulation indicates MYB and bHLH transcription factors, together with a WD40 co-regulator, form a complex which activates the target anthocyanin biosynthetic genes (Koes et al., 2005). The bHLH factor Lc is constitutively expressed in Lc petunia (Fig. 6A) and An11 (WD40) was shown to be expressed in leaves irrespective of light treatment (Fig. 6B). The involvement of the endogenous petunia bHLH factors Jaf13 and An1 is unlikely to determine the light-induced phenotype, as the related bHLH Lc transgene was expressed at high levels throughout the plant, and bHLH factors involved in anthocyanin production cannot bind DNA on their own and act through their binding partners (Sainz et al., 1997; Hernandez et al., 2004; Koes et al., 2005). Therefore, the possibility was investigated that MYB transcription factors may determine the light-induced anthocyanin biosynthesis in Lc petunia. Introduction of a known anthocyanin-regulating MYB transcription factor, ROSEA1, successfully complemented the requirement for high-light (Fig. 7B). It was hypothesized that endogenous MYB factors are induced by high light in vegetative tissues to control the production and distribution of anthocyanins to fulfil light-screening functions, and that the MYB(s) provide the specificity of target gene recognition. Mitchell petunia contains recessive alleles for the known anthocyanin-regulating MYB factors, an2– (null) and an4– (Quattrocchio et al., 1993, 1998; Koes et al., 2005), therefore, the hypothesized MYB determining light-induced vegetative pigmentation is a new MYB anthocyanin regulator.
Lc petunias grown under high-light conditions have higher transcript levels for CHS and CHI, as well as DFR and ANS, which suggests endogenous anthocyanin regulators may act upon both the early and late biosynthetic genes together with LC. The early anthocyanin biosynthetic genes are shared with multiple branches of the flavonoid pathway, including flavonols, indicating a requirement for multiple regulatory systems to act upon common genes to control the production of different flavonoid compounds. Such a system was demonstrated in Arabidopsis, where the promoters of the flavonoid genes CHS, CHI, and F3H contained multiple cis-elements conferring responsiveness to the anthocyanin regulators C1/SN (MYB/bHLH) or to light (light-regulated flavonol production) (Hartmann et al., 2005). These elements consisted of a MYB-recognition element (MRE), a bHLH recognition sequence named the R-response-element (RRE), and an ACGT containing element (ACE), which are thought to bind MYB, bHLH, and bZIP transcription factors, respectively. Light-regulated expression of the early genes required both the MRE and ACE, while activation by C1/SN required the MRE and RRE, suggesting that both the flavonol regulators and the anthocyanin regulators may act upon the shared early biosynthetic genes, differentially and independently to regulate the production of different flavonoids. The observation that myb11– myb12– myb11– triple mutants do not produce flavonols, yet still make anthocyanins is consistent with this (Stracke et al., 2007), suggesting that the anthocyanin regulators can act redundantly upon the early biosynthetic genes (CHS, CHI, F3H), allowing expression of these genes in the absence of the flavonol regulators.
Light-regulated flavonol production occurs by a system distinct from anthocyanin regulation in petunia. Flavonols were induced with increasing light intensity in Lc and Mitchell petunia leaves, which have previously been identified as various acylated kaempferol and quercetin-glycosides (Bloor et al., 1998). Transcripts for CHS, CHI, and FLS were induced by high light, in a manner that we believe is co-ordinated (Fig. 5). In Arabidopsis, co-ordinated expression of these genes is regulated by AtMYB11, AtMYB12, and AtMYB111 for flavonol production in different tissues throughout the plant (Mehrtens et al., 2005; Stracke et al., 2007). It is possible that homologous regulators to AtMYB12 regulate flavonol production in petunia, acting independently of anthocyanin regulators. Interestingly, FLS transcript levels were lower in high-light grown Lc petunia compared to Mitchell. It has been suggested that this was because the high level of anthocyanins in Lc petunia screened light from underlying cells, which reduced the light-induction of flavonol-related genes. A reduction in CHS and CHI transcripts was not observed, however, due to the activation of these genes by LC, together with endogenous anthocyanin regulators. Despite a reduction in the transcript levels for FLS in high-light Lc petunia, a reduction in flavonol content compared to Mitchell was not observed. FLS knock-down in Mitchell petunia flowers resulted in enhanced anthocyanin accumulation (Davies et al., 2003), suggesting FLS strongly competes with DFR for di-hydroflavonol substrates. Increased flux through the early steps of the flavonoid pathway in high-light-grown Lc petunia may have elevated the pool of di-hydroflavonols, providing more substrate for FLS, allowing flavonols to accumulate to similar levels to Mitchell.
Physiological impact of anthocyanin pigmentation upon photosynthesis
The light-induced anthocyanin phenotype observed in Lc petunia allowed a comparison of photosynthetic capacity to be made in pigmented and non-pigmented leaves. Although anthocyanin pigmentation did not reduce the maximum photosynthetic rate (Amax), a screening role for anthocyanins was demonstrated. Under low-light conditions, when anthocyanins were not induced, Mitchell and Lc petunia had similar photosynthetic assimilation rates, light saturation, and light compensation points. However, once anthocyanins were induced in Lc petunia, the light-saturation point increased by 46% compared with Mitchell plants grown under the same conditions. This suggests that the anthocyanins in Lc petunia leaves were screening light from the underlying photosynthetic tissues. A light-screening role for anthocyanins was also supported by analysis of the photosynthetic pigments in these plants, which showed that Lc petunia leaves containing high levels of anthocyanin also had higher chlorophyll and carotenoid levels and a reduced chlorophyll a/b ratio. These changes observed in the purple Lc plants are generally associated with shade leaves, whereas the Mitchell plants grown in high light had the typical sun leaf characteristics of a lower chlorophyll content and higher a/b ratios (Huner et al., 1998; Willows, 2004). Similar findings have been made in Quercus coccifera, comparing anthocyanic and acyanic leaves, where anthocyanin accumulation led to shade acclimation traits (Manetas et al., 2003). The presence of anthocyanins in leaves of Galax urceolata reduced the photoinhibition caused by exposure to strong white or green light, but not to red wavelengths, demonstrating an in vivo light attenuation and photoprotection role for anthocyanins (Hughes et al., 2005). While the absorption spectrum for anthocyanins peaks at around 550 nm in vivo, they also absorb wavelengths in the blue region, particularly when they accumulate to high levels (Merzlyak et al., 2008). Together with our own findings, this indicates that foliar anthocyanins do screen light from underlying tissues.
Conclusions
Lc and Mitchell petunia have provided a unique, near-isogenic experimental system to examine the mechanism for vegetative light-induced anthocyanin pigmentation and the effect of pigmentation upon photosynthesis, whilst removing the variability associated with plants of differing genetic backgrounds or developmental stages. The enhanced pigmentation of Lc petunia has allowed us to demonstrate that sub-epidermal anthocyanins do act as a light screen, but without affecting the maximum photosynthetic rate. The fact that intense foliar pigmentation in Lc petunia occurred only when grown under high light points to the existence of a light-induced regulatory factor responsible for the activation of the anthocyanin pathway. Given that both the bHLH (Lc) and WD40 (An11) were constitutively expressed in Lc petunia, and that a MYB anthocyanin-regulator Rosea1 was able to complement the high-light requirement, the endogenous regulatory factor appears likely to be a MYB protein.
Acknowledgments
We gratefully acknowledge the technical assistance of Steve Arathoon, Ian King, and Julie Ryan, as well as useful discussions and advice from Associate Professor Kevin Gould. We thank Duncan Hedderley for his assistance with the statistical analyses, International Flower Development Pty Ltd for the use of the petunia plasmid clones, and Drs Susan Wessler and Ronald Diamani for the pLc349 plasmid. NWA was supported by the Massey University Masterate Scholarship.
References
- Aida R, Yoshida K, Kondo T, Kishimoto S, Shibata M. Copigmentation gives bluer flowers on transgenic torenia plants with the antisense dihydroflavonol-4-reductase gene. Plant Science. 2000;160:49–56. doi: 10.1016/s0168-9452(00)00364-2. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Bahnsen K, Hanson M, Mitchell A, Smith HJ. Cell and tissue culture of haploid and diploid Petunia ‘Mitchell’. Plant Molecular Biology Newsletter. 1980;1:26–32. [Google Scholar]
- Bloor SJ, Bradley JM, Lewis DH, Davies KM. Identification of flavonol and anthocyanin metabolites in leaves of Petunia ‘Mitchell’ and its Lc transgenic. Phytochemistry. 1998;49:1427–1430. [Google Scholar]
- Boase MR, Bradley JM, Borst NK. Genetic transformation mediated by Agrobacterium tumefaciens of florists’ chrysanthemum (Dendranthema grandiflorum) cultivar ‘Peach Margaret’. In Vitro Cellular and Developmental Biology-Plant. 1998;34:46–51. [Google Scholar]
- Bovy A, de Vos R, Kemper M, et al. High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. The Plant Cell. 2002;14:2509–2526. doi: 10.1105/tpc.004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley JM, Davies KM, Deroles SC, Bloor SJ, Lewis DH. The maize Lc regulatory gene up-regulates the flavonoid biosynthetic pathway of Petunia. The Plant Journal. 1998;13:381–392. [Google Scholar]
- Bradley JM, Deroles SC, Boase MR, Bloor S, Swinny E, Davies KM. Variation in the ability of the maize Lc regulatory gene to upregulate flavonoid biosynthesis in heterologous systems. Plant Science. 1999;140:37–48. [Google Scholar]
- Chalker-Scott L. Environmental significance of anthocyanins in plant stress responses. Photochemistry and Photobiology. 1999;70:1–9. [Google Scholar]
- Davies KM, Schwinn KE. Transcriptional regulation of secondary metabolism. Functional Plant Biology. 2003;30:913–925. doi: 10.1071/FP03062. [DOI] [PubMed] [Google Scholar]
- Davies KM, Schwinn KE, Deroles SC, Manson DG, Lewis DH, Bloor SJ, Bradley JM. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica. 2003;131:259–268. [Google Scholar]
- Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. The Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feild T, Lee D, Holbrook N. Why leaves turn red in autumn. The role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiology. 2001;127:566–574. [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough AP, Tong Y, Yoder JI. Lc as a non-destructive visual reporter and transposition excision marker gene for tomato. The Plant Journal. 1996;9:927–933. [Google Scholar]
- Gould KS, Lister C. Flavonoid functions in plants. In: Andersen ØM, Markham KR, editors. Flavonoids: chemistry, biochemistry and applications. Boca Raton: CRC Press; 2006. pp. 397–411. [Google Scholar]
- Grotewold E. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Molecular Biology. 2005;57:155–171. doi: 10.1007/s11103-004-6910-0. [DOI] [PubMed] [Google Scholar]
- Haselhoff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences, USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatier J-HB, Gould K. Anthocyanin function in vegetative organs. In: Gould K, Davies K, Winefield C, editors. Anthocyanins: biosynthesis, functions, and applications. New York: Springer; 2009. pp. 1–20. [Google Scholar]
- Hernandez JM, Heine GF, Irani NG, Feller A, Kim MG, Matulnik T, Chandler VL, Grotewold E. Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. Journal of Biological Chemistry. 2004;279:48205–48213. doi: 10.1074/jbc.M407845200. [DOI] [PubMed] [Google Scholar]
- Hughes NM, Neufeld HS, Burkey KO. Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata. New Phytologist. 2005;168:575–587. doi: 10.1111/j.1469-8137.2005.01546.x. [DOI] [PubMed] [Google Scholar]
- Huner NPA, Öquist G, Sarhan F. Energy balance and acclimation to light and cold. Trends in Plant Science. 1998;3:224–230. [Google Scholar]
- Hunter DA, Steele BC, Reid MS. Identification of genes associated with perianth senescence in Daffodil (Narcissus pseudonarcissus L. ‘Dutch Master’) Plant Science. 2002;163:13–21. [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colourful model for the regulation and evolution of biochemical pathways. Trends in Plant Science. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee T-M, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UB-B irradiation. The Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AM, Walbot V, Davis RW. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science. 1992;258:1773–1775. doi: 10.1126/science.1465611. [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production encodes a protein similar to transcriptional activators and contains the Myc-homology region. Proceedings of the National Academy of Sciences, USA. 1989;86:7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetas Y, Petropoulou Y, Psaras GK, Driana A. Exposed red (anthocyanic) leaves of Quercus coccifera display shade characteristics. Functional Plant Biology. 2003;30:265–270. doi: 10.1071/FP02226. [DOI] [PubMed] [Google Scholar]
- Martin C, Jin H, Schwinn K. Mechanisms and applications of transcriptional control of phenylpropanoid metabolism. In: Romeo J, Saunders J, Matthews B, editors. Regulation of phytochemicals by molecular techniques. Oxford: Elsevier Science Ltd; 2001. pp. 155–170. [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiology. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak MN, Chivkunova OB, Solovchenko AE, Naqvi KR. Light absorption by anthocyanins in juvenile, stressed, and senescing leaves. Journal of Experimental Botany. 2008;59:3903–3911. doi: 10.1093/jxb/ern230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton EM, Teramura AH. The role of flavonol glycosides and carotenoids in protecting soybean from ultraviolet-B damage. Plant Physiology. 1993;103:741–752. doi: 10.1104/pp.103.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R. How genes paint flowers and seeds. Trends in Plant Science. 1998;3:212–217. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:743–747. [Google Scholar]
- Neill SO, Gould KS. Anthocyanins in leaves: light attenuators or antioxidants? Functional Plant Biology. 2003;30:865–873. doi: 10.1071/FP03118. [DOI] [PubMed] [Google Scholar]
- Paolocci F, Bovone T, Tosti N, Arcioni S, Damiani F. Light and an exogenous transcription factor qualitatively and quantitatively affect the biosynthetic pathway of condensed tannins in Lotus corniculatus leaves. Journal of Experimental Botany. 2005;56:1093–1103. doi: 10.1093/jxb/eri101. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. The Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JNM, Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. The Plant Journal. 1998;13:475–488. doi: 10.1046/j.1365-313x.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Ray H, Yu M, Auser P, Blahut-Beatty L, McKersie B, Bowley S, Westcott N, Coulman B, Lloyd A, Gruber MY. Expression of anthocyanins and proanthocyanidins after transformation of alfalfa with maize Lc. Plant Physiology. 2003;132:1448–1463. doi: 10.1104/pp.103.025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz M, Grotewold E, Chandler VL. Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. The Plant Cell. 1997;9:611–625. doi: 10.1105/tpc.9.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. The Plant Cell. 2006;18:831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso N, Sierra H, Carvajal L, Delpiano P, Günther G. Fast high performance liquid chromatography and ultraviolet-visible quantification of principal phenolic antioxidants in fresh rosemary. Journal of Chromatography. 2005;1100:20–25. doi: 10.1016/j.chroma.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Wellburn AR. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Journal of Plant Physiology. 1994;144:307–313. [Google Scholar]
- Willows RD. Chlorophylls. In: Davies K, editor. Plant pigments and their manipulation. Vol. 14. Cornwall: Blackwell Publishing Ltd; 2004. pp. 23–56. [Google Scholar]